Abstract
In the past decade, a growing set of immunotherapies including immune checkpoint blockade, chimeric antigen receptor T cells, and bispecific antibodies propelled the advancement of oncology therapeutics. Accumulating evidence demonstrates that immunotherapy could eliminate tumors better than traditional chemotherapy or radiotherapy with lower risk of adverse events in numerous cancer types. Unfortunately, a substantial proportion of patients eventually acquire resistance to immunotherapy. By analyzing the differences between immunotherapy-sensitive and immunotherapy-resistant populations, it was noticed that the composition of gut microbiota is closely related to treatment effect. Moreover, in xenograft models, interventional regulation of gut microbiota could effectively enhance efficacy and relieve resistance during immunotherapy. Thus, we believe that gut microbiota composition might be helpful to explain the heterogeneity of treatment effect, and manipulating gut microbiota could be a promising adjuvant treatment for cancer immunotherapy. In this mini review, we focus on the latest understanding of the cross-talk between gut microbiota and host immunity. Moreover, we highlight the role of gut microbiota in cancer immunotherapy including immune checkpoint inhibitor and adoptive cell transfer.
Keywords: gut microbiota, immunotherapy, PD-1, PD-L1, CTLA-4, adoptive cell transfer
Introduction
The gut microbiota contains a large number of microorganisms populating in gastrointestinal tract such as bacteria, fungi, protozoa, virus, phages, and archaea.1,2 It is generally believed that the gut flora consists of essential and opportunistic bacteria.3 The essential bacteria are beneficial to humans and participate in fermenting undigested carbohydrates and endogenous mucus, synthesizing short-chain fatty acids (SCFA) and vitamins, and defending against infection by pathogens.4-6 On the contrary, overgrowth of opportunistic bacteria could lead to infection.3 The imbalance between essential and opportunistic bacteria results in gut dysbiosis, which usually refers to the compositional and functional alteration in microbiota driven by environmental or host-associated factors.7 It has been well established that gut dysbiosis relates to some diseases including inflammatory bowel disease, nonalcoholic fatty liver disease, neurodegenerative disorders, and metabolic disease.8-13 Besides, gut dysbiosis is regarded as an important risk factor promoting tumor initiation and development.7 Some specific bacteria have been confirmed as carcinogens such as Helicobacter pylori for gastric cancer and Salmonella typhi for biliary cancer.14,15 The carcinogenic role of H pylori is related to its genotoxic effect, which further results in chronic inflammation and hyperactive proliferation signaling pathways in mucosal cells.14 Following long-term stimulation, H pylori could induce malignant transformation in gastric epithelia and mucosa-associated lymphoid tissues.14 Moreover, it has been verified that gut microbiota closely associate with host immunity.16,17 On the one hand, the gut microbiota participates in the development of the host immune system.18 On the other hand, the composition of gut microbiota is modulated by host immunity.19
Accumulating evidence demonstrates that gut microbiota could affect the therapeutic effect of multiple cancer treatments including chemotherapy, radiotherapy, as well as immunotherapy.20-22 The results of in vitro and in vivo studies showed that gut microbiota could regulate the efficacy of chemotherapy by multiple approaches, including (1) Translocation: bacteria cross chemotherapy-induced damaged gut mucosal barrier and enter peripheral lymph nodes; (2) Immunomodulation: gut microbiota promotes chemotherapy-related inflammation; (3) Metabolism and enzymatic degradation: gut microbiota could directly or indirectly modify the structure of pharmaceuticals, which might enhance or abrogate the efficacy of treatment and introduce toxic compounds; (4) Reduced diversity: chemotherapy tends to reduce to the diversity of gut microbiota and leads to the formation of pathogen-dominant gut flora and higher risk of gastrointestinal reactions.20 However, the exact mechanism by which gut microbiota modulates the efficacy of immunotherapy is still unclear.
Benefiting from the development of sequencing technology, it is now possible to analyze the composition of the microbiota.23 Commonly, 16S rRNA and metagenomic shotgun sequencing are adopted for taxonomic assignment.24 Taxonomic identification by 16S rRNA is based on the comparison between detection results and known database. Therefore, with 16S rRNA sequencing, it is difficult to identify unknown species.24 Compared with 16S rRNA sequencing, metagenomic shotgun sequencing could directly analyze the whole genomic context, which could be used for taxonomic identification and function analysis.24 Moreover, more and more microbiome studies utilize long-read sequencing that could overcome the limitations of next-generation sequencing such as identifying structural variants, repetitive regions, alleles, and highly homologous genomic regions. Given the vital role of gut microbiota in anticancer therapy, identifying efficacy-related bacteria provide a novel perspective to counteract drug resistance especially for immunotherapy.
The Cross-Talk Between Gut Microbiota and the Host Immune System
The cross-talk between gut microbiota and immunity is complicated. Host immunity not only sustains tolerance to symbiotic commensals and food antigens but also recognizes opportunistic bacteria and defends against pathogen infection.25 In the meantime, the influence of gut microbiota on the host immune system is multifaceted, from localized immune response to systemic innate or adaptive immunity.25 It was observed that mice that were bred and raised in a sterile environment (germ-free mice) were prone to harbor deficiencies in the development of gut-associated lymphoid tissues especially Peyer’s patches (PP) and isolated lymphoid follicles.26 Besides, depleting gut microbiota by broad-spectrum antibiotics inhibited murine bone marrow hematopoiesis and decreased the abundance of hematopoietic stem cells or multipotent progenitors.27
Gut Mucosal Immune System
The gut mucosal immune system contains organized lymphoid tissues located in the gut mucosal epithelium, lamina propria, and mesentery including PP, isolated lymphoid follicles, and mesenteric lymph node.28-30 Among them, the mucus layer and mucosal epithelium comprise the physical barrier of gut mucosal immunity.31 It is generally believed that the mucus is mainly produced by goblet cells.32 During mucus secretion, goblet cells in the small intestine can sense and sample luminal content.32 In a manner that has not been well studied, actively secreting goblet cells take up antigenic materials and deliver them to dendritic cells (DCs) in lamina propria.32 Notably, the mucus contains abundant antimicrobial peptides that effectively clear bacterial clones on gut epithelium. As a part of intestinal innate immunity, Paneth cells in the base of intestine crypts are main producers of antimicrobial peptides.33 Decreased antimicrobial peptides lead to elevated bacterial colonization and hyperactive adaptive immune response.34 Mucosal epithelial cells under the mucus layer not only directly isolate gut microbiota but also secrete cytokines and chemokines to regulate the mucosal immune system.31 In mucosal epithelium, innate lymphoid cells (ILCs) play an important role in regulating the magnitude of inflammation and maintaining intestinal homeostasis.31 By secreting interleukin (IL)-22, ILCs promote healing during infection and counteract the damaging effect of immune response.35 In the meantime, ILCs also stimulate the production of antimicrobial peptides to kill gram-positive bacteria.36
Peyer’s patches are the core component of gut-associated lymphoid tissue and are distributed throughout the small intestine.37 Distinguished from peripheral lymph organs, PP harbor some specialized structures.37 Notably, there are no afferent lymphatics in PP. Instead, PP are overlain by specialized microfold epithelial cells (termed M cells), which constantly sample and deliver antigens from the lumen into PP.38 A host of DCs are enriched in the area underneath M cells (subepithelial dome region), capturing and presenting antigens from M cells.38 Apart from antigen presentation, DCs in PP express retinol dehydrogenase that promotes the production of retinoic acid.28 Retinoic acid induces the homing of activated T or B cells to intestinal lamina propria by upregulating gut-imprinting molecules such as CCR9 and integrin α4β7 on lymphocytes.39-41 Apart from DCs in PP or isolated lymphoid follicles, it has been detected that a subset of DCs populate the gut mucosal epithelium, which are called “intraepithelial DCs.”42 Intraepithelial DCs are characterized by CX3CR1 expression and directly capture antigens from the intestinal lumen by their transepithelial dendrites.43 After activation, DCs traffic to mesenteric lymph nodes and induce the polarization of naïve CD4+ T cells toward inducible regulatory T cells (iTreg) or Th1/Th17 cells.25 After education (a process also known as imprinting, referring to how naïve T cells learn to express homing receptors for skin, gut, or other tissues) in mesenteric lymph node, most newly generated iTreg, Th17, and Th1 cells home to the gut by the guidance of gut-imprinting molecules, while a part of lymphocytes circulate systemically.25,44
The Regulatory Effect of Gut Microbiota on the Gut Mucosal Immune System
The gut microbiota and its metabolites have a broad and profound influence on multiple aspects of the host gut mucosal immune system.1 It has been reported that human commensal Bacteroides fragilis could induce the differentiation of CD4+ naïve T cell into Treg and enhance the secretion of anti-inflammatory cytokines (eg, IL-10).45 B fragilis–induced intestinal immune tolerance is dependent on polysaccharide A and toll-like receptor 2 signaling, favoring to maintain gut homeostasis.45 Similarly, Cebula et al found that most colonic Treg cells belonged to thymus-derived Tregs, which recognized the antigenic materials from bacteria such as Clostridiales, Bacteroides, and Lactobacillus.46 Simultaneously, the colonic Treg cells could maintain tolerance to these bacteria.46 It was notable that antibiotic-mediated alteration in gut microbiota composition (mainly reducing the members of Clostridiales) significantly decreased the abundance of colonic Treg cells and changed the T cell receptor (TCR) repertoire of these thymus-derived Tregs.46 Contrary to B fragilis, some commensals modify gut immunity toward a pro-inflammatory direction, such as commensal segmented filamentous bacteria and adherent invasive Escherichia coli.47-49 Segmented filamentous bacteria promote the development of Th17 and induce the production of IL-17 in RORγt+ CD4+ T cells.50
Bacterial metabolites have been documented as a vital regulator of gut immune response as well. SCFAs including acetate, propionate, butyrate, and isobutyrate are end products of the fermentation activity of intestinal microorganisms.51,52 A growing body of studies demonstrated that SCFAs enhanced the generation and immune inhibitory capability by counteracting the effect of histone deacetylase and promoting acetylation of Foxp3 locus.53 In addition, butyrate-mediated inhibition of histone deacetylase could interfere with some lipopolysaccharides-responsive signaling pathways in DC, further enhancing the conversion from naïve CD4+ T toward a Treg population.1
The Influence of Gut Microbiota on Host Systemic Immunity
The binding between pathogen-associated molecular patterns and pattern recognition receptors (eg, toll-like receptors) together with bacteria-derived metabolites (eg, SCFAs) influence the local immune response in gut.25 However, the regulatory effect of gut microbiota is not just limited to the localized mucosal immune system. Actually, gut microbiota have a substantial effect on host systemic immunity via cytokine secretion, cross-reactivation, lymphocyte homing, and recirculation.25 By consistent antigen sampling of interdigitation of DCs and M cells, the stimulation of pathogen-associated molecular patterns propel the maturation and activation of DCs.25 There are abundant draining lymph nodes in the mesentery of the small intestine and colon where the differentiation of naïve CD4+ T cells can be modulated by DCs.25 Apart from inducing CD4+ T cell differentiation (especially toward Tregs and Th17 cells), DCs might stimulate CD8+ T cells in mesenteric lymph nodes.25 Moreover, a subset of activated DCs in the gut enter into circulation and induce a broader immune response.25 Besides, as mentioned above, some primed lymphocytes in mesenteric lymph nodes could subsequently enter the circulation as well. Due to cross-reactivity, gut microbiota–specific lymphocytes recognize and attack distant tissues with similar antigenic epitopes.54,55 Moreover, cytokines afforded by gut mucosal immune response might be secreted into circulation and set immunological tone, promoting host immunity to robustly respond to pathogens and to sustain the tolerance to innocuous commensals.56
Anticancer Immune Response and Immunotherapy
During malignant transformation, accumulating mutations increase the immunogenicity of tumor cell by generating tumor-associated antigen or neoantigen.57-59 In the condition of intact immune surveillance, host immunity could recognize and clear these immunogenic materials.60 However, a proportion of cancer cells could escape from immune elimination via various manners such as losing immunogenic antigens, dysregulating antigen presentation machinery, activating immune checkpoint signaling pathway, recruiting pro-tumor immune cells, and transforming growth factor β signaling–mediated exclusion of CD8+ T cells by the tumor parenchyma.61-63 As a result, antitumor immune response is impaired and tumor cells proliferate uncontrollably.64 Immunotherapy is aimed at restoring robust immune surveillance through regulating the balance between immunosupportive factors and immunosuppressive factors.65 The efficacy of immunotherapy could be affected by various determinants such as antigen presentation, T cell priming and activation, T cell trafficking and infiltration, as well as cytotoxicity activity of tumor infiltrating lymphocytes (TILs).60,66-68 Therefore, interventions affecting any processes of the cancer-immunity cycle could influence the efficacy of immunotherapy.
Immune Checkpoint Inhibitor
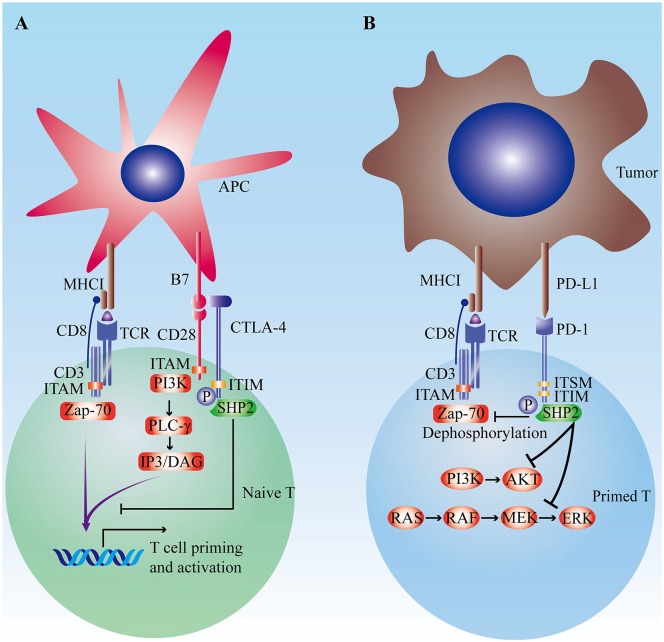
The activation of tumor-specific T cells needs 2 steps. First, TCR selectively binds to major histocompatibility complex I with anchored antigen peptides.69 Then, synergizing with co-stimulatory signals such as CD28, ICOS, and OX40, the activation signal of TCR/CD3 complex is further amplified and ultimately leads to the priming and activation of T cell.69 Contrarily, co-inhibitory signals (also known as immune checkpoints) including programmed cell death-1 (PD-1), cytotoxic T-lymphocyte–associated protein 4 (CTLA-4), T cell immunoglobulin domain and mucin domain-3 (TIM-3), and lymphocyte activation gene-3 (LAG-3) undermine T cell activation by intracellular immunoreceptor tyrosine–based inhibition motif (ITIM) to counteract TCR/CD3- or CD28-mediated tyrosine phosphorylation (Figure 1).70-72 Cancer cells tend to upregulate the activity of co-inhibitory signaling pathways to escape immune surveillance.73,74 Immune checkpoint inhibitors (ICIs) alleviate immune tolerance to tumor antigens and reinvigorate the antitumor response. Anti-PD-1/PD-L1 and anti-CTLA-4 have been successfully applied in multiple cancers.75-80 Nevertheless, there is a great potential to enhance the anticancer effect of ICI.
Figure 1.
The regulatory function of immune checkpoints. (A) The role of CTLA-4 in the priming and activation of naïve T cells. The activation of T cells is driven by stimulatory signals of TCR/CD3 complex and CD28. CTLA-4 could competitively antagonize co-stimulatory signal of CD28-B7 pathway and subsequently inhibits the T cells activation. (B) PD-1/PD-L1 signaling pathway. PD-1/PD-L1 signaling pathway to counteract CD3- or CD28-mediated tyrosine phosphorylation by ITIM and ITSM. Besides, PD-1 could disturb T cell proliferation and survival by inhibiting PI3K-AKT and Ras-Raf-MEK-ERK pathway.
Abbreviations: APC, antigen presentation cell; CTLA-4, cytotoxic T-lymphocyte–associated protein 4; ITIM, intracellular immunoreceptor tyrosine–based inhibition motif; ITSM, immunoreceptor tyrosine–based switch motif; MHC, major histocompatibility complex; PD-1, programmed cell death-1; TCR, T cell receptor.
Adoptive Cell Transfer
The therapeutic effect of ICI is highly dependent on preexisting tumor-specific immune cells.66 However, for some poorly immunogenic cancers, it is hard to eradicate cancer cells via ICI. In the context of an immune ignorant microenvironment, the injection of tumor-specific immune cells might be a reasonable strategy.81 Generally, adoptive cell transfer (ACT) could be deployed by 2 approaches: (1) expanding TILs in vitro, then reinfusing obtained TILs into patients; (2) isolating T cells from patients’ peripheral blood, genetically modifying T cells to express chimeric antigen receptor or specific TCR.82-85 ACT especially CAR-T exhibits potent anticancer effect in multiple hematological malignancies.86-88 However, the efficacy of ACT is limited to solid tumors, which is mainly attributed to unfavorable cytokine milieu, dysregulated Treg/T effector cell ratio, limited immune cell trafficking, as well as antigen heterogeneity.89 Interventions modulating the immune microenvironment and expanding T cell clones would be helpful to overcome the obstacles to ACT application in solid tumors.83,90
Gut Microbiota Modulates the Efficacy of Immunotherapy
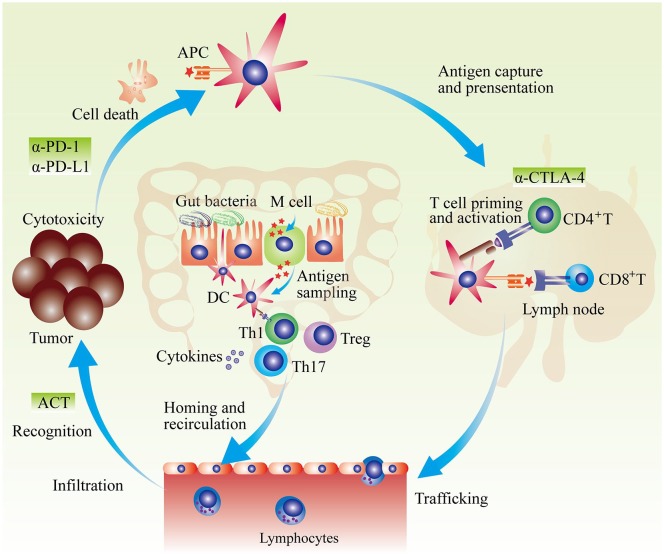
The gut microbiota possesses a broad range of regulatory effects on multiple immune effectors including the maturation of DCs, the differentiation of T cells, as well as the secretion of cytokines, which might regulate anticancer immunity (Figure 2).25,91 A series of studies indicated that gut microbiota composition is closely associated with the efficacy of cancer immunotherapy (summarized in Table 1).91
Figure 2.
Gut microbiota and anticancer immunotherapy. APCs capture and recognize dead tumor cell–derived antigens. Then, in peripheral lymphatic organs, APCs present possessed antigens and activate naïve T cells. Primed T cells migrate and infiltrate into tumor. After recognition of tumor antigen, activated T cells kill tumor cells. Factors interfering any producer of anticancer-immunity cycle could result in cancer immune escape. Generally, anti-PD-1/PD-L1 treatment mainly enhance tumor-killing activity; anti-CTLA-4 primarily promotes the priming and activation of T cells; and adoptive cell transfer mainly induces T cell clones recognizing tumor cells. Gut microbiota could affect anticancer immunotherapy by multiple manners. Gut microbiota–derived antigens could regulate the development and function of DC in gut, which further influences gut mucosa immunity. Induced immune response such as Th1-skweing immunity, Th17 polarization, Treg differentiation, and cytokines secretion could enter into circulation and influence the effect of systemic anticancer immunotherapy.
Abbreviations: ACT, adoptive cell transfer; APC, antigen presentation cell; α-CTLA-4, anti-cytotoxic T-lymphocyte–associated protein 4; α-PD-1, anti-programmed cell death-1; DC, dendritic cell; Treg, regulatory T cell.
Table1.
Regulatory Effect of Gut Microbiota on Cancer Immunotherapy.
| Bacterium | Regulatory Effect on Immunity | Influence on Immunotherapy | Author |
|---|---|---|---|
| Bifidobacterium | Enhancing the function of DC | Enhancing PD-1 blockade effect | Sivan et al99 |
| Upregulating tumor-specific CD8+ T | |||
| Increasing pro-inflammatory cytokine | |||
| Faecalibacterium | Increasing CD4+ and CD8+ T in circulation and in tumor | Enhancing PD-1 blockade effect | Gopalakrishnan et al100 |
| Bacteroidales | Upregulating systemic MDSC and Treg | Impeding PD-1 blockade effect | Gopalakrishnan et al100 |
| A group of bacteria including Bifidobacterium adolescentis, Bifidobacterium longum, and so on. | Elevating the secretion of IFN-γ | Enhancing PD-1 blockade effect | Matson et al101 |
| Increasing CD8+ tumor infiltrating T | |||
| Ruminococcus obeum and Roseburia intestinalis | Enriched in patients resistant to anti-PD-1 treatment | Impeding PD-1 blockade effect | Matson et al101 |
| Akkermansia muciniphila | Increasing CXCR3+CCR9+CD4+ T cell | Enhancing PD-1 blockade effect | Routy et al102 |
| Enhancing ability of DC and production of IL12 | |||
| Bacteroides fragilis | Inducing Th1 immune response and DC maturation | Enhancing CTLA-4 blockade | Vétizou et al103 |
| Faecalibacterium | Promoting development of Treg | Enhancing CTLA-4 blockade | Chaput et al104 |
| Upregulating ICOS expression of T cells; | |||
| Bacteroides | Leading to baseline systemic inflammation | Impeding CTLA-4 blockade effect | Chaput et al104 |
| Some species of Bacteroidetes | Decreasing DC and IL-12 | Impeding ACT effect | Uribe-Herranz et al105 |
| Inducing the formation of cold tumor |
Abbreviations: ACT, adoptive cell transfer; CTLA-4, cytotoxic T-lymphocyte–associated protein 4; DC, dendritic cell; ICOS, inducible T cell co-stimulator; MDSC, myeloid-derived suppressor cell; PD-1, programmed cell death-1; Treg, regulatory T cell.
The Role of Gut Microbiota in Anti-PD-1/PD-L1 Treatment
Anti-PD-1/PD-L1 treatment blocks the negative signal transduced by intracellular domains of PD-1 (ITIM and ITSM).92 PD-1/PD-L1 blockade not only promotes TCR/CD3- or CD28-mediated T cell activation but also enhances T cell survival and proliferation via upregulating Ras-Raf-MAPK and PI3K-AKT signaling pathways.93,94 Anti-PD-1/PD-L1 therapy has been approved for multiple types of cancers such as melanoma, non–small cell lung cancer, and kidney cell cancer.95-98 It has been verified that biomarkers including PD-L1 expression level, TIL status, and mismatch repair system deficiency highly correlate with the treatment effect of anti-PD-1/PD-L1.66 Besides these factors mentioned above, the gut microbiota contributes to the heterogeneity of therapeutic reaction as well.91
As early as 2015, Sivan et al noticed that the abundance of some special commensal bacteria was related to anti-PD-1 treatment effect in a mouse model.99 Researchers compared the efficacy of anti-PD-1 treatment in genetically similar mice (C57BL/6) from 2 different facilities (JAX and TAC) that harbored significantly different gut microbiota.99 The results showed that tumors grew more slowly and were more sensitive to anti-PD-1 therapy in JAX populations. This difference was attributed to enhanced antitumor immunity in JAX that could be transferred to TAC mice by cohousing or transplanting JAX fecal suspension to TAC.99 Based on the 16S rRNA sequencing technique, it was detected that markedly increased abundance of Bifidobacterium in JAX primarily led to elevated levels of TIL and better treatment response to anti-PD-1 therapy.99 Administration of commercially available Bifidobacterium including Bifidobacterium breve and Bifidobacterium longum significantly promoted tumor control especially combined with anti-PD-1 treatment.99 To interrogate the mechanism by which Bifidobacterium administration synergized with anti-PD-1 treatment, researchers monitored the abundance and function of tumor antigen–specific CD8+ T cell.99 It was observed that Bifidobacterium remarkably upregulated the level of tumor-specific CD8+ T cell and interferon (IFN)-γ secretion.99 In addition, in an in vitro experiment, DCs obtained from TAC receiving Bifidobacterium treatment showed improved capability to induce T cell priming and activation.99
Motivated by the encouraging finding in mouse models, a series of studies were deployed to explore the relationship between gut microbiota and anti-PD-1 treatment in cancer patients. Gopalakrishnan et al analyzed gut microbiota of melanoma patients undergoing anti-PD-1 treatment.100 The results demonstrated that gut microbial diversity was higher in responders, and the α-diversity (parameter reflecting bacterial community richness and evenness) of fecal samples was positively correlated to progression-free survival (PFS) time.100 Further analysis indicated that the level of Faecalibacterium (belonging to the Ruminococcaceae family, Clostridiales order) was higher in responders while Anaerotruncus colihominis, Bacteroides thetaiotaomicron (belonging to Bacteroidales order), and Escherichia coli were significantly enriched in nonresponders.100 In addition, it was found that the abundance of Faecalibacterium is positively correlated with the level of CD8+ TIL (R2 = 0.42, P < .01).100 The immune cell detection in circulation showed that increased gut Faecalibacterium accompanied elevated CD4+ or CD8+ T cells.100 Conversely, the level of systemic Bacteroidales positively related to the quantity of myeloid-derived suppressor cells and Tregs.100 Confirmed by fecal transplantation experiments in the mouse model, therapeutic benefit afforded by favorable bacteria was attributed to promoting the formation of hot tumor (according to the status of TILs, tumors can be classified as hot/T cell inflamed or cold/T cell non-inflamed tumors) via increasing local effector immune cells and decreasing suppressive immune cells.100
Similarly, Matson et al noticed the influence of gut microbiota on the efficacy of anti-PD-1 treatment in metastatic melanoma patients.101 Based on an integrative identification method (including 16S rRNA sequencing, metagenomics shotgun sequencing, and species-specific quantitative polymerase chain reaction), researchers observed that some bacteria such as Bifidobacterium adolescentis, B longum, Collinsella aerofaciens, Enterococcus faecium, Klebsiella pneumoniae, Lactobacillus species, Parabacteroides merdae, and Veillonella parvula were significantly enriched in responders, while Ruminococcus obeum and Roseburia intestinalis were remarkably abundant in nonresponders.101 Moreover, germ-free mice gavaged with fecal materials from responders had a markedly increased level of CD8+ TIL and secretion of IFN-γ, promoting the formation of an immunosupportive microenvironment.101
Around the same time, Routy et al reported the role of gut microbiota in anti-PD-1 treatment resistance.102 Researchers found that for cancer patients receiving anti-PD-1 treatment, additional oral antibiotic treatment (within 2 months before or 1 month after the start of anti-PD-1 treatment) significantly shortened overall survival (OS) and PFS time.102 To investigate the relationship between antibiotic-induced dysbiosis and impaired therapeutic effect, researchers compared the gut microbiota composition between responders and nonresponders.102 Among all bacteria overrepresented in responders, Akkermansia muciniphila was most significantly related to patients’ response rate (P = .007).102 Besides, immune reactivity of Tc1 or Th1 against A muciniphila correlated with improved survival data (P = .032).102 By fecal transplantation and antibiotics treatment in mice, researchers interrogated the influence of gut microbiota and oral antibiotic-induced dysbiosis on anti-PD-1 treatment effect.102 It was observed that mice receiving fecal transplantation from responders reacted better to PD-1 blockade with increased CXCL3+CD4+ TILs, while mice receiving fecal transplantation from nonresponders, in germ-free status, or undergoing antibiotic treatment were resistant to PD-1 blockade.102 Notably, the PD-1 resistance in germ-free or antibiotics-treated mice could be rescued by recolonization of A muciniphila and Enterococcus hirae.102 Further exploration showed that accumulated CXCR3+CCR9+CD4+ T cells and DC-IL-12 axis–mediated Th1-skewing priming might contribute to the enhanced therapeutic response to PD-1 blockade.102
The Effect of Gut Commensals on Anti-CTLA-4 Treatment
CTLA-4 blockade treatment mainly restores the activity of co-stimulatory signaling pathway (CD28-CD80/86) hijacked by CTLA-4. Exploring factors modulating anti-CTLA-4 treatment effect is helpful to enhance treatment response and relieve drug resistance. Vétizou et al found that some special gut bacteria supplements could enhance the effect of CTLA-4 blockade.103 Researchers noticed that broad-spectrum antibiotic treatment abrogated the antitumor effect of CTLA-4 blockade. Additionally, anti-CTLA-4 antibody could not effectively inhibit tumor progression in germ-free mice indicating that gut microbiota might participate in anti-CTLA-4 treatment.103 Recolonization of Bacteroides thetaiotaomicron, B fragilis, or Burkholderia cepacia in germ-free or antibiotic-treated mice rescued CTLA-4 blockade resistance.103 Further detection showed that oral gavage of B fragilis induced Th1 immune response and DC maturation in tumor-draining lymph node.103 Besides, adoptive B fragilis–specific Th1 cell transfer could partially restore sensitivity to CTLA-4 blockade in germ-free or antibiotic-treated mice.103 Apart from the enhanced CTLA-4 blockade effect, the recolonization of B fragilis and Burkholderia cepacia could alleviate treatment-induced colitis.103
Later in 2017, Chaput et al verified the regulatory effect of gut microbiota on CTLA-4 blockade in metastatic melanoma patients.104 In the recruited patients, baseline microbiota composition could herald prognostic status after undergoing CTLA-4 blockade treatment.104 Overrepresented Bacteroides at baseline predicted poor outcomes (P = .034), while increased Faecalibacterium at baseline indicated long-term benefits (P = .0092).104 Besides, all patients with survival time longer than 18 months could be screened out by gut microbiota composition harboring overrepresented Ruminococcus and Lachnospiraceae genus (belonging to the Firmicutes phylum).104 Contrary to the observations mentioned above, baseline antibiotic treatment could not disturb the composition of gut microbiota.104 Then, clustering analysis indicated that patients with gut microbiota containing Faecalibacterium or other bacteria belonging to the Firmicutes phylum (eg, unclassified Ruminococcaceae, Clostridium XIVa, and Blautia) tended to possess better clinical outcomes (PFS: P = .039; OS: P = .051).104 In line with enhanced CTLA-4 blockade effect, patients with overrepresented Faecalibacterium or other Firmicutes had increased risk of treatment-induced colitis, especially compared with patients with Bacteroides-dominant gut microbiota.104 To interrogate the mechanisms by which Faecalibacterium-dominant gut microbiota composition increased CTLA-4 blockade effect and corresponding adverse events, researchers monitored immune status–related parameters.104 It was found that patients with Faecalibacterium-dominant gut microbiota had lower CD4+/CD8+ T cells and systemic proinflammatory cytokine levels at baseline, as well as higher ICOS expression on CD4+ T after the start of anti-CTLA-4 treatment.104 Presumably, Faecalibacterium and other Firmicutes contributed to decreased systemic inflammation by inducing the development of Treg at baseline.104 However, as the primary target of anti-CTLA-4 treatment, increased Treg level endowed patients with elevated sensitivity to CTLA-4 blockade as well as decreased risk of treatment-induced colitis.104
Manipulating Gut Microbiota to Enhance Effect of ACT
The therapeutic effect of ACT is limited by peripheral tolerance and immune escape in the tumor microenvironment.105 Uribe-Herranz et al found that manipulating the composition of gut microbiota could modulate the effect of ACT.105 Researchers found that the treatment effect of ACT was different in genetically similar mice (C57BL/6) from 2 vendors (JAX and HAR). The 16S rRNA gene sequencing of stool samples distinguished gut microbiota composition between JAX and HAR: Bacteroidales S24-7 dominant commensals in JAX while a wide range of bacteria belonging to the Bacteroidetes phylum in HAR.105 After vancomycin treatment, bacteria belonging to the Bacteroidetes phylum were eliminated in JAX and HAR.105 Vancomycin treatment did not change the effect of ACT in JAX.105 However, this antibiotic intervention significantly enhanced the efficacy of ACT in HAR to an extent similar to the treatment effect in JAX.105 Besides, additional vancomycin administration remarkably increased the abundance and activity of tumor-specific TIL.105 This transformation to hot tumor was attributed to accumulated CD8α+ DC and IL-12 in peripheral circulation, as well as concurrent enhanced Th1-skewed immune response.105
Putative Mechanisms by Which the Gut Microbiota May Regulate the Effect of Anticancer Immunotherapy
Anticancer immunity is described by a model called the cancer-immunity cycle. Tumor-derived antigens initiate the immune response.60 After capture and presentation of antigen presentation cells, naïve T cells are primed and activated in peripheral lymphatic organs.60 Then, primed T cells migrate and infiltrate the tumor bed.60 Following the recognition of tumor antigens, activated T cells kill tumor cells.60 During tumor initiation and progression, one or more steps in cancer-immunity cycle are impaired.60 Anticancer immunotherapy is developed to unleash the exhausted T cells and restore anticancer immune response.106 Based on the cancer-immunity cycle, immunotherapy could compensate for one or multiple undermined anticancer immune procedures. However, as a cascade reaction, the actual effect of immunotherapy is limited by its upstream or downstream factors such as systemic cytokine repertoire, the cross-presentation of antigen presentation cell, as well as the inhibitory components in the tumor immune microenvironment.107 Gut microbiota could regulate a broad range of immune effectors, especially DC. As the core of antigen presentation and T cell activation, the function of DC is the determinant of immune surveillance and immune clearance. Some bacteria such as Bifidobacterium could enhance the function of DC by promoting DC maturation, upregulating cytokine secretion, stimulating DC-IL-12-Th1-skewing immune response, as well as facilitating the activation and survival of tumor-specific T cells.99 The cross-talk between gut microbiota and DC in PP not only induces local immune response in gut mucosa but also regulates systemic immune response by the peripheral circulation.25 Locally generated cytokines and active DCs enter into circulation that could provide a favorable immune tone and synergize with concurrent anticancer immunotherapy.25,91 Besides this nonspecific immune augmentation, partial bacteria antigen–loaded DCs might lead to molecular mimicry and eliminate tumor cells sharing similar antigen repertoire with gut microbiota.91
Clinical Application of Gut Microbiota in Immunotherapy
Motivated by the encouraging results of preclinical studies, multiple clinical trials investigating the influence of gut microbiota on immune cancer efficacy are ongoing. In 2019, Jin et al reported the data from non–small cell lung cancer patients undergoing nivolumab therapy (patients were enrolled from CheckMate 078 and CheckMate 870 studies).108 By analyzing the fecal samples of patients before and after anti-PD-1 therapy via 16S rRNA gene sequencing, researchers found that patients with higher diversity of gut microbiota possessed prolonged PFS compared with ones with lower diversity of gut microbiota.108 Moreover, patients responding to nivolumab therapy possessed higher diversity of gut microbiota at baseline, which sustained stable composition during treatment.108 Composition difference analysis between responder group and nonresponder group showed that bacteria such as Alistipes putredinis, B longum, and Prevotella copri were significantly enriched in responders, while unclassified Ruminococcus were enriched in nonresponders.108 Besides, given that antibiotics could reshape the composition of gut microbiota that further interferes the effect of immunotherapy, the relationship between antibiotic-associated dysbiosis and immunotherapy is another hot topic.109 Elkrief et al found that antibiotic treatment before immunotherapy such as anti-PD-1/PD-L1 and anti-CTLA-4 was an independent risk factor for worse PFS (hazard ratio = 0.32, 95% confidence interval = 0.13-0.83, P = .02).109 Patients receiving antibiotic treatment prior to immunotherapy exhibited lower possibility to effectively respond to immunotherapy (objective response rate of antibiotic group vs control group = 0% vs 34%) and improved prognosis (PFS of antibiotic group vs control group = 0.28, 95% confidence interval = 0.10-0.76, P = .01).109
Apart from utilizing gut microbiota to predict the efficacy of immunotherapy, some clinical studies focused on how to modulate the composition of gut microbiota to overcome anti-PD-1/PD-L1 resistance. NCT03341143 is a single-center phase 2 trial interrogating the efficacy of fecal microbiota transplant (FMT) plus pembrolizumab in melanoma patients resistant to anti-PD-1 therapy.110 In this phase 2 trial, FMT was conducted as following procedures: collecting stool from tested donors, mixing with saline or other solutions, then straining and infusing into colon by colonoscopy.110 NCT03341143 is ongoing, and the results of this study have not been reported.110 Meanwhile, NCT03595683 (phase 2 trial) evaluated the treatment effect of pembrolizumab with additional EDP1503 (an orally delivered monoclonal microbiota product).111 In this trial, 70 melanoma patients were involved and received pembrolizumab treatment (200 mg/3 weeks) and concurrent EDP1503 (≥15 × 1010 colony-forming units/day).111 Moreover, the treatment effect of combination therapy of other additional oral microbiome interventions such as SER-401 (NCT03817125) and ICI are under investigation.112
Although a myriad of preclinical studies demonstrated that gut microbiota regulated host systemic immune response, modulated immunotherapy efficacy, and affected treatment-induced adverse effects, the regulatory function of certain commensal bacteria still needs further investigation, especially for the extrapolation from the mouse model to humans. The results of these ongoing studies might provide more stable evidence to support the feasibility of enhancing immunotherapy effect by modulating gut microbiota composition. However, it is notable that original gut mucosa commensals interfere with the colonization of supplemental probiotics.113 The extent of resistance to probiotics colonization is heterogeneous among populations and could be influenced by baseline commensal status.113 Therefore, patient’s commensal background should be taken into consideration for manipulating gut microbiota by interventions such as fecal transplantation. Notably, in 2019, it has been reported that 2 patients receiving FMT treatment developed invasive infections caused by multidrug-resistant organisms and one of the patients died. It is necessary to keep alert to FMT therapy–induced adverse events in further clinical investigation.
Conclusion
Gut microbiota has a substantial influence on host immune response and modulates multiple steps of cancer-immunity cycle including antigen presentation, T cell priming, and activation. Manipulating gut microbiota to induce the formation of systemically immunologic tone is helpful to enhance effect and overcome resistance in immunotherapy. Identifying favorable bacteria and exploring feasible approaches to manipulating gut microbiota would be meaningful to cancer immunotherapy.
Footnotes
Author Contributions: MY performed the selection of literature, drafted the manuscript, and prepared the figures. DJ, SQ, and QC collected the related references and participated in discussion. KW and AL designed this review and revised the manuscript. All authors contributed to this manuscript. All authors read and approved the final manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (Nos. 81874120, 81572608, 81672984), and the Wuhan Science and Technology Bureau (No. 2017060201010170).
ORCID iD: Kongming Wu  https://orcid.org/0000-0003-2499-1032
https://orcid.org/0000-0003-2499-1032
References
- 1. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512-519. [DOI] [PubMed] [Google Scholar]
- 3. Sarkar SR, Banerjee S. Gut microbiota in neurodegenerative disorders. J Neuroimmunol. 2019;328:98-104. [DOI] [PubMed] [Google Scholar]
- 4. Srinivasan K, Buys EM. Insights into the role of bacteria in vitamin A biosynthesis: future research opportunities [published online January 13, 2019]. Crit Rev Food Sci Nutr. doi: 10.1080/10408398.2018.1546670 [DOI] [PubMed] [Google Scholar]
- 5. Le Roy CI, Woodward MJ, Ellis RJ, La Ragione RM, Claus SP. Antibiotic treatment triggers gut dysbiosis and modulates metabolism in a chicken model of gastro-intestinal infection. BMC Vet Res. 2019;15:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero R, et al. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients. 2018;11:E51. doi: 10.3390/nu11010051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219-232. [DOI] [PubMed] [Google Scholar]
- 8. Safari Z, Gérard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD). Cell Mol Life Sci. 2019;76:1541-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sugihara K, Morhardt TL, Kamada N. The role of dietary nutrients in inflammatory bowel disease. Front Immunol. 2019;9:3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quigley EMM. Microbiota-brain-gut axis and neurodegenerative diseases. Curr Neurol Neurosci Rep. 2017;17:94. [DOI] [PubMed] [Google Scholar]
- 11. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022-1023. [DOI] [PubMed] [Google Scholar]
- 12. Zhang L, Wu YN, Chen T, Ren CH, Li X, Liu GX. Relationship between intestinal microbial dysbiosis and primary liver cancer. Hepatobiliary Pancreat Dis Int. 2019;18:149-157. [DOI] [PubMed] [Google Scholar]
- 13. Lin C, Cai X, Zhang J, et al. Role of gut microbiota in the development and treatment of colorectal cancer. Digestion. 2019;100:72-78. [DOI] [PubMed] [Google Scholar]
- 14. Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014;345:196-202. [DOI] [PubMed] [Google Scholar]
- 15. Di Domenico EG, Cavallo I, Pontone M, Toma L, Ensoli F. Biofilm producing Salmonella typhi: chronic colonization and development of gallbladder cancer. Int J Mol Sci. 2017;18:E1887. doi: 10.3390/ijms18091887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. D’Amelio P, Sassi F. Gut microbiota, immune system, and bone. Calcif Tissue Int. 2018;102:415-425. [DOI] [PubMed] [Google Scholar]
- 17. Pronovost GN, Hsiao EY. Perinatal interactions between the microbiome, immunity, and neurodevelopment. Immunity. 2019;50:18-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swartwout B, Luo XM. Implications of probiotics on the maternal-neonatal interface: gut microbiota, immunomodulation, and autoimmunity. Front Immunol. 2018;9:2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cianci R, Franza L, Schinzari G, et al. The interplay between immunity and microbiota at intestinal immunological niche: the case of cancer. Int J Mol Sci. 2019;20:E501. doi: 10.3390/ijms20030501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14:356-365. [DOI] [PubMed] [Google Scholar]
- 21. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17:271-285. [DOI] [PubMed] [Google Scholar]
- 22. Yi M, Qin S, Chu Q, Wu K. The role of gut microbiota in immune checkpoint inhibitor therapy. Hepatobiliary Surg Nutr. 2018;7:481-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Ding X, Guan R, et al. Evaluation of different 16S rRNA gene V regions for exploring bacterial diversity in a eutrophic freshwater lake. Sci Total Environ. 2018;618:1254-1267. [DOI] [PubMed] [Google Scholar]
- 24. D’Argenio V. Human microbiome acquisition and bioinformatic challenges in metagenomic studies. Int J Mol Sci. 2018;19:E383. doi: 10.3390/ijms19020383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33:570-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spiljar M, Merkler D, Trajkovski M. The immune system bridges the gut microbiota with systemic energy homeostasis: focus on TLRs, mucosal barrier, and SCFAs. Front Immunol. 2017;8:1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Josefsdottir KS, Baldridge MT, Kadmon CS, King KY. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 2017;129:729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kunisawa J, Kurashima Y, Kiyono H. Gut-associated lymphoid tissues for the development of oral vaccines. Adv Drug Deliv Rev. 2012;64:523-530. [DOI] [PubMed] [Google Scholar]
- 29. Suzuki K, Kawamoto S, Maruya M, Fagarasan S. GALT: organization and dynamics leading to IgA synthesis. Adv Immunol. 2010;107:153-185. [DOI] [PubMed] [Google Scholar]
- 30. Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. 2010;28:243-273. [DOI] [PubMed] [Google Scholar]
- 31. Shi N, Li N, Duan X, Niu H. Interaction between the gut microbiome and mucosal immune system. Mil Med Res. 2017;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016;16:639-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ayabe T, Ashida T, Kohgo Y, Kono T. The role of Paneth cells and their antimicrobial peptides in innate host defense. Trends Microbiol. 2004;12:394-398. [DOI] [PubMed] [Google Scholar]
- 34. Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mukherjee S, Zheng H, Derebe MG, et al. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature. 2014;505:103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reboldi A, Cyster JG. Peyer’s patches: organizing B-cell responses at the intestinal frontier. Immunol Rev. 2016;271:230-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Foussat A, Balabanian K, Amara A, et al. Production of stromal cell-derived factor 1 by mesothelial cells and effects of this chemokine on peritoneal B lymphocytes. Eur J Immunol. 2001;31:350-359. [DOI] [PubMed] [Google Scholar]
- 39. Johansson-Lindbom B, Svensson M, Pabst O, et al. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. 2005;202:1063-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johansson-Lindbom B, Svensson M, Wurbel MA, Malissen B, Márquez G, Agace W. Selective generation of gut tropic T cells in gut-associated lymphoid tissue (GALT): requirement for GALT dendritic cells and adjuvant. J Exp Med. 2003;198:963-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mora JR, Bono MR, Manjunath N, et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 2003;424:88-93. [DOI] [PubMed] [Google Scholar]
- 42. Rescigno M, Urbano M, Valzasina B, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361-367. [DOI] [PubMed] [Google Scholar]
- 43. Niess JH, Brand S, Gu X, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254-258. [DOI] [PubMed] [Google Scholar]
- 44. Ferber D. Immunology. The education of T cells. Science. 2007;316:191-193. [DOI] [PubMed] [Google Scholar]
- 45. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204-12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cebula A, Seweryn M, Rempala GA, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677-689. [DOI] [PubMed] [Google Scholar]
- 49. Tomkovich S, Jobin C. Microbiota and host immune responses: a love-hate relationship. Immunology. 2016;147:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu J, Lin S, Zheng B, Cheung PCK. Short-chain fatty acids in control of energy metabolism. Crit Rev Food Sci Nutr. 2018;58:1243-1249. [DOI] [PubMed] [Google Scholar]
- 52. McNabney SM, Henagan TM. Short chain fatty acids in the colon and peripheral tissues: a focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients. 2017;9:E1348. doi: 10.3390/nu9121348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dehner C, Fine R, Kriegel MA. The microbiome in systemic autoimmune disease: mechanistic insights from recent studies. Curr Opin Rheumatol. 2019;31:201-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stewart LJ, Edgar JDM, Blakely G, Patrick S. Antigenic mimicry of ubiquitin by the gut bacterium Bacteroides fragilis: a potential link with autoimmune disease. Clin Exp Immunol. 2018;194:153-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017;46:562-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Capietto AH, Jhunjhunwala S, Delamarre L. Characterizing neoantigens for personalized cancer immunotherapy. Curr Opin Immunol. 2017;46:58-65. [DOI] [PubMed] [Google Scholar]
- 58. Wagner S, Mullins CS, Linnebacher M. Colorectal cancer vaccines: tumor-associated antigens vs neoantigens. World J Gastroenterol. 2018;24:5418-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yi M, Qin S, Zhao W, Yu S, Chu Q, Wu K. The role of neoantigen in immune checkpoint blockade therapy. Exp Hematol Oncol. 2018;7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1-10. [DOI] [PubMed] [Google Scholar]
- 61. Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol. 2016;27:1492-1504. [DOI] [PubMed] [Google Scholar]
- 62. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565-1570. [DOI] [PubMed] [Google Scholar]
- 63. Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res. 2015;21:687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest. 2015;125:3335-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yi M, Jiao D, Xu H, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li X, Shao C, Shi Y, Han W. Lessons learned from the blockade of immune checkpoints in cancer immunotherapy. J Hematol Oncol. 2018;11:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ramachandran M, Dimberg A, Essand M. The cancer-immunity cycle as rational design for synthetic cancer drugs: novel DC vaccines and CAR T-cells. Semin Cancer Biol. 2017;45:23-35. [DOI] [PubMed] [Google Scholar]
- 69. Kumar S, Leigh ND, Cao X. The role of co-stimulatory/co-inhibitory signals in graft-vs-host disease. Front Immunol. 2018;9:3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jiang X, Wang J, Deng X, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Long J, Lin J, Wang A, et al. PD-1/PD-L blockade in gastrointestinal cancers: lessons learned and the road toward precision immunotherapy. J Hematol Oncol. 2017;10:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Marin-Acevedo JA, Dholaria B, Soyano AE, Knutson KL, Chumsri S, Lou Y. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J Hematol Oncol. 2018;11:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dyck L, Mills KHG. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur J Immunol. 2017;47:765-779. [DOI] [PubMed] [Google Scholar]
- 74. Ren B, Cui M, Yang G, et al. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol Cancer. 2018;17:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Horn L, Mansfield AS, Szczesna A, et al. ; IMpower133 Study Group. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. 2018;379:2220-2229. [DOI] [PubMed] [Google Scholar]
- 76. Migden MR, Rischin D, Schmults CD, et al. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med. 2018;379:341-351. [DOI] [PubMed] [Google Scholar]
- 77. Eggermont AMM, Blank CU, Mandala M, et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N Engl J Med. 2018;378:1789-1801. [DOI] [PubMed] [Google Scholar]
- 78. Forde PM, Chaft JE, Smith KN, et al. Neoadjuvant PD-1 blockade in resectable lung cancer. N Engl J Med. 2018;378:1976-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Garassino MC, Cho BC, Kim JH, et al. ; ATLANTIC Investigators. Durvalumab as third-line or later treatment for advanced non-small-cell lung cancer (ATLANTIC): an open-label, single-arm, phase 2 study. Lancet Oncol. 2018;19:521-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xue S, Hu M, Iyer V, Yu J. Blocking the PD-1/PD-L1 pathway in glioma: a potential new treatment strategy. J Hematol Oncol. 2017;10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Met Ö, Jensen KM, Chamberlain CA, Donia M, Svane IM. Principles of adoptive T cell therapy in cancer. Semin Immunopathol. 2019;41:49-58. [DOI] [PubMed] [Google Scholar]
- 82. Choi BD, Maus MV, June CH, Sampson JH. Immunotherapy for glioblastoma: adoptive T-cell strategies. Clin Cancer Res. 2019;25:2042-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yu S, Li A, Liu Q, et al. Chimeric antigen receptor T cells: a novel therapy for solid tumors. J Hematol Oncol. 2017;10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fan J, Shang D, Han B, Song J, Chen H, Yang JM. Adoptive cell transfer: is it a promising immunotherapy for colorectal cancer? Theranostics. 2018;8:5784-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cogdill AP, Gaudreau PO, Arora R, Gopalakrishnan V, Wargo JA. The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol. 2018;39:900-920. [DOI] [PubMed] [Google Scholar]
- 86. Wang J, Chen S, Xiao W, et al. CAR-T cells targeting CLL-1 as an approach to treat acute myeloid leukemia. J Hematol Oncol. 2018;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Neelapu SS, Locke FL, Bartlett NL, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med. 2017;377:2531-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Maude SL, Laetsch TW, Buechner J, et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med. 2018;378:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Newick K, O’Brien S, Moon E, Albelda SM. CAR T cell therapy for solid tumors. Annu Rev Med. 2017;68:139-152. [DOI] [PubMed] [Google Scholar]
- 90. DeRenzo C, Gottschalk S. Genetic modification strategies to enhance CAR T cell persistence for patients with solid tumors. Front Immunol. 2019;10:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yi M, Yu S, Qin S, et al. Gut microbiome modulates efficacy of immune checkpoint inhibitors. J Hematol Oncol. 2018;11:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Parry RV, Chemnitz JM, Frauwirth KA, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol. 2005;25:9543-9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bardhan K, Anagnostou T, Boussiotis VA. The PD1:PD-L1/2 pathway from discovery to clinical implementation. Front Immunol. 2016;7:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Patsoukis N, Brown J, Petkova V, Liu F, Li L, Boussiotis VA. Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci Signal. 2012;5:ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Somasundaram A, Burns TF. The next generation of immunotherapy: keeping lung cancer in check. J Hematol Oncol. 2017;10:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ohaegbulam KC, Assal A, Lazar-Molnar E, Yao Y, Zang X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol Med. 2015;21:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liu SY, Wu YL. Ongoing clinical trials of PD-1 and PD-L1 inhibitors for lung cancer in China. J Hematol Oncol. 2017;10:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Massari F, Santoni M, Ciccarese C, et al. PD-1 blockade therapy in renal cell carcinoma: current studies and future promises. Cancer Treat Rev. 2015;41:114-121. [DOI] [PubMed] [Google Scholar]
- 99. Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-97. [DOI] [PubMed] [Google Scholar]
- 103. Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368-1379. [DOI] [PubMed] [Google Scholar]
- 105. Uribe-Herranz M, Bittinger K, Rafail S, et al. Gut microbiota modulates adoptive cell therapy via CD8α dendritic cells and IL-12. JCI Insight. 2018;3:94952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321-330. [DOI] [PubMed] [Google Scholar]
- 107. Li A, Yi M, Qin S, Song Y, Chu Q, Wu K. Activating cGAS-STING pathway for the optimal effect of cancer immunotherapy. J Hematol Oncol. 2019;12:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jin Y, Dong H, Xia L, et al. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in Chinese patients with NSCLC. J Thorac Oncol. 2019;14:1378-1389. [DOI] [PubMed] [Google Scholar]
- 109. Elkrief A, El Raichani L, Richard C, et al. Antibiotics are associated with decreased progression-free survival of advanced melanoma patients treated with immune checkpoint inhibitors. Oncoimmunology. 2019;8:e1568812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. ClinicalTrials.gov. Fecal microbiota transplant (FMT) in melanoma patients. https://www.clinicaltrials.gov/ct2/show/NCT03341143. Published November 14, 2017. Accessed August 30, 2019.
- 111. ClinicalTrials.gov. Pembrolizumab and EDP1503 in advanced melanoma. https://www.clinicaltrials.gov/ct2/show/NCT03595683. Published July 23, 2018. Accessed August 30, 2019.
- 112. ClinicalTrials.gov. Melanoma checkpoint and gut microbiome alteration with microbiome intervention (MCGRAW). https://www.clinicaltrials.gov/ct2/show/NCT03817125. Published January 25, 2019. Accessed August 30, 2019.
- 113. Zmora N, Zilberman-Schapira G, Suez J, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388-1405.e21. [DOI] [PubMed] [Google Scholar]