Gerstmann-Sträussler-Scheinker disease (GSS) is characterized by atypical N- and C-terminal cleaved protease-resistant PrPSc. Vanni et al. report that PrPSc oligomers purified from patients with GSS are authentic – but non-fibrillar – prions, thus uncovering the minimal prion protein domain able to encode infectious prions.
Keywords: prion disease, Gerstmann-Sträussler-Scheinker disease, PrPSc, infectivity, oligomers
Abstract
Prions are transmissible agents causing lethal neurodegenerative diseases that are composed of aggregates of misfolded cellular prion protein (PrPSc). Despite non-fibrillar oligomers having been proposed as the most infectious prion particles, prions purified from diseased brains usually consist of large and fibrillar PrPSc aggregates, whose protease-resistant core (PrPres) encompasses the whole C-terminus of PrP. In contrast, PrPSc from Gerstmann-Sträussler-Scheinker disease associated with alanine to valine substitution at position 117 (GSS-A117V) is characterized by a small protease-resistant core, which is devoid of the C-terminus. We thus aimed to investigate the role of this unusual PrPSc in terms of infectivity, strain characteristics, and structural features. We found, by titration in bank voles, that the infectivity of GSS-A117V is extremely high (109.3 ID50 U/g) and is resistant to treatment with proteinase K (109.0 ID50 U/g). We then purified the proteinase K-resistant GSS-A117V prions and determined the amount of infectivity and PrPres in the different fractions, alongside the morphological characteristics of purified PrPres aggregates by electron microscopy. Purified pellet fractions from GSS-A117V contained the expected N- and C-terminally cleaved 7 kDa PrPres, although the yield of PrPres was low. We found that this low yield depended on the low density/small size of GSS-A117V PrPres, as it was mainly retained in the last supernatant fraction. All fractions were highly infectious, thus confirming the infectious nature of the 7 kDa PrPres, with infectivity levels that directly correlated with the PrPres amount detected. Finally, electron microscopy analysis of these fractions showed no presence of amyloid fibrils, but only very small and indistinct, non-fibrillar PrPresparticles were detected and confirmed to contain PrP via immunogold labelling. Our study demonstrates that purified aggregates of 7 kDa PrPres, spanning residues ∼90–150, are highly infectious oligomers that encode the biochemical and biological strain features of the original sample. Overall, the autocatalytic behaviour of the prion oligomers reveals their role in the propagation of neurodegeneration in patients with Gerstmann-Sträussler-Scheinker disease and implies that the C-terminus of PrPSc is dispensable for infectivity and strain features for this prion strain, uncovering the central PrP domain as the minimal molecular component able to encode infectious prions. These findings are consistent with the hypothesis that non-fibrillar prion particles are highly efficient propagators of disease and provide new molecular and morphological constraints on the structure of infectious prions.
Introduction
Transmissible spongiform encephalopathies (TSEs), or prion diseases, are fatal neurodegenerative diseases, affecting humans and animals, that are associated with CNS accumulation of autocatalytically self-replicating aggregates of PrPSc, a misfolded pathological isoform of the host-encoded cellular prion protein (PrPC) (Prusiner, 1998). Although TSEs can occur as acquired, sporadic as well as inherited forms, they have been historically defined based on their experimental transmissibility. According to the protein-only hypothesis, PrPSc is considered the main, if not the sole, component of prions, the proteinaceous infectious agent of TSEs. Sporadic forms of human prion diseases include Creutzfeldt-Jakob disease (CJD) and the variably protease-sensitive prionopathy (VPSPr), while the acquired forms include variant CJD and Kuru. Genetic CJD, fatal familiar insomnia (FFI) and Gerstmann-Sträussler-Scheinker disease (GSS), are instead caused by pathogenic mutations in the prion protein PRNP gene which predisposes mutant PrPC to spontaneously convert to PrPSc.
For most human TSEs, the digestion of PrPSc with proteinase K (PK) results in PK-resistant PrP aggregates (PrPres) that are composed of N-terminally truncated, variably glycosylated C-terminal PrP fragments, with a molecular weight of ∼19–21 kDa for the non-glycosylated form (Parchi et al., 1996). By contrast, a PrPSc variant with biochemical features unique and distinctive from any other human prion disease characterizes GSS and VPSPr. Indeed, these diseases are characterized by an atypical PrPSc whose PK-resistant core is composed of a non-glycosylated, C- and N-terminally truncated PrP fragment, with a molecular weight that varies between 6 and 8 kDa depending on the specific PRNP mutation (Piccardo et al., 1998, 2001; Gambetti et al., 2011; Pirisinu et al., 2016). Among others, the GSS subtype associated with an alanine to valine mutation at the PRNP position 117 (GSS-A117V) is exclusively characterized by 6–7 kDa PrPres, composed of non-glycosylated PrP fragments spanning amino acids ∼90–150 (Tagliavini et al., 2001; Pirisinu et al., 2013; Cracco et al., 2019). As GSS cases exclusively associated with 6–8 kDa PrPres have been historically difficult to transmit in animal models, their nature as TSEs and the autocatalytic behaviour of their atypical PrPSc has been debated. However, recent evidence of their transmissibility to humanized mice (Asante et al., 2013) and bank voles (Pirisinu et al., 2016) indicated that these GSS subtypes are indeed true prion diseases.
A direct association between infectivity and PrPres had been reported for most TSEs characterized by classical PrPSc (Cronier et al., 2008; Lacroux et al., 2012; Miyazawa et al., 2015), and a recent study conducted on recombinant PrPSc confirmed and strengthened this association, demonstrating that the infectivity is encoded only within the structure of the PK-resistant, C-terminal, PrP fragment (Wang et al., 2018). Despite the latest findings on the transmissibility of prion diseases exclusively associated with 6–8 kDa PrPres, a comparable association between infectivity and atypical PrPres that lacks not only the N-terminus of the protein but also its C-terminus is still lacking. Here, we took advantage of the high susceptibility of bank voles to GSS and of a newly published purification method (Wenborn et al., 2015) to analyse the distribution of PrPres and infectivity during the purification of 7 kDa PrPres aggregates from brains of humans affected with GSS-A117V.
Our findings show that purified 7 kDa PrPres consists of small, non-fibrillar, autocatalytic aggregates composed of non-glycosylated and non-glycophosphatidylinositol (GPI) anchored PrP fragments spanning amino acids 90–150, which are solely responsible for the observed infectivity and encode the strain properties of GSS prions. In addition to demonstrating that aggregates composed of N- and C-terminally truncated PrP are truly infectious prions, our findings provide new important constraints regarding the molecular architecture that underlies the structure of infectious prions.
Material and methods
Prion isolates
Frozen brain samples from two GSS-A117V cases (samples GSS-A117V #1 and GSS-A117V #2) and from a sporadic CJD-MM1 case were provided by the National Prion Disease Pathology Surveillance Centre (Cleveland, USA) and the National Italian CJD Registry (Rome, Italy), respectively. The use of material was approved by UHCMC IRB #05-14-09, for the GSS cases, and by the Ethical Committee of the Istituto Superiore di Sanità for the sporadic CJD case. The PrP polymorphisms at position 129 were 129-VV and 129-MV for GSS-A117V #1 and GSS-A117V #2, respectively, and 129-MM for the sporadic CJD case. Bv109I-adapted CWD (Bv-CWD) and Bv109I-adapted Scrapie (Bv-Scr) samples used for the purification were from terminally affected Bv109I derived from previous studies (Di Bari et al., 2013). Frozen brain samples from Bv109I healthy voles (Bv-Neg) were also used for the purification as negative controls.
Preparation of brain homogenates for titration assays
Brain tissues were homogenized at 10% w/v in a phosphate-buffered saline (PBS) pH 7.4 and stored at −80°C. For each sample, two aliquots were taken. Aliquots were supplemented with 2% sarkosyl and incubated with gentle shaking at 37°C for 10 min. For each sample, an aliquot was added with 20 μg/ml PK (Sigma-Aldrich), while the untreated aliquot was added with an equal volume of PBS. Samples were then incubated for 1 h at 37°C with gentle shaking and the treatment was stopped with 10 mM PMSF (Sigma Aldrich). PK-treated and untreated brain homogenates from GSS-A117V, sCJD-MM1 and Bv-CWD were diluted to 0.4% w/v and then 10-fold serially diluted up to the dilution 4 × 10−8.
Prion purification
Purified prion preparations were prepared as described in Wenborn et al. (2015). Briefly, 200 µl aliquots of 10% w/v brain homogenates were treated for 30 min with Pronase E (PE) (Sigma-Aldrich) (100 μg/ml final concentration) with gentle shaking at 37°C. Digested samples were then added to 10 mM EDTA and incubated with an equal volume of sarkosyl 4% and Benzonase® (Merck) (50 U/ml final concentration) for 10 min at 37°C. Iodixanol 60% and NaPTA 4% were added to give final concentrations in the sample of 35% (w/v) and 0.3% (w/v), respectively, and then centrifuged at 16 100g for 90 min at 37°C. After carefully removing the upper flocculate fraction, the solubilized supernatant was filtered through a 0.45 μm membrane, mixed with an equal volume of 2% (w/v) sarkosyl in Dulbecco’s PBS containing 0.3% (w/v) NaPTA and then centrifuged at 16 100g for 90 min at 37°C to obtain the semi-purified P2 pellet and the SN2 supernatant fractions, which were used for western blot and bioassay analysis.
We also used a modified version of this protocol in which 200 µl aliquots of 10% brain homogenate were first incubated for 30 min with an equal volume of sarkosyl 4% with gentle shaking at 37°C and then treated with 50 μg/ml PK for 60 min at 55°C and gentle shaking. PK activity was blocked by adding 3 mM PMSF for 10 min at 4°C. The PK-treated samples were then subjected to the same steps of the Wenborn protocol that followed PE digestion, starting from the Benzonase® treatment as described above, obtaining again a semi-purified P2 pellet and the SN2 supernatant fractions that were then used for the western blot, electron microscopy and bioassay experiments. P2 pellets were resuspended in 20 µl PBS in the presence of 0.1% sarkosyl and both the P2 and SN2 fractions were kept at −20°C for western blotting and bioassay experiments. Considering the discarded fractions of the protocol, a single 20 µl P2 aliquot, as well as the corresponding SN2 fraction consisting of 1800 µl, corresponded to ∼150 µl of the starting brain homogenate (i.e. ∼15 mg brain equivalents).
With the PK protocol, P2 pellets were further purified to obtain highly enriched pure PrPres preparations. To this aim, P2 pellets were resuspended in a wash buffer composed of Dulbecco’s PBS, containing 17.5% (w/v) Iodixanol, 0.3% NaPTA and 0.1% (w/v) sarkosyl and centrifuged at 16 100g for 30 min. This step was repeated with the P3 pellets to obtain the final P4 pellets. Replica P4 pellets obtained after purification of GSS-A117V or Bv-Scr were resuspended either in 10 µl PBS in the presence of 0.1% sarkosyl when analysed for western blotting and bioassay analysis, or in 10 µl Tris HCl in the presence of 1 mM NaN3 and 0.1% sarkosyl for electron microscopy analysis. A single 10-µl P4 fraction corresponded to ∼150 µl of the starting brain homogenate.
To assess the infectivity of the purified fractions in relation to the original GSS-A117V inoculum, P2 and P4 fractions were diluted 180-fold and 360-fold in phosphate buffer, respectively, to obtain an inoculum comparable with the original GSS+PK 4 × 10−3 dilution, and then serially 10-fold diluted. SN2 supernatants, which were diluted 90-fold compared to the P2 fractions, were 2-fold diluted and then serially 10-fold diluted. Two dilutions were chosen for the titration assay: (i) the 4 × 10−4 dilution, to determine if the pellets containing only purified 7 kDa PrPres aggregates were still infectious; and (ii) the 4 × 10−6 dilution, to define the correlation between PrPres amount and infectivity.
Bioassay
All animal studies were carried out in derogated CL3 facilities. Bank voles were obtained from the breeding colony at the Istituto Superiore di Sanità (ISS). Inoculated voles were housed in autoclavable Eurostandard type II polysulphone cages (three voles/cage) with abundant wood chips as bedding material. Voles were kept under a 12-h light/dark cycle and given food and water ad libitum. Cohorts of voles (n = 6, 6–8 weeks of age and sex-matched) either expressing methionine (Bv109M, inoculated with sCJD-MM1) or isoleucine (Bv109I, for GSS-A117V and Bv-CWD) at PrP codon 109 were anaesthetized with ketamine (0.1 μg/g) and inoculated with 20 μl brain homogenates or purification fractions into the left cerebral hemisphere.
Inoculated voles were examined twice a week until the appearance of neurological signs, after which they were examined daily. Animals that died because of intercurrent disease were excluded from the study. Diseased animals were euthanized with CO2 at the terminal stage of the disease. The brain from each animal was removed and cut sagittally into two parts. One was stored at –80°C and one was fixed in formalin.
The research protocol, approved by the Italian Ministry of Health and performed under the supervision of the Service for Biotechnology and Animal Welfare of the ISS, adhered to the guidelines contained in the Italian legislative Decree 116/92, which transposed the European Directive 86/609/EEC on Laboratory Animal Protection, and then in the Legislative Decree 26/2014, which transposed the European Directive 2010/63/UE on Laboratory Animal Protection.
Neuropathology
Histology and immunohistochemistry were performed on formalin-fixed tissues as previously described (Nonno et al., 2006). Briefly, brains were trimmed at standard coronal levels, embedded in paraffin wax, cut at 6 µm and stained with haematoxylin and eosin. PrP immunolabelling in immunohistochemistry was carried out using a mix of SAF84 and 6C2 mAbs. For the construction of lesion profiles, vacuolar changes were scored in nine grey matter areas (Fraser and Dickinson, 1968).
SDS-PAGE, western blot and silver staining
Brain homogenates (10% w/v) were added with an equal volume of 100 mM Tris HCl containing 4% sarkosyl and incubated for 30 min at 37°C with gentle shaking. PK was added at a final concentration of 100 µg/ml and the samples were incubated for 60 min at 55°C with gentle shaking. PK was stopped with 3 mM PMSF. Samples were added with an equal volume of isopropanol/butanol (1:1 v/v), centrifuged for 10 min at 20 000g and the resulting pellets were resuspended in denaturing sample buffer (NuPAGE® LDS Sample Buffer and NuPAGE® Sample Reducing Agent, Invitrogen) and heated for 10 min at 95°C.
Purified pellets were either left untreated or digested with PK (10 µg/ml, 60 min at 55°C). PMSF (3 mM) was added to stop the digestion. Digested and untreated pellets were mixed with an equal volume of 2× denaturing sample buffer and heated at 95°C for 10 min. Aliquots of the supernatant fractions were added with an equal volume of isopropanol/butanol (1:1 v/v), centrifuged at 20 000g for 10 min and the pellets were resuspended in denaturing sample buffer and heated at 95°C for 10 min.
Electrophoresis, conducted with NuPAGE® MES SDS Running Buffer or NuPAGE® MOPS SDS Running Buffer (Invitrogen, Thermofisher) depending on the type of PrPSc analysed, and western blotting were carried as previously described (Pirisinu et al., 2018). The monoclonal antibodies used and their epitopes were as follow: SAF32 (octarepeat), 9A2 (bank vole sequence amino acids 99–101), SAF60 (human sequence amino acids 157–161) and SAF84 (bank vole sequence amino acids 163–169). Chemiluminescence signal was detected by ChemiDoc imaging system (Bio-Rad) and quantified using the Image Lab software (Bio-Rad).
For silver staining, SDS-PAGE gels were stained using the Pierce™ Silver Stain Kit (Thermofisher Scientific) following the manufacturer’s instructions, and visualized with the ChemiDoc Imaging system (Bio-Rad).
Electron microscopy
Negative staining of the purified samples was performed on carbon-coated, 200-mesh copper grids that were previously glow-discharged. P2 and P4 pellet fractions were resuspended in 10 µl Tris HCl (100 mM, pH 7.4) in the presence of 1 mM NaN3 and 0.1% sarkosyl. Whole SN2 fractions were added to an equal volume of isopropanol/butanol (1:1 v/v), centrifuged at 20 000g for 10 min, and the pellets were re-suspended in 10 µl Tris-HCl (100 mM, pH 7.4) in the presence of 1 mM NaN3 and 0.1% sarkosyl. Sample aliquots (5 µl) were loaded onto the grids and incubated for up to 5 min to allow proper adsorption of the protein sample. The grids were briefly washed with 0.1 M and 0.01 M ammonium acetate buffer, pH 7.4 to remove surplus, non-adsorbed protein sample and buffer and stained with two droplets (50 µl each) of freshly filtered 2% (w/v) uranyl acetate. After drying, the stained samples were viewed in a FEI Tecnai G2 TF20 electron microscope (FEI company) at an acceleration voltage of 200 kV. Each grid was analysed systematically using the tracking feature of the CompuStage (FEI Company) to determine the ultrastructure of the adsorbed sample. The recorded tracks allowed a wide-ranging, non-overlapping search pattern that covered most of the grid surface. This systematic surveying approach allowed us to ascertain the aggregation state of the adsorbed sample. Images were recorded on an Eagle 4k × 4k multiport CCD camera (FEI company) with a 4-port readout and a 14 × 14 µm2 physical pixel size.
Immunogold labelling
Fab 69 was selected from a phage display library for its ability to bind murine PrP (Senatore et al., 2020). For the immunogold labelling experiments Fab 69’s ability to recognize an epitope within residues 87GWGQGGGTHNQW98 of murine PrP was essential as this epitope is retained in the ∼7 kDa PrPres fragments. Fab 69 was expressed in Escherichia coli [BL21(DE3)] as a His-tagged, soluble, and functional protein. Upon lysis of the bacterial cells, Fab 69 was purified via IMAC chromatography, eluted using imidazole buffer, desalted, and characterized for its purity and activity (Rathod et al., unpublished data).
The immunogold labelling protocol was adapted from Wille et al. (2007): 2–3 μl samples were absorbed for 6 min onto glow-discharged formvar/carbon-coated nickel grids (Ted Pella, Inc.). The grids were then washed with 50 μl 0.1 M and 0.01 M ammonium acetate buffer pH 7.4 and treated with 50 μl of 3 M urea for 10 min, which served to increase the accessibility of the epitope that is recognized by Fab 69. After washing with 50 μl of the 0.1 M and 0.01 M ammonium acetate buffers, the grids were pre-stained with 2% phosphotungstate, pH 7.2 (PTA). The formvar/carbon-film surface was then blocked for 60–90 min with 0.3% bovine serum albumin (BSA) in Tris-buffered saline (TBS: 50 mM Tris HCl, pH 7.4; 150 mM NaCl), incubated with the primary antibody (Fab 69) for 3 h, rinsed five times with 0.1% BSA in TBS, incubated with a bridging goat F(ab′)2 anti-human IgG F(ab′)2 (Abcam ab98531) for 2 h, rinsed five times again with 0.1% BSA in TBS, incubated with a 5-nm gold-conjugated rabbit anti-goat IgG (Abcam ab202670) for 2 h, rinsed five times with 0.1% BSA in TBS, twice with TBS alone, and twice with water. Controls were treated identically, except that the primary Fab was omitted. Finally, the grids were counterstained twice with 50 μl 2% PTA and air-dried. The dried grids were analysed using a Tecnai G2 TF20 electron microscope (FEI company), as described. Gold particles that were observed within a radius of 25 nm from a structure of interest were considered specifically bound, since the length of a Fab molecule is 5 nm while that of an IgG is 10 nm (5 nm primary Fab + 5 nm bridging Fab + 10 nm IgG + 5 nm gold particle diameter = 25 nm). Gold particles that exceeded this 25 nm distance threshold were deemed non-specific background labelling.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.
Results
PK-treated GSS-A117V retains full infectivity
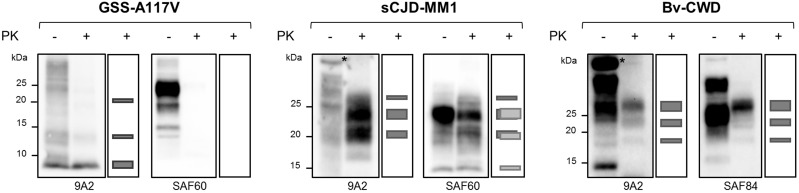
In a previous study, we demonstrated that GSS cases with only 7 kDa PrPres are experimentally transmissible in voles (Pirisinu et al., 2016), suggesting that infectivity could be associated with the 7 kDa PK-resistant core of PrPSc. However, those data could not exclude that other PrPSc conformers, sensitive to proteases, might have played a role in the observed phenotypes. To solve this issue, we sought to determine if prion infectivity associated with atypical PrPSc is resistant to PK, which is known to cleave both the N- and C-termini of atypical PrPSc. To this aim, we end-point titrated prion isolates from a GSS-A117V case (GSS-A117V #1) in bank voles, as a model for atypical PrPSc, and from two prion diseases associated with classical PrPSc, i.e. sCJD-MM1 and bank vole adapted CWD (Bv-CWD). GSS-A117V and Bv-CWD were chosen for their ease of transmission into bank voles carrying isoleucine at position 109 (Bv109I) (Di Bari et al., 2013; Pirisinu et al., 2016) and were thus titrated in Bv109I. In contrast, sCJD-MM1 was assayed in bank voles carrying methionine at codon 109 (Bv109M), which replicate this strain faster than Bv109I (Nonno et al., 2006; Rossi et al., 2017). Brain homogenates from the three cases were either left untreated or digested with PK at a concentration that allowed the complete degradation of PrPC, serially diluted 10-fold and inoculated in voles. Upon PK digestion, only PrPres species were detected in the inocula. The PK-treated GSS-A117V sample only showed N- and C-terminally truncated, unglycosylated PrPres of 7 kDa (Fig. 1). Faint PrPres fragments of 14 kDa and 21 kDa shared the same antibody reactivity as 7 kDa PrPres and likely represent its covalently linked multimers, as reported in previous studies (Pirisinu et al., 2013; Cracco et al., 2019). The N-terminally truncated, fully glycosylated PrPres types, with protease-resistant unglycosylated fragments of 19 kDa for Bv-CWD and of 21 and 14 kDa for sCJD-MM1, were detected in PK-treated Bv-CWD and sCJD samples instead (Fig. 1).
Figure 1.
PrP species in PK-treated and untreated inocula. Total PrP (first lane of each blot) and PrPres (second lane of each blot) in brain homogenates from GSS-A117V, sCJD-MM1 and Bv-CWD were analysed by western blot using monoclonal antibodies directed to the central region (9A2) or the C-terminus (SAF60 and SAF84 for human and bank vole PrP, respectively) of PrP. The second lane of each blot was duplicated as a schematic and added on the right of the blot to better illustrate the PrPres electrophoretic pattern of the PK-treated samples analysed; the size of each rectangle represents the amount of chemiluminescent signal of the respective PrPres fragment in the blot. The PK-treated GSS-A117V inoculum was characterized by 7 kDa PrPres devoid the C-terminus of the protein, as it is recognized by the internal antibody 9A2 but not by the C-terminal antibody SAF60 (larger rectangle in the 9A2 schematic box). Note the presence of multimers of 7 kDa PrPres (higher molecular weight bands of ∼14 and ∼21 kDa, represented by the thinner rectangles in the 9A2 schematic box), which share the same antibody reactivity of the 7 kDa PrPres, i.e. they are recognized by the internal antibody but not by the C-terminal one. The PK-treated sCJD-MM1 inoculum was characterized by a complex PrPres pattern, resulting from the overlapping of (i) type 1 PrPres, recognized by both the antibodies (three dark grey rectangles in both the schematic boxes), which is characterized by a dominance of the mono-glycosylated fragment, with an unglycosyilated fragment of 21 kDa in size; (ii) an additional C-terminal, fully glycosylated PrPres fragment that is recognized only by the C-terminal antibody (SAF60, three light grey rectangles in the SAF60 cartoon box), with an unglycosylated fragment of 14 kDa in size. The PrPres in the Bv-CWD inoculum was instead characterized by a classical PrPres pattern with an unglycosylated fragment of 19 kDa in size and a dominance of the di-glycosylated fragment (9A2 and SAF84, three dark grey rectangles in both the schematic boxes). The asterisks indicate non-specific bands detected in untreated brain homogenates by the antibody 9A2.
As expected, PK treatment did not substantially affect the infectivity titre of the two classical prion diseases (Supplementary Table 1). Indeed, sCJD-MM1 showed ID50 U/g values of 105.6 when untreated and 106.1 with PK treatment, while ID50 U/g values of 108.4 and 108.1 were observed for untreated and PK-treated Bv-CWD, respectively. Unexpectedly, GSS-A117V showed an extremely high infectious titre in voles, even higher than classical prions, with an ID50 U/g value of 109.3 for the GSS untreated inoculum. Moreover, the infectious titre was not substantially altered upon PK treatment, with the PK-treated GSS-A117V inoculum showing an ID50 U/g value of 109.0 (Supplementary Table 1). As previously reported (Nonno et al., 2006; Di Bari et al., 2013; Pirisinu et al., 2016), voles infected with GSS-A117V, sCJD-MM1 and Bv-CWD reproduced distinctive neuropathological patterns and PrPSc types, suggestive of the isolation of three distinct vole-adapted strains (Supplementary Figs 1 and 2). Most importantly, these phenotypes were unchanged in voles inoculated with PK-treated samples, suggesting that PK-resistant molecules retain the strain properties in these TSEs, including atypical GSS-A117V (Supplementary Figs 1 and 2). Interestingly, the severity of spongiosis in the cerebellum of voles infected with GSS-A117V varied according to the dilution of the inoculum, independently of the PK pretreatment of the inoculum (Supplementary Fig. 2).
Overall, these results establish that GSS-A117V contains PK-resistant and strain-specific prions, similar to prion diseases associated with classical PrPSc.
Purified 7 kDa PrPres aggregates retain prion infectivity and strain properties
The outcome of the previous experiment strongly suggested that the 7 kDa PrPres aggregates that remained after PK digestion are responsible for the infectivity and the strain features of GSS-A117V. To definitely demonstrate a direct association between infectivity and 7 kDa PrPres we decided to isolate these PrPres aggregates in order to obtain pure preparations of in vivo generated prions.
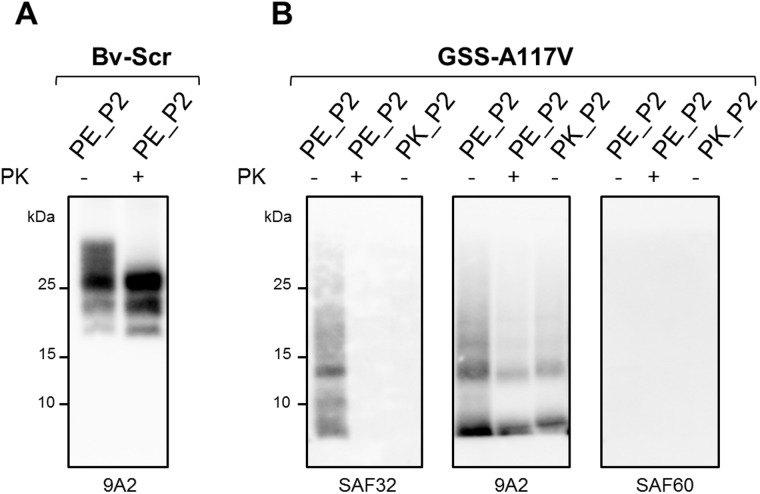
To this aim, we used a novel purification method for obtaining highly enriched pure prion preparations recently developed for mouse scrapie prions (Wenborn et al., 2015). In this method, an initial treatment with PE, which preserves the PK-sensitive N-terminus of classical PrPSc, allows the purification of full-length PrPSc that can then be N-terminally truncated into PrPres upon PK treatment.
We first used this method to purify classical PrPSc from the brains of scrapie affected bank voles (Bv-Scr). We obtained a first semi-purified pellet fraction, i.e. P2, containing full-length PrPSc, with intact C- and N-termini (Fig. 2). The semi-purified PrPSc obtained in P2 was partially resistant to PK, which cleaved the N-terminus leading to the typical three bands PrPres pattern of classical scrapie in western blot (Fig. 2). The PrPres amount obtained in the P2 pellet, determined by chemiluminescent signal, was about three times lower than the quantity of PrPres present in the starting brain homogenate (Supplementary Fig. 3). While attempting to purify PrPSc from GSS-A117V we found that, in contrast to classical PrPSc, the C-terminus of atypical PrPSc was PE-sensitive (Fig. 2), confirming that the C-terminus of atypical PrPSc has a distinct structure and is accessible to proteases. Thus, the semi-purified P2 fraction that we obtained from GSS-A117V #1, named PE_P2, did not contain full-length PrPSc but C-terminally cleaved PrPSc. Interestingly, the N-terminus of atypical PrPSc was partially preserved upon PE treatment, as confirmed by the weak signal detected in western blot with the antibody SAF32 (Fig. 2). Similar to classical PrPSc, semi-purified GSS-A117V PrPSc from the PE_P2 pellet was partially resistant to PK, which removed the remaining N-terminus thus resulting in the 7 kDa PrPres aggregates that we sought to purify.
Figure 2.
PrPSc in the purified fractions from scrapie and GSS-A117V. P2 pellets obtained from scrapie affected voles (Bv-Scr) (A) or GSS-A117V (B) with the purification methods using PE (PE_P2) or PK (PK_P2) as proteases, were analysed by western blot either untreated (–) or subjected to a further PK digestion (+). The PE_P2 fraction from Bv-Scr contained full-length PrPSc that upon PK digestion showed the typical three band PrPres pattern. In contrast, the GSS-A117V PE_P2 fraction contained low molecular weight C-terminal cleaved PrPSc fragments, not detected by the C-terminal antibody SAF60, but partially preserving the octarepeat region in the N-terminus (detected with SAF32). Upon PK digestion of PE_P2 from GSS-A117V, PrPres was only detected with antibody 9A2, revealing the expected 7 kDa PrPres profile, devoid of both the N- and C-terminus. The GSS-A117V PK_P2 pellet contained only 7 kDa PrPres, already devoid of N-terminal and C-terminal moieties, as it was only detected by the internal antibody 9A2.
However, the recovery of PrPres in the P2 fraction was lower than that observed with classical PrPSc, with PrPres in the PE_P2 pellet being ∼10 times less than in the original brain homogenate (Supplementary Fig. 3).
In an attempt to increase the yield of 7 kDa PrPres in the P2 fraction, we replaced PE treatment with PK treatment. We obtained a P2 fraction, i.e. PK_P2, containing only the 7 kDa N- and C-termini cleaved PrPres (Fig. 2). Again, the PK_P2 fraction contained ∼10 times less PrPres than the starting brain homogenate.
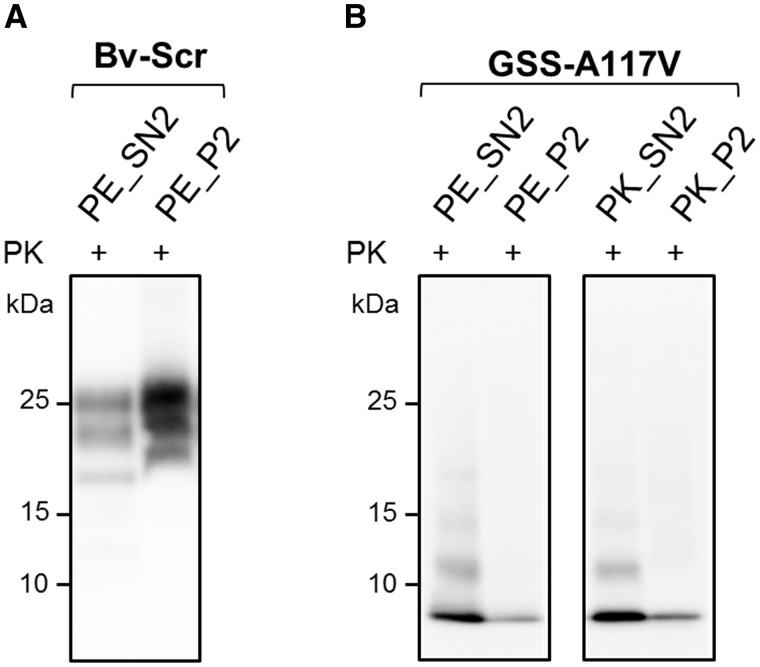
To investigate the reason for the low PrPres yield in the GSS-A117V P2 pellets, we analysed the amount of PrPres in all fractions of both Bv-Scr and GSS-A117V samples. Bv-Scr and GSS-A117V had a comparable recovery of PrPres after the first centrifugation, i.e. in the supernatant fraction SN1, but their behaviour was different during the centrifugation in 17.5% iodixanol, which allowed the separation of the sample into an insoluble pellet P2 and a soluble supernatant SN2. Indeed, as expected, classical PrPres was highly enriched in the P2 fraction and partitioned into SN2 and P2 at a ratio of ∼1:3, respectively, indicating that only a small amount of PrPres remained in the supernatant fraction. By contrast, most of the 7 kDa PrPres was retained in the SN2 fraction, with a SN2: P2 distribution of ∼2:1, indicating that under those conditions of centrifugation, the 7 kDa PrPres did not sediment efficiently, thus explaining the low amount of PrPres in the P2 fraction (Fig. 3).
Figure 3.
Comparison of PrPres in SN2 and P2 fractions. SN2 (PE_SN2) and P2 (PE_P2) fractions obtained from scrapie affected vole (Bv-Scr) (A) or from GSS-A117V (B) were treated with PK (+) and analysed by western blot with antibody 9A2. For GSS-A117V, SN2 and P2 fractions obtained with the purification protocol using PK as protease (PK_SN2 and PK_P2, respectively) were also analysed. PrPres was mainly enriched in the P2 fraction in Bv-Scr (A), while it was mainly detected in SN2 fractions from GSS-A117V (B). Note that no difference in terms of SN2/P2 partitioning was observed using PE or PK during the purification method. All membranes were detected with antibody 9A2.
The inefficient sedimentation observed with the 7 kDa PrPres means that the atypical PrPSc is characterized by sedimentation properties different from those of classical PrPSc, suggesting that this PrPSc type might predominantly consist of small-sized/low density aggregates.
We then aimed at evaluating the infectivity of the purified fractions containing 7 kDa PrPres. PE_P2 and PK_P2 GSS-A117V pellets were diluted 1800 and 180 000-fold in phosphate buffer, respectively, to obtain dilutions equivalent to the 4 × 10−4 g/ml and 4 × 10−6 g/ml of the brain homogenate dilutions used in the titration of the original GSS-A117V sample. These preparations were used to inoculate voles, along with the 4 × 10−4 g/ml dilutions of a P2 purified fraction derived from uninfected vole brain tissue as a negative control (Bv-Neg), and of the original untreated GSS-A117V sample as a positive control (Table 1).
Table 1.
Transmission in bank voles of purified fractions of PrPSd from GSS-A177V
| Inoculum | Status | Fraction |
4 × 10−4 g/ml
a
|
4 × 10−6 g/ml
a
|
||
|---|---|---|---|---|---|---|
| Attack rate, n/Nb | Survival time ± SD, days c | Attack rate, n/Nb | Survival time ± SD, days c | |||
| GSS-A117V | Supernatant | PE_SN2 | 5/5 | 112 ± 6 | 6/6 | 143 ± 39 |
| GSS-A117V | Pellet | PE_P2 | 6/6 | 136 ± 3 | 2/6 | 183 ± 7 |
| GSS-A117V | Pellet | PE_P2_PK | 6/6 | 124 ± 11 | nd | nd |
| GSS-A117V | Supernatant | PK_SN2 | 5/5 | 103 ± 4 | 6/6 | 113 ± 13 |
| GSS-A117V | Pellet | PK_P2 | 5/5 | 120 ± 11 | 3/6 | 142 ± 7 |
| GSS-A117V | Pellet | PK_P4 | 5/5 | 149 ± 12 | 0/7 | >350 |
| Bv-Neg | Pellet | PK_P2 | 0/4 | >350 | nd | nd |
| GSS-A117V | BHd | – | 4/4 | 112 ± 12 | nd | nd |
SD = standard deviation; n = number of voles that developed confirmed TSE disease; N = number of inoculated mice; nd = not done.
Purified fractions from GSS-A117V #1 were diluted to reach dilutions equivalent to the 4 × 10−4 g/ml and 4 × 10−6 g/ml dilutions of the starting brain homogenate, as reported in the ‘Materials and methods’ section.
Voles dead from intercurrent disease were excluded from the study.
When voles did not show clinical signs, the time of the last animal of the group tested negative was reported.
Untreated GSS-A117V #1 brain homogenate (BH) diluted and used as positive control.
Remarkably, all voles inoculated with the dilution 4 × 10−4 g/ml of the PE_P2 and PK_P2 pellets became clinically sick, confirming that the human-derived purified 7 kDa PrPres aggregates are fully infectious in voles. Infectivity was still detectable in the 4 × 10−6 g/ml dilution, i.e. after a >100 000-fold dilution of the P2 pellet fraction, although both PE_P2 and PK_P2 showed a partial attack rate at this dilution, comparable to the outcome of the 4 × 10−8 g/ml dilution of the PK-treated GSS-A117V brain homogenate (see Supplementary Table 1 for comparison). This allows us to conclude that the P2 pellets contained comparable levels of infectivity, being both ∼100-fold less infectious than the original GSS-A117V inoculum, in keeping with the low recovery of the 7 kDa PrPres during the purification. Interestingly, no difference in terms of infectivity was observed between the PE and PK-treated pellets, indicating that the octarepeat region of PrP that is partially preserved upon PE treatment does not play a significant role in prion infectivity. In agreement with these conclusions, the purified PE_P2 fraction preserved infectivity after a further PK treatment (referred to as PE_P2_PK in Table 1).
As SN2 fractions contained higher amounts of 7 kDa PrPres than the P2 fractions (Fig. 3), we aimed at determining if this correlated with higher levels of infectivity, i.e. if the distribution of infectivity in the SN2/P2 separation correlated with the distribution of PrPres, by inoculating voles with PE_SN2 and PK_SN2, both equivalently diluted to the 4 × 10−4 g/ml and 4 × 10−6 g/ml of the brain homogenate dilutions. Indeed, the two supernatants PE_SN2 and PK_SN2 contained higher levels of infectivity than the corresponding P2 fractions, resulting in 100% attack rates at both dilutions (Table 1).
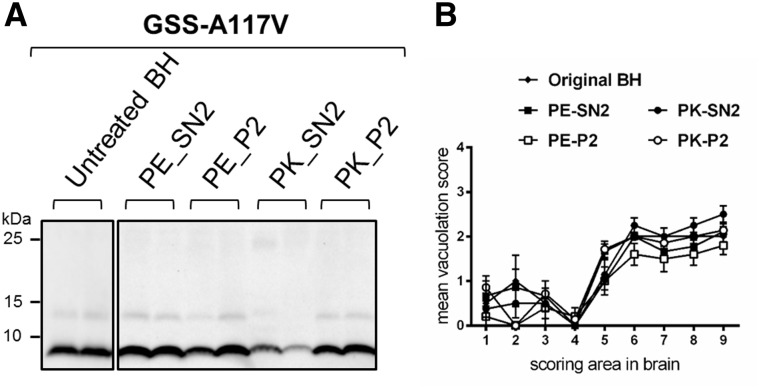
Finally, the analysis of the PrPres type and of the neuropathological phenotype in voles inoculated with the different fractions confirmed that the original GSS-A117V strain features were preserved in all fractions and were identical to those of the original inoculum (Fig. 4 and Supplementary Figs 4–6). The slight variation of the degree of spongiosis in the cerebellum of voles inoculated with SN2 and P2 fractions (Supplementary Fig. 6) was reminiscent of that observed in the titration experiment (Supplementary Fig. 2) and was probably due to the different dilutions and levels of infectivity in SN2 and P2 fractions.
Figure 4.
PrPSc and spongiform degeneration in voles inoculated with supernatant and pellet fractions from GSS-A117V. (A) Representative western blot of PrPres from voles inoculated with the original brain homogenate (untreated BH) or with supernatant (PE_SN2 or PK_SN2) and pellet (PE_P2 or PK_P2) fractions from GSS-A117V. Membranes were probed with antibody 9A2. (B) Patterns of neurodegeneration analysed by semiquantitative assessment of spongiform degeneration in different brain regions (lesion profiles) of voles inoculated with the untreated brain homogenate or with the different purification fractions from GSS-A117V, as indicated at the top of the graph. Brain scoring areas are: medulla (1), cerebellum (2), superior colliculus (3), hypothalamus (4), thalamus (5), hippocampus (6), septum (7), retrosplenial and adjacent motor cortex (8), cingulate and adjacent motor cortex (9). Each point represents the mean ± standard error of the mean (SEM) of at least four individual voles.
Overall, these findings imply that PrPres aggregates composed of only N- and C-terminally truncated 7 kDa PrPres, purified from a human brain affected with the prion disease GSS-A117V, are fully infectious prions and encode the strain features of the original full-length PrPSc molecule. Furthermore, our results clearly demonstrate that infectivity correlates directly with the 7 kDa PrPres, as it decreases at the same rate as the amount of PrPres in the inoculum.
Purified prions from GSS-A117V are small non-fibrillar particles
A crucial question in the prion field is the 3D structure of the infectious PrPSc conformer, but the experimental obstacles posed by the highly aggregated, glycosylated, and GPI-anchored conventional PrPSc conformers, and the difficulties of obtaining highly infectious synthetic prions, present formidable obstacles (Wille and Requena, 2018). In this context, purified 7 kDa PrPres aggregates, associated with high levels of infectivity and made up of short, non-glycosylated and non-GPI-anchored PrP fragments, might potentially overcome these obstacles and could be exploited for structural studies.
To obtain highly purified preparations void of contaminating proteins and suitable for analysis by electron microscopy, we added two further centrifugation steps to the whole purification method, resulting in pure P4 pellets. We thus generated purified 7 kDa PrPres samples from two unrelated GSS-A117V cases (named GSS-A117V #1, previously used for the titration assay and the first analysis of purified 7 kDa PrPres, and GSS-A117V #2) and equivalent 20 kDa PrPres from Bv-Scr. The final pure PK_P4 pellet from Bv-Scr contained similar amounts of PrPres than the PK_P2 fraction, with all major SDS-PAGE silver stained bands overlapping with the bands recognized by the anti-PrP antibody on western blot (Fig. 5). P4 pellets derived from GSS-A117V #1 and GSS-A117V #2 showed comparable or slightly lower (GSS-A117V #1) levels of PrPres than the corresponding P2 pellets, although unrelated proteins still present in the P2 fractions were absent in P4. Thus, the major SDS-PAGE silver stained band in GSS-A117V P4 pellet overlapped with the 7 kDa band recognized by the anti-PrP antibody on western blot (Fig. 5). Most importantly, infectivity co-segregated with PrPres aggregates, as P4 from Case 1 was still infectious after 3600-fold dilution (equivalent to the 4 × 10−4 g/ml of the brain homogenate) (Table 1). Still, the infectious titre was lower in P4 than in P2 (compare the 4 × 10−6 g/ml dilutions of PK_P2 and PK_P4 in Table 1), in keeping with the lower PrPres content in P4 (Fig. 5).
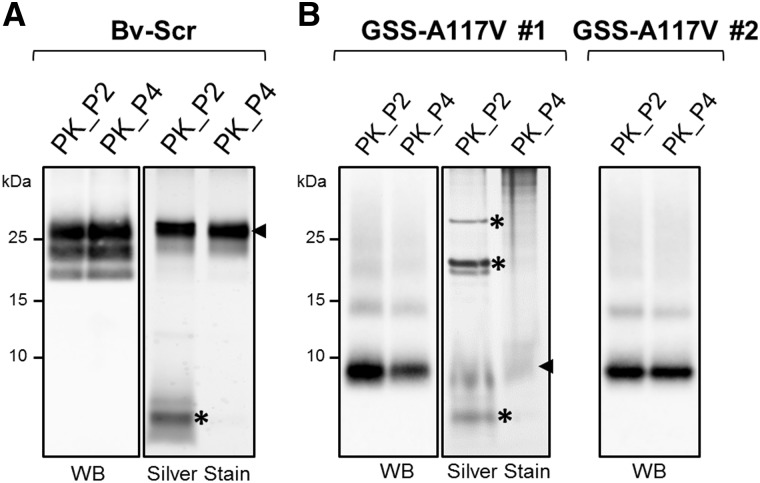
Figure 5.
Purified PrPres from Bv-Scr and human GSS-A117V isolates. P2 and P4 fractions obtained with the PK purification method from Bv-Scr (A) and from two GSS-A117V cases, indicated as GSS-A117V #1 and #2 (B), were analysed by western blot (WB) and silver staining to evaluate the presence and purity of PrPres. Fractions were loaded at 1.5 mg equivalent for western blot analysis and at 10 mg equivalent for silver staining. Blots were probed with antibody 9A2; SDS gels were used for the silver staining as indicated in the ‘Materials and methods’ section. Arrows on the right of silver-stained P4 fractions indicate the major SDS-PAGE silver-stained bands, which were consistent with PrPres detected with 9A2 by western blot. Asterisks indicate protein bands detected in the P2 fractions of both Bv-Scr and GSS-A117V that were absent in P4 pellets.
Ultrastructural analyses using negative stain electron microscopy of PK_P2 and PK_P4 samples from scrapie-affected vole brains that contain only classical PrPres revealed the presence of clumps of amyloid fibrils (Fig. 6B), morphologically similar to those observed in other classical prion strains (Wenborn et al., 2015). In sharp contrast, PK_P2 and PK_P4 pellets generated from the two GSS-A117V cases showed no fibrils at all; instead, they contained very small and structurally indistinct particles that covered the grid surface, with occasional lipid and amorphous structures (Fig. 6A). The alcohol precipitates of all samples contained only amorphous aggregates, as expected for alcohol-treated samples.
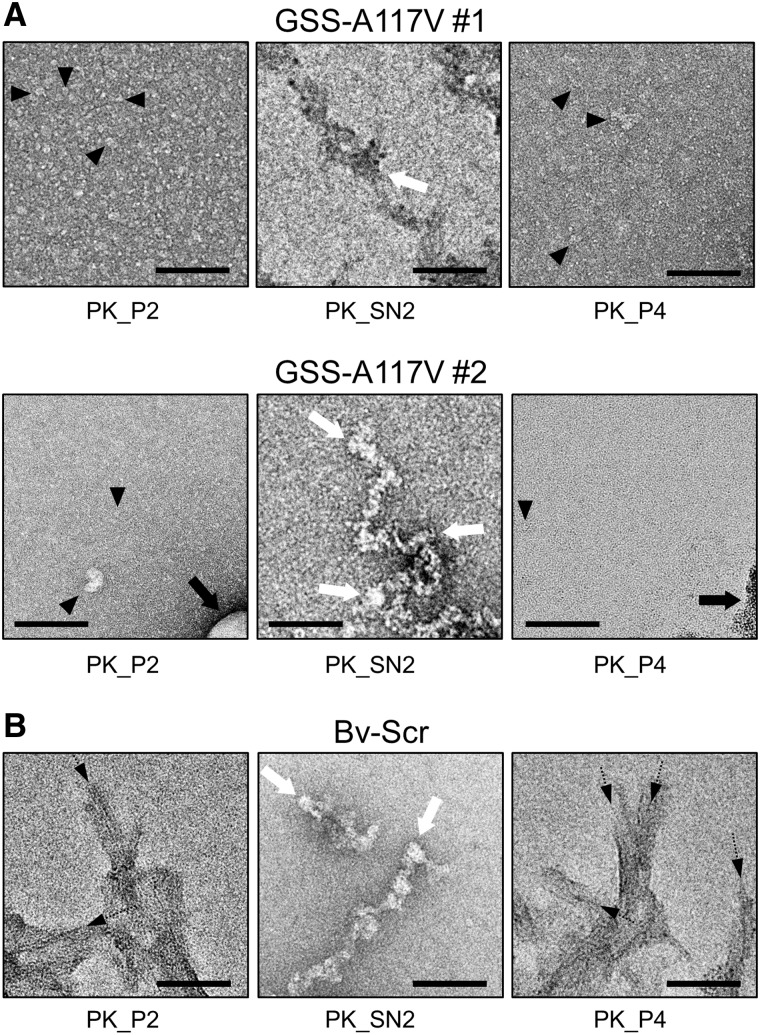
Figure 6.
Negative stain electron micrographs from human GSS-A117V and Bv-Scr isolates. The P2 and P4 fractions from the GSS-A117V isolates (A) and Bv-Scr (B) as well as the corresponding SN2 fractions (after alcohol precipitation) were negatively stained with uranyl acetate. The P2 and P4 fractions from the GSS-A117V isolates (A) contained a layer of small amorphous particles that covered the carbon film surface (filled arrowheads on select particles). No amyloid fibrils or larger protein aggregates were observed, occasional lipid structures and amorphous structures were visualized (black arrows). The SN2 precipitate was composed of amorphous aggregates only (white arrows), which is typical for alcohol precipitates. In contrast, the P2 and P4 fractions from Bv-Scr (B) contained substantial quantities of amyloid fibrils (black dashed arrows), which tended to aggregate into larger clumps, partially obscuring their ultrastructure. The SN2 precipitate again showed only amorphous structures (white arrows) because of the alcohol treatment. Scale bars = 100 nm.
To ascertain the nature of the structurally indistinct particles in the PK_P2 and PK_P4 pellets, we conducted immunogold labelling experiments with a recombinantly produced antibody fragment (Fab 69) that recognizes an epitope near the N-terminus of the 7 kDa fragment (Fig. 7). Using a binary system to detect bound Fab 69 we were able to decorate the small, 7 kDa fragment particles with 5 nm gold labels (Fig. 7A), while a control sample for which the primary Fab was omitted showed only a few gold particles as unspecific background labelling. Similarly, the amyloid fibrils in the Bv-Scr PK_P2 samples were recognized by Fab 69 and decorated with 5-nm gold labels (Fig. 7B), while the amyloid fibrils in the controls (no primary Fab) remained undecorated. Lastly, bank vole brains from uninfected animals that were subjected to the same purification procedure, resulting in PK_P2 control samples (Bv-Neg), revealed only a few, unspecific gold labels independent of the presence or absence of Fab 69 (Fig. 7C). Therefore, the immunogold labelling experiments confirmed that the observed, structurally indistinct oligomeric particles (Fig. 6A) contain a PrP epitope between residues 87 and 98.
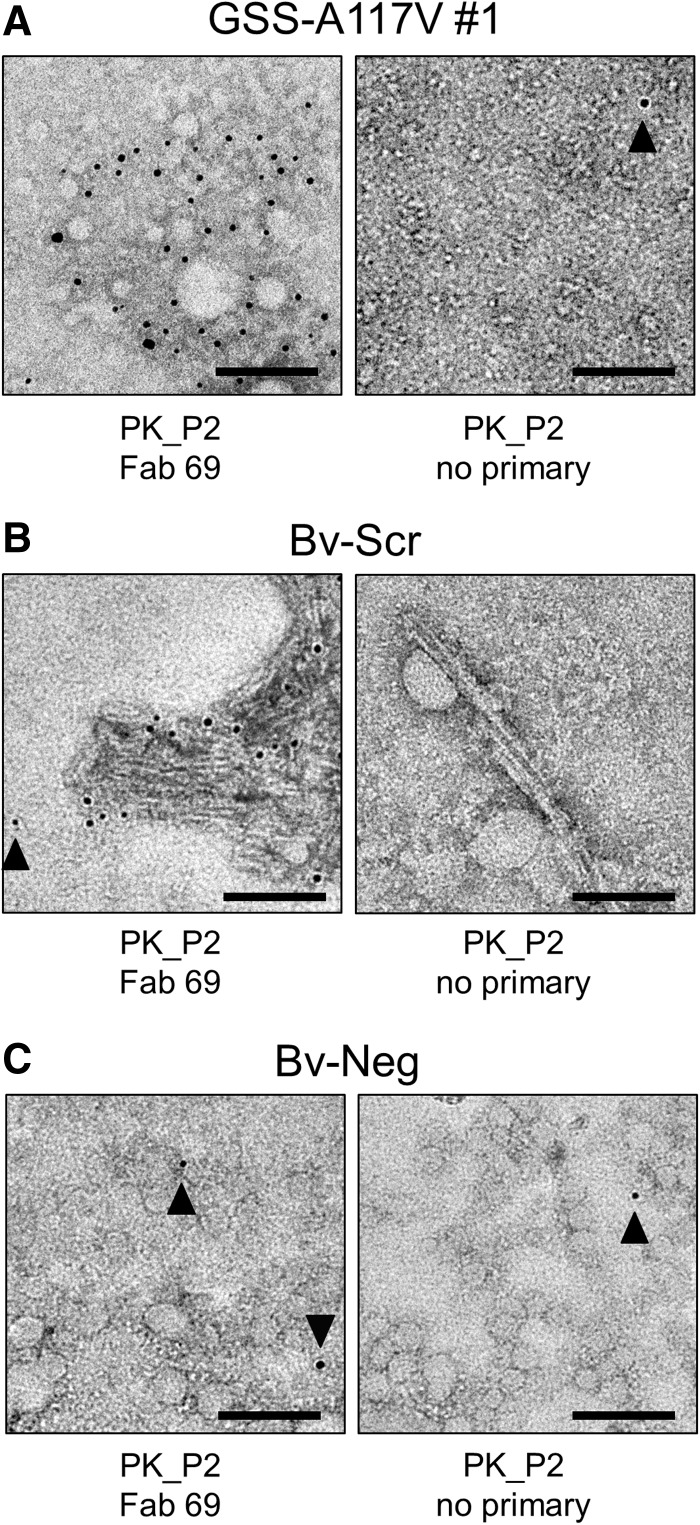
Figure 7.
Immunogold labelling of human GSS-A117V, Bv-Scr, and Bv-Neg isolates. The P2 fractions from GSS-A117V (A), Bv-Scr (B), as well as Bv-Neg isolates (C) were used for immunogold labelling experiments with an anti-PrP Fab (Fab 69), which recognizes an epitope between residues 87 and 98. The small amorphous particles in the GSS-A117V isolate were decorated with many 5 nm gold particles, while a sample that was treated identically except for the omission of the primary Fab showed only one gold particle (filled arrowhead). Because of the size and nature of the GSS-A117V particles, the attached gold labels cannot be assigned to specific particles. The amyloid fibrils in the Bv-Scr isolate (B) were decorated with gold particles indicating that they are composed of PrP, while in the control sample without primary Fab the fibrils remained undecorated. A gold particle that is too far from the amyloid fibrils represents non-specific background labelling (filled arrowhead). Bv-Neg samples (C) that lack PK-resistant PrP only showed low levels of background labelling as indicated by individual gold particles (filled arrowheads). Scale bars = 100 nm.
Discussion
Experimental evidence that GSS subtypes, which are exclusively associated with 6–8 kDa PrPres fragments, represent genuine prion diseases has recently been reported (Asante et al., 2013; Pirisinu et al., 2016). However, a direct association between low molecular weight, non-glycosylated, and non GPI-anchored PrPres fragments and infectivity was still missing. Indeed, even if 7 kDa PrP peptides had been reported as components of the amyloid plaques in the brains of GSS-A117V patients (Tagliavini et al., 2001), their role in the pathogenesis of the disease and in the propagation of infectious conformation remained elusive.
We discovered that GSS-A117V brain tissues are characterized by an extremely high infectious titre when inoculated in voles. Moreover, the PK-treated inoculum, which contained the 7 kDa PK-resistant PrP fragment, was characterized by the same level of infectivity as the untreated inoculum, which was composed of full-length PrPSc and PK-sensitive PrP isoforms. These findings allow us to conclude that, as for prion diseases associated with classical PrPSc, the infectivity of GSS-A117V is resistant to PK and is not associated with PK-sensitive forms of PrP. While this finding clearly suggested a correlation between 7 kDa PrPres aggregates and infectivity, a definitive proof of the infectious nature of the 7 kDa PrPres aggregates could only be achieved via purification of these atypical PrPres aggregates. Indeed, we found that purified, patient-derived 7 kDa PrPres aggregates are infectious in voles and faithfully transmit the strain features of the initial GSS-A117V brain inoculum. The specific infectious role of 7 kDa PrPres aggregates was demonstrated by the co-segregation of infectivity and PrPres in the purified P4 fraction and was further corroborated by the parallel decrease of infectivity and PrPres during purification, which followed the same rank order BH > SN2 > P2 > P4. Thus, PrPres aggregates that contain PrP-derived peptides encompassing residues ∼90–150 are sufficient as GSS prions and encode their strain properties. As similar ∼90–150 PrP peptides are formed in vivo and can be extracted from amyloid plaques of GSS-A117V patient brains (Tagliavini et al., 2001), it is conceivable that aggregates of misfolded ∼90–150 PrP fragments play a fundamental role in the autocatalytic propagation of the disease and could represent an important therapeutic target to slow the progression of the disease.
Besides the above-mentioned implications for better understanding the pathogenesis of GSS-A117V disease, our findings also provide new insights into the structural determinants of prion infectivity. We have demonstrated that human-derived small PrP peptides, devoid of Asn-linked glycans and GPI-anchor moieties, are able to interact with host PrPC and to catalyse its misfolding into a pathogenic and autocatalytic replicating conformation. These findings demonstrate that the misfolded conformation of ∼90–150 PrP peptides contains the key elements sufficient to encode for GSS-A117V prions and imply that the C-terminus of PrP, along with its Asn-linked glycans and the GPI-anchor, are dispensable for the structure of a prion. The latter conclusion is supported by several studies showing prion infectivity associated with in vitro misfolded recombinant PrP, which lacks all post-translational modifications (Legname et al., 2004; Wang et al., 2010), and with misfolded N- and/or C-terminally truncated recombinant PrP (Kaneko et al., 2000; Choi et al., 2016).
Although 7 kDa PrPres aggregates do not contain covalently linked glycans or a GPI anchor, our results do not exclude that other brain-derived molecules might be integral components of purified 7 kDa PrPres. The need for co-factors as an integral component of highly infectious prions is a matter of intense investigation (Wang et al., 2010; Deleault et al., 2012; Supattapone, 2014). Recent evidence suggests that pure preparations of recombinant PrPSc without co-factors may be infectious for wild-type mice (Choi et al., 2016) and bank voles (Fernández-Borges et al., 2018). It is noteworthy that, in both these studies, recombinant prions induced atypical, C-terminally truncated PrPres in the recipient rodents, similar to what was observed in the present study using Bv109I inoculated with purified GSS-A117V 7 kDa PrPres.
Usually, a PK-resistant core spanning residues ∼90–231 characterizes infectious PrPSc. Studies with recombinant PrP have shown that PrP conformers with a shorter, C-terminal PK-resistant core encompassing residues ∼160–231 are not infectious (Makarava et al., 2011; Zhang et al., 2014; Groveman et al., 2015). We have recently found that a brain-derived PrPSc conformer with an unusual C-terminal PrPres, which does not include the central PrP domain ∼90–150, behaves as a ‘defective prion’, competent for autocatalytic self-propagation in vitro but unable to propagate in vivo (Vanni et al., 2016), and PrPSc aggregates lacking the central polybasic domain showed comparable behaviour (Miller et al., 2011). Moreover, this central PrP domain contains the polybasic lysine cluster that has been indicated as a key structural determinant for PrPC-to-PrPSc conversion. Overall, these studies strongly suggest that the central ∼90–150 PrP domain might be an obligate component of the PK-resistant core of infectious prions. Conversely, an obvious consequence of the present study is that the C-terminal region of PrPSc, i.e. from residue ∼150 up to the C-terminus, is not necessary for infectivity. Taken together, these findings support the view that the PrP region spanning residues ∼90–150 is the minimal PrPSc component, which allows a level of structural complexity that is able to support self-replication of prions.
X-ray fibre diffraction of PrPSc and PrPres fibrils as well as cryo-electron microscopy analyses of GPI-anchorless PrPres fibrils independently indicated a four-rung β-solenoid structure as the key feature of the infectious conformer (Wille et al., 2009; Vázquez-Fernández et al., 2016). These findings suggested a more general role for the four-rung β-solenoid structure in the autocatalytic propagation mechanism that characterizes prions (Wille and Requena, 2018). However, the residue ∼90–150 minimal PrPSc component responsible for the infectivity of GSS-A117V is too small to account for a four-rung β-solenoid structure, thus suggesting that an alternative structural arrangement must account for the observed self-replication of the 7 kDa PrPres aggregates, which could entail a two-rung β-solenoid configuration. Indeed, it has been shown by X-ray fibre diffraction that the synthetic peptide PrP(89–143, P101L) supports both a two-rung solenoidal architecture and a stacked-sheet structure, depending on the conditions of amyloid formation (Wan et al., 2015). Finally, solid-state nuclear magnetic resonance studies of infectious PrP23-144 fibrils showed that its short β-sheet core (residues ∼112–139) was characterized by a parallel in-register organization of β-strands (Helmus et al., 2011).
The present study provides the biochemically simplest, highly infectious PrPSc prions ever purified from diseased brains, and our findings provide new constraints for studies aimed at investigating the structure of prions. The ability to unequivocally correlate infectivity with a known 3D structure is the necessary basis for understanding the molecular underpinnings of prion infectivity. Recent studies have shown that prions purified from multiple scrapie strains contain PrPSc that is assembled into rod-like assemblies, referred to as prion rods, with a common hierarchical assembly consisting of twisted pairs of short fibres with a repeating substructure (Terry et al., 2016; Vázquez-Fernández et al., 2016). In this scenario, one unexpected finding of our study was that GSS-A117V infectivity, although purified using the same purification protocol that allowed the direct association of infectivity with prion rods and fibrils (Wenborn et al., 2015; Terry et al., 2016), was instead associated with small oligomeric assemblies, and not with the commonly observed, larger fibrillar structures or prion rods.
Although unexpected, the non-fibrillar nature of GSS-A117V prion particles is in agreement with the relatively low amount of PrP monomers in our preparations compared to their high infectious titre, which correlates with the presence of numerous small oligomers versus only few prion rods. Moreover, this finding is also in agreement with the observed lower sedimentation rate of 7 kDa PrPres compared to the fibrillar PrPSc aggregates, which characterize classical scrapie prions. Lastly, these observations imply that a fibrillar quaternary structure is not an absolute requirement for prion infectivity, as had been seen with the complete lack of fibrillar assemblies in RML-infected FVB mice (Godsave et al., 2008, 2013; Levine et al., 2015).
Our findings are in line with a proposed key role of PrPSc oligomers in the propagation of prion disorders as it has been shown that prion infectivity distributes widely when fractionated in density gradients and that smaller particles appear to account for a substantial proportion of the total infectivity in brain homogenates (Silveira et al., 2005; Tixador et al., 2010; Kim et al., 2012; Laferrière et al., 2013). The fractionation of purified prions by sonication and detergents led to the identification of non-fibrillar oligomers of PrP, with masses equivalent to 14–28 PrP molecules, as the most infectious prion particles (Silveira et al., 2005). Both small and large PrPSc aggregates could contribute to disease pathogenesis, accounting in variable proportions for propagation and resistance in the host environment. Long-range interactions in large fibrillar infectious aggregates might confer high resistance in the host environment, while smaller oligomers are considered metastable intermediates to fibril formation or an off-pathway product of aggregation. Our findings challenge this view, in that we found only small amorphous PrPSc particles in GSS-A117V, but not larger fibrils, suggesting that these small particles are stable and could represent the best compromise between stability and infectivity of PrPSc in GSS-A117V. It could be hypothesized that the A117V pathogenic mutation in PrP confers stability to oligomers, while oligomers of wild-type human PrP would be only transient and progress towards a rapid structural transition to fibrils. Consistent with this hypothesis, molecular simulation studies with PrP model peptides showed that the A117V substitution enhanced the population of β-hairpin peptides that form oligomers and slowed the formation of fibrils once nucleation had occurred (Gill, 2014). An alternative hypothesis is that the absence of the C-terminus in 7 kDa PrPres aggregates plays a role in stabilizing PrPSc oligomers or in preventing their structural reorganization into fibrils.
Although purified amyloid proteins usually consist of large aggregates with a fibrillar quaternary structure, non-fibrillar oligomers have been observed for a number of amyloid proteins associated with neurodegeneration, such as amyloid-β, tau and α-synuclein. Accumulating evidence in the past two decades suggests that these oligomers are likely the most toxic species in Alzheimer’s, Parkinson’s and other neurodegenerative diseases (Haass et al., 2012; Kayed and Lasagna-Reeves, 2013; Forloni et al., 2016; Shafiei et al., 2017). The structural relationship between small-sized infectious particles in brain homogenate with that of the larger purified fibrils is still debated. A major obstacle to understanding this relationship has been the transient nature of oligomers, which prevented to date high resolution structural characterization of in vivo derived oligomers. Our findings that PrPSc oligomers can be purified from GSS-A117V provide a new tool to understand the molecular organization of PrPSc, also in light of the simplified biochemical composition of 7 kDa PrPres. The high resolution structure of the GSS-A117V PrPSc oligomers would thus provide new fundamental knowledge on the catalytic domain that underpins PrPSc infectivity, which could conceivably apply to larger and more common fibrillar structures.
Supplementary Material
Acknowledgements
We thank the National Prion Disease Pathology Surveillance Center, Cleveland, for providing us the GSS-A117V cases and Dr Maurizio Pocchiari (National CJD Registry) for the sCJD-MM1 case. Furthermore, we thank Geraldine Horny, Sylvie Fels, Nathalie George, Stefan Ewert and Thomas Pietzonka from Novartis (Basel, Switzerland) for providing the anti-mPrP Fab library that included Fab 69.
Funding
This work was supported by grants from the Ministero della Salute (RF-2009-1474624) to R.N. and the Alberta Prion Research Institute award (201600029) to H.W. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
The authors report no competing interests.
Glossary
- CJD =
Creutzfeldt-Jakob disease; GPI = glycophosphatidylinositol
- GSS =
Gerstmann-Sträussler-Scheinker disease
- PE =
Pronase E
- PK =
proteinase K
- PrPC =
cellular prion protein
- PrPSc =
misfolded pathological isoform of the host-encoded PrPC
- TSESc =
transmissible spongiform encephalopathy
References
- Asante EA, Linehan JM, Smidak M, Tomlinson A, Grimshaw A, Jeelani A, et al. Inherited prion disease A117V is not simply a proteinopathy but produces prions transmissible to transgenic mice expressing homologous prion protein. PLoS Pathog 2013; 9: e1003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Cali I, Surewicz Kong Q, Gambetti P, Surewicz WK.. Amyloid fibrils from the N-terminal prion protein fragment are infectious. Proc Natl Acad Sci USA 2016; 113: 13851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cracco L, Xiao X, Nemani SK, Lavrich J, Cali I, Ghetti B, et al. Gerstmann-Sträussler-Scheinker disease revisited: accumulation of covalently-linked multimers of internal prion protein fragments. Acta Neuropathol Commun 2019; 7: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronier S, Gros N, Tattum MH, Jackson GS, Clarke AR, Collinge J, et al. Detection and characterization of proteinase K-sensitive disease-related prion protein with thermolysin. Biochem J 2008; 416: 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, et al. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci USA 2012; 109: E1938–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bari MA, Nonno R, Castilla J, D’Agostino C, Pirisinu L, Riccardi G, et al. Chronic wasting disease in bank voles: characterisation of the shortest incubation time model for prion diseases. PLoS Pathog 2013; 9: e1003219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Borges N, Di Bari MA, Eraña H, Sánchez-Martín M, Pirisinu L, Parra B, et al. Cofactors influence the biological properties of infectious recombinant prions. Acta Neuropathol 2018; 135: 179–99. [DOI] [PubMed] [Google Scholar]
- Forloni G, Artuso V, La Vitola P, Balducci C.. Oligomeropathies and pathogenesis of Alzheimer and Parkinson’s diseases. Mov Disord 2016; 31: 771–81. [DOI] [PubMed] [Google Scholar]
- Fraser H, Dickinson AG.. The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol 1968; 78: 301–11. [DOI] [PubMed] [Google Scholar]
- Gambetti P, Puoti G, Zou WQ.. Variably protease-sensitive prionopathy: a novel disease of the prion protein. J Mol Neurosci 2011; 45: 422–4. [DOI] [PubMed] [Google Scholar]
- Gill AC. β-hairpin-mediated formation of structurally distinct multimers of neurotoxic prion peptides. PLoS One 2014; 9: e87354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsave SF, Wille H, Kujala P, Latawiec D, DeArmond SJ, Serban A, et al. Cryo-immunogold electron microscopy for prions: toward identification of a conversion site. J Neurosci 2008; 28: 12489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsave SF, Wille H, Pierson J, Prusiner SB, Peters PJ.. Plasma membrane invaginations containing clusters of full-length PrPSc are an early form of prion-associated neuropathology in vivo. Neurobiol Aging 2013; 34: 1621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groveman BR, Kraus A, Raymond LD, Dolan MA, Anson J, Dorward DW, et al. Charge neutralization of the central lysine cluster in prion protein (PrP) promotes PrP(Sc)-like folding of recombinant PrP amyloids. J Biol Chem 2015; 290: 1119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ.. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol 2012; 8: 101–12. [DOI] [PubMed] [Google Scholar]
- Helmus JJ, Surewicz K, Apostol MI, Surewicz WK, Jaroniec CP.. Intermolecular alignment in Y145Stop human prion protein amyloid fibrils probed by solid-state NMR spectroscopy. J Am Chem Soc 2011; 133: 13934–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K, Ball HL, Wille H, Zhang H, Groth D, Torchia M, et al. A synthetic peptide initiates Gerstmann-Sträussler-Scheinker (GSS) disease in transgenic mice. J Mol Biol 2000; 295: 997–1007. [DOI] [PubMed] [Google Scholar]
- Kayed R, Lasagna-Reeves CA.. Molecular mechanisms of amyloid oligomers toxicity. J Alzheimers Dis 2013; 33: S67–S78. [DOI] [PubMed] [Google Scholar]
- Kim C, Haldiman T, Surewicz K, Cohen Y, Chen W, Blevins J, et al. Small protease sensitive oligomers of PrPSc in distinct human prions determine conversion rate of PrP(C). PLoS Pathog 2012; 8: e1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroux C, Vilette D, Fernández-Borges N, Litaise C, Lugan S, Morel N, et al. Prionemia and leukocyte-platelet-associated infectivity in sheep transmissible spongiform encephalopathy models. J Virol 2012; 86: 2056–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferrière F, Tixador P, Moudjou M, Chapuis J, Sibille P, Herzog L, et al. Quaternary structure of pathological prion protein as a determining factor of strain-specific prion replication dynamics. PLoS Pathog 2013; 9: e1003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, et al. Synthetic mammalian prions. Science 2004; 305: 673–6. [DOI] [PubMed] [Google Scholar]
- Levine DJ, Stöhr J, Falese LE, Ollesch J, Wille H, Prusiner SB, et al. Mechanism of scrapie prion precipitation with phosphotungstate anions. ACS Chem Biol 2015; 10: 1269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Kovacs GC, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, et al. Genesis of mammalian prions: from non-infectious amyloid fibrils to a transmissible prion disease. PLoS Pathog 2011; 7: e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Geoghegan JC, Supattapone S.. Dissociation of infectivity from seeding ability in prions with alternate docking mechanism. PLoS Pathog 2011; 7: e1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa K, Okada H, Masujin K, Iwamaru Y, Yokoyama T.. Infectivity-associated PrP(Sc) and disease duration-associated PrP(Sc) of mouse BSE prions. Prion 2015; 9: 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonno R, Di Bari MA, Cardone F, Vaccari G, Fazzi P, Dell’Omo G, et al. Efficient transmission and characterization of Creutzfeldt-Jakob disease strains in bank voles. PLoS Pathog 2006; 2: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchi P, Castellani R, Capellari S, Ghetti B, Young K, Chen SG, et al. Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann Neurol 1996; 39: 767–78. [DOI] [PubMed] [Google Scholar]
- Piccardo P, Dlouhy SR, Lievens PM, Young K, Bird TD, Nochlin D, et al. Phenotypic variability of Gerstmann-Straussler-Scheinker disease is associated with prion protein heterogeneity. J Neuropathol Exp Neurol 1998; 57: 979–88. [DOI] [PubMed] [Google Scholar]
- Piccardo P, Liepnieks JJ, William A, Dlouhy SR, Farlow MR, Young K, et al. Prion proteins with different conformations accumulate in Gerstmann-Straussler-Scheinker disease caused by A117V and F198S mutations. Am J Pathol 2001; 158: 2201–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirisinu L, Di Bari MA, D’Agostino C, Marcon S, Riccardi G, Poleggi A, et al. Gerstmann-Sträussler-Scheinker disease subtypes efficiently transmit in bank voles as genuine prion diseases. Sci Rep 2016; 6: 20443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirisinu L, Nonno R, Esposito E, Benestad SL, Gambetti P, Agrimi U, et al. Small ruminant nor98 prions share biochemical features with human Gerstmann-Sträussler-Scheinker disease and variably protease-sensitive prionopathy. PLoS One 2013; 8: e66405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirisinu L, Tran L, Chiappini B, Vanni I, Di Bari MA, Vaccari G, et al. Novel type of chronic wasting disease detected in moose (Alces alces). Emerg Infect Dis 2018; 24: 2210–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci USA 1998; 95: 13363–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Saverioni D, Di Bari M, Baiardi S, Lemstra AW, Pirisinu L, et al. Atypical Creutzfeldt-Jakob disease with PrP-amyloid plaques in white matter: molecular characterization and transmission to bank voles show the M1 strain signature. Acta Neuropathol Commun 2017; 5: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore A, Frontzek K, Emmenegger M, Chincisan A, Losa M, Reimann R, et al. Protective anti-prion antibodies in human immunoglobulin repertoires. BioRxiv2020; doi: 10.1101/2020.02.05.933721. [DOI] [PMC free article] [PubMed]
- Shafiei SS, Guerrero-Muñoz MJ, Castillo-Carranza DL.. Tau oligomers: cytotoxicity, propagation, and mitochondrial damage. Front Aging Neurosci 2017; 9: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, et al. The most infectious prion protein particles. Nature 2005; 437: 257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supattapone S. Synthesis of high titer infectious prions with cofactor molecules. J Biol Chem 2014; 289: 19850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliavini F, Lievens PM, Tranchant C, Warter JM, Mohr M, Giaccone G, et al. A 7-kDa prion protein (PrP) fragment, an integral component of the PrP region required for infectivity, is the major amyloid protein in Gerstmann-Sträussler-Scheinker disease A117V. J Biol Chem 2001; 276: 6009–15. [DOI] [PubMed] [Google Scholar]
- Terry C, Wenborn A, Gros N, Sells J, Joiner S, Hosszu LL, et al. Ex vivo mammalian prions are formed of paired double helical prion protein fibrils. Open Biol 2016; 6: 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixador P, Herzog L, Reine F, Jaumain E, Chapuis J, Le Dur A, et al. The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog 2010; 6: e1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni I, Migliore S, Cosseddu GM, Di Bari MA, Pirisinu L, D’Agostino C, et al. Isolation of a defective prion mutant from natural scrapie. PLoS Pathog 2016; 12: e1006016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Fernández E, Vos MR, Afanasyev P, Cebey L, Sevillano AM, Vidal E, et al. The structural architecture of an infectious mammalian prion using electron cryomicroscopy. PLoS Pathog 2016; 12: e1005835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W, Wille H, Stöhr J, Kendall A, Bian W, McDonald M, et al. Structural studies of truncated forms of the prion protein PrP. Biophys J 2015; 108: 1548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang X, Abskharon R, Ma J.. Prion infectivity is encoded exclusively within the structure of proteinase K-resistant fragments of synthetically generated recombinant PrPSc. Acta Neuropathol Commun 2018; 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang X, Yuan CG, Ma J.. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327: 1132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenborn A, Terry C, Gros N, Joiner S, D’Castro L, Panico S, et al. A novel and rapid method for obtaining high titre intact prion strains from mammalian brain. Sci Rep 2015; 5: 10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Bian W, McDonald M, Kendall A, Colby W, Bloch L, et al. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci USA 2009; 106: 16990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Govaerts C, Borovinskiy AL, Latawiec D, Downing KH, Cohen FE, et al. Electron crystallography of the scrapie prion protein complexed with heavy metals. Arch Biochem Biophys 2007; 467: 239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Requena JR.. The structure of PrPSc prions. Pathogens 2018; 7: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang F, Wang X, Zhang Z, Xu Y, Yu G, et al. Comparison of 2 synthetically generated recombinant prions. Prion 2014; 8: 215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.