Abstract
Auchenorrhynchan insects (Hemiptera) generally depend on two bacterial symbionts for nutrition. These bacteria experience extreme genome reduction and loss of essential cell functions that require direct host support, or the replacement of failing symbionts with more capable ones. However, it remains unclear how hosts adapt to integrate symbionts into their systems, particularly when they are replaced. Here, we comparatively investigated the evolution of host-support mechanisms in the glassy-winged sharpshooter, Homalodisca vitripennis (GWSS), and the aster leafhopper, Macrosteles quadrilineatus (ALF). ALF harbors the ancestral co-symbionts of the Auchenorrhyncha that have tiny genomes, Sulcia (190 kb) and Nasuia (112 kb). In GWSS, Sulcia retains an expanded genome (245 kb), but Nasuia was replaced by the more capable Baumannia (686 kb). To support their symbionts, GWSS and ALF have evolved novel mechanisms via horizontal gene transfer, gene duplication, and co-option of mitochondrial support genes. However, GWSS has fewer support systems targeting essential bacterial processes. In particular, although both hosts use ancestral mechanisms to support Sulcia, GWSS does not encode all of the same support genes required to sustain Sulcia-ALF or Nasuia. Moreover, GWSS support of Baumannia is far more limited and tailored to its expanded capabilities. Our results demonstrate how symbiont replacements shape host genomes and the co-evolutionary process.
Subject terms: Symbiosis, Molecular evolution
Introduction
Many insects engage in obligate symbioses with bacteria that provide hosts with essential nutrition [1, 2]. Prominent instances occur in plant sap-feeding insects (Hemiptera), which generally rely on bacterial symbionts for essential amino acids (EAAs) and vitamins that are deficient in their phloem or xylem diets [3, 4]. These symbionts are confined to specialized organs (bacteriomes) and cells (bacteriocytes), and they are transmitted vertically within the matriline [3, 5, 6]. As a result, bacteria exhibit extensive and ongoing gene loss due to strong genetic drift, yielding some of the smallest known bacterial genomes (~100–1,000 kilobases [kb]) [7, 8]. Symbionts require genetic resources from their hosts in order to function as cellular entities and to provide essential nutrition. Recent studies have shown that hosts express 100–1,000s of genes in bacteriocytes that can complement the missing or incomplete cellular machinery and metabolisms of their symbionts [9–13]. A large number of these genes are derived from horizontal gene transfer, reassignment of mitochondrial (MT) support genes, and gene duplication [11, 13, 14]. Since symbionts continue to lose genes, and are even occasionally replaced, it remains unclear how hosts dynamically adapt these mechanisms to maintain degrading symbionts, or to establish new ones.
The genomic capabilities of a particular symbiont lineage can vary widely between different host species [15–18]. Thus, hosts must continually adapt their support mechanisms to maintain the distinct symbiont genotypes. However, in some cases, highly degenerate symbionts can also be completely replaced by more capable and reliable ones [19, 20]. Newly acquired bacteria often take up residence in novel bacteriomes and bacteriocytes specifically evolved to maintain them [19, 21]. The evolutionary origins of bacteriome tissues in the leafhoppers have not been investigated; however, they are diagnosable as discrete tissues differing by location in the body, pigmentation, cellular morphology, and number of nuclei [19, 22]. Recent work in pea aphids, which harbor the intracellular symbiont, Buchnera aphidicola, provides some insight into the mechanisms that may underly bacteriome evolution and development more generally. For example, aside from symbiont-tailored gene expression patterns [9, 23], bacteriocyte development is highly conserved across related species and an intrinsic part of embryogenesis [24]. Thus, when symbionts are lost or replaced, the intricately orchestrated support mechanisms that maintained replaced partners are likely to be lost, particularly since they are adapted to certain bacteria and delivered in a tissue-specific manner [10, 11, 13]. In order to integrate new symbionts into the symbiosis, hosts must either adapt existing mechanisms or evolve entirely new ones; they must also reprogram bacteriome tissues to deliver them. However, it is poorly understood how hosts evolve on a genomic level to reestablish complex symbioses with multiple partners after a replacement has occurred.
Symbioses in the Auchenorrhyncha (Hemiptera: suborder) are ancient and complex, reaching back over 300 million years (MY) [25–27]. Hosts typically harbor two companion symbionts that jointly supply the ten EAAs: “Candidatus Sulcia muelleri” (Bacteroidetes; hereafter Sulcia) and “Ca. Nasuia deltocephalinicola” (Betaproteobacteria; hereafter Nasuia). Both symbionts established in the common ancestor of the Auchenorrhyncha and have tiny genomes (~100 kb–250 kb) that are missing essential genes in every cellular process [26, 28, 29]. They are further segregated into distinct bacteriocytes that provide support of their limited cellular capabilities (Fig. 1; [13, 30]). It was recently shown in the aster leafhopper, Macrosteles quadrilineatus (ALF), that support mechanisms are complex, deriving from various combinations of gene duplications, horizontal gene acquisitions, and retargeting of MT support genes [13]. Despite this tightly integrated genomic coevolution between hosts and their symbionts, Nasuia has been occasionally replaced as occurred in the sharpshooter leafhoppers (Cicadellinae) (Fig. 1; [31, 32]). Sharpshooter species generally harbor “Ca. Baumannia cicadellinicola” (Gammaproteobacteria; hereafter Baumannia) that has converged to synthesize only the same two EAAs as Nasuia (methionine and histidine). However, its genome is much larger (686–759 kb), encoding many more independent functional capabilities [33–35]. Although some Baumannia cells co-occur with Sulcia, the host has also evolved a novel bacteriome organ that exclusively houses it [3, 19, 31, 36]. Baumannia in these tissues appears to have discrete nutritional responsibilities [37]. Thus, the new bacteriome is likely an adaptation to maintain and regulate Baumannia’s contributions to the symbiosis.
Fig. 1. Evolution of symbiotic associations in the Cicadellidae.
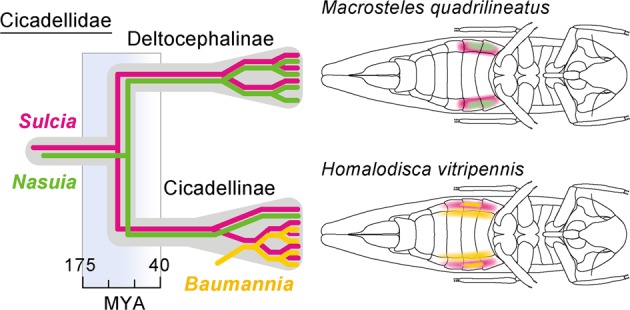
The estimated date of symbiont replacement that occurred in sharpshooter leafhoppers (Cicadellinae) is based on previous studies [25, 32]. Schematic illustration of bacteriome organs is adapted from previous fluorescence in situ hybridization studies [37, 39] (color figure online).
To address the questions of how hosts adapt to (i.) maintain symbiont species with varying capabilities, and (ii.) reestablish multipartner symbioses when members are replaced, we investigated host genome evolution in the glassy-winged sharpshooter, Homalodisca vitripennis (GWSS). The genome of Sulcia-GWSS (245 kb) has expanded gene sets involved in metabolic pathways and cellular functions relative to those of Sulcia-ALF (~190 kb) [38, 39]. Baumannia (~686 kb) has one of the largest known symbiont genomes from the Auchenorrhyncha [34]. It retains many additional metabolic capabilities that are lost from Nasuia (~112 kb). The GWSS host has two types of bacteriome tissues: a yellow bacteriome that harbors Sulcia-GWSS and Baumannia, and a red bacteriome that solely contains Baumannia [25, 31, 37]. The yellow bacteriome appears to be the only organ capable of vertically transmitting both symbionts, which may partially explain Baumannia’s occurrence there [3, 37]. Using comparative gene expression assays and host genomics, our results reveal that GWSS differentially expresses a wide range of genes between bacteriome types that deliver specific and distinct support of Sulcia-GWSS and Baumannia. To maintain a symbiosis with Baumannia, GWSS uses discretely evolved mechanisms to support its expanded and unique capabilities. Remarkably, many support mechanisms used to maintain the more degraded Sulcia lineage and Nasuia in ALF, are not present in the GWSS genome.
Materials and methods
Sample preparation and RNA sequencing
Insect collection, dissection, and RNA extraction were performed as described previously [37]. Strand-specific libraries were prepared with RNA ligation protocol (first biological replicate of body tissues) or deoxy-UTP protocol (all other libraries). Libraries were prepared and sequenced on an Illumina HiSeq 2500 at the UT Austin GSAF.
Transcriptome assembly, annotation, and differential expression analyses
Reads were quality-filtered and adapter-trimmed with Trimmomatic v0.32 (settings: illuminaclip:2:20:10:1:true leading:20 trailing:20 slidingwindow:4:15 minlen:50) [40]. Reads mapped to the Sulcia-GWSS and Baumannia genomes were removed in silico. Because one tissue replicate was prepared with an alternative strand-specific approach (see above), reads were reordered so that a combined de novo assembly could be done with the -SS_lib_type RF setting in Trinity v2.1.1 (all other settings were set to default) [41]. CD-HIT was used to reduce assembly redundancy by clustering at ≥95% identity [42]. Open reading frames were predicted for each transcript using EMBOSS’s getorf [43] and searched against the GenBank nr database with NCBI-BLASTP v2.2.30+ (e value 1e−6). Gene functions and metabolic pathways were determined with EcoCyc [44] and previous studies [34, 38]. Differentially expressed genes were determined by read mapping with Bowtie2 v2.2.7 and transcript abundance was estimated in RSEM v1.2.29 with Trinity’s align_and_estimate_abundance.pl script (TMM [trimmed mean of M values] normalized FPKM [fragments per kilobase per million]) [45, 46]. DE analysis (P ≤ 0.001 and fold change [FC] ≥2×) for all pair-wise tissue comparisons was conducted using edgeR v3.12.0 with the run_DE_analysis.pl script [47]. Since each tissue was deeply sequenced in triplicate replication, a qRT-PCR verification was not performed. Previous studies have demonstrated that the DE patterns are highly consistent between transcriptome sequencing and qRT-PCR assays [11–13].
Identification of symbiont support genes
Horizontally transferred genes (HTGs), MT support genes, and gene duplications in bacteriomes were identified as described previously [13]. However, the GWSS genome was obtained from GenBank (i5k project; Accession No. JJNS00000000). To detect if the GWSS genome retains gene residues of unique HTGs identified in the ALF genome, but that are not present in the GWSS transcriptome, we searched those HTGs against the GWSS genome using NCBI-BlastN v2.2.30+. We further identified the adjacent genes of each shared HTG between GWSS and ALF using NCBI-BlastX. Phylogenetic trees for HTGs and two MT support genes (elongation factor Ts, mt TSFM, and translation factor, mt GUF) were inferred using the maximum-likelihood (ML) and Bayesian approaches. Briefly, amino acid sequences of each gene were aligned with MAFFT using the L-INS-i model [48]. Models of amino acid substitutions for each gene were determined in Prottest3 v3.4.2 with the Akaike information criterion [49]. ML trees were reconstructed with RAxML-HPC2 via the CIPRES Science Gateway portal [50, 51]. A total of 1000 bootstrap replicates were run with an appropriate substitution model (see Fig. S1). Phylobayes v4.1 (settings: cat, gtr, dgam = 4) was used to reconstruct phylogenies in a Bayesian framework, which is more robust to issues of long-branch attraction [52]. Three chains for each gene were run 50,000 generation and convergence checked using bpcomp (maxdiff < 0.1) with 10% burn-in. We note that tree topologies should be interpreted cautiously as they are single gene phylogenies with taxa prone to long-branch attraction [52].
Proteomic verification of an HTG
Protein verification experiments were performed on the frr HTG in ALF since (i) it appears to be a conserved adaptation by leafhopper hosts to support Sulcia, and (ii) ALF is easily lab-reared in high numbers in contrast to GWSS. Full experimental procedures can be found in the Supplementary Materials and Methods. Briefly, immunoblot assays were performed on protein extractions from bacteriome and body tissues. A PVDF membrane was blocked and stained with rabbit anti-frr antibody and donkey anti-rabbit IgG. The reaction was visualized with a Supersignal West Dura extended duration substrate kit. Immunofluorescence microscopy experiments were further performed on dissected insect abdomens. Samples were dehydrated in EtOH, cleared in xylene, and embedded in paraffin. Sections were then slide deparaffinized, permeabilized, and treated with heat-induced antigen retrieval. Sections were incubated with frr antibody followed with Alexa Fluor 555 goat anti-rabbit IgG (H + L). Negative controls without either primary antibody or secondary antibody, or with excess amount of the frr antigen were conducted. Sections were visualized with a Zeiss LSM 880 laser confocal microscope.
Results and discussion
GWSS provides distinct and tailored support of its symbionts
In order to identify the genetic mechanisms that GWSS uses to support Sulcia-GWSS and Baumannia, we conducted a tissue-type specific differential RNA-seq experiment and host genomic analyses. To understand the evolutionary origins of symbiont support, we comparatively analyzed host-encoded symbiosis-associated genes with recent data from the ALF-Sulcia-Nasuia symbiosis [13]. RNA sequencing from three replicate dissections of the yellow and red bacteriomes and thoracic body tissues yielded 5.0–24.3 million quality-filtered, paired-end reads per library (Table S1). De novo assembly of the combined nine libraries generated 304,021 transcripts that represent 192,863 genes. CD-HIT reduced redundant contigs (similarity ≥95%) to 209,770 transcripts and 172,629 genes. Differential gene expression (DE) analysis showed that 3,595 and 7,598 genes are upregulated in the red and yellow bacteriomes relative to the body tissues, respectively (FC ≥ 2 and P ≤ 0.001; Fig. 2). Despite differences in sequencing depth, gene expression patterns are highly similar between tissue replicates (Fig. S2).
Fig. 2. Differential gene expression between symbiont-bearing bacteriomes and body tissues in GWSS.
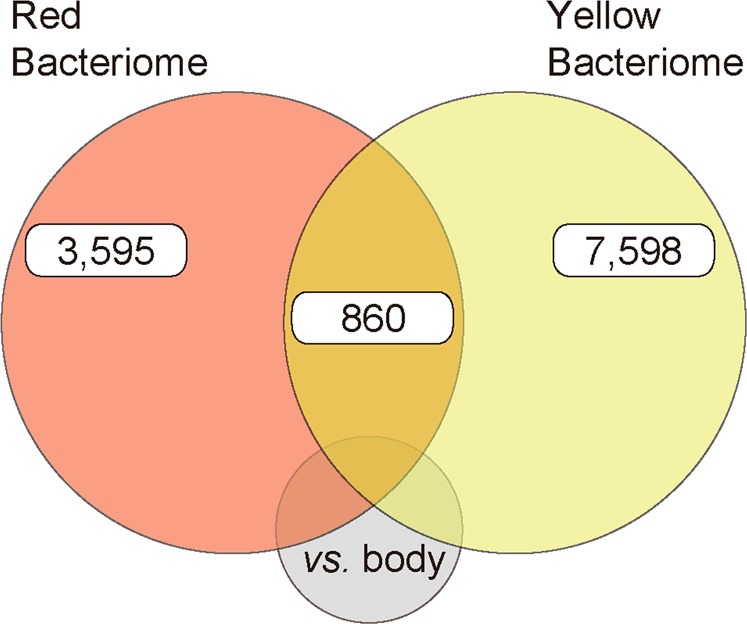
Highly expressed gene counts (edgeR; p ≤ 0.001 and fold change [FC] ≥ 2) in the yellow (harboring Sulcia and Baumannia) and red (harboring Baumannia) bacteriomes relative to insect body tissues (color figure online).
The overall DE patterns reflect the distinct cellular capabilities and requirements of Sulcia-GWSS and Baumannia. Similar to ALF and several other sap-feeding insect systems (e.g., psyllids, mealybugs, and whiteflies [10–13]), the GWSS lineage has evolved a range of novel traits to meet the specific needs of its symbionts via HTGs from other infecting bacteria, reassignment of MT support genes, and gene duplications with bacteriome-specific expression. GWSS support mechanisms that prop up incomplete cellular capabilities and nutritional pathways have been cobbled together with all three, sometimes in combination. While many support mechanisms appear to be conserved between ALF and GWSS leafhoppers—given the replacement of an ancient symbiont and the expanded genomic capabilities of Sulcia-GWSS and Baumannia—the evolution and deployment of these mechanisms in GWSS differ substantially from those in ALF.
The initial origins of bacteriocytes, and their supporting gene expression programs, remain uncertain for GWSS and ALF and also for other insect-bacterial symbioses. It has been proposed that symbionts may establish in tissues that already exhibit certain gene expression patterns that are suitable for symbiont residency (e.g., enriched nutrition; [53]). Nevertheless, the piecemeal acquisition of HTGs and gene duplications over evolutionary time, suggests that degrading symbiont genomes indeed drive the acquisition of novel genes and subsequent expression patterns in the bacteriocytes (discussed in detail below).
HTGs provide dynamic and tailored support of essential gene losses in specific symbionts
In GWSS, we identified fourteen genes horizontally transferred from infecting bacteria to the host genome. Seven HTGs appear to support the symbiosis and are more highly expressed in the yellow bacteriome. Strikingly, no HTGs are highly expressed in the red bacteriome (Table 1). This expression pattern suggests that Baumannia does not rely on HTGs to function. In contrast, the ALF genome encodes 27 HTGs that provided support of a range of cellular functions in Sulcia-ALF and Nasuia [13]. All identified HTGs in GWSS were further verified to be encoded in the host genome [54]. Six of them have the typical eukaryotic GT/AG intron boundary, as has been observed in other insect hosts. In order to determine whether HTGs are conserved in their genomic positions between ALF and GWSS, we further investigated local gene order. While two genes appear to maintain the same positions (cel and rnc; see Fig. S3), the analysis is inconclusive for others due to the fragmentary nature of both the ALF and GWSS genomes, relatively large noncoding tracts between genes, and abundance of poorly characterized hypothetical insect proteins. Nevertheless, phylogenetic analyses show that, in general, HTGs in GWSS group with their orthologs from ALF (Fig. 3a and Fig. S1), suggesting that they may have been acquired before the Deltocephalinae–Cicadellinae divergence.
Table 1.
Horizontally transferred genes in the Homalodisca vitripennis (GWSS) genome.
| Trinity ID | Gene | Product | Function | Predicted origin | FPKM (log2 FC) | ||
|---|---|---|---|---|---|---|---|
| Body | Red | Yellow | |||||
| DN151427_c3_g2 | rnc | Ribonuclease III | RNA processing | Wolbachiaa | 0 | 0.2 (-) | 371.2 (14.3) |
| DN152407_c0_g1 | ileSb | Isoleucine-tRNA ligase | Translation | Wolbachia | 0 | 0.1 (-) | 206 (13.9) |
| DN93430_c0_g1 | frr | Ribosome recycling factor | Translation | Alphaproteobacteria | 0 | 0.3 (-) | 152.9 (12.4) |
| DN157716_c1_g1 | ATPaseb | AAA-ATPase | ATPase activity | Firmicutesa | 1.5 | 1.4 (-) | 131.1 (6.6) |
| DN152668_c0_g5 | defb | Peptide deformylase | Translation factor | Rickettsia | 2.4 | 5.2 (-) | 226.6 (7.4) |
| DN145592_c1_g1 | yebC | Transcriptional regulator | Transcription | Midichloria | 0 | 0.3 (-) | 442.5 (14.5) |
| DN147782_c0_g1 | pel | Pectin lyase | Cell wall degradation | Pseudomonas | 39.1 | 0 (−8.5) | 0 (−8.9) |
| DN137942_c0_g1 | gh25-1b | Glycosyl hydrolase family 25 | Lysozyme activity | Pseudomonas | 63.3 | 123.7 (-) | 0.9 (−6.2) |
| DN145340_c0_g1 | gh25-2b | 2.2 | 1.1 (-) | 11.3 (2.4) | |||
| DN161451_c3_g1 | cel-1 | Cellulase | Cell wall degradation | Streptomyces | 84.2 | 0.1 (−10.1) | 0.1 (−9.7) |
| DN161451_c3_g3 | cel-2 | 329.5 | 0.4 (−9.5) | 0.1 (−10.7) | |||
| DN152450_c0_g1 | plc-1b | Phosphatidylinositol-specific phospholipase C domain | Lipid metabolism | Providencia | 5.8 | 0.3 (−3.9) | 0 (−8) |
| DN155959_c1_g1 | plc-2 | 29.3 | 2.8 (−3.3) | 0.5 (−5.6) | |||
| DN155959_c1_g2 | plc-3 | 127.4 | 19 (−2.8) | 2.3 (−5.8) | |||
FC is represented with “-” if p value is higher than 0.001.
FPKM fragments per kilobase per million, FC fold change in expression of bacteriomes relative to body tissues, Red red bacteriome, Yellow yellow bacteriome.
aThe origin is predicted based on the blast result.
bHTGs have typical GT/AG intron boundaries.
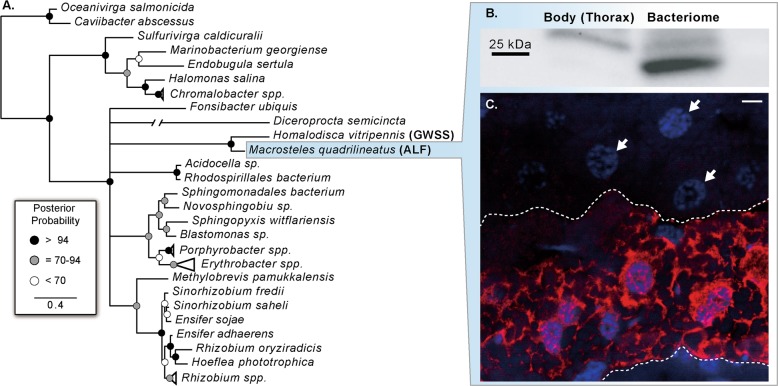
Fig. 3. Protein of an ancestral HTG frr in leafhoppers is specifically expressed in Sulcia-bearing bacteriocytes in ALF.
a Bayesian phylogeny of the horizontally transferred bacterial ribosomal recycling gene (frr; see methods and supplementary Fig. S1 for full phylogeny description). b Comparison of protein expression of frr between ALF bacteriome and body tissue using immunoblot approach. c Immunofluorescence localization of frr in Sulcia-ALF bacteriocytes with anti-frr primary antibody and Alexa Fluor 555 secondary antibody (red). DNA is counter-stained with DAPI (blue). The upper and lower dashed lines show the boundary between Sulcia and Nasuia bacteriocytes. White arrows indicate the nuclei of Nasuia-containing bacteriocytes. Scale bar is 5 µm (color figure online).
The differential occurrence of HTGs in GWSS relative to those in ALF, reveals why and when some HTGs were acquired in leafhoppers. HTGs in ALF that support Nasuia’s incomplete cellular information processing (CIP; e.g., rluA and dut, and specific copies of the duplicated HTGs) and vitamin synthesis (ribD) are not present in GWSS, and no gene residues of these HTGs were found in the GWSS genome. Their absence is likely explained by the fact that Baumannia still encodes and expresses these genes and does not require them [34, 37]. In contrast, HTGs that appear to fill the gaps of CIP systems (ileS, frr, and predicted ancestral copies of rnc, def, and yebC) in Sulcia-ALF are retained in GWSS. These genes further show correspondingly higher expression specifically in the Sulcia-containing bacteriocytes in both GWSS and ALF, indicating these HTG copies were acquired in a common ancestor to leafhoppers. Furthermore, some of these genes also appear to have undergone duplication, or repeated acquisition in ALF, to help shore-up convergent losses from Nasuia’s genome. For example, the rnc, def, and yebC genes are each represented by multiple copies that are differentially expressed between the Sulcia-ALF and Nasuia bacteriocytes [13]. In contrast, only a single copy of each of these genes is encoded in the GWSS genome (see Fig. S1), where they are required only to support Sulcia-GWSS [38]. Taken together, our results indicate that HTGs help hosts provide tailored support of their symbionts as they continue to lose one or more essential genes (see also [15]). Through the potential for duplication or repeated acquisition, HTGs further offer an adaptable mechanism to help facilitate symbioses with partner symbionts that also have highly degenerate genomes.
In order to validate HTG expression in their respective bacteriocytes, we conducted immunofluorescence microscopy and immunoblot assays on the frr HTG in ALF. Our results confirm that the frr gene is preferentially expressed in the bacteriocytes as a protein (Fig. 3b, c; [13]). Fluorescence microscopy demonstrated high expression of this protein within the bacteriocyte cytosol, but not in Sulcia-ALF cells. It is possible that this host-encoded frr gene functions to recycle Sulcia ribosomes from lysed cells [55]; however, its particular cellular roles remain to be confirmed. Nevertheless, both ALF and GWSS retain (see also Fig. S1), and significantly highly express, this gene in Sulcia harboring bacteriocytes. The horizontal acquisition of this gene is likely a conserved adaptation to support Sulcia in at least the common ancestor of the leafhoppers.
The acquisition of HTGs depends on the identity and needs of particular symbiont lineages. Few to no HTGs are shared between hosts that harbor distinct symbiont species (i.e., aphids, mealybugs, and leafhoppers; [13]). The establishment of HTGs likely goes through a three-step process. First, genes are initially acquired and maintain functional expression. Second, HTGs are then permitted to interact with certain symbiont functions via some element of the symbiont interface (e.g., directly inside bacteria, the symbiosomal membranes, or through indirect cellular processes). Finally, if these eukaryotic-encoded HTGs are more efficient than redundant bacterial orthologs, the symbiont gene will be lost. The long-term fate of established HTGs then depends on the evolutionary dynamics of the host and its symbiont community. For example, as in the case of ALF, HTGs may be duplicated with distinct copies supporting different symbiont species [13]. However, HTGs may also be lost from host genomes when younger symbionts replace older ones that depend on them. During this transition, HTGs can become superfluous as newly established symbionts are unlikely to require them, or be able to functionally interact with them. Indeed, variation in HTG composition among related hosts has been demonstrated in mealybug [15] and kleptoplastic dinoflagellate [56] species that have undergone symbiont replacements.
Repurposing of MT support genes is a conserved mechanism for providing tailored symbiont support in leafhoppers
Our recent study showed that the ALF host recruits 211 MT support genes to maintain Sulcia-ALF and Nasuia [13]. This evolutionary mechanism was also essential for the establishment of ancient symbioses between plants and cyanobacterial chloroplasts [57–59]. Thus, the reuse of MT support genes appears to offer a flexible set of genomic tools to help hosts to establish and stabilize obligate symbioses in a wide range of organisms. However, very little is known about how common the repurposing of MT support genes is in animal-bacterial symbioses.
In GWSS, we identified 94 MT support genes that are more highly expressed in either the yellow (83 genes) or the red (19 genes) bacteriomes (Table S2). These genes are able to support several critical bacterial functions, including nutritional metabolisms and nonnutritional cellular processes. For example, four MT aminoacyl-tRNA synthetases (aaRSs) capable of filling the gaps of missing genes in the translation system of Sulcia-GWSS show higher expression exclusively in the yellow bacteriome. In the red bacteriome, a MT ATP-binding cassette transporter that may be involved in heme biosynthesis retained in Baumannia [60] is highly expressed.
We further found that eight MT support genes have multiple copies with bacteriome-specific expression patterns in GWSS. In contrast, 33 highly expressed MT support genes in ALF have been duplicated [13]. Strikingly, most of the gene copies predicted to support Nasuia do not occur in the GWSS genome. For example, in ALF we identified two copies of MT translation factor GUF used in mitochondria to prevent the misincorporation of wrong amino acids under stress, which are differentially expressed in the Sulcia-ALF and Nasuia bacteriocytes [61]. GWSS has only one copy with a higher expression value in the yellow bacteriome. Phylogenetic analysis shows that this copy is likely orthologous with the one used to support Sulcia in ALF, suggesting that it is an ancestral support mechanism of Sulcia in leafhoppers (Fig. S1). These results indicate that, similar to HTGs, MT support genes are incorporated and occasionally duplicated to provide discrete maintenence of multiple bacterial symbionts with distinct requirements.
Widespread host-gene duplications support symbiont metabolite and energy requirements
Gene duplications are known to have contributed extensively to the integrated function and regulation of the pea aphid-Buchnera symbiont model system [14, 62–64]. For example, in order to exchange metabolites, transporter gene families have experienced widespread duplication and bacteriome-specific expression in aphids [62, 63]. In GWSS, we identified ~1,200 host genes (40–90% Blast identity and >80% sequence length coverage) that are derived from gene duplication events in the host genome with higher expression values in one or both bacteriomes. There are fewer duplicated genes in GWSS that support symbionts than there are in ALF (~1,600 genes). Similar to other sap-feeding insect systems, gene copies of amino acid transporters and sugar transporters are among the most widely duplicated gene sets in GWSS [13, 14, 62, 63, 65]. Many of the identified transporters appear to be involved in the trafficking of EAAs, EAA-associated metabolites, and resources important to other essential bacterial metabolisms.
We found further evidence that gene duplications are likely essential to the generation of bacterial energy. Baumannia maintains an expanded capability to perform glycolysis, which is generally lost from all known Sulcia and Nasuia lineages [20, 26, 28, 39, 66]. Consistent with Baumannia’s more capable energy metabolism, GWSS encodes a highly duplicated set of trehalose transporters Tret1 (n = 47) and two duplicate copies of the trehalase (TREH) gene. Twenty-three Tret1 copies and both of the TREH copies are upregulated in the bacteriomes (Table S3). Trehalose is the primary sugar in insect hemolymph and its transport into bacteriome tissues is essential for local energy production [67]. The TREH gene is further capable of hydrolyzing trehalose into glucose that can then be fed into Baumannia’s glycolysis pathway [34, 68]. Remarkably, the ALF host maintains fewer upregulated Tret1 transporters (n = 13) and no differentially expressed TREH genes, which is consistent with the fact that neither Sulcia-ALF, nor Nasuia, can perform glycolysis [13].
Hosts adapt to individually support bacterial metabolisms that provide essential nutrition
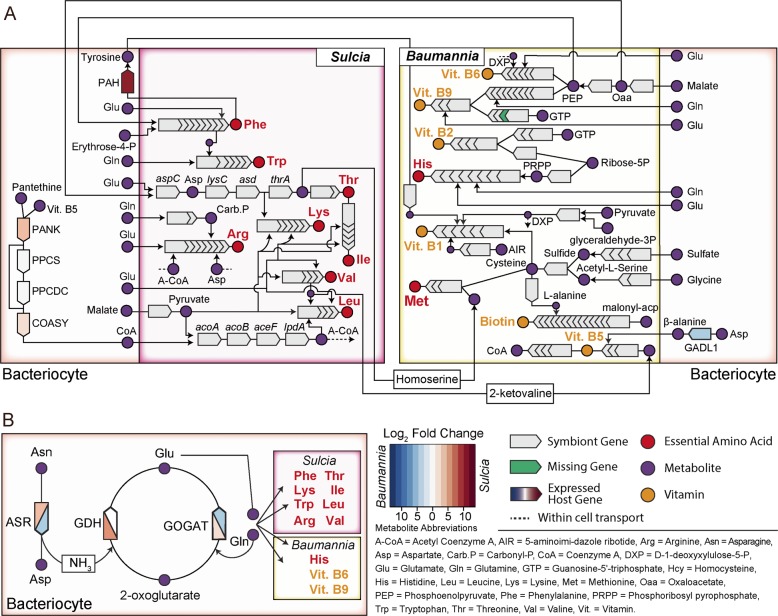
Sap-feeding insects generally supply metabolites to their obligate symbionts so that they can initiate and complete EAA synthesis [9–13, 69, 70]. In GWSS, both Sulcia-GWSS and Baumannia maintain relatively complete EAA synthesis pathways, but they require an array of metabolites in order to initiate and complete them. These requirements differ not only between Sulcia-GWSS and Baumannia, but also compared with the more ancient symbiont found in ALF. While GWSS appears to rely on some conserved mechanisms to provide metabolites, it has also evolved to match the needs of its symbionts. For example, Baumannia has converged to synthesize the same two EAAs, histidine and methionine, as does Nasuia [34, 39]. However, in contrast, Baumannia additionally retains the ability to make several required metabolites, including phosphoribosyl pyrophosphate and cysteine for histidine and methionine, respectively (Fig. 4a; [38, 70, 71]). Since Baumannia does not require these metabolites, GWSS does not highly express the relevant genes in its bacteriomes, as was previously found in ALF [13]. Although Baumannia requires homoserine to synthesize methionine, it has been previously predicted to acquire it from Sulcia-GWSS [38], which is further supported by our results.
Fig. 4. GWSS host support of nutritional metabolisms in Sulcia-GWSS and Baumannia.
a Inferred nutrition pathways in Sulcia-GWSS and Baumannia containing bacteriocytes. b The GS/GOGAT (glutamine synthetase/glutamate synthase) cycle and glutamate dehydrogenase (GDH) pathway for recycling NH3. More highly expressed host genes in bacteriocytes relative to body tissues are colored based on the log2 fold change ratio. See inset legend for additional details and explanation of metabolite abbreviations. See Table S3 for full names of gene products (color figure online).
The genome of Sulcia-GWSS, although much smaller than Baumannia’s, is responsible for the synthesis of eight EAAs. As such, it has expanded metabolite requirements of GWSS, including coenzyme A (CoA), oxaloacetate (Oaa), and phosphoenolpyruvate (PEP) (Fig. 4a). For CoA, GWSS appears to use the same alternative CoA synthesis pathway as ALF by more highly expressing pantothenate kinase and bifunctional CoA synthase in the yellow bacteriome (Fig. 4a and Table S3; [13, 72, 73]). Unlike ALF, however, we did not find highly expressed genes that would be required to synthesize Oaa or PEP in the yellow bacteriome. We predict that Sulcia-GWSS may instead acquire these from Baumannia, which has the capability to synthesize both (Fig. 4a; [34]). Baumannia further co-occurs in the yellow bacteriome where it could supply them directly to Sulcia-GWSS. In contrast, ALF more highly expresses pathways that can provide Oaa and PEP, since Nasuia is also unable to synthesize them and occurs in its own separate bacteriome tissue.
Both Baumannia and Sulcia-GWSS require the non-EAAs, glutamate (Glu) and glutamine (Gln), to synthesize nearly all EAAs (Fig. 4). Given that both symbionts have lost the ability to produce and transport these metabolites, it is essential that the host supplies all necessary mechanisms for their incorporation into symbiont metabolisms. Most examined phloem-feeding insect systems (e.g., pea aphids, mealybugs, psyllids, and ALF) use the GS/GOGAT (Gln synthetase/Glu synthase) pathway to recycle ammonia [9–11, 13, 69]. However, in GWSS, our results reveal that the host has evolved distinct ways of providing Glu and Gln between bacteriome types. In contrast to other symbioses [9–11, 13, 69], in GWSS only the GOGAT gene that produces Glu is more highly expressed in both bacteriomes (Fig. 4b and Table S3). It is feasible that the Gln producing activity of the GS gene is not required since the concentration of Gln is higher in xylem relative to phloem and the metabolite may be readily available in insect hemolymph [74]. In the yellow bacteriome, we further identified a more highly expressed MT support gene, Glu dehydrogenase (GDH; Fig. 4b and Table S3). The GDH pathway is an alternative to the GS/GOGAT pathway for recycling ammonia into Glu. It is widely employed by bacteria when ATP is limited, as may be the case for small genome symbionts that have lost most ATP synthesis capabilities [8, 75]. The incorporation of this gene to aid Baumannia and Sulcia-GWSS appears to be a unique adaptation by the GWSS lineage to support a novel symbiosis, as it has not yet been reported to play a role in other hemipteran systems [9–11, 13].
The GWSS lineage has uniquely evolved to support B vitamin synthesis by Baumannia
Plant saps are deficient in B vitamins that sap-feeding insects must acquire from exogenous sources, or from their symbionts [76]. Baumannia, unlike symbionts found in other auchenorrhynchan hosts, is capable of synthesizing a relatively comprehensive set of B vitamins: thiamine (B1), riboflavin (B2), pantothenate (B5), pyridoxine (B6), biotin (B7), and folic acid (B9) (see Fig. 4). Baumannia also retains the ability to synthesize some of the precursor metabolites required to initiate these pathways [34]. However, because Baumannia lacks all but two amino acid pathways, and B vitamin pathways require amino acids, they must be acquired from the host or from Sulcia-GWSS. For example, β-alanine and tyrosine are required for B5 and B1 pathways, respectively. An acidic amino acid decarboxylase (GADL1) host gene that can synthesize β-alanine from aspartate is highly expressed in the red bacteriome where B5 is predicted to be synthesized (Fig. 4a and Table S3; [37]). The synthesis of B5 also appears to require 2-ketovaline, which can be obtained from Sulcia-GWSS’s valine and leucine pathways, as was predicted previously (Fig. 4a; [38]). In contrast, B1 may be synthesized by Baumannia in both the yellow and red bacteriomes [37]. A host-encoded phenylalanine-4-hydroxylase gene capable of hydroxylating phenylalanine to tyrosine is highly expressed only in the yellow bacteriome, and tyrosine may be transported to Baumannia in both bacteriome types (Fig. 4a and Table S3).
Finally, since all symbiotic partners require B vitamins, they must be transported across tissues and cellular compartments. We identified nine highly expressed vitamin transporter genes (Table S3); three are more highly expressed in the yellow bacteriome, and seven are more highly expressed in the red bacteriome, where a bulk of B vitamin synthesis is predicted to occur [37]. In contrast, fewer vitamin transporters (n = 4) were identified in ALF, consistent with the fact that most of the vitamin pathways have been lost in Sulcia-ALF and Nasuia.
Sulcia, but not Baumannia, relies on their host to complete DNA replication, transcription, and translation
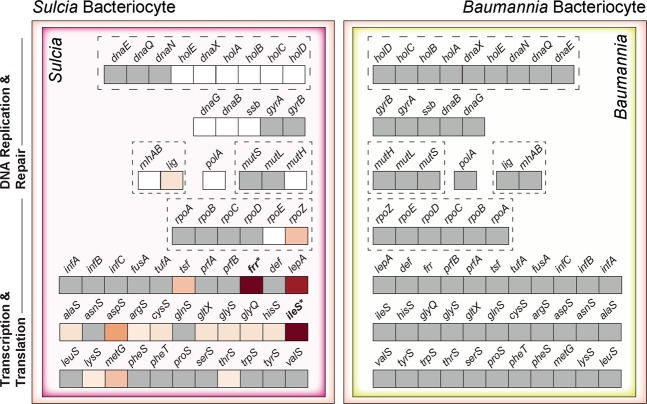
One of the most basic and essential functions of any cell is the ability to replicate, transcribe, and translate genetic information. However, nearly all tiny-genome symbionts have variably lost genes involved in each of these three core functions [7, 8, 77]. The symbionts of GWSS have differentially lost an array of genes involved in these cellular information processing (CIP) mechanisms and require discrete host inputs to function. Given Baumannia’s larger genome, it still encodes all genes predicted to be essential to CIP systems (Fig. 5). As such, no differentially expressed host genes involved in CIP systems were found in the red bacteriome, particularly in contrast to the 38 genes found to support Nasuia in ALF [13]. For example, six highly expressed MT aaRSs were reported to likely replace these missing genes in Nasuia’s genome, while none of their orthologs are more highly expressed in the red bacteriome. Furthermore, the HTGs predicted to support CIP functions in Nasuia are also absent in the GWSS genome [13]. For example, in Nasuia bacteriocytes, we identified four highly expressed copies of the def gene that is essential for the maturation of proteins [78], while GWSS has a single copy that is uniquely highly expressed in the yellow bacteriome (Table 1).
Fig. 5. The complete core cellular information processing (CIP) gene set in Baumannia and host compensation of missing CIP genes in Sulcia-GWSS.
Symbiont-encoded genes are colored in gray. Missing CIP genes in Sulcia-GWSS with no host complementary genes identified are shown in white. Horizontally transferred genes are shown in bold and labeled with an asterisk. More highly expressed host genes in bacteriocytes relative to body tissues are colored based on the log2 fold change ratio (see inset legend in Fig. 4). Genes grouped by dashed boxes are predicted to interact directly within the bacterial cytosol (color figure online).
In contrast to Baumannia, Sulcia-GWSS has lost 30 CIP genes. The GWSS host more highly expresses genes in the yellow bacteriome that can precisely fill most of these gaps (Fig. 5 and Table S3). Strikingly, however, we did not find highly expressed host genes that are able to replace several missing genes involved in DNA replication and repair (e.g., DNA polymerase genes holA-E). One possibility is that Baumannia cells co-occurring in the yellow bacteriome support DNA replication and repair in Sulcia-GWSS. Baumannia still encodes gene sets that could fill these gaps in Sulcia-GWSS’s CIP systems; however, this level of protein exchange between insect-bacterial symbionts remains uncertain. Compared with the Sulcia-ALF lineage, Sulcia-GWSS has a larger CIP gene set (29 vs. 22) [38, 39]. All missing CIP genes from the Sulcia-GWSS genome have also been lost in Sulcia-ALF, suggesting that they were purged in the common ancestor to leafhoppers. For most of those missing bacterial genes that are shared between Sulcia lineages, GWSS relies on the same support mechanisms as ALF. For example, we identified two differentially expressed HTGs, frr and ileS, in both GWSS and ALF that can replace the missing symbiont orthologs involved in translation in both Sulcia lineages (Fig. 5).
It is well known that symbiotic bacteria generally experience convergent gene losses. This appears to also be the case for Sulcia lineages. Some host compensatory mechanisms demonstrate independent origins to compensate for shared missing Sulcia genes. For example, we found that the DE host gene capable of complementing alaS (alanine-tRNA ligase) missing in both Sulcia lineages has different origins in ALF and GWSS hosts. GWSS has recruited a MT support ALAS gene, while ALF appears to use a more highly expressed cytoplasm-targeted ALAS gene. The use of two different ALAS genes to support its loss from Sulcia suggests that the two lineages independently lost the alaS gene after the two leafhopper groups diverged.
Conclusion
Symbionts of the Auchenorrhyncha have excessively small genomes and are segregated into distinct organs that support them [30, 70]. For Sulcia, one of the most ancient insect-associated symbionts known, our results show that a majority of host-derived support mechanisms are likely conserved, at least among leafhoppers. Most Sulcia lineages have genomes <250 kb with broadly similar cellular capabilities [28, 38, 39, 79], suggesting that some host support mechanisms for Sulcia may have evolved in an earlier auchenorrhynchan ancestor. However, all major Sulcia lineages differ in the identities of genes more recently lost and the demands that these losses place on their hosts. For example, as Sulcia slipped below a 200 kb genome in ALF and related deltocephalinae species, hosts incorporated a range of distinct mechanisms to maintain it. The genome of ALF reveals that hosts have integrated 17 additional HTGs into its genome to support Sulcia; some of these have been further duplicated to support its partner, Nasuia [13].
The replacement of an ancient symbiont has been posited as a way of escaping the limitations of highly degenerate ones [80]. Yet, newly acquired symbionts are still subject to the same genome degeneration process, requiring widespread and ongoing compensatory evolution by their hosts [81–84]. In the GWSS symbioses, it is clear that the acquisition of Baumannia still required sharpshooter hosts to evolve whole new support regimes. Our results demonstrate that indeed GWSS highly expresses a distinct set of genes to meet the metabolic and cellular needs of Baumannia. However, in contrast to more ancient symbionts in ALF, Baumannia retains extensive cellular capabilities. As such, we found fewer symbiont support mechansims overall and that several support mechanisms used by ALF are not present in the GWSS genome. In particular, although we found seven HTGs in GWSS devoted to symbiont support, none of them appear to aid Baumannia. With only two leafhopper host lineages to compare, it is not clear if these mechanisms were lost in sharpshooters, or if they never evolved. The split between leafhoppers and sharpshooters is relatively ancient (>40 MYA), making identification of gene remnants difficult. Regardless, the complexity of support mechanisms targeting Baumannia is likely to convergently increase as Baumannia’s genome inevitably shrinks, as has already occurred in other sharpshooter host species [33, 35, 85].
Supplementary information
Acknowledgements
We thank three anonymous reviewers for the helpful suggestions. We thank Dr Stephen Richards (University of California Davis), Adelaida Rhodes (Broad Institute), and the i5K team for help with accessing the GWSS genome. We thank Dr Nancy Moran (University of Texas Austin) and Dr Susan Ge (University of California Merced) for the use of laboratory resources. We also thank the UT Austin GSAF for sequencing. Dr Allen Yang and Dr Xiaoliang Liu provided valuable support with data analysis and immunoblot experiment. The data in this work were collected, in part, with a confocal microscope acquired through the National Science Foundation (NSF) MRI Award (DMR-1625733). This work was supported by a NSF Award (IOS1347116).
Data availability
Raw Illumina reads from GWSS were submitted to the Sequence Read Archive (SRA) database under the accession number PRJNA342859, and the assembled transcriptome was submitted to the Transcriptome Shotgun Assembly (TSA) database under the accession number GICT00000000.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Meng Mao, Gordon M. Bennett
Supplementary information
The online version of this article (10.1038/s41396-020-0616-4) contains supplementary material, which is available to authorized users.
References
- 1.Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–89. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- 2.Douglas AE. The microbial dimension in insect nutritional ecology. Funct Ecol. 2009;23:38–47. [Google Scholar]
- 3.Buchner P. Endosymbiosis of animals with plant microorganims. New York, NY: Interscience; 1965. [Google Scholar]
- 4.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–90. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 5.Koga R, Meng X-Y, Tsuchida T, Fukatsu T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte–embryo interface. Proc Natl Acad Sci USA. 2012;109:E1230–7. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luan J, Sun X, Fei Z, Douglas AE. Maternal inheritance of a single somatic animal cell displayed by the bacteriocyte in the whitefly Bemisia tabaci. Curr Biol. 2018;28:459–65. doi: 10.1016/j.cub.2017.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 8.Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 9.Hansen AK, Moran NA. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 2011;108:2849–54. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sloan DB, Nakabachi A, Richards S, Qu J, Murali SC, Gibbs RA, et al. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 2014;31:857–71. doi: 10.1093/molbev/msu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husnik F, Nikoh N, Koga R, Ross L, Duncan RP, Fujie M, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153:1567–78. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 12.Luan J-B, Chen W, Hasegawa DK, Simmons AM, Wintermantel WM, Ling K-S, et al. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol Evol. 2015;7:2635–47. doi: 10.1093/gbe/evv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao M, Yang X, Bennett GM. Evolution of host support for two ancient bacterial symbionts with differentially degraded genomes in a leafhopper host. Proc Natl Acad Sci USA. 2018;115:E11691–700. doi: 10.1073/pnas.1811932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan RP, Husnik F, Van Leuven JT, Gilbert DG, Dávalos LM, McCutcheon JP, et al. Dynamic recruitment of amino acid transporters to the insect/symbiont interface. Mol Ecol. 2014;23:1608–23. doi: 10.1111/mec.12627. [DOI] [PubMed] [Google Scholar]
- 15.Husnik F, McCutcheon JP. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci USA. 2016;113:5416–24. doi: 10.1073/pnas.1603910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong RA, Park H, Moran NA. Genome evolution of the obligate endosymbiont Buchnera aphidicola. Mol Biol Evol. 2019;36:1481–9. doi: 10.1093/molbev/msz082. [DOI] [PubMed] [Google Scholar]
- 17.Łukasik P, Nazario K, Van Leuven JT, Campbell MA, Meyer M, Michalik A, et al. Multiple origins of interdependent endosymbiotic complexes in a genus of cicadas. Proc Natl Acad Sci USA. 2018;115:E226–35. doi: 10.1073/pnas.1712321115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett GM, McCutcheon JP, McDonald BR, Moran NA. Lineage-specific patterns of genome deterioration in obligate symbionts of sharpshooter leafhoppers. Genome Biol Evol. 2015;8:296–301. doi: 10.1093/gbe/evv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koga R, Bennett GM, Cryan JR, Moran NA. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol. 2013;15:2073–81. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]
- 20.McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–18. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manzano‐Marín A, Szabó G, Simon JC, Horn M, Latorre A. Happens in the best of subfamilies: establishment and repeated replacements of co‐obligate secondary endosymbionts within Lachninae aphids. Environ Microbiol. 2017;19:393–408. doi: 10.1111/1462-2920.13633. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser B. Licht-und elektronenmikroskopische untersuchung der symbionten von Graphocephala coccinea forstier (Homoptera: Jassidae) Int J Insect Morphol Embryol. 1980;9:79–88. [Google Scholar]
- 23.Nakabachi A, Shigenobu S, Sakazume N, Shiraki T, Hayashizaki Y, Carninci P, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102:5477–82. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, Stern DL. Developmental origin and evolution of bacteriocytes in the aphid—Buchnera symbiosis. PLoS Biol. 2003;1:e21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–10. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett GM, Mao M. Comparative genomics of a quadripartite symbiosis in a planthopper host reveals the origins and rearranged nutritional responsibilities of anciently diverged bacterial lineages. Environ Microbiol. 2018;20:4461–72. doi: 10.1111/1462-2920.14367. [DOI] [PubMed] [Google Scholar]
- 27.Bell-Roberts L, Douglas AE, Werner GD. Match and mismatch between dietary switches and microbial partners in plant sap-feeding insects. Proc R Soc B. 2019;286:20190065. doi: 10.1098/rspb.2019.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao M, Yang X, Poff K, Bennett G. Comparative genomics of the dual-obligate symbionts from the treehopper, Entylia carinata (Hemiptera: Membracidae), provide insight into the origins and evolution of an ancient symbiosis. Genome Biol Evol. 2017;9:1803–15. doi: 10.1093/gbe/evx134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett GM, Abbà S, Kube M, Marzachì C. Complete genome sequences of the obligate symbionts “Candidatus Sulcia muelleri” and “Ca. Nasuia deltocephalinicola” from the pestiferous Leafhopper Macrosteles quadripunctulatus (Hemiptera: Cicadellidae) Genome Announc. 2016;4:e01604–15. doi: 10.1128/genomeA.01604-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Leuven JT, Mao M, Xing DD, Bennett GM, McCutcheon JP. Cicada endosymbionts have tRNAs that are correctly processed despite having genomes that do not encode all of the tRNA processing machinery. mBio. 2019;10:e01950–01918. doi: 10.1128/mBio.01950-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran NA, Dale C, Dunbar H, Smith WA, Ochman H. Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ Microbiol. 2003;5:116–26. doi: 10.1046/j.1462-2920.2003.00391.x. [DOI] [PubMed] [Google Scholar]
- 32.Takiya DM, Tran PL, Dietrich CH, Moran NA. Co‐cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol Ecol. 2006;15:4175–91. doi: 10.1111/j.1365-294X.2006.03071.x. [DOI] [PubMed] [Google Scholar]
- 33.Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. mBio. 2014;5:e01697–14. doi: 10.1128/mBio.01697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4:e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett GM, McCutcheon JP, McDonald BR, Moran NA. Lineage-specific patterns of genome deterioration in obligate symbionts of sharpshooter leafhoppers. Genome Biol Evol. 2016;8:296–301. doi: 10.1093/gbe/evv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobiałka M, Michalik A, Szwedo J, Szklarzewicz T. Diversity of symbiotic microbiota in Deltocephalinae leafhoppers (Insecta, Hemiptera, Cicadellidae) Arthropod Struct Dev. 2018;467:268–78. doi: 10.1016/j.asd.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Bennett GM, Chong RA. Genome-wide transcriptional d in the companion bacterial symbionts of the glassy-winged sharpshooter (Cicadellidae: Homalodisca vitripennis) reveal differential gene expression in bacteria occupying multiple host organs. G3 Genes, Genomes, Genet. 2017;7:3073–82. doi: 10.1534/g3.117.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–7. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett GM, Moran NA. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 2013;5:1675–88. doi: 10.1093/gbe/evt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 43.Rice Peter, Longden Ian, Bleasby Alan. EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 44.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–90. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katoh K, Misawa K. Kuma K-i, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). 2010; New Orleans, LA. p. 1–8.
- 51.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–5. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- 53.Wilson AC, Duncan RP. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc Natl Acad Sci USA. 2015;112:10255–61. doi: 10.1073/pnas.1423305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas GW, Dohmen E, Hughes DS, Murali SC, Poelchau M, Glastad K, et al. The genomic basis of arthropod diversity. BioRxiv. 2018. 10.1101/382945.
- 55.Janosi L, Shimizu I, Kaji A. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc Natl Acad Sci USA. 1994;91:4249–53. doi: 10.1073/pnas.91.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hehenberger E, Gast RJ, Keeling PJ. A kleptoplastidic dinoflagellate and the tipping point between transient and fully integrated plastid endosymbiosis. Proc Natl Acad Sci USA. 2019;116:17934–42. doi: 10.1073/pnas.1910121116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carrie C, Small I. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833:253–9. doi: 10.1016/j.bbamcr.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 58.Peeters N, Small I. Dual targeting to mitochondria and chloroplasts. Biochimica et Biochim Biophys Acta. 2001;1541:54–63. doi: 10.1016/s0167-4889(01)00146-x. [DOI] [PubMed] [Google Scholar]
- 59.Singer A, Poschmann G, Mühlich C, Valadez-Cano C, Hänsch S, Hüren V, et al. Massive protein import into the early-evolutionary-stage photosynthetic organelle of the amoeba Paulinella chromatophora. Curr Biol. 2017;27:2763–73. doi: 10.1016/j.cub.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 60.Bayeva M, Khechaduri A, Wu R, Burke MA, Wasserstrom JA, Singh N, et al. ATP-binding cassette B10 regulates early steps of heme synthesis. Circ Res. 2013;113:279–87. doi: 10.1161/CIRCRESAHA.113.301552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bauerschmitt H, Funes S, Herrmann JM. The membrane-bound GTPase Guf1 promotes mitochondrial protein synthesis under suboptimal conditions. J Biol Chem. 2008;283:17139–46. doi: 10.1074/jbc.M710037200. [DOI] [PubMed] [Google Scholar]
- 62.Price DR, Duncan RP, Shigenobu S, Wilson AC. Genome expansion and differential expression of amino acid transporters at the aphid/Buchnera symbiotic interface. Mol Biol Evol. 2011;28:3113–26. doi: 10.1093/molbev/msr140. [DOI] [PubMed] [Google Scholar]
- 63.Price DR, Tibbles K, Shigenobu S, Smertenko A, Russell CW, Douglas AE, et al. Sugar transporters of the major facilitator superfamily in aphids; from gene prediction to functional characterization. Insect Mol Biol. 2010;19:97–112. doi: 10.1111/j.1365-2583.2009.00918.x. [DOI] [PubMed] [Google Scholar]
- 64.Duncan RP, Nathanson L, Wilson AC. Novel male-biased expression in paralogs of the aphid slimfast nutrient amino acid transporter expansion. BMC Evol Biol. 2011;11:253. doi: 10.1186/1471-2148-11-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dahan RA, Duncan RP, Wilson AC, Dávalos LM. Amino acid transporter expansions associated with the evolution of obligate endosymbiosis in sap-feeding insects (Hemiptera: sternorrhyncha) BMC Evol Biol. 2015;15:52. doi: 10.1186/s12862-015-0315-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–9. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyatt G, Kalf G. The chemistry of insect hemolymph: II. trehalose and other carbohydrates. J gen physiol. 1957;40:833–47. doi: 10.1085/jgp.40.6.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mori H, Lee J-H, Okuyama M, Nishimoto M, Ohguchi M, Kim D, et al. Catalytic reaction mechanism based on α-secondary deuterium isotope effects in hydrolysis of trehalose by European honeybee trehalase. Biosci Biotechnol Biochem. 2009;73:2466–73. doi: 10.1271/bbb.90447. [DOI] [PubMed] [Google Scholar]
- 69.Poliakov A, Russell C, Ponnala L, Hoops H, Sun Q, Douglas A, et al. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol cell Proteom. 2011;10:M110.007039. doi: 10.1074/mcp.M110.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ankrah NY, Chouaia B, Douglas AE. The cost of metabolic interactions in symbioses between insects and bacteria with reduced genomes. mBio. 2018;9:e01433–18. doi: 10.1128/mBio.01433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Douglas AE. How multi-partner endosymbioses function. Nat Rev Microbiol. 2016;14:731–43. doi: 10.1038/nrmicro.2016.151. [DOI] [PubMed] [Google Scholar]
- 72.Balibar CJ, Hollis-Symynkywicz MF, Tao J. Pantethine rescues phosphopantothenoylcysteine synthetase and phosphopantothenoylcysteine decarboxylase deficiency in Escherichia coli but not in Pseudomonas aeruginosa. J Bacteriol. 2011;193:3304–12. doi: 10.1128/JB.00334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rana A, Seinen E, Siudeja K, Muntendam R, Srinivasan B, Van Der Want JJ, et al. Pantethine rescues a Drosophila model for pantothenate kinase–associated neurodegeneration. Proc Natl Acad Sci USA. 2010;107:6988–93. doi: 10.1073/pnas.0912105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andersen PC, Brodbeck BV, Mizell RF., III Feeding by the leafhopper, Homalodisca coagulata, in relation to xylem fluid chemistry and tension. J Insect Physiol. 1992;38:611–22. [Google Scholar]
- 75.Leigh JA, Dodsworth JA. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol. 2007;61:349–77. doi: 10.1146/annurev.micro.61.080706.093409. [DOI] [PubMed] [Google Scholar]
- 76.Douglas AE. The B vitamin nutrition of insects: the contributions of diet, microbiome and horizontally-acquired genes. Curr Opin Insect Sci. 2017;23:65–69. doi: 10.1016/j.cois.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 77.Klasson L, Andersson SG. Evolution of minimal-gene-sets in host-dependent bacteria. Trends Microbiol. 2004;12:37–43. doi: 10.1016/j.tim.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 78.Mazel D, Pochet S, Marliere P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–23. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsuura Y, Moriyama M, Łukasik P, Vanderpool D, Tanahashi M, Meng X-Y, et al. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc Natl Acad Sci USA. 2018;115:E5970–9. doi: 10.1073/pnas.1803245115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sudakaran S, Kost C, Kaltenpoth M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017;27:375–90. doi: 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 81.Bennett GM, Moran NA. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112:10169–76. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCutcheon JP, Boyd BM, Dale C. The life of an insect endosymbiont from the cradle to the grave. Curr Biol. 2019;29:R485–95. doi: 10.1016/j.cub.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 83.Wernegreen JJ. Endosymbiont evolution: predictions from theory and surprises from genomes. Ann N Y Acad Sci. 2015;1360:16–35. doi: 10.1111/nyas.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gil R, Latorre A. Unity makes strength: a review on mutualistic symbiosis in representative insect clades. Life. 2019;9:21. doi: 10.3390/life9010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sabater-Muñoz B, Toft C, Alvarez-Ponce D, Fares MA. Chance and necessity in the genome evolution of endosymbiotic bacteria of insects. ISME J. 2017;11:1291–304. doi: 10.1038/ismej.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw Illumina reads from GWSS were submitted to the Sequence Read Archive (SRA) database under the accession number PRJNA342859, and the assembled transcriptome was submitted to the Transcriptome Shotgun Assembly (TSA) database under the accession number GICT00000000.