The outer membrane is a major determinant of the intrinsic antibiotic resistance of Gram-negative bacteria. It is composed of both lipopolysaccharide (LPS) and phospholipid, and the synthesis of these lipid species must be balanced for the membrane to maintain its barrier function in blocking drug entry. In this study, we identified an essential protein of unknown function as a key new factor in modulating LPS synthesis in the model bacterium Escherichia coli. Our results provide novel insight into how this organism and most likely other Gram-negative bacteria maintain membrane homeostasis and their intrinsic resistance to antibiotics.
KEYWORDS: LpxC, YejM, lipid A, outer membrane, phospholipid
ABSTRACT
Gram-negative bacteria are surrounded by a complex cell envelope that includes two membranes. The outer membrane prevents many drugs from entering these cells and is thus a major determinant of their intrinsic antibiotic resistance. This barrier function is imparted by the asymmetric architecture of the membrane with lipopolysaccharide (LPS) in the outer leaflet and phospholipids in the inner leaflet. The LPS and phospholipid synthesis pathways share an intermediate. Proper membrane biogenesis therefore requires that the flux through each pathway be balanced. In Escherichia coli, a major control point in establishing this balance is the committed step of LPS synthesis mediated by LpxC. Levels of this enzyme are controlled through its degradation by the inner membrane protease FtsH and its presumed adapter protein LapB (YciM). How turnover of LpxC is controlled has remained unclear for many years. Here, we demonstrate that the essential protein of unknown function YejM (PbgA) participates in this regulatory pathway. Suppressors of YejM essentiality were identified in lpxC and lapB, and LpxC overproduction was shown to be sufficient to allow survival of ΔyejM mutants. Furthermore, the stability of LpxC was shown to be reduced in cells lacking YejM, and genetic and physical interactions between LapB and YejM were detected. Taken together, our results are consistent with a model in which YejM directly modulates LpxC turnover by FtsH-LapB to regulate LPS synthesis and maintain membrane homeostasis.
INTRODUCTION
The bacterial cell envelope is essential for maintaining cell shape, withstanding mechanical stress, and resisting osmotic pressure and is a bacterium’s first line of defense against antibiotics, bacteriophages, and immune cells (1, 2). In Gram-negative bacteria, the cell envelope includes a symmetric inner membrane (IM) composed of phospholipids and an asymmetric outer membrane (OM) consisting of phospholipids in the inner leaflet and lipopolysaccharide (LPS) in the outer leaflet (2). The cell wall is located in the periplasmic space between the IM and OM and is made from a cross-linked heteropolymer called peptidoglycan (PG). Biogenesis of the cell envelope is tightly coordinated with cellular growth and division (3), with all three layers having to expand and divide each cell cycle without compromising their structural integrity.
The phospholipid (PL) and LPS membrane components are synthesized in the cytosol and within the inner leaflet of the IM. They must be transported across the IM and through the periplasm to build the OM (for reviews, see references 1 and 4–6). LPS consists of three covalently attached groups: a glycolipid called lipid A, a core oligosaccharide, and a longer, variable O-antigen polysaccharide chain. O antigen is synthesized separately from the lipid A core, but the two components are joined by ligation while anchored in the outer leaflet of the IM. Mature LPS molecules are then transported from the IM to the OM by the Lpt system, which forms a protein bridge connecting the IM and OM (7). The mechanism by which PLs are transported to the OM remains unclear (8).
R-3-Hydroxymyristoyl-acyl carrier protein (ACP) serves as the acyl donor for the synthesis of both PL and LPS (Fig. 1A). In the PL synthesis pathway, it is a substrate of (3R)-hydroxymyristoyl-ACP dehydratase (FabZ) (9). It is also utilized by LpxA (UDP-N-acetylglucosamine acyltransferase) in the first step of lipid A synthesis (10). However, this reaction is reversible (11); the committed step for lipid A production is catalyzed by the second enzyme, UDP-3-O-acyl-N-acetylglucosamine deacetylase (LpxC) (12). Balanced synthesis of LPS and PL is required to prevent loss of membrane integrity and cell death (13). In Escherichia coli, this balance is in part maintained by the inner membrane-localized, ATP-dependent zinc metalloprotease FtsH (14). In conjunction with its presumed adapter protein LapB (also called YciM) (15, 16), FtsH degrades LpxC to regulate the flux of lipid precursors through the LPS pathway. However, how LpxC proteolysis is modulated in response to disruptions in LPS or PL synthesis to maintain homeostasis is unclear.
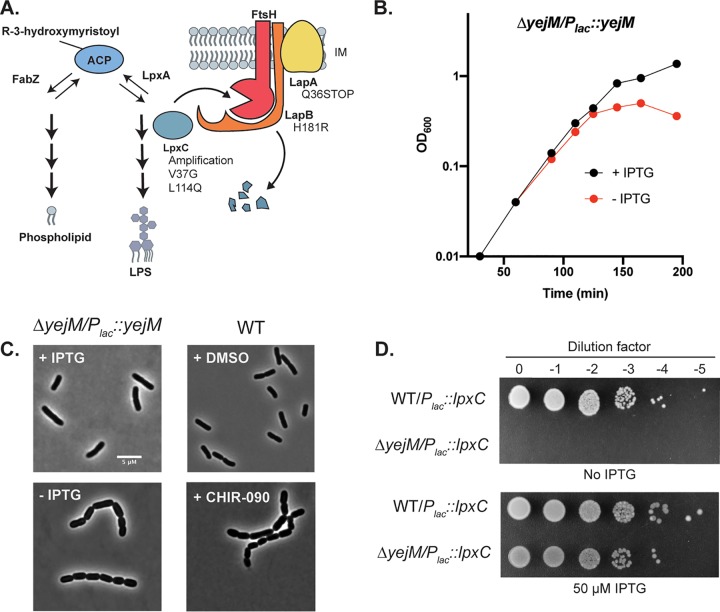
FIG 1.
The depletion of YejM leads to cell chaining and lysis. (A) Branched synthesis pathway leading from R-3-hydroxymyistoyl-ACP to either phospholipid or LPS. LpxC is turned over by FtsH. LapB is thought to serve as an adapter in this process. LapA is also shown, but its role in LpxC degradation is not clear. The identities of variants that suppress YejM essentiality are indicated in the diagram. (B) Growth curve following the depletion of YejM. Cells of strain EMF27 (ΔyejM/Plac::yejM) were grown without (red) or with (black) 50 μM IPTG to induce yejM expression, and growth was monitored by following the OD600 of the culture. (C) (Left) Micrographs of cells at the 125-min time point of the growth curve shown in panel B. (Right) Wild-type cells treated with DMSO (top) or 0.25 μg/ml CHIR-090 (bottom) for 2 h. Bar, 5 μm. (D) Serial dilutions of wild-type and ΔyejM cells harboring a Plac::lpxC plasmid were plated in the absence and presence of 50 μM IPTG.
Here, we identified the essential membrane protein of unknown function YejM (also called PbgA) as an inhibitor of LpxC turnover by FtsH-LapB in E. coli. Genetic suppressors of yejM essentiality first pointed us toward its potential role in maintaining sufficient LpxC levels for LPS synthesis. Subsequent analysis indicated that LpxC stability is reduced in the absence of YejM and that YejM interacts genetically and physically with LapB. Our results are therefore consistent with a model in which YejM interferes with LpxC proteolysis through its interaction with LapB. Complementary results that also support this model were reported while this paper was under review (17). We propose that the modulation of LpxC degradation by YejM is likely to be homeostatic and responsive to perturbations in the balance between LPS and PL synthesis.
RESULTS
Rationale.
In a genetic selection for suppressors of cell morphogenesis defects in E. coli, we identified mutations in yejM. These suppressors will be reported as part of a separate study, but they prompted us to further investigate YejM (PbgA) function, especially because it has remained one of the few essential genes in E. coli without a well-characterized activity. YejM is an IM protein with a five-transmembrane-domain N terminus that is essential for growth and a nonessential C-terminal periplasmic domain (18, 19). Nonsense mutations in yejM leading to the truncation of the periplasmic domain have previously been found to cause phenotypes consistent with defects in OM assembly (18–21), including reduced LPS/PL ratio, vancomycin sensitivity, temperature sensitivity, and leakage of periplasmic proteins. YejM shares structural similarities to LtaS, the enzyme that synthesizes lipoteichoic acids in many Gram-positive bacteria (22). Like LtaS, YejM has a hydrophobic binding pocket in the periplasmic domain that is important for protein function (23). However, the crystal structure of this domain of YejM revealed that it lacks residues that are important for LtaS catalytic activity, indicating that is unlikely to have a similar enzymatic function (23). Although previous studies have implicated YejM in the transport of cardiolipin to the OM in Salmonella enterica serovar Typhimurium (24, 25) and Shigella flexneri (26), a recent study suggested that it plays a broader yet ill-defined role in envelope assembly (27). We therefore thought that further study of the function of this essential protein was warranted.
Overproduction of LpxC suppresses the essentiality of YejM.
To begin investigating the essential function of YejM, we examined the terminal morphological phenotype induced upon its depletion. The native yejM gene was deleted in a strain harboring a plasmid that expressed yejM from an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible lac promoter (Plac). In the presence of inducer, these cells grew and divided normally (Fig. 1B). However, as observed previously (18, 19, 24), YejM depletion in the absence of IPTG led to slowed growth followed by partial lysis of the culture (Fig. 1B). Prior to lysing, the YejM-depleted cells formed cell chains (Fig. 1C; also, see Fig. S1 in the supplemental material), indicating a failure to complete cell separation. This morphology is reminiscent of cells defective in envelope biogenesis, including mutants defective in LPS assembly (28–30) and cells treated with the LpxC inhibitor CHIR-090 (Fig. 1C; also, see Fig. S1).
Quantification of cell chaining in the absence of YejM or following treatment with LpxC inhibitor CHIR-090. Cell chaining (Fig. 1C) was quantified by counting the number of cellular units per cell following the depletion of YejM (n = 20) (A) or treatment with 0.25 μg/ml CHIR-090 for 2 h (n =45) (B). The data were then plotted in Graphpad Prism. ****, P < 0.0001 (unpaired t-test). Download FIG S1, TIF file, 0.6 MB (618.9KB, tif) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next turned to suppressor analysis to identify mutations that bypass the essentiality of YejM as a means to understand its function. To this end, the yejM gene was cloned under the control of the lactose promoter (Plac) in a plasmid backbone designed by the de Boer laboratory to select for suppressors of ftsN essentiality (31). The vector encodes a temperature-sensitive replication protein [RepA(Ts)] and the restriction endonuclease I-SceI under the control of a temperature-sensitive lambda repressor (cI857), which is paired with an I-SceI cutting site in the vector backbone. To select for suppressors of YejM essentiality, a ΔyejM strain harboring the yejM suicide vector was plated at the nonpermissive temperature (37°C), where the plasmid will cease to replicate and be cleaved by expressed I-SceI. Under these conditions, suppressors arose at a frequency of ∼9 × 10−4. To increase the stringency of the selection, suppressors were also isolated on LB plates containing 1% sodium dodecyl sulfate (SDS), which reduced the suppressor frequency to ∼1 × 10−5. Twelve surviving colonies (5 selected on LB and 7 selected on LB containing 1% SDS) were isolated, and their mutations were mapped by whole-genome sequencing (Fig. 1A; also, see Table S1). Five of the suppressors contained genomic amplifications in a region containing lpxC. Two additional suppressors had missense mutations in lpxC, including one encoding a V37G substitution that has been demonstrated to lead to increased LpxC abundance in Klebsiella pneumoniae, suggesting increased stability (32). Three of the suppressors had mutations either in lapB, in its neighbor lapA, or in the region upstream of the lapAB operon. Overall, 10 of 12 suppressors had mutations predicted to increase cellular LpxC levels.
ΔyejM suppressors. Mutations mapped in each suppressor are listed. Download Table S1, PDF file, 0.1 MB (150.7KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The suppressor analysis suggested that an LpxC deficiency underpinned the lethality of a ΔyejM mutation. To investigate this possibility further, lpxC was cloned under Plac control on a multicopy plasmid. A strain harboring this vector was used as a recipient in a transduction using P1 phage grown on a ΔyejM::kanr donor strain. Kanamycin-resistant transductants were readily obtained on agar containing IPTG to induce lpxC expression from the plasmid, and these strains were found to be inducer-dependent for growth (Fig. 1D). Notably, the only previously reported suppressor of a ΔyejM mutation was a multicopy vector encoding holo-ACP synthase 2 (AcpT) (18–20), and catalytic activity of the synthase was found not to be required for suppression (18). Given our results, we wondered whether overproduction of AcpT also acts by promoting the accumulation of LpxC. Cells overexpressing acpT from a plasmid were indeed found to have elevated LpxC levels (see Fig. S2), suggesting that elevated AcpT may be an FtsH substrate and overwhelm the proteolytic machinery to allow LpxC accumulation. Taken together, our results thus far indicate that LpxC overproduction renders the normally essential YejM protein dispensable for growth.
The overexpression of acpT increases LpxC levels. Immunoblot detection of LpxC from extracts of wild-type cells harboring plasmids pPR111 (Plac::lpxC), pPR66 (Plac::empty), or pEMF43 (Plac::acpT) grown in the presence or absence of IPTG (50 μM [lane 2] or 1 mM [lanes 4 and 6]), as indicated. Download FIG S2, TIF file, 0.6 MB (639.3KB, tif) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LpxC is aberrantly degraded in the absence of YejM.
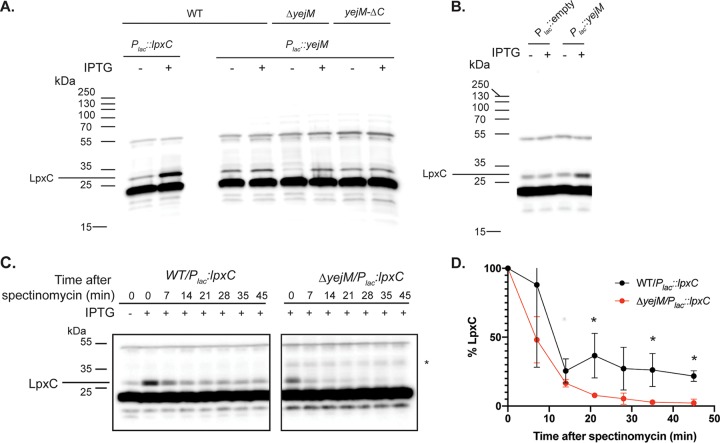
To further investigate the connection between YejM and LpxC, we investigated the effect of YejM inactivation on the steady-state levels of LpxC. Immunoblotting indicated that LpxC levels were unaffected in wild-type cells with or without moderate induction of yejM expression from a plasmid (Fig. 2A). However, depletion of YejM in cells with a deletion of the native gene led to a dramatic reduction in LpxC levels, to the point where it was barely detectable (Fig. 2A). Notably, cells harboring a yejM allele (yejM-ΔC) at its native locus in which a stop codon was introduced at residue 192 maintained normal LpxC levels whether or not expression of the full-length gene was induced from a plasmid (Fig. 2A), indicating that the N-terminal transmembrane domain of YejM is sufficient to prevent aberrant degradation of LpxC. Furthermore, increased expression of yejM from a plasmid with higher concentrations of inducer led to a striking increase in LpxC accumulation in otherwise wild-type cells (Fig. 2B). Based on these results, we hypothesized that LpxC is aberrantly degraded in the absence of YejM. To test this possibility, we took advantage of our ability to delete yejM in the presence of a plasmid overproducing lpxC. Wild-type and ΔyejM cells expressing lpxC from the plasmid were treated with spectinomycin to block translation, and the fate of previously translated LpxC was monitored by immunoblotting. Because LpxC is initially overproduced, rapid degradation of the protein is observed in both cell types in the first 7 min, which presumably reflects the degradation system attempting to return the concentration of LpxC to endogenous (uninduced) levels. After the initial phase of degradation, the LpxC concentration plateaued in wild-type cells at about 25% of the initial concentration and was maintained at this level for the rest of the time course (Fig. 2C). In contrast, LpxC levels decayed rapidly in the ΔyejM cells to undetectable levels without an observable plateau (Fig. 2C). Based on these results, we conclude that YejM is required to prevent excessive turnover of LpxC.
FIG 2.
Changes in yejM expression affect LpxC accumulation. (A) Immunoblot using anti-LpxC primary antibody. Lanes 1 and 2, extracts from wild-type cells with a Plac::lpxC (pPR111) plasmid grown in the absence (lane 1) or presence (lane 2) of 50 μM IPTG to serve as a marker for LpxC; lanes 3 and 4, wild-type cells with a Plac::yejM (pEMF17) plasmid grown for 195 min in the absence (lane 3) or presence (lane 4) of 50 μM IPTG; lanes 5 and 6, extracts from ΔyejM cells with a Plac::yejM (pEMF17) plasmid grown for 195 min in the absence (lane 5) or presence (lane 6) of 50 μM IPTG; lanes 7 and 8, extracts from yejM-ΔC cells with a Plac::yejM (pEMF17) plasmid grown for 195 min in the absence (lane 7) or presence (lane 8) of 50 μM IPTG. (B) Immunoblot detecting LpxC from extracts of wild-type cells with a Plac::empty (pPR66) or a Plac::artificialRBS_yejM (pEMF33) plasmid grown in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 1 mM IPTG. Note that the yejM in this plasmid has a strong artificial ribosome-binding site relative to the construct used for panel A. (C) Wild-type and ΔyejM cells harboring a Plac::lpxC plasmid (pPR111) were grown in the presence of 50 μM IPTG at 30°C in minimal medium to an OD600 of 0.5. Spectinomycin (300 μg/ml) was then added, and samples were taken at the indicated time points for the preparation of extracts and the detection of LpxC by immunoblotting. The asterisk denotes a nonspecific band that appeared sporadically in blots of the ΔyejM samples. Blots are representative of three independent experiments. Note that the first lane of the left blot shows the LpxC level in uninduced cells. (D) Quantification of LpxC levels following spectinomycin treatment in three independent experiments. Levels of LpxC are presented as a percentage of the initial LpxC concentration at time zero. The asterisk indicates a P value of <0.05 (unpaired t test). Error bars denote standard deviations (SD).
YejM interacts genetically and physically with LapB.
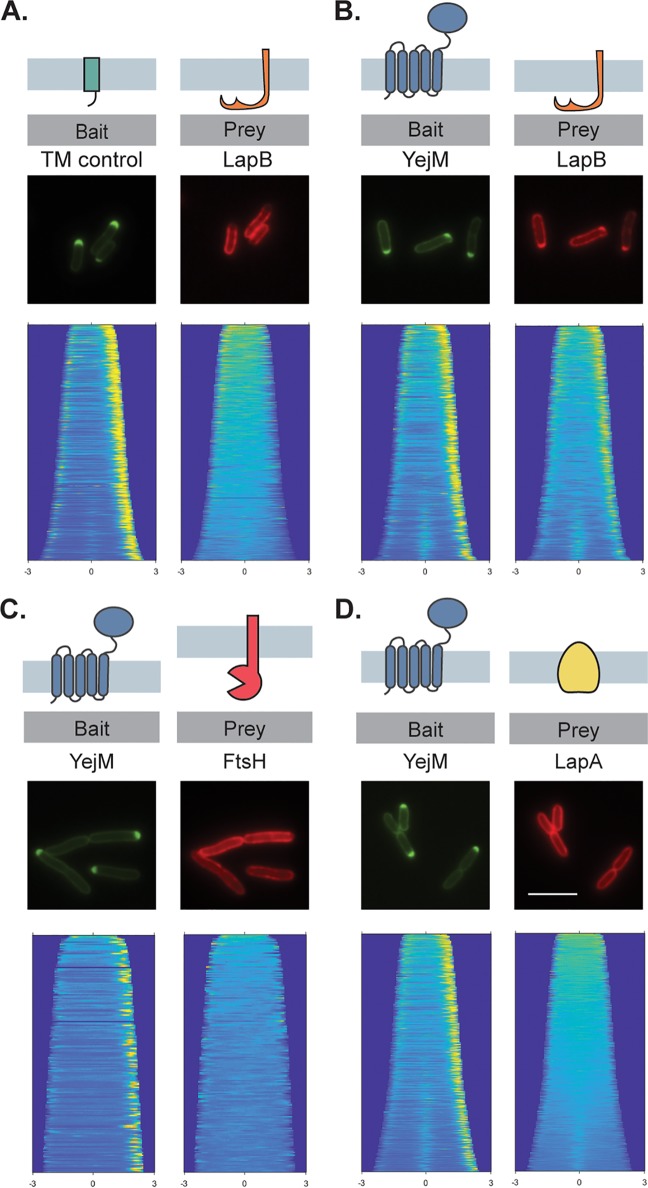
Our results thus far suggested a model in which YejM promotes LpxC accumulation by protecting it from the FtsH-LapB proteolytic system. We therefore investigated whether YejM interacts with any of the components of this system using our recently developed POLAR (PopZ-linked apical recruitment) two-hybrid assay (33). The POLAR assay takes advantage of the ability of the PopZ protein from Caulobacter crescentus to spontaneously form foci at the poles of E. coli cells. Therefore, protein-protein interactions can be assessed by fusing a “bait” protein to a PopZ-interaction domain called H3H4 along with green fluorescent protein (GFP) and then monitoring whether it can recruit a “prey” protein of interest fused to mScarlet to the cell pole. Using YejM as a bait, we found that it was able to recruit a LapB prey fusion to the pole (Fig. 3B), but not an FtsH or a LapA prey fusion (Fig. 3C and D). The recruitment of LapB prey to the pole was specific for YejM, as it was not recruited by a transmembrane control bait (Fig. 3A). However, we noticed that cells expressing LapB prey constructs formed chains and appeared to lyse when they were paired with the control bait (see Fig. S3A) but not with the YejM bait. Therefore, for the control in Fig. 3A we used a control bait construct in which yejM was silently expressed downstream of the control bait sequence, which eliminated the adverse morphological effects observed upon production of the LapB prey fusion. We also observed a positive interaction between YejM-ΔC and LapB (Fig. S3B), but unlike in assays with the full-length YejM bait and LapB prey, cell chaining was observed. Thus, some residual toxicity of the LapB prey construct was likely maintained when it was paired with the YejM-ΔC bait. Based on the POLAR results, we conclude that YejM interacts with the LapB component of the FtsH-LapB proteolytic system and that the N-terminal transmembrane domain of YejM is sufficient for this interaction.
FIG 3.
YejM interacts with LapB. The POLAR assay was used to assess protein-protein interactions (see the text for details). Shown are representative micrographs of cells expressing the indicated bait and prey proteins. Cells were transformed with plasmids producing the control bait, which consists of a single transmembrane domain derived from residues 2 to 55 of Pseudomonas aeruginosa PBP1b fused to PopZ-H3H4-GFP (pEMF55) (A) or PopZ-H3H4-GFP-YejM (pEMF35) (B to D). Note that the control bait construct also expresses unlabeled yejM in order to reduce LapB toxicity. Prey constructs produce C-terminal mScarlet fusions to the indicated proteins from plasmid vectors integrated into the chromosome. Bar, 5 μm.
The LapB prey construct does not interact with the control bait construct in the POLAR assay. Shown are representative micrographs of cells expressing the indicated bait and prey proteins. (A) Cells were transformed with plasmids expressing the control bait, which consists of a single transmembrane domain derived from residues 2 to 55 of Pseudomonas aeruginosa PBP1b fused to PopZ-H3H4-GFP (pHCL149). The LapB prey construct consists of a C-terminal mScarlet fusion to LapB and was expressed from an integrated plasmid (pHCL147 derivative pEMF36) under the control of the Plac promoter. The experimental setup is the same as for Fig. 3A, except that unlabeled YejM is not produced from the bait construct in this case. Note that without expressing the unlabeled YejM, cells begin to form chains, most likely due to the toxic effects of LapB overproduction. (B) The LapB prey construct is coexpressed with the YejM-ΔC bait construct (pEMF65). Bar, 5 μm. Download FIG S3, TIF file, 1.0 MB (1,005.6KB, tif) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
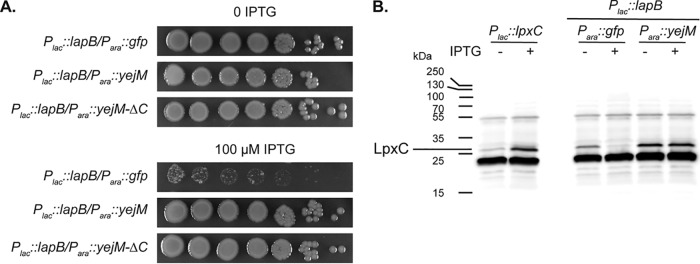
The effects of the LapB prey fusion observed on cell growth during the POLAR analysis suggested that LapB overproduction is toxic and that this toxicity can be overcome by coexpression of yejM. To investigate this possibility further, we monitored the effect of overproducing untagged LapB on cell growth when either full-length YejM, YejM-ΔC, or a GFP control was simultaneously overproduced from a compatible plasmid. Cells containing the lapB expression plasmid grew normally in the absence of inducer regardless of whether they overproduced YejM or GFP (Fig. 4A). When lapB expression was induced, plating efficiency was dramatically reduced for cells co-producing GFP (Fig. 4A). However, cells coproducing full-length YejM or YejM-ΔC were protected and plated at normal efficiency (Fig. 4A). To investigate whether the toxicity of LapB and its antagonism by YejM were related to LpxC turnover, we monitored LpxC abundance in cells overproducing LapB. Cells in which LapB and GFP were coproduced had reduced LpxC levels, whereas cells overexpressing yejM had elevated levels of LpxC and no apparent reduction in LpxC levels upon LapB overproduction (Fig. 4B). We conclude not only that YejM interacts with LapB but also that it likely serves as an antagonist of LapB activity to prevent LpxC degradation by the FtsH-LapB proteolytic machinery.
FIG 4.
YejM protects cells from LapB toxicity. (A) Serial dilutions of wild-type cells with the integrated lapB overexpression plasmid pEMF53 (Plac::lapB) and either pEMF57 (Para::gfp), pEMF54 (Para::yejM), or pEMF68 (Para::yejM-ΔC) were plated on LB agar with 0.2% arabinose and additionally supplemented with 100 μM IPTG, as indicated. Note that lapB-induced toxicity is relieved by yejM coexpression. (B) LpxC immunoblot. Extracts from WT cells harboring a Plac::lpxC plasmid (pPR111) grown with (lane 2) or without (lane 1) IPTG were used as a marker for the LpxC band. LpxC was also detected in extracts of MG1655(attHKEMF53) [WT(Plac::lapB)] cells harboring either pEMF57 (Para::gfp) (lanes 4 and 5) or pEMF54 (Para::yejM) (lanes 6 and 7) grown in LB containing 0.2% arabinose without (lanes 4 and 6) or with (lanes 5 and 7) 100 μM IPTG to induce lapB expression.
DISCUSSION
YejM (PbgA) has remained one of the few essential proteins of unknown function in E. coli. With the exception of a multicopy suppressor selection (18, 19), prior genetic studies primarily examined the phenotype of mutants encoding a truncated YejM protein lacking the nonessential C-terminal periplasmic domain (18–21, 24, 27). Cells producing these YejM-ΔC variants displayed a range of OM permeability barrier defects, suggesting an important yet ill-defined role for YejM in envelope biogenesis.
A more specific role was assigned to the YejM homolog from S. Typhimurium (PbgA), which was identified as a factor required for OM barrier function in cells activated for the two-component system PhoPQ (24). This regulatory system induces changes in the OM that help protect S. Typhimurium from the assaults it encounters in the phagosomes of host cells during infection (34). One of the observed changes in OM composition during PhoPQ induction is an increase in the content of the phospholipid cardiolipin (CL) (24, 35). This increase in CL levels in the OM was not observed in PhoPQ-activated cells producing a YejM-ΔC variant. This observation combined with the detection of CL binding by the YejM periplasmic domain in vitro led to the original proposal that the protein functions as a transporter that shuttles CL from the IM to the OM (24). A structure of full-length YejM was recently reported in which the protein was premixed with cardiolipin prior to crystallization (25). Two cardiolipin binding sites were observed in the structure, but they were positioned near the membrane facing surface of the transmembrane domain where any phospholipid species might be expected to interact with YejM when it is situated in the IM (25). Thus, the new structural information does not help explain how YejM might function as a cardiolipin transporter.
In another recent follow-up to the original S. Typhimurium study (24), mice were infected with S. Typhimurium producing a YejM-ΔC variant, and following growth in the host, suppressors that restored OM integrity to the YejM-ΔC cells were isolated in lpxC, lapB, and ftsH (27). It was also found that deletion of the C-terminal domain of YejM resulted in changes in the LPS and phospholipid composition of S. Typhimurium that could be at least partially rescued by the suppressors (27). As a result of this analysis, a variety of functions were proposed for YejM, including a general role in LPS assembly. It was also proposed that the periplasmic domain of YejM somehow facilitates lipid trafficking during stress and that the transmembrane region performs an undefined essential activity involving phospholipids (27).
A simplified picture of YejM function emerges from our study of a complete deletion allele of yejM in E. coli. Similar to the host-induced suppressors of the S. Typhimurium YejM-ΔC defect, we identified suppressors of YejM essentiality in the lpxC and lapB genes. We further showed that overexpression of lpxC was sufficient to allow complete deletion of yejM. Subsequent analysis demonstrated that levels of LpxC were strongly dependent on YejM. Inactivation of YejM led to aberrant degradation of LpxC and reduced levels of the enzyme, whereas overproduction of YejM promoted the hyperaccumulation of LpxC. We then identified an interaction between YejM and LapB, likely mediated through the N-terminal transmembrane domain of YejM, and discovered that the toxicity of LapB overproduction can be blocked by co-overproduction of either full-length YejM or YejM-ΔC. Overall, our results are consistent with a model in which YejM opposes LapB function to inhibit LpxC degradation by the FtsH protease (Fig. 5).
FIG 5.
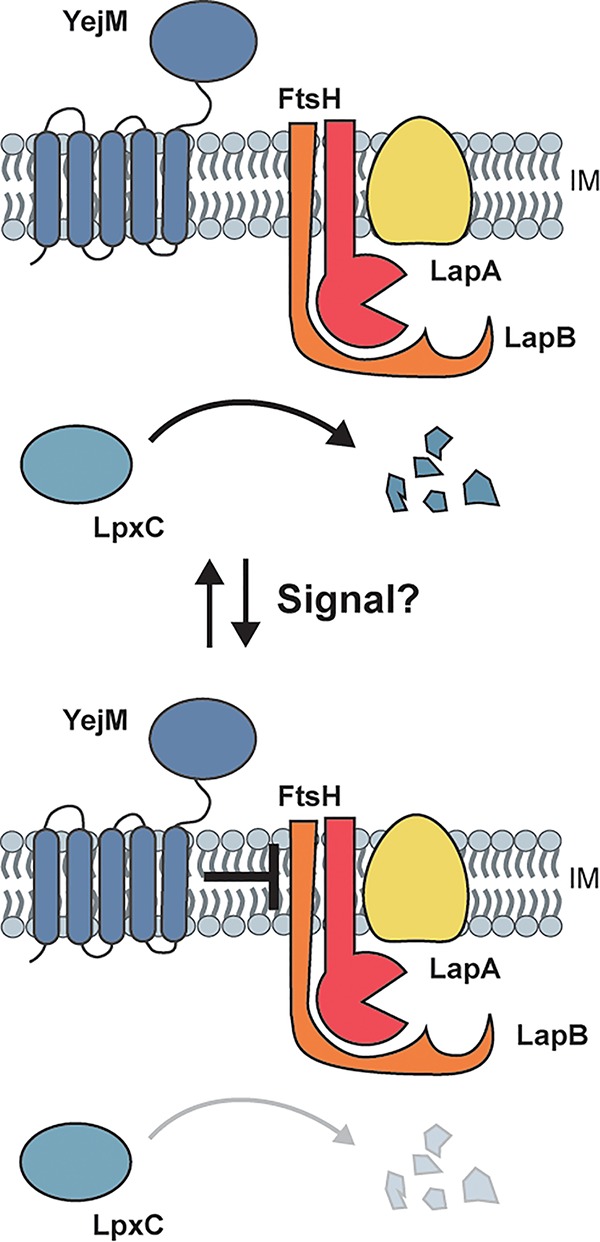
Model for the modulation of LpxC turnover by YejM. LpxC is degraded by FtsH in a LapB-dependent manner. The role of LapA is unclear. We propose that in response to some signal, potentially the buildup of a lipid molecule in the inner membrane, YejM blocks LpxC degradation by FtsH-LapB. This inhibition is most likely mediated by an interaction between YejM and LapB and may be used to help balance LPS and phospholipid synthesis in response to perturbations or fluctuations.
The ability of YejM to antagonize LpxC degradation makes it an attractive candidate for a factor that modulates the stability of LpxC in response to disruptions in LPS and phospholipid synthesis homeostasis. It has been known for some time that inhibition of LpxC or overexpression of fabZ, both of which presumably shift flux of lipid precursors away from the LPS synthesis pathway, leads to the stabilization of LpxC (9, 32, 36–38). Recently, it was also shown that LpxC turnover is reduced in cells with elevated activity of phospholipase A (PldA), which cleaves phospholipids in the outer leaflet of the OM, another marker for defects in LPS and phospholipid synthesis homeostasis (39). In each of these cases, the precise molecular signal(s) that modulates LpxC degradation by FtsH-LapB remains to be determined. However, lipids like acyl coenzyme A (acyl-CoA), LPS, and phospholipids, or intermediates in the synthesis of these molecules, are likely candidates (36, 39). Notably, YejM is related to enzymes like LtaS and EptA that use phospholipid substrates either to polymerize lipoteichoic acids or to modify lipid A with a lipid head group, respectively (22, 40–43). Even though YejM lacks amino acids predicted to be required for enzymatic activity, it is conceivable that it retains the ability to bind LPS, phospholipids, or both, and that such binding events modulate its ability to interfere with LpxC degradation by FtsH-LapB. The C terminus of YejM is likely involved in recognizing any potential signaling molecules despite its dispensability for interacting with LapB or in maintaining normal steady-state levels of LpxC. We therefore suspect that most of the OM defects observed previously for cells producing YejM-ΔC variants, including reduced cardiolipin content in the OM of S. Typhimurium, ultimately stem from the improper control of LpxC turnover in response to stress and that a lipid transport function for YejM is unlikely. Although further studies are required to test these possibilities, the identification of YejM as an essential component in the pathway controlling LpxC levels represents an important step toward a mechanistic understanding of how Gram-negative bacteria balance phospholipid and LPS synthesis to properly assemble the OM layer that defines them.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used and generated here are derivatives of MG1655. Strains were cultured in LB medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) or minimal M9 medium (44) supplemented with 0.2% Casamino Acids and 0.2% glucose, arabinose, or maltose, as indicated in the figure legends.
Antibiotic concentrations are as follows (unless otherwise indicated): 25 μg/ml chloramphenicol (Cam), 25 μg/ml kanamycin (Kan), 5 μg/ml tetracycline (Tet), and 50 μg/ml spectinomycin (Spec). Strains EMF27 [ΔyejM::kanr/Plac::yejM] and EMF30 [ΔyejM::kanr/Plac::lpxC] were maintained on medium supplemented with 50 μM IPTG unless otherwise stated. All strains, plasmids, and primers used in this study are listed in Table S2, Table S3, and Table S4, respectively. Methods used to construct the ΔyejM strains and expression constructs used in this study are detailed in the supplemental material.
Strains used in this study. Download Table S2, PDF file, 0.04 MB (40.1KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S3, PDF file, 0.1 MB (99.8KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S4, PDF file, 0.03 MB (29.4KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Selection of suppressors of YejM essentiality.
The suppressor strain plasmid (pEMF20) was cloned via Gibson assembly in strain JLB45, which expresses cI857, in order to prevent zygotic induction of I-SceI. The plasmid was then transformed into MG1655 Chung competent cells. The ΔyejM::kanr allele was transduced into MG1655/pEMF20 via P1 transduction and confirmed via PCR (see Text S1 in the supplemental material), generating strain EMF37. Suppressors of ΔyejM were selected by growing EMF37 cells at 37°C on LB plates (including 1% SDS, where indicated). An overnight culture of EMF37 was prepared under the permissive condition (30°C; LB containing Kan, Spec, and 100 μM IPTG). For suppressors EMF40 to -44, the EMF37 overnight culture was serially diluted, plated on LB plates, and incubated at 37°C. For suppressors EMF45 to -51, the overnight culture of EMF37 was back diluted 1:50 and grown under permissive conditions until an optical density at 600 nm (OD600) of 0.42 was reached. Cells were then pelleted, washed, and resuspended in LB and allowed to grow for an additional hour at 37°C (nonpermissive conditions). Cells were then plated on LB + 1% SDS and incubated overnight at 37°C. The loss of plasmid pEMF20 was confirmed by screening for spectinomycin sensitivity. Overnight cultures of the suppressor strains were prepared, and 5 ml of each culture was pelleted and stored at −20°C. Genomic DNA (gDNA) was isolated from each pellet using the Wizard genomic DNA purification kit (Promega) and further purified using the genomic DNA Clean & Concentrate kit (Zymo Research). Whole-genome sequencing was performed as described previously (45) with some modifications (46) (Nextera DNA sample preparation kit). The concentration of the DNA in the samples was determined using the Qubit dsDNA HS assay kit, and the sizes of the products following tagmentation were determined using a high-sensitivity D1000 screen tape run on an Agilent 4200 TapeStation. The sequencing was carried out using a MiSeq reagent kit v3 (Illumina). The data were analyzed using the CLC Genomics Workbench software (Qiagen).
Supplemental methods for plasmid and strain construction. Download Text S1, PDF file, 0.1 MB (76.2KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Immunoblotting.
Cell pellets were collected and resuspended in water and 2× Laemmli sample buffer (100 mM Tris-HCl, pH 6.8; 2% SDS; 0.1% bromophenol blue; 20% glycerol) at a 1:1 ratio. Samples were boiled for 10 min and sonicated (Qsonica tip sonicator; amplification, 25%; time, 1 min) two or three times. Protein concentration was measured using the noninterfering (NI) protein assay (with bovine serum albumin [BSA] protein standard) (G Biosciences catalog no. 786-005) and was normalized using 1× sample buffer. Samples were run on a 15% polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was then rinsed in phosphate-buffered saline containing 0.1% Tween (PBS-T) (10% 10× PBS-T buffer, pH 7.4 [Sigma-Aldrich]) and blocked in 5% milk in PBS-T for 1.5 h. The membrane was then incubated in a primary antibody solution of 1% milk in PBS-T containing rabbit anti-lpxC antibody (a generous gift from the Doerrler lab) at a 1:10,000 dilution for approximately 16 h at 4°C. The membrane was then washed four times in PBS-T (once quickly and three times for 10 min each). The membrane was then incubated in secondary antibody solution (horseradish peroxidase [HRP]-conjugated anti-rabbit IgG; 1:1,000 dilution; Rockland no. 18–8816-33) in 0.2% milk in PBS-T for 2 h. Following 5 washes with PBS-T, the membrane was developed using SuperSignal West Pico Plus chemiluminescent substrate (Thermo Fisher Scientific catalog no. 34577) and imaged using the c600 Azure Biosystems platform.
LpxC degradation assay.
MG1655/pPR111 (wild type [WT]/Plac::lpxC] and EMF30 [ΔyejM/Plac::lpxC] cells were incubated overnight in 4 ml of LB containing Cam without or with 50 μM IPTG, respectively. Overnight cultures were diluted to an OD600 of 0.025 in 5 ml of LB containing Cam with (MG1655/pPR111 and EMF30) or without (MG1655/pPR111) 50 μM IPTG. Cultures were incubated at 37°C with shaking to an OD600 of ∼0.3 to 0.4. Cell concentrations were normalized, and cultures were back diluted again at a ratio of 1:50 in 100 ml M9 minimal medium containing 50 μg/ml chloramphenicol and 50 μM IPTG (except for the MG1655/pPR111 control without IPTG). Cells were grown at 30°C with shaking to an OD600 of 0.5. Then, 300 μg/ml spectinomycin was added to the cultures to inhibit protein synthesis. Samples were taken at 0, 7, 14, 21, 28, 35, and 45 min following the addition of spectinomycin and analyzed via immunoblotting for LpxC. The amount of LpxC was quantified by measuring the intensity of the LpxC band at each time point and normalizing the intensity to a nonspecific band used as a loading control. The level of LpxC at each time point was then presented as a percentage of protein present at time zero for each strain. These data were plotted in GraphPad Prism. The results represent three independent experiments; error bars in the figures show standard deviations (SD).
Monitoring growth during YejM depletion.
Cultures of TB28/pEMF17 [ΔlacIZYA::frt/Plac::yejM] and EMF27 [ΔlacIZYA::frt ΔyejM/Plac::yejM] were grown overnight at 37°C in 4 ml of LB containing Cam plus 50 μM IPTG. Cultures were then diluted to an OD600 of 0.025 in 6 ml of LB containing Cam plus 50 μM IPTG and grown at 37°C with shaking to an OD600 of 0.3. Then, 3 ml of culture was pelleted for 2 min at 5,000 rpm, washed once in LB containing Cam, and resuspended in 3 ml LB containing Cam. The concentration of the cultures was then normalized, and the samples were diluted at a ratio of 1:50 in 50 LB containing Cam with or without 50 μM IPTG. Cultures were incubated at 37°C with shaking. Samples were taken every 15 to 30 min, as indicated in Fig. 2C. The culture density was measured, and samples of cells were removed for fixation. In order to quantify cell chaining, the number of cellular units per chain (indicated by partial constriction between units) was counted per cell. All data were plotted in GraphPad Prism.
Effect of CHIR-090 on cell morphology.
MG1655 (WT) cells were grown overnight in LB at 37°C and back diluted to an OD600 of 0.05 in 5 ml of LB and grown to an OD600 of 0.4. Cells were back diluted again to an OD600 of 0.1 in 5 ml LB with dimethyl sulfoxide (DMSO) or 0.25 μg/ml CHIR-090 (Cayman Chemical Company catalog no. 728865-23-4). Cells were grown for 2 h and fixed before being visualized by phase-contrast microscopy. The extent of cell chaining was quantified as described above.
Phase-contrast microscopy.
For the phase-contrast micrographs in Fig. 1, cells were fixed in 2.6% in formaldehyde and 0.04% glutaraldehyde at room temperature for 1 h and stored at 4°C for up to 3 days. Prior to imaging, samples were immobilized on 2% agarose pads on 1-mm glass slides, and number 1.5 coverslips were used. Samples were imaged on a Nikon TE2000 inverted microscope using Nikon Elements Acquisition software AR 3.2. Cropping was done and additional adjustments were made using Fiji software.
POLAR analysis.
Cells were prepared for imaging as described previously (33). Briefly, cells from a single colony were grown in LB medium supplemented with Tet and Cam for 2 h at 37˚C. Cells were back diluted in M9 containing 0.2% arabinose and 100 μM IPTG and incubated for 2 h at 37˚C to induce expression of bait and prey protein fusions. Cells were immobilized on agarose pads as described above for imaging. Micrographs were taken on a Nikon Ti inverted microscope with a Plan APO lambda 100×/1.45 oil Ph3 DM lens objective, Lumencore SpectraX LED illumination, Chroma ET filter cubes for GFP (49002) and mCherry (49008), an Andor Zyla 4.2 Plus sCMOS camera, and Nikon Elements 4.30 acquisition software. The microscope slide was kept at 30°C using an environmental control chamber. Demographs showing polar localization were generated using a custom MATLAB code as described previously (33). Cells were aligned by length and oriented so that the pole with greater bait intensity was located on the right. The corresponding demograph of the prey signal was generated using the same orientation.
ACKNOWLEDGMENTS
We thank all the members of the Bernhardt and Rudner labs for their thoughtful discussions and advice throughout this project, especially Patty Rohs for her insight and guidance in the early stages of this project. We also acknowledge the consultation, support, and services provided by Paula Montero Llopis and the other members of the MicRoN (Microscopy Resources on the North Quad) team at HMS. We thank Bill Doerrler for the generous gift of the anti-LpxC antibody.
This work was supported by the National Institutes of Health (AI083365 to T.G.B.) and Investigator funds from the Howard Hughes Medical Institute. E.M.F. was supported in part by the T32 Bacteriology PhD Training Program (AI132120-02) awarded to the Harvard Graduate Program in Bacteriology.
Footnotes
This article is a direct contribution from Thomas G. Bernhardt, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Natividad Ruiz, The Ohio State University, and M. Stephen Trent, University of Georgia.
Citation Fivenson EM, Bernhardt TG. 2020. An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli. mBio 11:e00939-20. https://doi.org/10.1128/mBio.00939-20.
REFERENCES
- 1.May KL, Silhavy TJ. 2017. Making a membrane on the other side of the wall. Biochim Biophys Acta Mol Cell Biol Lipids 1862:1386–1393. doi: 10.1016/j.bbalip.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos MP, Robert V, Tommassen J. 2007. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol 61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 5.Henderson JC, Zimmerman SM, Crofts AA, Boll JM, Kuhns LG, Herrera CM, Trent MS. 2016. The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu Rev Microbiol 70:255–278. doi: 10.1146/annurev-micro-102215-095308. [DOI] [PubMed] [Google Scholar]
- 6.Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. doi: 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherman DJ, Xie R, Taylor RJ, George AH, Okuda S, Foster PJ, Needleman DJ, Kahne D. 2018. Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359:798–801. doi: 10.1126/science.aar1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powers MJ, Trent MS. 2019. Intermembrane transport: glycerophospholipid homeostasis of the Gram-negative cell envelope. Proc Natl Acad Sci U S A 116:17147–17155. doi: 10.1073/pnas.1902026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohan S, Kelly TM, Eveland SS, Raetz C, Anderson MS. 1994. An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J Biol Chem 269:32896–32903. [PubMed] [Google Scholar]
- 10.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson MS, Bull HG, Galloway SM, Kelly TM, Mohan S, Radika K, Raetz CR. 1993. UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J Biol Chem 268:19858–19865. [PubMed] [Google Scholar]
- 12.Anderson MS, Raetz C. 1987. Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J Biol Chem 262:5159–5169. [PubMed] [Google Scholar]
- 13.Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su LH, Fierke CA, Jackman JE, Raetz CR, Coleman J, Tomoyasu T, Matsuzawa H. 1999. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol 31:833–844. doi: 10.1046/j.1365-2958.1999.01221.x. [DOI] [PubMed] [Google Scholar]
- 14.Ito K, Akiyama Y. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol 59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- 15.Klein G, Kobylak N, Lindner B, Stupak A, Raina S. 2014. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J Biol Chem 289:14829–14853. doi: 10.1074/jbc.M113.539494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahalakshmi S, Sunayana M, SaiSree L, Reddy M. 2014. yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol Microbiol 91:145–157. doi: 10.1111/mmi.12452. [DOI] [PubMed] [Google Scholar]
- 17.Guest RL, Samé Guerra D, Wissler M, Grimm J, Silhavy TJ. 2020. YejM modulates activity of the YciM/FtsH protease complex to prevent lethal accumulation of lipopolysaccharide. mBio 11:e00598-20. doi: 10.1128/mBio.00598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Lay NR, Cronan JE. 2008. Genetic interaction between the Escherichia coli AcpT phosphopantetheinyl transferase and the YejM inner membrane protein. Genetics 178:1327–1337. doi: 10.1534/genetics.107.081836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirvas L, Nurminen M, Helander IM, Vuorio R, Vaara M. 1997. The lipid A biosynthesis deficiency of the Escherichia coli antibiotic-supersensitive mutant LH530 is suppressed by a novel locus, ORF195. Microbiology 143:73–81. doi: 10.1099/00221287-143-1-73. [DOI] [PubMed] [Google Scholar]
- 20.Nurminen M, Hirvas L, Vaara M. 1997. The outer membrane of lipid A-deficient Escherichia coli mutant LH530 has reduced levels of OmpF and leaks periplasmic enzymes. Microbiology 143:1533–1537. doi: 10.1099/00221287-143-5-1533. [DOI] [PubMed] [Google Scholar]
- 21.Qiu N, Misra R. 2019. Overcoming iron deficiency of an Escherichia coli tonB mutant by increasing outer membrane permeability. J Bacteriol 201:e00340-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu D, Wörmann ME, Zhang X, Schneewind O, Gründling A, Freemont PS. 2009. Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc Natl Acad Sci U S A 106:1584–1589. doi: 10.1073/pnas.0809020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong H, Zhang Z, Tang X, Huang S, Li H, Peng B, Dong C. 2016. Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria. Sci Rep 6:30815. doi: 10.1038/srep30815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ressl S, Kulasekara BR, Blanc M-P, Miller SI. 2015. Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17:441–451. doi: 10.1016/j.chom.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan J, Petersen EM, Hinds TR, Zheng N, Miller SI. 2020. Structure of an inner membrane protein required for PhoPQ-regulated increases in outer membrane cardiolipin. mBio 11:e03277-19. doi: 10.1128/mBio.03277-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi RM, Yum L, Agaisse H, Payne SM. 2017. Cardiolipin synthesis and outer membrane localization are required for Shigella flexneri virulence. mBio 8:e01199-17. doi: 10.1128/mBio.01199-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cian MB, Giordano NP, Masilamani R, Minor KE, Dalebroux ZD. 2019. Salmonella enterica serovar Typhimurium use PbgA/YejM to regulate lipopolysaccharide assembly during bacteremia. Infect Immun 88:e00758-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Normark S. 1969. Transduction and dominance studies of the envA gene present in a chain-forming mutant of Escherichia coli K 12. J Gen Microbiol 57. [PubMed] [Google Scholar]
- 29.Beall B, Lutkenhaus J. 1987. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol 169:5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grundström T, Normark S, Magnusson K. 1980. Overproduction of outer membrane protein suppresses envA-induced hyperpermeability. J Bacteriol 144:884–890. doi: 10.1128/JB.144.3.884-890.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu B, Persons L, Lee L, de Boer PA. 2015. Roles for both FtsA and the FtsBLQ subcomplex in FtsN‐stimulated cell constriction in Escherichia coli. Mol Microbiol 95:945–970. doi: 10.1111/mmi.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mostafavi M, Wang L, Xie L, Takeoka KT, Richie DL, Casey F, Ruzin A, Sawyer WS, Rath CM, Wei J-R, Dean CR. 2018. Interplay of Klebsiella pneumoniae fabZ and lpxC mutations leads to LpxC inhibitor-dependent growth resulting from loss of membrane homeostasis. mSphere 3:e00508-18. doi: 10.1128/mSphere.00508-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim HC, Bernhardt TG. 2019. A PopZ‐linked apical recruitment assay for studying protein–protein interactions in the bacterial cell envelope. Mol Microbiol 112:1757–1768. doi: 10.1111/mmi.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalebroux ZD, Miller SI. 2014. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol 17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. 2014. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc Natl Acad Sci U S A 111:1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emiola A, Andrews SS, Heller C, George J. 2016. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc Natl Acad Sci U S A 113:3108–3113. doi: 10.1073/pnas.1521168113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng D, Zhao J, Chung HS, Guan Z, Raetz CRH, Zhou P. 2013. Mutants resistant to LpxC inhibitors by rebalancing cellular homeostasis. J Biol Chem 288:5475–5486. doi: 10.1074/jbc.M112.447607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomanek N, Arends J, Lindemann C, Barkovits K, Meyer HE, Marcus K, Narberhaus F. 2018. Intricate crosstalk between lipopolysaccharide, phospholipid and fatty acid metabolism in Escherichia coli modulates proteolysis of LpxC. Front Microbiol 9:3285. doi: 10.3389/fmicb.2018.03285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May KL, Silhavy TJ. 2018. The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. mBio 9:e00379-18. doi: 10.1128/mBio.00379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H, Hsu F-F, Turk J, Groisman EA. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol 186:4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrera CM, Hankins JV, Trent MS. 2010. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol Microbiol 76:1444–1460. doi: 10.1111/j.1365-2958.2010.07150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anandan A, Evans GL, Condic-Jurkic K, O'Mara ML, John CM, Phillips NJ, Jarvis GA, Wills SS, Stubbs KA, Moraes I, Kahler CM, Vrielink A. 2017. Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc Natl Acad Sci U S A 114:2218–2223. doi: 10.1073/pnas.1612927114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Wei W, Lei S, Lin J, Srinivas S, Feng Y. 2018. An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance. mBio 9:e02317-17. doi: 10.1128/mBio.02317-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 45.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohs PDA, Buss J, Sim SI, Squyres GR, Srisuknimit V, Smith M, Cho H, Sjodt M, Kruse AC, Garner EC, Walker S, Kahne DE, Bernhardt TG. 2018. A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet 14:e1007726. doi: 10.1371/journal.pgen.1007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of cell chaining in the absence of YejM or following treatment with LpxC inhibitor CHIR-090. Cell chaining (Fig. 1C) was quantified by counting the number of cellular units per cell following the depletion of YejM (n = 20) (A) or treatment with 0.25 μg/ml CHIR-090 for 2 h (n =45) (B). The data were then plotted in Graphpad Prism. ****, P < 0.0001 (unpaired t-test). Download FIG S1, TIF file, 0.6 MB (618.9KB, tif) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ΔyejM suppressors. Mutations mapped in each suppressor are listed. Download Table S1, PDF file, 0.1 MB (150.7KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The overexpression of acpT increases LpxC levels. Immunoblot detection of LpxC from extracts of wild-type cells harboring plasmids pPR111 (Plac::lpxC), pPR66 (Plac::empty), or pEMF43 (Plac::acpT) grown in the presence or absence of IPTG (50 μM [lane 2] or 1 mM [lanes 4 and 6]), as indicated. Download FIG S2, TIF file, 0.6 MB (639.3KB, tif) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The LapB prey construct does not interact with the control bait construct in the POLAR assay. Shown are representative micrographs of cells expressing the indicated bait and prey proteins. (A) Cells were transformed with plasmids expressing the control bait, which consists of a single transmembrane domain derived from residues 2 to 55 of Pseudomonas aeruginosa PBP1b fused to PopZ-H3H4-GFP (pHCL149). The LapB prey construct consists of a C-terminal mScarlet fusion to LapB and was expressed from an integrated plasmid (pHCL147 derivative pEMF36) under the control of the Plac promoter. The experimental setup is the same as for Fig. 3A, except that unlabeled YejM is not produced from the bait construct in this case. Note that without expressing the unlabeled YejM, cells begin to form chains, most likely due to the toxic effects of LapB overproduction. (B) The LapB prey construct is coexpressed with the YejM-ΔC bait construct (pEMF65). Bar, 5 μm. Download FIG S3, TIF file, 1.0 MB (1,005.6KB, tif) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains used in this study. Download Table S2, PDF file, 0.04 MB (40.1KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S3, PDF file, 0.1 MB (99.8KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S4, PDF file, 0.03 MB (29.4KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental methods for plasmid and strain construction. Download Text S1, PDF file, 0.1 MB (76.2KB, pdf) .
Copyright © 2020 Fivenson and Bernhardt.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.