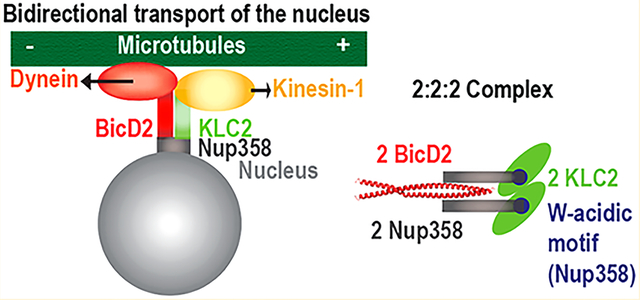
Abstract
Nup358 is a protein subunit of the nuclear pore complex that recruits the opposing microtubule motors kinesin-1 and dynein [via the dynein adaptor Bicaudal D2 (BicD2)] to the nuclear envelope. This pathway is important for positioning of the nucleus during the early steps of mitotic spindle assembly and also essential for an important process in brain development. It is unknown whether dynein and kinesin-1 interact with Nup358 simultaneously or whether they compete. Here, we have reconstituted and characterized a minimal complex of kinesin-1 light chain 2 (KLC2) and Nup358. The proteins interact through a W-acidic motif in Nup358, which is highly conserved among vertebrates but absent in insects. While Nup358 and KLC2 form predominantly monomers, their interaction results in the formation of 2:2 complexes, and the W-acidic motif is required for the oligomerization. In active motor complexes, BicD2 and KLC2 each form dimers. Notably, we show that the dynein adaptor BicD2 and KLC2 interact simultaneously with Nup358, resulting in the formation of 2:2:2 complexes. Mutation of the W-acidic motif results in the formation of 1:1:1 complexes. On the basis of our data, we propose that Nup358 recruits simultaneously one kinesin-1 motor and one dynein motor via BicD2 to the nucleus. We hypothesize that the binding sites are close enough to promote direct interactions between these motor recognition domains, which may be important for the regulation of the motility of these opposing motors. Our data provide important insights into a nuclear positioning pathway that is crucial for brain development and faithful chromosome segregation.
Graphical Abstract
The nucleus is bidirectionally transported along microtubules by the opposing motors cytoplasmic dynein and kinesin-1.1 Nuclear positioning facilitates fundamental processes during muscle and brain development,2–4 which is underscored by the fact that mutations in relevant proteins cause neuromuscular and neurodegenerative diseases, including distinct cases of spinal muscular atrophy (SMA). In infants, SMA is the most prevalent genetic cause of death.5–8
However, despite the significance of nuclear positioning, it is unknown how teams of opposing motors collaborate to achieve the correct timing, directionality, and velocity of transport. Notably, the interactions of opposing motors with cargoes in general have not been characterized extensively by biochemical methods. One of the proteins that recruit dynein and kinesin-1 to the nucleus is Nup358 (also known as RanBP2), a 358 kDa subunit of the nuclear pore complex that is the main constituent of the cytoplasmic filaments (Figure 1).1,11,12 Large portions of Nup358 are intrinsically disordered and contain docking sites for a variety of proteins.12 It has not been established whether Nup358 recruits dynein and kinesin-1 at the same time or whether these motors compete for binding. Furthermore, it is unknown how many dynein and kinesin-1 motors are recruited to Nup358, even though this information would be important to understand how the transport of the cell nucleus via Nup358 is facilitated.
Figure 1.
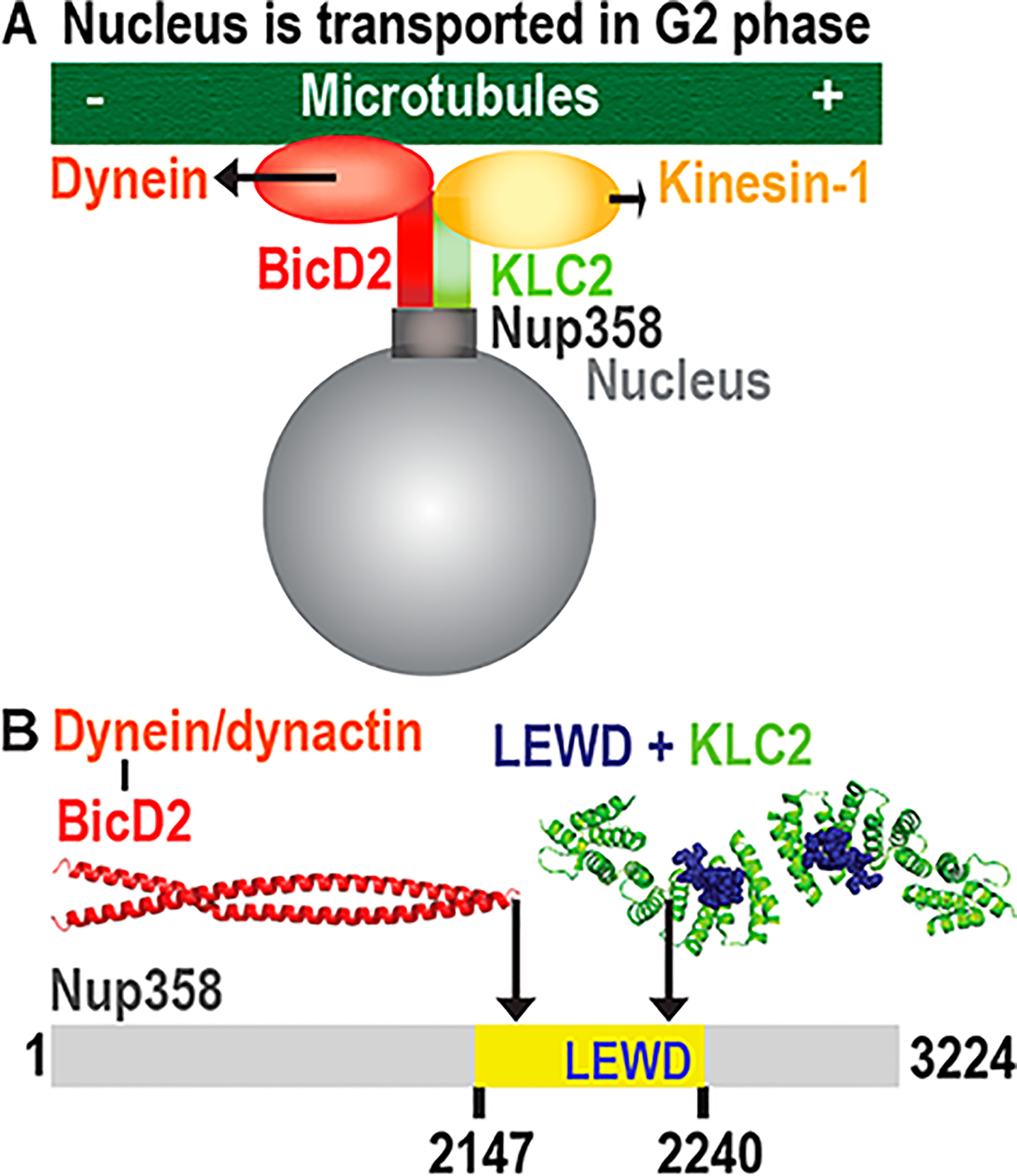
Bidirectional positioning of the nucleus by opposing microtubule motors dynein and kinesin-1. (A) Kinesin-1 and the dynein adaptor BicD2 are recruited to Nup358 during the G2 phase. BicD2 recruits dynein to the nucleus via this pathway.1 (B) Schematic representation of the minimal binding site for BicD2 on Nup358 (yellow). The structure of the cargo/adaptor interacting domain of BicD2 is shown (red).9 This region of BicD2 binds to the cargo adaptor Nup358. A KLC2-binding W-acidic motif (with the sequence LEWD) is located in the mapped minimal BicD2-binding site. The structure of the KLC2-TPR domain that has the W-acidic motif with the sequence LEWD bound is shown.10
Cytoplasmic dynein mediates the transport of almost all substrates that are trafficked to the minus end of microtubules.13 Cargo is recognized by adaptors, e.g., Bicaudal D2 (BicD2),9,13–21 which also connect it to the motor. Cargo loading is coupled to activation of dynein for processive motion, and dynein adaptor/cargo adaptor complexes such as BicD2/Nup358 are required for this process.9,13–26 A motor that opposes dynein is kinesin-1, which transports substrates to the plus end of microtubules.27 In the absence of cargo, kinesin-1 is autoinhibited. Kinesin-1 is a tetramer that is composed of two light chain (KLC) and two two heavy chain (KHC) subunits. KHCs have three isoforms in mammals (Kif5A–C), and KLCs have four isoforms (KLC1–4). The motor domains are located in the KHCs, whereas the KLCs are important for cargo recognition and autoinhibition.
For cargo recognition and for activation of kinesin-1 for processive transport, short sequence motifs (e.g., the sequence LEWD) are sufficient, which are termed W-acidic motifs and are recognized by the KLC2-TPR domain (tetratricopeptide repeat-containing domain).10,28–31 A structure of this KLC2 domain with a fused W-acidic motif peptide has provided insight into cargo recognition.10 W-acidic motifs consist of a tryptophan (W) residue that is sandwiched between two acidic residues and in addition often contain a leucine or methionine residue in the first position.28 Fusion of the specific W-acidic motif LEWD to a cargo is sufficient to initiate its transport by kinesin-1, and a mutation of this motif to LEAA prevents the association.28 However, full motor activity is achieved only if additional cargo/KLC and cargo/KHC interactions are formed.32–34
Many cargoes interact with both KHCs and KLCs through multiple sites. Interestingly, some W-acidic binding motifs interact with both the KLCs and the KHCs. However, the W-acidic motif alone is not sufficient for an interaction with the KHCs, and additional regions are required.32 Furthermore, the KLCs bind first and are required to prime access for the KHCs, which can bind to the W-acidic binding motif only after autoinhibition is relieved.32
Kinesin-1 is autoinhibited in the absence of cargo, which is facilitated by an intramolecular interaction within the KHCs that prevents ATP hydrolysis in the motor domains. In addition, in the autoinhibited state, a leucine-phenylalanine-proline (LFP) motif in a flexible region of KLC2 blocks access of the W-acidic motif to the binding site.29 Binding of a W-acidic motif displaces the LFP motif, resulting in conformational changes that activate the motor.29 This proposed mechanism is specific for cargoes with W-acidic binding motifs. Other sequence motifs, such as the Y-acidic motifs, bind at a different site on KLC2 and activate kinesin-1 by a distinct mechanism that also engages additional protein factors.10,29,35,36 A compelling hypothesis is that binding of cargo to the KLCs induces their dimerization, and the resulting 2:2 complex could be required to activate autoinhibited kinesin-1.36 However, while both W-acidic binding motifs and Y-acidic binding motifs induce dimerization of KLCs in the available crystal structures10,35,36 and while kinesin-1 motors contain two copies of the KLCs, this hypothesis remains to be confirmed by functional studies with intact motors and in context of additional interaction partners.
Kinesin-1 is recruited to the nucleus by residues 2134–2264 of human Nup358.11,37,38 A direct interaction of the KHC isoforms Kif5B and Kif5C with Nup358 was shown in vivo and in vitro, and the KLCs were also present in this complex.11 In addition, in the same region of Nup358, a W-acidic motif was identified, which is recognized by KLC2. Initial co-immunoprecipitation experiments in COS7 cells confirmed an interaction with KLC2 (see Figure S5B of ref 38). However, the interaction was not reconstituted from recombinant proteins, and no additional analysis was performed to confirm the interaction.
The same domain of Nup358 also recruits the dynein adaptor BicD2 (Figure 1).1 BicD2 in turn recruits dynein together with its activator dynactin.1 This Nup358/BicD2 pathway for nuclear positioning is active in the G2 phase1 and regulated in a cell cycle specific manner.2,19,39 During early steps of mitotic spindle assembly, the Nup358/BicD2 pathway facilitates in all cell types positioning of the nucleus with respect to the centrosome.1 The integrity of this pathway is important in order to time mitotic entry correctly.1 In brain progenitor cells, this pathway is in addition essential for apical nuclear migration, which is required for these progenitors to enter mitosis and thus to differentiate to neurons and other cell types.3
Kinesin-1 also has an active role in nuclear positioning in the G2 phase1 and is recruited to the same domain of Nup358 as BicD2 in the context of cells11 (Figure 1), but it is unknown if BicD2 and kinesin-1 bind simultaneously to Nup358. While dynein is the main motor responsible for nuclear positioning in the G2 phase, overall motility is also regulated by kinesin-1, which antagonizes dynein activity.1,11 The arrangement of opposing motors acting together may represent an important property of microtubule motor systems, likely needed for the spatiotemporal regulation of transport.1,4,33,38,40–47 The spatial proximity of binding sites of opposing motors is common,4,41 which has inspired the hypothesis that these motors compete for cargo.33 The alternative tug-of-war model proposes that the team of motors that produces the greater force determines the direction of transport.40,48,49 Finally, in the coordination model, opposing motors bind simultaneously to cargo and are tightly coordinated for unidirectional transport by regulatory mechanisms, such as adaptor or scaffolding proteins.44,50,51 Which of these models applies to nuclear positioning via Nup358 is unknown, and whether kinesin-1 and dynein bind simultaneously to Nup358 or whether they compete for binding is also unknown.
Here, we have reconstituted and characterized a minimal complex of Nup358 and KLC2. While the individual proteins form predominantly monomers, the resulting complex has a 2:2 stoichiometry, suggesting that the binding of the cargo Nup358 to KLC2 induces oligomerization. Intriguingly, we also show that both BicD2 and KLC2 interact with Nup358 simultaneously and form 2:2:2 complexes. We hypothesize that the binding sites are close enough to promote direct interactions between these motor recognition domains, which may be important for the regulation of the motility of these opposing motors.
MATERIALS AND METHODS
Protein Expression and Purification.
Expression constructs for the minimal interacting domains termed BicD2-CTD (residues 715–804 of human BicD2),39 Nup358-min (residues 2147–2240 of human Nup358),9 KLC2TPR (residues 196–480 of mouse kinesin-1 light chain 2 isoform 1),29 and KLC2TPR-trunc (residues 218–480)10 were previously described. Expression constructs for the fusion proteins and mutants were generated by commercial gene synthesis of the codonoptimized insert sequences with restriction sites added at each end, which were cloned into the pGEX-6P1 expression plasmid (GE Healthcare) by the company Genscript, using the restriction sites for BamHI and XhoI. To create the expression plasmids for the Nup358-KLC2-fusion protein and the Nup358/W2224A/D2225A-KLC2-fusion protein mutant, the sequence for residues 2147–2240 of Homo sapiens Nup358 (NCBI entry XM_005264002.2) was fused to the N-terminus of residues 196–480 of kinesin-1 light chain 2 isoform 1 from Mus musculus (NCBI entry NP_001356289.1).
Recombinant protein expression in Escherichia coli and protein purification were performed as described previously.9,10,29,39,52 The Nup358-KLC2-fusion protein and the W2224A/D2225A mutant were expressed at 37 °C as described for Nup358-min,9 but the E. coli RIL (DE3)pLysS expression strain was used.
Proteins were purified as described previously.9,29,39,52 His6-tagged BicD2-CTD was purified using Ni-NTA affinity chromatography. The affinity tag was cleaved off by thrombin, and the protein was then purified by another step of Ni-NTA affinity chromatography as described previously.39 GST-tagged Nup358-min and the Nup358-KLC2-fusion protein (WT and W2224A/D2225A mutant) were purified using glutathione affinity chromatography and eluted by proteolytic cleavage of the GST tag by PreScission protease, as described for Nup358-min.9 All proteins were subsequently injected onto a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) that was equilibrated with 20 mM HEPES (pH 7.5), 150 mM NaCl, and 0.5 mM TCEP and purified by gel filtration chromatography.9,39,52 His6-tagged KLC2TPR was purified using Ni-NTA affinity chromatography, with a binding and wash buffer of 25 mM HEPES (pH 7.5), 500 mM NaCl, 5 mM β-mercaptoethanol (BME), and 20 mM imidazole. For the elution buffer, the imidazole concentration was increased to 250 mM in the same buffer. The eluate was filtered (pore size of 0.02 um) and immediately purified by gel filtration chromatography using the following buffer: 20 mM HEPES (pH 7.5), 500 mM NaCl, 0.5 mM TCEP, and 5 mM BME. KLC2TPR was concentrated to 0.5 mg/mL but had a higher solubility when mixed with Nup358-min (5 mg/mL). The concentrations of proteins (c) were determined by absorbance (A) measurements: c (mg/mL) = (A228.5 – A234)/3.14. Proteins were flash-frozen in liquid nitrogen and stored at −80 °C.52
Analytical Size Exclusion Chromatography.
For analytical size exclusion chromatography, proteins were mixed in a 1:1:1 equimolar ratio (0.1 mg of Nup358-min, 0.1 mg of BicD2-CTD, and 0.25 mg of KLC2TPR in a volume of 610 μL). Samples were filtered (pore size of 0.02 μm) and centrifuged in a microcentrifuge (25 min, 21700g, 4 °C); 500 μL of these samples was injected onto a GE Healthcare Superdex 200 Increase 10/300 GL size exclusion chromatography column, which had been equilibrated with 20 mM HEPES (pH 7.5), 150 mM NaCl, and 0.5 mM TCEP.52,53 Elution fractions were loaded onto 16% acrylamide gels and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Proteins were detected by Coomassie Blue stain.
Size Exclusion Chromatography Coupled to Multi-angle Light Scattering (SEC–MALS).
Prior to analysis, purified proteins were incubated for 30 min at 4 °C, filtered (pore size of 0.02 μm), and centrifuged in a microcentrifuge (4 °C, 21700g, 20 min). Purified proteins (volume of 100 μL) were injected onto a GE Healthcare Superdex 200 Increase 10/300 GL gel filtration column at ambient room temperature. The column had been equilibrated with 20 mM HEPES (pH 7.5), 150 mM NaCl, and 0.5 mM TCEP.39,54–56 The column was connected to a Wyatt Technology DAWN 8+ multiangle light scattering detector and an Optilab TrEX refractive index detector. ASTRA 6.1 (Wyatt Technology) was used to determine weight-averaged molar masses as described previously.54,55 Elution peak boundaries were placed at half-peak height; the molar masses were determined across the selected peak area, and these molar masses of the selected peak area are plotted on a secondary axis in all SEC–MALS graphs. Representative SEC–MALS experiments are shown. Masses from two or three experiments were averaged. Standard deviations were calculated for all experiments, but because they were in all cases much lower compared to the experimental error of the method (which is 5%), the error was calculated at 5% for all molar masses. Therefore, the reproducibility of these experiments is much better than the error of mass determination.
Sequence Alignments and Figures.
Sequences were aligned with XCED,57 Jalview,58 and T-Coffee.59 Because human Nup358 includes 3224 residues, which is above the size limit that the sequence alignment programs can handle, Nup358 sequences were truncated by 0–400 residues at the N-terminus and 0–350 residues at the C-terminus prior to alignment. Structure figures were created with PyMOL (The PyMOL Molecular Graphics System, version 2.0, Schrödinger, LLC). Figures were created with Adobe Photoshop and Illustrator Creative Cloud 2017 (Adobe).
RESULTS
A Minimal Complex of Nup358 and KLC2 Was Reconstituted and Characterized.
The sequence of Nup358 contains a four-amino acid W-acidic motif with the sequence LEWD (residues 2222–2225), which can act as a recruitment site for KLC2 (Figure 1). Intriguingly, the W-acidic motif is located within the mapped minimal BicD2-binding site (Nup358-min, residues 2147–2240),9 which recruits the opposing motor dynein (Figure 1).
Because interactions of Nup358 with BicD2 and kinesin-1 have so far been described for only human and mouse proteins, we analyzed the sequence conservation for this domain of Nup358 to assess if these pathways are conserved among species.
The sequence alignment of Nup358 shows that the W-acidic motif with the sequence LEWD is highly conserved among all vertebrates, including mammals, birds, reptiles, amphibians, and fish (Figure 2). Key amino acids E and W of the motif are present in all sequences analyzed, and replacement of the amino acids L and D is found in only three of 30 sequences. In addition, the sequence region adjacent to the W-acidic motif is also highly conserved and may contribute additional interactions to recruit KLC2. Another conserved region is found in the N-terminal part of the domain, whereas a middle portion of the domain is less well conserved. Therefore, the Nup358-binding sites for BicD2 and the kinesin-1 recruiting W-acidic motif are highly conserved among vertebrates. The nuclear pore protein Nup358 is also found in insects but absent in lower organisms such as yeast. The W-acidic LEWD motif is however missing in Nup358 of insects such as Drosophila melanogaster (Dm) (Figure S1), and it therefore remains to be established whether Dm Nup358 recruits KLC2.
Figure 2.
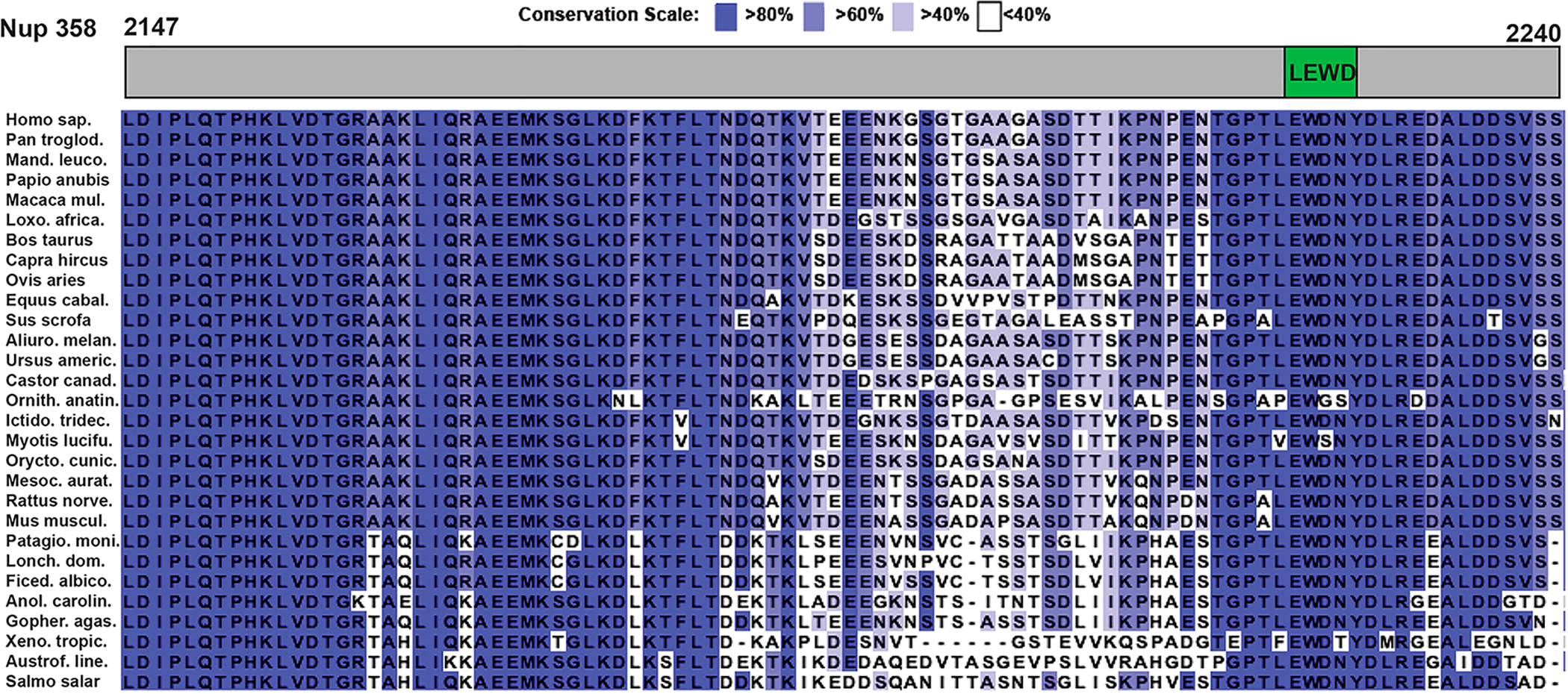
W-Acidic motif with the sequence LEWD conserved in vertebrate Nup358. Sequences of Nup358 from vertebrates were aligned. The sequence region homologous to human Nup358-min (residues 2147–2240) is shown. The location of the W-acidic motif is schematically indicated in a top panel. Residues are colored in different shades of purple, according to their degree of conservation (see the scale). Species names are abbreviated. Sequences from the following species are shown (in alphabetical order): Ailuropoda melanoleura, Anolis carolinensis, Astrofundulus limneaus, Bos taurus, Capra hircus, Castor canadensis, Equus caballus, Ficedula albicollis, Gopherus agassizii, Homo sapiens, Ictidomys tridecemilneatus, Lonchura striata domestica, Loxodonta africana, Macaca mulatta, Mandrillus leucophaeus, Mesocricetus auratus, Mus musculus, Myotis lucifugus, Ornithorhynchus anatinus, Oryctolagus cuniculus, Ovis aries, Pan troglodytes, Papio anubis, Patagioenas fasciata monilis, Rattus norvegicus, Salmo salar, Sus scrofa, Urgus americanus, and Xenopus tropicalis. The same sequence alignment is also shown for a larger region of Nup358 in Figure S2.
A W-acidic motif with the sequence LEWD is sufficient to recruit KLC2 to a cargo and to activate kinesin-1 for processive transport,10,28–31 provided it is accessible for the interaction.
Therefore, to assess if Nup358 recruits the KLC2, we performed a binding assay with the minimal interacting domains. A schematic representation of all minimal interacting domains that were purified for this study is shown in Figure 3, and an SDS–PAGE analysis of all purified proteins is shown in Figure S3. For our binding assay, we used the mapped minimal BicD2-interacting domain of Nup358 [Nup358-min (Figure 3A), residues 2147–2240] and the TPR domain of kinesin-1 light chain 2 [termed KLC2TPR throughout the paper (Figure 3B), residues 196–480]. KLC2TPR is sufficient to recognize the W-acidic motif, and a structure of the slightly truncated TPR domain with the W-acidic motif bound is available.10 We mixed purified Nup358-min and KLC2TPR and separated the mixture by analytical size exclusion chromatography (Figure 4). The elution fractions were analyzed by SDS–PAGE. The individual proteins were analyzed separately. In this experiment, Nup358-min and KLC2TPR co-elute and the elution peaks are shifted toward higher molecular weights compared to the individual proteins (Figure 4A,C,D). These data confirm that Nup358-min and KLC2TPR interact.
Figure 3.
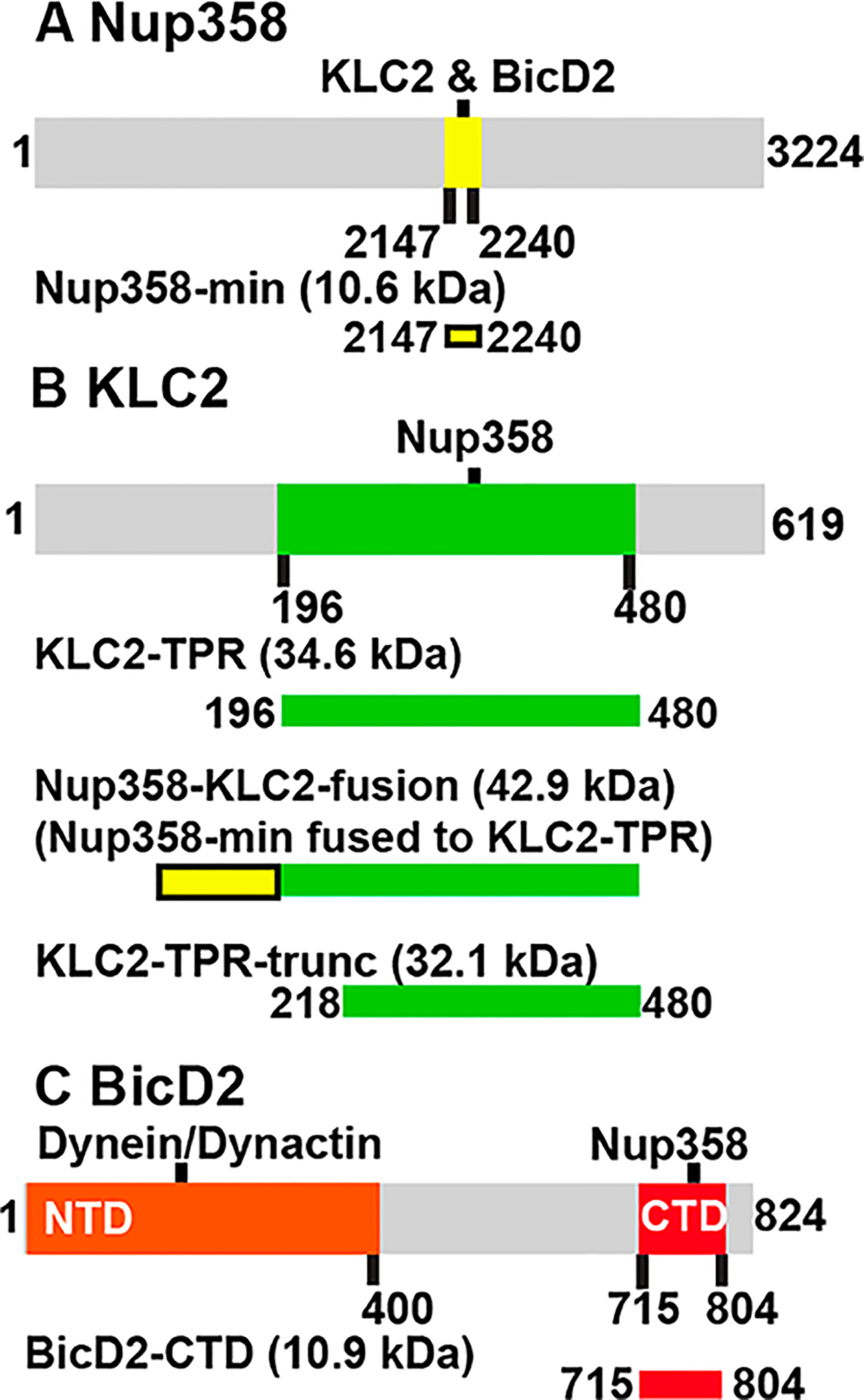
Schematic representation of the full-length proteins Nup358, KLC2, and BicD2 (gray bars) with mapped protein–protein interaction sites (colored). Purified interacting domains and fusion proteins used in this study are shown as bars below the full-length proteins. Calculated molar masses (in kilodaltons) of the protein fragments are indicated: (A) Nup358-min (yellow), (B) KLC2TPR (green), the Nup358-KLC2-fusion protein, and KLC2TPR-trunc, and (C) BicD2-CTD (red).
Figure 4.
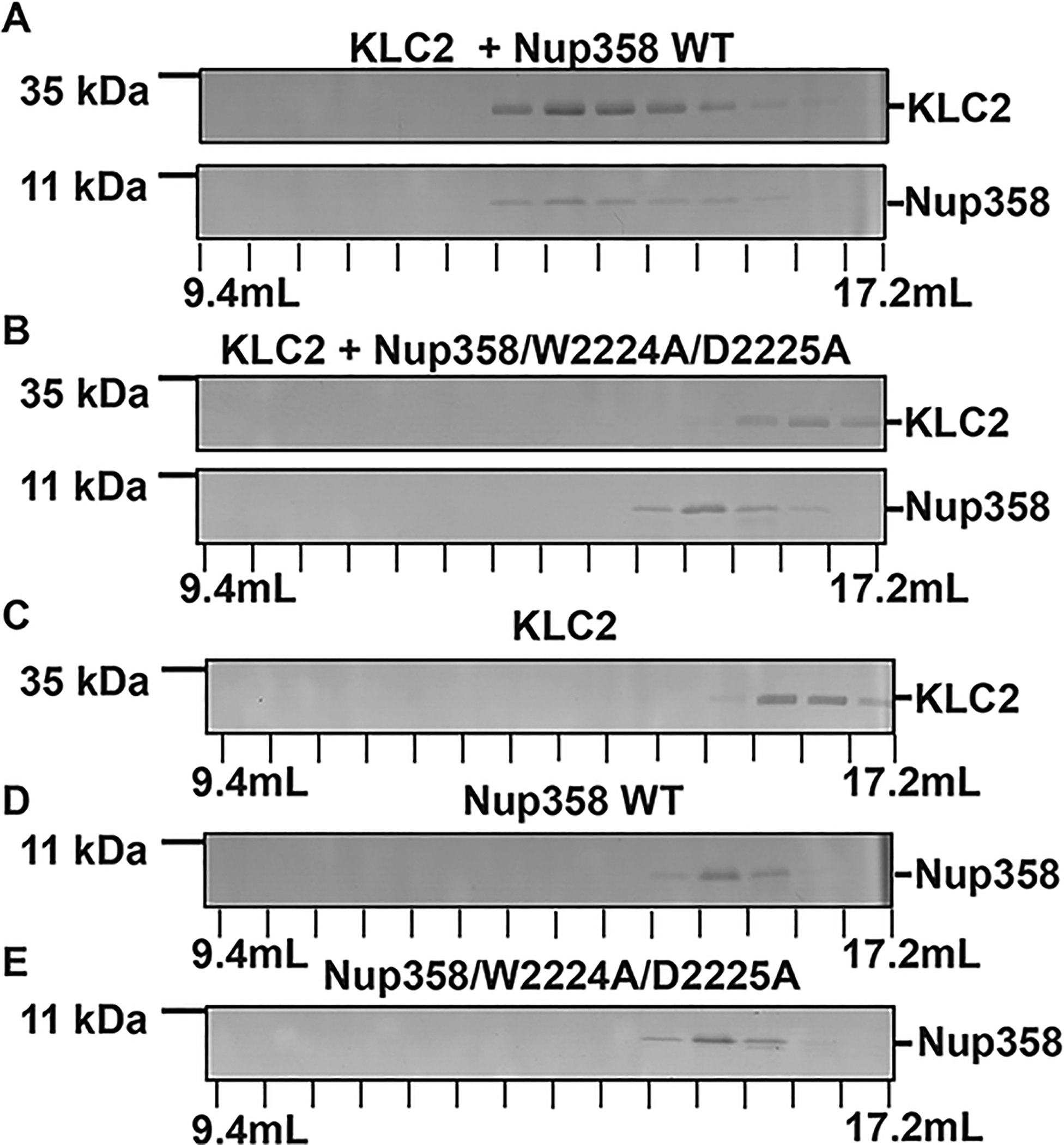
Nup358-min interacts with KLC2 through its W-acidic motif. (A–E) To assess binding, purified proteins were combined (in a 1:1 molar ratio) and then separated by gel filtration chromatography. The elution fractions were analyzed using SDS–PAGE. The individual proteins were also analyzed. Elution volumes are indicated on the bottom; each tick mark represents an elution fraction with a volume of 0.6 mL. The molecular weights of standard proteins are denoted on the left. (A) KLC2TPR and Nup358-min, (B) KLC2TPR and Nup358-min/W2224A/D2225A mutant, (C) KLC2TPR, (D) Nup358-min, and (E) Nup358-min/W2224A/D2225A mutant. SDS–PAGE analyses of the purified proteins and the loaded samples are shown in Figures S3 and S4, respectively. The experiments were repeated, and very similar results were obtained. The numbers of replicates for experiments shown in each figure panel were (A) 4, (B) 2, (C) 3, (D) 4, and (E) 2.
It should be noted that when individually analyzed, Nup358-min (10.6 kDa) elutes earlier than KLC2TPR (34.6 kDa), which indicates that Nup358-min has a hydrodynamic radius that is larger than that of KLC2TPR. Nup358-min is an intrinsically disordered protein, and therefore, the hydrodynamic radius is expected to be much larger compared to that of a globular protein of the same size. KLC2TPR is a TPR domain with a compact structure (see Figure 1B),10 and therefore, its hydrodynamic radius is expected to be in line with that of a globular protein at the same size.
It is established that mutation of the W-acidic motif with the sequence LEWD to the amino acid sequence LEAA abolishes the interaction between kinesin-1 and its cargoes.10,28,30,31 To confirm that the W-acidic motif is required for the interaction, we created a mutant of Nup358-min, in which tryptophan 2224 and aspartate 2225 of the LEWD motif were replaced by alanine residues (Nup358-min/W2224A/D2225A mutant). We then assessed the interaction of purified Nup358-min/W2224A/D2225A and KLC2TPR by analytical size exclusion chromatography (Figure 4B). When mixed, Nup358-min/W2224A/D2225A and KLC2TPR essentially elute at the same elution volume as the individual proteins (Figure 4C,E), confirming that the interaction is virtually abolished by the mutation. However, when a very large excess of Nup358/W2224A/D2225A is used, a very small portion of the KLC2TPR elution peak is shifted slightly toward a higher molecular weight, indicating that a very weak interaction may still be present (Figure S5).
In conclusion, we have reconstituted a minimal complex of KLC2 and Nup358, which confirms that these proteins indeed interact directly. For this interaction, a W-acidic sequence motif in Nup358 is required, which is conserved in vertebrates but not in insects.
The W-Acidic Motif Is Required for the Formation of 2:2 Complexes of KLC2 and Cargo.
Transport-active kinesin-1 motors are heterotetramers of two KLCs and to KHCs. In line with the oligomeric state of the active motor, KLC2TPR forms 2:2 complexes with cargoes in X-ray structures.10,35,36 Therefore, we determined the oligomeric state of the Nup358-min/KLC2TPR complex by SEC–MALS, a method that is applied to determine molar masses across all elution peaks with very high accuracy (5% error).
First, we analyzed the oligomeric state of the individual purified proteins by SEC–MALS. The molar mass (MW) of Nup358-min was determined to be 10.6 ± 0.5 kDa, matching closely the mass of a monomer (10.6 kDa) (Figure 5A, Table 1, and Table S1). The molar mass was not concentration-dependent (in the range of 1–5 mg/mL), and a very similar molar mass was also obtained for the Nup358-min/W2224A/D2225A mutant (10.0 ± 0.5 kDa at 5 mg/mL) (Figure 5B). These data suggest that Nup358-min forms monomers.
Figure 5.
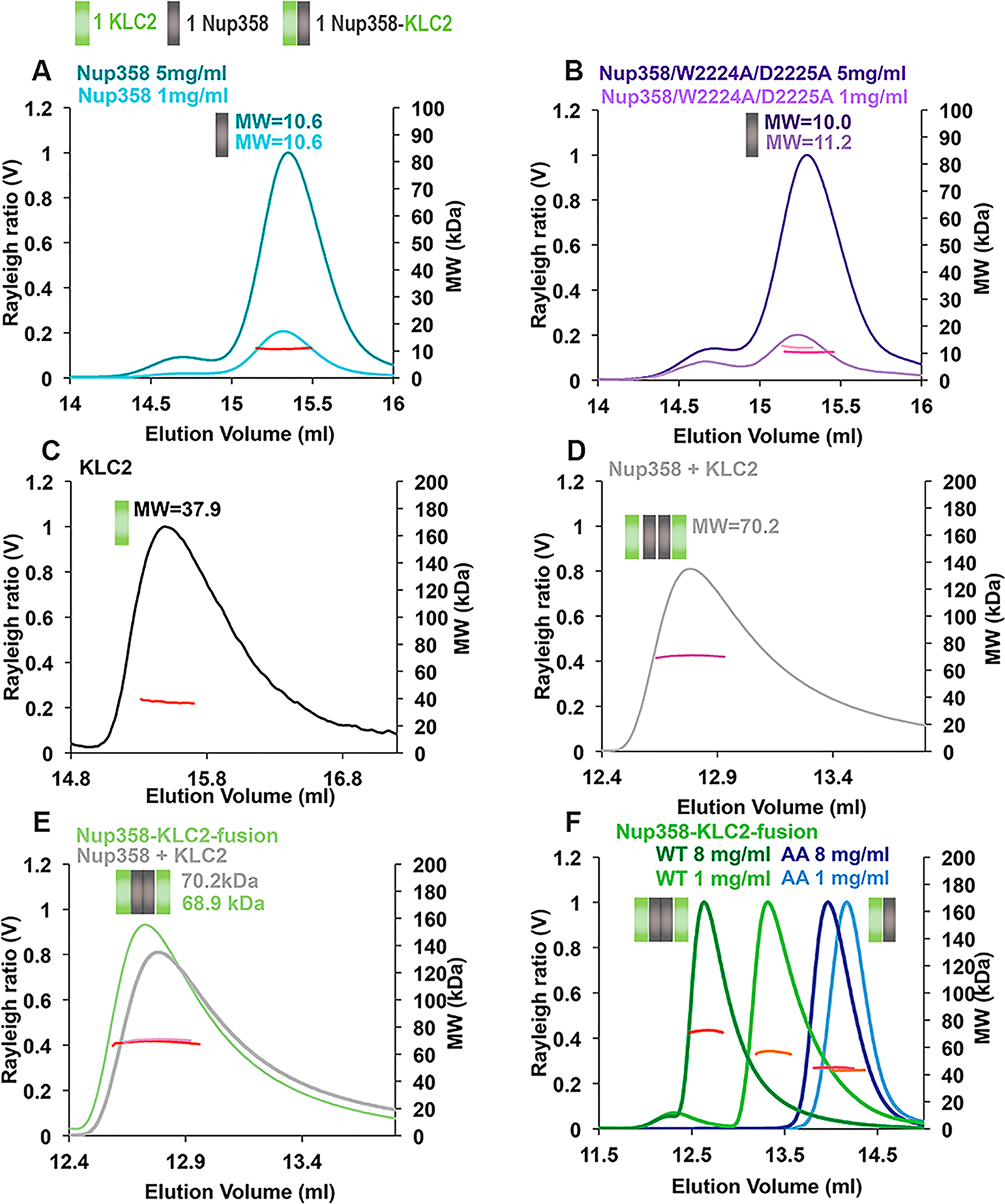
W-Acidic motif required for the formation of 2:2 complexes of Nup358-min and KLC2TPR, suggesting a role of cargo in dimerization of the KLC2s. Purified proteins were analyzed by size exclusion chromatography coupled to multiangle light scattering (SEC–MALS). The Rayleigh ratio (distinct colors) and weight-averaged molar masses (MW in kilodaltons) (colored in shades of red, orange, and pink) are plotted vs the elution volume. The oligomeric state that matches most closely the data is indicated in a schematic manner above each elution peak; each Nup358-min or KLC2TPR protomer is represented by a gray or green square, respectively. Elution peak boundaries were placed at half-peak height. Molar masses were determined across the selected peak area and plotted on a secondary axis in all SEC–MALS graphs. Average MWs are also shown. (A) Nup358-min. (B) Nup358-min/W2224A/D2225A mutant. (C) KLC2TPR-trunc. (D) Nup358-min and KLC2TPR mixed in a 1:1 molar ratio and concentrated to 4 mg/mL before analysis. (E) Overlay of the Nup358-min/KLCTPR complex that was assembled from individual proteins (from panel D) with the SEC–MALS profile of the Nup358-KLC2-fusion protein at the same concentration (4 mg/mL). (F) Nup358-KLC2-fusion protein (WT, green) and its W2224A/D2225A mutant (AA, blue). Representative experiments are shown, and molar masses were averaged from the following numbers of replicates: (A) 3 and 3, (B) 3 and 2, (C) 2, (D) 4, (E) 3 and 3, and (F) 2 and 2, and 2 and 3 (see Table S1). MWs, errors, and protein concentrations at distinct elution peaks are listed in Table S1. The standard deviations of the MWs were calculated, but because they were in all cases much below the experimental error of 5%, the errors of all MWs were calculated at 5%.
Table 1.
Calculated Molar Masses (MWs) of the Monomers
| protein domain | MW (kDa) |
|---|---|
| Nup358-min | 10.6 |
| BicD2-CTD | 10.9 |
| KLC2TPR (residues 196–480) | 34.6 |
| KLC2TPR-trunc (residues 218–480) | 32.1 |
| Nup358-KLC2-fusion | 42.9 |
The molar mass of KLC2TPR was also determined by SEC–MALS. For these experiments, a slightly shortened fragment was used, because without addition of a binding partner the solubility of the original fragment was too low for analysis (termed KLC2TPR-trunc, containing residues 218–480,10 compared to KLC2TPR, which contained residues 196–48029). The molar mass of KLC2TPR-trunc was determined to be 37.9 ± 1.9 kDa, which suggests that it forms predominantly monomers (the calculated mass of the monomer is 32.1 kDa) (Figure 5C).
To determine the molar mass of the Nup358-min/KLC2TPR complex by SEC–MALS, we mixed the individual proteins. The complex eluted in a single peak that had a molar mass of 70.2 ± 3.5 kDa (Figure 5D), suggesting formation of a 2:2 complex [calculated MW of 90.4 kDa (Table 1)], as well as some oligomers with a lower molar mass. The latter could include KLC2TPR and Nup358-min monomers (calculated molar masses of 34.6 and 10.6 kDa, respectively) or 1:1 or 2:1 complexes.
To conclude, our data suggest that Nup358-min and KLC2TPR form monomers. Binding of the cargo Nup358-min to KLC2TPR induces oligomerization and results in formation of a 2:2 complex of the KLC2TPR with its cargo Nup358, along with oligomers with lower molar masses.
To facilitate the analysis of oligomeric states, we also designed an N-terminal fusion protein of Nup358-min and KLC2TPR [Nup358-KLC2-fusion (Figure 3B)]. A fusion protein has the advantage that only one reactant is present, and only homo-oligomers are formed rather than a mixture of homo- and hetero-oligomers, which are more difficult to analyze. Within the fusion protein, the W-acidic motif of Nup358 present in the C-terminal portion of Nup358-min is expected to interact with KLC2TPR, resulting in a loop-type structure, whereas the N-terminus of Nup358-min is available for binding of BicD2. This construct was designed on the basis of the fusion protein that allowed the structure of KLC2TPR with a W-acidic motif bound to be determined.10,29 In the original construct, the W-acidic motif with the sequence LEWD was N-terminally fused via a flexible linker of 16 amino acids. Nup358-min is intrinsically disordered; therefore, we fused Nup358-min directly to KLC2TPR, because the W-acidic motif with the sequence LEWD is followed by 16 C-terminal amino acids that act as a flexible linker with a length similar to that of the original construct. Notably, the resulting Nup358-KLC2-fusion protein has a much higher solubility (9 mg/mL) compared to that of KLC2TPR (0.5 mg/mL) or a complex of KLC2TPR with its binding partner, Nup358-min (5 mg/mL). Because the fusion protein leads to a locally increased concentration of the binding partners, the interaction is expected to be stronger compared to that of a complex formed from the individual proteins.
We determined the oligomeric state of the Nup358-KLC2-fusion protein by SEC–MALS to assess if it forms similar oligomers as the individual protein domains Nup358-min and KLC2TPR. The molar mass of the Nup358-KLC2-fusion protein was determined to be 68.9 ± 3.4 kDa, which suggests formation of dimers, as well as a small population of monomers [calculated dimer MW of 85.8 kDa (Table 1)] (Figure 5E and Table S1). Notably, this matches closely to the molar mass of the complex formed by the individual Nup358-min and KLC2TPR proteins at the same protein concentration (MW = 70.2 ± 3.5 kDa). Furthermore, the SEC–MALS elution profiles of the complex and the fusion protein look very similar (Figure 5E). This suggests that the Nup358-KLC2-fusion protein forms similar oligomers as the complex assembled from the two individual proteins. It also suggests that the W-acidic motif of Nup358 is bound to KLC2TPR within the fusion protein, because the individual protein components form monomers. Furthermore, the Nup358-KLC2-fusion protein is highly soluble (9 mg/mL), even more than the Nup358-min/KLC2TPR complex (5 mg/mL), whereas KLC2TPR in the absence of cargo has a much lower solubility, likely because the hydrophobic pocket for the W-acidic motif is solvent-exposed.
To assess whether dimerization of the Nup358-KLC2-fusion protein is dependent on concentration, we performed SEC–MALS analyses at a range of concentrations between 1 and 8 mg/mL. Molar masses ranged from 56.3 ± 2.8 kDa (1 mg/mL) to 72.1 ± 3.6 kDa (8 mg/mL), suggesting that the fusion protein exists in a dimer–monomer equilibrium and that dimerization is favored at higher protein concentrations (Figure 5F).
We also created a W2224A/D2225A mutant of the Nup358-KLC2-fusion protein. In the resulting fusion protein, the internal interaction of Nup358 and KLC2 is expected to be strongly diminished. The molar mass of the Nup358/W2224A/D2225A-KLC2-fusion protein was determined to be 45.1 ± 2.3 kDa at a protein concentration of 8 mg/mL and 43.2 ± 2.2 kDa at 1 mg/mL, closely matching the molar mass of the monomer (42.9 kDa) (Figure 5F). These data suggest that the interaction between the W-acidic motif of Nup358 and KLC2 is required to form 2:2 complexes, because the W2224A/D2225A mutation results in formation of monomers.
To conclude, the minimal interacting domains Nup358-min and KLC2TPR form monomers in solution. Notably, formation of a complex between the two induces oligomerization and results in the formation of 2:2 complexes. A fusion protein of these two domains, in which the W-acidic motif is expected to interact with KLC2, also dimerizes. Notably, if this internal interaction is disrupted by a W2224A/D2225A mutation of the W-acidic motif, the mutated fusion protein forms monomers. These data suggest that the interaction of the W-acidic motif of the cargo Nup358 is required to form 2:2 complexes with KLC2. It should be noted that we cannot exclude formation of some additional oligomeric states, including 2:1 complexes; however, the oligomeric states of both the mixed individual proteins and the fusion protein (which enforces a 1:1 ratio of reactants and eliminates the presence of individual proteins) are similar, which supports the fact that a 2:2 complex is indeed formed, as observed in the crystal structures of KLC2/cargo complexes.10
BicD2 and KLC2 Interact with Nup358 Simultaneously and Form 2:2:2 Complexes.
Because KLC2 binds to the same domain of Nup358 as BicD2, we investigated if these proteins bind to Nup358 simultaneously or whether they compete. Therefore, we designed a competition assay, in which purified minimal interacting domains of all three proteins were mixed and analyzed by size exclusion chromatography (Figure 6). The C-terminal domain of BicD2 [BicD2-CTD, consisting of residues 715–804 (Figure 3C)] has been previously fine-mapped as the minimal interacting domain for Nup358-min.1,9,22,39
Figure 6.
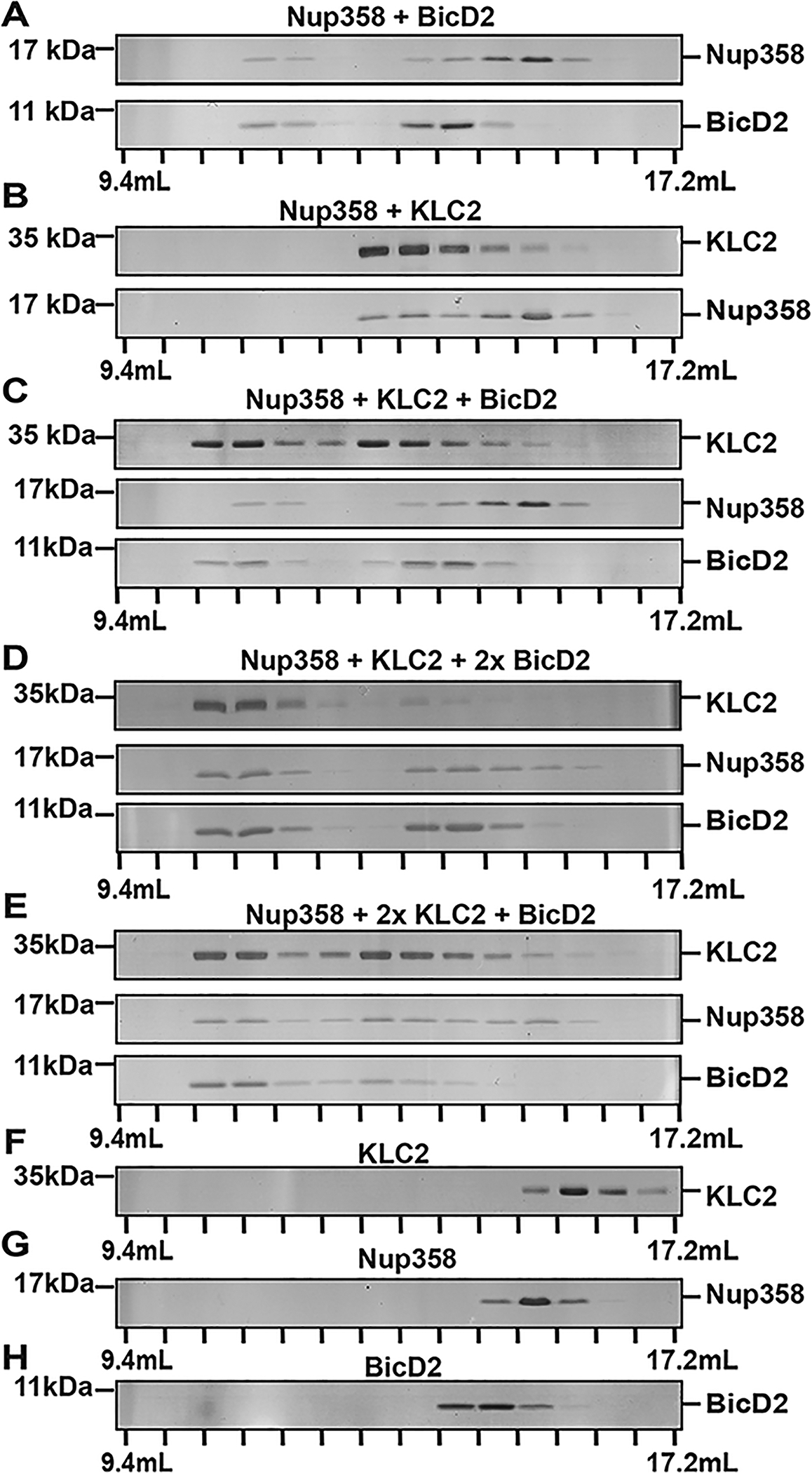
BicD2-CTD and KLC2 are capable of binding simultaneously to Nup358-min. (A–H) Purified BicD2-CTD, KLC2TPR, and Nup358-min were combined in an equimolar ratio and analyzed by gel filtration chromatography. SDS–PAGE gels of the elution fractions are shown. (A) Nup358-min and BicD2-CTD. (B) Nup358-min and KLC2TPR. (C) KLC2TPR, Nup358-min, and BicD2-CTD. (D and E) Same as panel C. 2x denotes a 2-fold molar excess of BicD2-CTD (D) or KLC2TPR (E). Samples in panels D and E were concentrated after mixing prior to analysis. (F) KLC2TPR. (G) Nup358-min. (H) BicD2-CTD. SDS–PAGE analyses of the purified proteins and of the loaded samples are shown in Figures S3 and S4, respectively. The experiments were repeated, and very similar results were obtained. The numbers of replicates for experiments shown in each figure panel were (A) 3, (B) 4, (C) 2, (D) 2, (E) 2, (F) 3, (G) 4, and (H) 5.
First, we mixed Nup358-min and BicD2-CTD and separated the mixture by gel filtration chromatography. These experiments suggested complex formation, because Nup358-min and BicD2-CTD co-elute and the elution peak is shifted toward a higher molecular weight when compared to the individual proteins (Figure 6A,G,H). An interaction was also confirmed for the mixture of Nup358-min and KLC2 (Figure 6B,F,G).
Next, we analyzed a mixture of all three proteins (Nup358-min, KLC2TPR, and BicD2-CTD) in an equimolar ratio. All three proteins co-elute, and the elution peak is shifted toward a higher molecular weight (Figure 6C) when compared to the binary complexes (Figure 6A,B), suggesting formation of a triple complex. We also repeated the experiment with either a 2-fold molar excess of BicD2-CTD (Figure 6D) or a 2-fold excess of KLC2TPR (Figure 6E). Adding an excess of one binding partner does not affect the interactions within the triple complex, suggesting that BicD2 and KLC2 likely do not compete for binding of Nup358-min.
Furthermore, in the absence of Nup358-min, no interaction was observed between BicD2-CTD and KLC2TPR (Figure S6). To conclude, our data confirm that BicD2-CTD and KLC2TPR interact simultaneously with Nup358-min. There is no indication from our data that these binding partners compete.
Highly processive dynein motors can be reconstituted from dynein, the N-terminal domain of BicD2 and dynactin.15–17 Notably, BicD2 fragments interact as dimers with dynein and dynactin in the structure of a minimal complex.18,60 Transport-active kinesin-1 motors are heterotetramers of two KLCs and to KHCs. In line with the oligomeric states observed for the active motors, KLC2TPR forms 2:2 complexes with cargoes in X-ray structures10,35,36 and BicD2 forms 2:2 complexes with cargoes in solution.39 Furthermore, BicD2-CTD also forms homodimers in solution.9,22,39 Therefore, we analyzed the oligomeric state of the Nup358-min/BicD2-CTD/KLC2TPR triple complex by SEC–MALS. The complex formed a broad elution peak that is likely composed of a mixture of oligomers. The molar mass of the first elution peak with the highest molar mass was 125.1 ± 6.3 kDa (Figure 7A and Table S2), suggesting formation of a 2:2:2 complex [calculated MW of 112.2 kDa (Table 1)]. For comparison, the molar mass of the binary KLC2TPR/Nup358-min complex (i.e., without BicD2) is 70.2 ± 3.5 kDa (Figure 7A); therefore, addition of BicD2 shifts the equilibrium even further toward the formation of 2:2:2 complexes, although other oligomeric states may be formed, as well.
Figure 7.
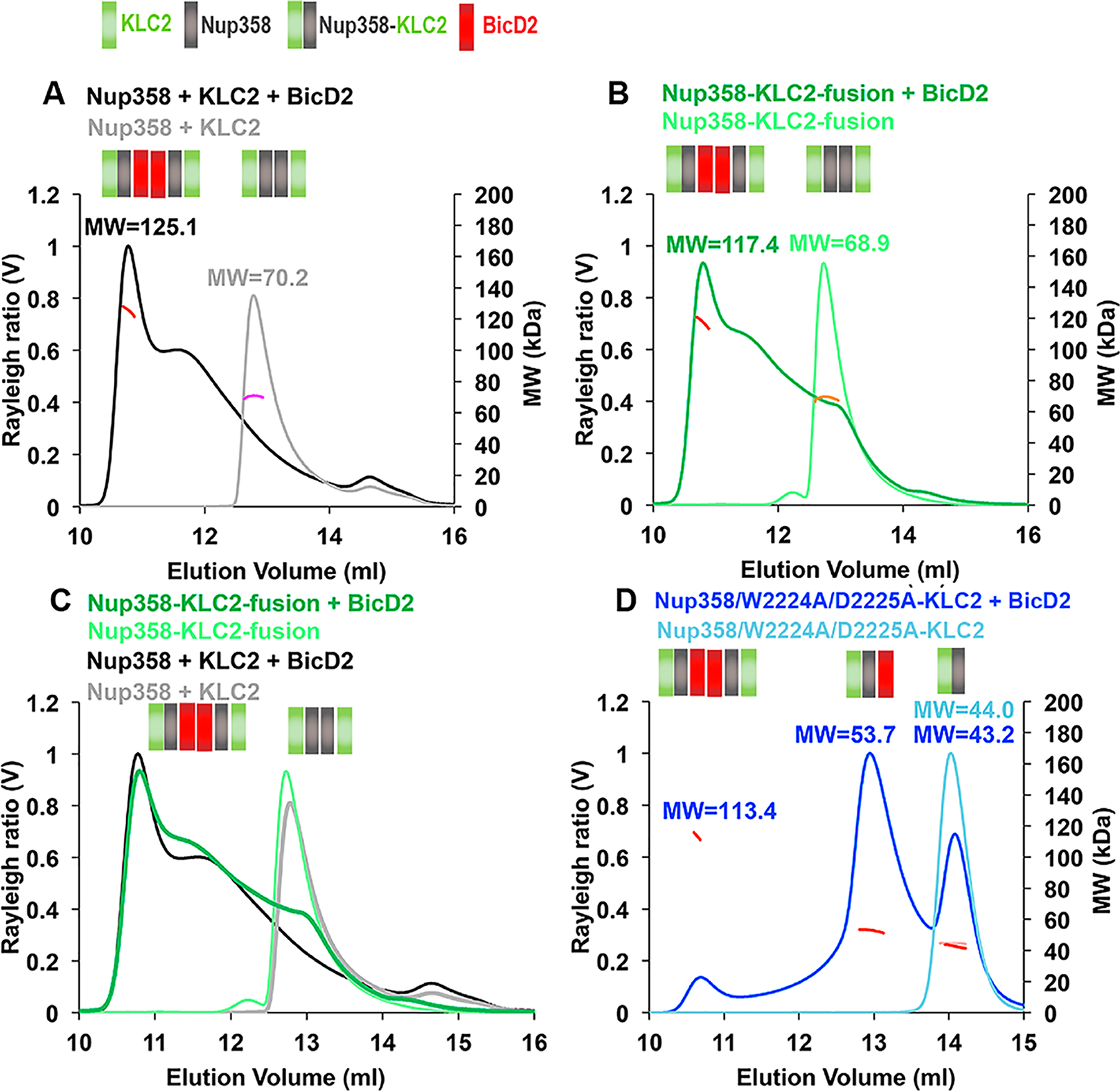
BicD2-CTD, Nup358-min, and KLC2TPR form a triple complex with a 2:2:2 stoichiometry. Purified proteins were analyzed by SEC–MALS. The Rayleigh ratio (distinct colors) and weight-averaged molar mass MW (in kilodaltons, colored in shades of red, orange, and pink) vs the elution volume are shown. MWs were averaged from two to three experiments (see Table S2 for errors and exact numbers of replicates) and are indicated. The oligomeric state that matches most closely to the data is indicated in a schematic manner above each elution peak; each Nup358-min, KLC2TPR, or BicD2-CTD protomer is represented by a gray, green, or red square, respectively. The same protein concentrations (4 mg/mL) were used for the Nup358-min/KLC2TPR complex (see Figure 5) as well as the Nup358-KLC2-fusion proteins (WT or W2224A/D2225A mutant). BicD2 was added at a fusion protein concentration of 1–4 mg/mL to achieve a 1:1 molar ratio of the reactants. The triple complex was assembled by mixing Nup358-min, KLC2TPR, and BicD2-CTD in an equimolar ratio, and the resulting triple complex was concentrated to 5 mg/mL before analysis. (A) Overlay of SEC–MALS elution profiles of Nup358-min and KLC2TPR (gray) and the Nup358-min/KLC2TPR/BicD2 triple complex (black). (B) Overlay of SEC–MALS elution profiles of the Nup358-KLC fusion protein and the Nup358-KLC fusion protein with BicD2-CTD. (C) Overlay of SEC–MALS elution profiles from panels A and B. (D) Overlay of SEC–MALS profiles of Nup358/W2224A/D2225A-KLC2-fusion protein and Nup358/W2224A/D2225A-KLC2-fusion protein with BicD2-CTD. Numbers of replicates for each experiment: (A) 3 and 3, (B) 3 and 3, (C) repeated from panels A and B, and (D) 2 and 2 (see Table S2).
We also performed analysis of the Nup358-KLC2-fusion protein with BicD2 bound, which reduces the triple complex to a two-component system, as it is expected to be more homogeneous. The SEC–MALS elution profile of the Nup358-KLC2-fusion protein mixed with BicD2-CTD over-lays very well with the SEC–MALS elution profile of the individual proteins Nup358-min, KLC2TPR, and BicD2-CTD at the same concentration (Figure 7B,C). The molar mass of the first elution peak of the Nup358-KLC2-fusion protein mixed with BicD2-CTD was determined to be 117.4 ± 5.9 kDa, which is quite similar to the molar mass of the mixture of the three individual proteins (125.1 ± 6.3 kDa) (Figure 7A,B). These data suggest that the Nup358-KLC2-fusion protein forms oligomers with BicD2-CTD similar to those of the individual proteins. It also confirms the formation of 2:2:2 complex for the triple complex and establishes the stoichiometry of these three proteins. We cannot exclude the formation of additional oligomeric states such as higher-order oligomers or possibly 2:1 complexes of fusion proteins and BicD2-CTD; however, it should be noted that the separate experiments analyzing fusion proteins and mixtures of Nup358-min and KLC2 with and without BicD2-CTD yield consistent results, supporting the formation of 2:2:2 complexes of BicD2, KLC2, and Nup358 among small amounts of other oligomeric states. These data establish an upper limit of the number of dynein and kinesin-1 motors that can be recruited to Nup358.
A binding assay suggested that the W-acidic motif is dispensable for the interaction between Nup358 and BicD2 and that the Nup358-min/W2224A/D2225A mutant interacts as strongly as the WT with BicD2-CTD (Figure S7). We were intrigued by the fact that the W2224A/D2225A mutant of the Nup358-KLC2-fusion protein forms monomers (rather than dimers, as observed in the WT); therefore, we investigated whether addition of BicD2-CTD affected the oligomeric state of the fusion protein/W2224A/D2225A mutant. The W2224A/D2225A double mutation is expected to disrupt the internal interaction between Nup358-min and KLC2TPR in the fusion protein. We focused on analysis of the highly soluble fusion protein (soluble to 8 mg/mL), because a mixture of the individual KLC2TPR and Nup358-min/W2224A/D2225A proteins had a very low solubility.
In the SEC–MALS elution profile of the Nup358/W2224A/D2225A-KLC2-fusion protein mutant and the BicD2-CTD, three elution peaks were observed for the complex (Figure 7D). The first elution peak with the highest mass is very small (~1/8 of the peak height of the WT) and has a molar mass of 113.4 ± 5.7 kDa, similar to that of the WT (117.4 kDa). The second elution peak is the highest and has a molar mass of 53.7 ± 2.7 kDa, which matches closely to a 1:1 complex [calculated MW of 53.8 kDa (Table 1)]. The third elution peak is quite large and has a molar mass of 43.2 ± 2.2 kDa, which matches closely the mass of a fusion protein monomer (calculated MW of 42.9 kDa). To conclude, the Nup358/W2224A/D2225A-KLC2-fusion protein forms monomers and can form 1:1 complexes with the BicD2-CTD. A very small amount of 2:2 complexes are formed, as well. Therefore, while the W-acidic motif is dispensable for binding of BicD2 to Nup358, the interaction of Nup358-min and KLC2TPR via the W-acidic motif is important for the formation of 2:2:2 complexes with BicD2. BicD2 is a dimer when bound to dynein/dynactin; therefore, it is conceivable that a disruption of oligomerization in the triple complex has mechanistic consequences.
To compare whether BicD2 remains stably associated with the WT and W2224A/D2225A mutant Nup358-KLC2-fusion proteins, we assembled a complex of the fusion proteins with the BicD2-CTD and isolated the preassembled complex by gel filtration. This approach allowed individual proteins to be removed, resulting in a more homogeneous sample (Figure 8).
Figure 8.
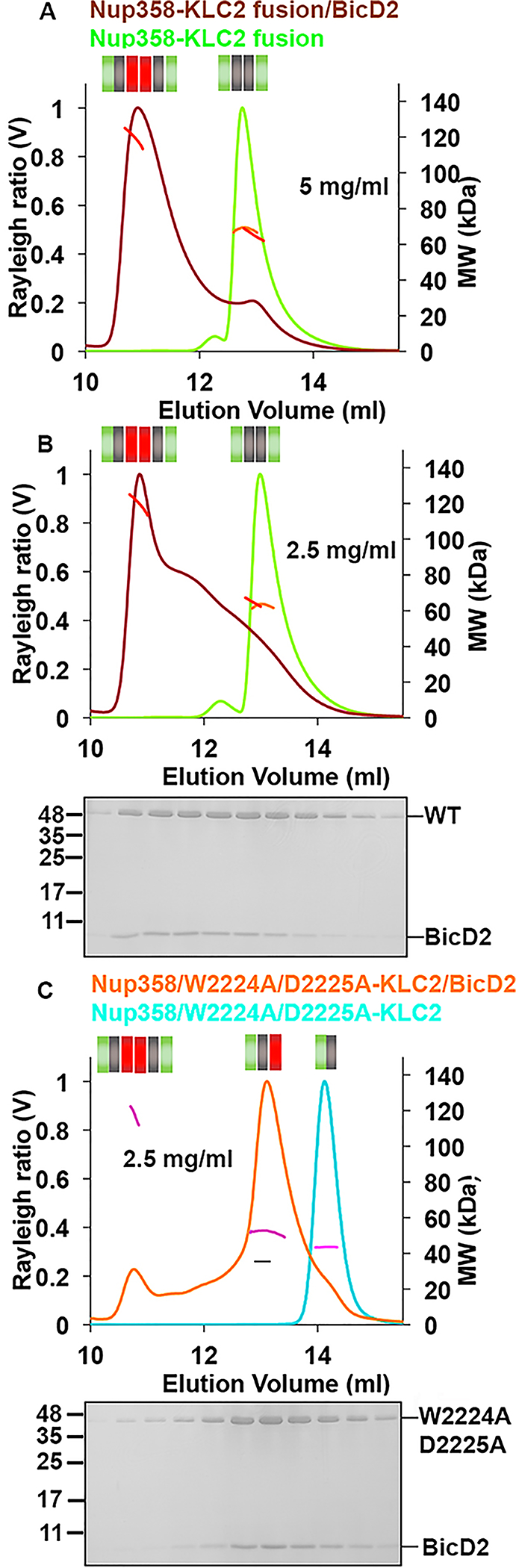
SEC–MALS analysis of purified Nup358-KLC2/BicD2-CTD complexes. The Nup358-KLC2-fusion protein (WT or W2224A/D2225A mutant) was mixed with the BicD2-CTD in a 1:1 molar ratio, and the complex was isolated by size exclusion chromatography to remove unbound reactants. These complexes were subsequently analyzed by SEC–MALS. The Rayleigh ratio (distinct colors) and molar mass MW (colored in shades of red, orange, and pink) are plotted vs elution volume (see also Table S3). The SDS–PAGE of the elution fractions is shown beneath the SEC–MALS profiles of the complexes. (A) The Nup358-KLC2-fusion protein and the purified Nup358-KLC2-fusion protein/BicD2-CTD complex (5 mg/mL) were analyzed. (B) Same experiment as in panel A, at 2.5 mg/mL. (C) Same experiment as in panel A, with the Nup358/W2224A/D2225A-KLC2-fusion protein. Numbers of replicates for each experiment: (A) 2 and 3, (B) 4 and 2, and (C) 3 and 3 (see Table S3).
The SEC–MALS elution profile of the purified Nup358-KLC2-fusion protein/BicD2-CTD WT complex at 2.5 mg/mL is similar to that observed via SEC–MALS analysis of the mixed proteins, and the molar mass is comparable (Figure 8B). SDS–PAGE of the elution fractions shows that BicD2 and the fusion protein form a stoichiometric complex. We also analyzed the purified complex at 5 mg/mL by SEC–MALS (Figure 8A). At the higher concentration, most of the protein elutes in the first elution peak with a higher molar mass [118.4 ± 5.9 kDa (Table S3)], which contains the 2:2 complex. The second peak is reduced to a small shoulder. Therefore, at higher protein concentrations, the 2:2 complex is the predominant oligomeric state.
We also analyzed the purified BicD2-CTD/Nup358/W2224A/D2225A-KLC2-fusion protein complex by SEC–MALS at a concentration of 2.5 mg/mL. The first elution peak with the highest mass that contains the 2:2 complex is rather small, similar to that observed via SEC–MALS analysis of the mixed proteins, and the molar mass is comparable. The second peak also has a similar height and molar mass as observed for the mixed individual proteins. However, the third elution peak that contained individual Nup358/W2224A/D2225A-KLC2-fusion protein in the SEC–MALS profile of the mixed proteins is not present in the SEC–MALS elution profile of the purified BicD2-CTD/Nup358/W2224A/D2225A-KLC2-fusion protein complex, and SDS–PAGE analysis of the elution fractions confirms that it forms a stoichiometric complex in both peaks (Figure 8C). These data suggest that the BicD2-CTD remains stably associated with the WT and W2224A/D2225A mutant fusion proteins (Figure 8).
To conclude, our data suggest that the dynein adaptor BicD2-CTD and KLC2TPR interact simultaneously with Nup358-min and do not compete for binding. The W-acidic motif of Nup358, which is required to recruit KLC2, is dispensable for the interaction with BicD2. However, the W-acidic motif is required for the formation of a 2:2 complex of Nup358-min and KLC2TPR and for formation of 2:2:2 complexes upon addition of BicD2. Disruption of the interaction of Nup358-min and KLC2TPR by a W2224A/D2225A mutation results in the formation of monomers and upon addition of BicD2 in the formation of 1:1:1 complexes. These results suggest a role of KLC2/cargo interactions and especially the W-acidic motif with the sequence LEWD in oligomerization. Both KLC2 and BicD2 dimerize within active motor complexes; therefore, cargo-induced dimerization of KLC2 may potentially be important for the activation mechanism of these motor adaptors.
DISCUSSION
Here, we have reconstituted and characterized a minimal complex of Nup358 and KLC2. While Nup358 and KLC2 form predominantly monomers, their interaction results in the formation of 2:2 complexes, and the W-acidic motif is required for the oligomerization. Furthermore, we show that the dynein adaptor BicD2 and KLC2 both interact simultaneously with Nup358 and form 2:2:2 complexes.
The fact that we were able to reconstitute a complex of KLC2TPR and Nup358-min is an important step as it confirms that the interaction is specific and that these proteins interact directly. While our study focuses on minimal recombinant interaction domains, the interaction of Nup358 with KLC2 was also confirmed in the context of cells.11 When the GST-tagged Nup358 kinesin-1-binding domain was incubated with retinal extracts, the KHC isoforms Kif5B and Kif5C were pulled down, and the complex also contained KLCs.11 In addition, initial immunoprecipitations in CO7 cells confirmed the interaction of this Nup358 domain with KLC2 (see Figure S5B of ref 38). Prior to our study, it was not clear if these proteins interacted directly. The significance of this interaction is further highlighted by the very high degree of sequence conservation of the W-acidic motif with the sequence LEWD and the BicD2-binding site among all vertebrates, and therefore, these pathways for the recruitment of dynein and kinesin-1 to the nucleus are likely conserved among vertebrates. The W-acidic motif is however missing in the Nup358 sequence of insects, and therefore, an interaction of Nup358 and KLC2 is likely not formed in insects, although the possibility cannot be excluded.61
Both KHCs and KLC2 bind to the same domain of Nup358.11,38 A combination of spatially close KHC-and KLC-binding sites is common, as full transport activity requires the formation of multiple cargo/KLC and cargo/KHC interactions.32–34 While the binding site of the KHCs on Nup358 has not been finely mapped, it has been shown that KHCs recognize a binding site that includes the W-acidic binding motif of the cargo SKIP.32 However, this interaction was formed only when autoinhibition was relieved, suggesting that the KLCs need to bind first, prior to the KHCs.32 Future studies will investigate if the KHCs also recognize the W-acidic motif of Nup358.
It has been previously proposed that the interaction of KLC2TPR with cargo results in the formation of 2:2 complexes, which could be a general mechanism for activation of autoinhibited kinesin-1 upon cargo binding.36 It should be noted that this hypothesis remains to be confirmed by functional assays with intact motors and additional interaction partners. In line with this idea, our results suggest that binding of Nup358 to KLC2 induces formation of 2:2 complexes, whereas the individual Nup358 and KLC2 domains form monomers. Several structures of KLC2/cargo complexes confirm that these generally form 2:2 complexes.10,29,35,36 However, the TPR domain of KLC2 forms monomers in solution,62 even though in crystal structures a dimer is observed.29,62,63
Notably, we show that the W-acidic motif, which promotes interaction between Nup358 and KLC2, is required for cargo-induced dimerization. Disruption of the internal interaction in the Nup358-KLC2-fusion protein by a W2224A/D2225A mutation of the W-acidic motif results in the formation of monomers. Our fusion proteins form similar oligomers as a mixture of the individual proteins but have higher solubility. Therefore, they are valuable tools for investigating the oligomeric state.
In the wild-type Nup358-KLC2-fusion protein dimer, two different conformations are possible, because the W-acidic motif is tethered to KLC2TPR via a flexible linker. The W-acidic motifs could bind to the KLC2s present in the same fusion protein molecule (internal interaction), or the W-acidic motifs could bind to the KLC2s of the second fusion protein molecule of the dimer (domain-swapped interaction). A domain swap would link the two wild-type fusion protein molecules in the dimer and thus would be expected to increase the degree of dimerization compared to those of the individually mixed proteins. We show that the wild-type fusion protein forms oligomeric states very similar to those of the individually mixed proteins (Nup358-min and KLC2TPR), and the degree of dimerization is similar and not increased. Furthermore, when the fusion protein is mixed with BicD2, very similar oligomeric states are formed compared to the three individually mixed proteins. Therefore, any potential conformational changes resulting from fusing Nup358-min to KLC2TPR are unlikely to affect oligomerization.
Notably, our data show that Nup358-min interacts simultaneously with BicD2-CTD and KLC2TPR, and that they do not compete. On the basis of our data, we hypothesize that dynein and kinesin-1 are simultaneously recruited to Nup358, and therefore, a competition model33 in which opposing motors compete for binding of cargo is likely not applicable for this transport pathway. However, our data are compatible with other models, such as the tug-of-war model,40,48,49 in which the team of motors that produces the greater force determines the direction of transport, or the coordination model,44,50,51 in which opposing motors bind simultaneously to cargo and are tightly coordinated for unidirectional transport by regulatory mechanisms.
While it is possible that both motors bind independently, the spatial proximity of the binding sites of BicD2 and KLC2 on Nup358 suggests that these motor recognition domains interact and regulate each other’s transport function. This Nup358 domain is an intrinsically disordered protein, which may undergo structural rearrangement upon binding of the BicD2-CTD coiled coil and the TPR domain of KLC2. It remains to be established if such structural changes occur and whether they result in cooperativity.
Both BicD2 and KLCs form dimers in kinesin-1 and dynein/dynactin/BicD2 motor complexes.18,60 Interestingly, the triple complex of Nup358, BicD2, and KLC2 has a 2:2:2 stoichiometry, and the W-acidic motif is required for oligomerization. A W2224A/D2225A double mutation disrupts the internal interaction between KLC2 and Nup358 and results in the formation of 1:1:1 complexes of BicD2, Nup358, and KLC2. Because the oligomerization state is important for the function of KLC2 and BicD2, the fact that a disruption of the KLC2/Nup358 interaction also affects the oligomeric state of BicD2 does indeed suggest that there is crosstalk between these adaptors. Future studies will test these hypotheses in vivo in the context of the full-length proteins.
For a 2:2:2 complex of kinesin-1, BicD2/dynein, and Nup358 to be formed in vivo, two copies of Nup358 need to be in spatial proximity within the NPC. Recently, a structure of the symmetric NPC core scaffold was determined by an interdisciplinary approach; however, Nup358 was not placed in the cryo-electron microscopy map.64–66 An electron density in the cytoplasmic filaments of the NPC remained unassigned, which may possibly be consistent with two or more spatially close molecules of Nup358, as the electron density was diminished upon RNAi knockdown of Nup358.65,67 However, it is not clear if the entire electron density represents Nup358, and thus, the exact copy number of Nup358 in the NPC remains to be established. The number of NPCs in each nucleus is known.68 Assuming 16 Nup358 molecules in each NPC,65,67 each nucleus contains an estimated ~32000 molecules of Nup358,39 which doubles at the onset of mitosis.68 These numbers provide a rough estimate of the motor docking sites that are provided by Nup358.
Our data also establish the stoichiometry of opposing motors that bind to Nup358. A Nup358 dimer can recruit one (heterotetrameric) kinesin-1 motor and one (dimeric) dynein motor via a BicD2 dimer. BicD2 is a dynein adapter that predominantly recruits one dynein motor, but a small population of BicD2 molecules can also recruit a second dynein motor.60 The forces of kinesin-1 and dynein 1 are comparable (6 and 4.3 pN, respectively).48 While dynein is the major motor responsible for nuclear positioning in the G2 phase, the overall motility is also regulated by kinesin-1, which antagonizes dynein activity.1 Teams of individual motors are sufficient to bypass obstacles, which poses the question of why both motors are needed.69 The arrangement of opposing microtubule motors acting together is likely required for spatiotemporal regulation of transport and may represent a fundamental property of these motor systems.1,4,33,38,40–47
CONCLUSIONS
We have reconstituted a minimal triple complex of Nup358 with the motor recognition domains of the opposing motors dynein and kinesin-1 bound. This purified complex can be used as a valuable tool for future studies of bidirectional transport along microtubules.
While Nup358 and KLC2 form predominantly monomers, their interaction results in the formation of 2:2 complexes, and the W-acidic motif is required for the oligomerization. This is in line with the idea that cargo binding induces oligomerization to 2:2 complexes, which may be a general mechanism for activation of the KLCs upon cargo binding.36
On the basis of our data, we propose that one dynein motor and one kinesin-1 motor are recruited simultaneously to Nup358. There is no indication in our data that the recognition domains of these motors compete for binding. The spatial proximity of the binding sites suggests that these motor recognition domains interact and there may be crosstalk between them. Notably, the interaction of the W-acidic motif of Nup358 with KLC2 is required for the formation of 2:2:2 complexes of Nup358, BicD2, and KLC2, as a W2224A/D2225A double mutation of the W-acidic motif results in 1:1:1 complexes. BicD2 and KLC2 both form dimers in active motor complexes, and a change in the oligomerization state may have consequences for the function of these adaptors. These data provide important insights into a nuclear positioning pathway that is crucial for brain development and control of the cell cycle stage.1,3
Supplementary Material
ACKNOWLEDGMENTS
The authors thank A. Akhmanova (Utrecht University, Utrecht, The Netherlands) for plasmids encoding DNA sequences of Nup358 and BicD2,1 M. P. Dodding for an expression construct of the TPR domain of KLC2,10,29 and D. King (Howard Hughes Medical Institute, University of California, Berkeley, CA) for mass spectrometry analysis. Furthermore, the authors thank B. Callahan and S. Bane (Binghamton University) for access to equipment and D. Moraga for critical reading of the manuscript.
Funding
This work was supported by the National Institutes of Health (NIH) via National Institute of General Medical Sciences (NIGMS) Grant 1R15GM128119-01 awarded to S.R.S. The Department of Chemistry of Binghamton University and the Research Foundation of the State University of New York provided additional funding.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.9b00907.
Seven figures and three tables (PDF)
Accession Codes
Nup358, NCBI XM_005264002.2, kinesin-1 light chain 2 isoform 1, NCBI NP_001356289.1, Bicaudal D2, NCBI NM_029791.4.
The authors declare no competing financial interest.
REFERENCES
- 1.Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, Demmers J, Fornerod M, Melchior F, Hoogenraad CC, Medema RH, and Akhmanova A (2010) Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 8, No. e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baffet AD, Hu DJ, and Vallee RB (2015) Cdk1 activates pre-mitotic nuclear envelope dynein recruitment and apical nuclear migration in neural stem cells. Dev. Cell 33, 703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu DJ, Baffet AD, Nayak T, Akhmanova A, Doye V, and Vallee RB (2013) Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell 154, 1300–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson MH, and Holzbaur ELF (2012) Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell Sci 125, 4158–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neveling K, Martinez-Carrera LA, Holker I, Heister A, Verrips A, Hosseini-Barkooie SM, Gilissen C, Vermeer S, Pennings M, Meijer R, te Riele M, Frijns CJM, Suchowersky O, MacLaren L, Rudnik-Schoneborn S, Sinke RJ, Zerres K, Lowry RB, Lemmink HH, Garbes L, Veltman JA, Schelhaas HJ, Scheffer H, and Wirth B (2013) Mutations in BICD2, which encodes a golgin and important motor adaptor, cause congenital autosomal-dominant spinal muscular atrophy. Am. J. Hum. Genet 92, 946–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peeters K, Litvinenko I, Asselbergh B, Almeida-Souza L, Chamova T, Geuens T, Ydens E, Zimon M, Irobi J, De Vriendt E, De Winter V, Ooms T, Timmerman V, Tournev I, and Jordanova A (2013) Molecular defects in the motor adaptor BICD2 cause proximal spinal muscular atrophy with autosomal-dominant inheritance. Am. J. Hum. Genet 92, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Synofzik M, Martinez-Carrera LA, Lindig T, Schöls L, and Wirth B (2014) Dominant spinal muscular atrophy due to BICD2: a novel mutation refines the phenotype. J. Neurol., Neurosurg. Psychiatry 85, 590–592. [DOI] [PubMed] [Google Scholar]
- 8.Oates EC, Rossor AM, Hafezparast M, Gonzalez M, Speziani F, MacArthur DG, Lek M, Cottenie E, Scoto M, Foley AR, Hurles M, Houlden H, Greensmith L, Auer-Grumbach M, Pieber TR, Strom TM, Schule R, Herrmann DN, Sowden JE, Acsadi G, Menezes MP, Clarke NF, Zuchner S, Muntoni F, North KN, and Reilly MM (2013) Mutations in BICD2 cause dominant congenital spinal muscular atrophy and hereditary spastic paraplegia. Am. J. Hum. Genet 92, 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noell CR, Loh JY, Debler EW, Loftus KM, Cui H, Russ BB, Zhang K, Goyal P, and Solmaz SR (2019) Role of Coiled-Coil Registry Shifts in the Activation of Human Bicaudal D2 for Dynein Recruitment upon Cargo Binding. J. Phys. Chem. Lett 10, 4362–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pernigo S, Lamprecht A, Steiner RA, and Dodding MP (2013) Structural basis for kinesin-1:cargo recognition. Science 340, 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Y, Singh BB, Aslanukov A, Zhao H, and Ferreira PA (2001) The Docking of Kinesins, KIF5B and KIF5C, to Ran-binding Protein 2 (RanBP2) Is Mediated via a Novel RanBP2 Domain. J. Biol. Chem 276, 41594–41602. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Matunis MJ, Kraemer D, Blobel G, and Coutavas E (1995) Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, RanGTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem 270, 14209–14213. [DOI] [PubMed] [Google Scholar]
- 13.Cianfrocco MA, DeSantis ME, Leschziner AE, and Reck-Peterson SL (2015) Mechanism and regulation of cytoplasmic dynein. Annu. Rev. Cell Dev. Biol 31, 83–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA, Hoogenraad CC, King SJ, and Akhmanova A (2012) BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell 23, 4226–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlager MA, Serra-Marques A, Grigoriev I, Gumy LF, Esteves da Silva M, Wulf PS, Akhmanova A, and Hoogenraad CC (2014) Bicaudal D family adaptor proteins control the velocity of dynein-based movements. Cell Rep 8, 1248–1256. [DOI] [PubMed] [Google Scholar]
- 16.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, and Carter AP (2014) In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J 33, 1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, and Vale RD (2014) Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, and Carter AP (2015) The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui H, Trybus K, Ali M, Goyal P, Aura X, Loh J, and Solmaz S (2019) Coiled-coil registry shifts in the F684I mutant of Bicaudal result in cargo-independent dynein recruitment. bioRxiv, DOI: 10.1101/776187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, Willemsen R, Visser P, Grosveld F, and Galjart N (2001) Mammalian Golgi associated Bicaudal D2 functions in the dynein dynactin pathway by interacting with these complexes. EMBO J 20, 4041–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoogenraad CC, Wulf P, Schiefermeier N, Stepanova T, Galjart N, Small JV, Grosveld F, de Zeeuw CI, and Akhmanova A (2003) Bicaudal D induces selective dynein mediated microtubule minus end directed transport. EMBO J 22, 6004–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Salter HK, Holding AN, Johnson CM, Stephens E, Lukavsky PJ, Walshaw J, and Bullock SL (2013) Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev 27, 1233–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terawaki S, Yoshikane A, Higuchi Y, and Wakamatsu K (2015) Structural basis for cargo binding and autoinhibition of Bicaudal-D1 by a parallel coiled-coil with homotypic registry. Biochem. Biophys. Res. Commun 460, 451–456. [DOI] [PubMed] [Google Scholar]
- 24.McClintock MA, Dix CI, Johnson CM, McLaughlin SH, Maizels RJ, Hoang HT, and Bullock SL (2018) RNA-directed activation of cytoplasmic dynein-1 in reconstituted transport RNPs. eLife 7, No. e36312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sladewski TE, Billington N, Ali MY, Bookwalter CS, Lu H, Krementsova EB, Schroer TA, and Trybus KM (2018) Recruitment of two dyneins to an mRNA-dependent Bicaudal D transport complex. eLife 7, No. e36306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury S, Ketcham SA, Schroer TA, and Lander GC (2015) Structural organization of the dynein-dynactin complex bound to microtubules. Nat. Struct. Mol. Biol 22, 345–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vale RD, Reese TS, and Sheetz MP (1985) Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodding MP, Mitter R, Humphries AC, and Way M (2011) A kinesin-1 binding motif in vaccinia virus that is widespread throughout the human genome. EMBO J 30, 4523–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yip YY, Pernigo S, Sanger A, Xu M, Parsons M, Steiner RA, and Dodding MP (2016) The light chains of kinesin-1 are autoinhibited. Proc. Natl. Acad. Sci. U. S. A 113, 2418–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Araki Y, Kawano T, Taru H, Saito Y, Wada S, Miyamoto K, Kobayashi H, Ishikawa HO, Ohsugi Y, Yamamoto T, Matsuno K, Kinjo M, and Suzuki T (2007) The novel cargo Alcadein induces vesicle association of kinesin-1 motor components and activates axonal transport. EMBO J 26, 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konecna A, Frischknecht R, Kinter J, Ludwig A, Steuble M, Meskenaite V, Indermühle M, Engel M, Cen C, Mateos J-M, Streit P, and Sonderegger P (2006) Calsyntenin-1 docks vesicular cargo to kinesin-1. Mol. Biol. Cell 17, 3651–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanger A, Yip YY, Randall TS, Pernigo S, Steiner RA, and Dodding MP (2017) SKIP controls lysosome positioning using a composite kinesin-1 heavy and light chain-binding domain. J. Cell Sci 130, 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu M-M, and Holzbaur ELF (2014) Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol 24, 564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blasius TL, Cai D, Jih GT, Toret CP, and Verhey KJ (2007) Two binding partners cooperate to activate the molecular motor Kinesin-1. J. Cell Biol 176, 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pernigo S, Chegkazi MS, Yip YY, Treacy C, Glorani G, Hansen K, Politis A, Bui S, Dodding MP, and Steiner RA (2018) Structural basis for isoform-specific kinesin-1 recognition of Y-acidic cargo adaptors. eLife 7, No. e38362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cockburn JJB, Hesketh SJ, Mulhair P, Thomsen M, O’Connell MJ, and Way M (2018) Insights into Kinesin-1 Activation from the Crystal Structure of KLC2 Bound to JIP3. Structure 26, e1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho K. i., Cai Y, Yi H, Yeh A, Aslanukov A, and Ferreira PA (2007) Association of the Kinesin-Binding Domain of RanBP2 to KIF5B and KIF5C Determines Mitochondria Localization and Function. Traffic 8, 1722–1735. [DOI] [PubMed] [Google Scholar]
- 38.Wilson MH, and Holzbaur ELF (2015) Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 142, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noell CR, Loftus KM, Cui H, Grewer CT, Kizer M, Debler EW, and Solmaz SR (2018) A quantitative model for BicD2/cargo interactions. Biochemistry 57, 6538–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendricks AG, Perlson E, Ross JL, Schroeder HW, Tokito M, and Holzbaur ELF (2010) Motor Coordination Via Tug-Of-War Mechanism Drives Bidirectional Vesicle Transport. Curr. Biol 20, 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu R, Antoku S, and Gundersen GG (2017) Centrifugal Displacement of Nuclei Reveals Multiple LINC Complex Mechanisms for Homeostatic Nuclear Positioning. Curr. Biol 27, 3097–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, and Doye V (2011) A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J. Cell Biol 192, 855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris RL, and Hollenbeck PJ (1993) The regulation of bidirectional mitochondrial transport is coordinated with axonal outgrowth. J. Cell Sci 104, 917–927. [DOI] [PubMed] [Google Scholar]
- 44.Encalada SE, Szpankowski L, Xia C-H, and Goldstein LSB (2011) Stable Kinesin and Dynein Assemblies Drive the Axonal Transport of Mammalian Prion Protein Vesicles. Cell 144, 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soppina V, Rai AK, Ramaiya AJ, Barak P, and Mallik R (2009) Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc. Natl. Acad. Sci. U. S. A 106, 19381–19386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dharan A, and Campbell EM (2018) Role of Microtubules and Microtubule-Associated Proteins in HIV-1 Infection. J. Virol 92, e00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dharan A, Opp S, Abdel-Rahim O, Keceli SK, Imam S, Diaz-Griffero F, and Campbell EM (2017) Bicaudal D2 facilitates the cytoplasmic trafficking and nuclear import of HIV-1 genomes during infection. Proc. Natl. Acad. Sci. U. S. A 114, E10707–E10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belyy V, Schlager MA, Foster H, Reimer AE, Carter AP, and Yildiz A (2016) The mammalian dynein–dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat. Cell Biol 18, 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller MJI, Klumpp S, and Lipowsky R (2008) Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc. Natl. Acad. Sci. U. S. A 105, 4609–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ally S, Larson AG, Barlan K, Rice SE, and Gelfand VI (2009) Opposite-polarity motors activate one another to trigger cargo transport in live cells. J. Cell Biol 187, 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gross SP, Guo Y, Martinez JE, and Welte MA (2003) A Determinant for Directionality of Organelle Transport in Drosophila Embryos. Curr. Biol 13, 1660–1668. [DOI] [PubMed] [Google Scholar]
- 52.Cui H, Loftus K, Noell C, and Solmaz S (2018) Identification of cyclin-dependent kinase 1 specific phosphorylation sites by an in vitro kinase assay. J. Visualized Exp, DOI: 10.3791/57674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loftus KM, Coutavas E, Cui H, King D, Ceravolo A, Pereiras D, and Solmaz S (2017) Mechanism for G2 phase-specific nuclear export of the kinetochore protein CENP-F. Cell Cycle 16, 1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma A, Solmaz SR, Blobel G, and Melcak I (2015) Ordered regions of channel nucleoporins Nup62, Nup54, and Nup58 form dynamic complexes in solution. J. Biol. Chem 290, 18370–18378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Solmaz SR, Blobel G, and Melcak I (2013) Ring cycle for dilating and constricting the nuclear pore. Proc. Natl. Acad. Sci. U. S. A 110, 5858–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solmaz SR, Chauhan R, Blobel G, and Melcak I (2011) Molecular architecture of the transport channel of the nuclear pore complex. Cell 147, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katoh K, Misawa K, Kuma K. i., and Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waterhouse AM, Procter JB, Martin DMA, Clamp M, and Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Notredame C, Higgins DG, and Heringa J (2000) T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 60.Urnavicius L, Lau CK, Elshenawy MM, Morales-Rios E, Motz C, Yildiz A, and Carter AP (2018) Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hampoelz B, Schwarz A, Ronchi P, Bragulat-Teixidor H, Tischer C, Gaspar I, Ephrussi A, Schwab Y, and Beck M (2019) Nuclear Pores Assemble from Nucleoporin Condensates During Oogenesis. Cell 179, 671–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen TQ, Chenon M, Vilela F, Velours C, Aumont-Nicaise M, Andreani J, Varela PF, Llinas P, and Ménétrey J (2017) Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain. PLoS One 12, e0186354–e0186354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu H, Lee HY, Tong Y, Hong B-S, Kim K-P, Shen Y, Lim KJ, Mackenzie F, Tempel W, and Park H-W (2012) Crystal structures of the tetratricopeptide repeat domains of kinesin light chains: insight into cargo recognition mechanisms. PLoS One 7, e33943–e33943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJH, Briggs JAG, Glavy JS, Hurt E, and Beck M (2016) Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull M-T, Banterle N, Parca L, Kastritis P, Buczak K, Mosalaganti S, Hagen W, Andres-Pons A, Lemke EA, Bork P, Antonin W, Glavy JS, Bui KH, and Beck M (2015) In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, Davenport AM, Jeon YE, and Hoelz A (2016) Architecture of the symmetric core of the nuclear pore. Science 352, aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ori A, Banterle N, Iskar M, Andrés-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, Lemke EA, and Beck M (2013) Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol 9, 648–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, Hsu BY-L, and Borun TW (1972) Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol 55, 433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferro LS, Can S, Turner MA, ElShenawy MM, and Yildiz A (2019) Kinesin and dynein use distinct mechanisms to bypass obstacles. eLife 8, No. e48629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


