Abstract
The amino acid derivative 4-borono-L-phenylalanine (BPA) has been used in the radiation medicine technique boron neutron capture therapy (BNCT). Here we have characterized its interaction with DNA when incorporated into a positively charged hexa-L-arginine peptide. This ligand binds strongly to DNA and induces its condensation, an effect which is attenuated at higher ionic strengths. The use of an additional tetra-L-arginine ligand enables the preparation of a DNA condensate in the presence of a negligible concentration of unbound boron. Under these conditions, Monte Carlo simulation indicates that >85% of energy deposition events resulting from thermal neutron irradiation derive from boron fission. The combination of experimental model systems and simulations that we describe here provides a valuable tool for accurate track structure modeling of the DNA damage produced by the high LET particles involved in BNCT.
Keywords: DNA damage, DNA condensation, Neutron irradiation, Boron, Monte Carlo simulation
1. Introduction
Ionizing radiation damages DNA in biological targets (Sage and Harrison, 2011). The chemical modifications to DNA result from two different pathways, the direct and indirect effects. With direct-type effects, the DNA and approximately 10–12 tightly bound waters of solvation are ionized. In the indirect effect, ionization of the aqueous solvent produces highly reactive intermediates some of which react with the DNA. The hydroxyl radical is the species mainly responsible for the indirect effect (von Sonntag, 2006). Irradiating a target such as a mammalian cell nucleus, scavenging data show that the direct effect and indirect effects both make a significant contribution to the biological effects of irradiation (Chapman et al., 1973).
Experimental investigation of the chemistry of the indirect effect in dilute solution is straightforward, because the contribution of the direct effect is negligible. Modeling the direct effect in aqueous solution is challenging because it requires attenuating the dominant indirect effect. Conditions required to achieve this employ cryogenic temperatures, dehydration, or very large concentrations of scavengers (Swarts et al., 1996; Sharma et al., 2008; Adhikary et al., 2009; Yokoya et al., 2009). The intention is to limit the formation or diffusion of the reactive intermediates responsible for the indirect DNA damage.
By the usual standards of biochemistry, these experimental conditions are unusually harsh. They also interfere with important further reactions of damaged DNA, for example with oxygen and water (von Sonntag, 2006). These limitations can be addressed by using condensed DNA in room temperature aqueous solution as the irradiation target (Ly et al., 2006). Condensed DNA assumes a highly aggregated conformation that is not accessible from the bulk solution. This DNA condensation can be replicated using peptides to model the close association of DNA with amino acid residues in cellular chromatin (van Holde and Slatanova, 2007; Perisic and Schlick, 2017). Peptides as the aggregating agents, can achieve the desired attenuation of the indirect effect. We wished to explore the suitability of this model system for the DNA damage produced under conditions used in boron neutron capture therapy (BNCT). Monte Carlo track structure calculations for BNCT seek to correlate their results with biological effects (Baiocco et al., 2016), but there is little chemical data available to calibrate these biophysical models.
Therapeutic applications of isotopes with large cross sections for neutron capture (Locher, 1936) were proposed only a few years after the discovery of the neutron itself (Chadwick, 1932). Attention concentrated on boron-10 (Sweet, 1951; Javid et al., 1952), because of its combination of reasonable natural abundance (ca. 20%), unusually efficient neutron capture (Sears, 1992), and prompt alpha particle emission. In the 10B(n,α)7Li reaction, neutron capture by boron-10 forms helium-4 (α-particle) and lithium-7 (Figure 1). Disintegration products are 1.5 MeV helium-4 and 0.84 MeV lithium-7 ions plus a 0.48 MeV γ-ray 94% of the time; or 1.8 MeV helium-4 and 1.0 MeV lithium-7 ions 6% of the time (Yue et al., 2018). The ions carry most of the 2.8 MeV Q-value, and comprise a pair of high linear energy transfer (LET) particles with ranges in tissue of below 10 μm (Kempe et al., 2007); which is the length scale of a mammalian cell. The dose deposited in a cell by a single α-particle track is ca. 0.5 Gy (Lorimore et al., 1998). Thus, several of these light ion tracks are required to inflict enough damage to be lethal (Raju et al., 1991; Fournier et al., 2012).
Figure 1.
Energy distribution of products from the 10B(n,α)7Li reaction.
Neutron capture by boron offers the possibility of major benefits compared to conventional radiation therapy with low-LET X-rays because it involves the formation of short-range high- LET particles (Wagner et al., 2012). The short range confers an advantage by mitigating lethal radiation effects to nearby healthy tissues (Prasanna et al., 2012). The high LET nature of the particles overcomes the radioresistance of hypoxic tumor cells (Tinganelli et al., 2013). In practice, there are substantial pharmacological challenges in targeting boron delivery agents to the desired biological location in sufficiently high concentrations (Hawthorne and Lee, 2003) as well as difficulties with the dosimetry of short-range particles (Ono et al., 2019). Nevertheless, there is continuing interest in BNCT (Moss, 2014; Barth et al., 2018a).
The most commonly used agent in BNCT is 4-dihydroxyboryl-L-phenylalanine, also called 4- borono-L-phenylalanine and abbreviated as BPA (Barth et al., 2018b). In this work, we linked BPA to a cationic peptide, which binds strongly to DNA. By adding a second boron-free peptide, which binds to DNA less strongly (because it has fewer positive charges (Mascotti and Lohman, 1997)), it is possible to produce a DNA condensate with very low levels of unbound boron. Thereby, we created a room-temperature aqueous model system for characterizing the interaction of DNA with the high LET particle tracks of BNCT. Importantly, we defined the conditions required for this condensate to be formed for irradiation, as well as the conditions for re-dissolution necessary for subsequent assays of DNA damage.
2. Materials and Methods
2.1. Biochemicals
The ligands N-acetyl-4-dihydroxyboryl-L-phenylalaninyl-hexa-L-arginine amide (Ac-BPA-Arg6- NH2) and N-acetyl-tetra-L-arginine amide (Ac-Arg4-NH2) were obtained commercially (Biosynthesis, Lewisville, TX) as trifluoroacetate salts. Structural formulas are shown in Figure 2. Sheared salmon sperm DNA (< 2 kb) was from Invitrogen (Waltham, MA). The fluorescent dyes thiazole orange and BOBO-3 were obtained from Sigma-Aldrich (St. Louis, MO) and Thermo-Fisher (Waltham, MA) respectively.
Figure 2.
Chemical structures of the oligo-arginine ligands Ac-Arg4-NH2 (left) and Ac-BPA- Arg6-NH2 (right). See Experimental section.
2.2. Solutions
Solutions contained sodium phosphate (10−2 mol L−1, pH 7.0), sodium perchlorate (0, or 1×10−2 to 1 mol L−1), DNA (10 μ g mL−1, equivalent to 3×10−5 mol L−1 nucleotide residues or 1.5×10−5 mol L−1 base pairs), the ligands Ac-BPA-Arg6-NH2 (0, or 1×10−7 to 2×10−4 mol L−1) and Ac-Arg4-NH2 (0, or 1×10−6 to 5×10−5 mol L−1), and thiazole orange (0 or 5×10−7 mol L−1) and BOBO-3 (0 or 2×10−7 mol L−1).
2.3. Pelleting
In some cases, the UV-visible light absorption, static light scattering (SLS) intensity, and fluorescence intensity of the supernatant were examined after centrifugation at 1.5×104 × g for 10 min.
2.4. Static light scattering
The intensity of SLS (Wyatt, 1993) was quantified (500 μ L sample volume, excitation path length 10 mm, emission path length 2 mm) at 90° with a PTI fluorescence spectrometer (Horiba, Piscataway, NJ). Both excitation and emission wavelengths were 400 nm. The biochemical reagents (ligands and DNA) show no significant absorption at this wavelength.
2.5. Dynamic light scattering
DLS measurements were made using a model Z3000 (Nicomp, Port Richey, FL) size analyzer. The cylindrical sample (500 μ L) was laser-illuminated (658 nm) and the scattered light collected at 90°. Brownian motion produces a time-dependent variation in the scattered coherent light intensity (Bloomfield, 2000). This was fitted with a proprietary auto-correlation function where the diffusion coefficient distribution was calculated using an inverse Laplace transform. The hydrodynamic diameter is calculated from the diffusion coefficient, which assumes water values for the refractive index and viscosity.
2.6. Absorbance spectroscopy
Absorption in the UV-visible region of DNA, thiazole orange, and BOBO-3 was quantified (500 μ L, 10 mm path length) with a Cary model 300 spectrometer (Varian, Palo Alto, CA).
2.7. Atomic force microscopy (AFM)
Samples were prepared by depositing an aliquot (1 μL) of the solution onto a freshly cleaved mica surface (1 cm diameter), rinsing with water (100 μ L), and drying in air. Topographic (intensity corresponds to height) data were collected over a 5×5 μm area with a Multi-mode 8 instrument (Bruker, Santa Barbara, CA) using the peak force tapping mode and a Scanasyst- Fluid nitride probe (20 nm tip radius, 0.7 N m−1 force constant, 150 kHz resonant frequency, Bruker, Santa Barbara, CA). The vertical piezo elements were modulated at ca. 2 kHz for each pixel of the image. The peak interaction force was used as the feedback signal to estimate the topography. The scan speed was 2 seconds for each row of 1024 pixels. Minor image processing was used to attenuate horizontal scanning artifacts and background curvature, and to adjust contrast. The images provide a poor estimate of dimensions in the horizontal plane, which are less than the nominal tip radius of 20 nm. However, measurements of vertical distances of the diameter of double stranded DNA (ca. 2 nm) are reliable.
2.8. Fluorescence spectroscopy
Fluorescence emission intensities at a nominal 90° angle were measured (500 μ L, 10 mm excitation path length, emission path length 2 mm) with a PTI instrument (Horiba, Piscataway, NJ). The excitation and emission wavelengths were 480 nm and 526 nm (thiazole orange), or 570 nm and 605 nm (BOBO-3).
2.9. Modeling of energy deposition by neutrons
We used the TOPAS Monte Carlo tool (Perl et al., 2012) version 3.2 layered on top of the Geant4 toolkit (Agostinelli et al., 2003) version 10.5.p01. The simulation volume consisted of a square prism of 1 cm × 1 cm × 1 μm, which was sufficient to allow one or two neutron interactions. The prism was embedded in a vacuum and irradiated with a 0.025 eV (thermal) neutron point source having zero divergence (the initial momentum of the neutrons was perpendicular to the target prism). The physics list used for the TOPAS simulation consisted of the so-called “high precision models of Geant4” for the interaction of thermal neutrons. These models use the cross-section data from ENDF/B-VI (ENDF, 1991), previously benchmarked with Geant4 elsewhere (Tran et al., 2018; van der Ende et al., 2016). For each secondary particle produced by neutron interactions, the relative contribution to the total energy deposit in the square prism was obtained by averaging ten simulation runs, 108 primary neutrons per run, using different random seeds.
The elemental composition of the material of the square prism was derived in the following manner. The experimental DNA target has 8 base pairs (charge Z = −8 + −8 = −16) bound by 2× Ac-BPA-Arg6-NH2 and 1× Ac-Arg4-NH2 ligands (Z = 6 + 6 + 4 = +16) (see Discussion). From the strand break yield in plasmid DNA condensed with a hexa-arginine ligand (Perry et al., 2011), the hydroxyl radical diffusion distance is ca. 5 nm. This defines a cylinder with a radius of 5 nm and a height of 8 base pairs (8 × 0.34 nm), having a volume of 210 nm3. This volume contains 10−21 / 18 mol nm−3 water, equivalent to 6×1023 × 10−21/18 = 33 molecules nm−3, for a total of 33 × 210 = 7×103 water molecules, or 2 × 7×103 = 1.4×104 1H atoms (ignoring the minor DNA and peptide contributions). The boron concentration in this target volume is two boron atoms per 7×103 water molecules, which is equivalent to 2×11 / 7×103 × 18 = 170 ppm (35 ppm of the isotope boron-10) by mass. Therefore, the simulation used 35 ppm boron-10. The 8 base pairs are assumed to consist of 4 GC (8× 14N atoms) and 4 AT (7× 14N atoms) pairs, which contain a total of 32 + 28 = 60 14N atoms. The peptides add another 69 14N atoms for a total of 129. Therefore, the material of the square prism had the following mass weight fractions: 0.8755 of oxygen, 0.1103 of hydrogen, and 0.0142 of nitrogen with aggregated 35 ppm of boron-10. The density of the material was assumed equal to that of water (1 g cm−3).
3. Results
3.1. Ligand structure
Two different cationic ligands were used to minimize the fraction of boron that was not bound to the DNA. The boron-containing ligand Ac-BPA-Arg6-NH2 (charge Z = +6) was more highly positively charged than the boron-free ligand Ac-Arg4-NH2 (charge Z = +4). The chemical structures of these ligands are shown in Figure 2. More highly positively charged oligo-arginine ligands have significantly larger DNA binding constants than those with fewer positive charges (Mascotti and Lohman, 1997). To model the highly scavenged environment of a mammalian cell nucleus, we used condensed DNA. Condensation of the polyanionic DNA by positively charged ligands involves its extensive neutralization (Yoo and Aksimentiev, 2016). The experimental conditions required to achieve this necessarily involve an excess of the ligand. The resulting presence of a small fraction of unbound boron would complicate track structure modeling of DNA damage after neutron irradiation. To mitigate this undesirable effect, we made use of the combination of a limiting concentration of a boron-containing ligand having a larger binding constant and an excess of a boron-free ligand with a smaller binding constant. We have described a related approach in the case of a fluorescent hydroxyl radical probe (Perry et al., 2011).
3.2. Ligand acidity
Because of the issue of ligand charge (see Introduction), we wished to characterize the acidity of the boron-containing ligand (Ac-BPA-Arg6-NH2), although it was not expected to deprotonate at pH values in the physiological range (Hall, 2011; Achilli et al., 2013). By examining the decrease in absorption at Δ = 243 nm with increasing pH (Soundararajan et al., 1989; Marinaro et al., 2012), the acid dissociation constant of the boronic acid in Ac-BPA-Arg6-NH2 was quantified as pKa = 10.1 (Figure 3). This is consistent with the value of pKa = 9.79 reported for borono-L- phenylalanine (BPA) itself (Watanabe et al., 2016).
Figure 3.
Acidity dependence of the protonation state of the boronic acid group in Ac-BPA- Arg6-NH2. The data are fitted with the Henderson-Hasselbalch equation for a pKa value of 10.1.
3.3. Sedimentation, light scattering, and AFM
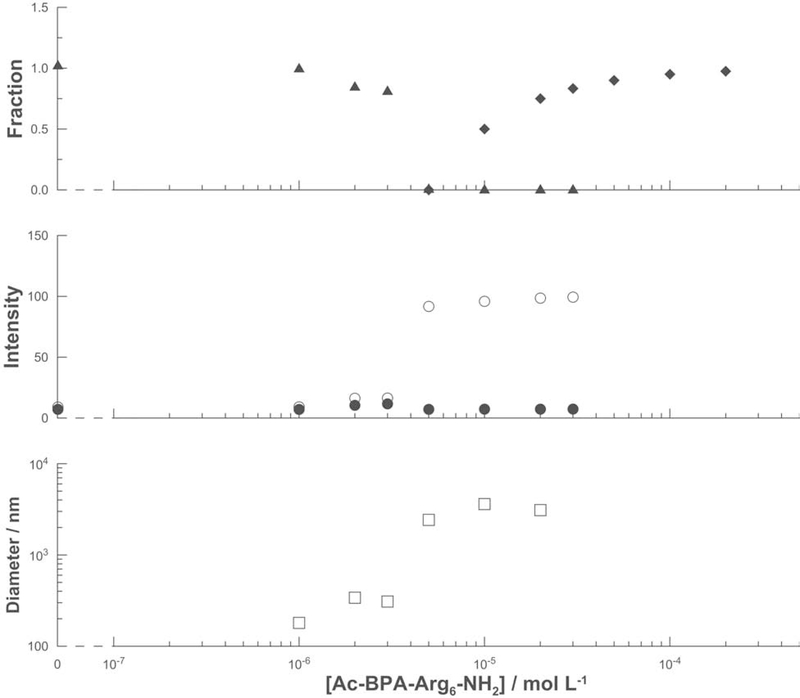
The effect of the ligand Ac-BPA-Arg6-NH2 on DNA (10 μ g mL−1) was examined using sedimentation, light scattering (both SLS and DLS), and atomic force microscopic (AFM) imaging (Figures 4, 5A, and 5B). There is a large increase in the sedimentation rate (Figure 4, upper panel) of the DNA at ligand concentrations above a sharply defined value of ca. 5×10−6 mol L−1, since after centrifugation the DNA is not detectable in the UV absorption spectrum of the supernatant solution. For ligand concentrations above 5×10−5 mol L−1, increasing amounts of the ligand remain in the supernatant. There is a sharp ca. 10-fold increase in the SLS intensity at the same threshold ligand concentration of ca. 5×10−6 mol L−1, but ligand concentrations greater than this value do not produce any further increase in scattering (Figure 4, middle panel). An abrupt increase in the DLS measurement of hydrodynamic radius from several hundred nanometers to several microns was also observed, again at the same threshold ligand concentration of 5×10−6 mol L−1 (Figure 4, lower panel). The AFM image of DNA in the presence of 1×10−6 mol L−1 Ac-BPA-Arg6-NH2 (Figure 5A) reveals filamentous objects with lengths in the micron range and heights of a few nanometers. These dimensions are typical of individual DNA strands (van Holde and Zlatanova, 2007). In the presence of 1×10−5 mol L−1 Ac- BPA-Arg6-NH2 (Figure 5B), significantly more compact structures are visible, which are consistent with the increase in light scattering. Note however that these images are of dried samples and unrepresentative of particle sizes in aqueous solution.
Figure 4.
Effect of the concentration of the ligand Ac-BPA-Arg6-NH2. Upper panel: fraction of DNA (triangle) and of the ligand (rhombus) remaining in solution after centrifugation. Middle panel: SLS intensity (arbitrary units) before (open circle) and after (closed circle) centrifugation. Lower panel: hydrodynamic radius from DLS (square).
Figure 5A.
Atomic force microscopy (AFM) image of DNA in the presence of 1×10−6 mol L−1 Ac-BPA-Arg6-NH2. The horizontal dimensions are 3×3 μ m. The vertical scale is 5 nm.
Figure 5B.
Atomic force microscopy (AFM) images of DNA in the presence of 1×10−5 mol L−1 Ac-BPA-Arg6-NH2. The horizontal dimensions are 3×3 μ m. The vertical scale is 5 nm.
3.4. Attenuation of thiazole orange and BOBO-3 fluorescence
The effect of the hexa-arginine ligand on DNA determined by using the intercalating fluorescent dyes thiazole orange and BOBO-3 (Figure 6). The dye (5×10−7 mol L−1 thiazole orange or 2×10−7 mol L−1 BOBO-3) was pre-bound to the DNA before addition of the ligand. At the DNA base pair concentration of 1.5×10−5 mol L−1 (see Experimental) the dye to base pair ratio was 1/30 for thiazole orange and 1/75 for BOBO-3. We quantified the fluorescence intensity of the dye before centrifugation, and its concentration was determined after centrifugation from its visible absorption spectrum. For both dyes, the fluorescence intensity decreases slightly (by less than 2- fold) at ligand concentrations less than 5×10−6 mol L−1, but extensively (to a residual ca. 10–20%) at ligand concentrations greater than 5×10−6 mol L−1. At ligand concentrations greater than 5×10−6 mol L−1, ca. 50% of the thiazole orange but only ca. 10% of the BOBO-3 is present in the supernatant after centrifugation. The rest of the dyes remains bound to the DNA and is pelleted. The fluorescence of the unbound dyes is negligible (Nygren et al., 1998; Ruedas-Rama et al., 2010). So the effect of ligand concentrations above 5×10−6 mol L−1 is to attenuate by ca. 5-fold the emission intensity from the dyes which remain bound to the DNA. Similar fluorescence quenching was reported for DNA binding dyes Hoechst 33258 (Saito et al., 2004) and YOYO-1 (Johnson et al., 2011).
Figure 6.
Effect of the concentration of the ligand Ac-BPA-Arg6-NH2. Upper panel: intensity (arbitrary units) of thiazole orange (open circle) or BOBO-3 (open square) fluorescence before centrifugation. Lower panel: fraction of thiazole orange (closed circle) or BOBO-3 (closed square) remaining in solution after centrifugation.
3.5. Ionic strength
We examined the influence of sodium ions on the interaction of DNA with 2×10−5 mol L−1 Ac- BPA-Arg6-NH2 (Figure 7), a ligand concentration 4-fold greater than the threshold value of 5×10−6 mol L−1 observed in Figure 4. In the absence of any added sodium perchlorate (the only contribution to the sodium ion concentration was from the buffer components), the SLS intensity was large, and DNA was undetectable in the supernatant after pelleting. Very similar effects were observed up to a sodium ion concentration of ca. 7×10−2 mol L−1. At sodium ion concentrations greater than ca. 0.2 mol L−1, no SLS signal was observed and the vast majority of the DNA remained in solution after centrifugation.
Figure 7.
Effect of sodium perchlorate at 2×10−5 mol L−1 Ac-BPA-Arg6-NH2. Upper panel: Fraction of DNA (triangle) remaining in solution after centrifugation. Lower panel: SLS intensity (arbitrary units) before (open circle) and after (closed circle) centrifugation.
3.6. Combination of both ligands
At concentrations of the hexa-arginine ligand Ac-BPA-Arg6-NH2 slightly less than the threshold value of 5×10−6 mol L−1 (see Figures 4 and 6), a ca. 7-fold increase in SLS intensity was detectable in the additional presence of the tetra-arginine ligand Ac-Arg4-NH2 (Figure 8). The combination of 1×10−6 mol L−1 Ac-BPA-Arg6-NH2 and 1.5×10−5 mol L−1 Ac-R4-NH2 produced an increased SLS intensity similar to that with 2×10−6 mol L−1 Ac-BPA-Arg6-NH2 and 2×10−6 mol L− 1 Ac-R4-NH2.
Figure 8.
Effect of Ac-Arg4-NH2 on the SLS intensity in the presence of 1×10−6 mol L−1 (square) or 2×10−6 mol L−1 (circle) Ac-BPA-Arg6-NH2.
3.7. Energy deposition by neutrons
The main interactions of thermal neutrons that compete with capture by boron-10 (cross section of 3840 barn × 19.8% isotopic abundance = 760 barn) are the 1H(n,γ)2H, 14N(n,p)14C, and 14N(n,γ)15N reactions (Young and Foster, 1972; Goorley et al., 2002; Hayashi et al., 2015), with cross sections of 0.33, 1.9, and 0.075 barn respectively (Young and Foster, 1972; Sears, 1992). This is based on the elemental composition of the target (see Materials and Methods).
The TOPAS results (Table 1) of the contributions of the side reactions show that energy deposition is mainly from the 10B(n,α)7Li reaction (87.3% ± 2.8%). Protons and recoil 14C ions from the 14N(n,p)14C reaction each contribute 6.1% ± 1.1% and 3.5% ± 0.5%, respectively. Recoil deuterons from the 1H(n,γ)2H reaction contribute 2.5% ± 0.2%. Energy depositions by products of other reactions are negligible.
Table 1.
Relative contributions to the total energy deposited by secondary particles produced by simulated neutron irradiation of a boron-containing target (see Experimental). Errors shown are one standard deviation of the mean.
| Particle | Source | Contribution |
|---|---|---|
| 7Li | 10B(n,α)7Li | 46.8% ± 1.9% |
| 4He | 10B(n,α)7Li | 40.4% ± 2.1% |
| 1H | 14N(n,p)14C | 6.6% ± 1.1% |
| 14C | 14N(n,p)14C | 3.5% ± 0.5% |
| 2H | 1H(n,γ)2H | 2.5% ± 0.2% |
| 15N | 14N(n,γ)15N | < 0.1% |
| Gamma | 1H(n,γ)2H | < 0.1% |
4. Discussion
The pKa of the boronic acid group in Ac-BPA-Arg6-NH2 was found by spectrophotometric titration to be 10.1 (Figure 3). Therefore its anionic form (a tetrahedral hydroxide adduct (Cammidge et al., 2006; Hall, 2011)) makes a negligible (0.1%) contribution under the conditions (pH 7.0) we used to examine DNA binding. Thus we assume that this ligand carries a charge of Z = +6.
The ligand Ac-BPA-Arg6-NH2 displaces thiazole orange from DNA (Figure 6), suggesting that it binds to DNA, in common with other oligo-arginines (Mascotti and Lohman, 1997; Perry et al., 2011). It is however unable to displace BOBO-3. The DNA binding constants for these dyes are (in the presence of 0.1 mol L−1 sodium chloride) 3×105 L mol−1 and 8×105 L mol−1 respectively (Nygren et al., 1998; Ruedas-Rama et al., 2010). Under similar ionic conditions (0.1 mol L−1 potassium acetate) a value of 7×104 L mol−1 has been reported for the binding of a hexa-cationic oligo-arginine peptide to polyU (Mascotti and Lohman, 1997). It is difficult to estimate the values applicable at the lower ionic strength we used, because the binding constants of more highly charged compounds exhibit a much larger ionic strength dependence.
The increase in SLS intensity is a typical consequence of mixing polyanions (including DNA) with different types of cations such as linear polyamines and basic proteins (Keller et al., 2002; Saccardo et al., 2009) to create a highly compact conformation. In the case of DNA, this process is described as condensation (Bloomfield, 1996; DeRouchey et al., 2013; Li et al., 2017; Kang et al., 2018). The observation of a sharp increase to a constant value indicates an essentially complete condensation of the DNA by the hexa-arginine ligand. This is supported by the quenching of the fluorescence of the DNA binding dyes (Saito et al., 2004; Johnson et al., 2011), and by AFM imaging. The pelleting and DLS assays both reveal a large increase in particle size caused by aggregation of the DNA. This effect is distinguishable from DNA condensation, since some condensing agents (e.g. poly-L-lysine) do not produce aggregation (Nayvelt et al., 2007).
At ligand concentrations greater than a sharply defined threshold value of ca. 5×10−6 mol L−1, the DNA forms micron sized aggregates, which can be pelleted by centrifugation. The remaining solution is depleted in both the DNA and ligand. This behavior is characteristic of the associative phase separation of oppositely charged polyelectrolytes (Spruijt et al., 2010; Jha et al., 2014). Assuming a mean molecular weight of 325 g mol−1 per nucleotide residue, a DNA concentration of 10 μg mL−1 is equivalent to 10×10−6 × 1000 / 325 = 3×10−5 mol L−1 nucleotide residues. The threshold ligand concentration of 5×10−6 mol L−1 corresponds to 6 × 5×10−6 = 3×10−5 mol L−1 arginine residues. DNA condensation by cations typically involves the neutralization of ca. 90% of its negative charges (Bloomfield 1996), so this close agreement indicates a near quantitative binding between the peptide ligand and DNA. It is also consistent with the levels of the ligand remaining in solution after pelleting the aggregated DNA from solutions containing ligand concentrations in excess of the 5×10−6 mol L−1 threshold value.
The combination of a limiting concentration of the strongly-binding boron-containing ligand Ac- BPA-Arg6-NH2 and a small excess of the weaker binding Ac-Arg4-NH2 is able to condense and aggregate the DNA (Figure 8). Under very similar conditions we have reported that only ca. 1% of the hexa-arginine ligand remains unbound (Perry et al., 2011), by using a fluorescent coumarin probe instead of BPA. Therefore, we are able to produce a boron-containing condensed DNA target with the 35 ppm boron concentration that is achievable in BNCT (Barth et al., 2018a). The condensate can be disassociated after irradiation by increasing the ionic strength. This frees the DNA for subsequent assays of the radiation damage it has sustained.
The TOPAS simulation of energy deposition in our DNA-peptide model indicates that under the experimental conditions needed to form the condensate, >85% of energy depositions in the DNA target volume are derived from boron-10 neutron capture (Table 1). The second-largest contributor (ca. 10%) is from neutron capture on 14N, although it is less important (<1.4% at 10 ppm boron-10) in vivo (Ono et al., 2019). In our system, this contribution could be reduced by replacing arginine with lysine, but the nucleophilic character of the latter would be expected to result in unwanted chemical reactions after irradiation (Xu et al., 2008; Konigsfeld et al., 2012).
5. Conclusion
The intention here was to design and characterize an experimental biochemical model system with which to assay DNA damage by the (n,μ) reaction of boron-10. The experimental model we have described is suitable to simulate BNCT conditions for four reasons. First, cellular chromatin is far better modeled by condensed DNA than a naked plasmid (Daban, 2003). Second, the boron concentration matches that of BNCT. Third, the quantitative binding of the boron-containing ligand to produce condensed DNA ensures that the vast majority of DNA damage results from neutron capture on boron. Fourth, the electrostatic binding of the boron- containing ligand is easily reversed by increasing the ionic strength. This releases the DNA for subsequent assays of its damage.
The experimental approach described here is able to model BNCT and can provide data essential for the calibration of biophysical track structure codes such as PARTRAC (Friedland et al., 2019) and Geant4-DNA (Incerti et al., 2019) (TOPAS-nBio is a wrapper for the latter). The combination of experimental data and Monte Carlo simulation has the capacity to address the complex dosimetry inherent in BNCT.
Highlights.
Boron-containing cationic peptide that binds strongly to DNA.
Peptide condensed DNA models cellular chromatin.
Boron concentration in condensed DNA within the therapeutic range.
Vast majority of energy deposition is from neutron capture on boron.
Acknowledgements
Supported by Loma Linda University School of Medicine. Jose Ramos-Méndez was supported by National Cancer Institute grant U24CA215123.
Footnotes
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achilli C, Ciana A, Fagnoni M, Balduini C, Minetti G (2013) Susceptibility to hydrolysis of phenylboronic pinacol esters at physiological pH. Cent Eur J Chem 11: 137–139. [Google Scholar]
- Adhikary A, Khanduri D, Sevilla MD (2009) Direct observation of the hole protonation state and hole localization site in DNA-oligomers. J Am Chem Soc 131: 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostinelli S, et al. (2003) GEANT4 - A simulation toolkit. Nucl Instruments Methods Phys Res Sect A Accel Spectrometers Detect Assoc Equip 506: 250–303. [Google Scholar]
- Baiocco G, Barbieri S, Babini G, Morini J, Alloni D, Friedland W, Kundrat P, Schmitt E, Puchalska M, Sihver L, Ottolenghi A (2016) The origin of neutron biological effectiveness as a function of energy. Sci Rep 6: 34033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth RF, Zhang Z, Liu T (2018a) A realistic appraisal of boron neutron capture therapy as a cancer treatment modality. Cancer Commun 38: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth RF, Mi P, Yang W (2018b) Boron delivery agents for neutron capture therapy of cancer. Cancer Commun (Lond) 38: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield VA (1996) DNA condensation. Curr Opin Struct Biol 6: 334–341. [DOI] [PubMed] [Google Scholar]
- Bloomfield VA (2000) Static and dynamic light scattering from aggregating particles. Biopolymers 54: 168–172. [DOI] [PubMed] [Google Scholar]
- Cammidge AN, Goddard VHM, Gopee H, Harrison NL, Hughes DL, Schubert CJ, Sutton BM, Watts GC, and Whitehead AJ (2006) Aryl trihydroxyborates: easily isolated discrete species convenient for direct application in coupling reactions. Org Lett 8: 4071–4074. [DOI] [PubMed] [Google Scholar]
- Chadwick J (1932) Existence of a neutron. Proc Royal Soc 136: 629–708. [Google Scholar]
- Chapman JD, Reuvers AP, Borsa J, Greenstock CL (1973) Chemical radioprotection and radiosensitization of mammalian cells growing in vitro. Radiat Res 56: 291–306. [PubMed] [Google Scholar]
- Daban JR (2003) High concentration of DNA in condensed chromatin. Biochem Cell Biol 81:91–99. [DOI] [PubMed] [Google Scholar]
- DeRouchey J, Hoover B, Rau DC (2013) A comparison of DNA compaction by arginine and lysine peptides: a physical basis for arginine rich protamines. Biochemistry 52: 3000–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ende BM, Atanackovic J, Erlandson A, Bentoumi G (2016) Use of GEANT4 vs. MCNPX for the characterization of a boron-lined neutron detector. Nucl Instruments Methods Phys Res Sect A Accel Spectrometers Detect Assoc Equip 820: 40–47. [Google Scholar]
- ENDF/B-VI: Cross Section Evaluation Working Group, ENDF/B-VI Summary Document, Report BNL-NCS-17541 (ENDF-201) (1991) edited by Rose PF, National Nuclear Data Center, Brookhaven National Laboratory, Upton, NY, USA. [Google Scholar]
- Fournier C, Zahnreich S, Kraft D, Friedrich T, Voss KO, Durante M, Ritter S (2012) The fate of a normal human cell traversed by a single charged particle. Sci Rep 2: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland W, Schmitt E, Kundrat P, Baiocco G, Ottolenghi A (2019) Track-structure simulations of energy deposition patterns to mitochondria and damage to their DNA. Int J Radiat Biol 95: 3–11. [DOI] [PubMed] [Google Scholar]
- Goorley JT, Kiger WS 3rd, Zamenhof RG (2002) Reference dosimetry calculations for neutron capture therapy with comparison of analytical and voxel models. Med Phys 29: 145–156. [DOI] [PubMed] [Google Scholar]
- Hall DG (2011) Structure, properties, and preparation of boronic acid derivatives. Overview of their reactions and applications In “Boronic Acids. Preparation and Applications in Organic Synthesis, Medicine, and Materials,” second edition, pp 1–133. Hall DG, ed. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- Hawthorne MF, Lee MW (2003) A critical assessment of boron target compounds for boron neutron capture therapy. J Neurooncol 62: 33–45. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Sakurai Y, Uchida R, Suzuki M, Usui S, Tominaga T (2015) Preliminary study of MAGAT polymer gel dosimetry for boron neutron capture therapy. J Phys Conf Ser 573: 012074. [Google Scholar]
- van Holde K, Zlatanova J (2007) Chromatin fiber structure: Where is the problem now? Semin Cell Dev Biol 18: 651–658. [DOI] [PubMed] [Google Scholar]
- Incerti S, Kyriakou I, Bernal MA, Bordage MC, Francis Z, Guatelli S, Ivanchenko V, Karamitros M, Lampe N, Lee SB, Meylan S, Min CH, Shin WG, Nieminen P, Sakata D, Tang N, Villagrasa C, Tran HN and Brown JMC (2018) Geant4-DNA example applications for track structure simulations in liquid water: A report from the Geant4-DNA. Project. Med. Phys. 45: e722–739. [DOI] [PubMed] [Google Scholar]
- Javid M, Brownell GL, Sweet WH (1952) The possible use of neutron-capturing isotopes such as boron 10 in the treatment of neoplasms. II. Computation of the radiation energies and estimates of effects in normal and neoplastic brain. J Clin Invest 31: 604–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha PK, Desai PS, Li J, Larson RG (2014) pH and salt effects on the associative phase separation of oppositely charged polyelectrolytes. Polymers 6: 1414–1436. [Google Scholar]
- Johnson RN, Chu DS, Shi J, Schellinger JG, Carlson PM, Pun SH (2011) HPMA-oligolysine copolymers for gene delivery: optimization of peptide length and polymer molecular weight. J Control Release 155:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Yoo J, Sohn B, Lee S, Lee HS, Ma W, Kee J, Aksimentiev A, Kim H (2018) Sequence- dependent DNA condensation as a driving force of DNA phase separation. Nucleic Acids Res, 46: 9401–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Tagawa T, Preuss M, Miller AD (2002) Biophysical characterization of the DNA binding and condensing properties of adenoviral core peptide mu. Biochemistry 41: 652–659. [DOI] [PubMed] [Google Scholar]
- Kempe J, Gudowska I, Brahme A (2007) Depth absorbed dose and LET distributions of therapeutic 1H, 4He, 7Li, and 12C beams. Med Phys 34: 183–192. [DOI] [PubMed] [Google Scholar]
- Konigsfeld KM, Lee M, Urata SM, Aguilera JA, Milligan JR (2012) Free terminal amines in DNA-binding peptides alter the product distribution from guanine radicals produced by single electron oxidation. Int J Radiat Biol 88: 230–238. [DOI] [PubMed] [Google Scholar]
- Li M, Matkovic M, Piantanida I, Schmuck C (2017) Incorporation of arginine mimetic residue into peptides for recognition of double stranded nucleic acid structure: binding and aggregation studies. Bioorg Med Chem 25: 1875–1880. [DOI] [PubMed] [Google Scholar]
- Locher GL (1936) Biological effects and therapeutic possibilities of neutrons. Am J Roentgenol 36: 1–13. [Google Scholar]
- Lorimore SA, Kadhim MA, Pocock DA, Papworth D, Stevens DL, Goodhead DT, Wright EG (1998) Chromosomal instability in the descendants of unirradiated surviving cells after alpha-particle irradiation. Proc Natl Acad Sci USA 95: 5730–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A, Bullick S, Won JH, Milligan JR (2006) Cationic peptides containing tyrosine protect against radiation-induced oxidative DNA damage. Int J Radiat Biol 82: 421–433. [DOI] [PubMed] [Google Scholar]
- Marinaro WA, Schieber LJ, Munson EJ, Day VW, Stella VJ (2012) Properties of a model aryl boronic acid and its boroxine. J Pharm Sci 101: 3190–3198. [DOI] [PubMed] [Google Scholar]
- Mascotti DP, Lohman TM (1997) Thermodynamics of oligoarginines binding to RNA and DNA. Biochemistry 36: 7272–7279. [DOI] [PubMed] [Google Scholar]
- Milligan JR, Aguilera JA, Ward JF (1993) Variation of single-strand break yield with scavenger concentration for plasmid DNA irradiated in aqueous solution. Radiat Res 133: 151–157. [PubMed] [Google Scholar]
- Moss RL (2014) Critical review, with an optimistic outlook, on boron neutron capture therapy (BNCT). Appl Radiat Isot 88: 2–11. [DOI] [PubMed] [Google Scholar]
- Nayvelt I, Thomas T, Thomas TJ (2007) Mechanistic differences in DNA nanoparticle formation in the presence ofoligolysines and poly-L-lysine. Biomacromolecules 8: 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren J, Svanvik N, Kubista M (1998) The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers 46: 39–51. [DOI] [PubMed] [Google Scholar]
- Ono K, Tanaka H, Tamari Y, Watanabe T, Suzuki M, Masunaga SI (2019) Proposal for determining absolute biological effectiveness of boron neutron capture therapy - the effect of 10B(n,α)7Li dose can be predicted from the nucleocytoplasmic ratio or the cell size. J Radiat Res 60: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic O, Schlick T (2017) Dependence of the linker histone and chromatin condensation on the nucleosome environment. J Phys Chem B 121: 7823–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl J, Shin J, Schümann J, Faddegon B, Paganetti H (2012) TOPAS: An innovative proton Monte Carlo platform for research and clinical applications. Med Phys 39: 6818–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry CC, Tang VJ, Konigsfeld KM, Aguilera JA, Milligan JR (2011) Use of a coumarin-labeled hexa-arginine peptide as a fluorescent hydroxyl radical probe in a nanoparticulate plasmid DNA condensate. J Phys Chem B 115: 9889–9897. [DOI] [PubMed] [Google Scholar]
- Prasanna PG, Stone HB, Wong RS, Capala J, Bernhard EJ, Vikram B, Coleman CN (2012) Normal tissue protection for improving radiotherapy. Where are the gaps? Transl Cancer Res 1: 35–48. [PMC free article] [PubMed] [Google Scholar]
- Raju MR, Eisen Y, Carpenter S, Inkret WC (1991) Radiobiology of α-particles. III. Cell inactivation by α-particle traversals of the cell nucleus. Radiat Res 128: 204–209. [PubMed] [Google Scholar]
- Ruedas-Rama MJ, Orte A, Crovetto L, Talavera EM, Alvarez-Pez JM (2010) Photophysics and binding constant determination of the homodimeric dye BOBO-3 and DNA oligonucleotides. J Phys Chem B 114:1094–1103. [DOI] [PubMed] [Google Scholar]
- Saccardo P, Villaverde A, Gonzalez-Montalban N (2009) Peptide-mediated DNA condensation for non-viral gene therapy. Biotechnol Adv 27: 432–438. [DOI] [PubMed] [Google Scholar]
- Sage E, Harrison L (2011) Clustered DNA lesion repair in eukaryotes: relevance to mutagenesis and cell survival. Mutat Res 711: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Kobayashi M, Iwabuchi S, Morita Y, Takamura Y, Tamiya E (2004) DNA condensation monitoring after interaction with Hoechst 33258 by atomic force microscopy and fluorescence spectroscopy. J Biochem 136:813–823. [DOI] [PubMed] [Google Scholar]
- Sears VF (1992) Neutron scattering lengths and cross sections. Neutron News 3: 26–37. [Google Scholar]
- Sharma KK, Milligan JR, Bernhard WA (2008) Multiplicity of DNA single-strand breaks produced in pUC18 exposed to the direct effects of ionizing radiation. Radiat Res 170: 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Sonntag C (2006) Free radical induced DNA damage and its repair. A chemical perspective. Springer, New York. [Google Scholar]
- Soundararajan S, Badawi M, Kohlrust CM, Hageman JH (1989) Boronic acids for affinity chromatography: spectral methods for determinations of ionization and diol-binding constants. Anal Biochem 178: 125–134. [DOI] [PubMed] [Google Scholar]
- Spruijt E, Westphal AH, Borst JW, Stuart MAC, van der Gucht J (2010) Binodal compositions of polyelectrolyte complexes. Macromolecules 43: 6476–6484. [Google Scholar]
- Swarts SG, Becker D, Sevilla M, Wheeler KT (1996) Radiation-induced DNA damage as a function of hydration. II. Base damage from electron-loss centers. Radiat Res 145: 304–314. [PubMed] [Google Scholar]
- Sweet WH (1951) The use of nuclear disintegration in the diagnosis and treatment of brain tumor. N Engl J Med 245: 875–878. [DOI] [PubMed] [Google Scholar]
- Tinganelli W, Ma NY, von Neubeck C, Maier A, Schicker C, Kraft-Weyrather W, Durante M (2013) Influence of acute hypoxia and radiation quality on cell survival. J Radiat Res 54: 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HN, Marchix A, Letourneau A, Darpentigny J, Menelle A, Ott F, Schwindling J, Chauvin N (2018) Comparison of the thermal neutron scattering treatment in MCNP6 and GEANT4 codes. Nucl Instruments Methods Phys Res Sect A Accel Spectrometers Detect Assoc Equip 893: 84–94. [Google Scholar]
- Wagner FM, Loeper-Kabasakal B, Breitkreutz H (2012) Neutron medical treatment of tumours - a survey of facilities. J Inst 7: C03041. [Google Scholar]
- Watanabe T, Hattori Y, Ohta Y, Ishimura M, Nakagawa Y, Sanada Y, Tanaka H, Fukutani S, Masunaga S, Hiraoka M, Ono K, Suzuki M, Kirihata M (2016) Comparison of the pharmacokinetics between L-BPA and L-FBPA using the same administration dose and protocol: a validation study for the theranostic approach using [18F]-L-FBPA positron emission tomography in boron neutron capture therapy. BMC Cancer 16: 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt PJ (1993) Light scattering and the absolute characterization of macromolecules. Analytica Chimica Acta 272: 1–40. [Google Scholar]
- Xu X, Muller JG, Ye Y, Burrows CJ (2008) DNA-protein cross-links between guanine and lysine depend on the mechanism of oxidation for formation of C5 vs C8 guanosine adducts. J Am Chem Soc 130: 703–709. [DOI] [PubMed] [Google Scholar]
- Yokoya A, Cunniffe SM, Watanabe R, Kobayashi K, O’Neill P (2009) Induction of DNA strand breaks, base lesions and clustered damage sites in hydrated plasmid DNA films by ultrasoft X-rays around the phosphorus K edge. Radiat Res 172: 296–305. [DOI] [PubMed] [Google Scholar]
- Yoo J, Aksimentiev A (2016) The structure and intermolecular forces of DNA condensates. Nucleic Acids Res 44: 2036–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue AT, Anderson ES, Dewey MS, Gilliam DM, Greene GL, Laptev AB, Nico JS, Snow WM (2018) Precision determination of absolute neutron flux. Metrologia 55: 460–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PG, Foster DG Jr (1972) An evaluation of the neutron and gamma-ray production cross sections for nitrogen. Los Alamos National Laboratory Publication LA-4725 (ENDF- 173). Los Alamos, NM, USA. [Google Scholar]