Abstract
Biofilms are structured microbial communities adhered to surfaces that cause many human infections. The study of oral biofilms has revealed complex composition, spatial organization, and phenotypic/genotypic diversity of the resident microbiota at the various sites in the mouth. Yet, knowledge about the spatial arrangement, positioning, and function of the polymicrobial community across the intact biofilm architecture remains sparse. Using multiple length scale imaging and computational analysis, we discovered unique spatial designs comprising mixed interbacterial species and interkingdom communities within intact biofilms formed on teeth of toddlers with caries. Intriguing structural patterns ranging from intermixed communities with extensive coaggregation (including bacterial-fungal clustering) to spatially segregated species forming a multilayered architecture were found. Among them, a distinctive 3-dimensional structure exhibited densely clustered cariogenic pathogens that were surrounded by outer layers of mixed bacterial communities in juxtaposition, forming a highly ordered spatial organization. These findings are particularly relevant as we approach the postmicrobiome era whereby studying the spatial structure of the pathogen and commensal microbiota may be important for understanding the microbiome function at the infection site to coordinate the disease process in situ.
Keywords: microbiome, biogeography, Streptococcus mutans, Candida albicans, dental caries, early childhood caries
Introduction
Polymicrobial biofilms are frequently observed throughout the human body, modulating the transition from health to disease, whereby different species or specific pathogens act within the microbial community, often enmeshed in a protective extracellular matrix, to modulate virulence (Stacy et al. 2016; Koo et al. 2017; Bowen et al. 2018). Molecular sequencing technologies and bioinformatics have advanced our understanding of the polymicrobial composition of the oral microbiome (Dewhirst et al. 2010). Conversely, multiple imaging modalities revealed that the different microbial species form spatially structured communities across the biofilm (Zijnge et al. 2010; Mark Welch et al. 2016). Thus, the microbial community “structure” can be defined by “microbial composition” (based on sequence read abundance over a range of taxonomy) and “spatial organization” (based on how microbes are physically associated to each other) as remarked recently (Mark Welch et al. 2019; Valm 2019).
Dental caries is a prime example of polymicrobial biofilm-induced disease that affects 3.9 billion people globally, costing >$290 billion/y (Listl et al. 2015; Vos et al. 2017). In particular, early childhood caries (ECC) is the most prevalent biofilm infection in preschoolers, constituting a major public health problem (Dye et al. 2015; Peres et al. 2019). Sequencing- and culturing-based studies on severe ECC revealed a polymicrobial community associated with the disease, including Streptococcus mutans (a cariogenic pathogen), acidogenic-aciduric bacteria, and even fungi such as Candida albicans (Hajishengallis et al. 2017; Xiao, Grier, et al. 2018). However, the spatiostructural organization of polymicrobial communities on diseased tooth surfaces remains unresolved.
Intact Biofilms and Spatial Design Approach
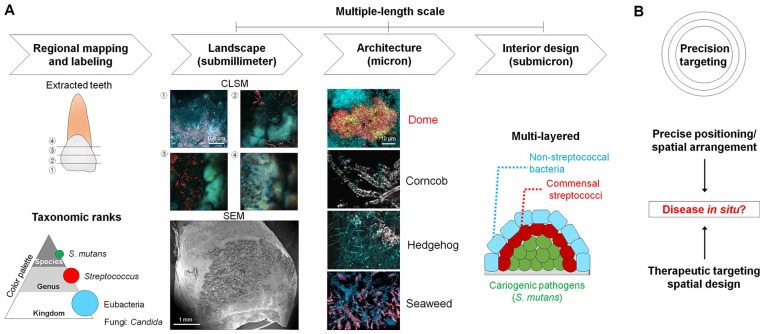
Rather than a homogeneous mixture of different species randomly distributed on the surface, the microbes within biofilms are arranged nonrandomly, forming spatially structured polymicrobial communities (Nadell et al. 2016; Bowen et al. 2018). Despite significant advances to understand the biofilm community structure, most of the knowledge has been generated from disrupted samples collected at various sites with few exceptions (Wood et al. 2002; Zijnge et al. 2010; Mark Welch et al. 2016). Imaging of undisturbed biofilm structure remains challenging as it requires sample collection from clinical sites and further processing that disrupts the original 3-dimensional (3D) architecture. To address this, we removed infected teeth from patients with severe ECC without perturbing the biofilm to keep its native structure intact. Primary maxillary anterior teeth with mesial or distal carious lesions were extracted with the use of a pediatric maxillary anterior forceps adapted to grasp mesiodistally. To assess the intact biofilm architecture, we examined the spatial organization of the microbial communities on the extracted primary teeth from children (ages between 36 and 72 mo) diagnosed with severe early childhood caries (S-ECC). Once collected, we examined the spatial structure of the naturally formed biofilm using confocal microscopy after fluorescent labeling of the microorganisms (Fig. 1A). For imaging purposes, we excluded the sample if over 30% of the enamel surface was cavitated or missing; the biofilm images were acquired from the noncavitated tooth surface. Since Streptococcus genus is one of major taxa in the mouth, particularly in supragingival plaque (Dewhirst et al. 2010; Eren et al. 2014; Johansson et al. 2016), and previous sequence-based analyses have revealed relatively high proportions of Streptococcus and S. mutans in S-ECC (Johansson et al. 2016), we employed taxa-based fluorescence in situ hybridization (FISH) probes (16S ribosomal RNA targeted), including eubacteria (EUB338-Cy3), Streptococcus (STR405-Cy5), and S. mutans (MUT590–Alexa Fluor 488), following the taxonomic ranks (kingdom-genus-species) (Fig. 1). This taxa-specific labeling allowed fluorescence subtraction through classification of a set of elements (kingdom [EUB]–genus [STR]–species [SMU]) (Fig. 2). As a targeted approach, we used a fluorescence subtraction method to analyze the spatial organization and composition of pathogens and commensal microbiota in the different phylogenetic scale (e.g., S. mutans [SMU], non-mutans streptococci [NSMU], nonstreptococcal bacteria [NSTR]), as well as multiple length scale from submicron to submillimeter across the intact biofilm (Ladau and Eloe-Fadrosh 2019) (Figs. 1 and 2).
Figure 1.
Intact biofilm imaging and spatial design concepts. (A) Schematic diagram of intact biofilm imaging consisted of the following steps: 1) mapping of tooth surface and taxa-specific labeling for confocal imaging, 2) low magnification or tile image acquisition for landscape of intact biofilms (submillimeter scale: 0.624 × 0.624 [x, y] mm2), 3) high-magnification imaging for biofilm architecture (micron scale; 2 × zoom, 156 × 156 µm2), and 4) assessment of interior design in spatially organized biofilm via further zoom-in imaging (4 × zoom, 78 × 78 µm2) and computational analysis (submicron scale). Diseased teeth (n = 10) were extracted from children (aged between 36 and 72 mo) diagnosed with severe early childhood caries (S-ECC). In the case of teeth extraction for healthy individuals (n = 2), only caries-free children were involved who had deciduous (primary) teeth that needed to be extracted clinically. Ethical approval of the study was obtained from the Institutional Review Board (IRB) at the University of Pennsylvania (IRB#824243). (B) Precise positioning of pathogens arranged with other microbes may coordinate the disease process in situ. This spatial design of the polymicrobial community could be a therapeutic target (precision targeting).
Figure 2.
Taxa-specific labeling and computational imaging. (A) Differentiation of bacterial group via taxa-specific labeling in planktonic cells. The cell mixture was prepared at an equal proportion of Streptococcus mutans, Streptococcus oralis, and Actinomyces naeslundii (1:1:1 ratio for each of the bacterial suspension at ~108 colony-forming units [CFU]/mL). For taxa-specific labeling, fluorescence in situ hybridization (FISH) probes (16S ribosomal RNA targeted) including all bacteria (EUB), Streptococcus (STR), and S. mutans (SMU) following the taxonomic ranks (kingdom-genus-species) were used (MUT590, 5′-ACTCCAGACTTTCCTGAC-3′ with Alexa Fluor 488 for S. mutans; STR405, 5′-TAGCCGTCCCTTTCTGGT-3′ with Cy5 for Streptococcus; EUB338, 5′-GCTGCCTCCCGTAGGAGT-3′ with Cy3 for all bacteria at a final concentration of 1 µM). Bacterial groups were differentiated by a specific coding based on the classification of fluorescent labeling: S. mutans was labeled by MUT590, STR405, and EUB338 (coded as SMU); S. oralis was labeled by STR405 and EUB338 (coded as NSMU for non-mutans streptococci); and A. naeslundii was labeled by EUB338 only (coded as NSTR for nonstreptococcal bacteria). If cells were labeled by SMU and EUB without STR labeling, it was considered nonspecific labeling. The images were acquired using an LSM 800 (Carl Zeiss) equipped with a 20× (1.0 numerical aperture [NA]) water immersion objective or with a 40× (1.2 NA) oil immersion objective. (B) A fluorescence subtraction method for assessing polymicrobial community organization in intact plaque biofilm. To analyze the positioning of S. mutans within the polymicrobial biofilm structure, fluorescence subtraction was applied using Image Calculator of ImageJ (National Institutes of Health): S. mutans (SMU); Streptococcus (STR405) – SMU = non-mutans streptococci (NSMU); all bacteria (EUB338) – STR405 = nonstreptococcal bacteria (NSTR). For computational image processing, acquired images with similar signal intensity from each taxon channel were used.
Given the multiscale nature of biofilm spatial organization, we applied the “spatial design” principle, whereby the landscape, architecture, and interior design converge for assessing the “living space.” In this context, the overall “landscape” of biofilm through the spatial organization can be assessed to identify unique “architectures” and dissect its “interior designs” to determine specific multicellular arrangements or bacterial positioning associated with disease (Fig. 1).
Spatial Design of Bacterial Communities on Human Tooth Surface
Based on the spatial design concept, the overall biofilm “landscape” revealed 3 common architectures on the teeth from both healthy individuals and patients with disease, such as corncob-, hedgehog-, and seaweed-like structures (Fig. 3A). Corncob- and hedgehog-like structures comprised Streptococcus associated with filamentous bacteria (Fig. 3A and Appendix Fig. 1), whereby filaments were located at the periphery of a spiny-like cluster in hedgehog-like structures, while corncob-like structures were characterized by cocci surrounding the central filament, consistent with previous findings (Mark Welch et al. 2016). The seaweed-like structure comprised a bacterial cluster organized with a stem-like arrangement. All these microbial organizations showed a polybacterial intermixing and relatively low abundance of S. mutans (Fig. 3A). However, we found a distinctive dome-like architecture on the diseased tooth surface. This structure displayed a multilayer cellular arrangement harboring a dense accumulation of S. mutans, contrasting from other architectures (Fig. 3A).
Figure 3.
Spatial design of bacterial community organization on tooth surface. (A) Landscape and architecture of intact biofilms on the teeth. Distinct biogeography of biofilms from healthy individuals and patients with disease. Biofilm architecture includes dome-, corncob-, hedgehog-, and seaweed-like structures. (B) Interior design of intermixed and multilayered dome-like structures. Cells were differentiated according to taxonomic ranks (kingdom-genus-species). EUB338, eubacteria (blue); STR405, Streptococcus (red); MUT590, Streptococcus mutans (green). (C) Spatial structure of pathogens and commensal microbiota in distinct architectures. NSMU, non-mutans streptococci, NSTR, nonstreptococcal bacteria; SMU, S. mutans.
We further investigated the interior design of the dome-shape community using taxa-specific labeling (kingdom-genus-species) and fluorescence subtraction method. The imaging analyses revealed a polymicrobial assembly composed of a bacterial cluster dominated by S. mutans (SMU) in the inner core that was spatially segregated from outer layers of other bacteria (Fig. 3B, C). In the context of S-ECC, the observation of such a densely packed S. mutans core in the 3D dome shape provides evidence of spatial positioning and arrangement of this pathogen within the polymicrobial community, which remained elusive despite its clinical association with the disease. Conversely, other structural organizations revealed relatively high abundance of non–S. mutans bacteria (Fig. 3B, C). Hence, the combination of imaging and computational methodologies employed here can unveil the spatial structuring of complex and highly heterogenous polymicrobial biofilm communities in 3D across multiple length scales and depth.
Cross-Kingdom Community Organization on Human Tooth Surface
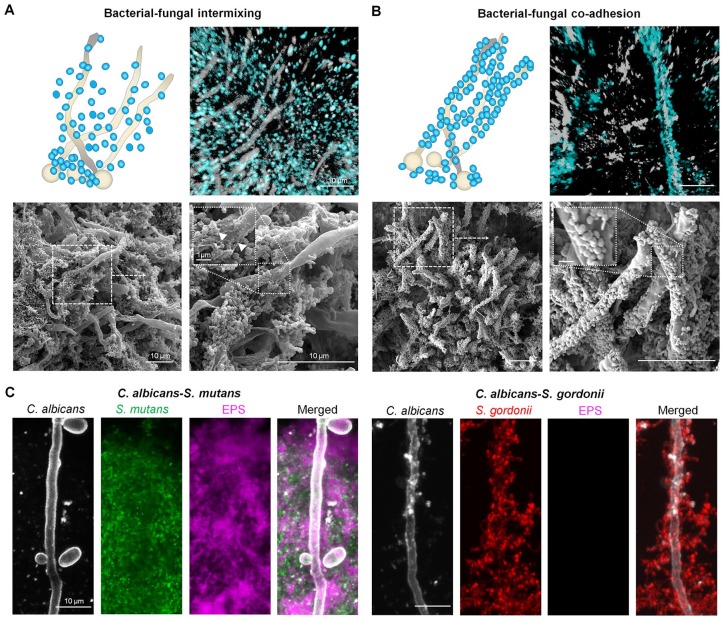
Although S-ECC has been associated with acidogenic/aciduric bacteria (Palmer et al. 2010; Tanner et al. 2011; Johansson et al. 2016), Candida albicans (an opportunistic fungal pathogen) has been frequently isolated in plaque biofilms from children with S-ECC and its presence correlated with the disease based on culturing and microbiome studies (Yang et al. 2012; Xiao et al. 2016; Jean et al. 2018; Xiao, Huang, et al. 2018). The confocal imaging also revealed the presence of C. albicans colocalized with oral streptococci (Fig. 4). For identification of C. albicans in the clinical sample, we tested the FISH probe for specificity of C. albicans, especially compared to other Candida ssp. (e.g., Candida dubliniensis, Candida tropicalis, and Candida glabrata) (Appendix Fig. 2), which can also be detected in the plaque and oral mucosa (Silva et al. 2012; Al-Ahmad et al. 2016; Xiao et al. 2016; Xiao, Grier, et al. 2018).
Figure 4.
Cross-kingdom communal organization of intact biofilms. Bacteria and Candida albicans association in vivo revealed bacterial-fungal intermixing and bacterial clustering (A) and cell-cell coadhesion (B). In confocal images, C. albicans is labeled in white and bacteria is depicted in blue. Fluorescence in situ hybridization (FISH) oligonucleotide probes used to label eubacteria and C. albicans: EUB338, 5′-GCTGCTCCCGTAGGATG-3′ with Cy3 for eubacteria; CAAL, 5′-GCCAAGGCTTATACTCGCT-3′ with FITC for C. albicans. Electron micrographs were acquired from the same sample as used in confocal imaging. (C) Assembly of cross-kingdom communal arrangements in vitro using coculture of Streptococcus mutans or Streptococcus gordonii with C. albicans in the presence of sucrose. For visualization of in vitro biofilm assembly, S. mutans and S. gordonii were stained with 2.5 µM Syto9 green-fluorescent nucleic acid stain while C. albicans cells were labeled with concanavalin A conjugated with tetramethylrhodamine. The extracellular polysaccharides (EPS) were labeled with 1 μM Alexa Fluor 647–dextran conjugate. For classification of S. mutans and non-mutans streptococci (NSMU) in cross-kingdom interactions, S. mutans (SMU) is depicted in green and S. gordonii (one of NSMU) is shown in red.
We found 2 different patterns of cross-kingdom spatial organization: 1) C. albicans intermixed with bacteria (with a high abundance of S. mutans; Fig. 4A and Appendix Fig. 3) but maintaining a certain spatial distance from each other and 2)C. albicans physically coadhering with non-mutans streptococci, forming a “cross-kingdom corncob” (Fig. 4B and Appendix Fig. 3). The observation of C. albicans intermixed with S. mutans is consistent with a recent clinical study showing that the presence of C. albicans alters the oral bacteriome, particularly by enhancing the levels of S. mutans in plaque biofilms in S-ECC (Xiao, Grier, et al. 2018). On the other hand, the bacterial-fungal “corncob” was previously observed in the supragingival biofilms from periodontitis patients and in cavitated surfaces via an embedding-decalcification-sectioning process (Zijnge et al. 2010; Dige and Nyvad 2019).
We were particularly intrigued with the presence of extracellular material (Fig. 4A, inset) on the fungal surface in the C. albicans and S. mutans mixed community. Previous studies have shown that S. mutans–derived glucosyltransferases (Gtfs) can bind avidly to the C. albicans cell wall and produce extracellular glucans in situ in the presence of sucrose using in vitro and rodent models (Gregoire et al. 2011; Falsetta et al. 2014; Hwang et al. 2017; Kim et al. 2018). Using a glucan-specific labeling technique based on Gtfs activity (Klein et al. 2009), we were able to assess localized production of glucans using the clinical samples. We detected functional Gtf activity bound to the C. albicans surface, which produced copious amounts of glucans surrounding the filamentous fungal cells (Appendix Fig. 4A, C). Interestingly, enzymatically active Gtfs were distributed across the depth, indicating that Gtfs were also present at the biofilm-tooth interface (Appendix Fig. 4B).
We recapitulated these cross-kingdom organizations using experimental mixed biofilms on the apatitic surface whereby C. albicans cells were cocultured with either S. mutans or Streptococcus gordonii (Fig. 4C). We found that S. mutans was intermixed with C. albicans in which extracellular polysaccharides (EPS) glucans interspersed and surrounded the bacterial-fungal cluster. In contrast, S. gordonii directly coadhered with C. albicans. C. albicans interacts with commensal (viridans) streptococci such as S. gordonii and Streptococcus oralis via well-characterized cell wall proteins and receptors on both organisms (Xu et al. 2014; Nobbs and Jenkinson 2015). Conversely, S. mutans forms a cross-kingdom association with C. albicans via a glucan-dependent mechanism whereby glucans produced in situ provide binding sites for S. mutans through glucan-binding proteins (Gregoire et al. 2011; Falsetta et al. 2014).
Concluding Remarks and Future Perspectives
Altogether, the spatial design of polymicrobial communities and mapping the pathogen positioning in their native diseased state revealed unique structural organization across multiple scales and taxonomic ranks. We found a 3D dome-like architecture composed of multiple species with a multilayer arrangement on the infected teeth associated with severe childhood tooth decay. Further analysis showed dense clustering of a cariogenic pathogen (S. mutans) forming an inner core surrounded by outer layers of other bacteria in a highly ordered manner. In addition, cross-kingdom communal organizations were also detected on the enamel surface, whereby C. albicans were physically associated with S. mutans or non-mutans streptococci. Notably, we found a functional bacterial exoenzyme (Gtf) on the surface of the Candida fungal cells within intact clinical plaque samples, which was capable of producing EPS in situ. The reasons for these polymicrobial positionings and spatial organization and how these structured communities can cause caries remain unknown, requiring further mechanistic studies.
At the same time, we emphasize the limitations of this study, including the small sample size, the limited number of species-specific labeling, and further validation of the C. albicans as well as S. mutans probes against other genotypes and closely related species (such as Streptococcus sobrinus). We are currently expanding the sample size and conducting additional enamel surface and quantitative imaging analyses to determine the association of the spatial organization and its virulence potential (i.e., acid dissolution of the tooth enamel). Furthermore, a more comprehensive multispecies (including uncultivable microbes, other mutans streptococci species and genotypes) and EPS (other polymeric matrices such as extracellular DNA, proteins) labeling can be achieved using a combination of fluorescence imaging methods, which could reveal additional spatial designs not observed here.
Future studies shall elucidate the impact of the multiscale spatial structuring and positioning of the pathogen and commensal microbiota in modulating the disease longitudinally across different dental sites and individuals. It would be interesting to elucidate how changes in the global microbiome composition and activity affect the spatial structuring of the biofilm community. Furthermore, technological advances using real-time microscopy-spectroscopy methods combined with an integrative multiomics approach (Lloyd-Price et al. 2019; Zhou et al. 2019) provide exciting opportunities to precisely assess the function of the structured community and how it coordinates the disease process spatiotemporally. In summary, our findings provide new insights about the spatial and structural organization of the polymicrobial biofilm communities on intact human teeth, which may be important for understanding the microbiome function and virulence at the infection site. Investigations in this area of research may reveal more precise ways to identify community structure associated with severe childhood caries and help develop enhanced approaches to prevent this highly prevalent and costly pediatric disease.
Author Contributions
D. Kim, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; H. Koo, contributed to conception, design, and data interpretation, drafted and critically revised the manuscript. Both authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520909313 for Spatial Design of Polymicrobial Oral Biofilm in Its Native Disease State by D. Kim and H. Koo in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
This work was supported in part by the National Institute for Dental and Craniofacial Research (NIDCR) grant DE025220 (H.K.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Al-Ahmad A, Auschill TM, Dakhel R, Wittmer A, Pelz K, Heumann C, Hellwig E, Arweiler NB. 2016. Prevalence of Candida albicans and Candida dubliniensis in caries-free and caries-active children in relation to the oral microbiota—a clinical study. Clin Oral Investig. 20(8):1963–1971. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Burne RA, Wu H, Koo H. 2018. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26(3):229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol. 192(19):5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dige I, Nyvad B. 2019. Candida species in intact in vivo biofilm from carious lesions. Arch Oral Biol. 101:142–146. [DOI] [PubMed] [Google Scholar]
- Dye BA, Thornton-Evans G, Li X, Iafolla TJ. 2015. Dental caries and sealant prevalence in children and adolescents in the United States, 2011–2012. NCHS Data Brief. 2015(191):1–8. [PubMed] [Google Scholar]
- Eren AM, Borisy GG, Huse SM, Mark Welch JL. 2014. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA. 111(28):E2875–E2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai C-H, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, et al. 2014. Symbiotic relationship between Streptococcus mutans and candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 82(5):1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI, Ambatipudi KS, Rosalen PL, Bauserman R, Waugh RE, et al. 2011. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl Environ Microbiol. 77(18):6357–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis E, Parsaei Y, Klein MI, Koo H. 2017. Advances in the microbial etiology and pathogenesis of early childhood caries. Mol Oral Microbiol. 32(1):24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G, Liu Y, Kim D, Li Y, Krysan DJ, Koo H. 2017. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 13(6):1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean J, Goldberg S, Khare R, Bailey LC, Forrest CB, Hajishengallis E, Koo H. 2018. Retrospective analysis of Candida-related conditions in infancy and early childhood caries. Pediatr Dent. 40(2):131–135. [PMC free article] [PubMed] [Google Scholar]
- Johansson I, Witkowska E, Kaveh B, Lif Holgerson P, Tanner ACR. 2016. The microbiome in populations with a low and high prevalence of caries. J Dent Res. 95(1):80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Liu Y, Benhamou RI, Sanchez H, Simón-Soro Á, Li Y, Hwang G, Fridman M, Andes DR, Koo H. 2018. Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J. 12(6):1427–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MI, Duarte S, Xiao J, Mitra S, Foster TH, Koo H. 2009. Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilm development. Appl Environ Microbiol. 75(3):837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. 2017. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol. 15(12):740–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladau J, Eloe-Fadrosh EA. 2019. Spatial, temporal, and phylogenetic scales of microbial ecology. Trends Microbiol. 27(8):662–669. [DOI] [PubMed] [Google Scholar]
- Listl S, Galloway J, Mossey PA, Marcenes W. 2015. Global economic impact of dental diseases. J Dent Res. 94(10):1355–1361. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. 2019. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 569(7758):655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Dewhirst FE, Borisy GG. 2019. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu Rev Microbiol. 73(1):335–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. 2016. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 113(6):E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Drescher K, Foster KR. 2016. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 14(9):589–600. [DOI] [PubMed] [Google Scholar]
- Nobbs AH, Jenkinson HF. 2015. Interkingdom networking within the oral microbiome. Microbes Infect. 17(7):484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CA, Kent R, Jr, Loo CY, Hughes CV, Stutius E, Pradhan N, Dahlan M, Kanasi E, Arevalo Vasquez SS, Tanner AC. 2010. Diet and caries-associated bacteria in severe early childhood caries. J Dent Res. 89(11):1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres MA, Macpherson LMD, Weyant RJ, Daly B, Venturelli R, Mathur MR, Listl S, Celeste RK, Guarnizo-Herreño CC, Kearns C, et al. 2019. Oral diseases: a global public health challenge. Lancet. 394(10194):249–260. Erratum in: Lancet. 394(10203):1010. [DOI] [PubMed] [Google Scholar]
- Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. 2012. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 36(2):288–305. [DOI] [PubMed] [Google Scholar]
- Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. 2016. The biogeography of polymicrobial infection. Nat Rev Microbiol. 14(2):93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner ACR, Mathney JMJ, Kent RL, Chalmers NI, Hughes C V, Loo CY, Pradhan N, Kanasi E, Hwang J, Dahlan MA, et al. 2011. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol. 49(4):1464–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM. 2019. The structure of dental plaque microbial communities in the transition from health to dental caries and periodontal disease. J Mol Biol. 431(16):2957–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos T, Abajobir AA, Abate KH, Abbafati C, Abbas KM, Abd-Allah F, Abdulkader RS, Abdulle AM, Abebo TA, Abera SF, et al. 2017. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 390(10100):1211–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood SR, Kirkham J, Shore RC, Brookes SJ, Robinson C. 2002. Changes in the structure and density of oral plaque biofilms with increasing plaque age. FEMS Microbiol Ecol. 39(3):239–244. [DOI] [PubMed] [Google Scholar]
- Xiao J, Grier A, Faustoferri RC, Alzoubi S, Gill AL, Feng C, Liu Y, Quivey RG, Kopycka-Kedzierawski DT, Koo H, et al. 2018. Association between oral Candida and bacteriome in children with severe ECC. J Dent Res. 97(13):1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Huang X, Alkhers N, Alzamil H, Alzoubi S, Wu TT, Castillo DA, Campbell F, Davis J, Herzog K, et al. 2018. Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res. 52(1–2):102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Moon Y, Li L, Rustchenko E, Wakabayashi H, Zhao X, Feng C, Gill SR, McLaren S, Malmstrom H, et al. 2016. Candida albicans carriage in children with severe early childhood caries (S-ECC) and maternal relatedness. PLoS One. 11(10):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jenkinson HF, Dongari-Bagtzoglou A. 2014. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 29(3):99–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XQ, Zhang Q, Lu LY, Yang R, Liu Y, Zou J. 2012. Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Arch Oral Biol. 57(8):1048–1053. [DOI] [PubMed] [Google Scholar]
- Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SR, Zhang MJ, Rao V, Avina M, Mishra T, et al. 2019. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature. 569(7758):663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, Gmür R., Harmsen HJ. 2010. Oral biofilm architecture on natural teeth. PLoS One. 5(2):e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520909313 for Spatial Design of Polymicrobial Oral Biofilm in Its Native Disease State by D. Kim and H. Koo in Journal of Dental Research