Abstract
Well-designed micropatterns present in native tissues and organs involve changes in extracellular matrix compositions, cell types and mechanical properties to reflect complex biological functions. However, the design and fabrication of these micropatterns in vitro to meet task-specific biomedical applications remains a challenge. A de novo design strategy to code and synthesize functional micropatterns is presented to engineer cell alignment through the integration of aqueous-peptide inkjet printing and site-specific biomineralization. The inkjet printing provides direct writing of macroscopic biosilica selective peptide-R5 patterns with micrometer-scale resolution on the surface of a biopolymer (silk) hydrogel. This is combined with in situ biomineralization of the R5 peptide for site-specific growth of silica nanoparticles on the micropatterns, avoiding the use of harsh chemicals or complex processing. The functional micropatterned systems are used to align human mesenchymal stem cells and bovine serum albumin. This combination of peptide printing and site-specific biomineralization provides a new route for developing cost-effective micropatterns, with implications for broader materials designs.
Coding cell micropatterns through peptide inkjet printing for arbitrary biomineralized architectures is demonstrated here. The functional micropatterned systems are used to align human mesenchymal stem cells and bovine serum albumin in vitro, avoiding the use of harsh chemicals or complex processing, while providing potential applications in developing cost-effective micropatterns to meet task-specific biomedical applications.
Keywords: inkjet printing, site-specific biomineralization, biosilica, micropattern, cell alignment
1. Introduction
Micropatterns are characteristic and intriguing features widely found in native tissues and organs, such as diatoms, plant cell walls, arthropod exoskeletons and bones, among many other systems.[1,2] These functional micropatterns enhance mechanical performance (such as strength and toughness) of tissues and organs, and also support biological functions, including structural support, defense and prey capture, yet formed from a limited set of components (e.g., proteins, polysaccharides, minerals).[3,4] For example, the highly ordered periodic hexagonal or plywood-like micropatterns found in diatoms and arthropod exoskeletons enhances toughness thousands of times greater than the pristine minerals, and the iridescence derived from these features (known as structural color) provides adaptive biological functions such as mating, signaling or camouflage.[1,5–7] Axially aligned cell micropatterns in human muscle tissues provide structural anisotropy to maintain contraction and tension resistance for tendons to facilitate mechanical functions, motility and survival.[8] The spatial oriented micropatterns of collagen as parallel fiber bundles in tendons, concentric waves in bone and oriented fibrils in cartilage, are always accompanied with mechanical benefits and support cellular functions to these tissues and organs.[9–11]
A variety of biomineralization strategies have been applied to mimic the components, structure and function of natural micropatterned tissues, including photolithography, microcontact printing and laser patterning, among others.[4,8,12–14] The resultant micropatterns suggest promising applications in regenerative medicine, microelectronics, drug screening, and optical and biomedical sensors.[8,15] In particular, silica-based micropatterns play a critical role in many biomedical fields, including tissue engineering, controlled drug delivery, cell transplantation, optics, electronics as well as in vitro tissue models, due to the biocompatibility, high versatility and osteoinductive properties of silica.[12,15–18] However, most of these silica patterns are created in harsh conditions, such as extremely high temperature and/or pressures, strongly acidic solutions, or complex processing techniques.[19] Moreover, the silica patterns generated by these methods usually exhibit weak substrate bonding to rigid substrates, thus can be damaged easily due to the inherent brittleness of silica.[15] These features reduce the broader utility of these systems due to costs or material weak points, and certainly limit direct processing and integration with biological components such as cell or bioactive therapeutics.
In contrast, organisms such as diatoms, which are known for their complex and intricate silica architectures, construct silica micro-/nano-patterns with precise control through bottom-up growth under physiological and ambient conditions without additional chemical treatments.[20] For instance, biosilica morphogenesis in the diatom cell wall consists of a matrix of organic macromolecules, i.e., native silaffin-1A (natSil-1A) protein for silica formation and native silaffin-2 (natSil-2) protein for biosilica template organization. During in vivo silicification, natSil-1A triggers and mediates the formation of the silica particles, while natSil-2 moderates the activities of natSil-1A to form organized patterns through molecular self-assembly and phase-separation processing.[20,21] Inspired by the route that nature uses to synthesize micropatterns, in the present study, we present a de novo strategy to code and synthesize functional micropatterns to engineer cell alignment. A series of previous studies have confirmed that micropatterns play a dominant role in controlling cell-to-cell interactions, the microenvironment of individual cells and in turn the decisions related to cell differentiation.[8,22,23]
2. Results and Discussion
The core methodology for this design is the synergistic integration of aqueous-peptide inkjet printing and site-specific biomineralization. The inkjet printing allows for non-contact, maskless, direct arbitrary coding peptide micropatterns with versertality and repeatability, and more importantly, can be scaled-up to accelerate material assembly processes.[24,25] Piezo-electric driven actuator dispenses droplets at different volume in a drop-on-demand (DOD) inkjet printer, which can accept a wider range of ink[26] and cause less damage to bioactive substances as compared to thermal actuation.[24,27,28] When in combination with functional inks and/or active substrates, this technique can be utilized in biofabrication for various applications including biosensors, therapeutics, and regenerative medicine.[28–30] Nature typically constructs patterns during growth phases, which is usually a slow process that takes place over months to years or even hundreds of years. This low production efficiency can not match the requirements of commercial production. In contrast, site-specific biomineralization (in situ biosilicification) is able to form well-organized silica structures on the biosilica selective peptide-R5 (a bioinspired analog derived from the silaffin peptides for silica synthesis in Cylindrotheca fusiformis[21]) micropatterns while avoiding the use of harsh chemicals or complex processing. Furthermore, the silk fibroin (hereinafter referred to as silk) protein substrates offer biocompatibility, degradability and tunable mechanical properties to meet different requirements related to biomaterials, tissue engineering and regenerative medicine depending on the specific goal of a project.[31,32] Through these synergistic approaches, we obtained macroscopic silica patterns with micrometer resolution. The dimensions of these micropatterns can be initiated at small scale, while also scalable to sub-meters to match the dimension of human tissues (e.g., centimeter to meter scale).[25] Finally, in vitro cell culture experiments confirmed that human mesenchymal stem cells (hMSCs) successfully aligned with the printed silica micropatterns on the silk hydrogels. Fluorescence microscopy showed that bovine serum albumin (BSA) was aligned to the R5/biosilica micropatterns. This combination of peptide printing and site-specific biomineralization provides a new route to develop cost-effective micropatterns, with implications for broader materials designs.
2.1. De novo design strategy to code micropatterns
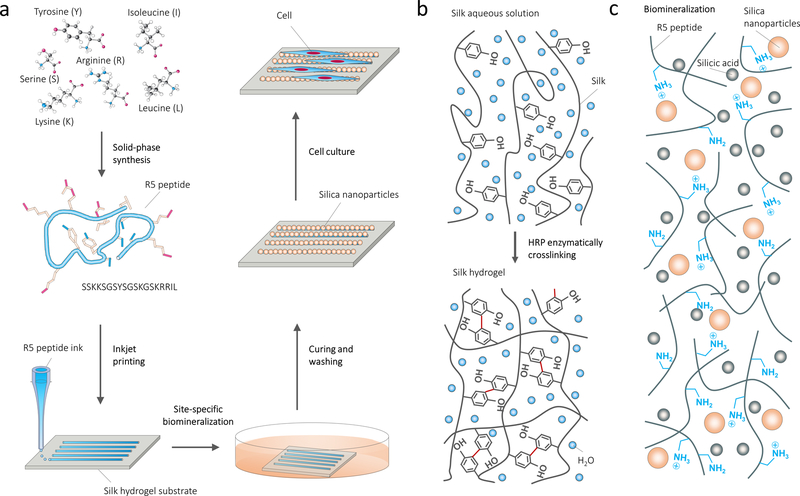
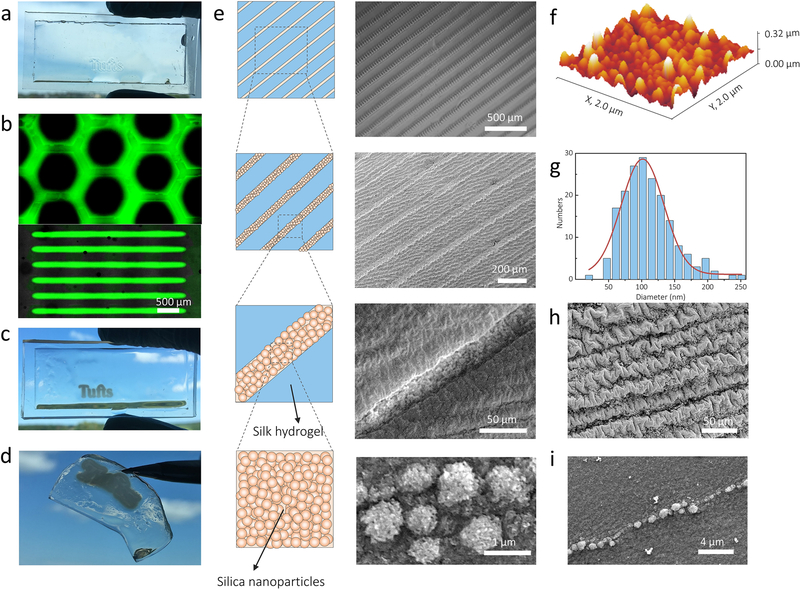
Figure 1a summarizes the bottom-up route to form biosilica micropatterns on protein substrates. The starting steps were to prepare the silk hydrogel substrates and the aqueous peptide ink. The enzymatically crosslinked silk hydrogels were used as protein substrates due to their biocompatibility, degradability and tunable mechanical properties (Figure 1b). The biosilica selective peptide-R5, a bioinspired analog derived from the silaffin peptides that are used for silicification in Cylindrotheca fusiformis,[21] was selected for the peptide ink since it can directly serve as a trapping agent to trigger the site-specific in vitro silicification (Figure 1c). Our previous studies indicated that enzymatic cross-linking between the silk and R5 molecular chains allowed for tight bonding between the substrate and printed peptide patterns.[31] A freshly printed 1.2 cm-wide peptide logo with micrometer resolution was visually discernable from the transparent underlying silk hydrogel (Figure 2a). The logo was well bonded with the hydrogels and deformed adaptively with deformation of the hydrogel. Through inkjet printing of fluorescein isothiocyanate (FITC)-labeled R5 peptide, well-defined peptide patterns on the substrates were evidenced by fluorescence images (Figure 2b). At the second stage, these all-protein micropatterns were flooded with a silicifying medium consisting of 3 vol% prehydrolyzed TEOS solution in buffer to induce biosilica deposition (in vitro silicification) on the micropattern surfaces. As a result, macroscopic peptide-biosilica patterns with 1.2 cm width were recognized from the transparent silk hydrogel (Figure 2c). The whole free-standing protein system can be removed from the container and used for further post-processing (Figure 2d). Our previous studies revealed that the positively charged amino acids dominated the silicification process, while the silk is negative charged.[31] As a result, the silica nanoparticles grew selectively on the R5 peptide patterned surface, represented by the schematic drawing and confirmed by optical imaging and scanning electron microscopy (SEM) (Figure 2e). The silica nanoparticles covered the printed R5 peptide lines with individual diameters of 50–200 nm measured from atomic force microscopy (AFM) topography images (Figure 2f, g). The diameter of these nanoparticles appeared as a sharp Gaussian distribution with a center value of ~100 nm. In contrast, no silica nanoparticles were observed on the silk hydrogel alone surfaces. We further reduced both the pattern linewidth and the gap distance between each line by adjusting the drop volume and line spacing during printing to investigate the spatial resolution of this method, with a gap distance between two lines of 20 μm and linewidth of 1 μm reached (Figure 2h, i). Careful examination of the silica nanoparticles on the micropatterns found that the silica nanoparticles remained bound to the pattern surfaces and even conformed to the wrinkled patterns that were caused by post-processing (ethanol treatment or freeze-drying). Energy dispersive spectrometer (EDS) mapping of Si and corresponding SEM image confirmed the presence of Si on the biosilica micropatterns (Figure S1).
Figure 1.
Design strategy and processing diagram of coding micropatterns. (a) De novo design to create silica micropatterns on protein substrates by in situ mineralization of silica-binding peptides and inkjet printing. (b) Silk hydrogelation mechanism. (c) Biomineralization of biosilica particles catalyzed by the biosilica selective peptide R5.
Figure 2.
Arrays of biosynthesized silica micropatterns on the hydrogel substrates. (a) Visual image shows freshly printed peptide logo with 1.2 cm width on the transparent silk hydrogel. (b) Fluorescence images of FITC-labeled R5 patterns on the silk hydrogels. (c) Visual image shows mineralized peptide logo with 1.2 cm width on the transparent silk hydrogel. (d) The whole free-standing protein system can be easily removed from the container used for gelation. (e) Schematic representation (left) and SEM images (right) show the linear array of mineralized micropatterns printed with different gap spacing between lines on the silk hydrogels. (f) AFM topography image of the silica particles on the mineralized micropatterns scanned over a 2 μm × 2 μm area. (g) The corresponding graph of the nanoparticle diameter distribution appears a sharp Gaussian distribution with a center value of ~100 nm, in a range of 50–200 nm. (h, i) SEM images show the spatial resolution of the gap spacing between two lines decreased to 20 μm and the linewidth reached ~1 μm.
2.2. Fourier transform infrared (FTIR) mapping elucidated the structural details of the micropatterns
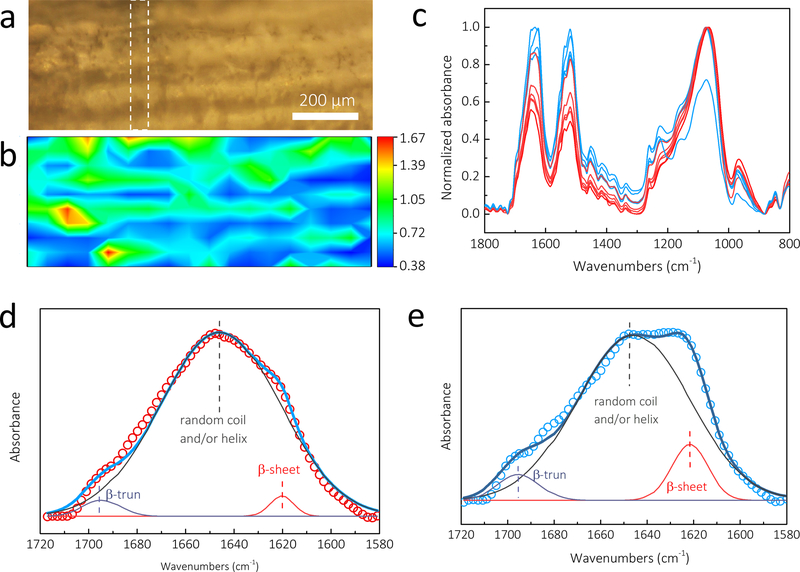
FTIR spectrometry coupled with microscope detection, supports mapping of spatial and spectral information of the samples simultaneously, enabling chemical visualization of the microscale patterns.[33–35] Previous studies demonstrated that FTIR mapping can provide useful information on the composition and conformations of proteins at defined locations with a spatial resolution of ~5–50 μm.[33–35] In this study, a 1032 × 397 μm rectangle micropattern was mapped with the pixel size (spatial resolution) of 57 × 40 μm (Figure 3a). To chemically image a particular component, the non-overlapping characteristic FTIR absorption for each element was required. Here, Si-O stretching bond (1300–1000 cm−1) and amide I band (1600–1700 cm−1) were chosen as representative peaks of silica micropattern and silk, respectively. To eliminate thickness inhomogeneities in the samples, the absorbance intensity ratios ASilica/ASilk (using A1060/A1625) were integrated for all pixel spectra to obtain chemical images of the micropatterns. Accordingly, in the resulting contour map (Figure 3b), the red/green and blue represent the strong and weak absorption of silica nanoparticles. It can be found that the strong (red/green) and weak (blue) silica absorption alternate, and the red/green lines in Figure 3b overlapped with the visual patterns in Figure 3a, confirming that the silica nanoparticles grew selectively only on the printed patterns.
Figure 3.
FTIR mapping showed the structural details of the biosilica micropatterns on the silk hydrogels. (a) Microscopic image of a 1032 × 397 μm rectangle micropattern. (b) The contour map with the red/green and blue represent the strong and weak absorption of silica nanoparticles. (c) The single pixel spectra extracted from the white dashed line range in FTIR images. Compared to the blue spectra from the blue region of the silk substrate, the red/green spectra from the red/green region of silica micropattern present significant weaker absorption in amide I and II bands. (d) The red/green spectra dominate a sharp peak at 1647 cm−1, which contributed to the random coil and/or helix structure. (e) The blue spectra show the strongest absorption at 1624 cm−1 and a shoulder at 1695 cm−1, which were assigned to β-sheets and β-turns, respectively. FTIR, Fourier transform infrared spectroscopy.
In addition, FTIR mapping can be used to monitor the secondary structure of the silk in a specific position. Figure 3c shows the single pixel spectra extracted from the white dashed line range in the FTIR images. The blue and red/green FTIR spectra corresponded to the spectra obtained from the blue (silk substrate) and red/green (silica micropattern) regions, respectively (Figure 3b). Compared to the spectra in the blue region, the spectra in the red/green region present significantly weaker absorption in the amide I and II bands, agreeing with the trend observed in the contour map. More dramatically, the red/green and blue spectra feature distinct maximum absorbance in the amide I band, a useful band to estimate the secondary structure of the silk protein. The red/green spectra dominated a sharp peak at 1647 cm−1, which contributed to the random coil and/or helix structure (Figure 3d). In comparison, the blue spectra showed the strongest absorption at 1624 cm−1 and a shoulder at 1695 cm−1, which were assigned to β-sheets and β-turns of the hairpin-folded antiparallel β-sheet structure of silk, respectively (Figure 3e). The deconvolution of the amide I band provided an estimation of β-sheet (crystalline) content in the blue region (silk substrates) of 11±3%, while that of the red/green part (micropatterns) was 3±2%. These results revealed that the micropatterns had a lower content of β-sheet. Our recent studies on the secondary structure of the R5 peptide before and after silicification characterized the R5 peptide as a random coil structure.[31] Thus, amide I absorption in the micropatterns was mainly contributed by the R5 peptide instead of the silk amide I band at the blue regions. In addition, the β-sheet content of the silk substrate was significantly lower than that of the silk hydrogel obtained by ethanol treatment (25±3%). As a result, theses enzymatically cross-linked hydrogels were transparent due to the limited β-sheet content, while the dityrosine bonds and limited β-sheets acted as crosslink points to form interlocking protein chains to keep the silk hydrogels stable in water.
2.3. Cell alignment on the micropatterns
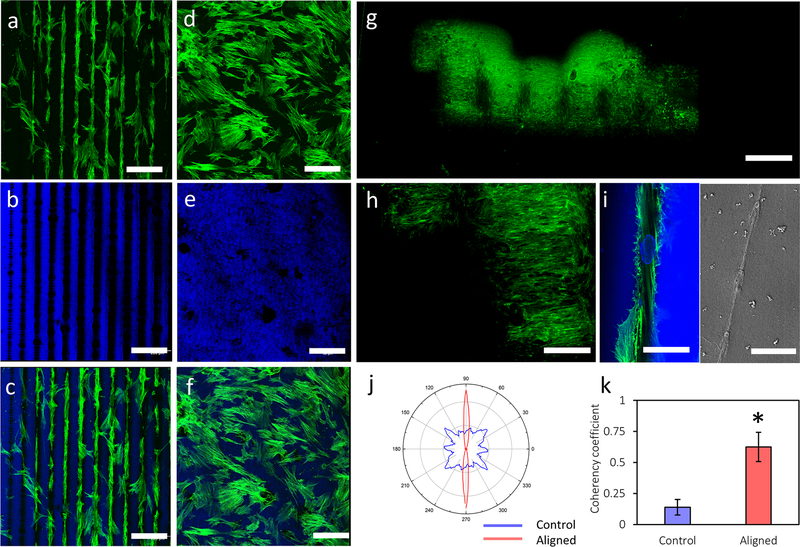
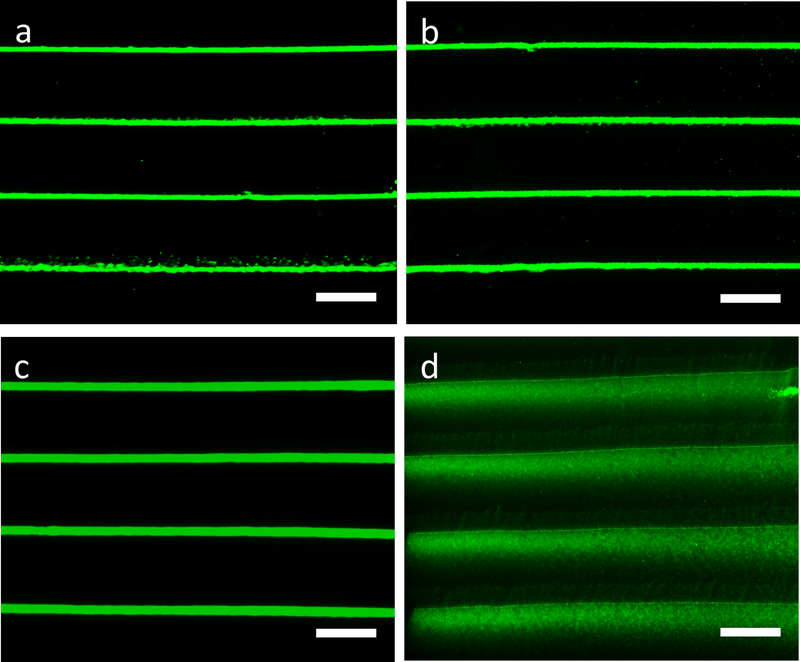
To test whether the patterned cues could be used for cell alignment, hMSCs were cultured on the micropatterned hydrogels with line spacings of 100 μm, in the range of the micropatterns in human tissues and organs.[4] hMSCs were choosen because of their utility as self-renewal and multipotential cells to differentiate to various lineages, including adipocytes, chondrocytes and osteoblasts.[36–40] Aligned hMSCs are important in tissue regeneration as cell micropatterns result in highly organized tissue architectures (e.g., bone, cartilage, cornea).[8,41] However, previous topographical micropatterning strategies often showed limited guidance of hMSC orientation on rigid substrates consisting of synthetic materials in relatively short culture periods.[8] In sharp contrast, this work demonstrated cell alignment extending the width of the printed micropatterns (e.g., 1 mm) on the hydrogel substrates after 4-day of cell culture, as shown in confocal micrographs of hMSCs on the silk hydrogels stained for F-actin (green) and counterstained for nuclei and silk[42] (blue, Figure 4a, b, c). In comparison, cells randomly distributed on the unpatterned control hydrogels demonstrated typical cell morphology over the culture period (Figure 4d, e, f). Furthermore, after 14 days of culture on the micropatterned “Tufts” logo of 1.2 cm width, fluorescence images of hMSCs stained for F-actin (green) showed that cells fully covered the logo, aligned to the biosilica micropatterns on the logo, and formed a “Tufts” logo by themselves (Figure 4g). The fluorescent intensity of F-actin stained hMSCs on the micropatterns was about two-fold stronger than that of the surrounding silk control surface (Figure 4h), providing further evidence for specific cell attachment and alignment to the biosilcia micropatterns.
Figure 4.
hMSC alignment on the biosilica micropatterned and control hydrogels. (a, b, c) hMSCs aligned to the biosilica micropatterns on the silk hydrogels; (d, e, f) hMSCs on the unpatterned control silk hydrogels demonstrated typical morphology. Confocal micrographs of hMSCs on the silk hydrogels stained for F-actin (green, a, d), counterstained for nuclei and silk (blue, b, e) and overlay images (c, f). (g) Fluorescence images of hMSCs stained for F-actin (green) on the micropatterned “Tufts” logo. (h) Partial enlargement of Figure g. (i) Zoomed view (left) and SEM image (right) of hMSCs on the biosilica micropatterns. (j) Graphical representation of cellular alignment quantified via ImageJ software and an OrientationJ plug-in. (k) Coherency coefficients indicating cellular distribution on the micropatterned and control hydrogels. Scale bars, a-f, 200 μm; g, 2 mm; h, 700 μm; i, 50 μm (left), 10 μm (right).
Interestingly, the hMSCs appeared to favor the silica particles and showed specific attachment to the silica patterns. Confocal micrographs of the stained hMSCs showed that cell nuclei also aligned to the biosilica micropatterns (Figure 4i, left). SEM images provided further evidence that a single hMSC attached to and spread its actin filaments on the silica particles (Figure 4i, right). The F-actin of hMSCs aligned parallelly with line spacing of 100 μm, following the printed micropattern. Quantification of cell alignment was performed on the confocal micrographs of F-actin stained hMSCs on the patterned and control silk hydrogels using ImageJ software equipped with a OrientationJ plug-in. Graphical representation of the cellular distribution showed that the hMSC filaments strongly oriented in the direction of the ellipse long axis, indicating significantly greater cellular alignment compared to controls (Figure 4j). The calculated coherency coefficient (a measure of cell alignment in the predominant direction) of the hMSCs on the micropatterns was 0.62, four times greater than that of the unpatterned controls (0.14, Figure 4k). These quantitative analyses further confirmed that cell orientation followed the peptide-silica micropatterns.
hMSCs were cultured on, and proliferated on, both aligned and unaligned hydrogel surfaces (Figure S2). On the unaligned samples, a random orientation of cells was observed with a characteristic fibroblast-like morphology. On the aligned samples, cells obtained an elongated morphology, aligned to the micropattern and mainly located on the microstrips (Figure S2, Figure 4a). The empty space between the microstrips may facilitate cell migration and proliferation.[43] A significantly greater number of cells was measured after 7 days of seeding (Figure S3), indicating the peptide/silica micropattern did not compromise hMSC proliferation. Moreover, our recent work on hMSC proliferation on silicified silk/R5 composites demonstrated cytocompatibility.[31]
2.4. Protein alignment on the micropatterns
The efficacy of this design to micropattern protein on the silk hydrogels was demonstrated by fluorescence microscopy (Figure 5). The FITC-R5 peptide was added into the R5 peptide solution at a molar ratio of 1/400 to generate ink for printing. The biomineralization process was initiated by flooding the micropatterned system with a silicifying medium consisting of 3 vol% prehydrolyzed TEOS solution in buffer (Figure S4). Inkjet printing of FITC-labeled BSA and R5 peptide followed by silicification resulted in FITC-BSA immobilized and aligned exclusively along the R5/biosilica micropatterns (Figure 5a). Fluorescence microscopy confirmed that after 6 days of incubation in PBS, the FITC-BSA was mainly limited to the biosilica regions, with FITC-BSA stripes distinctive along the R5/biosilica micropatterns (Figure 5b). Inkjet printing FITC-labeled BSA alone, followed by silicification, also resulted in FITC-BSA stripes (Figure 5c). However, after 6 days of incubation in PBS, the area covered by FITC-BSA extended over the hydrogel surface, with adjacent lines merged together (Figure 5d). The stability of the FITC-BSA/R5/silica micropatterns revealed spatial control of protein alignment using this design, probably due to BSA immobilization with the silica particles induced by the R5 peptide.
Figure 5.
BSA alignment on the biosilica micropatterns. (a, b) Fluorescence microscopy images of micropatterns containing FITC-labeled BSA and R5 peptide followed by silicification; (c, d) fluorescence microscopy images of micropatterns containing FITC-labeled BSA alone followed by silicification. Fluorescence microscopy images of micropatterns in PBS at day 1 (a, c), day 6 (b, d). The distinctive FITC-BSA stripes in the FITC-BSA/R5/silica micropatterns reveal the spatial control of protein alignment using this design, in comparison to the extended distribution of the control FITC-BSA micropatterns. Scale bars: a, b, c, d, 400 μm.
Aligned micropattern cues are crucial for cell proliferation, migration and differentiation,[43] and also result in the secretion and deposition of anistropic extracellular matrix (ECM) specific to tissue types.[4] Our recent work on hMSC differentiation in silicified silk/R5 composites demonstrated that this composite material promoted osteogenic differentiation of hMSCs in an osteoinductive environment in vitro, as compared to the silk plain controls.[31] Patterned silk films also supported osteogenic differentiation of hMSCs while inducing lamellar alignment of cells and ECM orientation.[44] Moreover, hMSCs cultured on 1,000 μm2 micropatterns, while prevented from spreading and flattening, upregulated chondrogenic genes and differentiated into chondrocytes in response to transforming growth factor beta (TGF-β) stimulation.[45] Pattern cues in cartilage play an important role in building mechanical strength and tissue functions, where interterritorial fibers are aligned perpendicular to the joint surface in the middle zone, while being parallel in the superficial zone.[46] Given the highly ordered mineral and collagen orientation in bone and cartilage, this directed mineral micropattern is crucial in improving bone and cartilage formation in vivo. This study emphasized the role of mineral micropatterns in osteochondral tissue engineering as cells reciprocally interact with ECM to adapt to structures and topography in the local environment.[47]
Inkjet printing of functional inks on protein substrates offers opportunities to micropattern a variety of substances, such as peptides, growth factors and biomacromolecules. Although inkjet printing has been used as a typical bioprinting approach to pattern protein and cells,[29,48] this work further extends its application by introducing immobolization and alignment of functional substances in well-organized silica micropatterns on protein substrates. Surface modifications of the inert biosilica particles allow for controllable encapsulation and sustained release of a library of bioactive molecules and substances (e.g., proteins, enzymes, and drugs), which are sensitive to heat or caustic chemicals.[49] The silk protein substrates offer biocompatiblility, degradablity and tunable mechanical properties to provide three-dimensional extracellular microenvironments and extrinsic factor reservoirs for applications in tissue engineering and regenerative medicine.[50–52] Geometric cues, in combination with bioactive factors, have a strong influence on hMSCs differentiation. For example, hMSCs were highly elongated and aligned in parallel along micropatterned fibronectin strips (20 μm width separated by 40 μm wide grooves) and gene expression was upregulated towards neurogenesis and myogenesis.[53] Directing hMSCs on the R5/silica micropatterned hydrogels, in combination with cell adhesion mediating molecules, could enable cell alignment for applications like cardiac muscle tissue engineering.[54] Micropatterning nerve growth factor (NGF) has potential to align Schwann cells in neurons with applications towards nerve tissue regeneration.[55] A variety of polycationic peptides, e.g., silaffin and long chain polyamines which are able to precipitate silica nanospheres individually, can be printed on the biopolymer substrates leading to functional material designs with different mineral morphologies.[56]
3. Conclusion
A de novo strategy to design and synthesize functional micropatterns to engineer cell alignment through the integration of aqueous-peptide inkjet printing and site-specific biomineralization was demonstrated. The resulting micropatterned systems supported the alignment of hMSCs along and in micropattern directions and space. In addtion, the micropatterns generated by inkjet printing of biosilicification domains on the biocompatible protein substrates under mild (peptide-mediated) conditions provide other potential applications. Inkjet printing of functional inks on protein substrates offers opportunities to micropattern a variety of substances, such as peptides, growth factors and biomacromolecules. Overall, this de novo design strategy to create functional micropatterns meets a broad range of needs in biomaterials and regenerative medicine involving interfacial tissue engineering, cell transplatation, cell-based sensors, and drug screening platforms,[22,57] as well as providing a system to improve the understanding of fundamental of cellular functions.[58]
4. Experimental Section
Materials:
All chemical reagents used for making the micropatterned hydrogels were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. All materials and reagents used for cell culture and analysis were purchased from Thermo Fisher Scientific (Waltham, MA) unless otherwise specified.
Preparation of aqueous silk fibroin solution:
Silk fibroin solutions (referred to as silk in the paper) were prepared using our previously established procedures.[59] Briefly, ten grams of silk cocoons were cut into pieces and boiled in sodium carbonate solution (4 L of 0.02 m) for 30 min to remove the coating of sericin protein. Degummed fibers were rinsed with deionized water three times and dried in air overnight. Five grams of dried fibers were dissolved in lithium bromide solution (20 ml of 9.3 m) at 60°C for 4 h. The silk solution was then dialyzed against deionized water using a dialysis cassette (Pierce 3.5 kDa MWCO; Fisher Scientific, PA) for 2 days. The solubilized silk solution was then centrifuged twice at 9000 RPM, 4°C for 20 min to remove insoluble particulates. Protein concentration was determined by drying a known mass of the silk solution at 60°C for 12 h and measuring the mass of the remaining solid.
Inkjet printing of the R5 peptide on the silk protein substrate:
The biosilica selective peptide R5 (H-SSKKSGSYSGSKGSKRRIL-OH) with a purity of 95% was synthesized by GenScript (Piscataway, NJ). Lyophilized horseradish peroxidase (HRP, type VI) powder and the R5 peptide powder were dissolved in deionized water to form a stock solution of 1000 U/mL and 100 mg/mL, respectively. To initiate the silk gelation, HRP (6 μL of 1kU ml−1) and H2O2 solution (5 μL of 1%) were mixed with silk solution (1 mL of 6 wt%) in a pre-made polydimethylsiloxane (PDMS) mold (size L×W×T: 60×24×0.6 mm), and left at 37℃ for 8 minutes before printing the R5 peptide.[60] Before complete gelation of the silk substrates, the R5 peptide ink was printed on the substrates at ambient conditions by a Dimatix Material Printer DMP-2830 equipped with Dimatix Materials Cartridges (DMS-11610 21 μm nozzle diameter, Fujifilm, Santa Clara, CA). A custom designed jetting waveform was created, and the R5 peptide solution was jetted at firing voltages of 40V and 25 V to achieve different drop volume. Gap spacing between lines were 20 μm, 50 μm, 100 μm or 200 μm. Drop spacing was 20 μm. The distance between the print head and the substrate was 300 μm. After printing, the silk hydrogel was left in a humidified chamber at room temperature for 2 hours to allow sufficient bonding between the silk hydrogel and the R5 peptide.
In vitro silicification:
Biosilica deposition was introduced into the patterned hydrogels post printing. Pre-hydrolyzed tetraethoxysilane (TEOS) solution was prepared by mixing TEOS solution (223 μl) with ethanol/water solution (766 μl of 50%), hydrogen chloride (HCl, 10 μl of 1 M) and left at room temperature for 15 min. The patterned hydrogel was then covered by silicifying medium (1 ml) consisting of prehydrolyzed TEOS solution (30 μl) and buffer solution (970 μl of 65 μl 1 M bis-tris propane, 35 μl 1 M citric acid solution and 870 μl deionized water) and allowed for silicification at room temperature overnight, respectively. After silicification, the patterned hydrogels were rinsed with DI water to remove excess reagents and stored at 4°C for further characterization.
Characterization:
The morphologies of the micropatterns were characterized by scanning electron microscopy (SEM, Ultra 55 field emission scanning electron microscopy, Carl Zeiss, AG) at an acceleration voltage of 5 kV. To prevent electrical charging, all specimens were coated with a 5 nm-thick Au layer before observation. The samples used for morphology characterization were treated by critical point drying (Tousimis 931 GL, MD). The structures of the biosilica nanoparticles were characterized by Atomic Force Microscopy (AFM) on a MFP-3D-BIO AFM instrument (Asylum Research, CA) in tapping mode. NP-O10 SiN probes were used (k = 0.12 N m−1, Bruker Corporation, MA). Fourier transform infrared (FTIR) spectroscopy mapping was performed using a Bruker LUMOS FTIR microscope system (Bruker, MA), equipped with a liquid nitrogen cooled mercury cadmium telluride (MCT) detector. The images of samples were obtained in ATR mode with an individual pixel size about 57 × 40 μm over a field of view of 1032 × 397 μm. For FTIR mapping, spectra were collected in the mid-infrared (MIR) range of 800–3600 cm−1 at a resolution of 4 cm−1 with 26 co-added scans. All the data collection and processing were performed using OPUS 6.5 (Bruker, MA) at Harvard University Center for Nanoscale Systems.
Fluorescence imaging of the fluorescent-labeled micropatterns:
The fluorescein isothiocyanate (FITC)-labeled R5 peptide (FITC-SSKKSGSYSGSKGSKRRIL-OH, GenScript, NJ) was added into the R5 peptide solution in a molar ratio of 1/400. The peptide mixture was used to create the ink for printing, following the same silicification process as described earlier. The FITC-labeled micropatterns on the silk hydrogels were imaged using the BZ-X700 series microscope (Keyence, IL, USA).
Cell Culture:
Human mesenchymal stem cells (hMSCs) were isolated from fresh bone marrow aspirate (Lonza, NJ) as previously described[61] and used in the cell alignment study. hMSCs were cultured in expansion medium containing Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% non-essential amino acids, 1% antibiotic/antimycotic (100 U ml−1 penicillin, 100 μg ml−1 streptomycin, 0.25 μg ml−1 fungizone) and 1 ng ml−1 fibroblast growth factor-basic (bFGF). Patterned hydrogels were sterilized in 70% ethanol for 5 hours prior to cell seeding. Cells at passage 2–3 were seeded onto the patterned and unpatterned control hydrogels from suspension in culture medium at a concentration of 2 × 104 cells cm−2. Cultures were maintained in growth media containing DMEM supplemented with 10% FBS, 1% non-essential amino acids and 1% antibiotic/antimycotic. All cell cultures were incubated at 37°C supplemented with 5% CO2. The actin expression of hMSCs in three-dimensional (3D) cell culture was visualized by fluorescence F-actin staining and counterstained by DAPI following the manufacturer’s instructions. Unpatterned plain silk HRP hydrogels served as controls.
Microscopy and Analysis:
Cell samples were rinsed in PBS and fixed in 4% paraformaldehyde (PFA) for 2 hours. For SEM analysis, samples were serial dehydrated in ethanol, critical point dried under CO2, Au sputter-coated, and imaged. For confocal microscopy, stained cells were imaged with a Leica DMRX microscope (Leica Microsystems Inc., IL). hMSC alignment was quantified using ImageJ software (NIH, Bethesda, MD) with a plug-in OrientationJ to calculate the directional coherency coefficient of F-actin filaments. The coherency coefficient ranges from 0 to 1. A coherency coeffient close to 1 represented as a slender ellipse, indicating the filaments strongly oriented in the direction of the ellipse long axis and greater cellular alignment.[62]
Protein alignment on the micropatterned hydrogels:
Fluorescein isothiocyanate (FITC)-labeled bovine serum albumin was added to the R5 peptide solution in a molar ratio of 1/1000. The peptide mixture was used to create the ink for printing, followed by the same printing setup and site-specific silicification process as described earlier. FITC-labeled BSA alone served as a control. The FITC-labeled micropatterns on the silk hydrogels were imaged using the BZ-X700 series microscope (Keyence, IL, USA).
Statistical analysis:
Statistical analysis was performed by paired or unpaired Student t-tests, as appropriate, to determine statistical differences between groups. One-way ANOVA was used when comparing features between groups. In all cases, n = 6 were used for data sets (p < 0.05 or 0.01). GraphPad software (GraphPad Prism software, CA) was utilized.
Supplementary Material
Acknowledgements
J. Guo, S. Ling and W. Li contributed equally to this work. This work was supported by the National Institutes of Health (R01DE016525, R01AR068048, U01EB014976) and the Air Force Office of Scientific Research (FA9550-17-1-0333). F. G. Omenetto would like to thank partial support from the ONR (Award N000141310596). The authors acknowledge use of the facilities at the Harvard Center for Nanoscale Systems.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Jin Guo, Department of Biomedical Engineering, Tufts University, MA 02155, USA; Department of Chemical and Biological Engineering, Tufts University, MA 02155, USA.
Shengjie Ling, Department of Biomedical Engineering, Tufts University, MA 02155, USA; School of Physical Science and Technology, Shanghai Tech University, Shanghai 201210, China; Department of Civil and Environmental Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Wenyi Li, Department of Biomedical Engineering, Tufts University, MA 02155, USA.
Ying Chen, Department of Biomedical Engineering, Tufts University, MA 02155, USA.
Chunmei Li, Department of Biomedical Engineering, Tufts University, MA 02155, USA.
Fiorenzo G. Omenetto, Department of Biomedical Engineering, Tufts University, MA 02155, USA
David L. Kaplan, Department of Biomedical Engineering, Tufts University, MA 02155, USA
References
- [1].Hamm CE, Merkel R, Springer O, Jurkojc P, Maier C, Prechtel K, Smetacek V, Nature 2003, 421, 841. [DOI] [PubMed] [Google Scholar]
- [2].Wegst UGK, Bai H, Saiz E, Tomsia AP, Ritchie RO, Nat. Mater 2014, 14, 23. [DOI] [PubMed] [Google Scholar]
- [3].Meyers MA, Chen P-Y, Lin AY-M, Seki Y, Prog. Mater. Sci 2008, 53, 1. [Google Scholar]
- [4].Thery M, J. Cell Sci 2010, 123, 4201. [DOI] [PubMed] [Google Scholar]
- [5].Chen P-Y, Lin AY-M, McKittrick J, Meyers MA, Acta Biomater 2008, 4, 587. [DOI] [PubMed] [Google Scholar]
- [6].Gordon R, Losic D, Tiffany MA, Nagy SS, Sterrenburg FAS, Trends Biotechnol 2009, 27, 116. [DOI] [PubMed] [Google Scholar]
- [7].Noyes J, Sumper M, Vukusic P, J. Mater. Res 2008, 23, 3229. [Google Scholar]
- [8].Li Y, Huang G, Zhang X, Wang L, Du Y, Lu TJ, Xu F, Biotechnol. Adv 2014, 32, 347. [DOI] [PubMed] [Google Scholar]
- [9].ELLIOTT DH, Biol. Rev. Camb. Philos. Soc 1965, 40, 392. [DOI] [PubMed] [Google Scholar]
- [10].Temenoff JS, Mikos AG, Biomaterials 2000, 21, 431. [DOI] [PubMed] [Google Scholar]
- [11].Weiner S, Traub W, FASEB J 1992, 6, 879. [PubMed] [Google Scholar]
- [12].Otzen D, Scientifica (Cairo) 2012, 2012, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Alom Ruiz S, Chen CS, Soft Matter 2007, 3, 168. [DOI] [PubMed] [Google Scholar]
- [14].Tamerler C, Sarikaya M, Philos. Trans. R. Soc. A Math. Phys. Eng. Sci 2009, 367, 1705. [DOI] [PubMed] [Google Scholar]
- [15].Hench LL, Wilson J, Science 1984, 226, 630. [DOI] [PubMed] [Google Scholar]
- [16].Cerruti M, Sahai N, Rev. Mineral. Geochemistry 2006, 64, 283. [Google Scholar]
- [17].Lebold T, Jung C, Michaelis J, Bräuchle C, Nano Lett 2009, 9, 2877. [DOI] [PubMed] [Google Scholar]
- [18].Schröder HC, Wang X, Schloßmacher U, Wiens M, Müller WEG, 2013; pp. 197–234.
- [19].Iler RK, The Chemistry of Silica: Solubility, Polymerization, Colloid and Surface Properties and Biochemistry of Silica; 1979. [Google Scholar]
- [20].Poulsen N, Sumper M, Kroger N, Proc. Natl. Acad. Sci 2003, 100, 12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sumper M, Brunner E, Adv. Funct. Mater 2006, 16, 17. [Google Scholar]
- [22].Chen CS, Science (80-.) 1997, 276, 1425. [DOI] [PubMed] [Google Scholar]
- [23].Kawaguchi K, Kageyama R, Sano M, Nature 2017, 545, 327. [DOI] [PubMed] [Google Scholar]
- [24].V Murphy S, Atala A, Nat. Biotechnol 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [25].Singh M, Haverinen HM, Dhagat P, Jabbour GE, Adv. Mater 2010, 22, 673. [DOI] [PubMed] [Google Scholar]
- [26].Li J, Rossignol F, Macdonald J, Lab Chip 2015, 15, 2538. [DOI] [PubMed] [Google Scholar]
- [27].Derby B, J. Mater. Chem 2008, 18, 5717. [Google Scholar]
- [28].Tao H, Marelli B, Yang M, An B, Onses MS, Rogers JA, Kaplan DL, Omenetto FG, Adv. Mater 2015, 27, 4273. [DOI] [PubMed] [Google Scholar]
- [29].Tasoglu S, Demirci U, Trends Biotechnol 2013, 31, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yoo S-S, Polio S, In Cell and Organ Printing; Springer; Netherlands: Dordrecht, 2010; pp. 3–19. [Google Scholar]
- [31].Guo J, Li C, Ling S, Huang W, Chen Y, Kaplan DL, Biomaterials 2017, 145, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vepari C, Kaplan DL, Prog. Polym. Sci 2007, 32, 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ling S, Qi Z, Knight DP, Shao Z, Chen X, Polym. Chem 2013, 4, 5401. [Google Scholar]
- [34].Ling S, Qi Z, Shao Z, Chen X, J. Mater. Chem. B 2015, 3, 834. [DOI] [PubMed] [Google Scholar]
- [35].Ling S, Qi Z, Watts B, Shao Z, Chen X, Phys. Chem. Chem. Phys 2014, 16, 7741. [DOI] [PubMed] [Google Scholar]
- [36].Mauney JR, Volloch V, Kaplan DL, Tissue Eng 2005, 11, 787. [DOI] [PubMed] [Google Scholar]
- [37].Peng R, Yao X, Cao B, Tang J, Ding J, Biomaterials 2012, 33, 6008. [DOI] [PubMed] [Google Scholar]
- [38].Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR, Science 1999, 284, 143. [DOI] [PubMed] [Google Scholar]
- [39].Prockop DJ, Science 1997, 276, 71. [DOI] [PubMed] [Google Scholar]
- [40].Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, Du J, Aldrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM, Nature 2002, 418, 41. [DOI] [PubMed] [Google Scholar]
- [41].Gong T, Xie J, Liao J, Zhang T, Lin S, Lin Y, Bone Res 2015, 3, 15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Han H, Ning H, Liu S, Lu Q, Fan Z, Lu H, Lu G, Kaplan DL, Adv. Funct. Mater 2016, 26, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhong H, Xuan L, Wang D, Zhou J, Li Y, Jiang Q, RSC Adv 2017, 7, 21837. [Google Scholar]
- [44].Tien LW, Gil ES, Park S-H, Mandal BB, Kaplan DL, Macromol. Biosci 2012, 12, 1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gao L, McBeath R, Chen CS, Stem Cells 2010, N/A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sophia Fox AJ, Bedi A, Rodeo SA, Sport. Heal. A Multidiscip. Approach 2009, 1, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liao S, Chan CK, Ramakrishna S, Mater. Sci. Eng. C 2008, 28, 1189. [Google Scholar]
- [48].Phillippi JA, Miller E, Weiss L, Huard J, Waggoner A, Campbell P, Stem Cells 2008, 26, 127. [DOI] [PubMed] [Google Scholar]
- [49].Luckarift HR, Spain JC, Naik RR, Stone MO, Nat. Biotechnol 2004, 22, 211. [DOI] [PubMed] [Google Scholar]
- [50].ILKHANIZADEH S, TEIXEIRA A, HERMANSON O, Biomaterials 2007, 28, 3936. [DOI] [PubMed] [Google Scholar]
- [51].Lutolf MP, Hubbell JA, Nat. Biotechnol 2005, 23, 47. [DOI] [PubMed] [Google Scholar]
- [52].Lutolf MP, Gilbert PM, Blau HM, Nature 2009, 462, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tay CY, Yu H, Pal M, Leong WS, Tan NS, Ng KW, Leong DT, Tan LP, Exp. Cell Res 2010, 316, 1159. [DOI] [PubMed] [Google Scholar]
- [54].Song W, Kawazoe N, Chen G, Nanomater J. 2011, 2011, 1. [Google Scholar]
- [55].Recknor JB, Sakaguchi DS, Mallapragada SK, Biomaterials 2006, 27, 4098. [DOI] [PubMed] [Google Scholar]
- [56].Rodríguez F, Glawe DD, Naik RR, Hallinan KP, Stone MO, Biomacromolecules 2004, 5, 261. [DOI] [PubMed] [Google Scholar]
- [57].Lee CJ, Huie P, Leng T, Peterman MC, Marmor MF, Blumenkranz MS, Bent SF, Fishman HA, Arch. Ophthalmol. (Chicago, Ill. 1960) 2002, 120, 1714. [DOI] [PubMed] [Google Scholar]
- [58].Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE, Science 1994, 264, 696. [DOI] [PubMed] [Google Scholar]
- [59].Rockwood DN, Preda RC, Yücel T, Wang X, Lovett ML, Kaplan DL, Nat. Protoc 2011, 6, 1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Partlow BP, Hanna CW, Rnjak-Kovacina J, Moreau JE, Applegate MB, Burke KA, Marelli B, Mitropoulos AN, Omenetto FG, Kaplan DL, Adv. Funct. Mater 2014, 24, 4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li C, Hotz B, Ling S, Guo J, Haas DS, Marelli B, Omenetto F, Lin SJ, Kaplan DL, Biomaterials 2016, 110, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fonck E, Feigl GG, Fasel J, Sage D, Unser M, Rufenacht DA, Stergiopulos N, Stroke 2009, 40, 2552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.