Abstract
Pulmonary arterial hypertension (PAH) is a rare disease with high mortality despite therapeutic advances. Clinical management of children with PAH is particularly challenging because of increased complexity of disease etiology and clinical presentation, and the lack of data from pediatric-specific clinical trials. In children, PAH often develops in association with congenital heart disease and other developmental disorders. Emerging data from genetic studies of pediatric-onset PAH indicate that the genetic basis is different than that of adults. There is a greater genetic burden in children, with rare genetic factors contributing to at least 35% of pediatric-onset idiopathic PAH (IPAH) compared with approximately 11% of adult-onset IPAH. De novo variants are the most frequent monogenetic cause of PAH in children, likely contributing to approximately 15% of all cases. Rare deleterious variants in BMPR2 contribute to pediatric-onset IPAH and familial PAH with similar frequency as adult-onset disease but rarely explain cases of PAH associated with other diseases. Rare deleterious variants in developmental genes—including TBX4, SOX17, and other genes requiring confirmation in larger cohorts—are emerging as important contributors to pediatric-onset disease. Because each causal gene contributes to only a small number of cases, large cohorts of pediatric-onset PAH are needed to further identify the unique etiologic differences of PAH in children. We propose a genetics-first approach followed by focused phenotyping of pediatric patients grouped by genetic diagnosis to define endophenotypes that can be used to improve risk stratification and treatment.
Key Words: genomics, lung disease, pediatrics
Abbreviations: APAH-CHD, PAH associated with congenital heart disease; FPAH, familial PAH; IPAH, idiopathic PAH; LGD, likely gene-disrupting; PAH, pulmonary arterial hypertension; PPHN, persistent pulmonary hypertension of the newborn
Pulmonary arterial hypertension (PAH) is a rare disease with high mortality despite therapeutic advances. Endothelial dysfunction, aberrant cell proliferation, and vasoconstriction give rise to increased pulmonary vascular pressures, increased vascular resistance, heart failure, and premature death. The disease is caused by genetic, epigenetic, and environmental factors, and gene × environment interactions wherein genetic contributions to disease risk are modified by environmental exposures. Most of our understanding of PAH etiology and treatment is based on studies in adults. However, emerging clinical and genetic data indicate that there are fundamental differences between pediatric- and adult-onset disease. Here, we highlight the need for dedicated studies of children with PAH because etiologies differ by age.
PAH usually manifests in early to midlife with an estimated prevalence of 4.8 to 8.1 cases per million people for pediatric-onset disease1 and 15 to 50 cases per million people for adult-onset disease.2 Pediatric PAH differs from adult-onset disease in several important aspects including sex bias, disease etiology, clinical presentation, and response to therapy.3, 4, 5 The three- to fourfold higher disease prevalence among women compared with men in adult-onset PAH is not observed in pediatric-onset disease, suggesting less dependence on sex-specific interacting factors in children.6, 7, 8 Etiologically, pediatric-onset PAH has a higher proportion of idiopathic PAH (IPAH), PAH associated with congenital heart disease (APAH-CHD), and developmental lung diseases (including persistent pulmonary hypertension of the newborn [PPHN]) than adult-onset disease.3,4,6,8 Data from the National Biological Sample and Data Repository for Pulmonary Arterial Hypertension (aka PAH Biobank, N = 2,572) indicate that children present with slightly higher mean pulmonary arterial pressure, decreased cardiac output, and increased pulmonary vascular resistance than adults at diagnosis (Table 1).8 Because of the lack of pediatric clinical trial data, fewer therapeutic options are indicated for use in children with PAH. In practice, therapeutic regimens are based on experiences of individual centers9 or statements of consensus guidelines.10,11 The management of pediatric PAH remains challenging because of the paucity of data regarding the natural history, mechanisms of disease, and treatment response.
Table 1.
Clinical Characteristics of Child- vs Adult-Onset Pulmonary Arterial Hypertension Cases at Diagnosis
| Group | Age at Diagnosis (y) | MPAP (mm Hg) | CO, Fick (L/min) | PVR (Woods Units) |
|---|---|---|---|---|
| Child (n = 226) | 8 ± 6 (226) | 55 ± 18 (225) | 3.3 ± 1.6 (168) | 17.7 ± 11.6 (164) |
| Adult (n = 2,345) | 52 ± 19 (2,345) | 50 ± 14 (2,293) | 4.6 ± 1.7 (1,630) | 10.0 ± 5.9 (1,579) |
| P value | < .0001 | < .0001 | < .0001 | < .0001 |
Data from the National Biological Sample and Data Repository for Pulmonary Arterial Hypertension (N = 2,572). Values are mean ± SD (No.) or as otherwise indicated. Child-onset = < 18 y of age at diagnosis; CO = cardiac output; MPAP = mean pulmonary artery pressure; PVR = pulmonary vascular resistance.
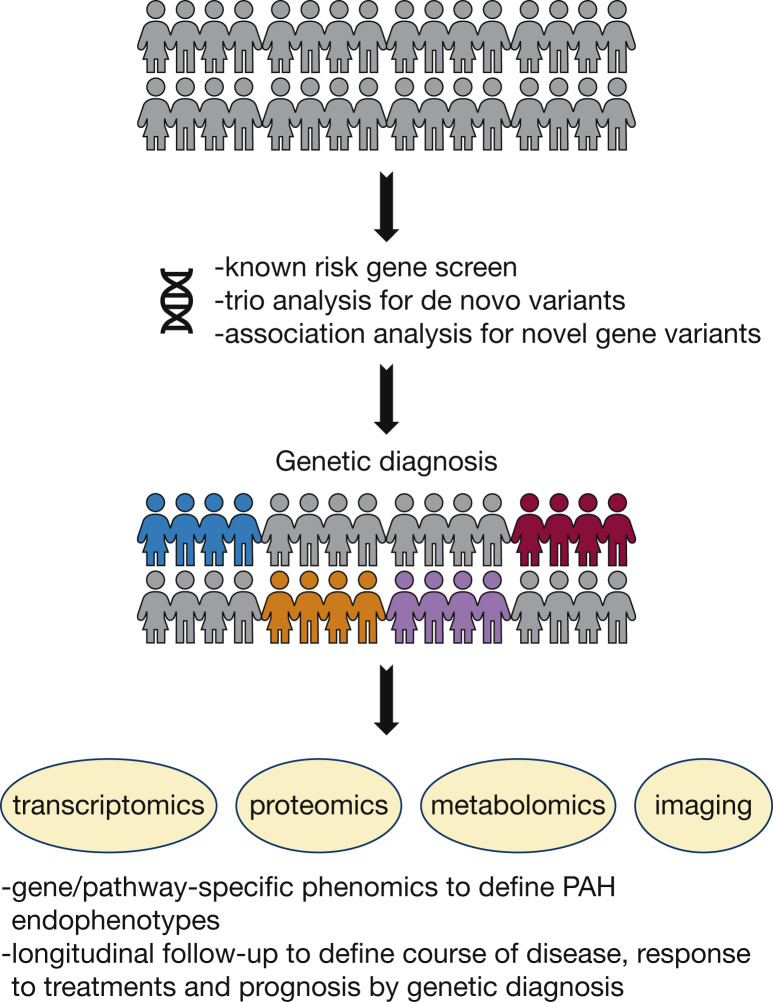
Identification of molecular subtypes of PAH has been proposed as a means to improve risk stratification, treatment, and outcomes. Multiple high-throughput omics technologies are available to provide data for molecular classification including genomics, transcriptomics, proteomics, and metabolomics. The National Heart, Lung, and Blood Institute-funded Pulmonary Vascular Disease Phenomics program is an example of such an effort, having enrolled > 1,000 patients for deep phenotyping.12 However, Pulmonary Vascular Disease Phenomics and most other cohorts are adult-centric with modest to minimal enrollment of pediatric patients. In addition, phenotyping requires biological sampling, imaging, and functional testing which may not be indicated for young children, especially those with comorbidities. The Pediatric Pulmonary Hypertension Network (www.pphnet.org) has been established to recruit and focus research efforts on pediatric patients. We propose a genetics-first approach for pediatric cohorts followed by detailed phenotyping and longitudinal follow-up of patients grouped by genetic diagnosis (Fig 1). A small-volume blood sample or saliva sample is all that is needed for whole exome or whole genome sequencing. Causal gene variant identification is straightforward for known risk genes and dependent on cohort size and number of family trios for the identification of novel genes. Comparison of clinical phenotypes among children carrying variants in specific genes, along with known biological functions of the genes, would then guide more detailed phenotypic analyses including pulmonary and cardiac imaging and assessment of systemic or extracardiopulmonary features. Comparison of metabolomic, proteomic, and transcriptomic data within and across genetic subtypes may enable identification of molecular phenocopies to prioritize rare variants and identify additional genetic subtypes. Longitudinal study of patients grouped by genetic diagnosis would provide invaluable data regarding natural history of disease, response to treatments, and prognosis. Large-scale pediatric-enriched cohorts are more likely to identify novel highly penetrant genes than adult cohorts, and families are more motivated to participate after a genetic diagnosis has been made in a child.
Figure 1.
A genetics-first approach toward better understanding and treatment of pediatric PAH. PAH = pulmonary arterial hypertension.
Genetics of Pediatric PAH
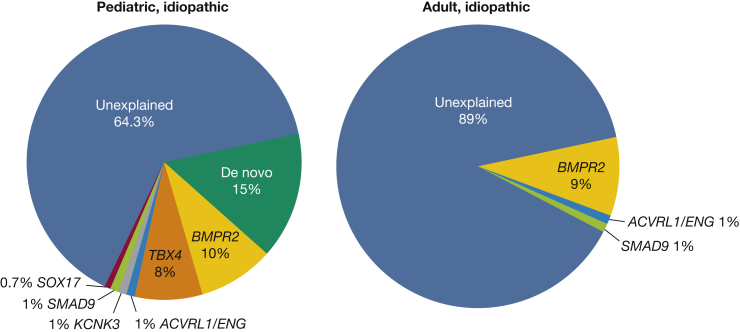
Knowledge of genetic factors specifically contributing to pediatric-onset PAH are starting to emerge. De novo genetic variants not inherited from parents contribute to a significant proportion of pediatric PAH (Fig 2).7 This is perhaps not unexpected because de novo variants have been shown to be important in other pediatric developmental diseases including congenital heart disease13, 14, 15 and congenital diaphragmatic hernia.16, 17, 18 In addition, rare heritable variants in at least three developmental pathways and/or transcription factors have been implicated in pediatric PAH: BMPR2, TBX4, and SOX17 (Figs 2, 3). Other known PAH risk genes such as ACVRL1, ENG, KCNK3, and SMAD9 rarely contribute to pediatric PAH7 and in aggregate account for approximately 3% of cases (Fig 2). In addition, two variants of uncertain significance were identified in each of BMPR1B19 and NOTCH320 by candidate gene screening. No effect of sex on risk allele frequencies has been identified to date.
Figure 2.
Relative contributions of de novo mutations and 11 established PAH risk genes in idiopathic pediatric- and adult-onset PAH in a cohort of 412 cases (130 with pediatric idiopathic PAH and 178 with adult idiopathic PAH). Risk genes included BMPR2, ACVRL1, BMPR1A, BMPR1B, CAV1, EIF2AK4, ENG, KCNK3, SMAD4, SMAD9, and TBX4. See Figure 1 legend for expansion of abbreviation.
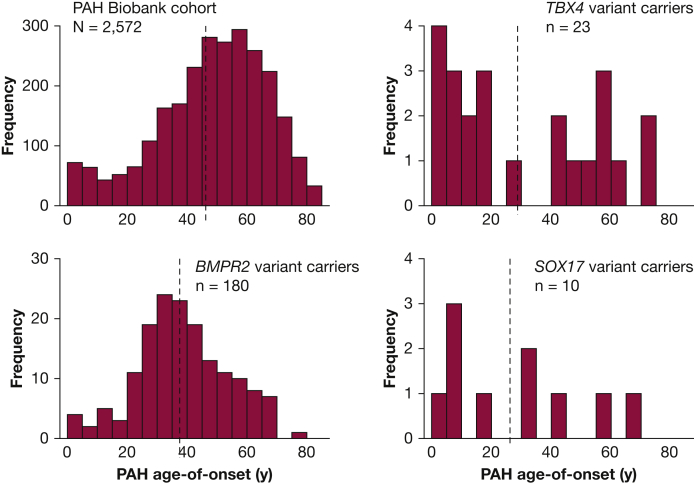
Figure 3.
Age distributions for cases with PAH from the National Biological Sample and Data Repository for Pulmonary Arterial Hypertension (N = 2,572). BMPR2, TBX4, and SOX17 variant carriers have younger mean age of onset than the whole cohort with significant enrichment of pediatric-onset cases among TBX4 variant carriers compared with the whole cohort. Dotted vertical lines indicate the group means. See Figure 1 legend for expansion of abbreviation.
The PAH genetics community has introduced the term hereditary PAH to include familial PAH (PAH that occurs in two or more family members) and PAH without a family history of PAH when there is a clear genetic diagnosis. Therefore, patients classified as IPAH at the time of diagnosis should be classified as hereditary PAH when a causal genetic variant is identified.
Role of De Novo Variants
De novo variants have emerged as an important class of genetic factors underlying rare diseases, especially early onset severe or lethal conditions14,18,21,22 because of strong negative selection decreasing reproductive fitness.23 In a cohort of 34 pediatric-onset IPAH cases for which we had samples from unaffected parents, we demonstrated a twofold enrichment of rare de novo variants in cases compared with an estimated background mutation rate (Table 2).7 The variants included both missense variants that change an amino acid with strong predictions of deleterious protein function and likely gene-disrupting (LGD) variants (frameshift, stop-gain, and canonical splice site), often causing protein truncation and nonsense-mediated decay leading to a state of haploinsufficiency (ie, loss of one functional copy of the gene). Among genes highly expressed in developing heart and lung, the enrichment was increased fourfold. All six of the LGD variants were identified in patients with an age of onset of < 5 years (Table 3).7 We now have additional data confirming the relative contribution of de novo variants in an expanded cohort of 124 trios with pediatric PAH probands, including both IPAH and PAH associated with other diseases (primarily APAH-CHD). The estimated fraction of pediatric PAH explained by de novo variants is approximately 15%. Some of the de novo variants occur in known risk genes (three in TBX4, two in BMPR2, one in ACVRL1, and one in ABCC8). However, the others occur in genes not previously implicated in PAH. Notably, some of the novel de novo variants occur in candidate genes with known or plausible roles in lung/vascular development. For example, AMOT encodes an angiostatin-binding protein involved in embryonic endothelial cell migration and tube formation and endothelial cell tight junctions and angiogenesis.24, 25, 26 KEAP1 regulates oxidative stress and apoptosis through protein interactions with NRF2 in murine vascular cells,27 and endothelial-specific deletion of NRF2 reduces endothelial cell sprouting in vivo.28 MAPK6 encodes ERK3, and mice carrying null alleles exhibit intrauterine pulmonary hypoplasia and early neonatal death.29 Although our data implicate a role for de novo variants in approximately 15% of pediatric PAH cases, larger trio cohorts will be required to develop a comprehensive list of PAH genes and confirm the role of individual genes with replication.
Table 2.
Enrichment of Rare De Novo Variants Among Pediatric-Onset Idiopathic Pulmonary Arterial Hypertension Trios (n = 36) Compared With an Estimated Background Mutation Rate
| Variant Type | No. Observed | No. Expected by Chance | Enrichment | P Value |
|---|---|---|---|---|
| SYN | 11 | 11.1 | 0.99 | 1 |
| Mis | 29 | 24.6 | 1.18 | .36 |
| D-Mis | 11 | 4.7 | 2.35 | .009 |
| LGD | 6 | 3.4 | 1.75 | .46 |
| D-Mis + LGD | 17 | 8.0 | 2.11 | .004 |
Bold font indicates statistically significant data. D-Mis = damaging missense predicted by MetaSVM; LGD = likely gene-disrupting (frameshift, stop-gain, and canonical splice site); Mis = all missense; SYN = synonymous.
Table 3.
Enrichment of De Novo LGD Variants in Very Early Onset (≤ 5 Years of Age) Compared With Older (> 5-18 Years of Age) Pediatric Patients
| Variant Type | Age ≤ 5 y (n = 16) | Age > 5-18 y (n = 18) | Enrichment | P Value |
|---|---|---|---|---|
| SYN | 2 | 8 | 3.6 | .12 |
| Mis | 10 | 16 | 1.4 | .44 |
| D-Mis | 3 | 6 | 1.8 | .51 |
| LGD | 6 | 0 | NA | .01 |
Bold font indicates statistically significant data. NA = not applicable. See Table 2 legend for expansion of other abbreviations.
BMPR2
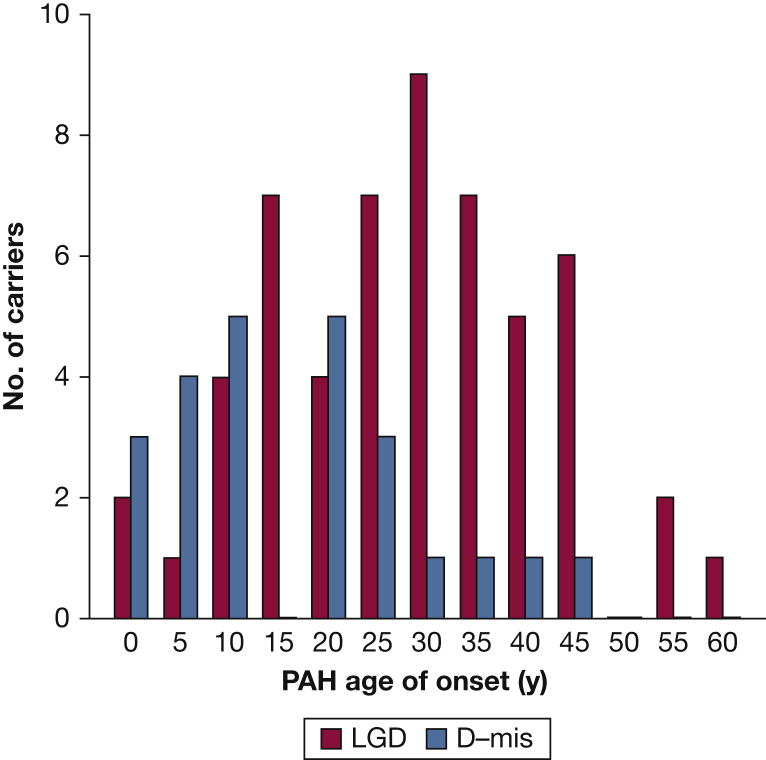
BMPR2 is a member of the transforming growth factor beta superfamily including transforming growth factor beta/bone morphogenetic protein ligands, receptors, accessory proteins, activins, and downstream signaling mediators (including mothers against decapentaplegic [SMAD] genes and NOTCH3). Rare deleterious variants in BMPR2 underlie approximately 70% of familial PAH (FPAH) and 10% to 20% of IPAH cases, with similar frequencies of BMPR2 variants in pediatric- vs adult-onset disease for both FPAH and IPAH.7,8 Overall, BMPR2 variant carriers have a younger mean age of onset and more severe PAH than noncarrier patients, at least in part because of impaired response to oxidative stress.30 However, BMPR2 variants are less frequent causes of APAH-CHD and have not been observed in PPHN. In a cohort of 258 APAH-CHD cases, only 7 cases (2.7%) carried rare deleterious BMPR2 variants.31 In the PAH Biobank—comprised of 4% FPAH, 43% IPAH, 48% PAH associated with other diseases, and 5% other PAH—only 12 of 119 of all BMPR2 carriers (10%) had PAH diagnoses other than FPAH/IPAH.8 Among 88 infants with PPHN, no rare deleterious variants in BMPR2 were identified.32 The wide distribution of deleterious variant locations across the BMPR2 gene has limited genotype-phenotype analyses with the sample sizes studied to date because of small numbers of individuals carrying any one variant. However, an analysis of mutation type—truncating or missense—showed earlier age of onset and decreased survival among carriers of missense compared with truncating variants.33 These data suggest that the presence of mutant/dysfunctional BMPR2 proteins arising from missense variants are more deleterious than haploinsufficiency of normal BMPR2 proteins. Similarly, mice carrying a Bmpr2 extracellular domain missense mutation developed more severe pulmonary hypertension in response to hypoxia or hypoxia with vascular endothelial growth factor inhibition than mice heterozygous for a Bmpr2 null allele.34 In the cohort of 412 pediatric- and adult-onset patients with PAH, there was a significant enrichment of deleterious missense variants in BMPR2 in patients with younger age of onset compared with LGD variant carriers (Fig 4),7 providing independent confirmation of the importance of missense variants in early onset disease. Therefore, in pediatric PAH, rare deleterious BMPR2 variants contribute primarily to FPAH/IPAH with deleterious missense variants associated with increased disease severity.
Figure 4.
Age distributions of BMPR2 LGD or predicted D-mis variant carriers from a cohort of 412 patients with pediatric- and adult-onset PAH. There was significant enrichment of D-mis variants among patients with younger age of onset compared with LGD variant carriers (83 total variant carriers). D-mis = damaging missense; LGD = likely gene-disrupting. See Figure 1 legend for expansion of other abbreviation.
TBX4
TBX4 is a transcription factor in the T-box gene family expressed in the developing atrium of the heart, limb buds, and mesenchyme of lung and trachea, with important roles in limb development and lung growth and branching.35 Rare deleterious TBX4 variants have been associated with small patella syndrome36 and PAH.7,37 TBX4 was first suggested as a candidate PAH risk gene because of its location on chromosome 17q23.1-23.2, where microdeletions were associated with severe neurodevelopmental delays and pulmonary hypertension.38,39 Kerstjens-Frederikse et al37 sequenced TBX2 and TBX4, both located within the 17q23.1-23.2 deletion, and identified three rare TBX4 variants among 49 adults with PAH and no rare variants in TBX2. In a small European cohort of 66 pediatric PAH cases, three of 40 FPAH/IPAH cases carried rare deleterious TBX4 variants40 compared with three of 136 adult carriers in a Spanish PAH cohort.41 In a larger cohort of 412 pediatric- and adult-onset FPAH/IPAH cases, we reported rare deleterious TBX4 variants in 13 cases with a significant enrichment of variants among pediatric-onset (12 of 155) compared with adult-onset (one of 257) patients.7 Furthermore, the mean age of onset was 20 years younger for TBX4 variant carriers compared with BMPR2 carriers. In the PAH Biobank, variants in TBX4 were the second most common genetic cause of PAH and accounted for approximately 1% of cases.8 Although 13 of 23 TBX4 variant carriers had a diagnosis of IPAH, the other diagnoses included APAH-CHD, PAH associated with connective tissue disease, FPAH, and PAH because of dietary toxin exposure. Age of onset for the TBX4 carriers exhibited a bimodal distribution with significant enrichment of variants among pediatric-onset cases (Fig 2). Interestingly, a clinical and histologic analysis of 19 children carrying rare deleterious TBX4 variants revealed a high frequency of severe developmental defects of the lung, skeleton, and heart.42 Ten of the infants presented with PPHN which resolved; however, the children were subsequently diagnosed with PAH later in infancy or childhood. This evidence indicates that TBX4 is especially important in pediatric-onset PAH, with rare deleterious variants causing a variety of disease subclasses and predicting disease recurrence in young patients initially diagnosed with PPHN.
SOX17
SOX17 is a highly constrained gene encoding a transcription factor involved in Wnt/β-catenin and Notch signaling during development.43 Genetic studies in mice show that Sox17 is required for correct development and function of the pulmonary vascular tree. Endothelial-specific inactivation of Sox17 leads to impaired arterial specification and embryonic death or, with conditional postnatal inactivation, arterial-venous malformations.44 Deletion in mesenchymal progenitor cells causes abnormal pulmonary vascular morphogenesis resulting in postnatal cardiopulmonary dysfunction and juvenile death.45 Moreover, in an elegant endothelial lineage tracing study in mice, Liu et al46 recently demonstrated that transcriptional activation of Sox17 via hypoxia-induced factor 1α, leads to upregulation of cyclin-E1 and endothelial regeneration in response to lung injury. Therefore, there are multiple mechanisms through which defective or deficient SOX17 could result in developmental cardiopulmonary defects or impaired response to hypoxic injury.
We identified SOX17 as a candidate risk gene for PAH using exome sequencing data in a cohort of 256 patients with APAH-CHD.31 Using a case-control gene-based association test, SOX17 was the only gene, out of approximately 18,000 genes, to reach genome-wide significance. Three of the top associated genes (BZW2, FTSJ3, and BAZ1B) were putative SOX17 transcriptional targets,47 and enrichment analysis of a gene set including 1,947 putative SOX17 target genes revealed enrichment of rare missense variants in the patient cohort, suggesting that multiple genes in the SOX17 pathway way be important in APAH-CHD. Most of these genes are expressed in pulmonary arterial endothelial cells or developing heart, and 28% (42 of 149) are expressed in the top quartile in both tissues/cell types. Pathway enrichment analysis showed that the SOX17 target genes with deleterious variants are overrepresented in developmental processes, transmembrane transport of small molecules, ion homeostasis, and extracellular matrix interactions. The association signal for SOX17 was because of LGD and deleterious missense variants carried by 10 patients with APAH-CHD, and seven of 10 of these patients had pediatric-onset disease. Screening of the separate cohort of 412 patients with FPAH/IPAH identified an additional three carriers (two pediatric carriers and one adult carrier).31 Rare deleterious variants have been identified in two additional IPAH cohorts, comprised mostly of adults. Genome sequencing data from the UK NIHR BioResource – Rare Diseases PAH Study identified nine of 1,038 IPAH SOX17 variant carriers48 and a candidate gene analysis of a Japanese cohort found four (three unrelated) of 140 FPAH/IPAH SOX17 variant carriers.49 In the PAH Biobank, rare deleterious SOX17 variants were identified in 10 of 2,572 patients (six with IPAH, two with APAH-CHD, one with PAH associated with portopulmonary disease, and one with PAH because of dietary toxin exposure).8 The mean age of onset for carriers in the PAH Biobank was 26 years, markedly younger than the overall cohort (48 years) of BMPR2 variant carriers (38 years) (Fig 3). Notably, 15 of 18 rare missense variants carried by patients from all five cohorts are located in the high mobility group (DNA binding) domain of the SOX17 protein. The high mobility group domain is evolutionally conserved, down to unicellular yeast species, and is essential for target-specific transcriptional control.43,50 Based on these data, we estimate that rare deleterious variants in SOX17 contribute to approximately 7% of pediatric-onset PAH (19 of 273), especially APAH-CHD, compared with approximately 0.4% (13 of 3,455) of adult-onset PAH. In addition, common single nucleotide polymorphisms in a putative endothelial-acting enhancer region of SOX17 have been associated with PAH,51 suggesting that variation in SOX17 gene expression may increase the risk for developing PAH or other vascular endothelium-related diseases more commonly.
Genetic Ancestry
The role of specific genes in PAH is likely heterogeneous across genetic ancestries. The results of genetic studies predominantly in individuals of European ancestry may not be generalizable to all other populations. A pediatric study of an Asian cohort revealed a higher carrier frequency of ALK1/ACVRLI variants (7 of 54, 12.9%)52 compared with studies of predominantly Europeans. A recent association analysis involving 331 IPAH cases and 10,508 control subjects of Asian ancestry identified BMP9/GDF2 as a significant risk gene in this population, second in frequency to BMPR2.53 Among 22 carriers of rare deleterious GDF2 variants, three had pediatric-onset disease accounting for 5.2% of the 57 pediatric cases. In a case study of a 5-year-old boy of Hispanic ancestry, a homozygous loss of function BMP9 variant, c.76C>T;p.Gln26Ter, was identified.54 The gnomAD population database (gnomad.broadinstitute.org) contains only two heterozygous counts of this allele, both of Latino ancestry, suggesting that this might be an ancestry-specific allele. Clearly, larger studies of children with greater diversity are needed to define ancestral-specific genetic factors and their overall role in pediatric-onset PAH.
PAH in the Setting of Other Developmental Diseases
Pediatric PAH is intrinsically linked with lung vascular development and growth and, as such, is frequently associated with other developmental diseases including Down syndrome, congenital heart disease, and congenital diaphragmatic hernia.55,56 Histopathologic studies have identified abnormal lung development and lung hypoplasia as common features of these diseases.55,57 Although the mechanism(s) for impaired lung development are not known, increased expression of antiangiogenic genes may contribute in Down syndrome.58,59 Decreased lung vascular and alveolar growth predispose to vascular injury during susceptible periods of growth and adaptation. A retrospective study of pediatric patients with PAH suggested that patients with PAH with Down syndrome may be less responsive to PAH treatments than patients without Down syndrome.60 Individuals with de novo monogenic causes of congenital diaphragmatic hernia have a higher risk of PAH (W. K. C., unpublished data, October 2019). With future studies, we should be able to learn more about which genes drive the phenotype and what the pleiotropic effects of these genes are on lung development beyond the impact of compression of the developing lung secondary to having abdominal contents in the chest. Important questions regarding risk remain unanswered. Which patients with associated developmental disorders are at risk for developing PAH? How much of the PAH clinical phenotype is reversible and for how long? Would early intervention prevent PAH in these patients? Genetic identification of individuals with de novo or rare inherited pathogenic variants with longitudinal follow-up of patients grouped by genetic etiology may provide answers to these questions.
Role of Other Omics in PAH
In addition to DNA sequencing to identify genetic etiologies, other omics including RNA sequencing, metabolomics, and proteomics can provide valuable information to improve predictions of who is at risk for disease, define endophenotypes, and guide effective therapies.61 For example, West et al62 performed RNA sequencing of peripheral blood lymphocytes derived from BMPR2 variant carriers with and without PAH to identify transcript patterns relevant to disease penetrance. Rhodes et al63 used metabolomics to identify circulating metabolites that distinguish PAH cases from healthy control subjects, predict outcomes among PAH cases, and monitor metabolite levels over time to determine whether correction could affect outcomes. Hemnes et al64 used transcriptomics to identify RNA expression patterns predictive of vasodilator-responsiveness among patients with PAH. These studies highlight the promise of other omics in improving predictions of PAH risk, diagnosis, classification, drug responsiveness, and prognosis. However, such studies have not yet been conducted in children. Delineation of genetic diagnoses followed by detailed omic phenotyping of patients grouped by diagnosis may limit the amount of biologic sampling required of pediatric patients.
Summary
Pediatric-onset PAH differs from adult-onset PAH in many important aspects including clinical presentation, genetic burden, and specific genes involved. Rare genetic factors contribute to approximately 35% of pediatric-onset IPAH compared with approximately 11% of adult-onset IPAH. De novo variants and rare deleterious variants in BMPR2, TBX4, and SOX17 currently explain most of the known genetic burden in pediatric PAH. Because of these differences, genetic information relevant to pediatric PAH cannot be extrapolated from adults. Large cohorts of patients with pediatric-onset PAH are necessary to identify the unique etiologic genes for PAH in children, and the natural history and response to therapy. Inclusion of diverse populations will allow the identification of ancestry-specific genetic factors to improve diagnostic yield and guide patient care. The yield of gene discovery efforts will be much greater in children, and the identification of novel genes will provide the framework for better understanding of pulmonary vascular biology and PAH. We propose a genetics-first approach to stratify pediatric PAH followed by molecular phenomics to define endophenotypes that can be clustered with known genetic etiologies to identify additional novel genetic etiologies. Once the molecular genetic subtypes are defined, detailed clinical phenotyping and longitudinal studies including differential response to treatment and prognosis will help to inform future clinical management. Identification of new causal genes will illuminate underlying disease pathophysiology and mechanisms for therapeutic targets for children and adults. Given the increased burden of genetic etiologies in children, genomic studies in children with a trio design to identify inherited and de novo variants will yield many more novel targets than similarly powered studies in adults. Although data have begun to emerge from studies of children with PAH, there is still much more to be learned.
Acknowledgments
Financial/nonfinancial disclosures: None declared.
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Footnotes
FUNDING/SUPPORT: This study was funded by the JPB Foundation[Grant U01HL125218] to Dr Chung.
References
- 1.Li L., Jick S., Breitenstein S., Hernandez G., Michel A., Vizcaya D. Pulmonary arterial hypertension in the USA: an epidemiological study in a large insured pediatric population. Pulm Circ. 2017;7(1):126–136. doi: 10.1086/690007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humbert M., Sitbon O., Chaouat A. Pulmonary arterial hypertension in France: results from a national registry. Am J Respir Crit Care Med. 2006;173(9):1023–1030. doi: 10.1164/rccm.200510-1668OC. [DOI] [PubMed] [Google Scholar]
- 3.Hansmann G. Pulmonary Hypertension in infants, children, and young adults. J Am Coll Cardiol. 2017;69(20):2551–2569. doi: 10.1016/j.jacc.2017.03.575. [DOI] [PubMed] [Google Scholar]
- 4.Rosenzweig E.B., Abman S.H., Adatia I. Paediatric pulmonary arterial hypertension: updates on definition, classification, diagnostics and management. Eur Respir J. 2019;53(1) doi: 10.1183/13993003.01916-2018. pii: 1801916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saji T. Update on pediatric pulmonary arterial hypertension. Differences and similarities to adult disease. Circ J. 2013;77(11):2639–2650. doi: 10.1253/circj.cj-13-1180. [DOI] [PubMed] [Google Scholar]
- 6.Barst R.J., McGoon M.D., Elliott C.G., Foreman A.J., Miller D.P., Ivy D.D. Survival in childhood pulmonary arterial hypertension: insights from the registry to evaluate early and long-term pulmonary arterial hypertension disease management. Circulation. 2012;125(1):113–122. doi: 10.1161/CIRCULATIONAHA.111.026591. [DOI] [PubMed] [Google Scholar]
- 7.Zhu N., Gonzaga-Jauregui C., Welch C.L. Exome sequencing in children with pulmonary arterial hypertension demonstrates differences compared with adults. Circ Genom Precis Med. 2018;11(4) doi: 10.1161/CIRCGEN.117.001887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N., Pauciulo M.W., Welch C.L. Novel risk genes and mechanisms implicated by exome sequencing of 2,572 individuals with pulmonary arterial hypertension. Genome Med. 2019;11(1):69–85. doi: 10.1186/s13073-019-0685-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humpl T., Berger R.M.F., Austin E.D. Treatment initiation in paediatric pulmonary hypertension: insights from a multinational registry. Cardiol Young. 2017;27(6):1123–1132. doi: 10.1017/S1047951116002493. [DOI] [PubMed] [Google Scholar]
- 10.Abman S.H., Hansmann G., Archer S.L. Pediatric pulmonary hypertension: guidelines from the American Heart Association and American Thoracic Society. Circulation. 2015;132(21):2037–2099. doi: 10.1161/CIR.0000000000000329. [DOI] [PubMed] [Google Scholar]
- 11.Hansmann G., Apitz C., Abdul-Khaliq H. Executive summary. Expert consensus statement on the diagnosis and treatment of paediatric pulmonary hypertension. The European Paediatric Pulmonary Vascular Disease Network, endorsed by ISHLT and DGPK. Heart. 2016;102(suppl 2):ii86–ii100. doi: 10.1136/heartjnl-2015-309132. [DOI] [PubMed] [Google Scholar]
- 12.Hemnes A.R., Beck G.J., Newman J.H. PVDOMICS: a multi-center study to improve understanding of pulmonary vascular disease through phenomics. Circ Res. 2017;121(10):1136–1139. doi: 10.1161/CIRCRESAHA.117.311737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaidi S., Choi M., Wakimoto H. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homsy J., Zaidi S., Shen Y. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350(6265):1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin S.C., Homsy J., Zaidi S. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu L., Sawle A.D., Wynn J. Increased burden of de novo predicted deleterious variants in complex congenital diaphragmatic hernia. Hum Mol Genet. 2015;24(16):4764–4773. doi: 10.1093/hmg/ddv196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longoni M., High F.A., Qi H. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum Genet. 2017;136(6):679–691. doi: 10.1007/s00439-017-1774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi H., Yu L., Zhou X. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS Genet. 2018;14(12) doi: 10.1371/journal.pgen.1007822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chida A., Shintani M., Nakayama T. Missense mutations of the BMPR1B (ALK6) gene in childhood idiopathic pulmonary arterial hypertension. Circ J. 2012;76(6):1501–1508. doi: 10.1253/circj.cj-11-1281. [DOI] [PubMed] [Google Scholar]
- 20.Chida A., Shintani M., Matsushita Y. Mutations of NOTCH3 in childhood pulmonary arterial hypertension. Mol Genet Genomic Med. 2014;2(3):229–239. doi: 10.1002/mgg3.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epi4K Consortium; Epilepsy Phenome/Genome Project. Allen A.S. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin Z.B., Wu J., Huang X.F. Trio-based exome sequencing arrests de novo mutations in early-onset high myopia. Proc Natl Acad Sci U S A. 2017;114(16):4219–4224. doi: 10.1073/pnas.1615970114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veltman J.A., Brunner H.G. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13(8):565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 24.Troyanovsky B., Levchenko T., Mansson G., Matvijenko O., Holmgren L. Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J Cell Biol. 2001;152(6):1247–1254. doi: 10.1083/jcb.152.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmgren L., Ambrosino E., Birot O. A DNA vaccine targeting angiomotin inhibits angiogenesis and suppresses tumor growth. Proc Natl Acad Sci U S A. 2006;103(24):9208–9213. doi: 10.1073/pnas.0603110103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y., Vertuani S., Nystrom S. Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ Res. 2009;105(3):260–270. doi: 10.1161/CIRCRESAHA.109.195156. [DOI] [PubMed] [Google Scholar]
- 27.Ungvari Z., Bagi Z., Feher A. Resveratrol confers endothelial protection via activation of the antioxidant transcription factor Nrf2. Am J Physiol Heart Circ Physiol. 2010;299(1):H18–H24. doi: 10.1152/ajpheart.00260.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei Y., Gong J., Thimmulappa R.K., Kosmider B., Biswal S., Duh E.J. Nrf2 acts cell-autonomously in endothelium to regulate tip cell formation and vascular branching. Proc Natl Acad Sci U S A. 2013;110(41):E3910–E3918. doi: 10.1073/pnas.1309276110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klinger S., Turgeon B., Levesque K., Wood G.A., Aagaard-Tillery K.M., Meloche S. Loss of Erk3 function in mice leads to intrauterine growth restriction, pulmonary immaturity, and neonatal lethality. Proc Natl Acad Sci U S A. 2009;106(39):16710–16715. doi: 10.1073/pnas.0900919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyer L.A., Pi X., Patterson C. The role of BMPs in endothelial cell function and dysfunction. Trends Endocrinol Metab. 2014;25(9):472–480. doi: 10.1016/j.tem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu N., Welch C.L., Wang J. Rare variants in SOX17 are associated with pulmonary arterial hypertension with congenital heart disease. Genome Med. 2018;10(1):56. doi: 10.1186/s13073-018-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byers H.M., Dagle J.M., Klein J.M. Variations in CRHR1 are associated with persistent pulmonary hypertension of the newborn. Pediatr Res. 2012;71(2):162–167. doi: 10.1038/pr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Austin E.D., Phillips J.A., Cogan J.D. Truncating and missense BMPR2 mutations differentially affect the severity of heritable pulmonary arterial hypertension. Respir Res. 2009;10:87. doi: 10.1186/1465-9921-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frump A.L., Datta A., Ghose S., West J., de Caestecker M.P. Genotype-phenotype effects of Bmpr2 mutations on disease severity in mouse models of pulmonary hypertension. Pulm Circ. 2016;6(4):597–607. doi: 10.1086/688930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arora R., Metzger R.J., Papaioannou V.E. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8(8) doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bongers E.M., Duijf P.H., van Beersum S.E. Mutations in the human TBX4 gene cause small patella syndrome. Am J Hum Genet. 2004;74(6):1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerstjens-Frederikse W.S., Bongers E.M.H.F., Roofthooft M.T.R. TBX4 mutations (small patella syndrome) are associated with childhood-onset pulmonary arterial hypertension. J Med Genet. 2013;50(8):500–506. doi: 10.1136/jmedgenet-2012-101152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballif B.C., Theisen A., Rosenfeld J.A. Identification of a recurrent microdeletion at 17q23.1q23.2 flanked by segmental duplications associated with heart defects and limb abnormalities. Am J Hum Genet. 2010;86(3):454–461. doi: 10.1016/j.ajhg.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nimmakayalu M., Major H., Sheffield V. Microdeletion of 17q22q23.2 encompassing TBX2 and TBX4 in a patient with congenital microcephaly, thyroid duct cyst, sensorineural hearing loss, and pulmonary hypertension. Am J Med Genet A. 2011;155A(2):418–423. doi: 10.1002/ajmg.a.33827. [DOI] [PubMed] [Google Scholar]
- 40.Levy M., Eyries M., Szezepanski I. Genetic analyses in a cohort of children with pulmonary hypertension. Eur Respir J. 2016;48(4):1118–1126. doi: 10.1183/13993003.00211-2016. [DOI] [PubMed] [Google Scholar]
- 41.Navas P., Tenorio J., Quezada C.A. Molecular analysis of BMPR2, TBX4, and KCNK3 and genotype-phenotype correlations in Spanish patients and families with idiopathic and hereditary pulmonary arterial hypertension. Rev Esp Cardiol (Engl Ed) 2016;69(11):1011–1019. doi: 10.1016/j.rec.2016.03.029. [DOI] [PubMed] [Google Scholar]
- 42.Galambos C., Mullen M.P., Shieh J.T. Phenotype characterisation of TBX4 mutation and deletion carriers with neonatal and pediatric pulmonary hypertension. Eur Respir J. 2019;54(2) doi: 10.1183/13993003.01965-2018. pii: 1801965. [DOI] [PubMed] [Google Scholar]
- 43.Francois M., Koopman P., Beltrame M. SoxF genes: key players in the development of the cardio-vascular system. Int J Biochem Cell Biol. 2010;42(3):445–448. doi: 10.1016/j.biocel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 44.Corada M., Orsenigo F., Morini M.F. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat Commun. 2013;4:2609. doi: 10.1038/ncomms3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lange A.W., Haitchi H.M., LeCras T.D. Sox17 is required for normal pulmonary vascular morphogenesis. Dev Biol. 2014;387(1):109–120. doi: 10.1016/j.ydbio.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu M., Zhang L., Marsboom G. Sox17 is required for endothelial regeneration following inflammation-induced vascular injury. Nat Commun. 2019;10(1):2126. doi: 10.1038/s41467-019-10134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lachmann A., Xu H., Krishnan J., Berger S.I., Mazloom A.R., Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graf S., Haimel M., Bleda M. Identification of rare sequence variation underlying heritable pulmonary arterial hypertension. Nat Commun. 2018;9(1):1416. doi: 10.1038/s41467-018-03672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hiraide T., Kataoka M., Suzuki H. SOX17 mutations in Japanese patients with pulmonary arterial hypertension. Am J Respir Crit Care Med. 2018;198(9):1231–1233. doi: 10.1164/rccm.201804-0766LE. [DOI] [PubMed] [Google Scholar]
- 50.Hou L., Srivastava Y., Jauch R. Molecular basis for the genome engagement by Sox proteins. Semin Cell Dev Biol. 2017;63:2–12. doi: 10.1016/j.semcdb.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Rhodes C.J., Batai K., Bleda M. Genetic determinants of risk in pulmonary arterial hypertension: international genome-wide association studies and meta-analysis. Lancet Respir Med. 2019;7(3):227–238. doi: 10.1016/S2213-2600(18)30409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chida A., Shintani M., Yagi H. Outcomes of childhood pulmonary arterial hypertension in BMPR2 and ALK1 mutation carriers. Am J Cardiol. 2012;110(6):586–593. doi: 10.1016/j.amjcard.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 53.Wang X.J., Lian T.Y., Jiang X. Germline BMP9 mutation causes idiopathic pulmonary arterial hypertension. Eur Respir J. 2019;53(3) doi: 10.1183/13993003.01609-2018. pii: 1801609. [DOI] [PubMed] [Google Scholar]
- 54.Wang G., Fan R., Ji R. Novel homozygous BMP9 nonsense mutation causes pulmonary arterial hypertension: a case report. BMC Pulm Med. 2016;16:17. doi: 10.1186/s12890-016-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abman S.H., Baker C., Gien J., Mourani P., Galambos C. The Robyn Barst Memorial Lecture: differences between the fetal, newborn, and adult pulmonary circulations: relevance for age-specific therapies (2013 Grover Conference series) Pulm Circ. 2014;4(3):424–440. doi: 10.1086/677371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bush D., Galambos C., Ivy D.D., Abman S.H., Wolter-Warmerdam K., Hickey F. Clinical characteristics and risk factors for developing pulmonary hypertension in children with Down syndrome. J Pediatr. 2018;202:212–219.e212. doi: 10.1016/j.jpeds.2018.06.031. [DOI] [PubMed] [Google Scholar]
- 57.Bush D., Abman S.H., Galambos C. Prominent intrapulmonary bronchopulmonary anastomoses and abnormal lung development in infants and children with Down syndrome. J Pediatr. 2017;180:156–162.e151. doi: 10.1016/j.jpeds.2016.08.063. [DOI] [PubMed] [Google Scholar]
- 58.Sanchez O., Dominguez C., Ruiz A. Angiogenic gene expression in Down syndrome fetal hearts. Fetal Diagn Ther. 2016;40(1):21–27. doi: 10.1159/000441356. [DOI] [PubMed] [Google Scholar]
- 59.Galambos C., Minic A.D., Bush D. Increased lung expression of anti-angiogenic factors in Down syndrome: potential role in abnormal lung vascular growth and the risk for pulmonary hypertension. PLoS One. 2016;11(8) doi: 10.1371/journal.pone.0159005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beghetti M., Rudzinski A., Zhang M. Efficacy and safety of oral sildenafil in children with Down syndrome and pulmonary hypertension. BMC Cardiovasc Disord. 2017;17(1):177. doi: 10.1186/s12872-017-0569-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hemnes A.R. Using omics to understand and treat pulmonary vascular disease. Front Med (Lausanne) 2018;5:157. doi: 10.3389/fmed.2018.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West J., Cogan J., Geraci M. Gene expression in BMPR2 mutation carriers with and without evidence of pulmonary arterial hypertension suggests pathways relevant to disease penetrance. BMC Med Genomics. 2008;1:45. doi: 10.1186/1755-8794-1-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rhodes C.J., Ghataorhe P., Wharton J. Plasma metabolomics implicates modified transfer RNAs and altered bioenergetics in the outcomes of pulmonary arterial hypertension. Circulation. 2017;135(5):460–475. doi: 10.1161/CIRCULATIONAHA.116.024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hemnes A.R., Trammell A.W., Archer S.L. Peripheral blood signature of vasodilator-responsive pulmonary arterial hypertension. Circulation. 2015;131(4):401–409. doi: 10.1161/CIRCULATIONAHA.114.013317. [DOI] [PMC free article] [PubMed] [Google Scholar]