Like commensal bacteria, commensal protozoa are an integral part of the vertebrate intestinal microbiome. How protozoa integrate into a commensal bacterium-enriched ecosystem remains poorly studied. Here, using the murine commensal Tritrichomonas musculis as a proof of concept, we studied potential factors involved in shaping the intestinal protozoal-bacterial community. Understanding the rules by which microbes form a multispecies community is crucial to prevent or correct microbial community dysfunctions in order to promote the host’s health or to treat diseases.
KEYWORDS: microbiota, protozoa, dietary fiber, cross-kingdom communication, intestinal colonization
ABSTRACT
Our current understanding of the host-microbiota interaction in the gut is dominated by studies focused primarily on prokaryotic bacterial communities. However, there is an underappreciated symbiotic eukaryotic protistic community that is an integral part of mammalian microbiota. How commensal protozoan bacteria might interact to form a stable microbial community remains poorly understood. Here, we describe a murine protistic commensal, phylogenetically assigned as Tritrichomonas musculis, whose colonization in the gut resulted in a reduction of gut bacterial abundance and diversity in wild-type C57BL/6 mice. Meanwhile, dietary nutrient and commensal bacteria also influenced the protozoan’s intestinal colonization and stability. While mice fed a normal chow diet had abundant T. musculis organisms, switching to a Western-type high-fat diet led to the diminishment of the protozoan from the gut. Supplementation of inulin as a dietary fiber to the high-fat diet partially restored the protozoan’s colonization. In addition, a cocktail of broad-spectrum antibiotics rendered permissive engraftment of T. musculis even under a high-fat, low-fiber diet. Furthermore, oral administration of Bifidobacterium spp. together with dietary supplementation of inulin in the high-fat diet impacted the protozoan’s intestinal engraftment in a bifidobacterial species-dependent manner. Overall, our study described an example of dietary-nutrient-dependent murine commensal protozoan-bacterium cross talk as an important modulator of the host intestinal microbiome.
IMPORTANCE Like commensal bacteria, commensal protozoa are an integral part of the vertebrate intestinal microbiome. How protozoa integrate into a commensal bacterium-enriched ecosystem remains poorly studied. Here, using the murine commensal Tritrichomonas musculis as a proof of concept, we studied potential factors involved in shaping the intestinal protozoal-bacterial community. Understanding the rules by which microbes form a multispecies community is crucial to prevent or correct microbial community dysfunctions in order to promote the host’s health or to treat diseases.
INTRODUCTION
The mammalian gut microbiome consists of a wide consortium of microbes from diverse phyla, including viruses, prokaryotic bacteria and archaea, and eukaryotic microbes. The different microorganisms in the gut are thought to be able to attain a homeostatic equilibrium state via functional interactions that contribute to the health and diseases of the host (1). Eukaryotic protists can be an integral part of the mammalian gut microbiome (2, 3), but how the protozoal species interact with other microbial community members in the gut remains poorly defined.
Trichomonads are flagellated anaerobic protozoa that belong to the phylum Parabasalia and that often have symbiotic relationships with their hosts (4). Recently, the finding of multiple Tritrichomonas species from the order Trichomonadida as commensal microbes in healthy rodents across many animal facilities inspired research to identify their importance in the host-microbe cross talk (3, 5–8). Merad and colleagues identified a murine commensal protozoan, Tritrichomonas musculis, that can enhance antibacterial defenses and increase intestinal inflammation via triggering inflammasome activation in the gut epithelial cell, promoting interleukin-18 (IL-18) production and Th1/Th17 cell differentiation (3). However, some Tritrichomonas species have instead been found to be able to trigger a type 2 immune response by producing succinate, which directly engages the succinate receptor that is expressed on intestinal tuft cells to produce cytokine IL-25 (5–8). Currently, we do not know the actual phylogenetic relationships among these multiple reported Tritrichomonas species and what distinguishes their induction of a type 1 versus type 2 response. Whether it is related to potential gut bacterial microbiota differences originating from different animal facilities is unclear. Furthermore, we still lack fundamental knowledge of how commensal protozoa interact with other microbial community members in general.
In this study, we described a critical contribution of the rodent commensal protist parabasalid T. musculis in shaping the ecology of the gut bacterial community. Meanwhile, T. musculis colonization and stability in the gut were greatly influenced by dietary nutrients and commensal bacteria. These results uncovered the involvement of a previously underappreciated cross-kingdom interaction between a protist and commensal bacteria in shaping the murine intestinal microbial ecosystem.
RESULTS
Identification of a gut eukaryotic symbiont in mice.
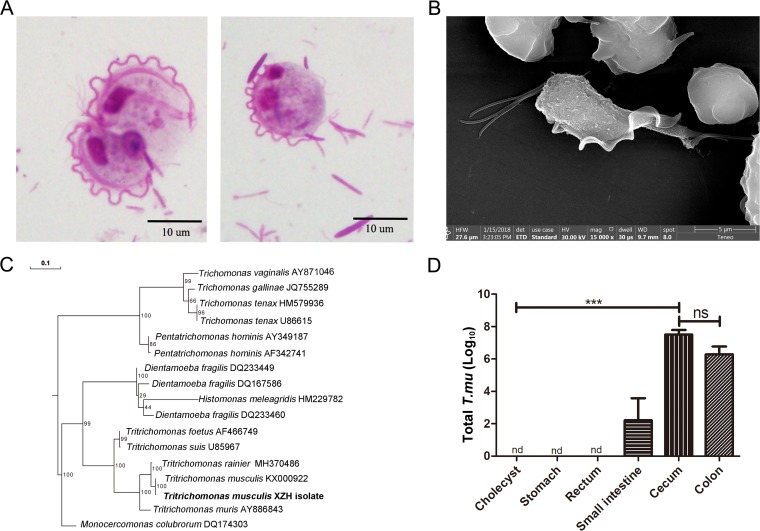
We screened cecal materials obtained from our in-house mouse colonies and microscopically detected the presence of unicellular flagellated microbes (Fig. 1A). We purified and imaged them by scanning electron microscopy (SEM) and identified them as tritrichomonads (Fig. 1B). Internal transcribed spacer (ITS) and 18S rRNA genomic DNA sequencing confirmed them to be a Tritrichomonas sp. phylogenetically identical to T. musculis (>99% homology) (Fig. 1C and supplemental data).
FIG 1.
Identification of a gut protozoan symbiont. (A) Representative microscopic image of the cecal content from protozoan-bearing mice, stained by Giemsa stain; scale bar, 10 μm. (B) Representative SEM image of purified protozoa. (C) Phylogenetic tree analysis according to the DNA sequences derived from the ITS rRNA region. The individual protozoal ITS sequence obtained from the NCBI GenBank database is indicated by its original species name followed by its accession number. The protozoan isolate that we found is in bold type. The evolutionary history was inferred using the RAxML method. Branch supports were computed out of 100 bootstrap trees. (D) The total number of T. musculis protozoa in the indicated tissues. The experiments were performed 2 times. The error bars represent standard deviations (n = 5 in each group). ***, P < 0.001; ns, no statistical significance; nd, not detected; T. mu, T. musculis.
T. musculis was found mostly in the gastrointestinal tract, with the highest number in the cecum and colon. It was not present in the hepatobiliary system (Fig. 1D). In the intestine, it was mostly found inside the lumen without adherence to the intestinal epithelial cell surface. Neonates born to T. musculis-colonized dams were initially T. musculis free and became T. musculis positive after weaning (see Fig. S1 in the supplemental material). Furthermore, T. musculis-negative adult B6 mice from Beijing Vital River Laboratory Animal Technology became T. musculis positive 2 weeks after being cohoused with our in-house T. musculis-bearing mice. These results suggested that the transmission of T. musculis was horizontal and most likely via the fecal-oral route.
T. musculis colonization decreased gut bacterium abundance and diversity.
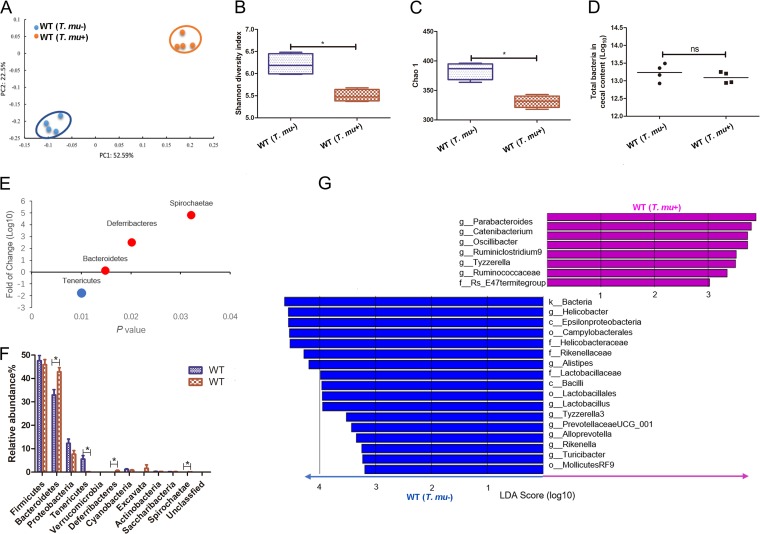
Although eukaryotic protozoa have been recognized as important members of the gut microbiota, the protozoan-commensal bacterium interactions remain largely unknown. We questioned how T. musculis colonization might influence the gut bacterial community. To this end, T. musculis-negative wild-type (WT) B6 mice from Beijing Vital River Laboratory Animal Technology were cohoused either with T. musculis-negative WT B6 mice, designated WT(T. musculis–), or with WT B6 mice that were orally administered with purified T. musculis, designated WT(T. musculis+). Four weeks after cohousing, the mice in the WT(T. musculis–) group remained T. musculis free, while the original T. musculis-negative mice in the WT(T. musculis+) group became T. musculis positive. We then collected the cecal contents from those mice, estimated the total number of bacteria in them using real-time quantitative PCR (RTqPCR) of the total bacterial 16S rRNA genes, and analyzed their bacterial 16S rRNA sequences using high-throughput DNA sequencing. Our results indicated that there were significant bacterial phylogenetic differences between T. musculis-colonized and -uncolonized mice based on unweighted UniFrac principal coordinate analysis (PCoA) of the bacterial 16S rRNA gene sequences (Fig. 2A). T. musculis colonization significantly reduced gut commensal bacterial abundance and diversity as indicated by Shannon and Chao 1 indices (Fig. 2B and C); however, the total number and concentrations of the cecal bacteria were not changed (Fig. 2D). At the phylum level, there was a relative decrease in Tenericutes and an increase of Bacteroidetes, Deferribacteres, and Spirochaetes (Fig. 2E and F). Based on linear discriminant analysis effect size (LEfSe), the family Rs-E47 termite group, which was previously found in higher termites (9), was determined to be significantly expanded following T. musculis colonization, and at the same time the families Helicobacteraceae, Rikenellaceae, and Lactobacillaceae were significantly reduced (Fig. 2G). These data suggested that there were potentially both cooperative and competitive cross-kingdom communications between gut bacteria and the protozoa.
FIG 2.
T. musculis colonization decreased gut bacterial abundance and diversity. (A) PCoA based on OTU abundance of indicated mice via cecal bacterial 16S rRNA sequencing. Each symbol represents one individual mouse. (B and C) The commensal bacterial α-diversity indicated by the Shannon index (B) and Chao1 index (C). (D) The total number of bacteria determined by quantitative real-time PCR. (E) The fold change of the indicated bacterial phyla’s abundance comparing T. musculis-bearing versus -nonbearing WT mice. (F) The relative bacterial abundance of the indicated taxonomic composition at the phylum level in the indicated mice. (G) LEfSe analysis to determine the alterations of the cecal bacteria after T. musculis colonization. In each group, n = 4 mice. The error bars represent standard deviations. *, P < 0.05; ns, no statistical significance; T. mu, T. musculis.
Western-type high-fat diet influence on T. musculis intestinal colonization.
Dietary changes are often accompanied by concomitant changes in the gut microbiota. Our modern Western-type high-fat low-fiber diet contributes to the loss of bacterial taxa and reduced commensal bacterial diversity in both humans and mice (10, 11). However, the impact of dietary shifts on the eukaryotic protozoan’s symbiotic membership in the context of a complex microbiota remains unclear. We observed that T. musculis-positive WT B6 mice that were initially fed a normal chow diet (ND) became T. musculis negative after switching to a high-fat diet (HFD) for 2 weeks. This suggests that an HFD can induce a loss of T. musculis.
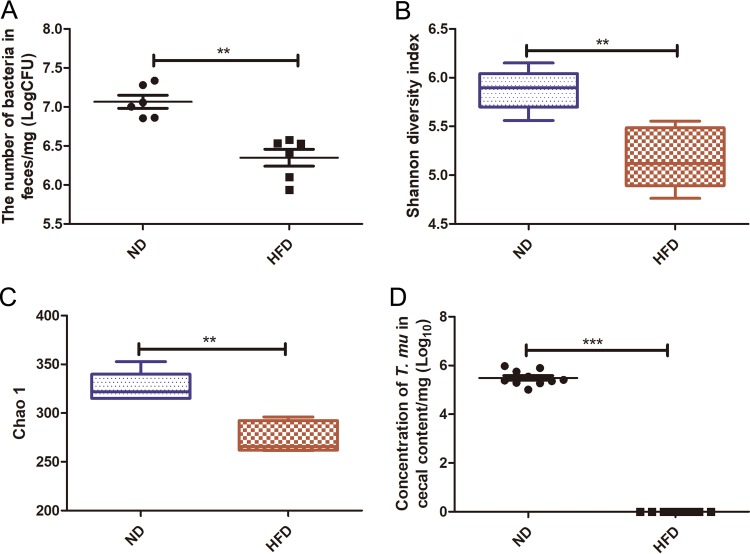
To further prove this notion, T. musculis-negative WT B6 mice fed ND were orally administered purified T. musculis (1 × 106 T. musculis protists/mouse) on day 0, and on the next day one group of mice was kept on ND and the other group was switched to HFD. Two weeks later, the cecal content was collected, the T. musculis presence was examined microscopically, and the bacterial community changes were determined using bacterial 16S rRNA gene sequencing. HFD feeding not only reduced the total bacterial number (Fig. 3A) but also significantly decreased the diversity and abundance of the bacterial community (Fig. 3B and C); meanwhile, the number of T. musculis organisms also dramatically declined (Fig. 3D). These results suggested that HFD reduced T. musculis colonization ability in addition to that of some commensal bacteria.
FIG 3.
HFD reduced T. musculis colonization ability. T. musculis-negative WT B6 mice fed ND were orally administered purified T. musculis on day 0, and on the next day the food was switched to the specific type of diet as indicated. Two weeks later, the cecal contents were collected. (A) The bacterial concentration in the cecal content. (B and C) The commensal bacterial α-diversity indicated by the Shannon index (B) and Chao1 index (C). (D) The concentration of T. musculis in the cecal content. The experiments were performed at least 2 times. The error bars represent standard deviations (n = 5 to 9 in each group). **, P < 0.01; ***, P < 0.001; T. mu, T. musculis.
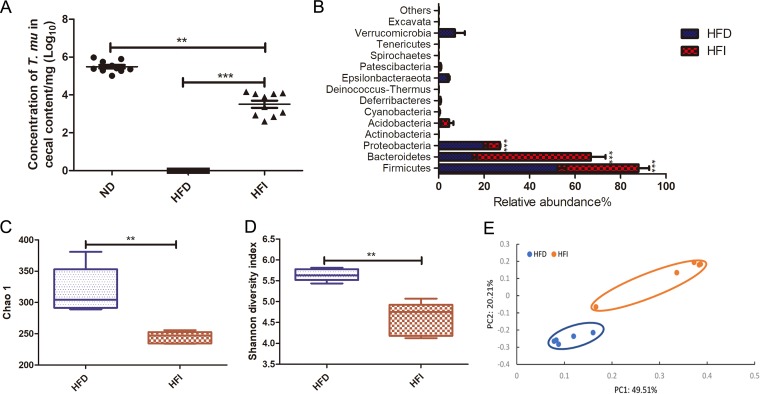
The Western-type high-fat diet has a relatively lower quantity of dietary fiber. An uncharacterized murine Tritrichomonas sp. has been reported to be able to utilize complex dietary fibers to establish colonization (8). To test whether the reduced amount of fiber in the HFD is also responsible for the diminishment of our T. musculis isolate, 10% fermentable oligofructan fiber inulin was supplemented to the HFD, and then the modified diet (HFD plus inulin [HFI]) was used to feed the mice. The T. musculis colonization ability was significantly improved when the mice were fed HFI relative to those fed HFD (Fig. 4A). Interestingly, inulin supplementation failed to restore the gut bacterial community (Fig. 4B to E). At the phylum level, mice fed HFI had more Bacteroidetes and fewer Firmicutes and Proteobacteria than the mice fed HFD (Fig. 4B), and the inulin supplementation further decreased the commensal bacterial community richness and diversity (Fig. 4C and D). These results together suggest that our T. musculis isolate requires a certain amount of dietary fermentable fiber for successful intestinal colonization. Too little dietary fiber in the food (e.g., HFD) cannot support T. musculis engraftment and can even endanger its stability in a complex microbial ecosystem.
FIG 4.
HFD-induced T. musculis colonization reduction was at least partially due to the limited amount of fermentable dietary fiber in the food. T. musculis-negative WT B6 mice fed ND were orally administered purified T. musculis on day 0, and on the next day, the food was switched to the specific type of diet as indicated. Two weeks later, the cecal contents were collected, and the presence of T. musculis was examined microscopically. (A) The concentration of T. musculis per milligram of cecal contents from the mice fed the indicated types of diet. The experiments were performed 3 times, and there was a total of 9 to 10 mice in each group. (B) The relative abundance of cecal bacterial phyla from the mice fed HFD or HFI. (C) Shannon index showing commensal bacterial community diversity. (D) Chao1 index showing commensal bacterial community richness. (E) PCoA based on OTU abundance of the cecal bacteria. Each symbol represents one individual mouse. In panels B to D, there were 5 mice in each group. The error bars represent standard deviations. **, P < 0.01; ***, P < 0.001; HFD, high-fat diet; HFI, HFD plus 10% inulin; T. mu, T. musculis.
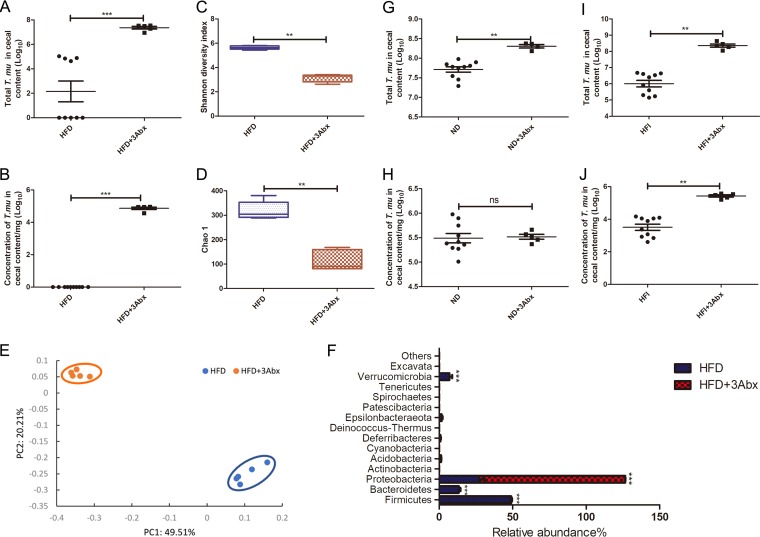
A cocktail of broad-spectrum antibiotics improves T. musculis colonization.
The balance of cooperation and competition both within and between microbial populations is a key determinant of microbial dynamics (12). To test the potential role of commensal bacteria in regulating T. musculis intestinal colonization, we treated the mice fed HFD with a cocktail of broad-spectrum antibiotics (vancomycin, neomycin-sulfate, and ampicillin) after administering a single dose of T. musculis via the oral route. The antibiotic treatment resulted in the reestablishment of T. musculis colonization in the gut under HFD conditions (Fig. 5A and B). The overall bacterial community richness and diversity both declined after antibiotic treatment (Fig. 5C and D), which was as expected. The antibiotic treatment diminished almost all the bacterial phyla except Proteobacteria (Fig. 5F). These data suggest that the reason for HFD-induced T. musculis diminishment is not solely a limitation of dietary fiber, and there is likely a competitive interaction between bacteria and protozoa to regulate T. musculis engraftment. Furthermore, the antibiotics’ effect of boosting T. musculis colonization is independent of the food type. A similar phenomenon was found in mice fed either ND (Fig. 5G and H) or HFI (Fig. 5I and J), suggesting a common mechanism of regulating T. musculis colonization.
FIG 5.
An antibiotic cocktail treatment improved T. musculis colonization. (A and B) The total number (A) and concentration (B) of T. musculis in the cecal content were quantified in WT B6 mice fed HFD with or without antibiotic treatment. (C and D) The α-diversity of gut commensal bacteria in the gut ecosystem with or without antibiotic treatment was indicated by the Shannon index (C) and Chao1 index (D). (E) PCoA based on OTU abundance of the cecal bacteria. Each symbol represents one individual mouse. (F) Relative abundance of the indicated taxonomic compositions at the phyla level in the WT mice with or without antibiotic treatment. (G and H) The total number (G) and concentration (H) of T. musculis in the cecal content from WT B6 mice fed ND with or without antibiotic treatment. (I and J) The total number (I) and concentration (J) of T. musculis in the cecal content from WT B6 mice fed HFI with or without antibiotic treatment. The experiments were repeated 2 times. A total of 5 to 10 mice were included in each group in panels A, B, and G to J, and a total of 5 mice in each group were included in panels C to F. The error bars represent the standard deviation. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no statistical significance; 3Abx, ampicillin plus vancomycin plus neomycin; ND, normal chow diet; HFD, high-fat diet; HFI, HFD plus 10% inulin; T. mu, T. musculis.
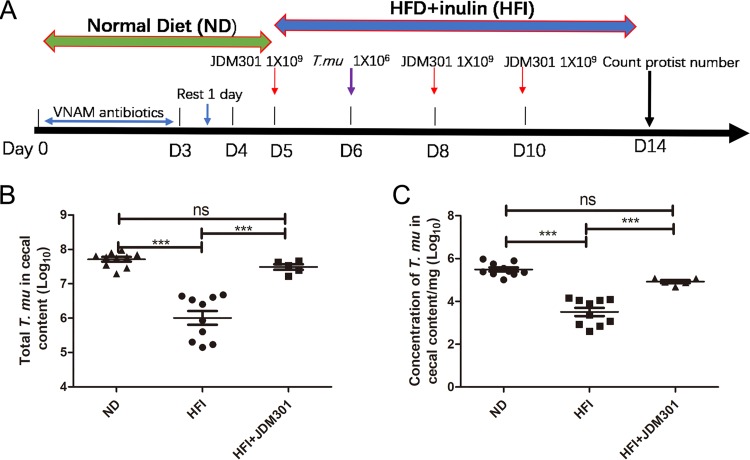
Some Bifidobacterium spp. can impact T. musculis colonization.
To further test the functional interactions between bacteria and T. musculis, we were interested in determining possible cross talk between specific bacterial species and T. musculis. Since inulin can sustain bifidobacterial growth and activity (13, 14), we tested the role of Bifidobacterium in T. musculis colonization in an inulin-enriched ecosystem. We orally administered Bifidobacterium longum JDM301 (1 × 109 CFU in 100 μl phosphate-buffered saline [PBS]/mouse) along with T. musculis to WT mice (Fig. 6A). B. longum JDM301 promoted T. musculis colonization under inulin-enriched HFD conditions (Fig. 6B and C). In contrast, administration of Bifidobacterium adolescentis (ATCC 15703) on a similar schedule inhibited T. musculis colonization (Fig. S2). Although the underlying mechanism is not clear, the data suggest that bacteria could impact T. musculis colonization, likely in a species- or even strain-specific manner, at least under certain dietary contexts.
FIG 6.
Bifidobacterium impacted T. musculis colonization in the inulin-enriched environment. (A) Schematic diagram of the experimental design. (B) The total number of T. musculis in the cecal content from the mice fed either ND, HFI, or HFI plus JDM301. (C) The T. musculis concentration in the cecal content. The experiments were performed twice. The error bars represent standard deviations (n = 5 to 10). ***, P < 0.001; ND, normal diet; HFI, high-fat diet plus 10% inulin; T. mu, T. musculis.
DISCUSSION
A lot of factors can influence gut microbiota diversity, composition, and function, including diet (11, 15), antibiotics (16), age (17), inflammation (18), and host genetics (19). In this study, we provided an example of cross-kingdom microbial communication as another force to drive gut microbiome dynamics. We showed that a murine commensal T. musculis colonization could strongly affect the gut prokaryotic bacterial community composition. Meanwhile, bacterial communities could also potentially provide constant competitive pressure on the protist. Antibiotic treatment could potentially release this pressure and favor the protozoan colonization. We also provided evidence of cooperative support of some bacterial spp. for T. musculis engraftment when the dietary nutrient conditions permitted it. These interkingdom commensal protozoan-bacterium interactions might be important in forming a functional microbiota that influences host physiopathology. In agreement with our notion, it has been shown that there is a direct association between the presence of Blastocystis and shifts in the gut bacterial and eukaryotic microbiome in people that do not have gastrointestinal disease or inflammation (20).
Multiple species of Tritrichomonas spp. have been isolated from different groups, with little to no knowledge of their actual taxonomic relationships, and they have been shown to be able to induce either type 1 or type 2 responses (3, 6, 8, 21). Although different Tritrichomonas spp. might function differently, uncharacterized potential gut bacterial microbiota differences raised by different animal facilities could also possibly influence which type of immune response to mount. Furthermore, Tritrichomonas and bacteria might interact and function as a whole. In accordance with this notion, it has been reported that the ability of helminths to modulate allergic inflammation is dependent on gut bacterial microbiota (22).
In our experiment, accompanied by T. musculis colonization, the abundances of Bacteroidetes, Deferribacteres, and Spirochaetes were increased. These positive correlations indicate potential cooperative relationships between these bacteria and Tritrichomonas. Interestingly, Bacteroidetes, Deferribacteres, and Spirochaetaes are characteristic members of the microbiota in the gut of wood-feeding termites and Cryptocercus cockroaches (23–25). Some members of the Bacteroidales in termite gut are symbionts of flagellated protozoa (26). Spirochete bacteria and wood-digesting protists were identified in the intestinal tissue of Miocene termites (27). In the lower termite gut, Spirochaetes are important in reductive acetogenesis to remove hydrogen produced by protistic symbionts (28). Together with our finding of expanded Bacteroidetes and Spirochaetaes in the murine gut after exposure to T. musculis, this indicates that there is a potentially very conservative rule of protozoan-bacterium interaction across various kinds of hosts. As these hosts have very different dietary habits, what determines this conservative symbiotic relationship requires further investigation.
It has been shown that the gut bacterial community rapidly and consistently responds to altered host diet (11, 29). Numerous studies have shown that a high-fat diet has strong effects on the abundance and diversity of the gut bacterial community, including an increase in the ratio of Firmicutes to Bacteroidetes and significantly decreased bacterial load in the host gut. Accordingly, the genetic composition and metabolic activity of gut microbiota changed when the host was exposed to HFD (30). Here, we showed that HFD could also dramatically influence the stability of the murine eukaryotic protist T. musculis in a complex commensal ecosystem. Thus, the modern Western-type diet can potentially change not only prokaryotic but also eukaryotic taxa. It will be interesting to further explore how these ecological changes in microbiota might influence host functions.
The reason for the instability of T. musculis under HFD conditions is related to at least two factors, dietary fiber and gut bacteria. We found that our T. musculis isolate required fermentable dietary fibers for its colonization and stability in the gut. This is similar to an uncharacterized Tritrichomonas sp. which Schneider described previously (8). Dietary fibers are the primary energy source of murine Tritrichomonas. However, there are additional factors to regulate Tritrichomonas colonization. By using antibiotics to remove bacteria, we found that T. musculis could engraft successfully even under food fiber-limited conditions. Thus, a potential cross-kingdom competitive interaction between commensal T. musculis and bacteria might be present, and this interaction might determine a dietary nutrient threshold for the protist to integrate into the gut community. Our data suggested that when the food fibers were scarce, T. musculis and commensal bacteria might compete for the limited resource for survival, and the net outcome was that T. musculis lost the war in a competitive ecosystem. However, we could not easily rule out other possibilities, since antibiotics have profound effects not only on bacteria but also on the host. Further work is needed to determine whether and which host factors impact T. musculis intestinal colonization.
Interestingly, when food fiber was provided in a large quantity, a bacterial species-specific interaction with T. musculis was revealed. B. longum JDM301 and B. adolescentis showed a competitive and cooperative effect, respectively. Thus, the growth of T. musculis in the gut ecosystem might be dependent on a delicate balance between competitive and cooperative relationships with other microbial community members. Bifidobacterium spp. in general can take up and degrade fructooligosaccharides (31), which could compete with T. musculis for the same food resources when food fiber is limited. Dietary fibers are the primary energy sources of many gut bacteria (32), and thus they could all be able to compete constantly with T. musculis for growth. Why B. longum JDM301 is supportive of T. musculis colonization under our experimental conditions is unclear. One possibility is that B. longum JDM301 might be somewhat unique in degrading inulin-type fructans. It has been reported that slow preferential degradation of the short-chain-length oligofructose intracellularly by B. adolescentis LMG 10734 enabled Faecalibacterium prausnitzii growth, while fast nonpreferential degradation of all chain-length fractions of oligofructose extracellularly and efficient degradation of the short-chain-length fractions of inulin by Bifidobacterium angulatum LMG 11039T and B. longum LMG 11047 inhibited F. prausnitzii growth (13). In any case, we propose that the relationships between bacteria and protozoa might be flexibly maintained in a context-dependent manner.
In previous reports, HFD significantly decreased Bifidobacterium abundance, while inulin supplementation increased Bifidobacterium and Anaerostipes abundances (33). In our study, when 10% inulin was added to HFD, no significant changes of Bifidobacterium and Anaerostipes abundances were found. It might be that 2 weeks of inulin supplementation is too short to restore Bifidobacterium and Anaerostipes abundances under the HFD diet. In addition, the supplementation of inulin to HFD induced a further reduction of the diversity of the intestinal bacterial community, which was consistent with a previous report (34). Thus, in contrast to T. musculis, the roles of complex dietary fibers in commensal bacteria are very sophisticated and less well understood. The dietary compositions, such as different types of fat and fibers, might all potentially be involved in regulating commensal bacterial community stabilities. This requires future investigation.
In summary, our data suggest the influence of dietary-nutrient-dependent functional protozoan-bacterium interactions in shaping a murine symbiotic commensal protozoan integration into gut microbiota. These results are the first step toward an understanding of the functional connections between commensal protists, bacteria, and the host.
MATERIALS AND METHODS
Animals and feeding conditions.
All animal experiments in this study were performed following the guidelines of the National Laboratory Animal Ethics Committee of China and were approved by the Animal Care and Use Committee of Xuzhou Medical University. C57BL/6 male mice (6 to 8 weeks old) free of T. musculis were purchased from Beijing Vital River Laboratory Animal Technology Co. Ltd., Beijing, China. The mice were fed irradiated food and autoclaved drinking water. All mice had free access to food and drinking water and were maintained under specific pathogen-free (SPF) conditions with a 12-h light phase and a 12-h dark phase. The high-fat diet (HFD), in which 60% of the energy is from fat, and HFD supplemented with 10% inulin (HFI) were purchased from Beijing Keao Xieli Feed Co. Ltd., Beijing, China. The HFD ingredient is as follows: casein, 200 g; l-cysteine, 3 g; maltodextrin, 125 g; sucrose, 68.8 g; cellulose, 50 g; soybean oil, 25 g; lard, 245 g; mineral mix S10026, 10 g; dicalcium phosphate, 13 g; calcium carbonate, 5.5 g; potassium citrate, 16.5 g; vitamin mix V1001, 10 g; choline bitartrate, 2 g; and FD&C blue dye no. 1, 0.05 g. When needed, a cocktail of broad-spectrum antibiotics (500 mg/liter ampicillin, 250 mg/liter vancomycin, and 500 mg/liter neomycin sulfate) was given to the mice in the drinking water for 2 weeks.
Purification of T. musculis from cecal content.
The cecal contents of T. musculis-containing mice were harvested into sterile PBS and filtered several times through a 70-μm-pore-size cell strainer. The suspension was spun at 1,000 rpm for 5 min at 4°C. The pellet was washed twice with PBS. T. musculis was further purified at the interface of a 40%/80% Percoll gradient. For in vivo administration, each mouse was inoculated with approximately 1 × 106 T. musculis protists via the oral route.
Bacterial strains and growth conditions.
Bifidobacterium longum JDM 301 was originally isolated from a commercial probiotic product from China (35). B. adolescentis (ATCC 15703) was purchased from the BeNa Culture Collection (http://www.bncc.org.cn). B. longum JDM301 and B. adolescentis were grown at 37°C in de Man, Rogosa, and Sharpe medium (MRS; BD Difco, NJ, USA) supplemented with 0.05% (wt/vol) l-cysteine-HCl (MRS-cys) or on MRS-cys containing agar (MRS-cys/agar) in an anaerobic workstation (Don Whitley Scientific, Bingley, UK). B. longum JDM301 and B. adolescentis in the exponential growth phase were harvested by centrifugation and washed three times with PBS. The pellets were resuspended in sterile phosphate saline buffer (PBS) (0.144 g KH2PO4/liter, 9 g NaCl/liter, 0.795 g Na2HPO4/liter, pH 7.5). The concentrations of B. longum and B. adolescentis cultures were adjusted to 1010 CFU/ml based on the turbidity. The bacterial counts were determined by plating serial dilutions of the cultures on MRS-cys/agar.
Scanning electron microscopy.
The cecal content containing T. musculis was collected and suspended in PBS. Then, the suspension was filtered through a 70-μm-pore-size cell strainer and spun and 1,500 rpm and 4°C for 5 min. After centrifugation, the supernatant was discarded, and the pellet was washed twice with PBS. After the final wash, the pellet was suspended and fixed with fixative (2.5% glutaraldehyde plus 4% paraformaldehyde) in phosphate buffer (PB) for 2 h at room temperature, followed by three washings of 5 min each with PB. Samples were dehydrated using the following series of ethanol-water mixtures: 25%, 50%, 70%, 80%, 90%, 100%, and 100% (7 steps). The replacement of the liquid medium was done in the wells of the plate. After treatment with 100% ethanol, the samples were incubated in isoamyl acetate twice for 15 min each time. The samples were dried in a critical-point dryer (HCP-2; Hitachi Ltd., Japan). Then, the samples were attached to the sample table, and gold coating was performed in a high-vacuum coating instrument (EM ACE600; Leica, Germany). After gold coating, the samples were observed using a Teneo VS scanning electron microscope (FEI, Hillsboro, OR, USA).
Quantification of T. musculis protozoa in the cecal content.
The cecum of mice was cut longitudinally and weighed. The harvested cecal content was suspended in PBS (10 μl/mg). The protist was counted using a hemocytometer as previously reported (3). The total number and concentration of T. musculis organisms per milligram were calculated accordingly.
Quantitative PCR (qPCR) was performed to determine the relative abundance of T. musculis in neonatal mice born to mothers with or without protozoan colonization. Total DNA was isolated from the cecal content. Autoclaved water was used as a protozoan-negative control. The primer sequences used to identify T. musculis were TTAGTAAGTGCGACCGAAGA (forward) and TAAGGCAGCATTCTCAAGC (reverse). qPCR was performed using the FastStart universal SYBR green master (Roche, Basel, Switzerland) and according to the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
Quantification of bacteria in the cecal content.
For the total bacterial quantification, DNA was isolated from the cecal content by using the QIAmp Fast DNA stool minikit (Qiagen, Hilden, Germany). The isolated DNA was used as the template for qPCR with the FastStart universal SYBR green master (Roche, Basel, Switzerland). The primers targeting the universal bacterial 16S rRNA gene were used according to a previous report (36). To convert qPCR threshold cycle (CT) values into bacterium numbers, we used the genomic DNA isolated from a known amount of B. longum JDM301 as a template to generate a standard curve by qPCR. The number of B. longum JDM301 bacteria was counted by plating serial dilutions of cultures on MRS-cys/agar (0.05% [wt/vol]) before extracting genomic DNA.
Phylogenetic analysis.
DNA was isolated using the QIAmp fast DNA stool minikit (Qiagen, Hilden, Germany) from the enriched protozoa from the cecal content with 40%/80% Percoll gradient centrifugation. Then, the 18S rRNA region was amplified using primers (forward [F]: TGCGCTACCTGGTTGATCCTGCC; reverse [R]: TGATCCTTCTGCAGGTTCACCTAC). The internal transcribed spacer (ITS) region was amplified using primers (F: TTAGTAAGTGCGACCGAAGA; R: TAAGGCAGCATTCTCAAGC). The PCR product was sequenced, and the sequence is presented in the supplemental data. Alignment and phylogenetic reconstructions were performed using the “build” function of ETE3 v3.1.1 (37) as implemented on GenomeNet (https://www.genome.jp/tools/ete/). The maximum likelihood (ML) tree was inferred using RAxML v8.1.20 run with the model GTRGAMMA and default parameters (38). Branch supports were computed out of 100 bootstrap trees.
Bacterial diversity analysis of the gut ecosystem.
The analysis was done by Nanjing Aurora Gene Technology Co. Ltd. (Nanjing, China). In brief, fresh cecal content was collected and weighed. The DNA of the cecal content was extracted using the E.Z.N.A. stool DNA kit (Omega Bio-Tek, Norcross, Georgia) according to the manufacturer’s instructions. The DNA of the cecal content was used as the template for PCR targeting the V3-V4 region of the bacterial 16S rRNA gene (39). The PCR was performed using the following conditions: 95°C for 3 min, followed by 27 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s, and finally ending at 72°C for 10 min. Reactions were performed in triplicate in a 20-μl mixture containing 4 μl of 5 × FastPfu buffer, 2 μl of 2.5 mM deoxynucleosides triphosphates (dNTPs), 0.8 μl of each primer (5 μM total), 0.4 μl of FastPfu polymerase, and 10 ng of template DNA. The PCR products were extracted from 2% agarose gels and cleaned with agarose gel purification. Purified PCR products were quantified using Qubit v3.0 (Life Invitrogen, California, USA), and all 24 amplicons whose barcodes were different were mixed equally. The pooled DNA product was used to construct the Illumina paired-end library following Illumina’s genomic DNA library preparation procedure. Then, the amplicon library was paired-end sequenced (2 × 250) on an Illumina HiSeq 2500 sequencing system according to the standard protocols.
Raw fastq files were demultiplexed and quality-filtered using QIIME v1.17 with the following criteria. (i) The 250-bp reads were truncated at any site receiving an average quality score of <20 over a 10-bp sliding window, and the truncated reads that were shorter than 50 bp were discarded. (ii) Reads were removed if they matched the exact barcode sequences, they had two nucleotide mismatches in primer matching, or they contained ambiguous characters. (iii) Only sequences that overlapped more than 10 bp were assembled according to their overlap sequence. Reads which could not be assembled were discarded. Sequences sharing 97% nucleotide sequence identity were binned into the same operational taxonomic units (OTUs) using UPARSE v7.1, and chimeric sequences were identified and removed using UCHIME in both reference mode (using the GOLD database) and de novo mode.
Sequences were deposited in the NCBI SRA database under accession number PRJNA542986. A taxonomical classification was performed using uclust against the SILVA (SSU123) 16S rRNA database (40). Based on the abundance data of OTUs, bacterial taxa summarization and rarefaction analyses of microbial diversity (dissimilarity value indicated by unweighted UniFrac distance) were then conducted, and PCoA plots were generated in QIIME.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism v5 software. Differences between multiple groups were compared using an analysis of variance (ANOVA) test with post hoc Bonferroni correction. Mann-Whitney’s test was used for comparisons between two groups. In microbiota analysis, the Wilcoxon signed rank test and Kruskal-Wallis (KW) sum-rank test were used. A P value of <0.05 was considered significant.
Data availability.
The raw bacterial 16S rRNA reads have been deposited in the SRA database under accession number PRJNA542986. Our T. musculis isolate’s ITS and 18S rRNA sequences are presented in the supplemental data.
Supplementary Material
ACKNOWLEDGMENTS
We declare no conflict of interest.
Project support was provided in part by the National Natural Science Foundation of China (grant no. 81770853), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) 2014 (grant no. KYLX14-1448), the Natural Science Foundation of the Higher Education Institutions of Jiangsu Province, China (grant no. 16KJB310016), and the Starting Foundation for Talents of Xuzhou Medical University (grant no. D2016029).
Y. Wang and Y. Wei designed the study; Y. Wei, J.G., Y.K., M.L., L.M., X.Z., H.S., and Y. Wang performed the experiments; Y. Wang, Z.L., and Y. Wei analyzed the data; and Y. Wang and Y. Wei wrote the paper. All of the authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. 2017. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol 15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 2.Lukeš J, Stensvold CR, Jirků-Pomajbíková K, Wegener Parfrey L. 2015. Are human intestinal eukaryotes beneficial or commensals? PLoS Pathog 11:e1005039. doi: 10.1371/journal.ppat.1005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, Remark R, Mogno I, Ng R, Gnjatic S, Amir ED, Solovyov A, Greenbaum B, Clemente J, Faith J, Belkaid Y, Grigg ME, Merad M. 2016. Host-protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell 167:444–456. doi: 10.1016/j.cell.2016.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maritz JM, Land KM, Carlton JM, Hirt RP. 2014. What is the importance of zoonotic trichomonads for human health? Trends Parasitol 30:333–341. doi: 10.1016/j.pt.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, Artis D, Garrett WS. 2016. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351:1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Fontana MF, Erle DJ, Anderson MS, Locksley RM, Raftery D, von Moltke J. 2018. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49:33–41.e37. doi: 10.1016/j.immuni.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei W, Ren W, Ohmoto M, Urban JF Jr, Matsumoto I, Margolskee RF, Jiang P. 2018. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci U S A 115:5552–5557. doi: 10.1073/pnas.1720758115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider C, O’Leary CE, von Moltke J, Liang H-E, Ang QY, Turnbaugh PJ, Radhakrishnan S, Pellizzon M, Ma A, Locksley RM. 2018. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell 174:271–284. doi: 10.1016/j.cell.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diouf M, Roy V, Mora P, Frechault S, Lefebvre T, Hervé V, Rouland-Lefèvre C, Miambi E. 2015. Profiling the succession of bacterial communities throughout the life stages of a higher termite Nasutitermes arborum (Termitidae, Nasutitermitinae) using 16S rRNA gene pyrosequencing. PLoS One 10:e0140014. doi: 10.1371/journal.pone.0140014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. 2016. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat Rev Microbiol 4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 13.Moens F, Weckx S, De Vuyst L. 2016. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int J Food Microbiol 231:76–85. doi: 10.1016/j.ijfoodmicro.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 15.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. 2014. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 17.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slingerland AE, Schwabkey Z, Wiesnoski DH, Jenq RR. 2017. Clinical evidence for the microbiome in inflammatory diseases. Front Immunol 8:400. doi: 10.3389/fimmu.2017.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. 2016. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieves-Ramirez ME, Partida-Rodriguez O, Laforest-Lapointe I, Reynolds LA, Brown EM, Valdez-Salazar A, Moran-Silva P, Rojas-Velazquez L, Morien E, Parfrey LW, Jin M, Walter J, Torres J, Arrieta MC, Ximenez-Garcia C, Finlay BB. 2018. Asymptomatic intestinal colonization with protist blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems 3:e00007-18. doi: 10.1128/mSystems.00007-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escalante NK, Lemire P, Cruz Tleugabulova M, Prescott D, Mortha A, Streutker CJ, Girardin SE, Philpott DJ, Mallevaey T. 2016. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J Exp Med 213:2841–2850. doi: 10.1084/jem.20161776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaiss MM, Rapin A, Lebon L, Dubey LK, Mosconi I, Sarter K, Piersigilli A, Menin L, Walker AW, Rougemont J, Paerewijck O, Geldhof P, McCoy KD, Macpherson AJ, Croese J, Giacomin PR, Loukas A, Junt T, Marsland BJ, Harris NL. 2015. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 43:998–1010. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai V, James ER, Nalepa CA, Scheffrahn RH, Perlman SJ, Keeling PJ. 2015. The role of host phylogeny varies in shaping microbial diversity in the hindguts of lower termites. Appl Environ Microbiol 81:1059–1070. doi: 10.1128/AEM.02945-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietrich C, Kohler T, Brune A. 2014. The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl Environ Microbiol 80:2261–2269. doi: 10.1128/AEM.04206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brune A, Dietrich C. 2015. The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu Rev Microbiol 69:145–166. doi: 10.1146/annurev-micro-092412-155715. [DOI] [PubMed] [Google Scholar]
- 26.Noda S, Inoue T, Hongoh Y, Kawai M, Nalepa CA, Vongkaluang C, Kudo T, Ohkuma M. 2006. Identification and characterization of ectosymbionts of distinct lineages in Bacteroidales attached to flagellated protists in the gut of termites and a wood-feeding cockroach. Environ Microbiol 8:11–20. doi: 10.1111/j.1462-2920.2005.00860.x. [DOI] [PubMed] [Google Scholar]
- 27.Wier A, Dolan M, Grimaldi D, Guerrero R, Wagensberg J, Margulis L. 2002. Spirochete and protist symbionts of a termite (Mastotermes electrodominicus) in Miocene amber. Proc Natl Acad Sci U S A 99:1410–1413. doi: 10.1073/pnas.022643899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brune A. 2014. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 29.Griffin NW, Ahern PP, Cheng J, Heath AC, Ilkayeva O, Newgard CB, Fontana L, Gordon JI. 2017. Prior dietary practices and connections to a human gut microbial metacommunity alter responses to diet interventions. Cell Host Microbe 21:84–96. doi: 10.1016/j.chom.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan Y, Wang F, Yuan J, Li J, Jiang D, Zhang J, Li H, Wang R, Tang J, Huang T, Zheng J, Sinclair AJ, Mann J, Li D. 2019. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: a 6-month randomised controlled-feeding trial. Gut 68:1417–1429. doi: 10.1136/gutjnl-2018-317609. [DOI] [PubMed] [Google Scholar]
- 31.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A 99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawicki CM, Livingston KA, Obin M, Roberts SB, Chung M, McKeown NM. 2017. Dietary fiber and the human gut microbiota: application of evidence mapping methodology. Nutrients 9:125. doi: 10.3390/nu9020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandeputte D, Falony G, Vieira-Silva S, Wang J, Sailer M, Theis S, Verbeke K, Raes J. 2017. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 66:1968–1974. doi: 10.1136/gutjnl-2016-313271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerezuela R, Fumanal M, Tapia-Paniagua ST, Meseguer J, Moriñigo MÁ, Esteban MÁ. 2013. Changes in intestinal morphology and microbiota caused by dietary administration of inulin and Bacillus subtilis in gilthead sea bream (Sparus aurata L.) specimens. Fish Shellfish Immunol 34:1063–1070. doi: 10.1016/j.fsi.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 35.Wei YX, Zhang ZY, Liu C, Zhu YZ, Zhu YQ, Zheng H, Zhao GP, Wang S, Guo XK. 2010. Complete genome sequence of Bifidobacterium longum JDM301. J Bacteriol 192:4076–4077. doi: 10.1128/JB.00538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA. 2009. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci U S A 106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay JM, Langella P, Xavier RJ, Sokol H. 2016. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M, Gillis M, Wilson BA, Nelson KE, White BA, Leigh SR. 2013. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J 7:1344–1353. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw bacterial 16S rRNA reads have been deposited in the SRA database under accession number PRJNA542986. Our T. musculis isolate’s ITS and 18S rRNA sequences are presented in the supplemental data.