Abstract
Respiratory viruses affect us throughout our lives, from infancy to old age, causing illnesses ranging from a common cold to severe pneumonia. They belong to several virus families, and although many features of infection with these diverse viruses are shared, some have unique characteristics. Here we explain what happens when we are infected by respiratory viruses, including SARS-CoV-2, which causes COVID-19.
Respiratory viruses affect us throughout our lives, from infancy to old age, causing illnesses ranging from a common cold to severe pneumonia. They belong to several virus families, and although several features of infection with these diverse viruses are shared, some have unique characteristics. Here we explain what happens when we are infected by respiratory viruses, including SARS-CoV-2, which causes COVID-19.
Main Text
Basic Concepts about Respiratory Viruses
Viral respiratory infections result when a virus infects the cells of the respiratory mucosa; this can occur when virus particles are inhaled or directly contact a mucosal surface of the nose or eyes. Infected individuals shed virus into the environment by coughing or sneezing or even during quiet breathing. Virus shed during coughing and sneezing is often present in large droplets that fall out of the air within a short distance. If the virus falls on a surface and survives, it can be transmitted when someone touches the infected surface and then touches their nose, eyes, or mouth. Virus is also spread by the airborne route in the form of small (<5 μm) droplet nuclei that can remain suspended for long periods of time and can be inhaled into the lower respiratory tract. The relative contribution of different particle sizes and of direct contact versus airborne transmission as a means of spread differs among the respiratory viruses. Influenza viruses are spread by contact as well as by airborne transmission, but the mode of transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is still being discussed. Some viruses are more fragile than others; for example, respiratory syncytial virus (RSV), the most important respiratory virus in early childhood, does not survive for long on inanimate surfaces, whereas coronaviruses are much more stable in the environment (van Doremalen et al., 2020). Infection control and prevention strategies are designed based on these features. Cough etiquette, hand hygiene, and surface decontamination are effective control measures for a virus that is spread by direct contact and large droplets. Preventing airborne spread via droplet nuclei requires use of special measures such as P2/N95 masks or respirators and special air handling.
What Happens When the Virus Reaches the Respiratory Mucosa?
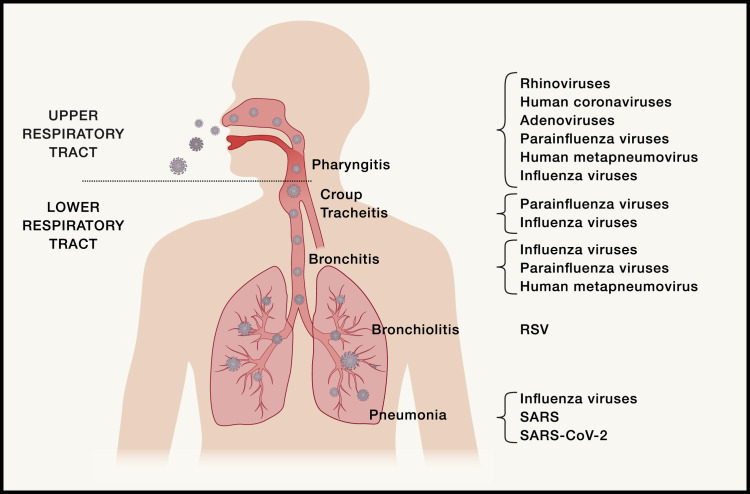
The respiratory epithelium is composed of a variety of cells that include ciliated and non-ciliated epithelial cells; goblet cells, which produce mucus that forms the first barrier for an incoming virus; and club cells, which produce proteases. Different respiratory viruses preferentially bind and infect ciliated or non-ciliated epithelial cells of the airways: pneumocytes lining the alveoli in the lungs and alveolar macrophages (Matrosovich et al., 2004). For example, avian influenza A viruses infect ciliated epithelial cells, whereas human influenza viruses infect non-ciliated epithelial cells. The presence of specific host cell molecules that are receptors for viral attachment and entry are the main determinants of which cells become infected. Human angiotensin-converting enzyme 2 (ACE2) is the receptor for SARS-CoV and SARS-CoV-2 (Zhou et al., 2020) as well as for human coronavirus NL63. The receptor for the Middle East respiratory syndrome (MERS) coronavirus is human dipeptidyl peptidase 4 (DPP4), and human coronavirus 229E uses aminopeptidase N as its receptor. All influenza viruses use sialic acid as their receptor, but there is added subtlety; human influenza viruses bind sialic acids with α2,6-linked oligosaccharides, whereas avian influenza viruses bind sialic acids with α2,3-linked oligosaccharides (reviewed in Paules and Subbarao, 2017). The presence of cells in the respiratory tract expressing the relevant viral receptor is critical for initiation of viral infection, and the clinical presentation depends on where these cells are situated in the respiratory tract. For example, if cells bearing the viral receptor are only present in the upper respiratory tract, then infection is likely to be limited to rhinitis (characterized by a runny nose and stuffiness) or pharyngitis (sore throat) (Figure 1 ). In contrast, if the viral receptor is present on cells in the lower respiratory tract (e.g., epithelial cells expressing α2,3-linked sialic acids beyond the respiratory bronchioles in the lungs), then the infecting virus (e.g., avian influenza A(H5N1) virus) causes lower respiratory tract infection.
Figure 1.
Schematic Illustration of the Human Respiratory Tract, Indicating the Clinical Presentations Associated with Different Respiratory Viruses that Infect Particular Parts of the Upper and Lower Respiratory Tract
After attachment to the receptor, the virus gains entry into the cell, and the viral genome is uncoated, releasing the viral genetic material, which is RNA in paramyxoviruses, orthomyxoviruses, and coronaviruses and DNA in adenoviruses. Viral transcription to produce viral proteins and viral replication to copy the viral genome are complex events unique to each viral family and occur in specific cellular compartments, although all viruses have crucial interactions with host cell proteins. Replication of paramyxoviruses, including RSV and parainfluenza viruses, occurs in the nucleus and cytoplasm. Influenza viruses are replicated entirely within the nucleus, and they utilize a unique strategy of “stealing” methylated capped ends of host cell messenger RNA (mRNA) as primers for viral mRNAs. The replication cycle of coronaviruses occurs entirely in the cytoplasm and involves generation of a series of subgenomic RNAs. Progeny virions are released from the infected cell into the respiratory tract, where they are shed by coughing and sneezing.
When host cells are infected by viruses, type I interferons (IFNs) and pro-inflammatory cytokines are expressed, cellular translation is suppressed, and an antiviral state is induced. However, many respiratory viruses are able to block IFN activation and/or signaling and inhibit apoptosis. This prevents the host from effectively clearing virally infected cells and also induces an antiviral state in neighboring cells, promoting viral replication in infected tissues that may contribute to the observed pathology.
What Are the Consequences of Infection in the Host?
When a person is infected with a respiratory virus, there is an incubation period before onset of clinical signs and symptoms. During the incubation period, the virus attaches to and infects cells, replicates its genome, and spreads to infect adjacent cells. The incubation period for influenza is short, typically 1–2 days, whereas for SARS-CoV-2, it is 4.5–5.8 days (Lauer et al., 2020). Productive viral infection of respiratory epithelial cells results in clinical symptoms and signs that depend on which part of the respiratory tract is infected (Figure 1). Infection of the nasal, nasopharyngeal, and oropharyngeal mucosa causes a runny nose, coughing, sneezing, and sore throat, whereas tracheobronchitis or croup presents with a characteristic barking seal-like cough. Bronchitis refers to inflammation of the bronchi and presents with a cough; bronchiolitis, involving the smaller distal airways, is characteristic of RSV infection in young infants and presents with wheezing. Pneumonia occurs when infection and inflammation involve the alveoli and lung parenchyma and is associated with a cough and shortness of breath. Rhinovirus, adenovirus, and human coronavirus infections are usually limited to the upper respiratory tract. Parainfluenza viruses cause croup, RSV causes bronchiolitis and influenza, and SARS, MERS, and SARS-CoV-2 can cause pneumonia (Figure 1). Sometimes respiratory viral infections are complicated by secondary bacterial infection, particularly in the middle ear (otitis media) or lungs (pneumonia). The best example of this is secondary bacterial infection caused by Streptococcus pneumoniae or Staphylococcus aureus following influenza virus infection. During the 1918 and 2009 influenza pandemics, a large proportion of the deaths resulted from secondary bacterial pneumonia.
The Immune Response to Respiratory Viruses
The immune system responds to viral infection with cellular and humoral (antibodies, complement, and antimicrobial peptides) responses. These responses are initiated by the innate immune system, which recognizes pathogens and induces production of proinflammatory cytokines and chemokines. This is followed by responses of the adaptive immune system, which consists of T cells, which can directly kill virus-infected cells, and B cells, which produce pathogen-specific antibodies in the serum and at mucosal surfaces (Chen and Subbarao, 2007).
Innate responses are activated when macrophages encounter damage-associated molecular patterns (DAMPs), such as intracellular contents released from dying cells or proteins released by tissue injury, or pathogen-associated molecular patterns (PAMPs), such as viral RNA or oxidized phospholipids from invading pathogens. DAMPs and PAMPs released during initial infection and lysis of pneumocytes (surface epithelial cells of the alveoli) activate multiple innate immune pathways through Toll-like receptors (TLRs), NLRP3 and/or inflammasome activation, or triggering of cytoplasmic DNA sensors such as RIG-I-MAVS and cGAS-STING (reviewed in Vardhana and Wolchok, 2020). The signal transduction that follows drives production of cytokines that activate antiviral gene expression programs in neighboring cells. They also recruit additional innate and adaptive immune cells with diverse roles in antiviral immunity, tissue repair, and homeostasis. Dysregulation of the innate immune response contributes to the cytokine storm seen in severe influenza A/H5N1 and SARS-CoV-2 infection.
Virus-encoded peptides stimulate activation, proliferation, and differentiation of naive CD8+ and CD4+ T cells. Effective viral clearance requires CD8+ effector T cell-mediated killing of virally infected cells as well as CD4+ T cell-dependent enhancement of CD8+ and B cell responses. Serum immunoglobulin G (IgG) is the dominant antibody involved in protection from respiratory viruses in the lower respiratory tract, whereas mucosal IgA plays an important role in immunity in the upper respiratory tract. Although cellular and humoral responses contribute to clearance of primary infection, neutralizing antibodies play a critical role in protection against re-infection. Immunological memory is maintained by T and B cell subsets (Sant and McMichael, 2012).
Persistence or excessive antigenic stimulation of T and B cells can induce a nonresponsive state known as T- or B cell “exhaustion.” This phenotype has been reported in many chronic viral infections and is frequently associated with lymphopenia. In these settings, lung-resident and circulating T cells upregulate specific markers of T cell exhaustion, such as PD-1, Tim-3, MKI67, and TYMS (Blank et al., 2019). Notably, lymphopenia and T cell exhaustion have been observed in recent viral epidemics caused by SARS and pandemic H1N1 influenza.
What Happens in COVID-19?
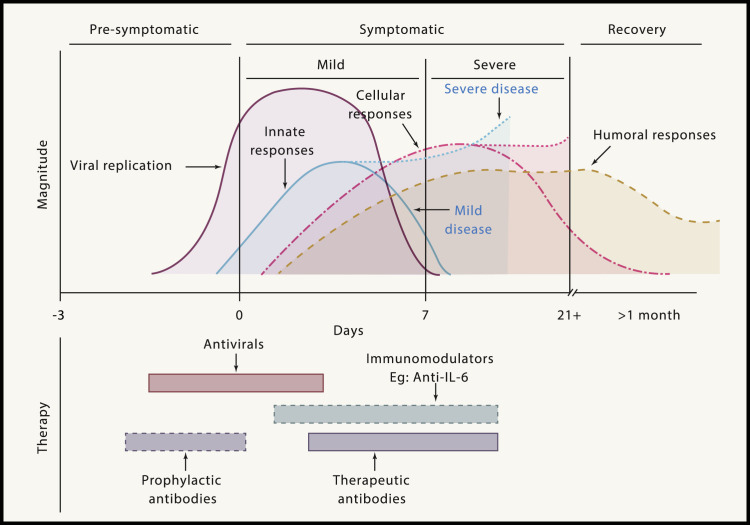
After an incubation period, infection progresses from the pre-symptomatic stage (1–3 days) through symptomatic infection (2–4 weeks) to a prolonged post-symptomatic or recovery stage (2–8 weeks) (Figure 2 ). The spectrum of disease with SARS-CoV-2 ranges from asymptomatic infection to severe, often fatal disease. Patients with mild disease (80%) have fever, cough, sore throat, loss of smell, headache, and body aches. Moderate illness is characterized by involvement of the lower respiratory tract. In influenza, the amount of infecting virus (inoculum) correlates with greater severity of infection. It is not yet known whether this is true for SARS-CoV-2; notably, asymptomatic patients with coronavirus disease 2019 (COVID-19; 10%–50% of infections) can excrete large quantities of virus (Vardhana and Wolchok, 2020).
Figure 2.
Schematic Illustration of the Phases of COVID-19, Showing the Kinetics of SARS-CoV-2 Replication and the Innate and Adaptive Immune Responses
The illness can be divided into three phases: pre-symptomatic, symptomatic, and recovery. Viral replication occurs during the late pre-symptomatic and early symptomatic phases. Innate immune responses follow, including activation of macrophages, natural killer (NK) cells, dendritic cells, and pro-inflammatory cytokine secretion. In severe disease, innate responses are exaggerated and remain elevated, contributing to tissue damage. Cellular immune responses with antigen-specific B and T cells are detected within the first week of infection in mild to moderate COVID-19 cases (Thevarajan et al., 2020). The bottom panel illustrates optimal timing of potential interventions based on viral and immune kinetics. Immunomodulators are genetically engineered or organism-derived proteins that target specific parts of the immune system.
Severe infection and complications occur more frequently in males and those with underlying medical conditions (called co-morbidities), including hypertension, diabetes, cardiovascular disease, and immunocompromised states. Findings associated with poor outcomes include an increase in white blood cell counts with lymphopenia, a prolonged prothrombin time (indicating coagulopathy), and elevated levels of liver enzymes, lactate dehydrogenase, D-dimer, interleukin-6, C-reactive protein, and procalcitonin (Gandhi et al., 2020).
The pathogenesis of severe SARS-CoV-2 infection is complex, and we are still learning about it at the time of this writing. The viral spike glycoprotein binds to the cell surface receptor ACE2 (Hoffmann et al., 2020), which plays an important regulatory role in the renin-angiotensin-aldosterone system (RAAS). The physiological role of ACE2 is in regulation of blood pressure; ACE2 metabolizes angiotensin II, a vasoconstrictor, to generate angiotensin 1-7, which is a vasodilator. Cells lining the mucosal surfaces of the nose and lungs are endowed with ACE2, which facilitates infection of the respiratory tract. However, ACE2 is also expressed on cells in many other tissues, including the endothelium, heart, gut, and kidneys, making these organs susceptible to infection by the virus.
A prominent feature of severe SARS-CoV-2 infections is the cytokine release syndrome. SARS-CoV-2 infection of respiratory epithelial cells activates monocytes, macrophages, and dendritic cells, resulting in secretion of a range of proinflammatory cytokines, including interleukin-6 (IL-6). IL-6 release instigates an amplification cascade that directly results in T helper 17 (Th17) differentiation, among other lymphocytic changes. Circulating IL-6 and soluble IL-6 receptor complexes indirectly activate many cell types, including endothelial cells, resulting in a flood of systemic cytokine production that contributes to hypotension and acute respiratory distress syndrome (ARDS). The key role of IL-6 in this cascade has prompted evaluation of IL-6 antagonists such as tocilizumab, sarilumab, and siltuximab for treatment of severe COVID-19 disease (Moore and June, 2020).
In addition to increased vascular permeability, the cytokine “storm” also leads to high levels of fibrinogen and activation of the coagulation cascade on endothelial surfaces of small blood vessels, signaled by very high levels of a fibrin breakdown product called D-dimer. Dysregulation of the RAAS because of competitive binding of ACE2 by the virus may also induce constriction of blood vessels. A puzzling phenomenon has been seen in COVID-19 pneumonia: some patients have extremely low blood oxygen levels, but they do not complain of breathlessness. It has been suggested that oxygen uptake in COVID-19 pneumonia is impeded because of clogged and constricted blood vessels in the lungs rather than because of congestion from accumulation of edema fluid in the alveoli, as seen in other viral pneumonias. The benefit of anticoagulants in the treatment of severe COVID-19 disease is therefore of great interest. It is becoming clear that the pathogenetic cascade triggered by SARS-CoV-2 infection damages many organs in the body, from the kidneys to the brain. Such pathophysiological changes are rarely seen with other respiratory virus infections.
Strategies for Treatment of SARS-CoV-2 Infection
There are no licensed drugs that specifically target SARS-CoV-2 (or other) coronaviruses. The scale of the COVID-19 pandemic, with more than 3 million cases and 230,000 deaths in 185 countries in a 4-month period, has driven an urgent search for drugs with antiviral activity against SARS-CoV-2. The fastest path to finding an effective treatment may be to repurpose licensed drugs that are approved for use in humans for other indications.
Drugs that target specific viral proteins or critical host cell processes have been selected for evaluation against SARS-CoV-2 in vitro and in clinical trials (Figure 2). More than 300 clinical trials have been registered around the world (listed on ClinicalTrials.gov), but not all are randomized controlled trials (RCTs), the gold standard in clinical research. In RCTs, participants are allocated by chance alone (randomly) to receive an intervention or drug or one among two or more drugs or interventions; randomization minimizes bias in measuring the effectiveness of a new intervention or treatment. Some of the drugs that were evaluated are the HIV protease inhibitor combination of lopinavir-ritonavir, chloroquine, hydroxychloroquine, and remdesivir. Lopinavir-ritonavir and hydroxychloroquine have not shown clear clinical benefits to date, although some RCTs are still ongoing. Clinical improvement regarding need for oxygen support and mechanical ventilation have been reported with use of remdesivir in a compassionate use study, where the drug was used in patients with severe disease outside of a clinical trial when no other treatments were available. A placebo-controlled RCT of remdesivir in China failed to demonstrate a statistically significant clinical benefit in treated patients (Wang et al., 2020), but an interim analysis from a large international clinical trial revealed that treatment with remdesivir reduced the duration of hospitalization by an average of 4 days. A preliminary report of the data is now available, and the drug has been approved for treatment of COVID-19 in the US (Beigel et al., 2020).
Immunomodulators may ameliorate severe COVID-19 and could be useful as adjuncts to antiviral agents (Figure 2). Cytokine release syndrome and the observation of elevated levels of serum IL-6 and other proinflammatory cytokines in patients with severe COVID-19 has prompted trials of drugs that inhibit the IL-6 pathway, such as tocilizumab and sarilumab. Antibodies that can block the infectivity of the virus can be derived from the plasma of recovered patients or from monoclonal antibodies generated from B cells from recovered patients or phage display libraries. Convalescent plasma has been used in SARS and severe influenza (Mair-Jenkins et al., 2015) and COVID-19 in non-randomized trials alone or as an adjunct to antiviral drugs for treatment of severe disease (Figure 2). Antibodies can also be administered to prevent infection in individuals who are known to be at risk of infection (Figure 2), referred to as immunoprophylaxis.
The optimal timing for administration of antiviral drugs, immunomodulators, and therapeutic antibodies should be guided by the phase of the disease and the kinetics of viral replication and immune response (Figure 2).
Recap
A diverse group of viruses, including coronaviruses, rhinoviruses, RSV, parainfluenza, and influenza viruses, target the respiratory tract, cause a range of infections from rhinitis to pneumonia, and represent a large global burden of disease. The viruses infect epithelial cells of the respiratory tract after binding distinct cell surface receptors, and infection rapidly triggers innate immune responses. However, viruses employ several strategies to inhibit antiviral immunity in the host. T cell responses are critical for viral clearance, and antibody responses directed against respiratory viruses can protect against re-infection, although reinfection occurs over the course of a lifetime with many respiratory viruses. COVID-19 refers to the respiratory illness caused by SARS-CoV-2, a zoonotic coronavirus that emerged in China in 2019 and has rapidly spread around the world in a pandemic. COVID-19 pneumonia differs from that caused by other respiratory viruses in the severity and duration of inflammation, which appears to be driven by an exuberant innate immune response, a flood of proinflammatory cytokines, and multiorgan dysfunction, driven by derangements in pulmonary and clotting physiology. As the pandemic evolves, we are gaining a better understanding of the complex pathophysiology underlying COVID-19 disease. Treatment strategies currently under study include antiviral agents, immunomodulators, antibodies, and adjunctive therapies.
Acknowledgments
The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health.
References
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., ACTT-1 Study Group Members Remdesivir for the Treatment of Covid-19 — Preliminary Report. The New England Journal of Medicine. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PubMed] [Google Scholar]
- Blank C.U., Haining W.N., Held W., Hogan P.G., Kallies A., Lugli E., Lynn R.C., Philip M., Rao A., Restifo N.P. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019;19:665–674. doi: 10.1038/s41577-019-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Subbarao K. The Immunobiology of SARS∗. Annu. Rev. Immunol. 2007;25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- Gandhi R.T., Lynch J.B., Del Rio C. Mild or Moderate Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMcp2009249. Published online April 24, 2020. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.M., Lim W.S., Makki S., Rooney K.D., Nguyen-Van-Tam J.S., Beck C.R., Convalescent Plasma Study Group The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M.N., Matrosovich T.Y., Gray T., Roberts N.A., Klenk H.D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Paules C., Subbarao K. Influenza. Lancet. 2017;390:697–708. doi: 10.1016/S0140-6736(17)30129-0. [DOI] [PubMed] [Google Scholar]
- Sant A.J., McMichael A. Revealing the role of CD4(+) T cells in viral immunity. J. Exp. Med. 2012;209:1391–1395. doi: 10.1084/jem.20121517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J. Exp. Med. 2020;217:e20200678. doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]