Abstract
Beyond being one the most widely used psychoactive drugs in the world, cannabis has been identified as an environmental risk factor for psychosis. Though the relationship between cannabis use and psychiatric disorders remains controversial, consistent association between early adolescent cannabis use and the subsequent risk of psychosis suggested adolescence may be a particularly vulnerable period. Previous findings on gene by environment interactions indicated that cannabis use may only increase the risk for psychosis in the subjects who have a specific genetic vulnerability. The type 1 cannabinoid receptor (CB1), encoded by the CNR1 gene, is a key component of the endocannabinoid system. As the primary endocannabinoid receptor in the brain, CB1 is the main molecular target of the endocannabinoid ligand, as well as tetrahydrocannabinol (THC), the principal psychoactive ingredient of cannabis. In this study, we have examined mRNA expression and DNA methylation of CNR1 in human prefrontal cortex (PFC), hippocampus, and caudate samples. The expression of CNR1 is higher in fetal PFC and hippocampus, then drops down dramatically after birth. The lifespan trajectory of CNR1 expression in the DLPFC differentially correlated with age by allelic variation at rs4680, a functional polymorphism in the COMT gene. Compared with COMT methionine158 carriers, Caucasian carriers of the COMT valine158 allele have a stronger negative correlation between the expression of CNR1 in DLPFC and age. In contrast, the methylation level of cg02498983, which is negatively correlated with the expression of CNR1 in PFC, showed the strongest positive correlation with age in PFC of Caucasian carriers of COMT valine158. Additionally, we have observed decreased mRNA expression of CNR1 in the DLPFC of patients with schizophrenia. Further analysis revealed a positive eQTL SNP, rs806368, which predicted the expression of a novel transcript of CNR1 in human DLPFC, hippocampus and caudate. This SNP has been associated with addiction and other psychiatric disorders. THC or ethanol are each significantly associated with dysregulated expression of CNR1 in the PFC of patients with affective disorder, and the expression of CNR1 is significantly upregulated in the PFC of schizophrenia patients who completed suicide. Our results support previous studies that have implicated the endocannabinoid system in the pathology of schizophrenia and provided additional insight into the mechanism of increasing risk for schizophrenia in the adolescent cannabis users.
Subject terms: Molecular neuroscience, Schizophrenia
Introduction
Cannabis is a widely used and increasingly popular psychoactive substance for both recreational and medicinal purposes1, especially in Western countries2. However, cannabis use is not entirely benign. There is increasing evidence that it is associated with increased risk for psychosis3 including the recurrence of manic symptoms in patients with bipolar disorder. Adolescent cannabis use has been associated with a 2.4-fold increase in the risk of schizophrenia4, and early adolescent-onset use is associated with a higher risk5. One intriguing study found an increased risk for schizophrenia with early adolescent heavy usage, especially if the subjects had the allele encoding for valine in the COMT Val158Met polymorphism (rs4680)6. Cannabis use disorder is a common comorbidity in patients with schizophrenia, exacerbating psychosis, reducing neuroleptic efficacy, and increasing relapse rates7–9. Nevertheless, the vast majority of individuals who use cannabis never develop psychosis, suggesting that there may be genetic vulnerabilities to this adverse side effect. Interestingly, a recent study reported that an acute effect of ∆9-THC on brain physiology measured with fMRI in normal subjects was modulated by COMT Val158Met10.
Cannabis contains about 400 bioactive molecules, more than 60 of which are cannabinoid compounds11. The most psychoactive of these is delta-9-tetrahydocannabinol (∆9-THC), acting via the cannabinoid receptor type 1 (CB1) in the central nervous system (CNS). Delta-9-THC, as an exogenous agonist of endocannabinoid receptors, has been classified by some as a hallucinogen due to its effect on perception, especially in high doses and in individuals at high risk for mental illness12. Identification of the psychoactive components of cannabis led to the discovery of an important neurotransmitter system, the endocannabinoid system (ECS), comprised of cannabinoid receptors (CB1 and CB2), endogenous ligands (anandamide and 2-arachadonylogycerol), and their metabolic enzymes (fatty acid amide hydrolase and monoacylglycerol lipase)13–18. This system is widely distributed in the central and peripheral nervous systems and is implicated in a wide variety of brain functions, such as memory, mood, motor control, and reward processing19–23.
Although CB1 and CB2 are both G-protein-coupled receptors, they have different localization. The CB1 receptor, encoded by the CNR1 gene, is expressed in both the central and peripheral nervous systems, especially on axon terminals in the cerebellum, hippocampus, basal ganglia, frontal cortex, amygdala, hypothalamus, and midbrain24,25. However, the CB2 receptor, encoded by the CNR2 gene, is mainly expressed in the periphery, on cells in the immune system such as monocytes, macrophages, B-lymphocytes, and T-lymphocytes13,26, making the ECS potentially associated with immunomodulation27,28. The CB2 receptor is also expressed in the CNS, but mainly in microglia and other immune cells of brain27,29,30. The CNR1 gene is composed of a single coding-exon and several alternative 5′ untranslated exons. The CB1 receptor, as expressed principally on presynaptic terminals, inhibits the release of excitatory and inhibitory neurotransmitters including acetylcholine, noradrenaline, 5-HT, GABA, glutamate, dopamine, D-aspartate, and cholecystokinin31–34. In summary, the CB1 receptor is an important component in the ECS in the nervous system, regulating synaptic transmission by modulating the release of neurotransmitters.
Genetic variants in CNR1 have been reported to influence the clinical presentation of schizophrenia in patients exposed to cannabis. Heavy cannabis use in the context of certain CNR1 genotypes may contribute to white matter volume deficits and cognitive impairment of patients with schizophrenia35. While some genetic studies have linked polymorphisms in the CNR1 gene with increased risk of schizophrenia36–38, other studies have failed to find this association39,40. A significant increase in the level of the endocannabinoids has been observed in the cerebrospinal fluid of patients with schizophrenia41. There are also numerous reports of the altered cannabinoid receptor protein levels and receptor binding in patients with schizophrenia42–44. On balance, there is a growing body of literature implicating the ECS in schizophrenia.
To further characterize the molecular correlates underlying clinical associations of the ECS with psychosis in general and schizophrenia in particular, we measured the mRNA expression of CNR1 across the human life span in the prefrontal cortex (PFC) and hippocampus and, based upon the Caspi study (2005)6, established expression profiles of the CNR1 transcripts based on age and COMT genotype. We have also compared the expression of the CNR1 transcripts in the PFC, hippocampus and caudate among controls, and individuals with either schizophrenia or affective disorders. To explore how epigenetics might affect the expression of CNR1 transcripts, we measured DNA methylation profiles of CNR1 in human PFC and hippocampus. By examining the relationship between the expression of CNR1, genetic variants and DNA methylation in a large human brain cohort across the lifespan and four different diagnostic groups, our study provides evidence of abnormalities in the ECS in association with schizophrenia, and defines expression quantitative trait loci and expression changes in this neuromodulatory system across the lifespan.
Materials and methods
Human postmortem brain tissue
Postmortem human brain tissue was collected at several sites for this study. A large number of samples were obtained at the Clinical Brain Disorders Branch (CBDB) at National Institute of Mental Health (NIMH) from the Northern Virginia and District of Columbia Medical Examiners’ Office, according to NIH Institutional Review Board guidelines (Protocol #90-M-0142). These samples were transferred to the Lieber Institute for Brain Development (LIBD) under an MTA with the NIMH. Additional samples were collected at the LIBD according to with a protocol approved by the Institutional Review Board of the State of Maryland Department of Health and Mental Hygiene (#12-24) and the Western Institutional Review Board (#20111080). Additional fetal, child, and adolescent brain tissue samples were provided by the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders via Material Transfer Agreements (NO1-HD-4-3368 and NO1-HD-4-3383) approved by Institutional Review Board of the University of Maryland. Audiotaped informed consent to study brain tissue was obtained for the legal next-of-kin on every case collected at NIMH and LIBD. Details of the donation process and specimens handling are described previously45,46. After next-of-kin provided audiotaped informed consent to brain donation, a standardized 36-item telephone screening interview was conducted, (the Lieber Institute for Brain Development Autopsy Questionnaire), to gather additional demographic, clinical, psychiatric history, substance abuse history, treatment, medical, and social history. A psychiatric narrative summary was written for every donor, to include data from multiple sources, including the Autopsy Questionnaire, medical examiner documents (investigative reports, autopsy reports, and toxicology testing), macroscopic and microscopic neuropathological examinations of the brain, as well as extensive psychiatric, detoxification, and medical record reviews, and/or supplemental family informant interviews using the MINI (Mini International Neuropsychiatric Interview). Two board-certified psychiatrists independently reviewed every case to arrive at DSM-5 lifetime psychiatric and substance use disorder diagnoses, including [schizophrenia and bipolar disorder, as well as substance abuse disorders], and if for any reason agreement was not reached between the two reviewers, a third board-certified psychiatrist was consulted. All donors were free from significant neuropathology, including cerebrovascular accidents and neurodegenerative diseases. Each subject was diagnosed retrospectively by two board-certified psychiatrists, according to the criteria in the DSM-IV. Brain specimens from the CBDB were transferred from the NIMH to the LIBD under a Material Transfer Agreement.
Available postmortem samples were selected based on RNA quality (RNA integrity number ≥ 5). In total, 703 DLPFC, 452 hippocampus, and 468 caudate postmortem brain samples were used in this study. The demographic data are summarized in Supplementary Table 1. In brief, the DLPFC cohort consists of 175 subjects with schizophrenia, 62 with bipolar disorder, 146 with major depression disorder (MDD), and 320 non-psychiatric controls. The hippocampus cohort includes 133 subjects with schizophrenia and 319 non-psychiatric controls. The caudate cohort contains 154 subjects with schizophrenia, 47 subjects with bipolar disorder and 273 non-psychiatric controls. The toxicological analysis was performed in each case. The non-psychiatric non-neurological controls had no known history of significant psychiatric or neurological illnesses, including substance abuse. Positive toxicology was exclusionary for control subjects but not for patients with psychiatric disorders.
RNA extraction, RNA sequencing, and real-time PCR
Gray matter in the DLPFC, hippocampus and caudate were dissected out, pulverized and stored at −80 °C. Briefly, brains were hemisected, and cut into 1.0–1.5-cm-thick coronal slabs, flash-frozen, and stored at −80 °C. DLPFC gray matter was dissected using a dental drill. For non-fetal cases, the DLPFC (Brodmann’s areas 9 and 46) was dissected from the middle frontal gyrus of the coronal slab, immediately anterior to the genu of the corpus callosum. For fetal cases, the PFC was obtained from the frontal cortex dissected at the dorsal convexity, midway between the frontal pole and anterior temporal pole. The hippocampus was dissected from the anterior tip posteriorly through to the mid body of the hippocampus at the level of the lateral geniculate nucleus. For the fetal cases, the dorsal-medial aspect of the temporal lobe was removed, including the adjacent cortex, medial to the hippocampal sulcus. In the rostral-caudal axis, the dissection was performed at the mid-point of the Sylvian fissure. Caudate tissue was dissected from the head of the caudate nucleus, at the level of the nucleus accumbens. The dissection was restricted to the dorsal third of the caudate at this level, to make certain that this tissue was distinct from the adjacent nucleus accumbens. Total RNA was extracted from 30 to 50 mg of pulverized tissue with RNeasy Lipid Tissue Mini Kit (Qiagen). The RNA was purified with RNeasy Mini Spin columns with on-column DNase digestion by RNase-free DNase set (Qiagen). The yield of total RNA was determined by Qubit RNA BR Assay Kit and Qubit Fluorometer (ThermoFisher Scientific). The RNA quality was assessed with high-resolution capillary electrophoresis on an Agilent Bioanalyzer 2100 (Agilent Technologies). Approximately 300 ng total RNA was applied to an RNA 6000 Nano LabChip without prior heating. RNA integrity numbers (RIN) were obtained from the entire electrophoretic trace with the RIN software algorithm and was used for the assessment of RNA quality.
Poly-A RNA-seq for DLPFC and RiboZero RNA-seq for hippocampus and caudate were performed as described previously47,48. In general, Poly-A-containing RNA was purified from 1 μg DNase treated total RNA. Sequencing libraries were constructed by the Illumina TruSeq RNA Sample Preparation V2 kit. An index (up to 12) was inserted into Illumina adapters allowing samples to be multiplexed across lanes in each flow cell. After purification and enrichment, final cDNA libraries were sequenced by an Illumina HiSeq 2000 with paired-end 2 × 100 bp reads. For RiboZero RNA-seq, paired-end strand-specific sequencing libraries were constructed from 300 ng RNA using Illumina TruSeq Stranded Total RNA Library Preparation kit with Ribo-Zero Gold ribosomal RNA depletion. The libraries were sequenced on an Illumina HiSeq 3000. The Illumina Real-Time Analysis (RTA) module was used to perform image analysis and base calling. The BCL converter (CASAVA v1.8.2) was used to generate FASTQ files containing the sequencing reads. The sequencing reads were aligned to the human genome (UCSC hg19 build) using splicing-read mapper TopHat249, STAR50 or GSNAP51, providing known transcripts from Ensembl Build GRCh37/hg19.
We have used real-time quantitative PCR (RT-PCR) to validate the mRNA expression of CNR1 in 662 post-mortem DLPFC samples, consisting of 164 subjects with schizophrenia, 55 subjects with Bipolar disorder, 131 subjects with MDD, and 312 non-psychiatric controls (Supplementary Table 2). In short, 4 μg of total RNA was reverse transcribed by SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). mRNA expression levels of CNR1 was measured by TaqMan Gene Expression Assay, Hs01038522_s1 (Applied Biosystems) using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). The mRNA expression level of CNR1 in DLPFC was normalized to geometric means of two constitutively expressed genes: β-actin (ACTB) and β-glucuronidase (GUSB).
To measure the mRNA expression of the CNR1 gene, we acquired the mapped reads covering the CNR1 genomic region from BrainSeq datasets48, corresponding to chr6:88,849,585-88,875,767 on genome build GRCh37/hg19. Reads mapping to the gene was used to quantify the expression CNR1 at the gene level. RPM (reads per million mapped reads in the targeted CNR1 locus) has been calculated to quantify gene-level expression and junction-level expression.
DNA extraction, genotyping, and DNA methylation
Genomic DNA was extracted from 100 mg of pulverized cerebellar tissue and DLPFC tissue with the phenol-chloroform method. SNP genotyping with HumanHap650Y, Human 1M-Duo BeadChips, HumanOmni5-Quad or HumanOmni2.5–8 (Illumina, San Diego, CA) was carried out according to the manufacturer’s instructions with DNA extracted from cerebellar tissue. The methylation and genotype data were extracted from a larger data set from one of our recent studies52. In brief, methylation of DNA extracted from DLPFC was assessed according to the manufacturer’s instruction using the Infinium HumanMethylation450 BeadChip Kit (Illumina), which measure DNA methylation of more than 485,000 CpG dinucleotides (cg) covering 99% RefSeq gene promoters, including the CNR1 gene. The methylation data were processed and normalized using the minfi Bioconductor package in R53 as previously described52.
Statistics
Statistical analyses were performed using the R package (Version3.5.2). The lifespan curve was generated using the LOESS fit (local polynomial regression fitting) by using an R package with default parameters. We controlled for heterogeneity of transcript feature expression resulting from potential latent factors by using principal component analysis (PCA). Expression principal components (PCs) were calculated using the log-transformed (log2 with an offset of 1) RPKM of all expressed transcript features mapping to the 2 Mb sequence flanking CNR1 gene. Genomic PCs, which can model quantitative ethnicity (ancestry), were also calculated based on the genomic variants across common linkage-disequilibrium independent variants across the genome. The first ten expression PCs and the first five genetic PCs were used as covariates in the following eQTL analysis. Statistical analyses of mRNA expression associated with genotypes were conducted in the patients with schizophrenia and unaffected controls as our previous study (http://eqtl.brainseq.org/phase1/eqtl/)47. Levene’s test indicated that the assumption of homogeneity of variance was met. Statistical models investigating diagnosis-related expression patterns in adults (age > 13) was conducted by the general linear model (GLM), adjusted for ten expression PCs. Comparisons within the different diagnostic groups for completed suicide, nicotine exposure, ethanol exposure, and psychotropic medications (toxicology screen results about antidepressants, antipsychotics, anticonvulsants, benzodiazepines and opiates), lifetime neuroleptic exposure, average daily neuroleptic dose, final neuroleptic dose and illicit substances were also conducted by GLM adjusted for ten expression PCs. Estimated of lifetime neuroleptic exposure, average daily dose and final neuroleptic dose were all converted to chlorpromazine equivalents for statistical comparisons. Cannabis exposure was determined by the clinical history from next of kin interviews and medical record review, and toxicology screen, which measured Delta-9 THC, Delta-9 Carboxy THC and 11-Hydroxy Delta-9 THC. For qPCR validation, comparisons between the patient groups and controls were proceeded by GLM, adjusting for age, sex, race, and RIN. Pearson correlation was used to calculate the correlation between expression level or methylation level and age. The correlation coefficients were compared by Fisher’s r-to-z transformation.
Results
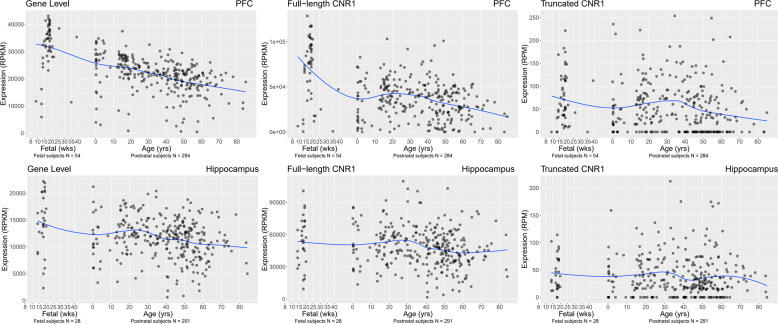
Based on NCBI Reference Sequence Database Release 109 (hg38), there are 5 previously identified alternative transcripts of CNR1 in human. These are predicted to produce only two different proteins: the previously identified full-length CB1 receptor and a truncated CB1 receptor. The truncated transcript is due to a 99 nt deletion located in the 5′ coding region of the last exon of CNR1, leading to a 439aa protein instead of the 472aa full-length CB1 receptor. We have used the reads aligning to the 99 nt deletion region and reads spanning the 99 nt deletion region to measure the expression of full-length and truncated CNR1 transcripts respectively (Supplementary Fig. 1). Lifespan expression was measured from gestational weeks 14 to 20 and from birth up to 85 years of age in the DLPFC and hippocampus, and from birth to 86 years of age in caudate. In both PFC and hippocampus, the expression of CNR1 at the gene level was the highest during the second-trimester fetal period, followed by decreased expression after birth (Fig. 1). The expression of the full-length and truncated CNR1 transcripts showed the similar lifespan patterns in PFC, but the flat life-span expression trajectories in hippocampus (Fig. 1). The expression of the CNR1 gene in PFC measured by RT-PCR also showed the same lifespan pattern (Supplementary Fig. 2). These findings suggest that the fetal brain may be susceptible to the effects of maternal cannabis consumption during pregnancy54–56.
Fig. 1. The mRNA expression lifetime trajectory of CNR1 gene and CNR1 transcripts in PFC and hippocampus of nonpsychiatric controls. The x-axis represents age.
The y-axis represents the relative expression at gene level or transcript level in the DLPFC and hippocampus. Each dot represents a nonpsychiatric control subject.
mRNA expression of CNR1 in schizophrenia and affective disorders
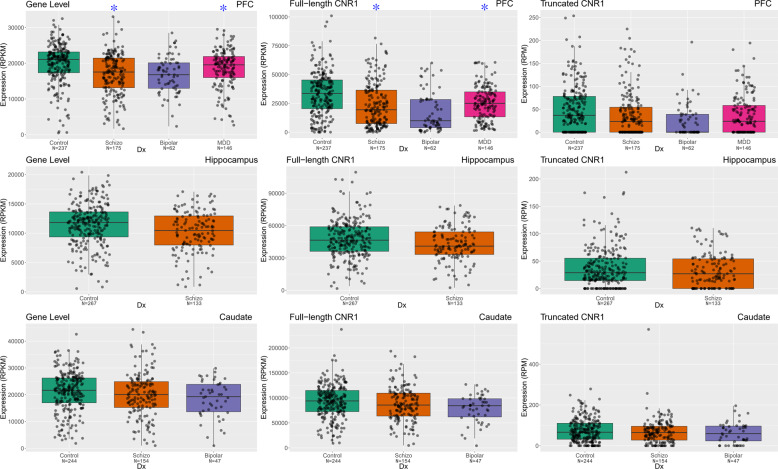
We then examined the relationship between CNR1 expression and diagnosis in neurotypical adult controls, and schizophrenia, bipolar, and major depression samples (age > 13). These analyses were conducted both at the gene and specific transcript level (full-length and truncated transcripts). In DLPFC, the expression of CNR1 at the gene level (Fig. 2) is significantly decreased in patients with schizophrenia (p = 3.31E−04, FDR = 9.92E−04) and patients with MDD (p = 1.67E−04, FDR = 7.50E−04) compared with non-psychiatric controls, but not in patients with bipolar disorder (p = 5.49E−02). At the transcript level, the expression of the full-length CNR1 transcript (Fig. 2) was downregulated significantly in the DLPFC of patients with schizophrenia (p = 8.86E−03, FDR = 0.02), and patients with MDD (p = 1.18E−04, FDR = 7.50E−04), but not in patients with bipolar disorder (p = 0.40). The expression of the truncated CNR1 transcript (Fig. 2) was not significantly altered in the DLPFC of patients with schizophrenia (p = 0.62), bipolar disorder (p = 0.67) and MDD (p = 0.09).
Fig. 2. The comparison of mRNA expression of CNR1 gene and CNR1 transcripts in human DLPFC, hippocampus and caudate.
The mRNA expression of CNR1 gene, full-length CNR1 transcript and 99 nt truncated CNR1 transcript was studied across four cohorts of subjects in DLPFC, two cohorts of subjects in hippocampus and three cohorts of subjects in caudate. The x-axis showed the different diagnostic groups: Control, nonpsychiatric control group; Schizo, patients with schizophrenia; Bipolar, patients with bipolar disorder; MDD, patients with major depression. The y-axis represents the relative expression quantities at gene level or transcript level in the DLPFC, hippocampus or caudate. Each dot represents a subject. Blue stars indicate patient groups with significantly differential expression compared with control group.
There is regional selectively in the relationship between diagnosis and CNR1 expression in the human brain. In the hippocampus, in contrast to the DLPFC, there is no significant difference in expression of the CNR1 gene and transcripts between patients with schizophrenia and non-psychiatric controls (p > 0.5). Also, there is no significant difference between the expression of the CNR1 gene and transcripts in caudate in the patients with schizophrenia or bipolar disorders, and non-psychiatric controls (p > 0.05).
To validate the expression difference revealed in DLPFC by RNA sequencing data, we performed TaqMan RT-PCR to measure the expression of the CNR1 gene in postmortem PFC samples. By linear regression, the expression of the CNR1 gene is marginally decreased in the DLPFC of the patients with schizophrenia (p = 1.61E−02, FDR = 4.83E−02), but not in patients with bipolar disorder (p = 5.19E−02) or MDD (p = 0.81) (Supplementary Fig. 3). The RT-PCR assay (Hs01038522_s1) binds on the last exon of CNR1. The RT-PCR results partly validated the RNA sequencing findings. The different statistical significance might due to different cohorts, different covariates, or different reference loci of quantifications between RT-PCR and RNA sequencing.
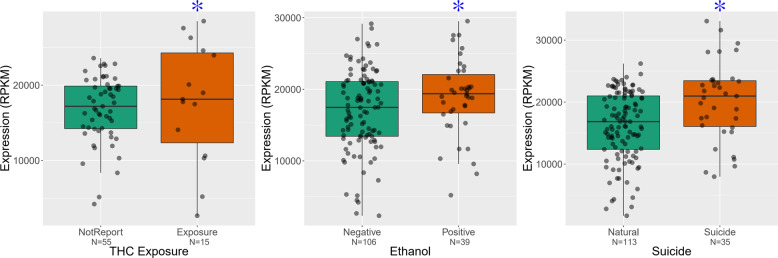
Based on toxicology screens at the time of death and clinical records, we tested for an association between CNR1 expression and death by completed suicide as well as illicit and legal drug exposure by GLM adjusted by the first ten expression PCs. Among the patients with affective disorders, the expression of CNR1 gene in DLPFC is significantly upregulated in subjects with THC exposure (p = 4.13E−03, FDR = 0.025) or ethanol exposure (p = 5.23E−03, FDR = 0.03). The expression of the CNR1 gene in DLPFC significantly increased in the patients with schizophrenia who had completed suicide (p = 0.01, FDR = 0.07) (Fig. 3). For a broad range of substances, such as nicotine, morphine, amphetamines, PCP, anticholinergics, anticonvulsants, antidepressants, antihistamines, anti-inflammatories, antipsychotics, CPZ equivalents, and lithium, there was no significant association with the expression of CNR1 gene in PFC, hippocampus and caudate.
Fig. 3. Expression of CNR1 gene in PFC correlated with substance abuse and suicide.
The y-axis represents the expression of CNR1 gene in DLPFC of patients with schizophrenia and affective disorders. Each black dot represents a subject. THC or Ethanol exposure associated with the increased expression of CNR1 gene in PFC with affective disorders, highlighted by blue stars. The x-axis showed the status of THC or ethanol exposure. The expression of CNR1 gene increased in the PFC of patients with schizophrenia who completed suicide, highlighted by blue star. The x-axis separates suicide verses unsuicide schizophrenia patients.
Differential mRNA expression of CNR1 in association with genetic variation in COMT
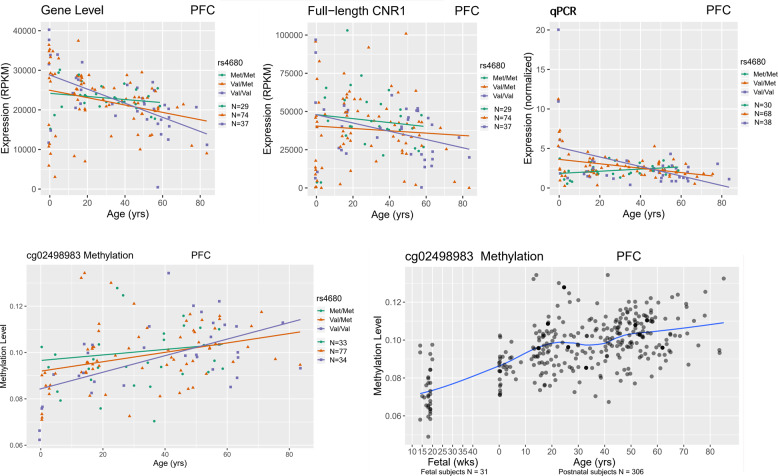
To explore the interaction between rs4680, a functional polymorphism in the COMT gene, and early THC exposure, we hypothesized that the expression of CNR1 in the human brain might have different developmental trajectories in association with rs4680 genotype. By Pearson’s correlation, the lifespan trajectory of CNR1 gene expression in PFC of Caucasian controls is differentially correlated with age by allelic variation at rs4680. Caucasian carriers of the COMT Val158 allele showed a significantly negative correlation between CNR1 expression in PFC and age. The correlation coefficients were significantly different between the Caucasian carriers of the Val158 homozygotes and Met158 homozygotes (p = 0.01, FDR = 0.015). The Val158 homozygotes showed the steepest slope of the decline in expression of CNR1 in PFC across the lifespan (COR = −0.55, p = 3.23E−04). The Val158Met heterozygotes also had a significantly negative slope of the expression of CNR1 in PFC across the lifespan (COR = −0.44, p = 5.04E−05) (Fig. 4). At the transcript level, we observed similar correlations between the full-length CNR1 transcript and age by rs4680 genotype (Fig. 4). In addition, the RT-PCR data validated that the Caucasian carriers of COMT Val158Val and Val158Met have negative correlations between the expression of full-length CNR1 in DLPFC and age (COR = −0.42, p = 0.008; COR = −0.25, p = 0.004). This assay also validated the finding that Caucasian carriers with Val158 homozygote showed the steepest slope of expression of CNR1 in PFC across the lifespan (COR = −0.42, p = 0.008) (Fig. 4), and the correlation coefficients were significantly different between the Caucasian carriers who were Val158 homozygotes compared with Met158 homozygotes (p = 6.11E−08, FDR = 1.45E−07).
Fig. 4. The mRNA expression of CNR1 gene and methylation level of cg02498983 is differentially correlated with age in the PFC of Caucasian controls by rs4680 genotype.
In the PFC of Caucasian controls who are COMT Val158 homozygotes, the expression of CNR1 gene and full-length transcript is most negatively correlated with age. Our qPCR results also validated this finding. Interestingly, the methylation level of cg02498983 is most positively correlated with age in the PFC of Caucasian controls who are COMT Val158 homozygotes. Each dot represents a control subject: green line, rs4680 Met/Met genotype; orange line, rs4680 Val/Met genotype; purple line, rs4680 Val/Val genotype. Blue line represents the lifespan trajectory of the methylation level of cg02498983 in PFC.
By Pearson’s correlation, we have also observed that the lifespan trajectories of three methylation loci (cg19961480, cg08037684, and cg02498983) in the DLPFC of Caucasians are correlated with age by allelic variation at rs4680. Caucasian carriers of the COMT Val158 allele showed significantly positive correlations between methylation levels of these three loci in DLPFC and age (Table 1). Correlation coefficient comparisons showed that only cg02498983 has a significant slope difference comparing Caucasian Val158 homozygote carriers to Met158 homozygotes (p = 0.0042, FDR = 0.038). The Val158 homozygote Caucasian subjects had the steepest slope of methylation levels in PFC across the lifespan (Fig. 4). In the PFC of control subjects, these three methylation loci showed increased methylation level by age across the lifespan (Fig. 4, Supplementary Fig. 4), and significantly (p < E−20) negative correlations with the expression of CNR1 gene and full-length CNR1.
Table 1.
Pearson’s correlation between the methylation level of three loci and age in the PFC of Caucasians by genotype at rs4680, a functional polymorphism in the COMT gene.
| cg19961480 | cg08037684 | cg02498983 | ||||
|---|---|---|---|---|---|---|
| rs4680 | CORa | p value | CORa | p value | CORa | p value |
| Met/Met | 0.16 | 8.83E−02 | 0.06 | 5.57E−01 | 0.23 | 1.73E−02 |
| Val/Met | 0.33 | 1.28E−06 | 0.29 | 2.62E−05 | 0.40 | 1.75E−09 |
| Val/Val | 0.36 | 4.71E−04 | 0.43 | 2.27E−05 | 0.60 | 3.35E−10 |
aCOR represents the values of the Pearson Correlation.
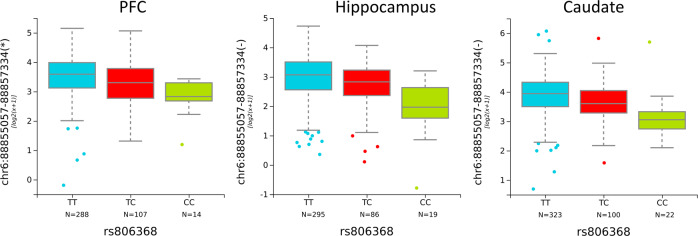
The association of genetic variation in CNR1 at rs806368 with transcript expression in PFC, hippocampus and caudate
For gene-level expression and full-length transcript expression of CNR1, we did not observe significant expression quantitative trait loci (eQTL). However, eQTL analyses for CNR1 revealed a positive SNP, rs806368, which associated with expression of an alternative 5′ junction of CNR1 (hg19: chr6:88855057-88857334). This unique junction links an alternative 5′ exon and the coding exon of CNR1, defining a novel CNR1 transcript. This transcript was not previously identified in the RefSeq. rs806368 is located at the 3′ UTR of CNR1. Genetic variation at this locus predicted the expression of a novel transcript of CNR1 in DLPFC (p = 8.42E−06, FDR = 9.75E−04) (Fig. 5), hippocampus (p = 4.46E−08, FDR = 1.87E−05) (Fig. 5) and caudate (p = 7.29E−08, FDR = 1.77E−05) (Fig. 5). In clinical studies, allelic variation at this SNP has been associated with increased risk for drug addiction, impulsivity, and nicotine addiction57. The clinically identified risk allele, C allele (minor allele), is associated with lower expression of the novel CNR1 transcript. This novel transcript is characterized by a unique junction between the last coding exon and an alternative 5’ adjacent exon, which is 48 nt longer than the 143 nt canonical exon. In addition to rs806368, we have observed additional SNPs with statistically significant eQTLs for this newly identified novel CNR1 transcript. However, these associations were found only in one or two brain regions, such as rs806371 in DLPFC (p = 3.61E−05, FDR = 6.19E−03), rs9450891 in hippocampus (p = 1.81E−05, FDR = 3.86E−03), or rs806371 in caudate (p = 5.45E−07, FDR = 1.09E−04) (Supplementary Table 3).
Fig. 5. Positive eQTLs for CNR1 with genetic variation at rs806368 in PFC, hippocampus, and caudate.
rs806368 genotype significantly associated with the expression of a novel transcript in PFC, hippocampus and caudate. The x-axis showed the genotypes for each SNP. The y-axis represents the relative expression quantities in PFC, hippocampus or caudate.
Discussion
Brain maturation continues during adolescence, especially in the limbic and neocortical regions involved in the regulation of complex behaviors and emotional responses. Multiple studies have suggested that changes in the ECS during adolescence contribute to the maturation of local corticocortical and corticolimbic circuit neurons58,59. Changes in the ECS during adolescence may produce long-lasting effects into adulthood. Caspi et al.6 presented evidence that cannabis exposure during adolescence may interact with genetic factors, such as COMT genotype at rs4680, to impact brain maturation and the risk for emergence of psychotic symptoms.
The CB1 receptor is the main cannabinoid receptor in the brain, and evidence in animal and human studies have suggested that dysregulation of the CB1 receptor in multiple brain regions, including the DLPFC and hippocampus, is involved in the development of schizophrenia by compromising complex circuits mediating cognition and memory60. In this study, we measured mRNA expression of the CNR1 gene, which encodes the CB1 receptor, across the human life span in the PFC and hippocampus, and established the expression profiles of CNR1 gene based on age and COMT genotype. Our results support previous studies that suggested a link between CNR1 and COMT genotype. In particular, the expression of CNR1 was the highest during the fetal period, followed by decreased expression after birth in human PFC and hippocampus. The correlations between CNR1 expression in the PFC of Caucasians and age differ by COMT genotype. Moreover, we have observed that the methylation levels at locus, cg02498983, is differentially correlated with age by COMT genotype in the PFC of Caucasians. In Caucasians, the COMT Val158Val carriers have the steepest negative slope of CNR1 expression and positive slope of methylation levels in PFC across lifespan compared with COMT Met158Val and Met158Met carriers. These results suggest that the levels of CNR1 protein might mediate the vulnerability towards dysmaturation of the PFC, secondary to cannabis exposure during adolescence. CNR1 expression levels in the PFC are modulated by rs4680 genotype in Caucasians, thereby altering signaling through the ECS. This in turn may lead to abnormal circuit formation between the PFC and other brain regions, including the hippocampal formation, the key elements for increasing risk of psychotic symptoms in adolescent cannabis users. During the development, CB1 receptor is critical to regulate neurotransmitter release and synaptic plasticity61, as well as neurodevelopmental programming for various of events such as proliferation, neurite growth and synaptogenesis62. Excessive stimulation of the cannabinoid system with exogenous cannabis during brain development, including in the adolescent period, may alter the biology and function of critical components of the maturing brain. This is especially important in the maturation of prefrontal regions of the brain as significant maturational events are not completed until early adulthood. It is not surprising that abnormal development and connectivity of the PFC as a result of cannabis exposure in genetically vulnerable individuals may lead to long-term effects on brain function later in life. Animal studies have suggested that maternal cannabis use affects the development of neural circuits, especially the dopaminergic system. In addition, animal model studies have found behavioral and cognitive deficits caused by cannabinoid exposure during early developmental periods63. These deficits are associated with changes of neurotransmission, especially in hippocampal and cortical excitatory circuits63. This disparity between different genotype groups is an example of different genetic backgrounds leading to a differential impact on gene expression and possibly differential susceptibility to the impact of environmental factors. We have also tested other genes they are important components of the cannabinoid signaling system, such as CNR2, FAAH and MGLL (Supplementary Fig. 5), but did not observe any positive associations between rs4680 genotype and gene expression in human postmortem brains.
The last exon of the CNR1 gene is transcribed to produce the CB1 receptor. Previous studies have reported several splicing variants of CNR1 with a truncated coding sequence in humans64,65, caused by several deletions inside the last exon of CNR1. Current RefSeq maps show one well-characterized splicing variant for CNR1 with a truncated coding sequence (NM_033181.3), caused by an in-exon 99 nt deletion. Splicing variants with truncated coding sequences are rare in the human PFC66, hippocampus and caudate. There are additional splicing events in the 5’ UTR of the CNR1 mRNA, but these do not change the protein-coding sequence. The longest 5′ UTR in CNR1 is about 500 nt and contains more than 600 potential transcription factor binding sites for 153 distinct transcription factors67. In this study, we have reported a novel junction in the 5’ UTR, which introduced an extra 48 nt sequence into the 5’ UTR. This novel extra 48 nt sequence contains an estimated 40 potential transcription factor binding sites (Supplementary Table 4). Three of them are unique and will bind Signal Transducer and Activator of Transcription proteins, whose biological activities ultimately regulate many critical aspects of cell growth, survival, and differentiation. The complexity of the 5′ UTR of CNR1 gene suggests that the expression of the CNR1 gene is regulated by multiple transcription factors in an “on-demand” fashion and this regulation may be important in aspects of brain development and function.
Our finding of decreased CNR1 expression in schizophrenia replicates and extends findings from previous studies. Egan et al. reported decreases in both CB1 mRNA and receptor levels, measured by in situ hybridization and immunocytochemistry, in the DLPFC of patients with schizophrenia68. A more recent study reported the lower availability of CB1 receptor in male patients with first-episode psychosis, greater reductions in CB1 receptor levels were associated with poor cognitive functioning and symptom severity69. Previously our group reported decreased expression of CB1 in layer 6 of the PFC of patients with schizophrenia, using in situ hybridization70. Consistent with our previous study70, here we have found decreased expression of the CNR1 gene in the DLPFC of patients with schizophrenia compared with non-psychiatric controls. In short, in examining three brain regions from one of the largest postmortem cohorts to date, our study has observed lower expression of CNR1 in DLPFC of patients with schizophrenia. To be certain, although CNR1 expression can be modulated pharmacologically by a large number of compounds, including atypical antipsychotic medications71, the results in this study are not clearly affected by those potential confounds. It is worth noting that, if we applied qSVA framework72 for removing confounding effects instead of expression PCs, there are no significant difference of the expression of CNR1 between the control group and case groups.
An important confound for postmortem human brain studies is comorbid substance use disorders. Comparisons of affective disorder patients with and without THC or ethanol exposure revealed upregulated expression of the CNR1 gene in the patients with mood disorders. In light of this finding we then examined a SNP at rs806368. Genotype at this locus in clinical studies has been associated with increased risk of cocaine dependence73,74, alcohol dependence75,76, nicotine dependence77, cannabis dependence78, impulsivity79, and drug dependence76. Here we have found that the minor allele of rs806368, a risk allele for substance dependence, is associated with lower expression of a novel CNR1 transcript in DLPFC. Our results provide further evidence that dysregulation of endocannabinoid signaling may be involved in substance abuse disorders, at least in patients with comorbid mood disorders. Since the novel transcript produces the same canonical CB1 receptor despite its unique 5’ UTR, it is more likely that this novel splicing event regulates the expression level of CB1 receptor in response to specific environmental stimuli, such as substance abuse. To understand the underlying mechanism of this positive eQTL, it is necessary to characterize this transcript at the cellular level across human development and to study this transcript in animal models of substance abuse and addiction.
There is abundant evidence from rodents and humans indicating that endocannabinoid signaling undergoes age-dependent changes80. CB1 receptor expression, its coupling to G proteins and the level of 2-Arachidonoylglycerol (2-AG) are reduced in the brain of older animals81–84. Fatty acid amide hydrolase (FAAH) is the primary catabolic enzyme of anandamide (AEA), which is one of two endogenous cannabinoid ligands85. The mRNA and protein levels and protein activity of FAAH are increased in late-onset (age > 65) Alzheimer’s disease (LOAD) patients86. Low doses of THC can reverse the age-related decline in cognitive performance in older animals, accompanied by enhanced expression of synaptic markers and increased hippocampal spine density87. In summary, the activity of the ECS declines during aging with decreased receptor levels and activity and decreased levels of an endogenous cannabinoid ligand because of increased levels of its catabolizing enzyme. In this study, we showed that the decreased lifespan trajectory of mRNA expression of CNR1 in PFC and hippocampus, as well as the increased lifespan trajectory the methylation level of cg02498983 in PFC. Our data suggested the decreasing signaling of cannabinoid system during the aging in the human brain, especially PFC.
There are high rates of recreational and illicit drug use among patients with schizophrenia and mood disorders88–92. Explanations include shared genetic vulnerabilities, self-medication to mitigate the side effects of medications, and/or psychosocial factors93,94. The self-medication hypothesis proposes that self-administered substances may reduce negative symptoms. As a consequence, positive symptoms appear to be more prominent among substance-abusing patients with schizophrenia. Genetic studies have observed positive associations between CNR1 gene and substance abuse, including cannabis, alcohol, nicotine, and cocaine73,76,77,95–97. The CB1 receptor regulates the release of neurotransmitters, modulating neuronal activity throughout the CNS98,99. In patients with schizophrenia, cigarette smokers require significantly higher neuroleptic doses, and smoking is associated with a significant reduction in measurements of medication-induced Parkinsonism100. Possibly, increasing activity of ECS normalizes neurotransmission in multiple neural networks in the CNS thereby decreasing both negative symptoms and the discomforting Parkinsonian side effects causing by antipsychotic medications. Our results suggest that allelic variation in the CNR1 gene is associated with differential gene expression, leading to differential vulnerability towards the development of psychiatric disorders. As shown by our findings, abnormalities in mRNA expression in postmortem human brains support the hypothesis that dysregulation of the ECS is involved in the pathology of schizophrenia and affective disorders. It follows that the ECS is a potential therapeutic target for both schizophrenia and affective disorders.
In conclusion, we have found differential expression of CNR1 in PFC and caudate of patients with psychosis, namely schizophrenia, bipolar disorder, and MDD. While CNR1 expression declines overall with aging, there is a differential effect of COMT genotype on age-related gene expression in Caucasians. We also have identified differences in methylation in CNR1 by COMT genotype, which represent the epigenetic contributions of the environment to CNR1 expression in the course of normal human brain development and psychosis. And last, but not least, we have identified a novel transcript in CNR1, whose expression is associated with a genetic variant that also is associated with increased risk for drug addiction, a major comorbidity in both schizophrenia and affective disorders.
Supplementary information
Acknowledgements
The authors would like to extend their appreciation to the Offices of the Chief Medical Examiner of Washington DC, Northern Virginia, and Maryland, along with the University of Maryland Brain and Tissue Bank for their provision of brain tissue used in this study. Appreciation is also extended to Dr. Llewellyn B. Bigelow and the LIBD diagnostic team for their work in assembling and curating the clinical and demographic information.
Conflict of interest
We have NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ran Tao, Chao Li
These authors jointly supervised this work: Thomas M. Hyde, Joel E. Kleinman
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-020-0832-8).
References
- 1.Caulkins JP, Hawken A, Kilmer B, Kleiman MAR. Marijuana Legalization: What Everyone Needs to Know. New York, NY: Oxford University Press,; 2012. [Google Scholar]
- 2.WDR-2009. http://www.unodc.org/unodc/en/data-and-analysis/WDR-2009.html (2009).
- 3.Skosnik PD, Spatz-Glenn L, Park S. Cannabis use is associated with schizotypy and attentional disinhibition. Schizophr. Res. 2001;48:83–92. doi: 10.1016/s0920-9964(00)00132-8. [DOI] [PubMed] [Google Scholar]
- 4.Andréasson S, Allebeck P, Engström A, Rydberg U. Cannabis and schizophrenia. A longitudinal study of Swedish conscripts. Lancet. 1987;2:1483–1486. doi: 10.1016/s0140-6736(87)92620-1. [DOI] [PubMed] [Google Scholar]
- 5.Arseneault L, et al. Cannabis use in adolescence and risk for adult psychosis: longitudinal prospective study. BMJ. 2002;325:1212–1213. doi: 10.1136/bmj.325.7374.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caspi A, et al. Moderation of the effect of adolescent-onset cannabis use on adult psychosis by a functional polymorphism in the catechol-O-methyltransferase gene: longitudinal evidence of a gene X environment interaction. Biol. Psychiatry. 2005;57:1117–1127. doi: 10.1016/j.biopsych.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 7.Schoeler T, et al. Continued versus discontinued cannabis use in patients with psychosis: a systematic review and meta-analysis. Lancet Psychiatry. 2016;3:215–225. doi: 10.1016/S2215-0366(15)00363-6. [DOI] [PubMed] [Google Scholar]
- 8.Hides L, Dawe S, Kavanagh DJ, Young RM. Psychotic symptom and cannabis relapse in recent-onset psychosis: prospective study. Br. J. Psychiatry. 2006;189:137–143. doi: 10.1192/bjp.bp.105.014308. [DOI] [PubMed] [Google Scholar]
- 9.Zammit S, et al. Effects of cannabis use on outcomes of psychotic disorders: systematic review. Br. J. Psychiatry J. Ment. Sci. 2008;193:357–363. doi: 10.1192/bjp.bp.107.046375. [DOI] [PubMed] [Google Scholar]
- 10.Bossong, M. G. et al. Acute effects of ∆9-tetrahydrocannabinol (THC) on resting state brain function and their modulation by COMT genotype. Eur. Neuropsychopharmacol.10.1016/j.euroneuro.2019.03.010 (2019). [DOI] [PubMed]
- 11.Atakan Z. Cannabis, a complex plant: different compounds and different effects on individuals. Ther. Adv. Psychopharmacol. 2012;2:241–254. doi: 10.1177/2045125312457586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben Amar M, Potvin S. Cannabis and psychosis: what is the link? J. Psychoactive Drugs. 2007;39:131–142. doi: 10.1080/02791072.2007.10399871. [DOI] [PubMed] [Google Scholar]
- 13.Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- 14.Felder CC, Glass M. Cannabinoid receptors and their endogenous agonists. Annu. Rev. Pharmacol. Toxicol. 1998;38:179–200. doi: 10.1146/annurev.pharmtox.38.1.179. [DOI] [PubMed] [Google Scholar]
- 15.Pertwee RG. Pharmacology of cannabinoid receptor ligands. Curr. Med. Chem. 1999;6:635–664. [PubMed] [Google Scholar]
- 16.Heifets BD, Castillo PE. Endocannabinoid signaling and long-term synaptic plasticity. Annu. Rev. Physiol. 2009;71:283–306. doi: 10.1146/annurev.physiol.010908.163149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kano M, Ohno-Shosaku T, Hashimotodani Y, Uchigashima M, Watanabe M. Endocannabinoid-mediated control of synaptic transmission. Physiol. Rev. 2009;89:309–380. doi: 10.1152/physrev.00019.2008. [DOI] [PubMed] [Google Scholar]
- 18.Katona I, Freund TF. Multiple functions of endocannabinoid signaling in the brain. Annu. Rev. Neurosci. 2012;35:529–558. doi: 10.1146/annurev-neuro-062111-150420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill MN, et al. Suppression of amygdalar endocannabinoid signaling by stress contributes to activation of the hypothalamic-pituitary-adrenal axis. Neuropsychopharmacology. 2009;34:2733–2745. doi: 10.1038/npp.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupica CR, Riegel AC, Hoffman AF. Marijuana and cannabinoid regulation of brain reward circuits. Br. J. Pharmacol. 2004;143:227–234. doi: 10.1038/sj.bjp.0705931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranganathan M, D’Souza DC. The acute effects of cannabinoids on memory in humans: a review. Psychopharmacology. 2006;188:425–444. doi: 10.1007/s00213-006-0508-y. [DOI] [PubMed] [Google Scholar]
- 22.Zanettini C, et al. Effects of endocannabinoid system modulation on cognitive and emotional behavior. Front. Behav. Neurosci. 2011;5:57. doi: 10.3389/fnbeh.2011.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuurman L, Ippel AE, Moin E, van Gerven JMA. Biomarkers for the effects of cannabis and THC in healthy volunteers. Br. J. Clin. Pharmacol. 2009;67:5–21. doi: 10.1111/j.1365-2125.2008.03329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glass M, Dragunow M, Faull RL. Cannabinoid receptors in the human brain: a detailed anatomical and quantitative autoradiographic study in the fetal, neonatal and adult human brain. Neuroscience. 1997;77:299–318. doi: 10.1016/s0306-4522(96)00428-9. [DOI] [PubMed] [Google Scholar]
- 25.Herkenham M, Groen BG, Lynn AB, De Costa BR, Richfield EK. Neuronal localization of cannabinoid receptors and second messengers in mutant mouse cerebellum. Brain Res. 1991;552:301–310. doi: 10.1016/0006-8993(91)90096-e. [DOI] [PubMed] [Google Scholar]
- 26.Galiègue S, et al. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur. J. Biochem. 1995;232:54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x. [DOI] [PubMed] [Google Scholar]
- 27.Onaivi ES, et al. Discovery of the presence and functional expression of cannabinoid CB2 receptors in brain. Ann. N Y Acad. Sci. 2006;1074:514–536. doi: 10.1196/annals.1369.052. [DOI] [PubMed] [Google Scholar]
- 28.Acharya N, et al. Endocannabinoid system acts as a regulator of immune homeostasis in the gut. Proc. Natl Acad. Sci USA. 2017;114:5005–5010. doi: 10.1073/pnas.1612177114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Sickle MD, et al. Identification and functional characterization of brainstem cannabinoid CB2 receptors. Science. 2005;310:329–332. doi: 10.1126/science.1115740. [DOI] [PubMed] [Google Scholar]
- 30.Gong J-P, et al. Cannabinoid CB2 receptors: immunohistochemical localization in rat brain. Brain Res. 2006;1071:10–23. doi: 10.1016/j.brainres.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 31.Szabo, B. & Schlicker, E. Effects of cannabinoids on neurotransmission. Handb. Exp. Pharmacol.168, 327–365 (2005). [DOI] [PubMed]
- 32.Pertwee RG, Ross RA. Cannabinoid receptors and their ligands. Prostaglandins Leukot. Essent. Fatty Acids. 2002;66:101–121. doi: 10.1054/plef.2001.0341. [DOI] [PubMed] [Google Scholar]
- 33.Howlett AC, et al. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol. Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- 34.Freund TF, Katona I, Piomelli D. Role of endogenous cannabinoids in synaptic signaling. Physiol. Rev. 2003;83:1017–1066. doi: 10.1152/physrev.00004.2003. [DOI] [PubMed] [Google Scholar]
- 35.Ho B-C, Wassink TH, Ziebell S, Andreasen NC. Cannabinoid receptor 1 gene polymorphisms and marijuana misuse interactions on white matter and cognitive deficits in schizophrenia. Schizophr. Res. 2011;128:66–75. doi: 10.1016/j.schres.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chavarría-Siles I, et al. Cannabinoid receptor 1 gene (CNR1) and susceptibility to a quantitative phenotype for hebephrenic schizophrenia. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2008;147:279–284. doi: 10.1002/ajmg.b.30592. [DOI] [PubMed] [Google Scholar]
- 37.Martínez-Gras, I. et al. (AAT)n repeat in the cannabinoid receptor gene, CNR1: association with schizophrenia in a Spanish population. Eur. Arch. Psychiatry Clin. Neurosci. 256, 437–441 (2006). [DOI] [PubMed]
- 38.Ujike H, et al. CNR1, central cannabinoid receptor gene, associated with susceptibility to hebephrenic schizophrenia. Mol. Psychiatry. 2002;7:515–518. doi: 10.1038/sj.mp.4001029. [DOI] [PubMed] [Google Scholar]
- 39.Tsai SJ, Wang YC, Hong CJ. Association study of a cannabinoid receptor gene (CNR1) polymorphism and schizophrenia. Psychiatr. Genet. 2000;10:149–151. doi: 10.1097/00041444-200010030-00008. [DOI] [PubMed] [Google Scholar]
- 40.Seifert J, Ossege S, Emrich HM, Schneider U, Stuhrmann M. No association of CNR1 gene variations with susceptibility to schizophrenia. Neurosci. Lett. 2007;426:29–33. doi: 10.1016/j.neulet.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Leweke FM, Giuffrida A, Wurster U, Emrich HM, Piomelli D. Elevated endogenous cannabinoids in schizophrenia. Neuroreport. 1999;10:1665–1669. doi: 10.1097/00001756-199906030-00008. [DOI] [PubMed] [Google Scholar]
- 42.Dean B, Sundram S, Bradbury R, Scarr E, Copolov D. Studies on [3H]CP-55940 binding in the human central nervous system: regional specific changes in density of cannabinoid-1 receptors associated with schizophrenia and cannabis use. Neuroscience. 2001;103:9–15. doi: 10.1016/s0306-4522(00)00552-2. [DOI] [PubMed] [Google Scholar]
- 43.Zavitsanou K, Garrick T, Huang XF. Selective antagonist [3H]SR141716A binding to cannabinoid CB1 receptors is increased in the anterior cingulate cortex in schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2004;28:355–360. doi: 10.1016/j.pnpbp.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Newell KA, Deng C, Huang X-F. Increased cannabinoid receptor density in the posterior cingulate cortex in schizophrenia. Exp .Brain Res. 2006;172:556–560. doi: 10.1007/s00221-006-0503-x. [DOI] [PubMed] [Google Scholar]
- 45.Deep-Soboslay A, et al. Reliability of psychiatric diagnosis in postmortem research. Biol. Psychiatry. 2005;57:96–101. doi: 10.1016/j.biopsych.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 46.Lipska BK, et al. Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol. Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 47.Jaffe AE, et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci. 2018;21:1117. doi: 10.1038/s41593-018-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.BrainSeq: A Human Brain Genomics Consortium. Electronic address: drweinberger@libd.org, BrainSeq: A Human Brain Genomics Consortium. BrainSeq: Neurogenomics to Drive Novel Target Discovery for Neuropsychiatric Disorders. Neuron. 2015;88:1078–1083. doi: 10.1016/j.neuron.2015.10.047. [DOI] [PubMed] [Google Scholar]
- 49.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaffe AE, et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 2016;19:40–47. doi: 10.1038/nn.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kooijman MN, et al. The Generation R Study: design and cohort update 2017. Eur. J. Epidemiol. 2016;31:1243–1264. doi: 10.1007/s10654-016-0224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fried PA. The Ottawa Prenatal Prospective Study (OPPS): methodological issues and findings—it’s easy to throw the baby out with the bath water. Life Sci. 1995;56:2159–2168. doi: 10.1016/0024-3205(95)00203-i. [DOI] [PubMed] [Google Scholar]
- 56.Day NL, Leech SL, Goldschmidt L. The effects of prenatal marijuana exposure on delinquent behaviors are mediated by measures of neurocognitive functioning. Neurotoxicol. Teratol. 2011;33:129–136. doi: 10.1016/j.ntt.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dinu IR, Popa S, Bicu M, Mota E, Mota M. The implication of CNR1 gene’s polymorphisms in the modulation of endocannabinoid system effects. Rom. J. Intern. Med. 2009;47:9–18. [PubMed] [Google Scholar]
- 58.Eiland L, Romeo RD. Stress and the developing adolescent brain. Neuroscience. 2013;249:162–171. doi: 10.1016/j.neuroscience.2012.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer, H. C., Lee, F. S. & Gee, D. G. The role of the endocannabinoid system and genetic variation in adolescent brain development. Neuropsychopharmacology10.1038/npp.2017.143 (2017). [DOI] [PMC free article] [PubMed]
- 60.Zamberletti E, Rubino T, Parolaro D. The endocannabinoid system and schizophrenia: integration of evidence. Curr. Pharm. Des. 2012;18:4980–4990. doi: 10.2174/138161212802884744. [DOI] [PubMed] [Google Scholar]
- 61.Katona I, Freund TF. Endocannabinoid signaling as a synaptic circuit breaker in neurological disease. Nat. Med. 2008;14:923–930. doi: 10.1038/nm.f.1869. [DOI] [PubMed] [Google Scholar]
- 62.Maccarrone M, Guzmán M, Mackie K, Doherty P, Harkany T. Programming of neural cells by (endo)cannabinoids: from physiological rules to emerging therapies. Nat. Rev. Neurosci. 2014;15:786–801. doi: 10.1038/nrn3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hurd YL, et al. Cannabis and the developing brain: insights into its long-lasting effects. J. Neurosci. 2019;39:8250–8258. doi: 10.1523/JNEUROSCI.1165-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shire D, et al. An amino-terminal variant of the central cannabinoid receptor resulting from alternative splicing. J. Biol. Chem. 1995;270:3726–3731. doi: 10.1074/jbc.270.8.3726. [DOI] [PubMed] [Google Scholar]
- 65.Ryberg E, et al. Identification and characterisation of a novel splice variant of the human CB1 receptor. FEBS Lett. 2005;579:259–264. doi: 10.1016/j.febslet.2004.11.085. [DOI] [PubMed] [Google Scholar]
- 66.Jaffe, A. E. et al. Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nat. Neurosci.18, 154–161 (2015). [DOI] [PMC free article] [PubMed]
- 67.Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- 68.Eggan SM, Hashimoto T, Lewis DA. Reduced cortical cannabinoid 1 receptor messenger RNA and protein expression in schizophrenia. Arch. Gen. Psychiatry. 2008;65:772–784. doi: 10.1001/archpsyc.65.7.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borgan, F. et al. In vivo availability of cannabinoid 1 receptor levels in patients with first-episode psychosis. JAMA Psychiatry.10.1001/jamapsychiatry.2019.1427 (2019). [DOI] [PMC free article] [PubMed]
- 70.Guillozet-Bongaarts AL, et al. Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol. Psychiatry. 2014;19:478–485. doi: 10.1038/mp.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laprairie R, Kelly M, Denovan-Wright E. The dynamic nature of type 1 cannabinoid receptor (CB1) gene transcription. Br. J. Pharmacol. 2012;167:1583–1595. doi: 10.1111/j.1476-5381.2012.02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jaffe AE, et al. qSVA framework for RNA quality correction in differential expression analysis. Proc. Natl Acad. Sci USA. 2017;114:7130–7135. doi: 10.1073/pnas.1617384114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clarke T-K, et al. Further evidence for association of polymorphisms in the CNR1 gene with cocaine addiction: confirmation in an independent sample and meta-analysis. Addict. Biol. 2013;18:702–708. doi: 10.1111/j.1369-1600.2011.00346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuo L, et al. Interaction between two independent CNR1 variants increases risk for cocaine dependence in European Americans: a replication study in family-based sample and population-based sample. Neuropsychopharmacology. 2009;34:1504–1513. doi: 10.1038/npp.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marcos M, et al. Cannabinoid receptor 1 gene is associated with alcohol dependence. Alcohol Clin. Exp. Res. 2012;36:267–271. doi: 10.1111/j.1530-0277.2011.01623.x. [DOI] [PubMed] [Google Scholar]
- 76.Zuo L, Kranzler HR, Luo X, Covault J, Gelernter J. CNR1 variation modulates risk for drug and alcohol dependence. Biol. Psychiatry. 2007;62:616–626. doi: 10.1016/j.biopsych.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 77.Chen X, et al. Cannabinoid receptor 1 gene association with nicotine dependence. Arch. Gen. Psychiatry. 2008;65:816–824. doi: 10.1001/archpsyc.65.7.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Agrawal A, et al. Evidence for association between polymorphisms in the cannabinoid receptor 1 (CNR1) gene and cannabis dependence. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2009;150B:736–740. doi: 10.1002/ajmg.b.30881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ehlers CL, Slutske WS, Lind PA, Wilhelmsen KC. Association between single nucleotide polymorphisms in the cannabinoid receptor gene (CNR1) and impulsivity in southwest California Indians. Twin Res. Hum. Genet. 2007;10:805–811. doi: 10.1375/twin.10.6.805. [DOI] [PubMed] [Google Scholar]
- 80.Marzo VD, Stella N, Zimmer A. Endocannabinoid signalling and the deteriorating brain. Nat. Rev. Neurosci. 2015;16:30–42. doi: 10.1038/nrn3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang L, Liu J, Harvey-White J, Zimmer A, Kunos G. Endocannabinoid signaling via cannabinoid receptor 1 is involved in ethanol preference and its age-dependent decline in mice. Proc. Natl Acad. Sci. USA. 2003;100:1393–1398. doi: 10.1073/pnas.0336351100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berrendero F, et al. Changes in cannabinoid receptor binding and mRNA levels in several brain regions of aged rats. Biochim. Biophys. Acta. 1998;1407:205–214. doi: 10.1016/s0925-4439(98)00042-8. [DOI] [PubMed] [Google Scholar]
- 83.Piyanova A, et al. Age-related changes in the endocannabinoid system in the mouse hippocampus. Mech. Ageing Dev. 2015;150:55–64. doi: 10.1016/j.mad.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 84.Romero J, et al. Loss of cannabinoid receptor binding and messenger RNA levels and cannabinoid agonist-stimulated [35S]guanylyl-5’O-(thio)-triphosphate binding in the basal ganglia of aged rats. Neuroscience. 1998;84:1075–1083. doi: 10.1016/s0306-4522(97)00552-6. [DOI] [PubMed] [Google Scholar]
- 85.Pertwee RG. The pharmacology of cannabinoid receptors and their ligands: an overview. Int. J. Obes. 2006;30(Suppl 1):S13–S18. doi: 10.1038/sj.ijo.0803272. [DOI] [PubMed] [Google Scholar]
- 86.D’Addario C, et al. Epigenetic regulation of fatty acid amide hydrolase in Alzheimer disease. PLoS ONE. 2012;7:e39186. doi: 10.1371/journal.pone.0039186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bilkei-Gorzo, A. et al. A chronic low dose of Δ9-tetrahydrocannabinol (THC) restores cognitive function in old mice. Nat. Med.10.1038/nm.4311 (2017). [DOI] [PubMed]
- 88.Fowler IL, Carr VJ, Carter NT, Lewin TJ. Patterns of current and lifetime substance use in schizophrenia. Schizophr. Bull. 1998;24:443–455. doi: 10.1093/oxfordjournals.schbul.a033339. [DOI] [PubMed] [Google Scholar]
- 89.Cantwell R, et al. Prevalence of substance misuse in first-episode psychosis. Br. J. Psychiatry J. Ment. Sci. 1999;174:150–153. doi: 10.1192/bjp.174.2.150. [DOI] [PubMed] [Google Scholar]
- 90.Quello SB, Brady KT, Sonne SC. Mood disorders and substance use disorder: a complex comorbidity. Sci. Pract. Perspect. 2005;3:13–21. doi: 10.1151/spp053113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davis L, Uezato A, Newell JM, Frazier E. Major depression and comorbid substance use disorders. Curr. Opin. Psychiatry. 2008;21:14–18. doi: 10.1097/YCO.0b013e3282f32408. [DOI] [PubMed] [Google Scholar]
- 92.Swartz MS, et al. Substance use and psychosocial functioning in schizophrenia among new enrollees in the NIMH CATIE study. Psychiatr. Serv. 2006;57:1110–1116. doi: 10.1176/ps.2006.57.8.1110. [DOI] [PubMed] [Google Scholar]
- 93.Kavanagh DJ, McGrath J, Saunders JB, Dore G, Clark D. Substance misuse in patients with schizophrenia: epidemiology and management. Drugs. 2002;62:743–755. doi: 10.2165/00003495-200262050-00003. [DOI] [PubMed] [Google Scholar]
- 94.Winklbaur B, Ebner N, Sachs G, Thau K, Fischer G. Substance abuse in patients with schizophrenia. Dialogues Clin. Neurosci. 2006;8:37–43. doi: 10.31887/DCNS.2006.8.1/bwinklbaur. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benyamina A, Kebir O, Blecha L, Reynaud M, Krebs M-O. CNR1 gene polymorphisms in addictive disorders: a systematic review and a meta-analysis. Addict. Biol. 2011;16:1–6. doi: 10.1111/j.1369-1600.2009.00198.x. [DOI] [PubMed] [Google Scholar]
- 96.Gerra MC, et al. Gene variants and educational attainment in cannabis use: mediating role of DNA methylation. Transl. Psychiatry. 2018;8:23. doi: 10.1038/s41398-017-0087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Evans DE, et al. Cannabinoid receptor 1 (CNR1) gene variant moderates neural index of cognitive disruption during nicotine withdrawal. Genes Brain. Behav. 2016;15:621–626. doi: 10.1111/gbb.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kendall, D. A. & Yudowski, G. A. Cannabinoid receptors in the central nervous system: their signaling and roles in disease. Front. Cell Neurosci.10.3389/fncel.2016.00294 (2017). [DOI] [PMC free article] [PubMed]
- 99.Zou, S. & Kumar, U. Cannabinoid receptors and the endocannabinoid system: signaling and function in the central nervous system. Int. J. Mol. Sci.10.3390/ijms19030833 (2018). [DOI] [PMC free article] [PubMed]
- 100.Goff DC, Henderson DC, Amico E. Cigarette smoking in schizophrenia: relationship to psychopathology and medication side effects. Am. J. Psychiatry. 1992;149:1189–1194. doi: 10.1176/ajp.149.9.1189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.