Abstract
Newly synthesized major histocompatibility complex (MHC) class I proteins are stabilized in the endoplasmic reticulum (ER) by binding 8–10-mer-long self-peptide antigens that are provided by transporter associated with antigen processing (TAP). These MHC class I:peptide complexes then exit the ER and reach the plasma membrane, serving to sustain the steady-state MHC class I expression on the cell surface. A novel subset of MHC class I molecules that preferentially bind lipid-containing ligands rather than conventional peptides was recently identified. The primate classical MHC class I allomorphs, Mamu-B*098 and Mamu-B*05104, are capable of binding the N-myristoylated 5-mer (C14-Gly-Gly-Ala-Ile-Ser) or 4-mer (C14-Gly-Gly-Ala-Ile) lipopeptides derived from the N-myristoylated SIV Nef protein, respectively, and of activating lipopeptide antigen-specific cytotoxic T lymphocytes. We herein demonstrate that Mamu-B*098 samples lysophosphatidylethanolamine and lysophosphatidylcholine containing up to a C20 fatty acid in the ER. The X-ray crystal structures of Mamu-B*098 and Mamu-B*05104 complexed with lysophospholipids at high resolution revealed that the B and D pockets in the antigen-binding grooves of these MHC class I molecules accommodate these lipids through a monoacylglycerol moiety. Consistent with the capacity to bind cellular lipid ligands, these two MHC class I molecules did not require TAP function for cell-surface expression. Collectively, these results indicate that peptide- and lipopeptide-presenting MHC class I subsets use distinct sources of endogenous ligands.
Keywords: major histocompatibility complex (MHC), MHC class I, lysophospholipid, antigen presentation, human immunodeficiency virus (HIV), structural biology, adaptive immunity, lipopeptide, Mamu-B, self-peptide, transporter associated with antigen processing (TAP)
Introduction
Major histocompatibility complex (MHC)2 class I molecules play a pivotal role in host defenses against viral infections by allowing cytotoxic T lymphocytes (CTLs) to monitor the cell-surface display of viral protein–derived peptide fragments and eliminate virus-infected cells without affecting uninfected cells (1). The sustained expression of self-peptide–bound MHC class I molecules is also critical for healthy cells because natural killer (NK) cells primarily target those with reduced MHC class I expression (2). Thus, the quality and quantity of cell surface–expressed MHC class I molecules are carefully controlled because these killer-type lymphocytes may be harmful to healthy cells. The biogenesis of MHC class I molecules occurs in the endoplasmic reticulum (ER), at which newly synthesized MHC class I heavy chains associate noncovalently with their partner, β2-microglobulin (β2m) (3–5). These unstable heterodimers are incorporated into the peptide-loading complex composed of tapasin, ERp57, calreticulin, and the ER membrane–associated ABC transporter, termed transporter associated with antigen processing (TAP) (6). MHC class I molecules are subsequently stabilized by binding peptide antigens (Ags), which are delivered from the cytosol through TAP. Only the fully assembled trimer complex exits the ER and reaches the cell surface, whereas free heavy chains as well as those associated with β2m, but still peptide-unbound, are retained in the ER (7). Thus, the cell-surface expression of MHC class I molecules is markedly impaired in cells defective in TAP (8, 9).
MHC class I molecules capture typically 8–10-mer-long peptides in their Ag-binding groove composed of six (A–F) pockets (10–12). The first, second, and C-terminal amino acid residues of the peptide ligand generally anchor at the A, B, and F pockets, respectively, by establishing mainly hydrogen bond interactions that serve to stabilize MHC class I complexes. This well-known molecular model depicting MHC class I–peptide interactions is applicable to most peptide-presenting MHC class I molecules; however, there may be some scope for further consideration because a distinct subset of MHC class I molecules has been identified that bind lipidated short peptides (lipopeptides) rather than long peptides (13–16). The Mamu-B*098 allomorph of the rhesus classical MHC class I family has been shown to bind the N-myristoylated 5-mer lipopeptide (C14-Gly-Gly-Ala-Ile-Ser) derived from the N-myristoylated retroviral Nef protein and present it to CTLs. The C14 fatty acid (myristic acid) anchored at the large, hydrophobic B pocket of Mamu-B*098 by establishing numerous intermolecular van der Waals (VDW) interactions, whereas the C-terminal Ser residue was accommodated in the small F pocket. On the other hand, the A pocket of Mamu-B*098 was partially collapsed by some bulky amino acid residues at the A/B pocket channel, leaving the A pocket dysfunctional (17). Collectively, these structural features indicate that Mamu-B*098 is unable to bind conventional long peptides in a similar manner to known MHC class I molecules and also that Mamu-B*098 samples a distinct chemical class of endogenous ligands rather than long peptides in the ER.
In the present study, we demonstrated that a soluble form of Mamu-B*098 expressed in mammalian cells associated with lysophospholipid species. We provide X-ray crystallographic evidence to show that lysophospholipids sit in the Ag-binding groove of Mamu-B*098 as ligands. The monoacylglycerol moiety of bound lysophospholipids appears to function as an anchor that is accommodated in the large, hydrophobic B pocket and shallow D pocket. Furthermore, an additional crystal structure of the second lipopeptide-presenting MHC class I allomorph, Mamu-B*05104, complexed with lysophosphatidylcholine demonstrated that Mamu-B*05104 is also capable of accommodating lysophospholipids through the B and D pockets, suggesting that lysophospholipids are shared endogenous ligands among lipopeptide-presenting MHC class I molecules. Consistent with these results, in contrast to conventional peptide-presenting MHC class I molecules, the surface expression of Mamu-B*098 as well as Mamu-B*05104 is completely unaffected in TAP-deficient cells. The ability to bind lysophospholipids and completely TAP-independent expression may be unique to lipopeptide-presenting MHC class I molecules and have never been reported for these presenting peptides.
Results
Identification of lysophospholipid species as cellular ligands for Mamu-B*098
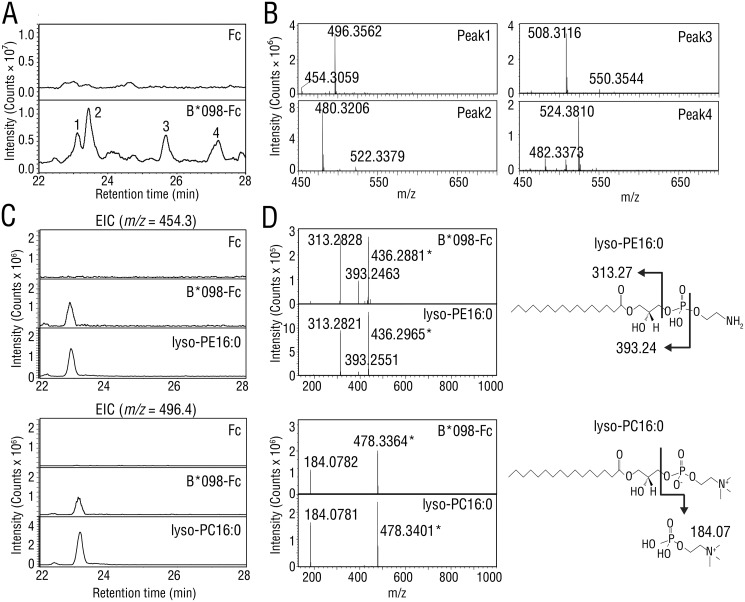
Mamu-B*098 is equipped with a voluminous, hydrophobic B pocket structure that appears to discriminate it from peptide-presenting MHC class I molecules. Due to the essential role of the B pocket in ligand binding (18), we predicted that Mamu-B*098 may favorably sample lipidic ligands in the ER. To search for cellular ligands with a potential to bind Mamu-B*098, the Mamu-B*098 ectodomain-IgG Fc fusion protein (B*098-Fc) as well as the control IgG Fc fragment (Fc) were expressed in Expi293F cells by transfection, and secreted recombinant proteins were purified. Ligands extracted in acid methanol from B*098-Fc and Fc proteins were analyzed by LC-MS/MS. Total ion chromatography of the B*098-Fc extract exhibited four specific peaks (Fig. 1A (bottom), indicated with Arabic numerals) that were undetectable with the Fc extract (Fig. 1A, top) at a retention time of 22–28 min. Mass spectra for each peak revealed ion species with m/z ranging between 450 and 550, and pairs of ion species with a 42 mass unit difference were detected for all peaks (Fig. 1B). By using lipid databases (19), we found that pairs of ion species were reasonably accounted for only by lysophosphatidylethanolamine (lyso-PE) and lysophosphatidylcholine (lyso-PC) with a given acyl chain, the exact mass of which differed by 42.05 (i.e. the m/z values of the ion pairs at peaks 1, 2, 3, and 4 strongly corresponded to lyso-PE and lyso-PC with C16:0, C18:1, C20:1, and C18:0 acyl chains, respectively). We compared the m/z 454.3 and m/z 496.4 signals at peak 1 with the standard preparation of lyso-PE16:0 and lyso-PC16:0, respectively, and found that these signals were identical to those of the standards for the retention time as well as MS/MS fragmentation, indicating that peak 1 was mainly composed of lyso-PE16:0 and lyso-PC16:0 (Fig. 1 (C and D), left panels). Furthermore, unique fragment ions of m/z 313.3 for lyso-PE16:0 and m/z 184.1 for lyso-PC16:0 were observed (Fig. 1D). These ions were presumably derived from the monoacylglycerol fragment of lyso-PE and the headgroup of lyso-PC, respectively, and the corresponding ions were also detected in peaks 2–4 (Fig. S1). This fragmentation is often regarded as a hallmark for the identification of lyso-PE and lyso-PC (Fig. 1D, right panels) (20–22). Therefore, lyso-PE and lyso-PC were novel ligand candidates of MHC class I molecules, to which the soluble form of Mamu-B*098 bound in the secretory pathway.
Figure 1.
LC-MS/MS analysis of bound ligands in soluble Mamu-B*098. A, samples extracted from the Fc protein (top) or B*098-Fc protein (bottom) were injected and separated in reversed-phase HPLC using a C18 column. Total ion chromatograms are shown at a retention time of 22–28 min. Four specific peaks that were undetectable with the Fc sample are indicated with Arabic numbers. B, MS spectra obtained at each peak in A are shown for the m/z range of 450–700. C and D, standard preparations of lyso-PE16:0 (top panels) and lyso-PC16:0 (bottom panels) were analyzed with Fc and B*098-Fc proteins for comparison. Extracted ion chromatograms (EIC) are shown at m/z 454.3 for lyso-PE16:0 and m/z 496.4 for lyso-PC16:0, respectively (C). MS/MS spectra at a retention time of 23.1 min are shown, and the structures and MS/MS fragmentation of lyso-PE16:0 and lyso-PC16:0 are also illustrated (D). Asterisks, dehydrated ions.
Lysophospholipids are anchored at B and D pockets through only a monoacylglycerol
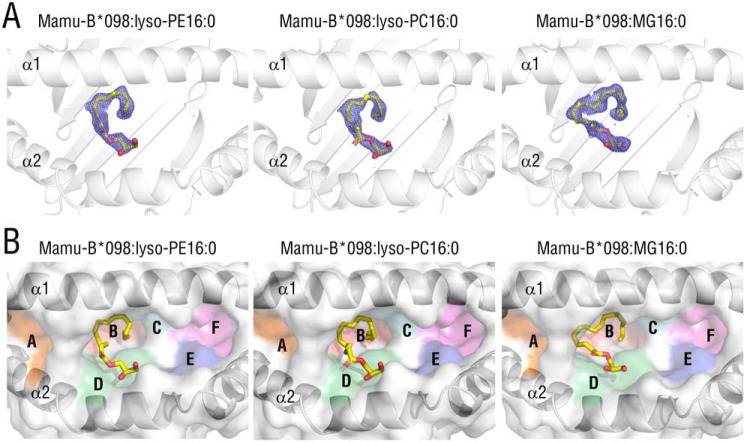
Because lysophospholipid species lack amino acid residues, these lipid ligands may be accommodated in the Ag-binding groove of Mamu-B*098 in a manner distinct from that of any known MHC class I:peptide or MHC class I:lipopeptide complexes. To confirm whether lysophospholipids are genuine ligands of Mamu-B*098 and clarify the binding mode of Mamu-B*098:lysophospholipid complexes, we produced the ectodomain of the Mamu-B*098 heavy chain and monkey β2m in Escherichia coli and purified them as inclusion bodies for in vitro refolding with lyso-PE16:0 or lyso-PC16:0. We elucidated X-ray crystal structures at a resolution of 1.80 Å for the Mamu-B*098:lyso-PE16:0 complex and 1.87 Å for the Mamu-B*098:lyso-PC16:0 complex (Table 1). The overall structures were composed of α1, α2, and α3 domains and noncovalently attached β2m and were nearly identical to that of the Mamu-B*098:C14-GGAIS lipopeptide complex (root mean square deviation (RMSD) of 0.374 Å for Mamu-B*098:lyso-PE16:0 and 0.375 Å for Mamu-B*098:lyso-PC16:0 over all Cα atoms in the α1 and α2 domains). In both structures, clear electron density maps were observed for only the monopalmitoylglycerol (MG16:0) moiety of the lysophospholipids in the Ag-binding cleft built up of the α1 and α2 domains of Mamu-B*098 (Fig. 2A (left two panels) and Fig. S2A). On the other hand, the ethanol amine (lyso-PE) and choline (lyso-PC) headgroups were not assigned in the electron maps, suggesting that these fluctuated and were disordered, and, thus, they were not included in the final structures. To verify that the maps obtained were derived from the MG16:0 moiety of lysophospholipids and to rule out the possibility of contamination by E. coli–derived unknown compounds, we elucidated the additional crystal structure of Mamu-B*098 complexed with MG16:0 itself (no headgroup) at 1.69 Å (Table 1). The whole structure also strongly resembled that of Mamu-B*098:C14-GGAIS (RMSD of 0.130 Å). A continuous electron map observed in the Ag-binding groove indeed corresponded to the structure of MG16:0 (Fig. 2A (right panels) and Fig. S2A). Bound MG16:0 was accommodated in the B and D pockets of Mamu-B*098, and this structural orientation was consistent with those of lyso-PE16:0 and lyso-PC16:0 (Fig. 2B), indicating that the MG16:0 backbone was sufficient to bind Mamu-B*098. Other pockets (A, C, E, and F pockets) were occupied by only water molecules or ethylene glycol, which was used as a cryoprotectant and did not interact with bound ligands (Fig. S2, B and C). Therefore, we concluded that lyso-PE and lyso-PC with a C16 palmitoyl group anchored at the B and D pockets of Mamu-B*098 through only a monoacylglycerol, whereas their headgroups appeared to be exposed to the solvent and not critically involved in the interaction between Mamu-B*098 and lysophospholipids.
Table 1.
Data collection and refinement statistics (molecular replacement)
| Mamu-B*098:lyso-PC16:0 | Mamu-B*098:lyso-PE16:0 | Mamu-B*098:MG16:0 | Mamu-B*05104:lyso-PC16:0 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | C2 | C2 | C2 | P21 21 21 |
| Cell dimensions | ||||
| a, b, c (Å) | 251.44, 46.44, 84.93 | 251.88, 46.73, 84.98 | 252.54, 46.90, 85.09 | 54.451, 81.028, 106.108 |
| α, β, γ (degrees) | 90.0, 90.9, 90.0 | 90.0, 90.8, 90.0 | 90.0, 90.8, 90.0 | 90, 90, 90 |
| Resolution (Å) | 50–1.87 (1.90–1.87)a | 50–1.87 (1.83–1.80) | 50–1.69 (1.73–1.69) | 50–2.15 (2.19–2.15) |
| Rmerge | 0.048 (0.362) | 0.054 (0.254) | 0.056 (0.403) | 0.061 (0.363) |
| I/σI | 33.4 (3.66) | 46.3 (5.94) | 39.0 (4.21) | 32.3 (4.83) |
| Completeness (%) | 99.5 (98.8) | 99.1 (98.0) | 98.5 (97.5) | 96.0 (92.5) |
| Redundancy | 3.9 (3.5) | 6.2 (5.7) | 4.4 (4.2) | 7.7 (7.7) |
| Refinement | ||||
| Resolution (Å) | 1.87 (1.90–1.87) | 1.80 (1.82–1.80) | 1.69 (1.71–1.69) | 2.150 (2.23−2.15) |
| No. of reflections | 76,941 (2451) | 86,953 (2687) | 103,819 (2992) | 25,216 (2417) |
| Rwork/Rfree (%) | 19.4 (26.3)/23.0 (34.4) | 19.1 (23.9)/22.9 (29.0) | 19.5 (27.1)/22.7 (33.6) | 18.19 (22.63)/23.15 (33.38) |
| No. of atoms | ||||
| Protein | 6219 | 6266 | 6377 | 3131 |
| EKG/metal ion/EDO/buffer molecule | 46/6/128/0 | 69/9/104/16 | 69/8/172/0 | 23/1/32/0 |
| Water | 412 | 505 | 587 | 209 |
| B-Factors (Å2) | ||||
| Protein | 33.8 | 29.1 | 28.5 | 35.51 |
| Ligand and ion | 42.1 | 38.1 | 39.9 | 47.16 |
| Water | 36.2 | 33.8 | 33.9 | 37.15 |
| RMSDs | ||||
| Bond lengths (Å) | 0.009 | 0.007 | 0.008 | 0.008 |
| Bond angles (degrees) | 1.012 | 0.937 | 1.201 | 0.96 |
| Ramachandran plot | ||||
| Favored (%) | 98.53 | 98.79 | 98.39 | 98.12 |
| Outliers (%) | 0.0 | 0.0 | 0 | 0.0 |
a The highest-resolution shell is shown in parentheses.
Figure 2.
Crystal structures of Mamu-B*098 complexed with lyso-PE16:0, lyso-PC16:0, and MG16:0. A, top views of the Ag-binding groove of Mamu-B*098 complexed with lyso-PE16:0 (left), lyso-PC16:0 (middle), and MG16:0 (right) are displayed with ribbon models of heavy chains and 2Fo − Fc maps (contoured at 0.8σ) for bound ligands (yellow sticks). The maps obtained for bound lysophospholipids only correspond to the structure of monopalmitoylglycerol, and, thus, the headgroup structures are not included. B, the surface of the antigen-binding groove is shown with pockets A–F colored, and the bound ligands are also displayed as yellow sticks.
Interactions between lysophospholipids and Mamu-B*098
The palmitoyl group of bound lyso-PE16:0 partially made a spiral shape and was accommodated in the large B pocket through a number of VDW interactions with the side chains of Tyr-7, Val-24, Arg-66, Val-67, Ala-70, Phe-74, Thr-97, and Ser-99 (Fig. 3A, top). These hydrophobic interactions were similar to those with the Mamu-B*098:C14-GGAIS lipopeptide, highlighting the broad capacity of the B pocket for binding an acyl chain (Fig. 3A, bottom). On the other hand, the glycerol moiety was laid on the shallow, hydrophobic cleft composed of Gln-155, Trp-156, and Tyr-159 in the D pocket. The glycerol moiety interacted with the side chains of Trp-156 and Tyr-159 through VDW forces, and was further wedged by a sole hydrogen bond interaction between the side chain and main chain of Gln-155 and two oxygen atoms at the sn-2 and sn-3 positions of glycerol (Fig. 3B, top). In contrast, interactions corresponding to those in the D pocket were not found in lipopeptide-bound Mamu-B*098 (Fig. 3B, bottom), indicating the unique role of the D pocket in accommodating lysophospholipids. Furthermore, the binding mode of lyso-PE16:0 to Mamu-B*098 was almost identical to that of lyso-PC16:0 (Fig. 3) and MG16:0 (Fig. S3). Thus, the headgroups of lysophospholipids did not appear to have any impact on binding to lipopeptide-presenting MHC class I proteins. For some structures, there was some variability for the density maps of bound ligands, presumably representing alternate conformations, whereas the binding mode was basically unaltered (Fig. S4).
Figure 3.
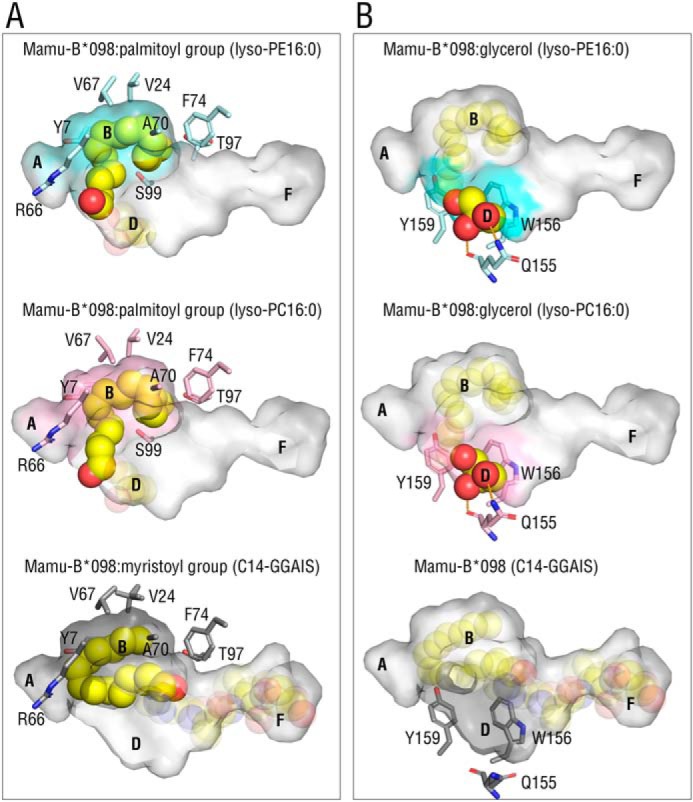
Interactions with monopalmitoylglycerol and Mamu-B*098. A and B, top view images of the palmitoyl group (A) and glycerol moiety (B) of bound ligands (yellow sphere) are displayed with the semitransparent antigen-binding groove of Mamu-B*098 complexed with lyso-PE16:0 (top, cyan), lyso-PC16:0 (middle, pink), and the C14-GGAIS lipopeptide (bottom, gray, 4ZFZ). The side chains of amino acid residues surrounding the palmitoyl group in the colored B pocket (A) and surrounding the glycerol moiety in the colored D pocket (B) are also shown. Hydrogen bonds are shown as dashed orange lines.
Crystallographic evidence of the capacity of Mamu-B*05104 to bind lysophospholipids
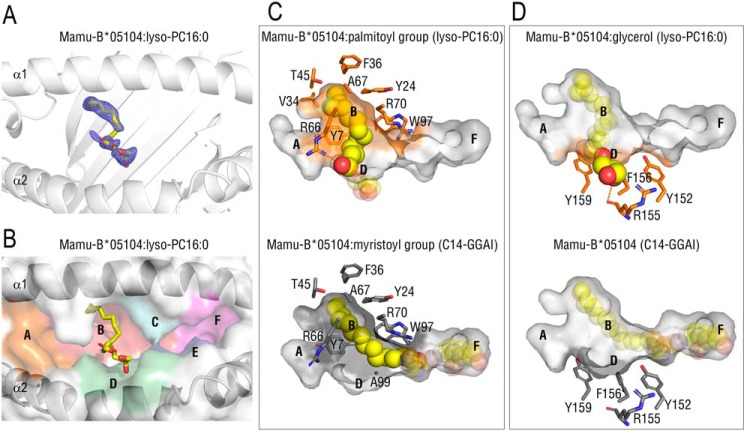
We recently identified the second lipopeptide-presenting MHC class I allomorph, Mamu-B*05104, which preferably presented N-myristoylated 4-mer lipopeptides to T cells and was only remotely related to Mamu-B*098. To identify shared features between Mamu-B*098 and Mamu-B*05104 and generalize our result showing that lysophospholipids were a novel ligand of MHC class I molecules, we refolded the recombinant Mamu-B*05104 protein in the presence of lyso-PC16:0. We then solved the X-ray crystal structure at a resolution of 2.1 Å (Table 1). An electron density map in the Ag-binding groove corresponded to the structure of MG16:0 of lyso-PC, whereas an obvious map for the chorine moiety was not observed (Fig. 4A and Fig. S2A), suggesting that the headgroup was disordered similar to Mamu-B*098:lysophospholid complexes. The palmitoyl group made a straight conformation and was accommodated in the large B pocket through a number of VDW interactions with the side chains of Tyr-7, Tyr-24, Val-34, Phe-36, Thr-45, Ala-67, Arg-66, Arg-70, and Trp-97 of Mamu-B*05104 (Fig. 4 (B and C), top panels). These interactions were mostly conserved in the Mamu-B*05104:lipopeptide complex (Fig. 4C, bottom). The carbonyl oxygen of the bound palmitoyl group was wedged by a hydrogen bond with the side chain of Arg-66 of Mamu-B*05104, whereas the myristoylated Gly residue was anchored by the side chain of Arg-70 for lipopeptide-bound Mamu-B*05104. On the other hand, the glycerol moiety was laid in the D pocket, which was not actively utilized to accommodate the lipopeptide (Fig. 4D). The glycerol moiety interacted with the side chains of Tyr-152, Arg-155, Phe-156, and Tyr-159 (VDW interactions) and the main chain of Arg-155 (a hydrogen bond) of Mamu-B*05104. No interactions existed between other pockets (A, C, E, and F) and lyso-PC16:0 (Fig. S2, B and C). This binding mode was equivalent to that of Mamu-B*098:lysophospholipid complexes (Fig. 2). The recombinant protein of the peptide-presenting MHC class I, HLA-B27, was not refolded in a buffer system by lyso-PC16:0, in contrast to lipopeptide-presenting MHC class I (Fig. S5), ruling out the nonspecific interaction of lysophospholipids with MHC class I molecules. Collectively, these results indicated that lysophospholipids were common cellular ligands among lipopeptide-presenting MHC class I molecules.
Figure 4.
Crystal structure of Mamu-B*05104 complexed with lyso-PC16:0. A and B, top view image of the Ag-binding groove of the Mamu-B*05104 complex with lyso-PC 16:0 with 2Fo − Fc maps (contoured at 0.8σ) (A) and the colored surface of the antigen-binding groove (B) are shown as in Fig. 2, A and B, respectively. The map obtained only corresponded to the structure of monopalmitoylglycerol, similar to Mamu-B*098:lysophospholipid complexes. C and D, top view images of the acyl chain (C) and glycerol moiety (D) of bound lyso-PC16:0 in the antigen-biding groove of Mamu-B*05104 (top, orange) are displayed with the side chains of amino acid residues interacting with bound ligands as in Fig. 3. The structure of Mamu-B05104 complexed with the C14-GGAI lipopeptide (bottom, 6IWG) is also shown for comparison. Hydrogen bonds are displayed as dashed orange lines.
TAP-independent cell-surface expression of lipopeptide-presenting MHC class I molecules
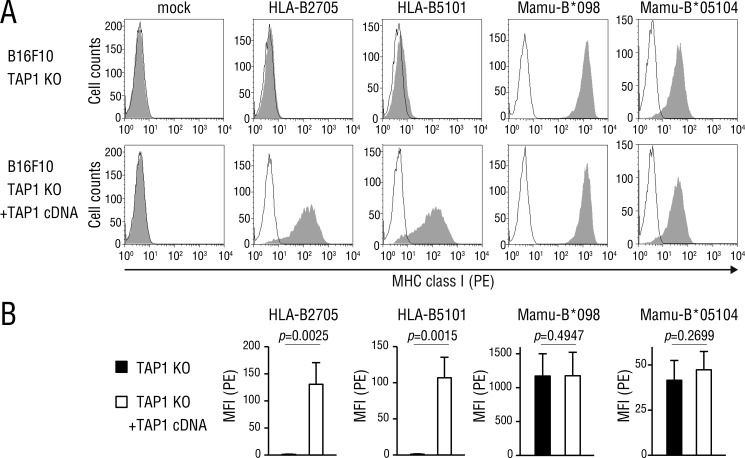
Because Mamu-B*098 and Mamu-B*05104 bound cellular lipid ligands, such as lysophospholipids, we assumed that the cell-surface expression of these allomorphs did not require TAP functions, in contrast to that of peptide-presenting MHC class I molecules. A flow cytometric analysis of TAP-deficient (Fig. 5A, top panels) and TAP-complemented (bottom panels) B16F10 cells transfected with HLA-B27 and HLA-B51 heavy chains confirmed that peptide-presenting MHC class I molecules failed to reach the plasma membrane without TAP functions. On the other hand, Mamu-B*098 and Mamu-B*05104 were expressed on the surface of TAP-deficient cells, and no significant difference was observed in its expression levels between TAP-deficient and TAP-complemented cells (Fig. 5B). Thus, we concluded that these two MHC class I molecules were expressed on the cell surface independently of TAP functions, which contrasted sharply with peptide-presenting MHC class I molecules.
Figure 5.
MHC class I expression in TAP-deficient cells. A, Mamu-B*098 and Mamu-B*05104 cDNA as well as those encoding the representative peptide-presenting MHC class I alleles, HLA-B2705 and HLA-B5101 were transiently transfected into TAP1-knockout (top panels) and TAP1-complemented (bottom panels) B16F10 cells. Cells were co-transfected with GFP cDNA, and GFP-positive cells were analyzed for MHC class I expression on the cell surface by flow cytometry. B, mean fluorescence intensity (MFI) representing MHC class I expression was assessed in three independent experiments and displayed as the mean with S.D. p values are also shown. PE, R-phycoerythrin.
Discussion
MHC class I molecules constantly sample 8–10-mer-long peptides provided by TAP transporters in the ER for their presentation to CTLs; however, the lipopeptide-presenting MHC class I molecules, Mamu-B*098 and Mamu-B*05104, clearly depart from this established paradigm. A LC-MS/MS analysis of bound ligands eluted from soluble Mamu-B*098 identified lyso-PE and lyso-PC species with up to a C20 fatty acid as a cellular ligand of MHC class I (Fig. 1). The unique anchoring of lyso-PC16:0 and lyso-PE16:0 via only a monoacylglycerol moiety was clearly demonstrated by the high resolution X-ray crystal structures of Mamu-B*098 complexed with lyso-PE16:0, lyso-PC16:0, and MG16:0 (Figs. 2 and 3) as well as the Mamu-B*05104:lyso-PC16:0 complex (Fig. 4). It is important to note that their binding manners closely resembled each other for the role of the B and D pockets in accommodating lysophospholipids. These results were further supported by evidence showing that their cell-surface expression was completely TAP-independent, in contrast to the conventional MHC class I molecules, HLA-B27 and HLA-B51 (Fig. 5). We cannot exclude the possibility that TAP-independent peptides, such as leader peptides, or lipid species other than lysophospholipids were also captured by Mamu-B*098 and Mamu-B*05104 in the ER. On the other hand, it was very unlikely for these MHC class I molecules to bind diacyl or triacyl lipids because they were equipped with only one pocket that was large enough to accommodate an acyl chain. Furthermore, lysophospholipids are the most abundant among monoacyl lipids in the ER (23). Therefore, these results suggest that lysophospholipids comprise a novel MHC class I endogenous ligand and were at least partially responsible for the TAP-independent expression of lipopeptide-presenting MHC class I molecules. Because Mamu-B*098 and Mamu-B*05104 were not closely related alleles, the capacity to bind lysophospholipids and TAP-independent expression may be shared features that are unique to lipopeptide-presenting MHC class I molecules. Collectively, the present results indicated that a lipopeptide-presenting MHC class I subset utilizes a unique source of endogenous ligands distinct from a peptide-presenting subset.
Lysophospholipids are mainly produced within cells by phospholipase A2 (PLA2), which cleaves and releases one fatty acid of diacyl phospholipids at the sn-2 position (24). Because members of cytosolic PLA2 (cPLA2) and Ca2+-independent PLA2 (iPLA2) localize at the ER membrane and produce lysophospholipids to induce the formation of the characteristic membrane tubules of the ER-Golgi network, cPLA2 and/or iPLA2 may serve as producers of lysophospholipids loaded on MHC class I molecules (25). However, our preliminary data showed that the surface expression of Mamu-B*098 as well as Mamu-B*05104 was unaffected by multiple PLA2 inhibitors, including arachidonyl trifluoromethyl ketone (cPLA2 and iPLA2 inhibitor) (26) or PLA2 gene overexpression, possibly due to the massive overlap in function among PLA2 gene family members (27). Thus, the members of PLA2 responsible and the extent to which PLA2 contributes to the production of lysophospholipids for a ligand supply to lipopeptide-presenting MHC class I molecules currently remain unclear.
In the endogenous ligand analysis of the B*098-Fc protein, we have so far failed to detect any signals corresponding to endogenous lipopeptides even though we easily observed lipopeptide-specific signals from an acid methanol eluate of recombinant MHC class I:lipopeptide complexes. In a steady state, the amount of N-myristoylated lipopeptide fragments present in cells may be markedly smaller than that of peptide fragments because only a fraction of proteins (estimated as ∼1% in a whole mammalian protein (28)) are N-myristoylated, and basically one N-myristoylated protein produces only one lipopeptide. Therefore, lipopeptides alone are not sufficient to sustain the expression of the lipopeptide-presenting MHC class I subset. We speculate that, in the evolutionary process, these presenting lipopeptides may need to acquire the capacity to bind lysophospholipids instead of self-peptides to sustain cell-surface expression. Sustained expression at a steady state may enable cells to avoid NK cell–mediated cytolysis and maintain the homeostasis of lipopeptide-specific T cells, similar to that of peptide-specific T cells (29). Upon viral infection, the cellular protein expression machinery, including N-myristoylation, is hijacked by viruses, and N-myristoylated viral proteins are vigorously produced. In this case, a massive amount of lipopeptide Ags may be generated and provided to lipopeptide-presenting MHC class I in the ER, followed by the replacement or binding competition of self-lysophospholipids with viral lipopeptides for their presentation to T cells.
Alternatively, lipopeptide Ags may be loaded on MHC class I molecules in a different compartment from the ER. Nonclassical MHC class I, CD1 molecules are equipped with hydrophobic pockets to accommodate various numbers of lipid Ags (30, 31). CD1 proteins also bind self-lipids, including lysophospholipids, as an endogenous ligand (32–35) and are expressed on the cell surface in a TAP-independent manner (36). In mycobacterial infections, self-lipids bound in CD1 proteins are considered to be replaced with nonself-lipid Ags derived from the bacteria in endosomal or lysosomal environments, followed by their presentation to lipid-specific T cells (37). A previous study reported that mouse CD1d was capable of binding 22-mer-long peptides and eliciting T-cell responses, implying a functional overlap between MHC class I and CD1 (38). Based on the structural and molecular similarities between lipopeptide-presenting MHC class I and CD1, a similar mechanism may exist for lipopeptide Ag presentation by utilizing lysophospholipids as a spacer ligand.
Based on the unaltered expression of lipopeptide-presenting MHC class I molecules in TAP-disrupted cells, lipopeptide-specific CTLs may play a critical role, particularly when pathogenic viruses utilize TAP inhibitors, such as the herpes simplex virus ICP47 protein, to escape from peptide-specific CTLs (39). Furthermore, the N-myristoylation motif is constituted within the N-terminal amino acid residues of proteins; difficulties are associated with introducing mutations into N-terminal lipopeptide Ag sequences without disrupting the N-myristoylation motif. Therefore, N-myristoylated lipopeptides may be an ideal target Ag by host CTLs. The molecular mechanisms underlying MHC class I–mediated lipopeptide antigen presentation are now being elucidated and may provide novel insights into host defenses against viral infections.
Experimental procedures
Preparation of Mamu-B*098-IgG Fc fusion proteins and a ligand analysis by MS
A cDNA sequence encoding the Mamu-B*098 ectodomain (from Gly-1 to Pro-276) was cloned in the pFUSE-hIgG1-Fc2 vector (InVivoGen, San Diego, CA). The cDNA construct was transfected into Expi293F cells (Thermo Fisher Scientific) with rhesus β2m cDNA in pcDNA3.1(+) to produce a soluble form of the Mamu-B*098-human IgG Fc fusion protein (B*098-Fc), whereas control transfection was performed using the empty pFUSE-hIgG1-Fc2 vector that encoded the human IgG Fc protein. After 5 days, cultured supernatants were collected and loaded onto HiTrap Protein G HP columns (GE Healthcare). The columns were washed sequentially with 10 ml of 20 mm phosphate buffer (pH 7.0), 10 ml of 0.5 m NaCl in 20 mm phosphate buffer (pH 7.0), and 3 ml of 2 mm phosphate buffer (pH 7.0), and column-bound B*098-Fc and Fc proteins were eluted in 5 ml of 0.2 n acetic acid. Ligands were released from purified protein preparations by adding 9 volumes of acid methanol (pH 4.0), followed by a 10-min sonication in a chilled water bath. Samples were filtered through the Millex-GV membrane with a pore size of 0.22 μm (Merck Millipore, Burlington, MA) and subjected to reversed-phase HPLC with an InertSustain C18 column (1.0 × 100 mm, particle size of 3 μm; GL Science, Tokyo, Japan). Elution was performed at a flow rate of 0.07 ml/min using a linear gradient from 25 to 100% acetonitrile in the presence of 0.1% formic acid, followed by MS/MS analysis with a scan range of 450–700 m/z in the positive ion mode using electrospray ionization-ion trap-TOF-MS (Shimadzu, Kyoto, Japan) (40). Collected data were analyzed by using LCMS solution software (Shimadzu).
Generation of lipid-bound MHC class I complexes
The ectodomain of MHC class I heavy chains and rhesus β2m were expressed in E. coli in the form of inclusion bodies as described previously (15). Purified inclusion bodies were dissolved in a buffer containing 6 m guanidine-HCl, and insoluble material was removed by centrifugation. The supernatant was treated with 50 mm DTT at 37 °C for 3 h, and aliquots were stored at −80 °C until used. To obtain ligand-bound MHC class I complexes, MHC class I heavy chains (0.5 μmol) and β2m (0.5 μmol) were refolded in the presence of a 20-fold molar excess of 1-hexadecanoyl-sn-glycero-3-phosphocholine (lyso-PC16:0) (Avanti Polar Lipids, Alabaster, AL), 1-hexadecanoyl-sn-glycero-3-phosphoethanolamine (lyso-PE16:0) (Avanti Polar Lipids), or 1-hexadecanoyl-sn-glycerol (MG16:0) (Tokyo Chemical Industry, Tokyo, Japan) by rapid dilution in 500 ml of refolding buffer (100 mm Tris-HCl, pH 8.3, 500 mm l-arginine, 2 mm EDTA, 0.5 mm oxidized GSH, and 5 mm reduced GSH). After dialysis against 10 mm Tris-HCl, pH 8.0, refolded proteins were purified by HiLoad 16/600 Superdex 200 pg (GE Healthcare) size-exclusion chromatography, followed by monoQ (GE Healthcare) anion-exchange chromatography.
X-ray crystallographic analysis
Crystals of lipid-bound MHC class I complexes were formed by a sitting-drop vapor-diffusion method as described previously (15). Briefly, 1 μl of a protein solution (10–14 mg/ml) was mixed at 20 °C with 1 μl of a mother liquid containing 2 mm ZnCl2, 13% PEG 6000, and 0.1 m Tris-HCl at pH 6.15 for Mamu-B*098:lyso-PE16:0, pH 6.4 for Mamu-B*098:lyso-PC16:0, pH 6.7 for Mamu-B*098:MG16:0, and 0.2 m sodium malonate, 20% PEG 3350, and 0.1 m Bistris propane at pH 7.5 for Mamu-B*05104:lyso-PC16:0, respectively. The crystals that formed were cryoprotected in 20% ethylene glycol. Diffraction data were collected at 100 K (in a cold nitrogen gas stream) on a Rigaku Saturn A200 detector (Rigaku, Woodlands, TX) with a wavelength of 1.0 Å. The resulting data sets were processed, merged, and scaled using HKL-2000 (HKL Research, Charlottesville, VA) (41). Structures were solved by molecular replacement with the lipopeptide-bound Mamu-B*098 (Protein Data Bank entry 4ZFZ) complex as a search model, as implemented in CCP4i software (42). The model was refined using REFMAC5 and PHENIX1.10 software (43). The structures were rebuilt using COOT 0.8.9 (44) and further modified based on σ-weighted (2|Fo| − |Fc|) and (|Fo| − |Fc|) electron density maps. Crystallographic images were depicted using PyMOL software. The Mamu-B*098 crystals contained two nearly identical trimer complexes in the asymmetric unit. Crystallographic structures were analyzed primarily for chain A, chain B, and the corresponding ligand.
Cell lines, transfection, and flow cytometry
TAP1-deficient cells were derived from the mouse melanoma cell line, B16F10, by utilizing a CRISPR/Cas9 system. Briefly, TAP1-targeting oligonucleotides (5′-CCG GGC ACG GCC GTG TGC ACA GAG-3′ and 5′-AAA CCT CTG TGC ACA CGG CCG TGC-3′) were annealed and cloned in pGuide-it (Clontech). The guide RNA expression construct was introduced into B16F10 by lipofection, and cloned cell transfectants were analyzed for mutations in the targeted third exon of the TAP1 gene. A mutant clone was selected and transfected with MHC class I and GFP cDNAs in pcDNA3.1(+) (Addgene, Watertown, MA) either with or without TAP1 cDNA in pEF6 (Thermo Fisher Scientific). Two days after transfection, cells were harvested and labeled with primary antibodies to MHC class I (clone W6/32), followed by phycoerythrin-conjugated donkey antibodies to mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). GFP-positive cells were analyzed for MHC class I expression by flow cytometry using FACS LSR Fortessa (BD Biosciences) and FlowJo software (Tree Star, Ashland, OR). Statistical analysis was performed, using Student's t test.
Data availability
Atomic coordinates and structural factors have been submitted to the Protein Data Bank under accession numbers 6LAM, 6LAH, 6LB2, and 6LT6. All remaining data are contained within the article.
Author contributions
Y. S., D. M., N. M., and B. M. data curation; Y. S. and D. M. validation; Y. S. and D. M. visualization; Y. S. and D. M. writing-original draft; D. M. and M. S. conceptualization; D. M., T. M., and M. S. supervision; D. M. and M. S. funding acquisition; D. M. and M. S. project administration; D. M., T. M., N. M., B. M., and M. S. writing-review and editing; N. M. and B. M. methodology.
Supplementary Material
Acknowledgments
We thank Ryu Nakata for advice and suggestions concerning the LC-MS/MS experiments. Diffraction data were collected at the BL26B1 stations of SPring-8 (Hyogo, Japan) with the approval of JASRI (proposal nos. 2017A2546, 2017A2547, 2018B2533, and 2018B2563).
This work was supported by Japan Society for the Promotion of Science KAKENHI Grants 17H05791, 18K19563, 18H02852, and 19H04805 (to M. S.) and 18K07172 (to D. M.) and by a grant from the Kato Memorial Bioscience Foundation (to D. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- MHC
- major histocompatibility complex
- CTL
- cytotoxic T lymphocyte
- ER
- endoplasmic reticulum
- β2m
- β2-microglobulin
- lyso-PE
- lysophosphatidylethanolamine
- lyso-PC
- lysophosphatidylcholine
- RMSD
- root mean-square deviation
- VDW
- van der Waals
- TAP
- transporter associated with antigen processing
- PLA2
- phospholipase A2
- cPLA2
- cytosolic PLA2
- iPLA2
- Ca2+-independent PLA2
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- NK
- natural killer
- Ag
- antigen.
References
- 1. Gotch F., Rothbard J., Howland K., Townsend A., and McMichael A. (1987) Cytotoxic T lymphocytes recognize a fragment of influenza virus matrix protein in association with HLA-A2. Nature 326, 881–882 10.1038/326881a0 [DOI] [PubMed] [Google Scholar]
- 2. Kumar V., and McNerney M. E. (2005) A new self: MHC-class-I-independent natural-killer-cell self-tolerance. Nat. Rev. Immunol. 5, 363–374 10.1038/nri1603 [DOI] [PubMed] [Google Scholar]
- 3. Nuchtern J. G., Bonifacino J. S., Biddison W. E., and Klausner R. D. (1989) Brefeldin A implicates egress from endoplasmic reticulum in class I restricted antigen presentation. Nature 339, 223–226 10.1038/339223a0 [DOI] [PubMed] [Google Scholar]
- 4. Rajagopalan S., and Brenner M. B. (1994) Calnexin retains unassembled major histocompatibility complex class I free heavy chains in the endoplasmic reticulum. J. Exp. Med. 180, 407–412 10.1084/jem.180.1.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sugita M., and Brenner M. B. (1994) An unstable β-2-microglobulin: major histocompatibility complex class I heavy chain intermediate dissociates from calnexin and then is stabilized by binding peptide. J. Exp. Med. 180, 2163–2171 10.1084/jem.180.6.2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neefjes J. J., Momburg F., and Hämmerling G. J. (1993) Selective and ATP-dependent translocation of peptides by the MHC-encoded transporter. Science 261, 769–771 10.1126/science.8342042 [DOI] [PubMed] [Google Scholar]
- 7. Neefjes J., Jongsma M. L., Paul P., and Bakke O. (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 10.1038/nri3084 [DOI] [PubMed] [Google Scholar]
- 8. Van Kaer L., Ashton-Rickardt P. G., Ploegh H. L., and Tonegawa S. (1992) TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4–8+ T cells. Cell 71, 1205–1214 10.1016/S0092-8674(05)80068-6 [DOI] [PubMed] [Google Scholar]
- 9. Cromme F. V., Airey J., Heemels M. T., Ploegh H. L., Keating P. J., Stern P. L., Meijer C. J. L. M., and Walboomers J. M. M. (1994) Loss of transporter protein, encoded by the TAP-1 gene, is highly correlated with loss of HLA expression in cervical carcinomas. J. Exp. Med. 179, 335–340 10.1084/jem.179.1.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Falk K., Rötzschke O., Stevanović S., Jung G., and Rammensee H. G. (1991) Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 10.1038/351290a0 [DOI] [PubMed] [Google Scholar]
- 11. Saper M. A., Bjorkman P. J., and Wiley D. C. (1991) Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 Å resolution. J. Mol. Biol. 219, 277–319 10.1016/0022-2836(91)90567-P [DOI] [PubMed] [Google Scholar]
- 12. Matsumura M., Fremont D. H., Peterson P. A., and Wilson I. A. (1992) Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science 257, 927–934 10.1126/science.1323878 [DOI] [PubMed] [Google Scholar]
- 13. Morita D., Igarashi T., Horiike M., Mori N., and Sugita M. (2011) Cutting edge: T cells monitor N-myristoylation of the Nef protein in simian immunodeficiency virus-infected monkeys. J. Immunol. 187, 608–612 10.4049/jimmunol.1101216 [DOI] [PubMed] [Google Scholar]
- 14. Morita D., Yamamoto Y., Suzuki J., Mori N., Igarashi T., and Sugita M. (2013) Molecular requirements for T cell recognition of N-myristoylated peptides derived from the simian immunodeficiency virus Nef protein. J. Virol. 87, 482–488 10.1128/JVI.02142-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morita D., Yamamoto Y., Mizutani T., Ishikawa T., Suzuki J., Igarashi T., Mori N., Shiina T., Inoko H., Fujita H., Iwai K., Tanaka Y., Mikami B., and Sugita M. (2016) Crystal structure of the N-myristoylated lipopeptide-bound MHC class I complex. Nat. Commun. 7, 10356 10.1038/ncomms10356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamamoto Y., Morita D., Shima Y., Midorikawa A., Mizutani T., Suzuki J., Mori N., Shiina T., Inoko H., Tanaka Y., Mikami B., and Sugita M. (2019) Identification and structure of an MHC class I-encoded protein with the potential to present N-myristoylated 4-mer peptides to T cells. J. Immunol. 202, 3349–3358 10.4049/jimmunol.1900087 [DOI] [PubMed] [Google Scholar]
- 17. Morita D., and Sugita M. (2016) Lipopeptides: a novel antigen repertoire presented by major histocompatibility complex class I molecules. Immunology 149, 139–145 10.1111/imm.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo H. C., Madden D. R., Silver M. L., Jardetzky T. S., Gorga J. C., Strominger J. L., and Wiley D. C. (1993) Comparison of the P2 specificity pocket in three human histocompatibility antigens: HLA-A*6801, HLA-A*0201, and HLA-B*2705. Proc. Natl. Acad. Sci. U.S.A. 90, 8053–8057 10.1073/pnas.90.17.8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sud M., Fahy E., Cotter D., Brown A., Dennis E. A., Glass C. K., Merrill A. H. Jr., Murphy R. C., Raetz C. R. H., Russell D. W., and Subramaniam S. (2007) LMSD: LIPID MAPS structure database. Nucleic Acids Res. 35, D527–D532 10.1093/nar/gkl838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liebisch G., Drobnik W., Lieser B., and Schmitz G. (2002) High-throughput quantification of lysophosphatidylcholine by electrospray ionization tandem mass spectrometry. Clin. Chem. 48, 2217–2224 10.1093/clinchem/48.12.2217 [DOI] [PubMed] [Google Scholar]
- 21. Davis B., Koster G., Douet L. J., Scigelova M., Woffendin G., Ward J. M., Smith A., Humphries J., Burnand K. G., Macphee C. H., and Postle A. D. (2008) Electrospray ionization mass spectrometry identifies substrates and products of lipoprotein-associated phospholipase A2 in oxidized human low density lipoprotein. J. Biol. Chem. 283, 6428–6437 10.1074/jbc.M709970200 [DOI] [PubMed] [Google Scholar]
- 22. Brügger B., Erben G., Sandhoff R., Wieland F. T., and Lehmann W. D. (1997) Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 94, 2339–2344 10.1073/pnas.94.6.2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schlager S. I., and Ohanian S. H. (1980) Tumor cell lipid composition and sensitivity to humoral immune killing. II. Influence of plasma membrane and intracellular lipid and fatty acid content. J. Immunol. 125, 508–517 [PubMed] [Google Scholar]
- 24. Dennis E. A. (1994) Diversity of group types, regulation, and function of phospholipase A2. J. Biol. Chem. 269, 13057–13060 [PubMed] [Google Scholar]
- 25. Bechler M. E., de Figueiredo P., and Brown W. J. (2012) A PLA1–2 punch regulates the Golgi complex. Trends Cell Biol. 22, 116–124 10.1016/j.tcb.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Balboa M. A., Pérez R., and Balsinde J. (2008) Calcium-independent phospholipase A2 mediates proliferation of human promonocytic U937 cells. FEBS J. 275, 1915–1924 10.1111/j.1742-4658.2008.06350.x [DOI] [PubMed] [Google Scholar]
- 27. Vasquez A. M., Mouchlis V. D., and Dennis E. A. (2018) Review of four major distinct types of human phospholipase A2. Adv. Biol. Regul. 67, 212–218 10.1016/j.jbior.2017.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thinon E., Serwa R. A., Broncel M., Brannigan J. A., Brassat U., Wright M. H., Heal W. P., Wilkinson A. J., Mann D. J., and Tate E. W. (2014) Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun. 5, 4919 10.1038/ncomms5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Markiewicz M. A., Girao C., Opferman J. T., Sun J., Hu Q., Agulnik A. A., Bishop C. E., Thompson C. B., and Ashton-Rickardt P. G. (1998) Long-term T cell memory requires the surface expression of self-peptide/major histocompatibility complex molecules. Proc. Natl. Acad. Sci. U.S.A. 95, 3065–3070 10.1073/pnas.95.6.3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Rhijn I., Godfrey D. I., Rossjohn J., and Moody D. B. (2015) Lipid and small-molecule display by CD1 and MR1. Nat. Rev. Immunol. 15, 643–654 10.1038/nri3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morita D., Hattori Y., Nakamura T., Igarashi T., Harashima H., and Sugita M. (2013) Major T cell response to a mycolyl glycolipid is mediated by CD1c molecules in rhesus macaques. Infect. Immun. 81, 311–316 10.1128/IAI.00871-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Birkinshaw R. W., Pellicci D. G., Cheng T. Y., Keller A. N., Sandoval-Romero M., Gras S., de Jong A., Uldrich A. P., Moody D. B., Godfrey D. I., and Rossjohn J. (2015) αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat. Immunol. 16, 258–266 10.1038/ni.3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lepore M., de Lalla C., Gundimeda S. R., Gsellinger H., Consonni M., Garavaglia C., Sansano S., Piccolo F., Scelfo A., Häussinger D., Montagna D., Locatelli F., Bonini C., Bondanza A., Forcina A., et al. (2014) A novel self-lipid antigen targets human T cells against CD1c+ leukemias. J. Exp. Med. 211, 1363–1377 10.1084/jem.20140410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cox D., Fox L., Tian R., Bardet W., Skaley M., Mojsilovic D., Gumperz J., and Hildebrand W. (2009) Determination of cellular lipids bound to human CD1d molecules. PLoS ONE 4, e5325 10.1371/journal.pone.0005325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. López-Sagaseta J., Sibener L. V., Kung J. E., Gumperz J., and Adams E. J. (2012) Lysophospholipid presentation by CD1d and recognition by a human natural killer T-cell receptor. EMBO J. 31, 2047–2059 10.1038/emboj.2012.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanau D., Fricker D., Bieber T., Esposito-Farese M. E., Bausinger H., Cazenave J. P., Donato L., Tongio M. M., and de la Salle H. (1994) CD1 expression is not affected by human peptide transporter deficiency. Hum. Immunol. 41, 61–68 10.1016/0198-8859(94)90086-8 [DOI] [PubMed] [Google Scholar]
- 37. Huang S., Cheng T. Y., Young D. C., Layre E., Madigan C. A., Shires J., Cerundolo V., Altman J. D., and Moody D. B. (2011) Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc. Natl. Acad. Sci. U.S.A. 108, 19335–19340 10.1073/pnas.1112969108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Girardi E., Wang J., and Zajonc D. M. (2016) Structure of an α-helical peptide and lipopeptide bound to the nonclassical major histocompatibility complex (MHC) class I molecule CD1d. J. Biol. Chem. 291, 10677–10683 10.1074/jbc.M115.702118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oldham M. L., Hite R. K., Steffen A. M., Damko E., Li Z., Walz T., and Chen J. (2016) A mechanism of viral immune evasion revealed by cryo-EM analysis of the TAP transporter. Nature 529, 537–540 10.1038/nature16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshinaga N., Aboshi T., Ishikawa C., Fukui M., Shimoda M., Nishida R., Lait C. G., Tumlinson J. H., and Mori N. (2007) Fatty acid amides, previously identified in caterpillars, found in the cricket Teleogryllus taiwanemma and fruit fly Drosophila melanogaster larvae. J. Chem. Ecol. 33, 1376–1381 10.1007/s10886-007-9321-2 [DOI] [PubMed] [Google Scholar]
- 41. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 42. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structural factors have been submitted to the Protein Data Bank under accession numbers 6LAM, 6LAH, 6LB2, and 6LT6. All remaining data are contained within the article.