The discovery of Asgard archaea has changed the existing ideas on the origins of eukaryotes. Researchers propose that eukaryotic cells evolved from Asgard archaea. This hypothesis partly stems from the presence of multiple eukaryotic signature proteins in Asgard archaea, including homologs of ESCRT proteins that are essential components of the endomembrane system in eukaryotes. However, structural and functional features of Asgard ESCRT remain unknown. Our study provides evidence that Asgard ESCRT is functionally comparable to the eukaryotic counterparts, suggesting that despite the apparent absence of endomembranes in archaea, eukaryotic ESCRT was inherited from an Asgard archaeal ancestor, alongside the emergence of endomembrane system during eukaryogenesis.
KEYWORDS: endomembrane system, Asgard archaea, ESCRT, eukaryogenesis, evolution
ABSTRACT
The emergence of the endomembrane system is a key step in the evolution of cellular complexity during eukaryogenesis. The endosomal sorting complex required for transport (ESCRT) machinery is essential and required for the endomembrane system functions in eukaryotic cells. Recently, genes encoding eukaryote-like ESCRT protein components have been identified in the genomes of Asgard archaea, a newly proposed archaeal superphylum that is thought to include the closest extant prokaryotic relatives of eukaryotes. However, structural and functional features of Asgard ESCRT remain uncharacterized. Here, we show that Vps4, Vps2/24/46, and Vps20/32/60, the core functional components of the Asgard ESCRT, coevolved eukaryote-like structural and functional features. Phylogenetic analysis shows that Asgard Vps4, Vps2/24/46, and Vps20/32/60 are closely related to their eukaryotic counterparts. Molecular dynamics simulation and biochemical assays indicate that Asgard Vps4 contains a eukaryote-like microtubule-interacting and transport (MIT) domain that binds the distinct type 1 MIT-interacting motif and type 2 MIT-interacting motif in Vps2/24/46 and Vps20/32/60, respectively. The Asgard Vps4 partly, but much more efficiently than homologs from other archaea, complements the vps4 null mutant of Saccharomyces cerevisiae, further supporting the functional similarity between the membrane remodeling machineries of Asgard archaea and eukaryotes. Thus, this work provides evidence that the ESCRT complexes from Asgard archaea and eukaryotes are evolutionarily related and functionally similar. Thus, despite the apparent absence of endomembranes in Asgard archaea, the eukaryotic ESCRT seems to have been directly inherited from an Asgard ancestor, to become a key component of the emerging endomembrane system.
INTRODUCTION
Eukaryogenesis is a major, long-standing puzzle in evolutionary biology because the specifics of the evolutionary process leading to the eukaryotic cellular complexity are far from clear. One of the key distinctions between the cells from eukaryotes and the cells of prokaryotes is the presence in the former of the sophisticated endomembrane system. Undoubtedly, the emergence of the endomembrane system was a milestone event in eukaryogenesis because it is a prerequisite of the intracellular compartmentalization which is a hallmark of eukaryotic cells (1). The endosomal sorting complex required for transport (ESCRT) machinery is an essential component of the eukaryotic endomembrane system that, as such, has been thought to be restricted to eukaryotic cells (2). For instance, Saccharomyces cerevisiae ESCRT consists of five main subcomplexes: ESCRT-0, -I, -II, -III, and Vps4 (3–5). Of the subcomplexes, the Vps4 and ESCRT-III subunits are central players in ESCRT function that mediate remodeling and scission of endomembranes (6, 7). The ESCRT-III subunits can be further divided into two classes, termed Vps2/24/46 and Vps20/32/60, and both participate in either directly or indirectly forming membrane-bound polymeric assemblies that sever membrane necks (8). On the other hand, Vps4, an ATPase, promotes ATP-dependent disassembly of the ESCRT-III polymers, thus ensuring that the ESCRT-III subunits turn over. Several studies have shown that the N-terminal microtubule-interacting and transport (MIT) domain of Vps4 recognizes and interacts with the type 1 MIT-interacting motif (MIM1) that is present in the Vps2/24/46 class ESCRT-III subunits and type 2 MIT-interacting motif (MIM2) present in the Vps20/32/60 subunits. These recognition models are essential for the biological function of ESCRT-III and Vps4 (9–11).
The cell division (Cdv) systems discovered in some archaeal orders, such as Sulfolobales and Desulfurococcales within the TACK (Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota) superphylum include a homolog of eukaryotic Vps4 (CdvC) and several homologs of eukaryotic ESCRT-III subunits (CdvBs) (12, 13). Given these homologies and because in eukaryotes, the MIT-MIM2 interactions occurred between CdvC and CdvB (12, 14, 15), the creanarchaeal Cdv system has been proposed to be the evolutionary ancestor of eukaryotic ESCRT (16). However, this evolutionary relationship remains uncertain. One reason for the uncertainty is that CdvBs lack the well-characterized MIM1, and the absence of the MIT-MIM1 interaction is likely to reflect major functional differences between crenarchaeal Cdv and eukaryotic ESCRT (12, 17). Such differences might indicate that, although the two systems consist of homologous subunits, the Cdv system is not the direct ancestor of eukaryotic ESCRT.
The recently discovered Asgard archaea (including Lokiarchaeota, Thorarchaeota, Heimdallarchaeota, Odinarchaeota, and Helarchaeota) have been proposed to include the closest archaeal relatives of eukaryotes. This proposition stems, partly, from the findings that the Asgard genomes encode a broad repertoire of eukaryotic signature proteins (ESPs) that are far more prevalent in Asgard than they are in other archaea (18–21). Among these ESPs are highly conserved homologs of eukaryotic ESCRT-I, -II, -III, and Vps4. Notably, the presence of these proteins in Asgard archaea that was originally demonstrated on metagenomic assemblies has been confirmed by analysis of the first closed Asgard genome, ruling out the possibility of a eukaryotic contamination (12, 18, 19, 22).
Here, we explore the phylogenetic relationships among the ESCRT-III components, reconstitute and biochemically characterize the Asgard Vps4, and test its potential biological function in the heterologous S. cerevisiae endomembrane system. The combined phylogenetic, genetic, and biochemical analyses reveal close relationships between the ESCRT-III subunits and Vps4 of Asgard archaea and eukaryotes, to the exclusion of other archaea.
RESULTS
Eukaryotic-like ESCRT-III subunits and Vps4 in Asgard archaea.
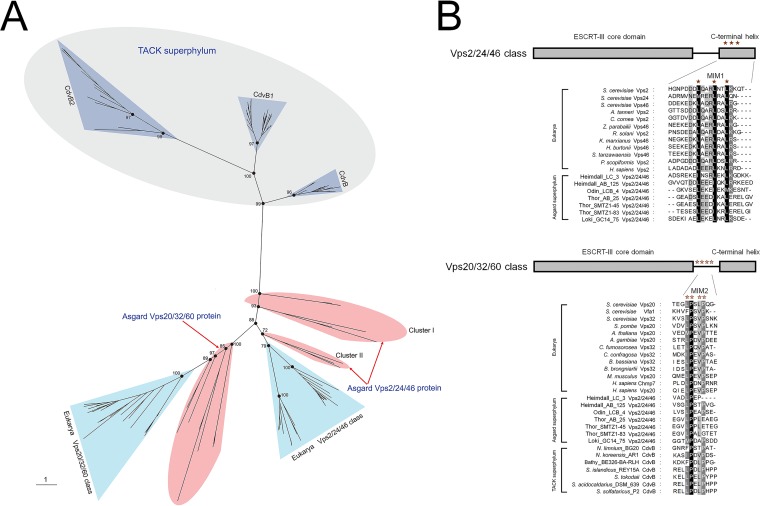
Given that the ESCRT-III subunits are tightly linked to the functional complexity of ESCRT (12), we first performed a detailed sequence comparison and phylogenetic analysis of the Vps2/24/46 and Vps20/32/60 as well as the Vps4 ATPase from Asgard archaea based on the available genomic data (18, 19). In the unrooted maximum likelihood phylogenetic tree Vps2/24/46 and Vps20/32/60, the Asgard proteins form a cluster with eukaryotic homologs that is separated from the archaeal (TACK) CdvB cluster by a long branch (Fig. 1A; see also Fig. S1 and Table S1 in the supplemental material), supporting the notion that Asgard archaea possess “eukaryote-like” ESCRT-III subunits. All the Asgard Vps20/32/60 proteins form a strongly supported clade with the eukaryotic Vps20/32/60 which is compatible with a direct ancestral relationship. The Asgard Vps2/24/46 proteins formed three clades, one of which (Odinarchaeota, Lokiarchaeota, and Thorarchaeota) clustered with the eukaryotic homologs, whereas the remaining two (Heimdallarchaeota) placed near the root of the Asgard-eukaryote branch (Fig. 1A). This tree topology probably resulted from the acceleration of evolution in Heimdallarchaeota.
FIG 1.
Phylogenetic and amino acid sequence analysis of the ESCRT-III-related subunits in archaea and eukarya. (A) Unrooted maximum likelihood phylogenetic tree of the ESCRT-III-related subunits in archaea and eukarya. Additional information on Asgard Vps2/24/46 and Vps20/32/60 can be found in Table S1 in the supplemental material. Bootstrap values are shown for some nodes. (B) Predicted MIM1 and MIM2 in Asgard Vps2/24/46 and Vps20/32/60, respectively. Additional information of proteins used here can be found in Table S1. The ESCRT-III core domain, C-terminal helix, and MIM1 and MIM2 are presented.
Phylogenetic analysis of ESCRT-III-related proteins in the eukarya and archaea. The tree was reconstructed by maximum likelihood analysis using 156 representative amino acid sequences based on the LG+G4 model (recommended by the “TESTONLY”), with the “-bb 1000” option, and the bootstrap values are shown on nodes. Download FIG S1, PDF file, 0.3 MB (316.2KB, pdf) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of proteins used in this study. Download Table S1, DOCX file, 0.02 MB (24.3KB, docx) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In addition to the phylogenetic results, we found that the Asgard Vps2/24/46 contained leucine-rich motifs located in the C-terminal helix and resembling the C-terminal MIM1 that are conserved in eukaryotes although some leucine residues were substituted by isoleucine in the Asgard homologs (Fig. 1B) (9). The C-terminal regions of the Asgard Vps20/32/60 contain proline-rich motifs that resemble MIM2, although they do not fully conform to the MIM2 consensus in eukaryotes and TACK archaea (10, 11). Taken together, the results of phylogenetic analysis and motif search for ESCRT-III subunits not only demonstrate the Asgard-eukaryote affinity but also show that the ancestors of the Vps2/24/46 and Vps20/32/60 groups had already diverged in Asgard archaea, antedating eukaryogenesis.
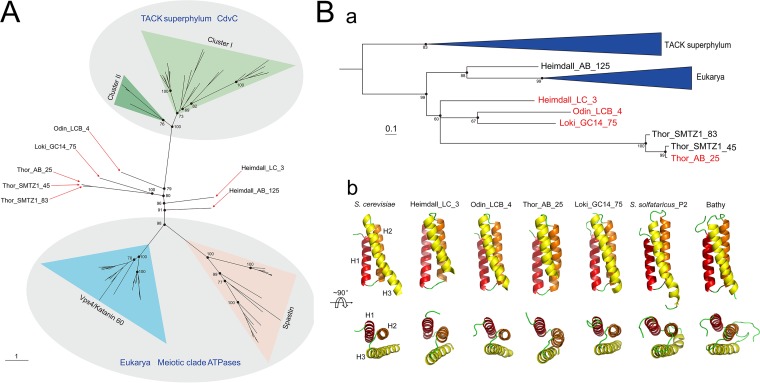
It appears likely that Vps4 structurally and functionally coevolved with ESCRT-III subunits in Asgard archaea. To explore the evolution of Vps4, an unrooted maximum likelihood phylogenetic tree was constructed for the group of ATPases, including CdvC from the TACK superphylum, Asgard Vps4, and the so-called eukaryotic “meiotic clade” comprised of Vps4, katanin 60, and spastin (23). As in the ESCRT-III subunit tree, the Asgard Vps4 formed a branch with the eukaryotic homolog that was separated by a long, strongly supported branch from the archaeal CdvC branch (Fig. 2A and Fig. S2). The Asgard Vps4 did not form a single clade, but rather four clades, all of which were located close to the root of the Asgard-eukaryote branch.
FIG 2.
Phylogenetic and structural analysis of the Asgard Vps4. (A) Unrooted maximum likelihood phylogenetic analysis of the Vps4-related in archaea and eukarya. Additional information on Asgard Vps4 can be found in Table S1. Bootstrap values are shown for some nodes. (B) Phylogenetic (a) and structural (b) analysis of the Asgard Vps4 MIT domain. The sequences of CdvC MIT domain are used as an outgroup to further confirm the phylogenetic relationship of the MIT domain in eukaryotic and Asgard Vps4. The antiparallel three-helix bundle of MIT domains is shown explicitly.
Phylogenetic analysis of Vps4-related proteins in eukarya and archaea. The tree was reconstructed by maximum likelihood analysis using 76 representative amino acid sequences based on the LG+I+G4 model (recommended by the “TESTONLY”), with the “-bb 1000” option, and the bootstrap values are shown on nodes. Download FIG S2, PDF file, 0.2 MB (257.2KB, pdf) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Despite their high divergence demonstrated by the lack of monophyly in the phylogenetic tree (Fig. 2A and Fig. S2), all Asgard Vps4 contain the eukaryotic-like “arginine collar” that consists of three conserved arginine residues (Fig. S3A). In eukaryotes, this motif is located in the pore loop 2 of Vps4 and is involved in the ESCRT-III filament translocation to the central pore of the Vps4 hexamer for disassembly (Fig. S3B) (24, 25).
Predicted “arginine collar” in Vps4 of Asgard archaea and eukarya. (A) The Walker A, Walker B, Sensor I, ARG finger, and Sensor II are conserved across all the indicated sequences and are shown to confirm the location of the “arginine collar”. Conserved arginine residues of the “arginine collar” are highlighted (red shading and red letters, respectively). The information of proteins used here can be found in Table S1. (B) The top and bottom views of the hexameric ring (gray) were constructed for Heimdall_LC_3 Vps4 (white) as an example to demonstrate the location of the “arginine collar” including R222, R231, and R232. Download FIG S3, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Because Vps4 recognizes ESCRT-III subunits via the MIT domain, we specifically analyzed the phylogeny of the MIT domains of the Vps4 proteins from Asgard archaea, eukaryotes, and TACK archaea. The tree demonstrates a clear affiliation of Asgard archaea with eukaryotes that, in this case, form a clade with one of the MIT domains from Heimdallarchaeota (Fig. 2B and Fig. S4). Affiliation with Heimdallarchaeota has been previously observed for many Asgard archaeal genes (19, 26, 27).
Phylogenetic analysis of the MIT domain in Vps4-related proteins. The tree was reconstructed by maximum likelihood analysis using 48 amino acid sequences based on theLG+I+G4 model (recommended by the “TESTONLY”), with the “-bb 1000” option, and the bootstrap values are shown on nodes. Download FIG S4, PDF file, 0.2 MB (230.1KB, pdf) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To further structurally characterize the MIT domain in Asgard Vps4, we constructed stable models of full-length Vps4 from Heimdallarchaeota_LC_3 (Heimdall_LC_3), Odinarchaeota_LCB_4 (Odin_LCB_4), Thorarchaeota_AB_25 (Thor_AB_25), and Lokiarchaeum_GC14_75 (Loki_GC14_75) using homology modeling and molecular dynamics simulation, and compared these with the S. cerevisiae Vps4 structure. For a control for the Asgard Vps4, we include CdvC from Sulfolobus solfataricus_P2 (cluster I in Fig. 2A) and Bathyarchaeota (cluster II in Fig. 2A). All MIT domains of Asgard Vps4 and TACK CdvC adopted a three-helix bundle structure that is closely similar to the S. cerevisiae MIT domain structure, although the helices in both the Asgard and TACK structures are somewhat shorter than in the S. cerevisiae structure (Fig. 2Bb and Fig. S5).
The number of Vps4 and CdvC MIT domain amino acid residues in alpha conformation of Asgard archaea, Saccharomyces cerevisiae, and TACK archaea during molecular dynamics simulations. The curves of the numbers of Vps4 MIT domain amino acid residues in the alpha conformation of Asgard and S. cerevisiae, which were calculated from the last 200-ns MD simulation trajectories, were plotted against simulation time. Download FIG S5, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taken together, the above data suggest that the evolution of Asgard Vps4, especially their MIT domain, was accompanied by the functional divergence of the ESCRT-III subunits. Thus, although the Asgard Vps4 proteins are highly diverged, the results of sequence comparison, phylogenetic analysis, and structural modeling are compatible with coevolution of Vps4 with ESCRT-III subunits and an ancestral relationship between the membrane remodeling machineries of Asgard and eukaryotes. Furthermore, it can be predicted that Asgard Vps2/24/46 and Vps20/32/60 form ESCRT-III-like filaments similar to those in eukaryotes.
Interactions between Asgard Vps4 and ESCRT-III subunits.
As previously described, unlike the CdvBs, Asgard Vps2/24/46 and Vps20/32/60 share the same eukaryotic ESCRT-III secondary structure, and these structures are probably responsible for their ability to bind to Vps4 like their eukaryotic counterparts (12). Isothermal titration calorimetry (ITC) was used to verify that the MIT domain of Asgard Vps4 binds to Vps2/24/46 and Vps20/32/60, respectively (Fig. S6). To characterize the interactions between Asgard Vps4 and ESCRT-III subunits, the respective binding free energies were estimated by molecular mechanics-generalized Born surface area (MM-GBSA) calculations (Table S2A) (28, 29). The binding free energies of Vps4-Vps2/24/46 in Heimdall_LC_3, Odin_LCB_4, Thor_AB_25, and Loki_GC14_75 were calculated as −39.02, −61.85, −71.81, and −72.24 kcal/mol, respectively. All these values, although compatible with stable binding, are lower than the binding free energy of Vps4-Vps2 (−82.98 kcal/mol) in S. cerevisiae, suggesting that the affinity of Asgard Vps4 for Vps2/24/46 is weaker than that of S. cerevisiae Vps4 for Vps2. The binding free energies for Asgard Vps4-Vps20/32/60 differed to a greater degree, indicating variation in the affinities (Table S2B). The Thor_AB_25 value of −119.97 kcal/mol was substantially greater than the binding free energy of the Vps4-Vps20 interaction in S. cerevisiae (−88.30 kcal/mol), the values for Heimdall_LC_3 (−81.43 kcal/mol) and Loki_GC14_75 (−88.89 kcal/mol) were comparable to those for S. cerevisiae, and the value for Odin_LCB_4 (−49.53 kcal/mol) was much lower than in S. cerevisiae.
ITC binding profiles of Asgard Vps4 MIT domain titrated with Asgard Vps2/24/46 and Vps20/32/60. (A) The curve of Heimdall_LC_3 TF-Vps4-MIT titrated with Heimdall_LC_3 TF-Vps2/24/46 was fit to Sequential Binding Sites with ΔH1 = −4.73 × 105 cal mol−1 and ΔH2 = 9.31 × 105 cal mol−1. The curve of Heimdall_LC_3-Vps4-MIT titrated with Heimdall_LC_3 TF-Vps20/32/60 was fit to Sequential Binding Sites with ΔH1 = 2.78 × 106 cal mol−1 and ΔH2 = −2.31 × 106 cal mol−1. (B) The curve of Odin_LCB_4 TF-Vps4-MIT titrated with Odin_LCB_4 Vps2/24/46 was fit to One Set of Sites with ΔH = 3.92 × 105 cal mol−1. The curve of Odin_LCB_4 TF-Vps4-MIT titrated with Odin_LCB_4 TF-Vps20/32/60 was fit to One Set of Sites with ΔH = 3.31 × 105 cal mol−1. (C) The curve of Thor_AB_25 TF-Vps4-MIT titrated with Thor_AB_25 TF-Vps2/24/46 was fit to Sequential Binding Sites with ΔH1 = 3.25 × 108 cal mol−1 and ΔH2 = 1.29 × 106 cal mol−1. The curve of Thor_AB_25 TF-Vps4-MIT titrated with Thor_AB_25 TF-Vps20/32/60 was fit to One Set of Sites with ΔH = 1.62 × 106 cal mol−1. (D) The curve of Loki_GC14_75 TF-Vps4-MIT titrated with Loki_GC14_75 TF-Vps2/24/46 was fit to Sequential Binding Sites with ΔH1 = −1.21 × 105 cal mol−1 and ΔH2 = 4.22 × 105 cal mol−1. The curve of Loki_GC14_75 TF-Vps4-MIT titrated with Loki_GC14_75 TF-Vps20/32/60 was fit to One Set of Sites with ΔH = 1.42 × 106 cal mol−1. Binding to a transcription factor (TF) control surface was negligible (not shown). Download FIG S6, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted binding free energies between Vps4 and ESCRT-III subunits (Vps2/24/46 [A] and Vps20/32/60 [B]). Download Table S2, DOCX file, 0.02 MB (18.9KB, docx) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
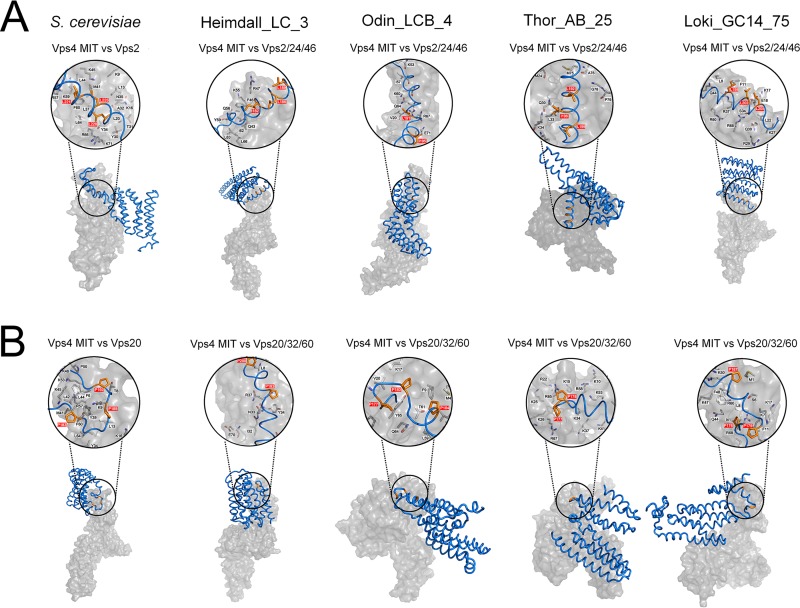
We further analyzed the structural basis for the MIT domain of Asgard Vps4 binding to the putative MIM1 and MIM2 of Vps2/24/46 and Vps20/32/60, respectively, by using MM-GBSA calculations (28). The key amino acids that contribute to the Vps4 MIT domain binding to the Vps2/24/46 MIM1 in Heimdall_LC_3, Odin_LCB_4, and Thor_AB_25 are mainly located in helix 2 and helix 3 of the MIT domain similar to the locations of MIM1-interacting residues in S. cerevisiae Vps4 (Fig. 3A and Table S3). These findings are consistent with the MIM1 peptide binding at the interface between helix 2 and helix 3 of the MIT domain as observed in eukaryotes (10, 30). In Loki_GC14_75, the key amino acid residues are located in helix 1 and helix 2, suggesting a distinct interaction mode.
FIG 3.
Comparison of the Vps4 (surface representation, gray) in complex with ESCRT-III subunits (ribbon representation, blue) in Asgard archaea. The MIM1 and MIM2 are shown in orange (stick representation, orange) and highlighted in red in close-up views (space-filling representation). The black letters indicate the main residues in the MIT domains that contribute to the interaction. The Vps4 MIT domain in complex with Vps2/24/46 (A) and Vps20/32/60 (B) subunits in S. cerevisiae, Heimdallarchaeota_LC_3 (Heimdall_LC_3), Odinarchaeota_LCB_4 (Odin_LCB_4), Thorarchaeota_AB_25 (Thor_AB_25), and Lokiarchaeum_GC14_75 (Loki_GC14_75) are indicated.
The dominant amino acid residues of Vps4 MIT domain involved in binding with Vps2/24/46 are listed. Download Table S3, DOCX file, 0.02 MB (17.7KB, docx) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The key residues involved in the MIT-Vps20/32/60 interactions are spread among helix 1, helix 2, and helix 3, in positions closely similar to those involved in the MIT-Vps20 interactions in S. cerevisiae (Fig. 3B and Table S4). Thus, the MIM2 peptide is predicted to bind the grooves formed by the three-helix bundle rather than helix 1 and helix 3 only as also observed for the eukaryotic ESCRT-III (10, 11). Taken together, these findings indicate that the interactions of the Asgard Vps4 MIT domain with the MIM1 (in Vps2/24/46) and MIM2 (in Vps20/32/60) closely resemble the corresponding interactions in eukaryotes.
The dominant amino acid residues of Vps4 MIT domain involved in binding with Vps20/32/60 are listed. Download Table S4, DOCX file, 0.02 MB (17.4KB, docx) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Asgard Vps4 phenotypically complements the vps4 null mutation in S. cerevisiae.
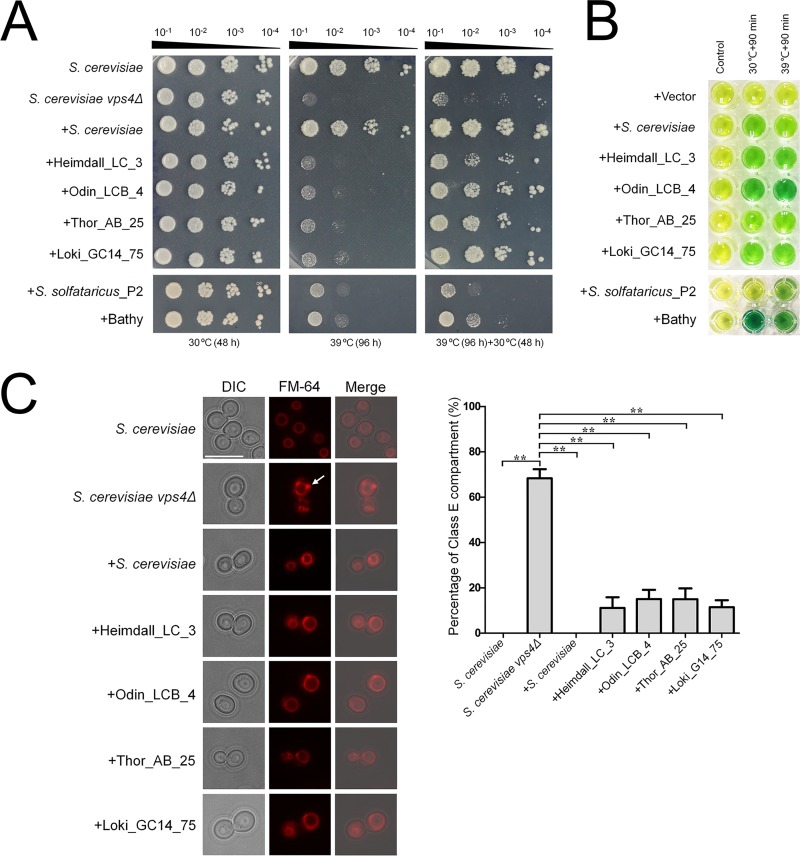
We further sought to determine whether the Asgard and eukaryotic Vps4 ATPases were functionally interchangeable. To this end, Heimdall_LC_3, Odin_LCB_4, Thor_AB_25, and Loki_GC14_75 were tested for the ability to complement the S. cerevisiae vps4 null mutation. For a control for the Asgard Vps4, we performed the complementation assays with CdvC from S. solfataricus_P2 and Bathyarchaeota. Briefly, we performed codon optimization again for the coding sequences of Asgard Vps4 and TACK CdvC for expression in S. cerevisiae and assembled the coding sequences into transcription units of the pPOT-RFP vector that contains a native promoter region of S. cerevisiae BY4741 vps4 (a 500-bp DNA sequence region upstream from the ATG start codon of this gene) and an S. cerevisiae cytochrome c isoform 1 (CYC1) terminator using the YeastFab Assembly method (31), respectively. The assembly products were transformed into an S. cerevisiae vps4Δ strain by the lithium acetate (LiAc)/polyethylene glycol (PEG) method (32). As previously described, in S. cerevisiae, vps4 null mutation resulted in temperature-sensitive growth defect, causing growth arrest at 39°C (33, 34). We found that the Asgard Vps4 could slightly suppress the growth defect of vps4Δ strain at 39°C (Fig. 4A). Remarkably, however, after incubation at 39°C for 96 h, the growth of a vps4Δ strain bearing Asgard Vps4 showed substantial, although variable, restoration at 30°C, in sharp contrast with a vps4Δ strain for which no restoration was observed (Fig. 4A). Nevertheless, both CdvCs showed only minimal growth restoration of the vps4Δ strain at 39°C and a limited enhancement of viability at 30°C; complementation with these proteins was substantially less efficient than that observed with their Asgard counterparts. Furthermore, the S. cerevisiae Vps4, Asgard Vps4, and CdvCs were re-codon-optimized, synthesized, and cloned into a pCold-TF vector (TaKaRa Bio Co. Ltd., Japan), respectively. After expression in Escherichia coli BL21, proteins were purified by Mag-Beads His-Tag Protein purification kit (BBI Co., Ltd., China). The biochemical experiments in vitro show that these purified proteins are active ATPases both at 30°C and 39°C (Fig. 4B). This observation eliminates the possibility that the poor complement result of CdvCs was due to the lack of ATPase activity at 39°C and is compatible with the involvement of the ATPase activity of Asgard Vps4 in sustaining the viability of the S. cerevisiae vps4Δ mutant under nonpermissive conditions.
FIG 4.
Functional complementation of Saccharomyces cerevisiae vps4 null mutants by Asgard Vps4. (A) Complementation of the high-temperature-sensitive growth defect of vps4 mutant cells. Five microliters of a series of 10-fold dilutions derived from a starting suspension of an OD600 of 10−1 was inoculated into SC-Ura medium. (B) The ATPase activity of S. cerevisiae Vps4, Asgard Vps4, and Cdvs at 30°C and 39°C were confirmed by a malachite green assay. The substrates would turn from golden to green owing to the inorganic phosphate released from ATP hydrolysis by Vps4 under the indicated condition. (C, left) The class E compartments in S. cerevisiae vps4 null mutants were largely abrogated by Asgard Vps4. The vacuolar morphologies in the indicated strains were visualized by fluorescence microscopy. The white arrow indicates the class E compartment in a vps4 null mutant. Bar = 10 μm. (Right) Quantification of class E compartment in the indicated strains. The results represented the means from three independent replicates (20 cells per experiment), and standard deviations are indicated by the error bars. Statistical significance was assessed by one-way analysis of variance with Bonferroni’s multiple-comparison test. **, P < 0.01.
As previously described, vps4 null mutation could induce formation of an aberrant prevacuolar compartment adjacent to the vacuoles, known as class E compartment, due to the block of intracellular protein trafficking (3, 30, 33). To further demonstrate that Asgard Vps4 is functionally analogous to its eukaryotic counterpart, we observed the vacuoles in the S. cerevisiae cells bearing Asgard Vps4. As expected, the characterized class E compartment vacuolar morphology was clearly observed in the S. cerevisiae vps4Δ cells, and this defect was almost completely rescued by S. cerevisiae Vps4 (Fig. 4C). We found that the Vps4 of Heimdall_LC_3, Odin_LCB_4, Thor_AB_25, and Loki_GC14_75 also partially complemented the aberrant vacuoles in the vps4 null mutant, with reduction of the class E compartment to about 80% of that observed with the native S. cerevisiae Vps4 (Fig. 4C). However, the enlarged vacuoles induced in the vps4Δ strain were not markedly eliminated by the Asgard Vps4. Taken together, these findings show that the Asgard Vps4 are functionally more similar to the eukaryotic homologs than homologs from other archaea.
DISCUSSION
In this work, we combined computational approaches, including sequence comparison, phylogenetic analysis, and structural modeling, with genetic and biochemical experiments to investigate the evolutionary and functional relationships between the ESCRT-III machineries of Asgard archaea and eukaryotes. Phylogenetic analyses of both the ESCRT-III subunits and Vps4 ATPase show a clear affinity between Asgard archaea and eukaryotes, to the exclusion of the other archaea. Moreover, the divergence of the two groups of ESCRT-III subunits already occurred in Asgard archaea.
The results of amino acid sequence analysis and structural modeling are best compatible with the coevolution of Vps4 with the ESCRT-III subunits. In particular, the interaction between the MIT domain of Vps4 and the MIM1- and MIM2-like of the ESCRT-III subunits appears to have evolved already in Asgard archaea.
The findings of the computational analysis are complemented by our experimental results. In particular, we show that Asgard Vps4 is capable of complementing the S. cerevisiae vps4 null mutant much more efficiently than homologs from Crenarchaeota and Bathyarchaeota. This enhanced functionality might be underpinned by the evolution of distinct, “eukaryote-like” structural features, such as the arginine collar that is involved in the disassembly of ESCRT-III polymers.
Taken together, all these findings are compatible with the direct origin of the eukaryotic ESCRT machinery from the Asgard ancestor. In a broader evolutionary context, the ESCRT complex likely evolved in the common ancestor of the TACK and Asgard superphyla, whereas its further elaboration occurred in the Asgard lineage. The key event apparently was the duplication of CdvB that seems to combine features of Vps2/24/46 and Vps20/32/60 (12), with subsequent functional diversification of the subunits and coevolution with Vps4.
An intriguing outstanding question is the function of the ESCRT machinery in the Asgard archaea. There is no indication that these (or any other) archaea possess intracellular membranes (22), so the ESCRT-III proteins and Vps4 are likely to be involved in cell division as demonstrated for the Cdv proteins of Crenarchaeota. However, the specialization of the ESCRT-III subunits might provide for the formation of eukaryote-like filaments that could be involved not only in the inside-out fission to produce membrane vesicles that have been observed in the MK-D1 strain, but also the outside-in fission that allows the Asgard archaea to engulf their bacterial metabolic partners. The latter capability is critical for the “Entangle-Engulf-Enslave model” of eukaryogenesis (22). Further molecular and cell biological study of the Asgard membrane remodeling apparatus, even if challenging due to the recalcitrance of these organisms to growth in culture, should shed light on the origin of the eukaryotic endomembrane system, one of the key aspects of eukaryogenesis.
MATERIALS AND METHODS
Bioinformatics analysis.
All the protein sequences were obtained either by NCBI accession number or by BLAST search (35) of the nonredundant protein sequences against the local Nr database. The protein sequences were aligned using MUSCLE (V3.8.1551) (36), trimmed with TrimAl (V1.4) (37) before construction of phylogenetic trees using IQ-Tree (V1.6.5) (38). The indicated functional domains of proteins were analyzed by Interpro (https://www.ebi.ac.uk/interpro/) and NCBI’s conserved domain database.
Homology modeling and docking study.
We searched the Vps4, Vps2/24/46, and Vps20/32/60 sequences belonging to S. cerevisiae, Lokiarchaeum_GC14_75, Thorarchaeota_AB_25, Heimdallarchaeota_LC_3, and Odinarchaeota_LCB_4 from the NCBI database and CdvC sequences belonging to Sulfolobus solfataricus_P2, and Bathyarchaeota (https://www.ncbi.nlm.nih.gov, NCBI:protein accession numbers KZV07689.1, P36108.2, and NP_013794.1; KKK42121.1, KKK42122.1, and KKK44605.1; OLS30569.1, OLS30568.1, and OLS30800.1; OLS27542.1, OLS27541.1, and OLS27540.1; OLS18192.1, OLS18193.1, OLS18194.1, AAK41192.1, and WP_119819537.1, respectively) to build the homology model. The cryo-electron microscopy (cryo-EM) structure of Vps4 (E233Q) hexamer belonging to S. cerevisiae was obtained from the Protein Data Bank (PDB accession number or code 5XMI) (39); subunit B was chosen for the modeling template, and the missing residues (1 to 118) were built at the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER). Sequence alignments and homology modelings of Vps4 for Lokiarchaeota, Thorarchaeota, Heimdallarchaeota, and Odinarchaeota with unknown structures were conducted using the MODELLER program (40), downloaded and installed from the salilab server (https://salilab.org/modeller/download_installation.html). The three-dimensional structures of Vps2/24/46 and Vps20/32/60 for S. cerevisiae and four Asgard archaea were also built at the I-TASSER server. Among several three-dimensional models generated using homology modeling and ab initio method, the best model was selected after a series of refining and minimization and molecular dynamics simulation employing ff14SB force fields parameters by AMBER 16.0 package (41). Then the complexes of Vps2/24/46 and Vps20/32/60 against Vps4 were simulated using the ZDOCK server (42). Ten top docking poses were generated.
Molecular dynamics simulation.
The parallel version of AMBER 16.0 package was used to prepare the complex files and conduct molecular dynamics (MD) simulations employing ff14SB force field parameters. The ionizable residue default protonation states in AMBER 16.0 were assigned. All MD simulations were conducted by applying cubic periodic boundary conditions (PBC) and in an explicit water box of TIP3P (transferable intermolecular potential with three points) water molecules (43) with a minimum distance of 10.0 Å between the complex surface and water box boundary. The Na+ or Cl− counterions were added in sufficient number to neutralize any net charges of the structures above. All of the chemical bond lengths of hydrogen-heavy atoms were restrained by the SHAKE algorithm (44). A cutoff radius of 10.0 Å was set for both nonbonded electrostatic and van der Waals interactions. Long-range electrostatic forces were taken into account using the particle mesh Ewald (PME) method (45). Langevin dynamics and Langevin piston methods were applied to keep the temperature (300 K) and pressure (1 bar) of the system constant, respectively. The time step was set at 2.0 fs.
The solvated systems were minimized using the PMEMD.CUDA module enabled with NVIDIA graphics processing units (GPUs) (46, 47) in three stages: (i) keeping the solute fixed and minimizing the positions of the water and counterions first with 100 kcal/(mol·Å2) restraints and (ii) reducing to 10 kcal/(mol·Å2), and (iii) last, for the entire system without any restraining force. Each stage was conducted with 10,000 steps of steepest descent algorithm, followed by 1,000 steps of conjugate gradient minimization to get rid of any unfavorable steric contacts for both solvent and protein molecules. Then, a NVT (amount of substance [N], volume [V], and temperature [T]) simulation was conducted to slowly heat the systems temperature from 0 K to 300 K over a period of 500 ps, and density was equilibrated for 2,000 ps with a weak restraint applied to the whole protein at 1 atm and 300 K. Finally, all restraints were removed, and production MD simulations were conducted at constant pressure (1 atm) and temperature (300 K) in the NPT (amount of substance [N], pressure [P], and temperature [T]) ensemble. For each system, MD simulation was performed for 500 ns and repeated thrice with different random number, and a total of 1.5-μs trajectory was analyzed by using a CPPTRAJ module (48).
Calculations of binding free energy.
The binding free energies of Vps2/24/46 and Vps20/32/60 against Vps4 were calculated by molecular mechanics-generalized Born surface area (MM-GBSA) method (28, 29). All energy components were calculated using 500 snapshots that were extracted every 200 ps during the last 100 ns of each MD simulation trajectory. The configurational entropy was not considered in the approach, as it is extremely time-consuming. Therefore, the binding free energy in the solvent environment can be expressed as follows:
The ΔEele, ΔEvdw, ΔGnp, and ΔGele represented electrostatic energy in the gas phase, van der Waals energy in the gas phase, nonpolar solvation energy, and polar solvation energy, respectively. All energy terms were calculated using MM-GBSA calculations, and the ΔGele is estimated by the generalized Born (GB) model (29), and the ΔGnp is calculated from the solvent-accessible surface area (SASA) of the molecules by molsur with the 0.00542 and 0.92 values for SURFTEN and SURFOFF, respectively (49). The decomposition of binding free energies were calculated at the residue pair level for a further investigation of the interactions between complexes using the MM-GBSA decomposition program (50, 51) implemented in AMBER 16.0.
Protein expression in Escherichia coli BL21 and purification for biochemical assays in vitro.
The Vps4, Vps2/24/46, and Vps20/32/60 coding sequences belonging to S. cerevisiae, Lokiarchaeum_GC14_75, Thorarchaeota_AB_25, Heimdallarchaeota_LC_3, and Odinarchaeota_LCB_4, and CdvC coding sequences belonging to Sulfolobus solfataricus_P2, and Bathyarchaeota from NCBI database (https://www.ncbi.nlm.nih.gov, NCBI:protein accession numbers KZV07689.1, P36108.2, and NP_013794.1; KKK42121.1, KKK42122.1, and KKK44605.1; OLS30569.1, OLS30568.1, and OLS30800.1; OLS27542.1, OLS27541.1, and OLS27540.1; OLS18192.1, OLS18193.1, OLS18194.1, AAK41192.1, and WP_119819537.1, respectively) were codon optimized by GeneDesign (http://54.235.254.95/gd/) for expression in E. coli BL21, synthesized (BGI Genomics Co., Ltd.), and respectively, cloned into a pCold-TF vector (TaKaRa Bio Co. Ltd., Japan) that includes an N-terminal His tag and a soluble trigger factor chaperone tag. The E. coli BL21 (TaKaRa Bio Co. Ltd., Japan) bearing the recombinant vectors were inoculated in LB medium containing 100 μg/ml carbenicillin, and incubated at 37°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8, and then isopropyl-d-1-thiogalactopyranodside was added at the final concentration of 0.5 mM, followed by incubation at 15°C for 18 to 24 h. The cell pellets were collected and resuspended in 20 ml binding buffer (20 mM phosphate buffer [pH 7.4], 500 mM NaCl, 50 mM imidazole, 1 mM dithiothreitol, 1 mM lysozyme, and 1 mM phenylmethylsulfonyl fluoride), followed by ultrasonic decomposition. Next, the target proteins were purified by Mag-Beads His-Tag Protein purification kit (BBI Co., Ltd., China) with wash buffer (20 mM phosphate buffer [pH 7.4], 500 mM NaCl, 100 mM imidazole, and 0.1% NP-40) and elution buffer (20 mM phosphate buffer [pH 7.4], 500 mM NaCl, and 500 mM imidazole). Finally, the purified proteins were concentrated to 1 to 2 ml in phosphate-buffered saline (PBS) (pH 7.4) by 30K Amicon Ultra-15 (Millipore Co. Ltd., USA). The concentrations of these proteins were determined by Bradford Protein assay kit (Beytotime Bio Co. Ltd., China). The purified Vps4, Vps2/24/46, and Vps20/32/60 belonging to S. cerevisiae, Lokiarchaeum_GC14_75, Thorarchaeota_AB_25, Heimdallarchaeota_LC_3, and Odinarchaeota_LCB_4 were used for isothermal titration calorimetry assay. The purified Vps4 belonging to S. cerevisiae, Lokiarchaeum_GC14_75, Thorarchaeota_AB_25, Heimdallarchaeota_LC_3, and Odinarchaeota_LCB_4, and Cdv belonging to Sulfolobus solfataricus_P2, and Bathyarchaeota were used for ATPase activity assay.
Isothermal titration calorimetry assay.
The isothermal titration calorimetry (ITC) assay was conducted at 25°C using an ITC200 system (MicroCal, USA). The Vps4 MIT domain (3 μM in PBS buffer) was placed in the cell and titrated with 19 injections of 10 μl of Vps2/24/46 or Vps20/32/60 (33 μM in PBS buffer) at 2-min intervals. The heat of ligand dilution into buffer was subtracted from the reaction heat, after removing the data of first injection. Data analysis was conducted using Origin 7.0 (MicroCal, USA).
ATPase activity assay.
The ATPase activity was determined by a slightly modified malachite green assay (52). In short, the purified proteins (4 μM) were incubated with reaction buffer (1 mM ATP, 20 mM HEPES [pH 7.4], 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol [DTT]) in a total volume of 50 μl at the indicated temperature for 90 min, and was immediately stopped by liquid nitrogen. Then, the reaction mixture was added with 100 μl of malachite green color buffer (14 mM ammonium molybdate, 1.3 M HCl, and 1.5 mM malachite green) and 50 μl of 21% (wt/vol) citric acid, followed by incubation at room temperature for 30 min. Finally, the reaction mixture that turned green was attributed to the free phosphate released by Vps4 ATP hydrolysis. Additionally, the control experiments were identical to the treatment group, except that the mixture of Vps4 and reaction buffer was immediately treated with liquid nitrogen before the addition of malachite green color buffer and citric acid, and these experiments were performed to eliminate the interference of irrelevant free phosphate. Also, the empty vector was used to prove that the ATP hydrolysis is due to Vps4.
S. cerevisiae strains and cultivation.
The S. cerevisiae strain BY4741 (MATa leu2Δ0 met15Δ0 ura3Δ0 his3Δ1) and its derivative vps4 null mutant strain YPR173Ca (designated S. cerevisiae vps4Δ in this study) were from S. cerevisiae deletion mutant library (53). S. cerevisiae cells were routinely cultured in YPD medium (10 g/liter yeast extract, 20 g/liter peptone, 20 g/liter glucose) or SC-Ura medium (synthetic complete medium lacking uracil) [6.7 g/liter yeast nitrogen base (YNB), 0.01 μmol/liter Fe(NH4)2(SO4)2, 20 g/liter glucose, and complete amino acids without uracil] at 30°C unless otherwise noted. The solid media were identical to those of YPD or SC-Ura except that agar was present.
Complementation assay.
The Vps4 coding sequences belonging to Lokiarchaeum_GC14_75, Thorarchaeota_AB_25, Heimdallarchaeota_LC_3, and Odinarchaeota_LCB_4, and CdvC coding sequences belonging to Sulfolobus solfataricus_P2, and Bathyarchaeota (NCBI:protein accession numbers KKK42121.1, OLS30569.1, OLS27542.1, OLS18192.1, AAK41192.1, and WP_119819537.1, respectively) were codon optimized by GeneDesign (http://54.235.254.95/gd/) for expression in S. cerevisiae, before synthesis by BGI Genomics Co., Ltd. (54). To eliminate the interference of transcriptional level factors, a native promoter region of S. cerevisiae BY4741 vps4 (a 500-bp DNA sequence region upstream from the ATG start codon of this gene) was used to drive the coding sequences. Then, we assembled the coding sequences, the S. cerevisiae vps4 native promoter, and an S. cerevisiae CYC1 (cytochrome c isoform 1) terminator into a pPOT-RFP vector according to a developed YeastFab Assembly protocol (31). In addition, the pPOT-RFP vector containing the entire S. cerevisiae BY4741 vps4 with its native promoter and the CYC1 terminator were transformed into the S. cerevisiae vps4 null mutant (32), and this reconstituted strain was designated the “+S. cerevisiae” strain shown in the figures. In this study, both the S. cerevisiae and S. cerevisiae vps4Δ were transformed with the pPOT-RFP vector as the control.
FM-64M staining.
S. cerevisiae cells of each strain were cultured in SC-Ura medium at 30°C and normalized to an OD600 of 0.5 to 0.8. Then, the S. cerevisiae cells were stained with 80 μM FM-64M (AAT Bioquest Co. Ltd., China) at 30°C for 20 min, and next cultured for 120 min after washes with medium. Finally, the S. cerevisiae cells were examined under an N-STORM fluorescence microscope (Nikon Co. Ltd., Japan).
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 91851105, 31622002, 31970105, 31725002, 21625302, and 31800615), the China Postdoctoral Science Foundation (grant 2018M643153), the Basic and Applied Basic Research of Guangdong Province (grant 2019A1515110089), the Shenzhen Science and Technology Program (grants JCYJ20170818091727570 and KQTD20180412181334790), Shenzhen Key Laboratory of Synthetic Genomics (ZDSYS201802061806209), Guangdong Provincial Key Laboratory of Synthetic Genomics (2019B030301006), and the Key Project of Department of Education of Guangdong Province (grant 2017KZDXM071). E.V.K. is supported by Intramural Research Program funds of the National Institutes of Health of the United States.
Zhongyi Lu and Meng Li conceived and designed the experiments. Zhongyi Lu, Tianyi Li, Siyu Zhang, and Jinquan Li performed the experiments. Huiying Chu and Guohui Li designed the molecular dynamics strategy. Ting Fu performed the simulations. Zhongyi Lu, Ting Fu, and Huiying Chu analyzed the data. Yang Liu and Junbiao Dai contributed reagents/materials/analysis tools. Zhongyi Lu, Ting Fu, Eugene Koonin, and Meng Li wrote the paper, and all authors edited and approved the paper.
We declare that we have no conflicts of interest.
Footnotes
Citation Lu Z, Fu T, Li T, Liu Y, Zhang S, Li J, Dai J, Koonin EV, Li G, Chu H, Li M. 2020. Coevolution of eukaryote-like Vps4 and ESCRT-III subunits in the Asgard archaea. mBio 11:e00417-20. https://doi.org/10.1128/mBio.00417-20.
REFERENCES
- 1.Dacks JB, Peden AA, Field MC. 2009. Evolution of specificity in the eukaryotic endomembrane system. Int J Biochem Cell Biol 41:330–340. doi: 10.1016/j.biocel.2008.08.041. [DOI] [PubMed] [Google Scholar]
- 2.Hartman H, Fedorov A. 2002. The origin of the eukaryotic cell: a genomic investigation. Proc Natl Acad Sci U S A 99:1420–1425. doi: 10.1073/pnas.032658599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babst M, Sato TK, Banta LM, Emr SD. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J 16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. 2004. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 5.Olmos Y, Carlton JG. 2016. The ESCRT machinery: new roles at new holes. Curr Opin Cell Biol 38:1–11. doi: 10.1016/j.ceb.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCullough J, Frost A, Sundquist WI. 2018. Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu Rev Cell Dev Biol 34:85–109. doi: 10.1146/annurev-cellbio-100616-060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leung KF, Dacks JB, Field MC. 2008. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9:1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- 8.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. 2009. Membrane scission by the ESCRT-III complex. Nature 458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 10.Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. 2008. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell 15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 11.Kojima R, Obita T, Onoue K, Mizuguchi M. 2016. Structural fine-tuning of MIT-interacting motif 2 (MIM2) and allosteric regulation of ESCRT-III by Vps4 in yeast. J Mol Biol 428:2392–2404. doi: 10.1016/j.jmb.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Caspi Y, Dekker C. 2018. Dividing the archaeal way: the ancient Cdv cell-division machinery. Front Microbiol 9:174. doi: 10.3389/fmicb.2018.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova KS, Yutin N, Bell SD, Koonin EV. 2010. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol 8:731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL, Williams RL, Bell SD. 2011. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell 41:186–196. doi: 10.1016/j.molcel.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. 2008. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A 105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samson RY, Dobro MJ, Jensen GJ, Bell SD. 2017. The structure, function and roles of the archaeal ESCRT apparatus. Subcell Biochem 84:357–377. doi: 10.1007/978-3-319-53047-5_12. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Gao R, Li C, Ni J, Yang Z, Zhang Q, Chen H, Shen Y. 2017. Functional assignment of multiple ESCRT-III homologs in cell division and budding in Sulfolobus islandicus. Mol Microbiol 105:540–553. doi: 10.1111/mmi.13716. [DOI] [PubMed] [Google Scholar]
- 18.Spang A, Saw JH, Jorgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema T. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Backstrom D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJ. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 20.Seitz KW, Dombrowski N, Eme L, Spang A, Lombard J, Sieber JR, Teske AP, Ettema TJG, Baker BJ. 2019. Asgard archaea capable of anaerobic hydrocarbon cycling. Nat Commun 10:1822. doi: 10.1038/s41467-019-09364-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Zhou Z, Pan J, Baker BJ, Gu JD, Li M. 2018. Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota. ISME J 12:1021–1031. doi: 10.1038/s41396-018-0060-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito Y, Sakai S, Song C, Tasumi E, Yamanaka Y, Yamaguchi T, Kamagata Y, Tamaki H, Takai K. 2020. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577:519–525. doi: 10.1038/s41586-019-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monroe N, Hill CP. 2016. Meiotic clade AAA ATPases: protein polymer disassembly machines. J Mol Biol 428:1897–1911. doi: 10.1016/j.jmb.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. 2008. Biochemical and structural studies of yeast Vps4 oligomerization. J Mol Biol 384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang B, Stjepanovic G, Shen Q, Martin A, Hurley JH. 2015. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat Struct Mol Biol 22:492–498. doi: 10.1038/nsmb.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Da Cunha V, Gaia M, Nasir A, Forterre P. 2018. Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genet 14:e1007215. doi: 10.1371/journal.pgen.1007215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spang A, Stairs CW, Dombrowski N, Eme L, Lombard J, Caceres EF, Greening C, Baker BJ, Ettema T. 2019. Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat Microbiol 4:1138–1148. doi: 10.1038/s41564-019-0406-9. [DOI] [PubMed] [Google Scholar]
- 28.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE III. 2000. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 29.Bashford D, Case DA. 2000. Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem 51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 30.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. 2007. ESCRT-III recognition by VPS4 ATPases. Nature 449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 31.Guo Y, Dong J, Zhou T, Auxillos J, Li T, Zhang W, Wang L, Shen Y, Luo Y, Zheng Y, Lin J, Chen GQ, Wu Q, Cai Y, Dai J. 2015. YeastFab: the design and construction of standard biological parts for metabolic engineering in Saccharomyces cerevisiae. Nucleic Acids Res 43:e88. doi: 10.1093/nar/gkv464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz RD, Schiestl RH. 2007. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:35–37. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- 33.Scheuring S, Röhricht RA, Schöning-Burkhardt B, Beyer A, Müller S, Abts HF, Köhrer K. 2001. Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. J Mol Biol 312:469–480. doi: 10.1006/jmbi.2001.4917. [DOI] [PubMed] [Google Scholar]
- 34.Scheuring S, Bodor O, Rohricht RA, Muller S, Beyer A, Kohrer K. 1999. Cloning, characterisation, and functional expression of the Mus musculus SKD1 gene in yeast demonstrates that the mouse SKD1 and the yeast VPS4 genes are orthologues and involved in intracellular protein trafficking. Gene 234:149–159. doi: 10.1016/s0378-1119(99)00163-8. [DOI] [PubMed] [Google Scholar]
- 35.Mount DW. 2007. Using the basic local alignment search tool (BLAST). CSH Protoc 2007:pdb.top17. doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- 36.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun S, Li L, Yang F, Wang X, Fan F, Yang M, Chen C, Li X, Wang HW, Sui SF. 2017. Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution. Nat Commun 8:16064. doi: 10.1038/ncomms16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez R, Sali A. 2000. Comparative protein structure modeling. Introduction and practical examples with modeller. Methods Mol Biol 143:97–129. doi: 10.1385/1-59259-368-2:97. [DOI] [PubMed] [Google Scholar]
- 41.Case DA, Betz RM, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Lin C, Luchko T, Luo R, Madej B, Mermelstein D, Merz KM, Monard G, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Sagui C, Simmerling CL, Botello-Smith WM, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Xiao L, Kollman PA. 2016. AMBER 2016. University of California, San Francisco, CA.
- 42.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. 2014. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 44.Ryckaert JP, Ciccotti G, Berendsen H. 1977. Numerical integration of Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 45.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. 1995. A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 46.Gotz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. 2012. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J Chem Theory Comput 8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, Walker RC. 2013. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput 9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 48.Roe DR, Cheatham TE III. 2013. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 49.Weiser J, Shenkin PS, Still WC. 1999. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J Comput Chem 20:217–230. doi:. [DOI] [Google Scholar]
- 50.Tsui V, Case DA. 2000. Theory and applications of the Generalized Born solvation model in macromolecular simulations. Biopolymers 56:275–291. doi:. [DOI] [PubMed] [Google Scholar]
- 51.Wang W, Donini O, Reyes CM, Kollman PA. 2001. Biomolecular simulations: recent developments in force fields, simulations of enzyme catalysis, protein-ligand, protein-protein, and protein-nucleic acid noncovalent interactions. Annu Rev Biophys Biomol Struct 30:211–243. doi: 10.1146/annurev.biophys.30.1.211. [DOI] [PubMed] [Google Scholar]
- 52.Merrill SA, Hanson PI. 2010. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J Biol Chem 285:35428–35438. doi: 10.1074/jbc.M110.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian K-D, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 54.Richardson SM, Wheelan SJ, Yarrington RM, Boeke JD. 2006. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res 16:550–556. doi: 10.1101/gr.4431306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analysis of ESCRT-III-related proteins in the eukarya and archaea. The tree was reconstructed by maximum likelihood analysis using 156 representative amino acid sequences based on the LG+G4 model (recommended by the “TESTONLY”), with the “-bb 1000” option, and the bootstrap values are shown on nodes. Download FIG S1, PDF file, 0.3 MB (316.2KB, pdf) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of proteins used in this study. Download Table S1, DOCX file, 0.02 MB (24.3KB, docx) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis of Vps4-related proteins in eukarya and archaea. The tree was reconstructed by maximum likelihood analysis using 76 representative amino acid sequences based on the LG+I+G4 model (recommended by the “TESTONLY”), with the “-bb 1000” option, and the bootstrap values are shown on nodes. Download FIG S2, PDF file, 0.2 MB (257.2KB, pdf) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted “arginine collar” in Vps4 of Asgard archaea and eukarya. (A) The Walker A, Walker B, Sensor I, ARG finger, and Sensor II are conserved across all the indicated sequences and are shown to confirm the location of the “arginine collar”. Conserved arginine residues of the “arginine collar” are highlighted (red shading and red letters, respectively). The information of proteins used here can be found in Table S1. (B) The top and bottom views of the hexameric ring (gray) were constructed for Heimdall_LC_3 Vps4 (white) as an example to demonstrate the location of the “arginine collar” including R222, R231, and R232. Download FIG S3, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis of the MIT domain in Vps4-related proteins. The tree was reconstructed by maximum likelihood analysis using 48 amino acid sequences based on theLG+I+G4 model (recommended by the “TESTONLY”), with the “-bb 1000” option, and the bootstrap values are shown on nodes. Download FIG S4, PDF file, 0.2 MB (230.1KB, pdf) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The number of Vps4 and CdvC MIT domain amino acid residues in alpha conformation of Asgard archaea, Saccharomyces cerevisiae, and TACK archaea during molecular dynamics simulations. The curves of the numbers of Vps4 MIT domain amino acid residues in the alpha conformation of Asgard and S. cerevisiae, which were calculated from the last 200-ns MD simulation trajectories, were plotted against simulation time. Download FIG S5, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ITC binding profiles of Asgard Vps4 MIT domain titrated with Asgard Vps2/24/46 and Vps20/32/60. (A) The curve of Heimdall_LC_3 TF-Vps4-MIT titrated with Heimdall_LC_3 TF-Vps2/24/46 was fit to Sequential Binding Sites with ΔH1 = −4.73 × 105 cal mol−1 and ΔH2 = 9.31 × 105 cal mol−1. The curve of Heimdall_LC_3-Vps4-MIT titrated with Heimdall_LC_3 TF-Vps20/32/60 was fit to Sequential Binding Sites with ΔH1 = 2.78 × 106 cal mol−1 and ΔH2 = −2.31 × 106 cal mol−1. (B) The curve of Odin_LCB_4 TF-Vps4-MIT titrated with Odin_LCB_4 Vps2/24/46 was fit to One Set of Sites with ΔH = 3.92 × 105 cal mol−1. The curve of Odin_LCB_4 TF-Vps4-MIT titrated with Odin_LCB_4 TF-Vps20/32/60 was fit to One Set of Sites with ΔH = 3.31 × 105 cal mol−1. (C) The curve of Thor_AB_25 TF-Vps4-MIT titrated with Thor_AB_25 TF-Vps2/24/46 was fit to Sequential Binding Sites with ΔH1 = 3.25 × 108 cal mol−1 and ΔH2 = 1.29 × 106 cal mol−1. The curve of Thor_AB_25 TF-Vps4-MIT titrated with Thor_AB_25 TF-Vps20/32/60 was fit to One Set of Sites with ΔH = 1.62 × 106 cal mol−1. (D) The curve of Loki_GC14_75 TF-Vps4-MIT titrated with Loki_GC14_75 TF-Vps2/24/46 was fit to Sequential Binding Sites with ΔH1 = −1.21 × 105 cal mol−1 and ΔH2 = 4.22 × 105 cal mol−1. The curve of Loki_GC14_75 TF-Vps4-MIT titrated with Loki_GC14_75 TF-Vps20/32/60 was fit to One Set of Sites with ΔH = 1.42 × 106 cal mol−1. Binding to a transcription factor (TF) control surface was negligible (not shown). Download FIG S6, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted binding free energies between Vps4 and ESCRT-III subunits (Vps2/24/46 [A] and Vps20/32/60 [B]). Download Table S2, DOCX file, 0.02 MB (18.9KB, docx) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The dominant amino acid residues of Vps4 MIT domain involved in binding with Vps2/24/46 are listed. Download Table S3, DOCX file, 0.02 MB (17.7KB, docx) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The dominant amino acid residues of Vps4 MIT domain involved in binding with Vps20/32/60 are listed. Download Table S4, DOCX file, 0.02 MB (17.4KB, docx) .
Copyright © 2020 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.