Abstract
Many fine-scale features of ribosomes have been explained in terms of function, revealing an elegant molecular machine optimized for error-correction, speed and control. Here we mathematically demonstrate that many less understood, larger-scale features of ribosomes – e.g. why a few rRNA molecules dominate the mass and why r-protein is divided into 55–80 small and similarly-sized segments – speed up their autocatalytic biogenesis.
Ribosomes are central to life by translating sequences of nucleic acids into sequences of amino acids [1]. Their features are therefore typically explained by how they affect translation [1]. However, in recent years it has become clear that ribosomes also are exceptional as customers of the translation machinery [2–4]. Not only do r-proteins make up a large fraction of the total protein content in many cells [5], but the autocatalytic nature of ribosome production introduces additional constraints. Specifically, the ribosome doubling time places a hard bound on the cell doubling time, since for every additional ribosome to share the translation burden there is also one more to make [2, 3]. Even for the smallest and fastest ribosomes known it takes at least ~6 min for one ribosome to make a new set of r-proteins (SI) and typically much longer, not accounting for the substantial time invested in the synthesis of ternary complexes [4]. This appears to explain the observed limits on bacterial growth given that ribosomes must also spend much of their time making other proteins [6–11] and shows that ribosomes are under tremendous selective pressure to minimize the time they spend reproducing themselves. Similar principles may also apply to some eukaryotes since ribosomes then are larger and slower [1, 12, 13] (SI). In fact, even organisms where cell doubling times are far from limited by ribosome doubling times would benefit from faster ribosome production, allowing ribosomes to spend more of their time to produce the rest of the proteome. This efficiency constraint was recently shown to have broad physiological consequences for cells [3, 4, 11, 14–16] and here we mathematically demonstrate that it may also explain many broader features of the ribosome itself (Fig. 1).
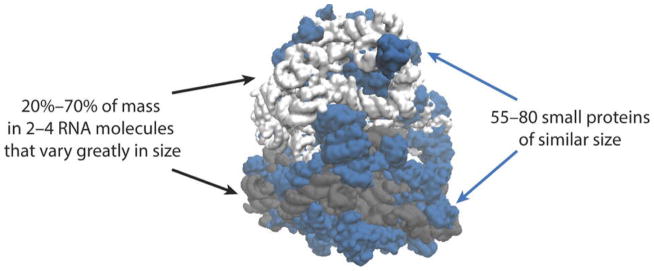
Figure 1.
Many unusual features of ribosomes are not well understood: With the exception of mitochondrial ribosomes, most of the mass is in a few RNA molecules that are very large on average but vary greatly in size (gray), whereas the proteins (blue) are unusually small, numerous and similar to each other in size.
Why do ribosomes have so many small r-proteins?
The total time τ it takes one ribosome to elongate a set of r-proteins for a new ribosome is proportional to their total length. However, the time that each ribosome on average must dedicate to that process also depends on how the total protein mass is divided up into individual segments. Specifically, because so many ribosomes produce r-proteins in parallel and because each ribosome consists of many r-proteins, complete sets of r-proteins will form by chance more quickly than individual ribosomes could elongate that amount of protein on their own. For example, if ribosomes consisted of two similarly sized r-proteins, complete pairs would start to form τ/2 time units after production initiated, and those newly made ribosomes could then share the translation burden. Similarly, if ribosomes contained n proteins of equal genomic length, the nascent peptides that cannot contribute to new ribosomes would be about n times shorter and mature r-proteins would be released n times faster (Fig. 2A). Increasing n thus reduces the fraction of time ϕ ribosomes must spend on their own production, according to (SI):
| (1) |
where Tgen is the cell generation time. This expression asymptotically reaches the previously known bound on growth [4, 10, 11] τln(2)/Tgen in the limit of high n, and shows that dividing the ribosomal protein content into a large number of small proteins can reduce the time to make new ribosomes by as much as 30% (Fig. 2B), but with diminishing returns at higher n. Because increasing n also has disadvantages not accounted for above, there should be an optimal number, nopt, of r-proteins.
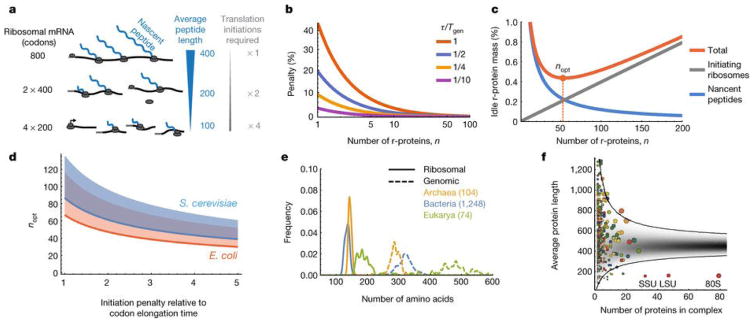
Figure 2.
A. Nascent r-protein peptides cannot participate in translation before their own translation is completed. Sub-dividing the protein mass into more and smaller pieces reduces the inactive fraction, but also increases the number of translation initiations required. B. The penalty, due to finite number of r-proteins, that is incurred on the minimal time fraction (from Eq. 1) that ribosomes must spend on their own production. Penalty is plotted as a function of n for different values of τ/Tgen (1 Red, 1/2 Blue, 1/4 Yellow, 1/10 Magenta), and is calculated relative to τln(2)/Tgen which is the asymptotic value achieved at high n, possibly reducing the penalty as much as 1/ln(2)≃1.44 fold. C. The fraction of dedicated r-protein mass that is elongationally idle, due to nascent r-protein peptides and ribosomes occupied in initiation, is minimized for an optimal number of r-proteins, nopt. D. The nopt from Eq. (2) and panel C for E. coli (orange) and S. cerevisiae (blue), in the range of the expected ratios between initiation and elongation rates (see main text). The shaded area above the lines would be accessible if ribosome could produce slightly less of the larger sub-unit, which only enters at the last step of initiation (SI). E. Across organisms, average lengths of proteins in ribosomes are 2–3 times smaller than in genomes [22]. F. The average length (amino acids) of proteins in S. cerevisiae multi-protein complexes vs. the number of proteins in the complex. Each colored circle corresponds to a known complex [23] (area is proportional to total number of amino acids). Gray represents the predicted probability density when drawing random genes from the genome. Most of the 403 complexes fall within the expected 99 % confidence interval, except the ribosome (80S, including both subunits SSU and LSU independently), that falls ~7 standard deviations below.
One disadvantage of dividing the ribosomal protein complement into many small segments is that the different proteins will not be made in exactly the same numbers in individual cells: Even if production rates for all r-proteins were perfectly matched on average, chance would inevitably create more of some r-proteins and less of others (Extended data Fig. 1). The assembly of complete ribosomes is then limited by the r-protein that by chance is present in the lowest number, creating a wasteful surplus pool of all other r-proteins and thereby reducing efficiency. Because each r-protein runs the risk of being under-produced, this surplus should increase with n. However, for the types of distributions often observed for gene expression, the average surplus is an exceedingly damped function of n, closely following (SI). Combining this diminishing disadvantage with the diminishing returns of shorter nascent peptides in Eq. (1) produces no effective upper limit on n : all n > nopt are virtually equally optimal (SI), even without expression control. E. coli cells also reduce this problem by negative feedback loops [17, 18] that prevent surplus accumulation, and by expressing the r-proteins in operons such that terminating ribosomes skip directly to the next start site [20], possibly ensuring that r-proteins in the same operon are expressed in almost identical numbers [17–19].
However, there is another countervailing selective force that limits n for a given total ribosome mass: For every additional r-protein one more translation initiation is required, sequestering ribosomes from the elongation process and creating another form of overhead (Fig. 2A). With τoh as the overhead time during which a ribosome is occupied making ribosomal proteins without actually elongating, and again τ as the time it takes a ribosome to elongate another set of r-proteins
| (2) |
where A ≃ 1 (SI). The square root reflects the fact that nopt minimizes the total idleness due to the length of nascent peptides which decreases as ~1/n, and due to ribosome sequestration from initiation which increases as nτoh/τ (Fig. 2C).
The exact initiation penalties – the times that ribosomes are sequestered to initiate translation – are not known in vivo. However, we only need to know how those times increase with n, and only relative to elongation rates. An estimate can then be determined in E. coli from an observed average spacing of about 15 codons [21] between initiating ribosomes. However, this may greatly exaggerate the relevant initiation penalty for several reasons. First, the previous ribosome must move away some distance from the initiation site before the next one can bind, but during that time the latter is free to bind other transcripts. In fact, if mRNA levels are abundant the previous ribosome can on average move away a longer distance before another ribosome binds the same transcript, increasing the spacing without decreasing efficiency. Second, the total time ribosomes must search the cytoplasm for mRNAs to produce n r-proteins should be roughly independent of n: producing twice as many r-proteins with half the size requires twice as many initiations, but also doubles the total concentration of the target mRNA for a given total investment in mRNA and thus approximately halves the time it takes each diffusing ribosome to find an r-protein mRNA. Thus, only the time the ribosome spends bound to the mRNA without elongating would be relevant here. Finally, as noted above most r-proteins in E. coli do not require typical initiations because terminating ribosomes continue directly to the next start site [20] in the operon. The relevant penalty due to initiation should thus be a small fraction of the time it takes to elongate 15 codons, possibly as low as one or two codon equivalents.
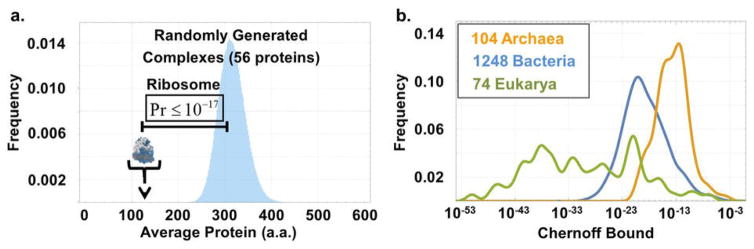
Due to the square root effect, initiation penalties in the broad range of 1–5 codons approximately predict 40 ≤ nopt ≤ 85 (Fig. 2D) – closely consistent with the 56 r-proteins observed in nature for E. coli even though the first-principles derivation is agnostic to the typical size of proteins or any structural properties. Formulated in terms of size, this means that r-proteins should be about 2–4 times smaller than average proteins. Indeed, the average r-protein length in E. coli is only ~130 amino acids compared to ~315 amino acids over the genome [22]. This is ~6.5 standard deviations below average, given that there are 56 r-proteins, and the probability of observing such low averages when drawing genes randomly from the genome is <10−17 (Extended data Fig. 2). Similar principles are observed across organisms (Fig. 2E, Extended data Fig. 2, SI) but not generally in multiprotein complexes (Fig. 2F).
Why are ribosomal proteins so similar in length?
The efficiency argument above also has direct consequences for how similar in length ribosomal proteins should be in order to maximize the efficiency of their own production. For example, if 99% of the ribosomal protein mass was in a single r-protein, it would not help to divide the rest into small pieces even if that would reduce the average length. Specifically, for a given production rate, the probability of finding a ribosome in the process of translating a particular r-protein is proportional to its length, as also shown experimentally [19], simply because longer r-proteins take proportionally longer time to translate. Thus, on average if one of the r-proteins is twice as long as another, it not only has twice as long nascent peptides but also occupies twice as many ribosomes, contributing four times more to the average length of nascent peptides. The average amino acid length over all r-protein nascent peptides in cells is then not simply half of their average genomic codon lengths, 〈L〉/2, as when the r-proteins have identical lengths, but approximately 〈L2〉/(2〈L〉), i.e., proportional to the average square of their genomic codon lengths. Because by definition of variances , the efficiency for a given average length is thus reduced by variation in length (Fig. 3A). Specifically, with CVL =σL/〈L〉 for the length distribution of r-proteins, Eq. (1) generalizes to (SI)
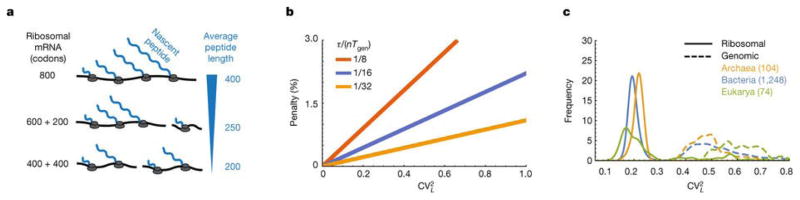
Figure 3.
A. Dividing up a long r-protein (e.g. 8oo codons) into two unequal parts (600 and 200 codons) does not reduce the average length of the nascent peptides as much as dividing it into equal parts (400 and 400 codons). B. The minimal time fraction that ribosomes must spend on their own production from Eq. 3, normalized by n(2τ/nTgen −1) from Eq. 1 and plotted as a function of for different values of τ/nTgen (1/8 red, 1/16 blue, 1/32 yellow) to show the percentage penalty that results from CVL > 0. C. Relative frequencies of the normalized variance ( from Eq. 3) of protein length distributions in ribosomes and genomes, showing substantially lower CV s for the former.
| (3) |
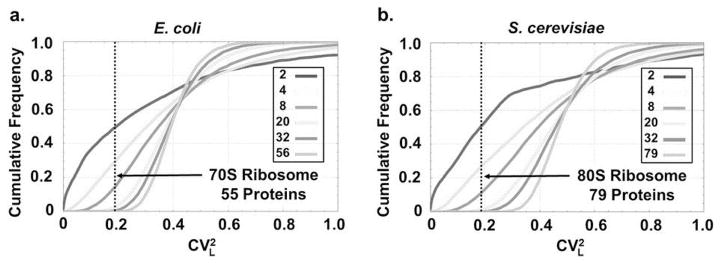
Inspecting this expression shows that to maximize the efficiency of their own production, the r-proteins should not only be short but also of similar lengths (Fig. 3B). Indeed we find that for a wide range of organisms, CVL is substantially lower for r-proteins than for the genome overall (Fig. 3C), whether or not we account for the fact that the r-proteins also are smaller on average (Extended data Fig. 3, SI). For bacteria similar results hold for gram-positives, while for gram-negatives the effect is also substantial except for the single large r-protein S1. However, S1 is only transiently associated with the ribosome and did not even appear in initial crystal structures[24]. It has also been shown to be involved in the selective initial binding to some mRNAs[25], and it is not produced in an operon with other r-proteins[17, 18]. We therefore propose, supported by some biochemical evidence[26], that cells use S1 to create specialized ribosome, e.g. with different balances between speed and accuracy. Regardless, this single significant exception to the length-variation rule clearly plays a different role in ribosomes.
Why are ribosomes so rich in RNA?
Another unusual feature of ribosomes is that so much of their mass is rRNA based [1, 12]. This could also be explained by selection to minimize the time each ribosome is occupied in self-production. With τpol as the translation time for the proteins of one RNA polymerase, each ribosome could produce Tgen/τpol new polymerases each generation Tgen, and each of those polymerases could produce rRNA for Tgen/τrRNA new ribosomes per generation where τrRNA is the time it takes to make one set of rRNA. The fold difference in the times that ribosomes must spend producing additional RNA polymerase (RNAP) for one set of rRNA versus the time to make one set of r-proteins is then (SI)
| (4) |
This ratio is typically very small despite rRNA dominating the ribosome mass, and even when accounting for inactive RNA polymerases (SI). For example, numbers for fast growing E. coli (Fig. 4A) suggest that the time ribosomes invest in r-protein synthesis could be two orders of magnitude higher than for an equivalent mass of rRNA (SI). Considering other auxiliary costs – e.g. making the proteins required to synthesize the nucleotides needed for the rRNA versus the proteins required to produce the nucleotides, mRNAs, amino acids, charged tRNAs, initiation and elongation factors that are needed for r-proteins – further supports these conclusions (SI).
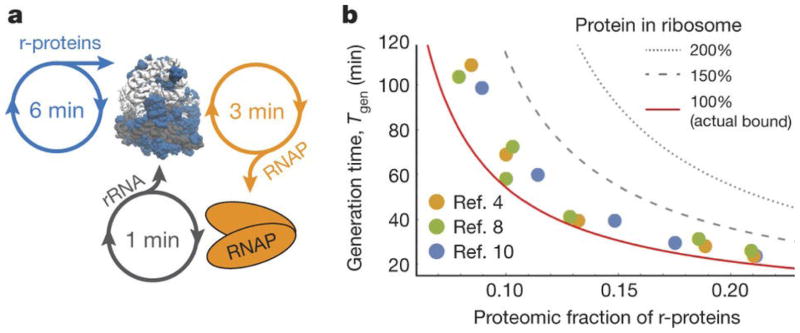
Figure 4.
A. Ribosome auto-production occupies ribosome time to make r-proteins and RNA polymerases for rRNA production. An RNA polymerase can be translated in half the time compared to a set of r-proteins, and in fast-growing E. coli, each polymerase can produce 20 sets of rRNAs per generation. B. Generation time vs. proteomic fraction of E. coli r-proteins. Circles are published experimental data: yellow [4], green [8], and blue[10]. Because most proteins in E. coli are stable, the proteomic fraction of r-proteins roughly equals the fraction of time, ϕ, ribosomes spend translating r-proteins. The solid line is the bound on generation time set by ribosome production, Tgen ≥ τln(2)/ϕ (rearranged Eq. 1 in high n limit). For the same ribosome speed, cells with hypothetical ribosomes replacing rRNA mass for r-protein (e.g., 150% or 200% the actual amount) could not divide as quickly.
The analysis above suggests a great efficiency advantage for using rRNA over protein whenever possible, and may thus explain why ribosomes defy the general rule that enzymes are mostly made of protein [27] (Fig. 1). That does not mean that the role of rRNA is merely to ensure appropriate overall dimensions of the ribosome. However, it does provide a fundamental reason for why proteins must be used sparingly in the ribosome, e.g. to increase accuracy or speed up translation, whereas rRNA should be used wherever possible without compromising function. If even a quarter of the rRNA mass was replaced with r-protein without increasing translation rates, many bacteria could not double as fast as they do (Fig. 4B).
Why are rRNAs so few, large and varying in size?
Considering the principles for r-proteins above, it may seem that rRNA should also be produced in small and uniformly sized pieces to maximize the efficiency of ribosome production. However, in most organisms, nascent rRNAs already participate in ribosome assembly by binding r-proteins during their transcription [28, 29], eliminating the need to release the rRNA in smaller pieces. In fact, producing many small rRNAs could greatly reduce efficiency because any differences in relative rRNA abundances would also waste the r-proteins bound. Because wasting r-protein has almost two orders of magnitude greater impact on ribosome efficiency than wasting rRNA, this should completely dominate over any gains in efficiency from producing rRNA in small pieces and suggests that cells should produce one single large rRNA. In E. coli the entire rRNA mass is indeed produced as a single transcript, which is then cut at various stages of assembly, creating three rRNAs for the two ribosomal subunits. This elegantly ensures that the rRNAs are made in strict stoichiometric proportions and minimizes the waste of r-proteins from binding to surplus rRNAs. Similar mechanisms occur broadly, from bacteria to mammalian cells, creating rRNAs that on average are very large. Such mechanisms also completely relieve the selective pressure for similar lengths identified for r-proteins above. Indeed the rRNAs vary greatly in length, much more than the genome-wide mRNAs and consistent with uniformly distributed random cuts. Though the theory does not specifically predict that rRNA sizes must be broadly distributed, the principle of minimizing the time for making new ribosomes is thus perfectly consistent with the rRNA having the opposite size characteristics of r-proteins.
Why do r-proteins differ between ribosomes?
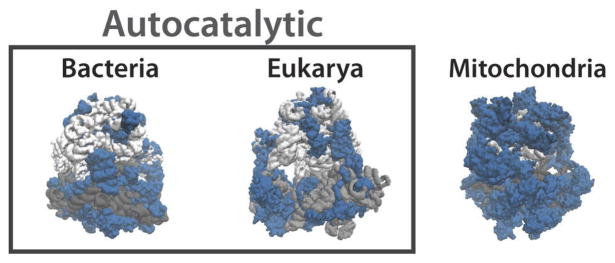
The efficiency perspective may also explain differences between ribosomes. For example, bacterial ribosomes – which are arguably under the most severe selective pressure for fast biogenesis, possibly along with archea for which less is known – are the smallest and fastest, with the shortest r-proteins and the largest percentage of rRNA mass, as much as 70%. Mitochondrial ribosomes – which are predicted to be at the opposite extreme and an exception to our theory since they are not present in high abundances and are made by cyto-ribosomes rather than by self-production – have the largest protein mass, the longest r-proteins and as little as 20% rRNA mass [1, 30–33] (Extended data Fig. 4). Interestingly, phylogenetic studies suggest that mitochondria originated from bacteria [34, 35], and that over evolutionary time, the rRNA fraction of the mito-ribosomes were gradually replaced by larger r-proteins. Though rRNA still may have originated in a hypothetical pre-protein RNA world [27] – the most common explanation for why ribosomes contain so much RNA – it is thus not necessary to invoke explanations based on evolutionary frozen accidents: In addition to its many specific roles [1], rRNA serves a fundamental purpose in ribosomes by providing appropriate dimensions and an assembly backbone at a two orders of magnitude lower ribosome sequestration cost than would be possible using protein. When this cost is less important, as it is to some extent for higher organisms but particularly for mitochondrial ribosomes, it seems that rRNA indeed is replaced by protein.
Cytosolic ribosomes in eukaryotes are between these extremes. Though the theory is agnostic about how much proteins vs. RNA they should contain, since that depends on the pressure for efficiency versus other functionality, it does make a clear prediction about the relative fold changes in the numbers and sizes of r-proteins. Specifically, it does not predict higher numbers for greater selective pressures, but rather an optimal number set by the relative initiation penalty and the total protein mass, which potentially could be reached by any organism since translational efficiency is always important. Though initiation complexes can build up slowly for some controlled genes in eukaryotes, elongation is also slower and observed distances between translating ribosomes could be as low as a few codons [36], strongly suggesting that the relative initiation penalty is similar to or lower than in E. coli, and likely equivalent to one or two elongation steps. Eq. (2) then predicts that any increase in the total ribosome protein mass should be achieved by similar fold changes in the numbers and sizes of the r-proteins, or a slightly larger increase in the numbers since the initiation penalty seems slightly lower. Indeed, eukaryotic ribosomes achieve a 1.7–1.8 fold higher protein content than bacteria by increasing the number of r-proteins by about 1.4 and the average length by 1.2–1.3. Though this near-perfect match could be coincidental, given the uncertainty in the estimates of initiation penalties, the theory certainly predicts a much higher number of r-proteins in eukaryotes given their larger size.
Outlook
Decades of structure-function analyses have shown how individual parts of the ribosome are optimized for translation. Here we expand that approach to consider functionality in a broader cellular context, and show how many features that previously were hard to explain – the unusually small, numerous and similarly sized r-proteins, and the high rRNA complement in a few large molecules – increase overall ribosome efficiency by reducing the time that ribosomes are sequestered for their own production. This may e.g. afford cells to use fewer ribosomes or ribosomes that trade speed for some other functionality. Rather than being mere relics of an evolutionary past, the unusual features of ribosomes thus seem to reflect an additional layer of functional optimization acting on collective properties of its parts.
Extended Data
Extended Data Figure 1.
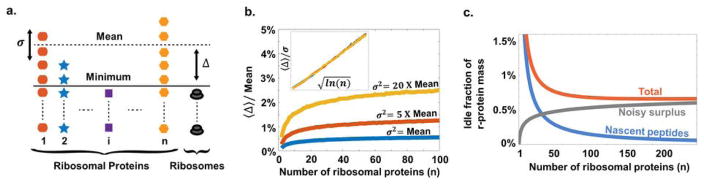
a. Cartoon of how stochastic gene expression will create temporary shortages of some r-proteins and surplus pools of others. The number of complete ribosomes assembled is then limited by the ribosomal protein in lowest abundance, and the average value of the difference between the minimum and the mean, 〈Δ〉, is the mean number of unmatched r-proteins. b. The relative mean free r-protein pool (from panel A) increases very slowly with the number of ribosomal proteins, n. For illustrative purposes we used negative binomial distributions with mean μ and different variances σ2 (SI), since this distribution has been observed and predicted in many studies of stochastic gene expression. The inset shows a curve collapse for . Similar results were obtained for more complete kinetic models. c. The fraction of the r-protein mass in the form of nascent peptides or free pools due to noisy expression (assuming Poisson noise for simplicity, but with similar results for many other noise models). Technically there is an optimal n but the curves around and above the optimum are exceedingly shallow meaning that there is no practical upper limit.
Extended Data Figure 2.
a. Distribution of the average protein length in 106 random samples of 56 proteins taken from E. coli’s genome, and the mark for the mean length of a protein in the bacterial ribosome. The probability to generate an average protein length as small as that seen in the ribosome is vanishingly small. b. The Chernoff upper bound (SI) on the probability that the average length of r-proteins could have arisen at random, i.e., with no size selection, is computed for 1424 different organisms. The value for E. coli is 10−17, but vanishingly small probabilities are attributed to all organisms we have examined.
Extended Data Figure 3.
Cumulative frequency of for randomly generated protein complexes composed of varying numbers of proteins (see SI for details). Only complexes in which the average protein length is identical ( ± 5 amino acids) to that seen in the ribosomes of E. coli (left) and S. cerevisiae (right) entered the statistics. As the number of proteins in a complex gradually increases from n = 2 to the number of proteins in the ribosome— n = 55 in E. coli (protein S1 was excluded see main text) and n = 79 in S. cerevisiae—the occurrence of coefficients of variation as low as those seen for the set of r-proteins becomes extremely rare.
Extended Data Figure 4.
Bacterial ribosomes contain about 55 r-proteins (blue) that average ~130 amino acids and make up 30–35% of the ribosome mass. Eukaryotic ribosomes contain about 80 r-proteins that average ~165 amino acids and make up about 45% of the ribosome mass. In both these cases ribosomes catalyse their own production. In contrast, in mitochondrial ribosomes which do not produce themselves rRNA (gray) is scarcer, and r-proteins are significantly larger and could make up as much as 80% of the ribosome mass.
Supplementary Material
Acknowledgments
S.R. was supported by a James S. McDonnell Foundation fellowship. S.R. and J.P. were supported by HFSP grant RGP0042, NSF-DMS grant PD127334, and NIH grant R01GM095784. We are grateful to R. Ward, A. Hilfinger, R. Milo, R. Jajoo and M. Landon for helpful discussions.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions S.R. and J.P. conceived the work, derived results and wrote the paper. M.E. contributed extensive advice and ideas.
The authors declare no competing financial interests.
Data Aviliability A list of genomes analyzed to support the findings of this study is appended to this paper. Genomes are available via UniProt at http://www.uniprot.org/proteomes/.
References
- 1.Rodnina MV, Wintermeyer W, Green R. Ribosomes Structure, Function, and Dynamics. Springer Science and Business Media; 2011. [Google Scholar]
- 2.Milo R, et al. BioNumbers – The database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38(suppl 1):D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dill KA, et al. Physical limits of cells and proteomes. Proc Natl Acad Sci USA. 2011;108(44):17876–17882. doi: 10.1073/pnas.1114477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klumpp S, et al. Molecular crowding limits translation and cell growth. Proc Natl Acad Sci USA. 2013;110(42):16754–16759. doi: 10.1073/pnas.1310377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liebermeister W, et al. Visual account of protein investment in cellular functions. Proc Natl Acad Sci USA. 2014;111:8488–8493. doi: 10.1073/pnas.1314810111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaechter M, et al. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 7.Maaloe O. An analysis of bacterial growth. Dev Biol. 1969;3(Suppl):33–58. [Google Scholar]
- 8.Forchhammer J, Lindahl L. Growth rate of polypeptide chains as a function of the cell growth rate in a mutant of Escherichia coli 15. J Mol Biol. 1971;55:563–568. doi: 10.1016/0022-2836(71)90337-8. [DOI] [PubMed] [Google Scholar]
- 9.Ehrenberg M, Kurland CG. Costs of accuracy determined by a maximal growth rate constraint. Q Rev Biophys. 1984;17(01):45–82. doi: 10.1017/s0033583500005254. [DOI] [PubMed] [Google Scholar]
- 10.Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus. 2008;3 doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- 11.Scott M, et al. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 12.Melnikov S, et al. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 13.See BNID 107871:http://bionumbers.hms.harvard.edu//bionumber.aspx?id=107871ver=3
- 14.Scott M, et al. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol. 2014;10(8):747. doi: 10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maitra A, Dill KA. Bacterial growth laws reflect the evolutionary importance of energy efficiency. Proc Natl Acad Sci USA. 2015;112(2):406–411. doi: 10.1073/pnas.1421138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maitra A, Dill KA. Modeling the overproduction of ribosomes when antibacterial drugs act on cells. Biophys J. 2016;110(3):743–748. doi: 10.1016/j.bpj.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nomura M, et al. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 18.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Progress in nucleic acid research and molecular biology. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 19.Li GW, et al. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157(3):624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto H, et al. 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc Natl Acad Sci USA. 2016;113:E1180–E1189. doi: 10.1073/pnas.1524554113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandt F, et al. The native 3D organization of bacterial polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 22.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pu S, et al. Up-to-date catalogues of yeast protein complexes. Nucleic acids research. 2009;37(3):825–831. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayati S, et al. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc Natl Acad Sci USA. 2001;98:11991–11996. doi: 10.1073/pnas.211266898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu X, et al. Ribosomal protein S1 unwinds double-stranded RNA in multiple steps. Proc Natl Acad Sci USA. 2012;109:14458–14463. doi: 10.1073/pnas.1208950109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauert M, et al. Heterogeneity of the translational machinery: Variations on a common theme. Biochimie. 2015;114:39–47. doi: 10.1016/j.biochi.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cech TR. Evolution of biological catalysis: ribozyme to RNP enzyme. Cold Spring Harb Symp Quant Biol. 2009;74:11–16. doi: 10.1101/sqb.2009.74.024. [DOI] [PubMed] [Google Scholar]
- 28.Shajani Z, et al. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 29.Woolford JL, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195(3):643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Brien TW. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene. 2002;286:73–79. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 31.Sharma MR, et al. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 32.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:6178, 1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greber BJ, et al. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348:303–308. doi: 10.1126/science.aaa3872. [DOI] [PubMed] [Google Scholar]
- 34.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 35.Andersson SG, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 36.Myasnikov AG, et al. The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes. Nat Commun. 2014;5 doi: 10.1038/ncomms6294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.