Summary
The application of machine learning toward DNA encoded library (DEL) technology is lacking despite obvious synergy between these two advancing technologies. Herein, a machine learning algorithm has been developed that predicts the conversion rate for the DNA-compatible reaction of a building block with a model DNA-conjugate. We exemplify the value of this technique with a challenging reaction, the Pictet-Spengler, where acidic conditions are normally required to achieve the desired cyclization between tryptophan and aldehydes to provide tryptolines. This is the first demonstration of using a machine learning algorithm to cull potential building blocks prior to their purchase and testing for DNA-encoded library synthesis. Importantly, this allows for a challenging reaction, with an otherwise very low building block pass rate in the test reaction, to still be used in DEL synthesis. Furthermore, because our protocol is solution phase it is directly applicable to standard plate-based DEL synthesis.
Subject Areas: Organic Chemistry, Organic Reaction, Artificial Intelligence
Graphical Abstract
Highlights
-
•
A mild solution-phase, plate applicable DNA-compatible Pictet-Spengler (PS) reaction
-
•
An efficient strategy for DNA-encoded diversified tryptoline libraries synthesis
-
•
A machine learning algorithm of building blocks filtering for DEL synthesis
-
•
An elegant application of machine learning for DNA-encoded library technology
Organic Chemistry; Organic Reaction; Artificial Intelligence
Introduction
DNA-encoded libraries (DELs) are collections of small molecules covalently linked to unique, structure-identifying DNA tags, which enable screens of a large pool of billions (even trillions) of library members for binders of disease-related biologically interesting targets (Clark et al., 2009, Ralph et al., 2011, Favalli et al., 2018, Neri and Lerner, 2018, Zhou et al., 2018, Faver et al., 2019, Reddavide et al., 2019, Dichson and Kodadek, 2019, Yuen et al., 2019). Compared with traditional combinatorial encoded methods, a distinctive and amplifiable DNA tag facilitates the decoding process and enables the screening of much larger libraries (trillions versus millions) (Buller et al., 2010, Encinas et al., 2014, Franzini and Randolph, 2016, Ottl et al., 2019). After affinity selection, the hit molecule's structural information is deciphered from the attached DNA via next-generation sequencing (Eidam and Satz, 2016, Roman et al., 2018).
High-quality DELs are the basis for the success of subsequent screening experiments, and quality includes high conversion rate for each building block (BB) used during library synthesis. Thousands of BBs are routinely reacted with a model DNA-conjugate to determine their appropriateness for use, in a particular reaction, prior to DNA-encoded library (DEL) synthesis. For all investigated reactions, a significant percentage of the acquired and tested BBs fail this validation step (generally a >50% conversion to desired product is required for a BB to “pass” the validation), greatly increasing reagent costs and library development time. Additionally, for particularly challenging reactions, the BB pass rate can be extremely low. Owing to the limited resources, it is not practical to pick high-conversion-rate BBs by experimentally determining the conversion rate of each commercially available BB, as a significant percentage of purchased BBs will fail to pass this validation step. To maximize the likelihood that purchased BBs will pass chemical validation, we envision the use of an informatics filter that could readily and inexpensively assess the likelihood of any particular BB to provide a high yield of desired product. Machine learning (ML) is a technology to build a mathematical model based on sample data, known as "training data," in order to make predictions or decisions without being explicitly programmed how to make the decision (Bishop, 2006). Great successes have been made with the method in the field of computer vision, natural language processing, and biological medicine (LeCun et al., 2015). However, no research regarding DEL reaction conversion rate ML prediction has been reported. Studies in traditional organic synthesis have used ML for the estimation of catalytic performance (Kite et al., 1994, Omata and Yamada, 2004) and reaction success (Skoraczynski et al., 2017, Raccuglia et al., 2016). More recently, a study has applied descriptors obtained by quantum chemical calculation to predict reaction yield (Ahneman et al., 2018). However, quantum chemical calculation is a time-consuming process, which is not practical when applied to DEL reaction yield prediction because tens of thousands of BBs are needed to be evaluated in a library constructing process.
Furthermore, applying ML for BB filtering is particularly valuable for challenging DNA-compatible reactions owing to the expected low BB passing rate. Although DEL is successful for hit identification and widely used throughout the academic and industrial small molecule drug discovery community, it still suffers from a limited number of DNA-compatible reactions and thus limited access to desirable drug-like chemical space (Satz et al., 2015, Malone and Paegel, 2016, Lu et al., 2017a, Lu et al., 2017b, Wang et al., 2018a, Wang et al., 2018b, Li et al., 2018, Flood et al., 2019, Wang et al., 2019, Du et al., 2019, Lerner et al., 2019, Liu et al., 2019, Škopic et al., 2019, Xu et al., 2019). More DNA-compatible organic transformations, especially the challenge but highly valuable ones, are strongly desired to improve the chemical diversity of DNA-encoded libraries. We envisioned develop a new challenge DNA-compatible reaction and applied ML for the BB filtering for DEL synthesis is a rational strategy for DEL chemical space expansion especially for valuable privileged scaffolds based DELs.
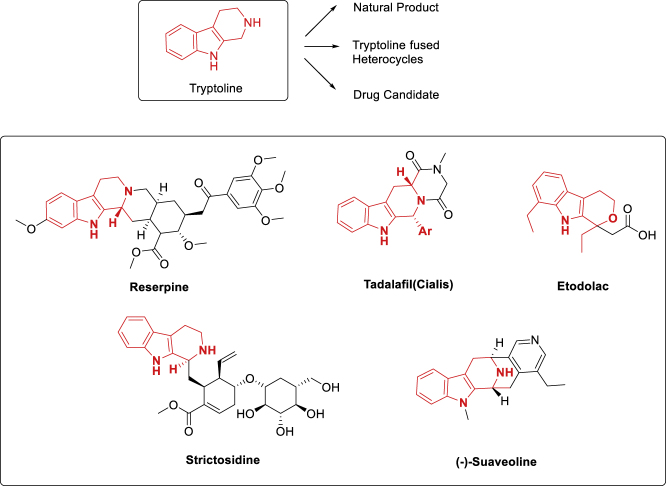
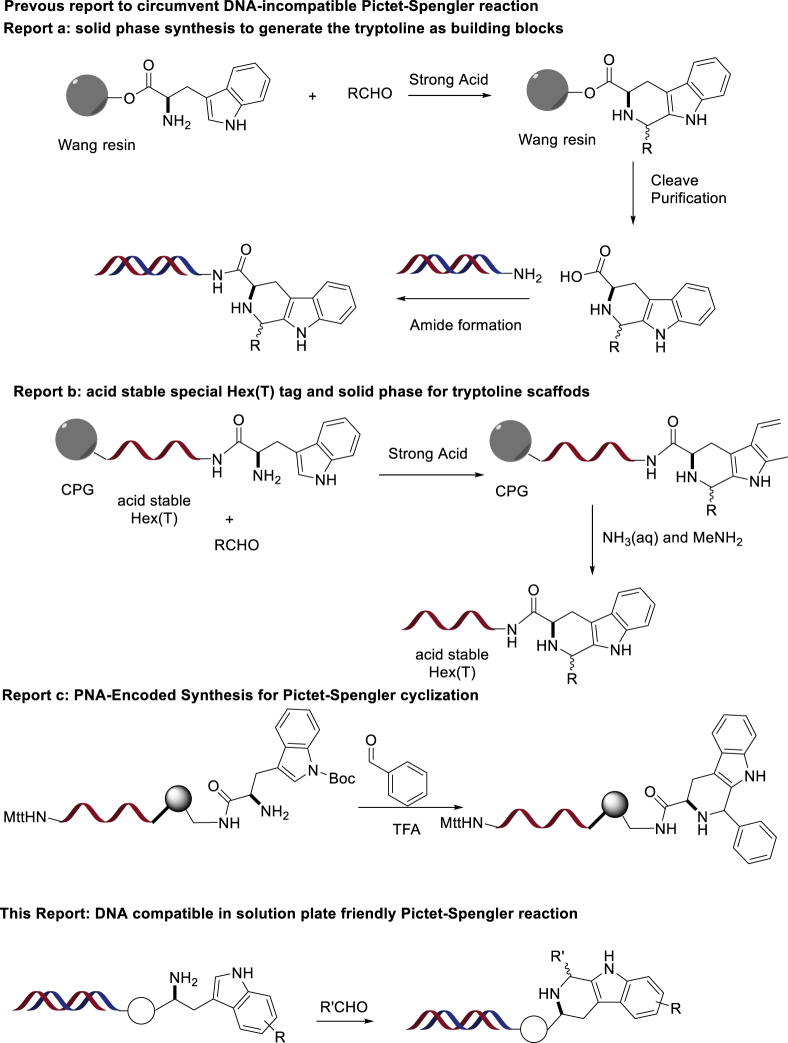
We decided to focus on the DNA-compatible cyclization of highly functionalized and rigid rings. (Note that our laboratory has previously discussed the design and synthesis of orthogonally protected heterocyclic scaffolds for use in DEL synthesis [Gong et al., 2017]). Poly-substituted optically active tryptoline derivatives classified as nonisoprenoids are common structural motifs in indole-based alkaloids. As depicted in Figure 1, functionalization of the C-1 position of tryptoline derivatives is generally observed in natural-product-based indole alkaloids and commercial drugs such as tadalafil (Cialis) (Yamamura et al., 2017) and etodolac (LaPlante et al., 2013, Maity et al., 2019). The World Drug Index contains over 200 listings of this distinctive heterocycle, which is usually assembled by the (Pictet-Spengler) PS reaction (Maity et al., 2019). Unfortunately, owing to the acidic conditions required for the PS reaction, there were no DEL PS reactions reported in the literature until recently. Importantly, both reports demonstrate proof of concept only and would require non-trivial deviation from existing protocols to actually synthesize a DEL. One report details a high-throughput solid phase methodology to synthesize tryptoline-containing BBs (Zambaldo et al., 2019). After release of the desired products from resin, the tryptoline products were then conjugated to DNA oligomers in a 96-well plate-based format. The second method demonstrated that short DNA oligomers attached to resins were protected from mildly acidic conditions, such that the PS reaction could be successfully accomplished in combination with electron-poor aldehydes (Figure 2) (Skopic et al., 2017). Besides, the preparation of PNA monomers with a protecting group combination (Mtt/Boc), which is orthogonal to Fmoc-based synthesis and compatible with PS reaction, was also elegantly demonstrated (Chouikhi et al., 2012). Despite the potential of the above methods, we believe there is still a clear need for a solution-phase PS reaction that is compatible with existing and proven DEL synthesis protocols (Figure 2).
Figure 1.
Bioactive C-1-Functionalized Tryptoline Derivatives through the Pictet-Spengler Cyclization Strategy
Figure 2.
In-Solution Plate Friendly DNA-Compatible Pictet-Spengler Reaction
Herein, we discuss the optimization (and reagent design) of a DNA-compatible and solution-phase PS reaction. The PS is a challenging reaction, as conditions that increase conversion rate often also increase DNA damage. Thus, under our optimized reaction conditions (which avoids DNA damage), a majority of randomly chosen aldehyde BBs fail to give high conversation to desired cyclized products. To better filter commercial BBs prior to their purchase, we trained a deep neural network (DNN) model (a type of ML model) to predict the conversion rate of BBs in our challenging PS reaction. We then purchased a subset of these BBs and compared our model's predictions with experimental results.
Results and Discussion
The Development of On-DNA PS Reaction
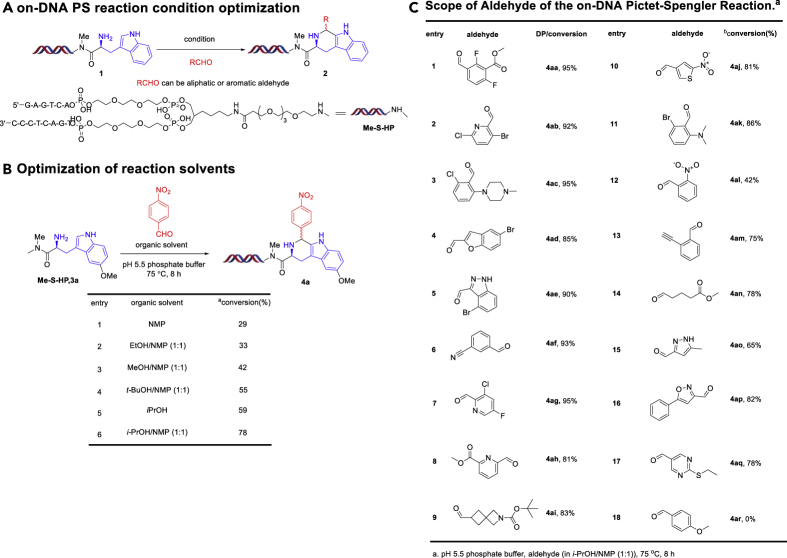
Developing a DNA-compatible solution-phase PS reaction using DNA-conjugated tryptamine substrates 1 and providing the desired products 2 (Figure 3A) (Zambaldo et al., 2019) is challenging since acidic conditions are typically required. To optimize a DNA-compatible reaction, we carried out parallel screening to test whether any acidic promoter could not only efficiently promote the PS reaction with simple aldehydes but also preserve the DNA without decomposition. Unfortunately, unlike reported literature procedures in traditional organic solvents, neither Lewis acids (Srinivasan and Ganesan, 2003) Sc(OTf)3, In(OTf)3, YbCl3, YCl3, Sm(OTf)3 nor Brønsted acids H3PO4, HCOOH appeared to promote the PS reaction, and only DNA damage was observed. Unsurprisingly, basic conditions such as adding NaOH or employing pH12 buffer also did not promote the reaction, and using I2 (Dipak and Mukut, 2008) also gave disappointing results.
Figure 3.
On-DNA PS Reaction Condition Optimization
(A) On-DNA PS reaction condition optimization.
(B) Optimization of reaction solvents.
(C) Scope of aldehyde of the on-DNA Pictet-Spengler Reaction. aConversions determined by liquid chromatography-mass spectrometry (LC-MS).
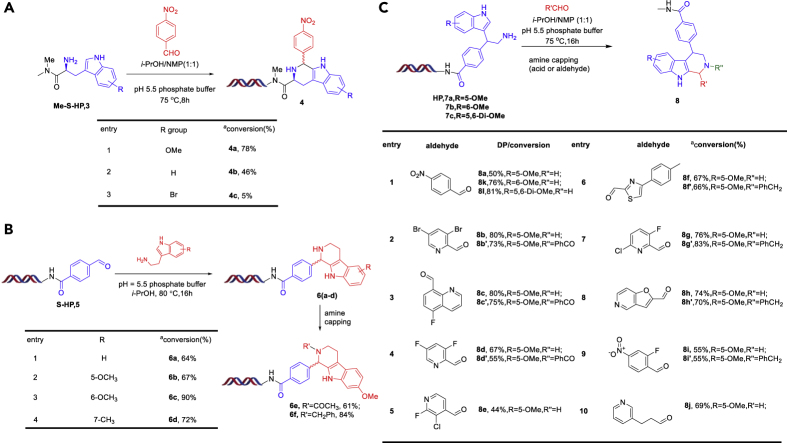
Drawing upon existing literature reports and the known mechanism of the PS reaction, we hypothesized that the combination of an electronic-rich tryptamine derivative and an electronic-deficient aldehyde may have better reactivity compared with the previous substrates 1. Thus, we chose to investigate a methoxy-substituted tryptamine-conjugated DNA substrate 3a. Employing a pH5.5 phosphate buffer to maintain a weakly acidic condition, we observed ∼29% conversion to the desired cyclized product 4a and no obvious signs of DNA damage. Next, we screened a series of solvents as tabulated in Figure 3B. The solvent i-PrOH led to an increase to 59% conversion, despite the test aldehyde 4-Nitrobenzaldehyde being poorly soluble under these conditions (Entry 2, Figure 3B). Thus, we chose a mixture of NMP and i-PrOH to improve solubility and observed a further increase in conversion to 78% (Entry 3, Figure 3B).
Next, we explored the scope of our optimized conditions and gratifyingly saw the PS reaction proceed smoothly with a broad spectrum of aldehydes. We found that our optimized conditions tolerated different functional groups including halides (Figure 3C, entries 1, 2, 3, 4, 5, 7, 11, 15), esters (entries 1, 8, 14), alkynes (entry 13), t-butyloxy carbonyls (entry 9), and nitriles (entry 6). Most of the heterocyclic aryl aldehydes gave moderate to excellent conversion (entries 4, 5, 15, 16, 17); however, an electron-rich (OCH3) aryl aldehyde even does not work (entry 18). And several aldehydes gave two different cyclization products because of stereo isomers (entries 1, 3, 5, 6, 7, 8, 10, 12, 16, 17) (for details see the Supplemental Information). To confirm that we were correctly assigning our DNA-conjugated products, we synthesized the corresponding off-DNA small molecule 4 as the free acid and then acylated this fully characterized cyclized molecule onto a DNA-conjugate. HPLC comparison of the two batches of 4 confirmed them to be the same (see the Supplemental Information).
DNN Model Construction and Validation
From the above exploration of aldehydes, 1,655 reaction records were collected (Table S1), based on which a DNN model was established (k-NearestNeighbor algorithm, KNN, a traditional machine learning methodwas also investigated as a baseline model, see Table S5). The extended connectivity fingerprints (ECPF4 [David and Mathew, 2010]) with a radius of two consecutive bonds and a length of 1,024 bits were used as input features, and conversion rates were used as the output task for learning (MACCS keys fingerprints were also tried; for details see Table S6).
To train the model, 20% of the data was randomly selected as an internal test dataset and the rest was selected as training dataset. Five-fold cross-validation was performed within the training dataset during the training process. In detail, training dataset was split into five folds and each fold is then used once as a validation, whereas the four remaining folds form the training set. For each fold, early stopping (Montavon et al., 2012) was applied and the training process was stopped when the mean square error (MSE) did not decrease for 50 epochs. The mean MSE of 5-fold cross-validation was chosen for searching the optimal k of KNN, and parameters of DNN and Adam optimizer (Kingma and Ba, 2014) were chosen for DNN parameters optimization (Table S2). In order to reduce the risk of overfitting, dropout (Srivastava et al., 2014) and weight decay (Kingma and Ba, 2014) were used for regularization. Bayesian optimization (BO) was also applied with pyGPGO (Jiménez and Ginebra, 2017) for DNN; however, no better hyper parameters were found. In the end, the DNN model with two hidden layers and the size 1,024 combined with ECFP4 showed the lowest mean MSE in 5-fold cross-validation and was chosen for further use.
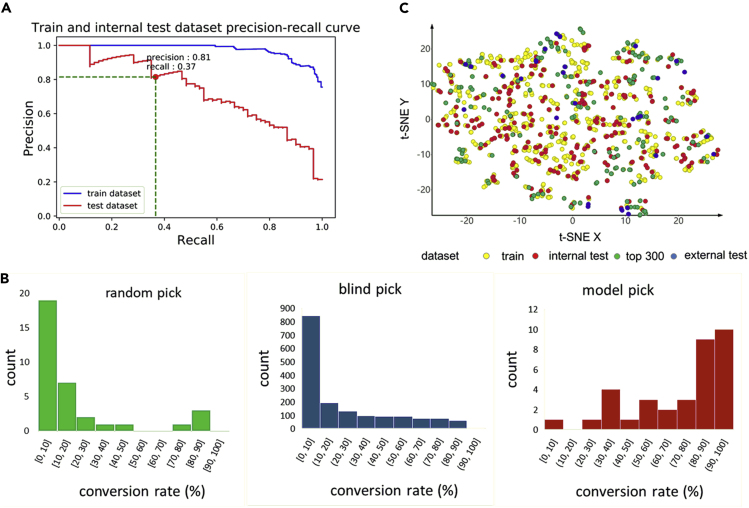
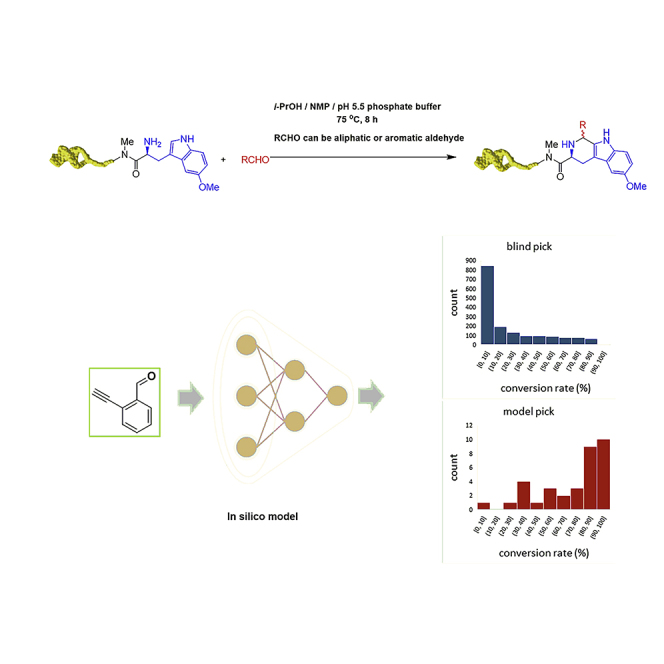
Generally, only BBs showing high conversion rate in a test PS reaction (i.e. the validation) will be selected for DEL construction and those with low conversion rate will be discarded. Thus, a useful model must correctly identify BBs that latter give high conversion rate in the test PS reaction. To better quantify the performance of the model, BBs with conversion rates over 50% are labeled 1 and others are labeled 0. Following this definition, a precision-recall curve can be plotted (Figure 4A). The results indicate a satisfactory performance for selecting BBs with high conversion rates (the precision is 0.81), although some positive samples might be missed (i.e., the recall is 0.37, but this is of less concern when picking a small subset of BBs from a large list of commercial reagents and clustering could be further used to make that the final selection of BBs to be as diverse as possible).
Figure 4.
Performance of DNN and Comparison with Random Pick and Blind Pick
(A) Model performance on train dataset and internal test dataset.
(B) Left: conversion rate distribution of 34 randomly picked BBs. Middle: conversion rate distribution of 1,655 blindly picked BBs. Right: conversion rate distribution of 34 model picked BBs.
(C) Structure distribution of train dataset, internal test dataset, top 300 and external test dataset shown as t-SNE plot.
We then carried out an external experimental validation of our ML model. All data containing aldehyde BB (Table S3) from the WuXi LabNetwork platform were collected, cleaned (remove ions), and evaluated by the model. The final predicted value of BB is the mean value of five best model in 5-fold cross-validation. In order to maximize the performance of the model, BBs were sorted in descending order according to predicted conversion rate and top 300 BBs were retained and those previously included in the train or internal test dataset were discarded (Table S4). After further filtering for price and real-time availability, 34 BBs were acquired for experimental evaluation (external test dataset, Table S4). The results show that the performance of the model on the external dataset was similar to that on the internal test dataset, with a precision for identifying BBs with conversion rate above 50% being 0.79 (0.81 for the internal test dataset). Compared with 1,655 blindly picked BBs, the model has a better performance to find high-conversion-rate BBs (high-conversion-rate BBs percentage: 18.4% versus 79.4%, Figure 4B). For parallel comparison of the performance of “random pick” and “model pick,” a random-pick test was carried out, where subsets with the size of 34 BBs were randomly selected from the WuXi LabNetwork platform (BBs already in train and external dataset were discarded), and their conversion rates distribution was depicted in Table S4 and Figure 4B. The results verified that the performance of “model pick” is better than that of “random pick.” In addition, t-SNE result (Figure 4C) and structure clustering analysis (Table S7) showed that diverse of structures have been included in model-recommended BBs (top 300 and external test) compared with “blind pick” (train and internal test). In practice, more rigorous clustering selection can be made to make picked BBs as diverse as possible if enough BBs are available. We believe our above-described model serves as a proof of concept in how BBs can be filtered for purchase, particularly in cases of challenging reactions with otherwise low validation pass rates.
The Scope of Pictet-Spengler Reaction
After exploring the scope of aldehydes reacting with DNA-conjugated tryptamine 3a, we further investigated differing DNA-conjugated tryptamines as shown in Figure 5A. We confirmed that the electronic effect of the substrate has an important influence on the reaction and unsurprisingly that the methoxy-substituted substrate (3a) gives the best result, whereas a bromosubstituted substrate gives almost no desired product. We then proceeded to investigate the reaction between a DNA-conjugated aryl aldehyde and different tryptamine substrates. At 80°C the reaction proceeded smoothly with good to excellent conversions (Figure 5B). For the on-DNA PS product 6c, we have also carried out the amine capping with acetic acid and benzaldehyde, and both reactions provided the desired amine-capped products with acceptable conversions. This result indirectly demonstrates formation of the desired DNA-conjugated tryptamine PS product and also illustrates an interesting potential library design. (Note that, in contrast, capping of DNA-conjugate 4 with carboxylic acids or aldehydes is extremely difficult.)
Figure 5.
Pictet-Spengler Reaction
(A) Electronic effect with Pictet-Spengler reaction.
(B) Pictet-Spengler reaction between on-DNA aldehyde and tryptamine substrates.
(C) Pictet-Spengler Reaction between DNA-conjugated indole-substituted amine and aldehyde.
In order to further explore the potential libraries employing this new on-DNA PS reaction condition, we designed DNA-conjugated indole amine 7. The on-DNA PS reaction between 7 and 10 different aldehydes are provided in Figure 5C. Again, the amine capping experiments were carried out employing both carboxylic acids and aldehydes; again, successful capping confirms the presence of the desired DNA-conjugated PS cyclized starting material and the ability to further diversity the scaffold during future library synthesis (Figure 5C).
DNA Damage Evaluation
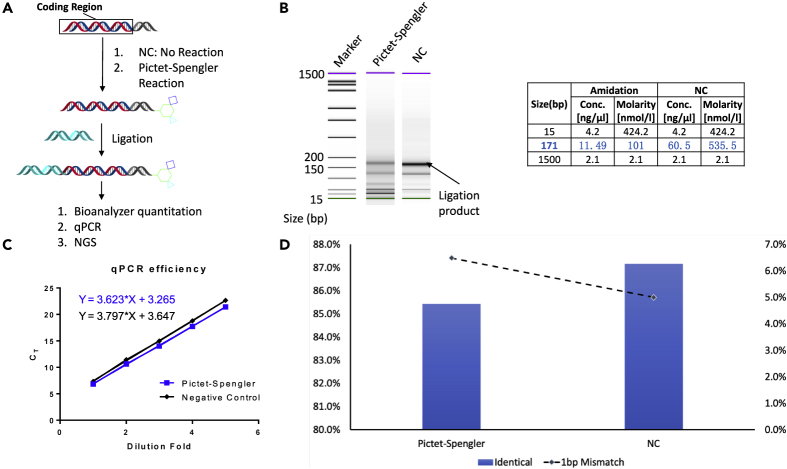
A PS reaction was performed with a DNA-conjugated compound containing a double-stranded DNA coding region to mimic the library component, and the product was then ligated to an oligonucleotide to generate a full-length DNA fragment for qPCR analysis and next-generation sequencing (NGS) to assess the DNA integrity (Figure 6A). Ligation without any chemical reaction was used as a negative control separately. The ligation product was first examined with capillary electrophoresis (Bioanalyzer 2100, Agilent) to assess the size and quantity of the DNA strain. The result showed no shift of PS reaction product compared with the reactant, indicating no change of DNA size by PS reaction (Figure 6B). Then, the amplification efficiency was analyzed by qPCR (QuantStudio 7, Thermo Fisher) and the result suggested no significant change of DNA amplification efficiency when compared with the negative control groups. This result indicates that the PS reaction did not introduce unknown variation to the nucleotide, otherwise the efficiency will probably change between the two comparative groups (Figure 6C). In order to further assess the nucleotide-level integrity of the DNA, the ligation product was amplified and sequenced by the NGS. The NGS results suggested no significant modification of nucleotide from the PS reaction compared with the negative control NC (Figure 6D). In summary, the comprehensive DNA integrity assessment study demonstrated no damage to DNA by the PS reaction and, thus, could potentially be used for the DNA-encoded library construction (for details see Supplemental Information 5).
Figure 6.
DNA Integrity was not Compromised by Pictet-Spengler Reaction
(A) Schematic illustration of the experiment design and workflow.
(B) Ligation and capillary electrophoresis, which suggest no DNA modification by Pictet-Spengler reaction.
(C) qPCR efficiency with CT as a function of dilution fold. No significant change of qPCR efficiency was observed on Pictet-Spengler reaction compared with the negative control groups.
(D) NGS data, suggesting no significant modification of DNA at the nucleotide level by Pictet-Spengler reaction.
Potential DEL Library Synthetic Route
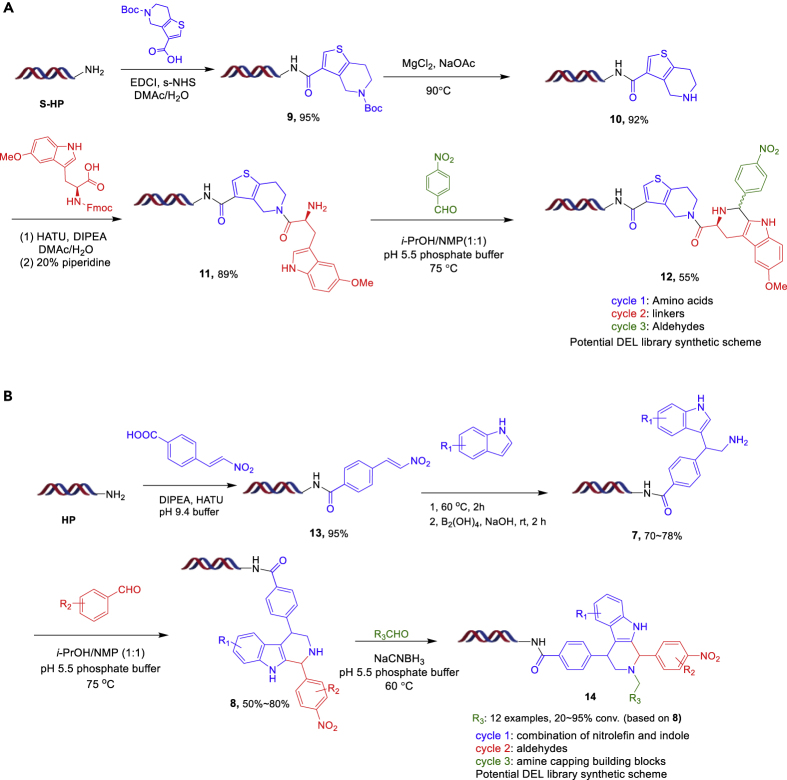
Lastly, we demonstrate proof-of-concept synthesis for two different three-cycle libraries (Figure 7A). For library 1, commercial aldehyde BBs would be purchased after filtering by our trained ML model. In library 2 (Figure 7B) we synthesize a different tryptoline scaffold, which is capable of being further diversified via acylation or alkylation. After on-DNA acylation to install the DNA-conjugated nitroalkene 13, the addition reaction followed by nitro reduction yields the DNA-conjugated indole substituted amine 7. The on-DNA PS reaction between 7 and a corresponding aldehyde provided the product 8 with good conversion (see the Supplemental Information). The amine capping of 8a then provides further diversification of the THBC scaffold (12 aldehydes were tested with 20%–95% conversions). These two diverse library designs demonstrate the potential of this novel on-DNA PS reaction.
Figure 7.
Potential DEL Library Synthetic Scheme
(A) Route for library 1.
(B) Route for library 2.
In summary, with the rational design of the on-DNA indole substrates, we have developed the first DNA-compatible PS reaction for a variety of aldehydes under the optimized reaction conditions. Besides, suitable reaction conditions were identified for various combinations of PS reaction coupling partners. Moreover, a DNN model has been developed to make the prediction of the reaction conversion rate for the BBs, which was the first example of applying ML for the BB selections of the corresponding on-DNA reactions for DNA-encoded library synthesis. The detailed library production applying this new developed on-DNA PS reaction and selection results of the interesting biological interesting targets will be reported in due time.
Limitations of the Study
This reaction has certain limitations on the indole substrates; for example, the presence of methoxy group can make the PS reaction proceed smoothly. Besides, in order to have better prediction, a large set of reaction records need to be available.
Resource Availability
Lead Contact
XiaojieLu.
Materials Availability
See details in supplemental information.
Data and Code Availability
All the data has been attached in Supplemental information.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file. Full details of synthesis and LC-MS/MS analysis are provided in the Supplemental Information.
Acknowledgments
We gratefully acknowledge financial support from the National Natural Science Foundation of China (81773634 to M.Z., 21877117 and 91953203 to X.L., 21907105 to Z.D.), National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program,” China (Number: 2018ZX09711002 to H.J., No. 2018ZX09711002-005 to X.L.), and “Personalized Medicines—Molecular Signature-based Drug Discovery and Development,” Strategic Priority Research Program of the Chinese Academy of Sciences (XDA12050201 to M.Z.; XDA12040330 and XDA12050405 to X.L.), Shanghai Commission of Science and Technology (18431907100 to X.L.).
Author Contributions
K.L., X. Liu, and S.L. contributed equally to this article. K.L. performed the PS reaction optimization, X. Liu. performed all the machine learning work, S.L. performed the validation and synthetic application; Y.A. and Y.S. performed the reaction optimization and validation; Q.S., X.S., W.S., and W.C. performed the DNA damage evaluation; Z.D. performed the synthetic application; L.K., H.Y., and A.S. guided the study and revised the manuscript; K.C. and H.J. guided the study; M.Z., X.P., and X. Lu conceived and designed the project and prepared the manuscript with feedback from all the authors.
Declaration of Interests
The authors declare no competing interests.
Published: June 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101142.
Contributor Information
Mingyue Zheng, Email: myzheng@simm.ac.cn.
Xuanjia Peng, Email: peng_xuanjia@wuxiapptec.com.
Xiaojie Lu, Email: xjlu@simm.ac.cn.
Supplemental Information
Sheet 1 contained structures and predicted conversion rate values of top 300 BBs. Sheet 2 contained structures, predicted and true conversion rate values of 34 BBs picked by model. Sheet 3 contained structures and true conversion rate values of 34 BBs picked by random selection.
References
- Ahneman D.T., Estrada J.G., Lin S., Dreher S.D., Doyle A.G. Predicting reaction performance in C–N cross-coupling using machine learning. Science. 2018;360:186–190. doi: 10.1126/science.aar5169. [DOI] [PubMed] [Google Scholar]
- Bishop C.M. Springer; 2006. Pattern Recognition and Machine Learning. [Google Scholar]
- Buller F., Mannocci L., Scheuermann J., Neri D. Drug discovery with DNA-encoded chemical libraries. Bioconjug. Chem. 2010;21:1571–1580. doi: 10.1021/bc1001483. [DOI] [PubMed] [Google Scholar]
- Chouikhi D., Ciobanu M., Zambaldo C., Duplan V., Barluenga S., Winssinger N. Expanding the scope of PNA-encoded synthesis (pes): Mtt-protected PNA fully orthogonal to Fmoc chemistry and a broad array of robust diversity generating reactions. Chem. Eur. J. 2012;18:12698–12704. doi: 10.1002/chem.201201337. [DOI] [PubMed] [Google Scholar]
- Clark M.A., Charya R.A., Arico-Muendel C.C., Belyanskaya S.L., Benjamin D.R., Carlson N.R., Centrella P.A., Chiu C.H., Creaser S.P., Cuozzo J.W. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- David R., Mathew H. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Dichson P., Kodadek T. Chemical composition of DNA-encoded libraries, past present and future. Org. Biomol. Chem. 2019;17:4676–4688. doi: 10.1039/c9ob00581a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipak P., Mukut G. Iodine-catalyzed highly effective Pictet–Spengler condensation: an efficient synthesis of tetrahydro-β-carbolines. Synth. Commun. 2008;38:4426–4433. [Google Scholar]
- Du H.C., Bangs M.C., Simmons N., Matzuk M.M. Multistep synthesis of 1,2,4-oxadiazoles via DNA-conjugated aryl nitrile substrates. Bioconjug. Chem. 2019;30:1304–1308. doi: 10.1021/acs.bioconjchem.9b00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam O., Satz A.L. Analysis of the productivity of DNA encoded libraries. Med. Chem. Commun. 2016;7:1323–1331. [Google Scholar]
- Encinas L., O'Keefe H., Neu M., Remuinan M.J., Patel A.M., Guardia A., Davie C.P., Perez-Macias N., Yang H., Convery M.A. Encoded library technology as a source of hits for the discovery and lead optimization of a potent and selective class of bactericidal direct inhibitors of Mycobacterium tuberculosis InhA. J. Med. Chem. 2014;57:1276–1288. doi: 10.1021/jm401326j. [DOI] [PubMed] [Google Scholar]
- Favalli N., Bassi G., Scheuermann J., Neri D. DNA-encoded chemical libraries-achievements and remaining challenges. FEBS Lett. 2018;592:2168–2180. doi: 10.1002/1873-3468.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faver J.C., Riehle K., Lancia D.R., Milbank J.B.J., Kollmann C.S., Simmons N. Quantitative comparison of enrichment from DNA-encoded chemical library selections. ACS Comb. Sci. 2019;21:75–82. doi: 10.1021/acscombsci.8b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood D.T., Asai S., Zhang X., Wang J., Yoon L., Adams Z.C., Dillingham B.C., Sanchez B.B., Vantourout J.C., Flanagan M.E. Expanding reactivity in DNA-encoded library synthesis via reversible binding of DNA to an Inert quaternary ammonium support. J. Am. Chem. Soc. 2019;141:9998–10006. doi: 10.1021/jacs.9b03774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini R.M., Randolph C. Chemical space of DNA-encoded libraries. J. Med. Chem. 2016;59:6629–6644. doi: 10.1021/acs.jmedchem.5b01874. [DOI] [PubMed] [Google Scholar]
- Gong Z., Hu G., Li Q., Liu Z., Wang F., Zhang X., Xiong J., Li P., Xu Y., Ma R. Compound libraries: recent advances and their applications in drug discovery. Curr. Drug Discov. Tech. 2017;14:216–228. doi: 10.2174/1570163814666170425155154. [DOI] [PubMed] [Google Scholar]
- Jiménez J., Ginebra J. PyGPGO: Bayesian optimization for python. J. Open Source Softw. 2017;2:431. [Google Scholar]
- Kingma D.P., Ba J. Adam: a method for stochastic optimization. arXiv e-prints. 2014 arXiv:1412.6980. [Google Scholar]
- Kite S., Hattorib T., Murakamib Y. Estimation of catalytic performance by neural network-product distribution in oxidative dehydrogenation of ethylbenzene. Appl. Catal. 1994;114:173–178. [Google Scholar]
- LaPlante S.R., Carson R., Gillard J., Aubry N., Coulombe R., Bordeleau S., Bonneau P., Little M., O’Meara J., Beaulieu P.L. Compound aggregation in drug discovery: implementing a practical NMR assay for medicinal chemists. J. Med. Chem. 2013;56:5142–5150. doi: 10.1021/jm400535b. [DOI] [PubMed] [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Lerner R.A., Ma P., Xu H., Li J., Lu F., Ma F., Wang S., Xiong H., Wang W., Buratto D. Functionality-independent DNA encoding of complex natural products. Angew. Chem. Int. Ed. 2019;58:9254–9261. doi: 10.1002/anie.201901485. [DOI] [PubMed] [Google Scholar]
- Li H., Sun Z., Wu W., Wang X., Zhang M., Lu X., Zhong W., Dai D. Inverse-Electron-Demand Diels−Alder reactions for the synthesis of pyridazines on DNA. Org. Lett. 2018;20:7186–7191. doi: 10.1021/acs.orglett.8b03114. [DOI] [PubMed] [Google Scholar]
- Liu F., Wang H., Li S., Bare G.A.L., Chen X., Wang C., Moses J.E., Wu P., Sharpless K.B. Biocompatible SuFEx click chemistry: thionyl tetrafluoride (SOF4)-derived connective hubs for bioconjugation to DNA and proteins. Angew. Chem. Int. Ed. 2019;58:8029–8033. doi: 10.1002/anie.201902489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Fan L., Phelps C.B., Davie C.P., Donahue C.P. Ruthenium promoted On-DNA ring-closing metathesis and cross-metathesis. Bioconjug. Chem. 2017;28:1625–1629. doi: 10.1021/acs.bioconjchem.7b00292. [DOI] [PubMed] [Google Scholar]
- Lu X., Roberts S., Franklin G.J., Davie C. On-DNA Pd and Cu promoted C–N cross-coupling reactions. Med. Chem. Commun. 2017;8:1614–1617. doi: 10.1039/c7md00289k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity P., Adhikari D., Jana A.K. An overview on synthetic entries to tetrahydro-β-carbolines. Tetrahedron. 2019;75:965–1028. [Google Scholar]
- Malone M.L., Paegel B.M. What is a “DNA-compatible” Reaction? ACS Comb. Sci. 2016;18:182–187. doi: 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon G., Orr G.B., Müller K.R. Second Edition. Springer; 2012. Neural Networks: Tricks of the Trade. [Google Scholar]
- Neri D., Lerner R.A. DNA-encoded chemical libraries: a selection system based on endowing organic compounds with amplifiable information. Annu. Rev. Biochem. 2018;87:479–502. doi: 10.1146/annurev-biochem-062917-012550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata K., Yamada M. Prediction of effective additives to a Ni/Active carbon catalyst for vapor-phase carbonylation of methanol by an artificial neural network. Ind. Eng. Chem. Res. 2004;43:6622–6625. [Google Scholar]
- Ottl J., Leder L., Schaefer J.V., Dumelin C.E. Encoded library technologies as Integrated lead finding platforms for drug discovery. Molecules. 2019;24:1629–1650. doi: 10.3390/molecules24081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raccuglia P., Elbert K.C., Adler P.D., Falk C., Wenny M.B., Mollo A., Zeller M., Friedler S.A., Schrier J., Norquist A.J. Machine-learning-assisted materials discovery using failed experiments. Nature. 2016;533:73–76. doi: 10.1038/nature17439. [DOI] [PubMed] [Google Scholar]
- Ralph E.K., Christoph E.D., David R.L. Small-molecule discovery from DNA-encoded chemical libraries. Chem. Soc. Rev. 2011;40:5707–5717. doi: 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddavide F.V., Cui M., Lin W., Fu N., Heiden S., Andrade H., Thompson M., Zhang Y. Second generation DNA-encoded dynamic combinatorial chemical libraries. Chem. Commun. 2019;55:3753–3756. doi: 10.1039/c9cc01429b. [DOI] [PubMed] [Google Scholar]
- Roman J.P., Haro R., Blas J.D., Jessop T.C., Castanon J. Design and development of a technology platform for DNA-encoded library production and affinity selection. SLAS Discov. 2018;23:387–396. doi: 10.1177/2472555217752091. [DOI] [PubMed] [Google Scholar]
- Satz A.L., Cai J., Chen Y., Goodnow R., Gruber F., Kowalczyk A., Petersen A., Naderi-Oboodi G., Orzechowski L., Strebel Q. DNA compatible multistep synthesis and applications to DNA encoded libraries. Bioconjug. Chem. 2015;26:1623–1632. doi: 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- Skopic M.K., Salamon H., Bugain O., Jung K., Gohla A., Doetsch L.J., Dos Santos D., Bhat A., Wagner B., Brunschweiger A. Acid- and Au(i)-mediated synthesis of hexathymidine-DNA-heterocycle chimeras, an efficient entry to DNA-encoded libraries inspired by drug structures. Chem. Sci. 2017;8:3356–3361. doi: 10.1039/c7sc00455a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoraczynski G., Dittwald P., Miasojedow B., Szymkuc S., Gajewska E.P., Grzybowski B.A., Gambin A. Ror2 signaling regulates golgi structure and transport through IFT20 for tumor invasiveness. Sci. Rep. 2017;7:3582. 3542. [Google Scholar]
- Srinivasan N., Ganesan A. A highly efficient Lewis acid-catalysed Pictet–Spengler reactions discovered by parallel screening. Chem. Commun. 2003;7:916–917. doi: 10.1039/b212063a. [DOI] [PubMed] [Google Scholar]
- Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- Škopic M.K., Götte K., Gramse C., Dieter M., Pospich S., Raunser S., Weberskirch R., Brunschweiger A. Micellar Bronsted acid mediated synthesis of DNA-tagged heterocycles. J. Am. Chem. Soc. 2019;141:10546–10555. doi: 10.1021/jacs.9b05696. [DOI] [PubMed] [Google Scholar]
- Wang J., Lundberg H., Asai S., Martin-Acosta P., Chen J.S., Brown S., Farrell W., Dushin R.G., O'Donnell C.J., Ratnayake A.S. Kinetically guided radical-based synthesis of C (sp3)-C (sp3) linkages on DNA. Proc. Natl. Acad. Sci. U S A. 2018;115:E6404–E6410. doi: 10.1073/pnas.1806900115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Sun H., Liu J., Dai D., Zhang M., Zhou H., Zhong W., Lu X. Ruthenium-promoted C−H activation reactions between DNA conjugated acrylamide and aromatic Acids. Org. Lett. 2018;20:4764–4768. doi: 10.1021/acs.orglett.8b01837. [DOI] [PubMed] [Google Scholar]
- Wang X., Sun H., Liu J., Dai D., Zhang M., Zhou H., Zhong W., Lu X. Palladium-promoted DNA-compatible Heck reaction. Org. Lett. 2019;21:719–723. doi: 10.1021/acs.orglett.8b03926. [DOI] [PubMed] [Google Scholar]
- Xu H., Ma F., Wang N., Hou W., Xiong H., Lu F., Li J., Wang S., Ma P., Yang G., Lerner R.A. DNA-Encoded Libraries: aryl fluorosulfonates as versatile electrophiles enabling facile On-DNA Suzuki, Sonogashira, and Buchwald reactions. Adv. Sci. 2019;23:1901551–1901556. doi: 10.1002/advs.201901551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura A., Fujitomi E., Ohara N., Tsukamoto K., Sato M., Yamamura H. Tadalafil induces antiproliferation, apoptosis, and phosphodiesterase type 5 downregulation in idiopathic pulmonary arterial hypertension in vitro. Eur. J. Pharm. 2017;810:44–50. doi: 10.1016/j.ejphar.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Yuen L.H., Dana S., Liu Y., Bloom S.I., Thorsell A.G., Neri D., Donato A.J., Kireev D.B., Schuler H., Franzini R.M. A focused DNA-encoded chemical library for the discovery of Inhibitors of NAD+-Dependent enzymes. J. Am. Chem. Soc. 2019;141:5169–5181. doi: 10.1021/jacs.8b08039. [DOI] [PubMed] [Google Scholar]
- Zambaldo C., Geigle S.N., Satz A.L. High-throughput solid-phase building block synthesis for DNA-encoded libraries. Org. Lett. 2019;21:9353–9357. doi: 10.1021/acs.orglett.9b03553. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Li C., Peng J., Xie L., Meng L., Li Q. DNA-encoded dynamic chemical library and its applications in Ligand Discovery. J. Am. Chem. Soc. 2018;140:15859–15867. doi: 10.1021/jacs.8b09277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet 1 contained structures and predicted conversion rate values of top 300 BBs. Sheet 2 contained structures, predicted and true conversion rate values of 34 BBs picked by model. Sheet 3 contained structures and true conversion rate values of 34 BBs picked by random selection.
Data Availability Statement
All the data has been attached in Supplemental information.