Abstract
Lung cancer remains one of the leading causes of cancer deaths around the world. Previous studies have shown that microRNAs have pivotal functions in tumorigenesis including lung cancer. It is reported that microRNA-195-5p acts as a tumor suppressor role in human cancers. However, the function and molecular mechanism of microRNA-195-5p in lung cancer progression is still unclear. In the present study, the results showed that the expression of microRNA-195-5p was downregulated both in lung cancer tissues and in lung cancer cell lines. Enhanced expression of microRNA-195-5p inhibited cell proliferation, migration, and invasion in lung cancer cells. Furthermore, Forkhead box k1 was identified as the direct target of microRNA-195-5p. Forkhead box k1 overexpression could restore the repressed cell proliferation and metastasis caused by microRNA-195-5p overexpression. Our results demonstrated that a functional mechanism of microRNA-195-5p in regulating lung cancer. It indicates that microRNA-195-5p may regulate lung cancer growth and metastasis through the regulation of Forkhead box k1, highlighting the potential application for the treatment of lung cancer in the future.
Keywords: miR-195-5p, FOXK1, lung cancer, proliferation, migration, invasion
Background
Lung cancer is one of the most common cancers and one of the leading causes of cancer deaths in the world.1,2 The dominant subtype of lung cancer is non-small cell lung cancer (NSCLC), which includes adenocarcinoma, squamous carcinoma, and large cell carcinoma accounting for almost 80% of all lung cancers. Despite treatment for lung cancer such as new drugs and therapeutic regimens has been made great improvement, to date, the overall 5-year survival rate of patients with NSCLC is still low.3 Thus, it is essential to investigate the underlying molecular mechanisms of NSCLC growth, development, and progression.
MicroRNAs (miRNAs) are a class of small, noncoding RNAs with 20 to 24 nucleotides in length, which regulate genes expression through binding to the 3′-untranslated region (3′-UTR) of targeted mRNAs.4-6 MicroRNAs play important roles in various physiological and developmental conditions. Some miRNAs are dysregulated in many human cancers, functioning as oncogene or tumor suppressor roles in tumorigenesis.7-11 MicroRNA-195-5p (miR-195-5p) has been reported to be a cancer regulator in multiple cancers such as colon cancer,12 gastric cancer,13 and so on, yet the function of miR-195-5p and the underlying mechanism in regulating lung cancer still remains to be investigated.
Forkhead box k1 (FOXK1), a member of the FOX transcription factor family,14 has been reported to be involved in tumorigenesis,15 including colorectal cancer,16 esophageal cancer,17 and hepatocellular carcinoma.18 However, it is not clear whether FOXK1 is directly associated with miR-195-5p in regulating NSCLC development.
In this study, we first examined the expression levels of miR-195-5p in both lung cancer tissues and lung cancer cell lines H1299 and A549. Then, the effects of miR-195-5p on lung cancer growth and metastasis were determined. Moreover, we explored whether FOXK1 was a direct target of miR-195-5p and performed restoration experiments. In sum, our results highlight the underlying mechanisms of miR-195-5p/FOXK1 regulation in lung cancer.
Materials and Methods
Clinical Samples and Cell Lines
Tissue samples of lung cancer and paired normal tissues were obtained from patients who received surgical treatment at the First Affiliated Hospital of Peking University. This study was approved by the Human Research Ethics Committee from the First Affiliated Hospital of Peking University (PKDYFS201706206). Informed consent was obtained from all patients. The human lung cell line BEAS-2B and lung cancer cell lines (H1299, A549) were obtained from Shanghai Institute of Cell Biology, China Academy of Sciences (Shanghai, China).
Cell Culture and Cell Transfection
All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Sigma, MO, USA) supplemented with 10% fetal bovine serum (Sigma, MO, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Pen/Strep; Sigma, MO, USA). Cells were incubated at 37°C in an atmosphere of 5% CO2. Cells were transfected with FOXK1 (Addgene), miR-195-5p mimic (Ribobio) or miR-NC using Lipofectamine 3000 reagent (Invitrogen).
Reverse Transcription and Quantitative Real-Time PCR
Total RNA was extracted from tissues or cultured cells with Trizol reagent according to the manufacture’s protocol (Invitrogen, USA). The quantitative real-time polymerase chain reaction (qRT-PCR) was performed on the ABI7300 system (Applied Biosystems, CA, USA), according to the manufacturer’s instructions. The mRNA levels were detected by using the SYBR Green PCR Master Mix and miR-195-5p levels were detected by using the TaqMan microRNA assay kit (Applied Biosystems, CA, USA). The relative expression of FOXK1 and miR-195-5p was normalized to the internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6, respectively. The relative expression was calculated using the 2−ΔΔCt method. The sequences of PCR primers were as follows: FOXK1 forward, 5′-ACACGTCTGGAGGAGACAGC-3′ and reverse, 5′-GAGAGGTTGTGCCGGATAGA-3′; miR-195-5p, forward 5′-GAATTCGCCTCAAGAGAACAAAGTGGAG-3′ and reverse 5′-AGATCTCCCATGGGGGCTCAGCCCCT-3′; U6, forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse 5′-CGCTTCACGAATTTGCGTGTCAT-3′; and GAPDH, forward 5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse 5′-ATGGCATGGACTGTGGTCAT-3′.
Cell Proliferation Assay
Cell proliferation was performed by using Cell Counting Kit-8 (CCK-8; Dojindo, Japan), according to the manufacturer’s protocols. H1299 and A549 cells were transfected with miR-NC, miR-195-5p then seeded in 96-well plates and cultured for 24, 48, 72 and 96 hours, respectively. Then, 10 µL CCK-8 reagent was added to each well and incubated for another 1 to 2 hours. The absorbance at OD450 was measured with the microplate reader. This experiment was performed in triplicate.
Transwell Migration and Invasion Assay
Cell migration assay was performed with migration chambers (BD Biosciences, CA, USA). Transfected cells (5 × 104 cells/well) suspended in serum-free DMEM were seeded into the upper chamber. The completed DMEM medium was placed into the lower chamber. After 24 hours, any unmigrated cells were removed and indicated cells were fixed and stained with 0.5% crystal violet (Sigma, MO, USA), followed by photographed and counted under the microscopy. Cell invasion assay was evaluated using Matrigel invasion chambers (BD Biosciences, CA, USA). The cells were treated referring to the migratory assay. After 24 hours of incubation, cells remaining on the upper membrane were removed carefully. The adherent to the bottom of the membrane was fixed and stained with crystal violet (Sigma, MO, USA). Then, they were measured under the microscopy.
Luciferase Reporter Assay
The wild type (WT) and mutant (MUT) 3′-UTR of FOXK1 containing the miR-195-5p binding site was amplified from human genomic DNA and validated by DNA sequencing. Indicated plasmids with miR-195-5p or miR-NC were transfected into H1299 or A549 cells in 24-well plates. After 48 hours, luciferase assays were detected by using the Dual Luciferase Reporter Assay System (Promega, Wisconsin) following the manufacturer’s protocol. Firefly luciferase was normalized to Renilla luciferase.
Western Blot
Total proteins from tissues and cultured cells were extracted with the RIPA lysis buffer with 3% proteinase inhibitor cocktail (Sigma, MO, USA), according to the manufacturer’s instruction. Cell lysates were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride (PVDF) membranes (Invitrogen, USA). After blocking with nonfat milk in TBST at room temperature for 1 hour, membranes were incubated with primary antibody against FOXK1 (1:1000, Abcam, Cambridge, Massachusetts) at 4°C overnight. Membranes were washed 3 times and incubated with horseradish peroxidase–conjugated secondary antibody for 2 hours at room temperature. Signals were detected by an enhanced chemiluminescence system (Amersham Biosciences).
Statistical Analysis
All data are shown as mean ± standard deviation. Student t test and 1-way analysis of variance analysis were used to estimate significant difference in different groups. P < .05 represented the difference was statistically significant.
Results
MicroRNA-195-5p Is Downregulated in Lung Cancer Tissues and Cell Lines
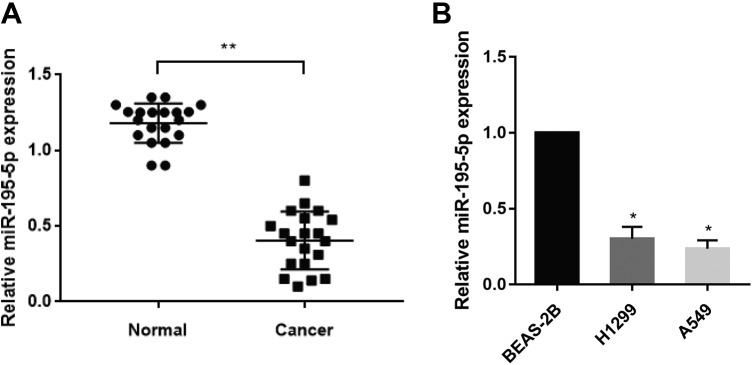
To investigate the role of miR-195-5p in lung adenocarcinoma, we first detected miR-195-5p expression in 20 pairs of lung cancer samples using qRT-PCR. The results showed that miR-195-5p expression was markedly downregulated in lung cancer tissues compared to adjacent normal tissues (Figure 1A). Consistently, miR-195-5p expression was also decreased in lung cancer cell lines (H1299, A549) than in the human lung epithelia cell line BEAS-2B (Figure 1B). These findings implied that miR-195-5p was downregulated in lung cancer tissues and cell lines.
Figure 1.
Micro-195-5p is downregulated in lung cancer tissues. A, Relative miR-195-5p expression levels were measured by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) in 20 pairs of human lung cancer tissues and adjacent normal tissues. U6 RNA level was used as an internal control. B, Relative miR-195-5p expression was analyzed by qRT-PCR in human lung cancer cell lines. Each experiment was repeated at least 3 times. *P < .05, **P < .01.
MicroRNA-195-5p Overexpression Suppresses Lung Cancer Cell Proliferation, Migration, and Invasion
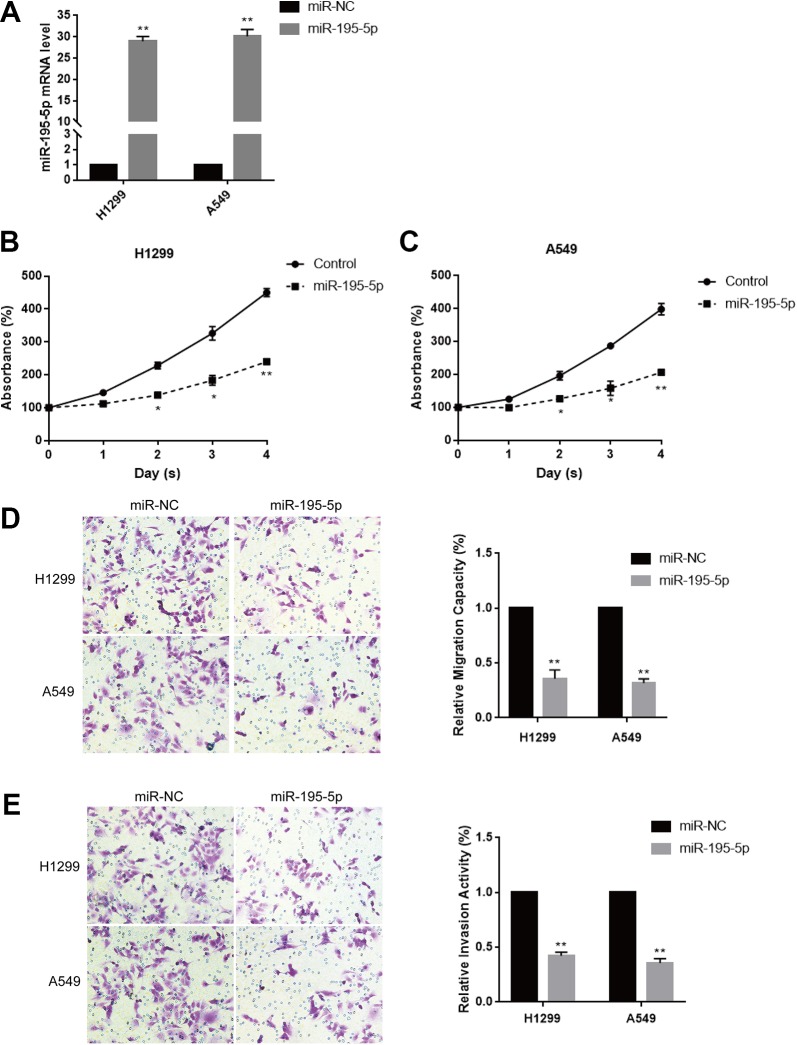
Since miR-195-5p expression was significantly lower in tumor tissues and A549 and H1299 cells than adjacent normal tissues and the normal cell line BEAS-2B, respectively, firstly we investigated these 3 tumorigenic activities between BEAS-2B normal cell line and A549/H1299 tumor cell lines. These results showed that the proliferative abilities, the migration and invasion activities of A549 and H1299 were obviously higher than that of BEAS-2B (Supplementary 1), indicating that miR-195-5p expression was inversely correlated with tumorigenic activities. To explore the function of miR-195-5p in lung cancer, we overexpressed miR-195-5p in H1299 and A549 cells by transfection with miR-195-5p mimic or negative control. The result showed that the miR-195-5p expression was significantly increased in H1299 and A549 cells after transfection determined by qRT-PCR (Figure 2A). We then used CCK-8 and transwell assays to analyze whether the miR-195-5p overexpression affected lung cancer cell proliferation and metastasis. Cell Counting Kit-8 assay showed that forced expression of miR-195-5p remarkably suppressed cell proliferation in both H1299 and A549 cells (Figure 2B and C). What’s more, transwell migration assay indicated that miR-195-5p overexpression significantly repressed H1299 and A549 cells migration (Figure 2D). Moreover, the transwell invasion assay was in accordance with the migration result, that was, increased miR-195-5p expression strikingly inhibited H1299 and A549 cells invasion (Figure 2E). Taken together, the above results suggested that the overexpression of miR-195-5p suppressed the proliferation, migration, and invasion of lung cancer cells.
Figure 2.
Overexpression of miR-195-5p inhibits lung cancer cell proliferation, migration, and invasion. A, The expression level of miR-195-5p in lung cancer cells H1299 and A549 transfected with miR-195-5p mimic or negative control was determined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). B and C, Cell Counting Kit-8 (CCK-8) assay was used to examine cell viability in H1299 and A549 cells transfected with miR-195-5p mimic and negative control, respectively, for 48 hours. D, Transwell migration assay was used to measure the migratory ability in H1299 and A549 cells transfected with miR-195-5p mimic and negative control, respectively, for 48 hours. E, Transwell-matrigel assay was used to measure the potential of invasion of H1299 and A549 cells transfected with miR-195-5p mimic and negative control, respectively, for 48 hours. Each experiment was repeated at least 3 times. *P < .05, **P < .01.
Forkhead Box k1 Is a Direct Target of miR-195-5p
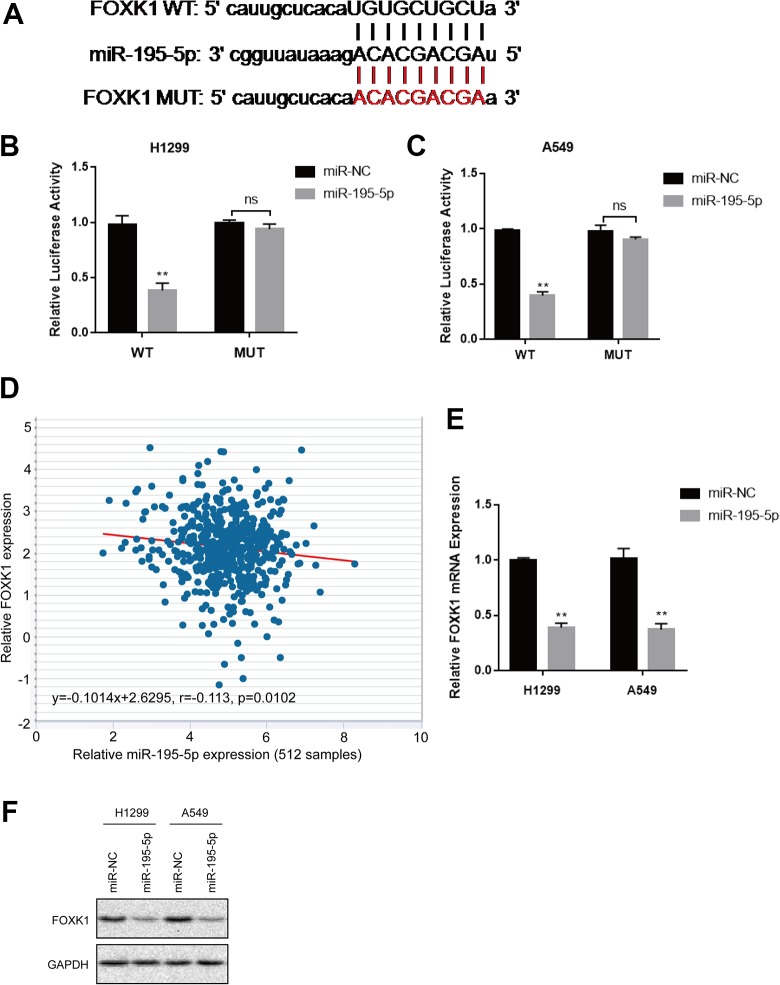
We then explored the underlying molecular mechanism of miR-195-5p by bioinformatics analysis using TargetScan (www.targetscan.org). Figure 3A showed that the 3′-UTR regions of FOXK1 contained binding site for the miR-195-5p seed region, indicating FOXK1 was the potential target. To confirm whether miR-195-5p directly binds to miR-195-5p, we performed luciferase reporter assay. The results displayed that the luciferase activity was inhibited in H1299 and A549 cells cotransfected with WT FOXK1 3′-UTR and miR-195-5p mimics whereas restored in cells cotransfected with MUT FOXK1 3′-UTR and miR-195-5p mimics (Figure 3B and 3C). Furthermore, Pearson rank correlation analysis showed that FOXK1 and miR-195-5p in lung cancer samples were inversely correlated (Figure 3D, Pearson correlation r = −0.133). In addition, the mRNA and protein level of FOXK1 was also decreased with the overexpression of miR-195-5p (Figure 3E and F). These data indicated that FOXK1 was a direct target of miR-195-5p in lung cancer cells.
Figure 3.
Forkhead box k1 (FOXK1) is a direct target of miR-195-5p. A, The schematic diagram showed the sequences of the miR-195-5p binding site within the human FOXK1 3′-untranslated region (3′-UTR), wild type FOXK1 3′-UTR and the mutant FOXK1 3′-UTR. The mutant nucleotides of the FOXK1 3′-UTR are labeled in red. B and C, Luciferase reporter assay on H1299 and A549 transfected with miR-195-5p or miR-NC together with wt FOXK1 3′-UTR or mut FOXK1 3′-UTR was measured 24 hours post-transfection. D, The correlation between relative FOXK1 mRNA levels and relative miR-195-5p expression in 512 lung cancer tissue samples was analyzed with Pearson rank correlation (r = −0.113, P = .0102). E, FOXK1 expression at mRNA level in transfected H1299 and A549 cells was measured by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). F, FOXK1 expression at protein level in transfected H1299 and A549 cells was examined by Western blot. Each experiment was repeated at least 3 times. *P < .05, **P < .01.
Loss of FOXK1 Suppressed Lung Cancer Cell Growth and Metastasis
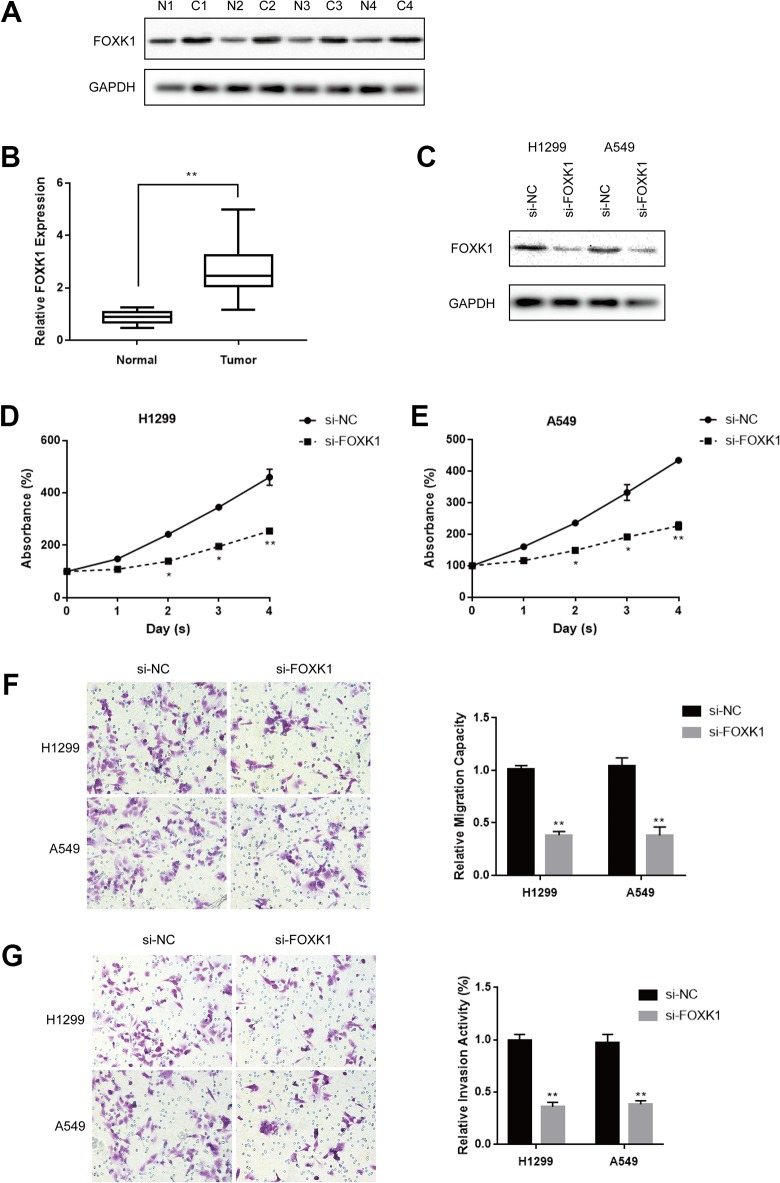
Next, we explored the cell function of FOXK1 in lung cancer cells, the relative expression of FOXK1 at protein level and mRNA level in lung cancer tissues was significantly higher compared to adjacent normal tissues as shown in Figure 4A and B. Given that FOXK1 was upregulated in lung cancer, we investigated the effects of FOXK1 on lung cancer cell proliferation, migration, and invasion with specific si-FOXK1. The knockdown efficiency of si-FOXK1 was validated by Western blot (Figure 4C). Cell Counting Kit-8 assay showed that FOXK1 depletion suppressed cell proliferation both in H1299 and in A549 cells (Figure 4D and E). Results of transwell assays showed that the migratory ability and invasive capability were significantly inhibited following FOXK1 knockdown (Figure 4F and G). In conclusion, the results suggested that FOXK1 knockdown suppressed lung cancer cell proliferation, migration, and invasion.
Figure 4.
Knockdown of Forkhead box k1 (FOXK1) suppressed lung cancer cell proliferation, migration, and invasion. A, Representative images of FOXK1 protein expression in lung cancer tissues (C) and normal tissues (N) were detected by Western blot. B, The mRNA levels of FOXK1 in lung cancer tissues and normal tissues were determined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). C, The knockdown efficiency of si-FOXK1 was measured by qRT-PCR. D and E, Cell Counting Kit-8 (CCK-8) assay was used to measure cell proliferation in H1299 and A549 transfected with si-NC and si-FOXK1, respectively, for 48 hours. Each experiment was repeated at least 3 times. *P < .05, **P < .01. F and G, Transwell migratory assay and transwell invasion assay were performed to measure cell migratory ability and invasive capacity in H1299 and A549 cells transfected with si-NC and si-FOXK1, respectively, for 48 hours. Each experiment was repeated at least 3 times. *P < .05, **P < .01.
MicroR-195-5p Exerts Its Role by Regulation of FOXK1 Expression in Lung Cancer
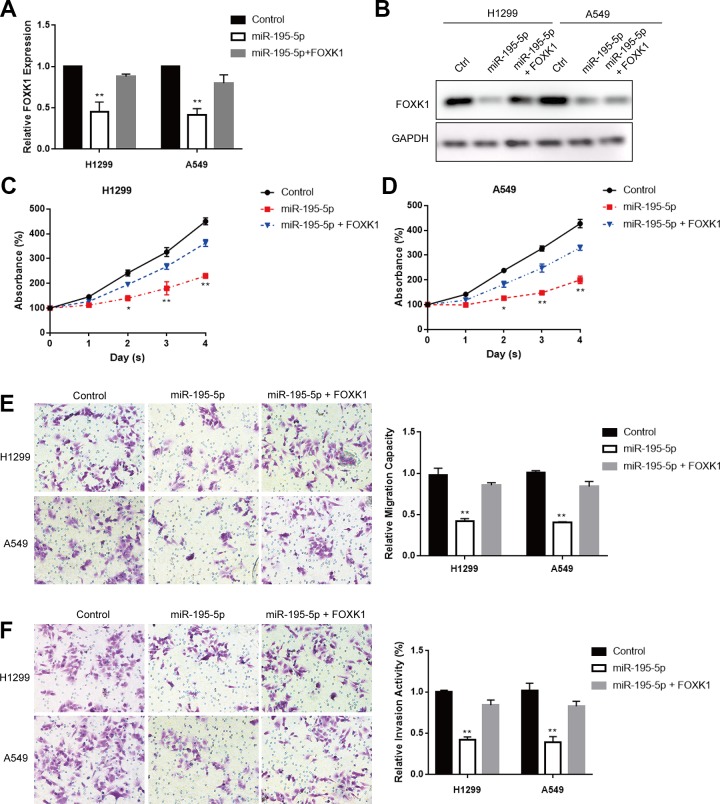
Based on the above results, we speculated that miR-195-5p might exert its effects by repressing FOXK1 expression. The expression of FOXK1 was reduced at mRNA level and protein level with miR-195-5p overexpression by qRT-PCR and Western blot (Figure 5A and B) while restored following FOXK1 overexpression (Figure 5A and B). Cell Counting Kit-8 results showed that the overexpression of FOXK1 could reverse the repressed cell proliferation induced by miR-195-5p overexpression in both H1299 and A549 cells (Figure 5C and D). Transwell migration assay and transwell invasion assay displayed that FOXK1 overexpression could abolished miR-195-5p-caused cell migratory ability and invasive capability suppression in both H1299 and A549 cells (Figure 5E and F). Taken together, these data demonstrated that miR-195-5p acted as a tumor suppressor role via the regulation of FOXK1 in lung cancer.
Figure 5.
Forkhead box k1 (FOXK1) overexpression abolished the suppressive effect of miR-195-5p in lung cancer. A and B, Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blot assays were used to analyze FOXK1 expression in H1299 and A549 cells transfected with miR-195-5p mimic and negative control RNA oligos, respectively, for 48 hours. The expression of FOXK1 was normalized to Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. C and D, Cell Counting Kit-8 (CCK-8) assay was sued to measure cell viability in H1299 and A549 cells transfected with miR-195-5p mimics or negative control together with FOXK1 overexpression, respectively, for 48 hours. E and F, Transwell migratory assay and invasion assay were used to determine the migratory ability and invasive capability of H1299 and A549 cells transfected with miR-195-5p mimic or negative control together with FOXK1 overexpression, respectively, for 48 hours. Each experiment was repeated at least 3 times. *P < .05, **P < .01.
Discussion
MicroRNAs play vital roles in tumorigenesis under a series of mechanisms in recent studies, which have been shown to be associated with clinical characteristics of multiple cancers and outcomes. The roles of miRNAs in lung cancer have also been studied. Such as, Yang et al reported that microRNA-218 functioned as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlated with poor prognosis.19 Liu et al showed that miR-330-3p controlled cell proliferation by targeting early growth response 2 in NSCLC.20 Liu et al studied that microRNA-4458 suppressed the proliferation of human lung cancer cells in vitro by directly targeting Lin28B,21 while Sun et al found that microRNA-346 facilitated cell growth and metastasis and suppresses cell apoptosis in human NSCLC by regulation of XPC/extracellular-signal regulated kinase (ERK)/Snail/E-cadherin pathway.10
Previous studies shown that miR-195-5p might act as a tumor suppressor role in several cancers. For example, it was reported that miR-195-5p suppressed the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14.22 It was shown that miR-497-5p, miR-195-5p, and miR-455-3p functioned as tumor suppressors by targeting human telomerase reverse transcriptase (hTERT) in melanoma A375 cells.23 We found in this study that miR-195-5p expression was low in lung cancer samples compared with normal control. The result was consistent in the lung cancer cell lines. Furthermore, miR-195-5p overexpression inhibited lung cancer cells proliferation, migration, and invasion. Thus, we demonstrated that miR-195-5p suppressed lung cancer growth and metastasis.
Forkhead box k1 has been identified to play an oncogenic role in the development of cancer. In the present research, FOXK1 was identified as the target of miR-195-5p. First, miR-195-5p repressed FOXK1 expression by binding to the 3′-UTR of FOXK1 by luciferase reporter assay. Second, the expression of FOXK1 in lung cancer cell lines was remarkably decreased with miR-195-5p expression. Third, FOXK1 was upregulated in lung cancer tissues compared to the adjacent normal tissues. What’s more, FOXK1 restoration treatment could abolish the suppressed effects caused by miR-195-5p overexpression. These results suggested that FOXK1 acted as an oncogene role in lung cancer development and miR-195-5p repressed lung cancer growth by targeting FOXK1.
In conclusion, this study revealed that miR-195-5p played an important role in lung cancer through regulation of FOXK1. Despite we identified that miR-195-5p could suppress progression of lung cancer by targeting FOXK1, there might be many other targets of miR-195-5p affecting carcinogenesis of lung cancer. Nevertheless, in our study, we confirmed that miR-195-5p functioned as a tumor suppressor through suppression of FOXK1. Further studies are required to investigate more molecular mechanisms of miR-195-5p and signaling pathway in lung cancer.
Conclusions
In summary, our study presents the regulatory mechanism of miR-195-5p in lung cancer cell growth directly through the target FOXK1 and implicates miR-195-5p as a potential target for lung cancer therapies and FOXK1 as a prospective prognosis factor.
Supplemental Material
Supplemental_1 for miR-195-5p Suppresses Lung Cancer Cell Proliferation, Migration, and Invasion Via FOXK1 by Zhiqiang Long and Yadong Wang in Technology in Cancer Research & Treatment
Abbreviations
- CCK-8
cell counting kit-8
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FOXK1
Forkhead box k1
- miR-195-5p
microRNA-195-5p
- miRNA
microRNAs
- MUT
mutant
- NSCLC
non-small cell lung cancer
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- 3′-UTR
3′-untranslated region
- WT
wild type.
Footnotes
Authors’ Note: Z.L. collected, analyzed the data, and draft the manuscript. Y.W. designed the project and revised the manuscript. All authors read and approved the final manuscript. All data generated in this study are included in this published article. All of the authors listed consented to publication. The research protocol was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Peking University (PKUFAHEC2017A-009023).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Yadong Wang  https://orcid.org/0000-0002-0417-9760
https://orcid.org/0000-0002-0417-9760
Supplemental Material: Supplemental material for this article is available online.
References
- 1. She J, Yang P, Hong Q, Bai C. Lung cancer in China: challenges and interventions. Chest. 2013;143(4):1117–1126. doi:10.1378/chest.11-2948. [DOI] [PubMed] [Google Scholar]
- 2. Li H, Chen S, Liu J, et al. Long non-coding RNA PVT1-5 promotes cell proliferation by regulating miR-126/SLC7A5 axis in lung cancer. Biochem Biophys Res Commun. 2018;495(3):2350–2355. doi:10.1016/j.bbrc.2017.12.114. [DOI] [PubMed] [Google Scholar]
- 3. Al-Shahrabani F, Vallböhmer D, Angenendt S, Knoefel WT. Surgical strategies in the therapy of non-small cell lung cancer. World J Clin Oncol. 2014;5(4):595–603. doi:10.5306/wjco.v5.i4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang R, Su B. Small but influential: the role of microRNAs on gene regulatory network and 3’UTR evolution. J Genet Genomics. 2009;36(1):1–6. doi:10.1016/S1673-8527(09)60001.-1. [DOI] [PubMed] [Google Scholar]
- 5. Mazière P, Enright AJ. Prediction of microRNA targets. Drug Discov Today. 2007;12(11-12):452–458. doi:10.1016/j.drudis.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 6. Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA. RNA. 2005;11(12):1753–1761. doi:10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104(2):235–240. doi:10.1038/sj.bjc.6606010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–677. doi:10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 9. Weeraratne SD, Amani V, Teider N, et al. Pleiotropic effects of miR-183∼96∼182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123(4):539–552. doi:10.1007/s00401-012-0969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun CC, Li SJ, Yuan ZP, Li DJ. MicroRNA-346 facilitates cell growth and metastasis, and suppresses cell apoptosis in human non-small cell lung cancer by regulation of XPC/ERK/Snail/E-cadherin pathway. Aging (Albany NY). 2016;8(10):2509–2524. doi:10.18632/aging.101080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Meng H, Wang K, Chen X, et al. MicroRNA-330-3p functions as an oncogene in human esophageal cancer by targeting programmed cell death 4. Am J Cancer Res. 2015;5(3):1062–1075. [PMC free article] [PubMed] [Google Scholar]
- 12. Feng C, Zhang L, Sun Y, et al. GDPD5, a target of miR-195-5p, is associated with metastasis and chemoresistance in colorectal cancer. Biomed Pharmacother. 2018;101:945–952. doi:10.1016/j.biopha.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 13. Wang J, Li L, Jiang M, Li Y. MicroRNA-195 inhibits human gastric cancer by directly targeting basic fibroblast growth factor. Clin Transl Oncol. 2014;19(11):1320–1328. doi:10.1007/s12094-017-1668-4. [DOI] [PubMed] [Google Scholar]
- 14. Katoh M, Katoh M. Human FOX gene family (Review). Int J Oncol. 2004;25(5):1495–1500. [PubMed] [Google Scholar]
- 15. Shi X, Wallis AM, Gerard RD, et al. Foxk1 promotes cell proliferation and represses myogenic differentiation by regulating Foxo4 and Mef2. J Cell Sci. 2012;125(pt 22):5329–5337. doi:10.1242/jcs.105239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu M, Wang J, Tang W, et al. FOXK1 interaction with FHL2 promotes proliferation, invasion and metastasis in colorectal cancer. Oncogenesis. 2016;5(11):e271 doi:10.1038/oncsis.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen D, Wang K, Li X, et al. FOXK1 plays an oncogenic role in the development of esophageal cancer. Biochem Biophys Res Commun. 2017;494(1-2):88–94. doi:10.1016/j.bbrc.2017.10.080. [DOI] [PubMed] [Google Scholar]
- 18. Li P, Yu Z, He L, et al. Knockdown of FOXK1 inhibited the proliferation, migration and invasion in hepatocellular carcinoma cells. Biomed Pharmacother. 2017;92:270–276. doi:10.1016/j.biopha.2017.05.087. [DOI] [PubMed] [Google Scholar]
- 19. Yang Y, Ding L, Hu Q, et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017;16(1):141 doi:10.1186/s12943-017-0710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu X, Shi H, Liu B, Li J, Liu Y, Yu B. MiR-330-3p controls cell proliferation by targeting early growth response 2 in non-small-cell lung cancer. Acta Biochim Biophys Sin (Shanghai). 2015;47(6):431–440. doi:10.1093/abbs/gmv032. [DOI] [PubMed] [Google Scholar]
- 21. Liu CH, Lv DS, Li M, Sun G, Zhang XF, Bai Y. MicroRNA-4458 suppresses the proliferation of human lung cancer cells in vitro by directly targeting Lin28B. Acta Pharmacol Sin. 2017;38(9):1297–1304. doi:10.1038/aps.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang T, Ren Y, Liu R, et al. MiR-195-5p Suppresses the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14. Biomed Res Int. 2017;2017:7378148 doi:10.1155/2017/7378148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chai L, Kang XJ, Sun ZZ, et al. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag Res. 2018;10:989–1003. doi:10.2147/CMAR.S163335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental_1 for miR-195-5p Suppresses Lung Cancer Cell Proliferation, Migration, and Invasion Via FOXK1 by Zhiqiang Long and Yadong Wang in Technology in Cancer Research & Treatment