Abstract
The cellular and molecular basis of metaplasia and declining neurogenesis in the aging olfactory epithelium (OE) remains unknown. The horizontal basal cell (HBC) is a dormant tissue-specific stem cell presumed to only be forced into self-renewal and differentiation by injury. Here we analyze male and female mice and show that HBCs also are activated with increasing age as well as non-cell-autonomously by increased expression of the retinoic acid-degrading enzyme CYP26B1. Activating stimuli induce HBCs throughout OE to acquire a rounded morphology and express IP3R3, which is an inositol-1,4,5-trisphosphate receptor constitutively expressed in stem cells of the adjacent respiratory epithelium. Odor/air stimulates CYP26B1 expression in olfactory sensory neurons mainly located in the dorsomedial OE, which is spatially inverse to ventrolateral constitutive expression of the retinoic acid-synthesizing enzyme (RALDH1) in supporting cells. In ventrolateral OE, HBCs express low p63 levels and preferentially differentiate instead of self-renewing when activated. When activated by chronic CYP26B1 expression, repeated injury, or old age, ventrolateral HBCs diminish in number and generate a novel type of metaplastic respiratory cell that is RALDH− and secretes a mucin-like mucus barrier protein (FcγBP). Conversely, in the dorsomedial OE, CYP26B1 inhibits injury-induced and age-related replacement of RALDH− supporting cells with RALDH1+ ciliated respiratory cells. Collectively, these results support the concept that inositol-1,4,5-trisphosphate type 3 receptor signaling in HBCs, together with altered retinoic acid metabolism within the niche, promote HBC lineage commitment toward two types of respiratory cells that will maintain epithelial barrier function once the capacity to regenerate OE cells ceases.
SIGNIFICANCE STATEMENT Little is known about signals that activate dormant stem cells to self-renew and regenerate odor-detecting neurons and other olfactory cell types after loss due to injury, infection, or toxin exposure in the nose. It is also unknown why the stem cells do not prevent age-dependent decline of odor-detecting neurons. We show that (1) stem cells are kept inactive by the vitamin A derivative retinoic acid, which is synthesized and degraded locally by olfactory cells; (2) old age as well as repeated injuries activate the stem cells and exhaust their potential to produce olfactory cells; and (3) exhausted stem cells alter the local retinoic acid metabolism and maintain the epithelial tissue barrier by generating airway cells instead of olfactory cells.
Keywords: aging; inositol-1,4,5-triphosphate; metaplasia; olfactory epithelium; retinoic acid; stem cells
Introduction
The horizontal basal cell (HBC) is a pluripotent and neurogenic stem cell in the adult mammalian olfactory epithelium (OE) that is dormant during normal tissue homeostasis, but becomes activated to proliferate and/or differentiate following extensive death of OE cells (Leung et al., 2007; Schwob et al., 2017). HBCs can be experimentally activated by genetic inactivation of the tumor protein 63 (p63; Fletcher et al., 2011; Packard et al., 2011; Schnittke et al., 2015; Fletcher et al., 2017) or by injury caused by methimazole, which is an olfactotoxin that kills all OE cell types except HBCs (Brittebo, 1995; Gadye et al., 2017). The adjacent respiratory epithelium (RE), which is not affected by methimazole, harbors basal stem cells with histochemical characteristics similar to those of HBCs (Rock et al., 2010).
Niche factors that regulate HBC activation in response to injury remain elusive. Neither is it known why HBCs cannot prevent age-related changes, such as decreased neurogenesis and appearance of respiratory metaplasia (Naessen, 1971; Nakashima et al., 1984; Loo et al., 1996; Genter and Ali, 1998; Kondo et al., 2009; Brann and Firestein, 2014; Child et al., 2018). Unexplored is also the regulation of HBC activation in relation to the functional organization of OE into overlapping zones with olfactory sensory neurons (OSNs) that express different odorant receptor genes, depending on their localization along the dorsomedial–ventrolateral nasal cavity axis (Ressler et al., 1993; Vassar et al., 1993; Mombaerts et al., 1996; Zapiec and Mombaerts, 2020). The dorsomedial OE zone (Z1) differs significantly from ventrolateral zones (Z2-4) with regard to gene expression (Alenius and Bohm, 1997; Gussing and Bohm, 2004), OSN turnover (Vedin et al., 2009; Login et al., 2015b), sensitivity to toxins, and age-related loss of OSNs (Loo et al., 1996; Genter and Ali, 1998; Vedin et al., 2004; Kondo et al., 2009). We and others have shown that dorsomedial–ventrolateral differences in all-trans-retinoic acid (RA) metabolism regulates olfactory progenitor cell fate and differentiation (Peluso et al., 2012; Paschaki et al., 2013; Login et al., 2015b). RA is a morphogen derived from vitamin A that regulates organogenesis and tissue regeneration and is widely used to induce differentiation of pluripotent stem cells cultures (Gudas and Wagner, 2011). RA bioavailability is determined by RA synthesizing retinaldehyde dehydrogenases (RALDHs) and members of the cytochrome P450 family 26 (CYP26) that inactivate RA (Duester, 2008; Isoherranen and Zhong, 2019). Local RA metabolism regulates the fate selection of stem cells in the embryonic gonadal niche and promotes hematopoietic stem cell dormancy in the bone marrow niche (Feng et al., 2014; Cabezas-Wallscheid et al., 2017). While a role for RA in the regulation of adult neurogenesis has been demonstrated (Wang et al., 2005; Jacobs et al., 2006; Goodman et al., 2012), less is known about the role of local RA metabolism in the regulation of adult neuronal stem cells. In the adult OE stem cell niche, nasal RA levels are controlled by non-neural RALDH+ cells and odor/air stimuli that induce CYP26B1 in such a way that an activity-dependent Z1high–Z4low spatial gradient of RA catabolic activity is formed (Niederreither et al., 1997; Öztokatli et al., 2012; Peluso et al., 2012; Login et al., 2015a,b).
We show that HBCs are activated not only by injury, but also naturally by old age as well as experimentally by increased CYP26B1 in OSNs. These three activating stimuli induce dormant HBCs to express the IP3 receptor and intracellular Ca2+ release channel (IP3R3). Thus, HBCs resemble RE stem cells, which have constitutive high IP3R3 levels. We find that HBCs in ventrolateral Z4 express relatively low levels of p63 and are biased to differentiate rather than to self-renew. Repeated injuries or chronically increased CYP26B1 cause metaplasia, which in the ventrolateral OE is of a previously unspecified secretory respiratory type with cells that express the mucin-like protein FcγBP. In Z1, CYP26B1 stimulates neurogenesis and inhibits a distinct type of injury-induced and age-related metaplasia with ciliated respiratory cells. Together, our results show that HBCs exhibit a limited self-renewal capacity and are more multipotent than previously thought, and that the balance of RA metabolizing enzymes in differentiated cell progeny of HBCs regulate HBC activation as well as differences in neurogenesis along the dorsomedial–ventrolateral axis.
Materials and Methods
Mice.
The OMP-CYP26B1 transgenic mouse line (herein referred to as OMP-Cyp) in which the OMP gene promoter drives Cyp26B1 overexpression selectively in OSNs has been previously described (Login et al., 2015a). Mice were housed with food and water available ad libitum in light- and temperature-controlled environment and killed by cervical dislocation followed by immediate exsanguination. All animal experiments were approved by the local ethics committee for animal research at the court of appeal for the upper northern area of Norrland (Umeå, Sweden).
Tissue preparation.
Nasal tissue was dissected, fixed for 4 h at 4°C in 4% (w/v) paraformaldehyde in PBS, pH 7.4, cryoprotected in 20% (w/v) sucrose at 4°C for 16–24 h, frozen in Tissue-Tek OCT compound (Sakura Finetek) and sectioned at 12 μm. Tissue from animals older than 3 weeks was treated with RDO Rapid Decalcifier (Apex Engineering).
Intranasal drug administration.
Two-week-old C57BL/6J mice were given carprofen (5 mg/kg, s.c.), and anesthesia was accomplished by injection of a mixture of medetomidine (0.25 mg/kg, s.c.), midazolam (2.5 mg/kg, s.c.), and fentanyl (0.025 mg/kg, s.c.; Henke and Erhardt, 2004). Antidotes atipamezole (1.25 mg/kg) and naloxone (0.6 mg/kg) were given subcutaneously. Control experiments using fluorescent beads added to the solutions assured that the technique used resulted in solutions spreading over the entire OE surface. Liarozole dihydrochloride (6-[(3-chlorophenyl)-imidazol-1-ylmethyl]-1H-benzimida-zole, Tocris Bioscience) was diluted to 1.5 mm in 7% dimethylsulfoxide (DMSO) for intranasal administration. 13-cis-RA (isotretinoin, Sigma-Aldrich) was administered at a concentration of 0.1 mm. Vehicle (7% DMSO) was administered (1 μl/g) to one naris. Two microliters of fluid was placed at the naris, timed with the inspiratory phase of the breathing cycle when the mouse was in supine position. One minute was allowed between administrations. The mouse was then turned 90° to right lateral recumbent position for the second dose, followed by the third dose given when in left lateral recumbent position. Drug or vehicle was administered once a day for 3 consecutive days, and mice were killed 24 h after the last dose. Liarozole, 13-cis-RA, and DMSO were administrated at nontoxic doses as assessed in pilot experiments by cleaved caspase-3 immunohistochemistry.
Methimazole treatment.
To ablate cells in OE, OMP-Cyp and littermate control mice received one, two, or three intraperitoneal injections of 75 mg/kg methimazole (Sigma-Aldrich) or physiological 0.9% NaCl saline vehicle (Brittebo, 1995; Suzukawa et al., 2011) with an interval of 3 weeks. Mice were killed 6 or 21 d postinjection (see Fig. 4O). Pilot experiments using 50, 75, or 100 mg/kg methimazole followed by analysis of tissue sections throughout the anterior–posterior axis of OE showed that 75 mg/kg injections resulted in a complete detachment of OE, leaving a single layer of HBCs in all zones. Mice intended for analysis of proliferating cells were injected intraperitoneally with 50 mg/kg bromodeoxyuridine (BrdU; Sigma-Aldrich) twice per day 6 h apart for 2 d [at 4 and 5 d postinjection (dpi) of methimazole] and were killed 24 h or 21 d later (representing 6 or 21 dpi of methimazole, respectively).
Figure 4.
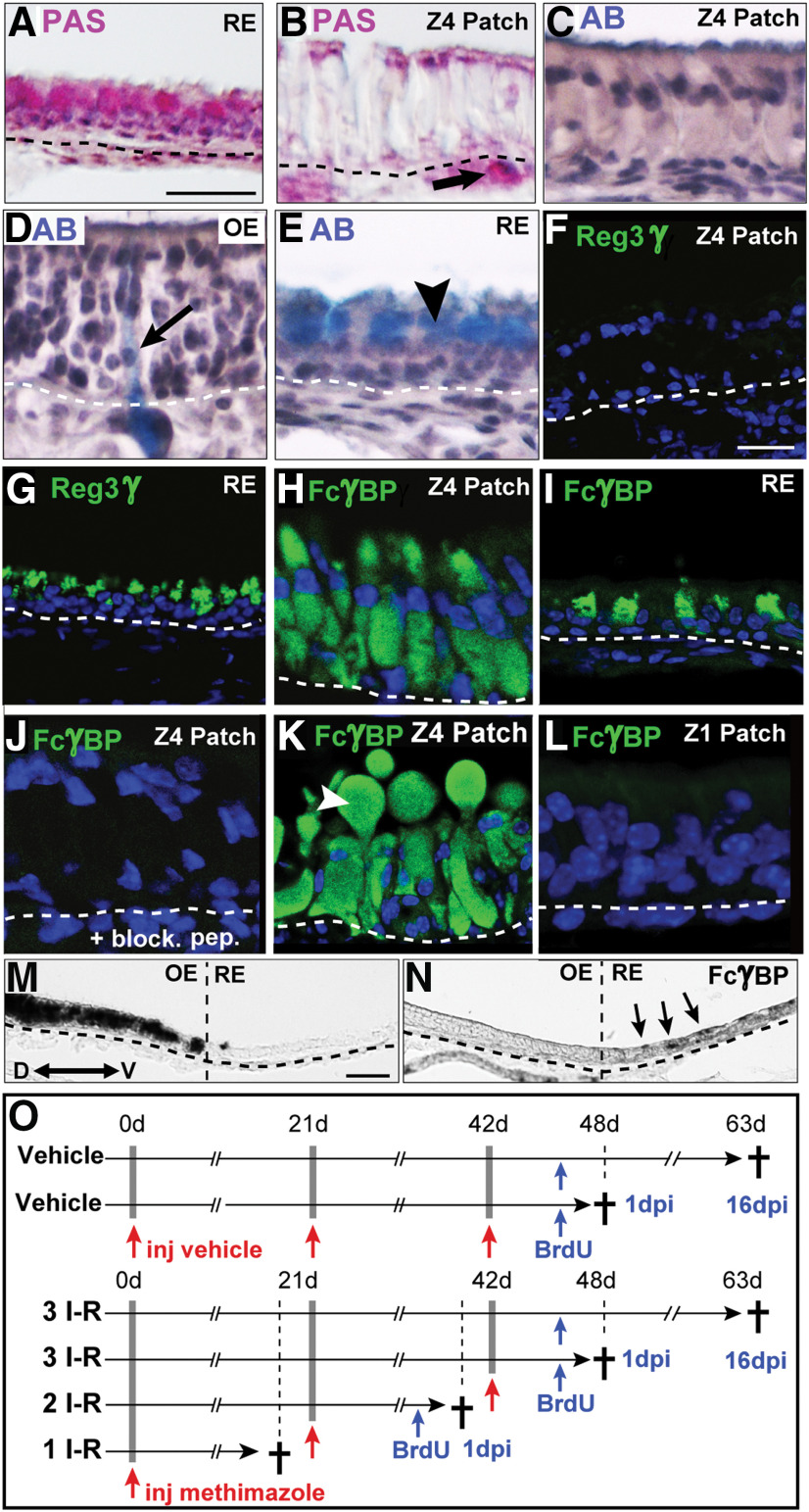
Identification of the metaplastic Z4 cell type. A–N, Analyses of OE, metaplastic patches in Z4 (Z4 patch), and RE of OMP-Cyp mice are shown. A, B, Shows PAS+ (pink) goblet cells in RE (A) and Bowman's glands in lamina propria of OE (B, arrow). Patch cells are negative (B). Nuclei are stained by hematoxylin (purple). C–E, Alcian Blue (AB) staining (light blue) shows negative patch cells (C) while Bowman's duct/glands in OE (D, arrow) and goblet cells in RE (E, arrowhead) stain positive. F, G, Immunofluorescence for Reg3γ (green) shows that Z4 patch cells are negative (F) while RE cells are positive (G). H, I, FcγBP in patch cells (H) and in RE cells (I). J, Shows lack of FcγBP immunofluorescence in Z4 patch when blocking with the immunizing peptide (block. pep.). K, Secreted FcγBP+ (green) extracellular globule (arrowhead) that is present in a fraction of the patches. L, Cells in Z1 patches do not stain for FcγBP. M, N, In situ hybridization analysis showing OMP mRNA+ in OE (M) and FcγBP mRNA+ cells (N, arrows) on serial sections of the nasal septum. O, Experimental time lines for vehicle treatments of control mice (control) and methimazole-induced I–R cycles of OE in control and OMP-Cyp mice. Two-month-old mice received one (1 I–R), two (2 I–R), or three (3 I–R) injections (inj.) of vehicle or methimazole (red arrows) 21 d apart. Mice were killed (†) at 6 or 21 dpi following methimazole injections and received BrdU (blue arrows) 1 or 16 d before being killed. Dashed lines indicate basal lamina. Scale bars: A–E, 25 µm; F–L, 12.5 µm; M, N, 50 µm.
Histology and immunodetection.
Before immunofluorescence analysis, the tissue sections were rinsed in PBS and background was blocked by incubation for 1 h in 3% normal donkey serum (Jackson ImmunoResearch) in PBS with 0.1% Triton X-100, followed by overnight incubation at 4°C in blocking solution with the primary antibody. Primary antibodies not requiring antigen retrieval were raised against the following: adenylate cyclase 3 (AC3; made in rabbit; dilution, 1:500; catalog #C-20, Santa Cruz Biotechnology); acetylated tubulin (mouse; 1:100; catalog #T7451, Sigma-Aldrich); CD36 (goat; 1:200; catalog #AF2519, R&D Systems); CYP26B1 (rabbit; 1:200; catalog #21 555-1-AP, ProteinTech); cytokeratin 5 (rabbit; 1:300; catalog #905501, BioLegend); ICAM1 (rabbit; 1:200; catalog #ab25375, Abcam); IP3R3 (rabbit; 1:500; catalog #AB9076, Merck); NAD(P)H dehydrogenase quinone 1 (NQO1; rabbit; 1:500; catalog #ab2346, Abcam); pS6 (rabbit; 1:2000; catalog #44-923G, Thermo Fisher Scientific); RALDH1/2 (mouse; 1:1000; catalog #SC-166362, Santa Cruz Biotechnology); REG3γ (rabbit; 1:500; catalog #ABIN1870299, Antibodies-online.com); STMN1 (rabbit; 1:1000; catalog #ab24445, Abcam); and cleaved caspase 3 (1:500; catalog #559565; BD Biosciences). Specific antigen retrieval protocols were necessary to improve the detection by some of the antibodies. Retrieval for BrdU (rat antibody; dilution, 1:500; catalog #NB500-169, Novus Biologicals) and SOX2 (mouse; 1:500; catalog #SC-365964, Santa Cruz Biotechnology) was with 2 m HCl at 37°C for 30 min. For cytokeratin 19 (rat; 1:10; catalog #AB_2133570, Developmental Studies Hybridoma Bank), FcγBP (rabbit; 1:200; catalog #NBP1-90 462, Novus Biologicals), FoxJ1 (mouse; 1:500; catalog #14996582, Thermo Fisher Scientific; rabbit; 1:100; catalog #NBP-87928, Novus Biologicals), IP3R3 (mouse; 1:50; catalog #610312, BD Biosciences), p63 (mouse; 1:100; catalog #4A4 CM163 ABC, Biocare Medical), and Ki67 (rabbit; 1:500; catalog #AB9076, Merck) heat retrieval in 10 mm sodium citrate buffer, pH 6 for 20 min, was used. FcγBP antibody competition was with FcγBP recombinant protein antigen (catalog #NBP1-90462PEP, Novus Biologicals) at a 10 m excess. A specific signal was detected by incubation with Alexa Fluor 488- or 546-conjugated donkey antibodies toward goat (1:500; catalog #A11055 and #A11056, Invitrogen), mouse (1:500, #A21202, Invitrogen), rabbit (1:500; Invitrogen catalog #A21206 and #A10040, Thermo Fisher Scientific), or rat IgG (1:500; Invitrogen catalog #A21208 and #A11081, Thermo Fisher Scientific). Biotin-conjugated donkey anti-mouse IgG (1:1000; catalog #AS10 1242, Agrisera) and anti-rabbit IgG (1:1000; catalog #AS101083, Agrisera) was used for 1 h at room temperature followed by 30 min of incubation with streptavidin DyLight 549 (1:750; catalog #SA-5549, Vector Laboratories). Sections were counterstained with Hoechst 33258 (Sigma-Aldrich) and mounted with fluorescent mounting medium (catalog #S3023, Dako). Images were captured with a Nikon Digital Eclipse C1 plus Confocal Microscope. For bright-field detection either ImmPACT DAB peroxidase or Vectastain Elite ABC HRP kits (Vector Laboratories) were used. For histologic hematoxylin and eosin staining, Mayer's hematoxylin (Histolab) was used for 5 min, sections were rinsed in tap water, followed by 0.2% eosin (Histolab) for 15 s, and rinsed. For Alcian Blue staining, sections were incubated with 3% acetic acid for 3 min, then 1% Alcian Blue 8GX (Sigma Aldrich) in 3% acetic acid, pH 2.5, for 30 min, rinsed in tap water, and counterstained with hematoxylin. For Periodic acid-Schiff (PAS) staining sections were incubated with 0.1% periodic acid (Merck) for 15 min, followed by 30 min in Schiff's reagent (Sigma-Aldrich), rinsed in tap water, and counterstained with hematoxylin. After histologic staining, sections were dehydrated in a 70%, 95%, and 100% ethanol series, cleared in xylene, and mounted with DPX media (BDH Laboratories).
In situ hybridization.
In situ hybridization analysis was performed as previously described (Vedin et al., 2009). Histone mRNA expression in S-phase was detected with a digoxygenin-labeled histone 3 cRNA probe made by using IMAGE clone no. 478445 with an insert corresponding to bases 27–480 of replication-dependent histone 3 (RefSeq accession no. NM_175653.2; Vedin et al., 2009). The FCγBP cRNA probe was generated using IMAGE clone 4209006 with an insert corresponding to bases 5922–7957 of RefSeq accession no. NM_001122603.1.
Experimental design and statistical analyses.
Comparisons among control, OMP-Cyp, and methimazole-treated mice were performed in groups of at least three littermate mice, with the same overall proportion of males and females. Quantifications of immunofluorescence and histone 3 mRNA+ cells were done from eight serial coronal hemisections (section interval, 90 μm) per mouse, collected from the middle part of OE in a region located 300–800 μm anterior to the olfactory bulb. Data for Z1, Z2, and Z4 were obtained from regions of 300 μm in length in each zone, per data point. Positive cells were assessed and counted directly after scanning with a Nikon C1confocal microscope. The numbers of histone 3 mRNA+ cells were determined by quantification of in situ hybridization signal in basal cells with a discernable nucleus. OE from regions 500 μm in length in Z4 (13-cis-RA) or Z1 and Z4 (liarozole) of 2-week-old mice (three mice per group) were analyzed using light field (in situ hybridization signal) and fluorescence (nuclear staining with Hoechst stain). Statistical significance of all datasets were determined by unpaired two-tailed t tests using Microsoft Excel and averaged data are presented as mean ± SEM. All quantifications were performed blinded across conditions. For a Krt5+ cell to be included in the count, the image had to include the nucleus.
Results
Effects of spatially delimited RA metabolic enzymes inadult OE
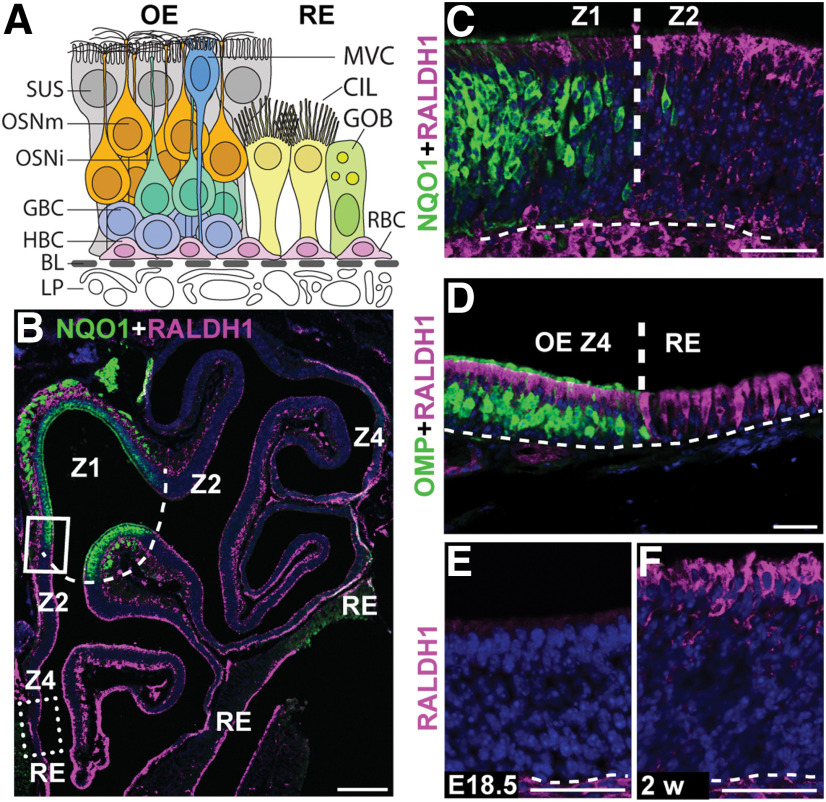
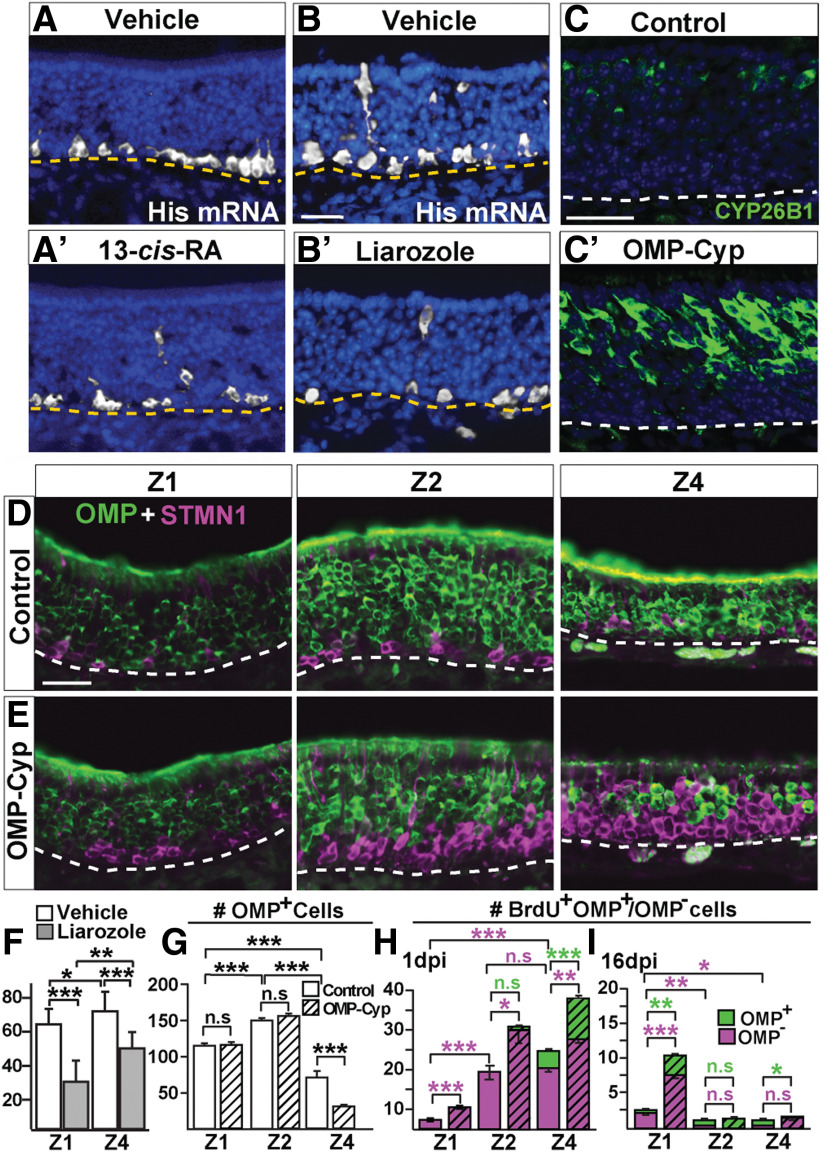
HBC cell progeny includes OSNs as well as supporting sustentacular (SUS) cells, globose basal cells (GBCs), Bowman's gland, and microvillous cells (MVCs; Fig. 1A). The principal cell types of RE are ciliated respiratory goblet cells and respiratory basal cells, the latter being the stem/progenitor cells of the RE. Double-immunofluorescence analyses with antibodies recognizing RALDH1 and the Z1 marker NQO1 (Gussing and Bohm, 2004), showed that SUS cells in Z2-4, but not in Z1, expressed the RA-synthesizing enzyme RALDH1 (Fig. 1B,C). RALDH1 was also in the RE adjacent to Z4 (Fig. 1B,D). Analysis of OE at embryonic day 18.5 and 2 weeks of age revealed that Z2-4 RALDH1+ SUS cells first appeared after birth (Fig. 1E,F). This finding, together with previous results showing RALDHs in lamina propria cells and the postnatal development of a Z1high to Z4low gradient of CYP26B1+ OSNs, make it highly likely that RA bioavailability in Z1, Z2, and Z4 becomes low, medium, and high, respectively, in the adult OE (Niederreither et al., 1997; Norlin et al., 2001; Peluso et al., 2012; Login et al., 2015a,b). To address the function of RA for adult neurogenesis, we administered 13-cis-RA (isotretinoin) intranasally to 2-week-old mice and analyzed them for cell proliferation (Fig. 2A–A′). 13-cis-RA is an endogenous agonist of retinoic acid receptors that is less efficiently cleared from tissue than RA (Isoherranen and Zhong, 2019). Cells in S‐phase have a very high level of histone mRNA that is rapidly degraded once genome replication is completed (Marzluff and Duronio, 2002; Vedin et al., 2009). The numbers of dividing basal cells per mm in vehicle (DMSO) and 13-cis-RA-treated mice were 66 ± 13 and 51 ± 7, respectively (p < 0.01, Student's t test). This indicated that transiently increased RA signaling in OE inhibited cell proliferation. To address the function of CYP26B1 for basal cell proliferation, mice were treated with the CYP26B1 inhibitor liarozole intranasally. Because of the Z1high to Z4low gradient of CYP26B1+ OSNs, we analyzed the effect in both Z1 and Z4. The result showed that liarozole, which conceivably resulted in increased RA due to CYP26B1 inhibition, decreased cell proliferation 1.9-fold and 1.6-fold in Z1 and Z4, respectively (Fig. 2B–B′,F). Together, these results indicated that a transient decrease of CYP26B1 activity inhibited cell proliferation in OE and that the effect was most pronounced in Z1, which had the most CYP26B1+ OSNs.
Figure 1.
Spatial expression of CYP26B1 and RALDH1 in the adult OE. A, Schematic illustration of cell types in the OE and RE. OE is a columnar pseudostratified neuroepithelium with underlying vascularized lamina propria (LP). Close to the basal lamina (BL; dashed line) are HBCs and GBCs. GBCs generate immature OSNs (OSNis), which mature into OMP+ OSNs (OSMm) with their cell bodies centrally in OE. In the apical layer are SUS cells and scattered MVCs, both of which are in contact with the BL by endfeet. Acinar and ductal cells of Bowman's glands are additional OE cells not included in the illustration. Main RE cell types are the ciliated respiratory cell (CIL), goblet cell (GOB), and respiratory basal cell (RBC). B, Immunofluorescence for NQO1 (green) in Z1 and RALDH1 (magenta) in Z2-4 at 2 months of age, showing the overall structure of the cartilaginous turbinates lined with OE in one nasal cavity. The dashed line represents the NQO1/RALDH1 border between Z1 and Z2. C, Close-up of region boxed in B showing the Z1 (NQO1)/Z2 (RALDH1) border. D, Region corresponding to the dashed box in B, showing the border between OMP+ OSNs and negative RE. Note that RALDH1 is in both OE SUS cells and RE cells. E, F, SUS cells are negative for RALDH1 at embryonic day 18.5 (EXVIII.5), but positive at 2 weeks (2 w) of age. Scale bars: B, 200 µm; C–F, 25 µm. Dashed line in C–F indicates BL. Nuclei are counterstained in blue.
Figure 2.
Altered proliferation of basal cells in OE resulting from liarozole and 13-cis-RA treatment and CYP26B1 expression. A–B′, In situ hybridization analysis showing a reduced number of histone mRNA+ S‐phase basal cells in OE after nasal instillation of 13-cis-RA (A, A′) and the CYP26B1 antagonist liarozole (B, B′) compared with vehicle (DMSO). C, C′, As expected, CYP26B1 (green) is increased in OSNs of transgenic OMP-Cyp (C′) compared with control mice (C). D, E, Double OMP (green, mature OSNs) and STMN1 (magenta, immature OSNs) immunofluorescence in Z1, Z2, and Z4 in 4-month-old OMP-Cyp mice and littermate controls, is shown. F, Quantification of basal cells positive for histone mRNA after nasal instillation of liarozole and vehicle shows that that there was significantly less proliferation in both Z1 and Z4 of liarozole-treated mice. G, Quantification of OMP+ OSNs in OMP-Cyp and controls is shown. H, I, The number of BrdU+/OMP− (magenta) and BrdU+/OMP+ (green) cells in OMP-Cyp mice (striped bars) and controls (open bars) after 1 (H) or 16 (I) dpi of BrdU. Graphs show N = 3 mice and n = 8 hemisections per mouse. Error bars represent the mean ± SEM. Two-tailed Student's t test: n.s. nonsignificant; *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars, 25 µm. Dashed line indicates basal lamina. Nuclei are counterstained in blue.
That RA normally appear to have a suppressive effect onproliferation is supported by the finding that a chronic vitamin A-deficient diet increases proliferation in adult rat OE (Asson-Batres et al., 2003). To analyze the long-term effect of chronically increased RA catabolism in OE, we analyzed mice (OMP-Cyp) that expressed a CYP26B1 transgene under the control of the OMP promoter (Fig. 2C–C′; Login et al., 2015a). As the OMP promoter drives high constitutive transcription selectively in OSNs, the expectation was that bioavailability of RA was continuously reduced throughout OE in OMP-Cyp mice compared with controls. Analysis of 4-month-old control mice demonstrated an innate zonal difference in neurogenesis as the immature STMN1+ OSN layer was thinner in Z1 compared with Z2-4 (Fig. 2D). This zonal difference was also found in the transgenics (Fig. 2E). Quantification revealed that Z2 had the greatest number of mature OMP+ OSNs per unit length compared with Z1 and Z4 (Fig. 2F). Compared with controls, there were, however, relatively fewer OMP+ OSNs in Z4 of OMP-Cyp mice (Fig. 2F), which was in line with our previous observation that OSN death is increased in Z4 of postnatal OMP-Cyp (Login et al., 2015b).
GBCs are a heterogeneous population of proliferating stem/progenitor cells that maintains normal tissue homeostasis in OE as OSNs turnover throughout life (Caggiano et al., 1994; Schwob et al., 2017). Quantification of S-phase GBC progeny 1 dpi of BrdU revealed higher proliferation in Z2 and Z4 than in Z1 in controls (Fig. 2G). The increased proliferation in response to CYP26B1 gain-of-function in OMP-Cyp mice was in keeping with the finding that proliferation decreased following intranasal administration of 13-cis-RA or the CYP26B1 inhibitor liarozole. Importantly, proliferation, as determined by BrdU+/OMP− cell numbers at 1 dpi, was increased in all zones of OMP-Cyp mice compared with controls (Fig. 2G). Analysis of the fraction of BrdU+/OMP+ double-positive cells suggested that the shortest time for transit from a proliferating BrdU-incorporating progenitor into a mature OMP+ OSN was in Z4 for both control and OMP-Cyp mice (Fig. 2G). As expected from previous studies (Graziadei and Graziadei, 1979; Vedin et al., 2009), the continuous cell turnover in adult OE led to clearance of most BrdU+ cells 16 d after BrdU labeling (Fig. 2H). However, in both control and OMP-Cyp mice, relatively more BrdU+ cells were retained in Z1 at 16 dpi, suggesting a slower cell turnover and a relatively high accumulation of BrdU+ GBCs in Z1 (Fig. 2H). Together, these results suggested that the Z1low, Z2medium, and Z4high RA bioavailability in controls correlated to low proliferation and low OSN turnover in Z1 compared with Z2 and Z4. Z4 differed from Z1 and Z2 by showing a higher rate of OSN differentiation. Moreover, a hallmark of Z1 was the relatively slow turnover of GBCs. Reduced RA bioavailability by high constitutive transgenic CYP26B1 expression exaggerated these zonal differences and, in addition, resulted in increased death of Z4 OSNs.
OMP-Cyp mice have patches in Z4 with HBCs, MVCs, and an unidentified cell type
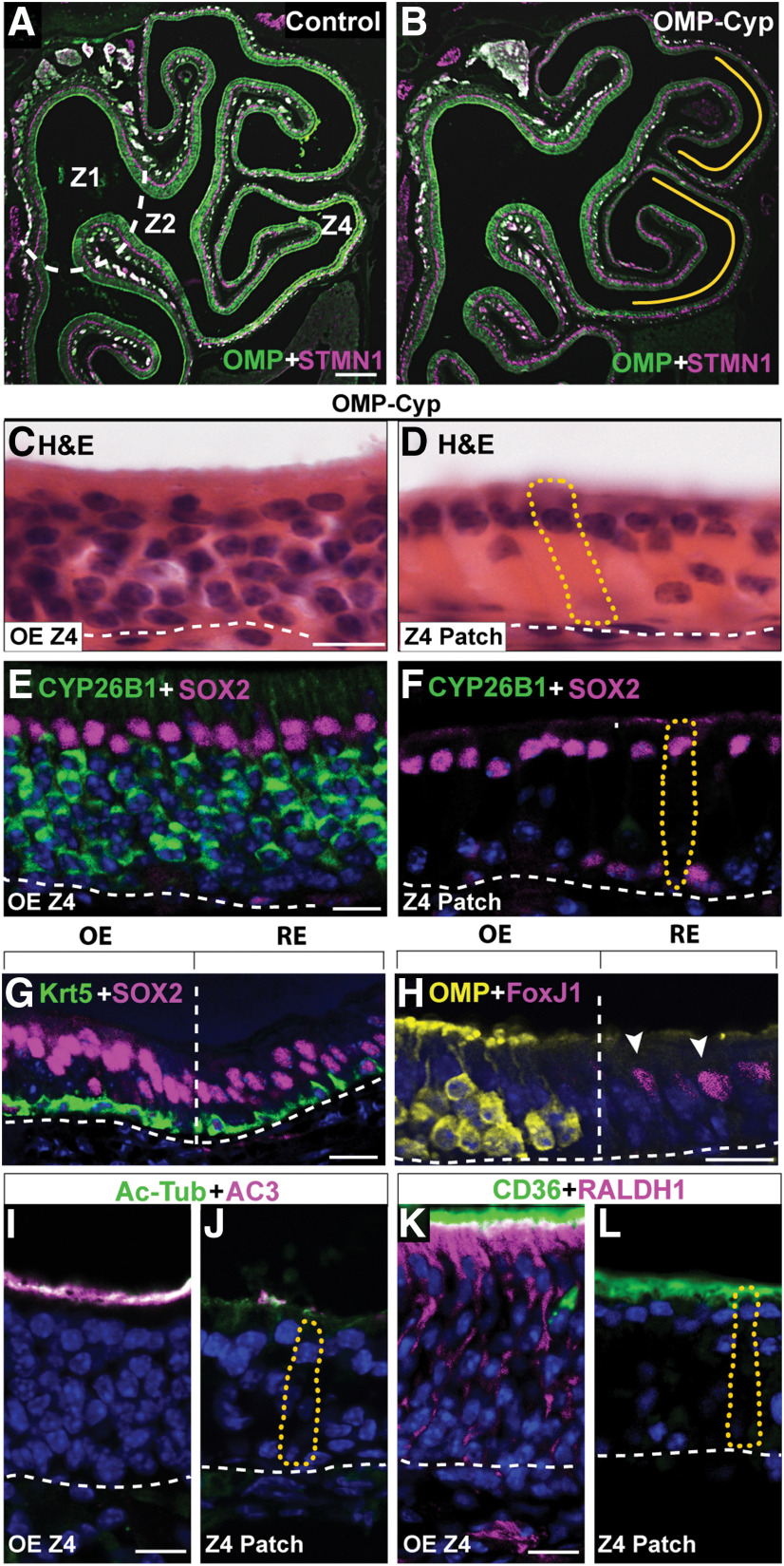
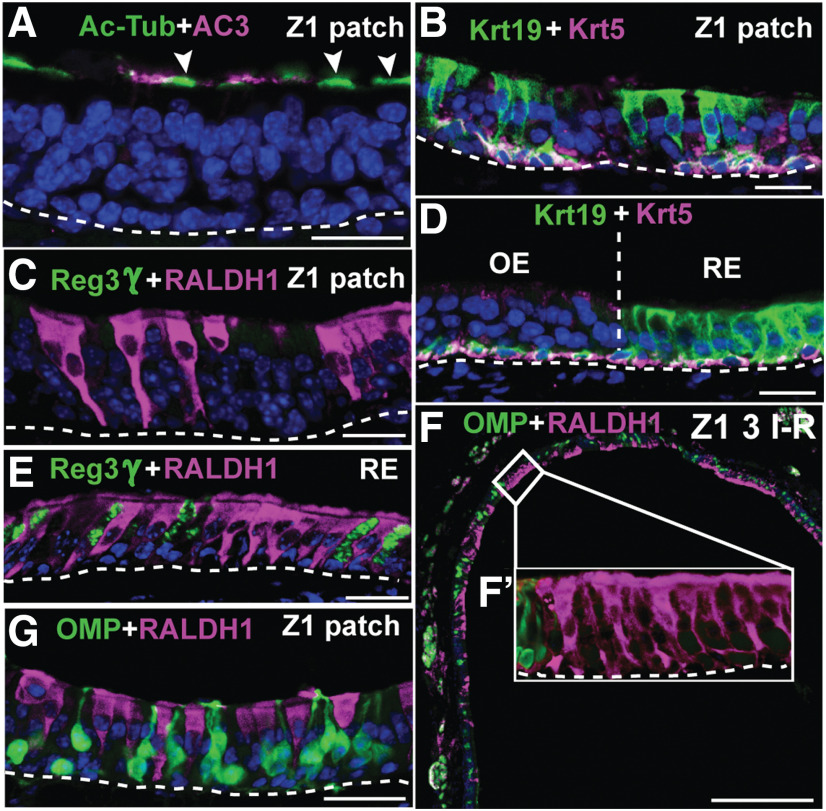
The persistently increased proliferation and increased OSN deaths in Z4 of OMP-Cyp mice correlated with the appearance of epithelial patches that were devoid of OSNs (Fig. 3A–D). This Z4 patch type was not found in 4-month-old controls and was thus caused by the increased CYP26B1 in OSNs. The established patches were maintained in a CYP26B1-independent way because, once established, these patches lacked CYP26B1+ OSNs (Fig. 3E,F). In addition to HBCs and some MVCs, the predominating unclassified cell type was a columnar cell with an apical nucleus (Fig. 3D,F). This cell type was voluminous, with distended basal cytoplasm and at first glance resembled a hypertrophic SUS cell. Similar to SUS and RE cells, the unclassified cell type showed nuclear SOX2 (Fig. 3F,G). Analyses for cilia markers acetylated-tubulin (Ac-Tub) and AC3 as well as the microvilli marker CD36 (Lee et al., 2015) showed that the patch cells, similar to SUS cells, had microvilli and not cilia as OSNs have (Fig. 3I–L). The histologic appearance of the patch also resembled goblet cell metaplasia of RE. However, patch cells were negative for PAS staining as well as Alcian Blue, which only stained goblet cells in RE and cells of Bowman's gland in OE (Fig. 4A–E). Additional experiments showed that the patch cells were negative for regenerating family member 3 γ (Reg3γ), which marks a large subclass of nasal RE cells (Sammeta and McClintock, 2010; Fig. 4F,G). A feature that distinguished the patch cells from both SUS and RE cells in Z4 was that they were RALDH1 negative (Fig. 3K,L). Instead patch cells stained intensely for FoxJ1, which is a marker for ciliated RE cells (Pardo-Saganta et al., 2015; Fig. 3H). However, in contrast to the nuclear staining of RE cells, patch cells showed cytoplasmic FoxJ1 staining (see Fig. 6E,F). A second unrelated anti-FoxJ1 antibody did not stain the Z4 patch cells, which indicated that the cytoplasmic staining first obtained was unspecific. Further irrelevant cytoplasmic immunofluorescence when using additional antibodies, raised the suspicion that the patch cells bound antibodies in a Fab-independent manner. We therefore examined expression of FcγBP, which is a secreted mucin-like glycoprotein that binds to the Fc portion of IgG (Harada et al., 1997; Kobayashi et al., 2002; Johansson et al., 2009). Anti-FcγBP antibodies reacted specifically with the cells in Z4 patches and surprisingly also a population of nasal RE cells (Fig. 4H,I). Antigen-specific blockade demonstrated that staining was specific to the Fab of the anti-FcγBP antibody (Fig. 4J). The patches in Z4 differed from RE as FcγBP+ cells in RE were smaller. In situ hybridization analysis confirmed that FcγBP+ cells were present in RE of control mice (Fig. 4M,N). In contrast to SUS cells, there was a fraction of patch cells associated with large secreted extracellular globules (Fig. 4K). Moreover, in contrast to RE, Z4 patches did not harbor ciliated respiratory or goblet cells. Altogether, these results strongly indicated that Z4 patches were composed of a metaplastic, possibly hypertrophic, secretory and FcγBP+/RALDH− respiratory cell type. It is likely that this cell type has been overlooked because of its histologic similarity to SUS cells, and it thus may have been referred to as “SUS-like,” “swollen SUS cells,” “globules-associated SUS cells,” or “SUS cells with greatly expanded cytoplasm” (Genter and Ali, 1998; Grubb et al., 2007; Kondo et al., 2009; Renne et al., 2009; Herrick et al., 2017). In addition, Jang et al. (2003) have reported a similar cell type that appears in the most ventrolateral OE following methyl bromide injury. Based on electron microscopy, the authors defined the cell as “similar to gland cells, having secretory vesicles in the cytoplasm.” The replacement of RALDH1+ SUS cells with RALDH−/FcγBP+ respiratory cells in OMP-Cyp transgenics conceivably lowered RA bioavailability even further and contributed to the maintenance of the metaplasia in Z4 once CYP26B1+ OSNs had disappeared (Fig. 3F).
Figure 3.
Increased expression of CYP26B1 results in patches without OSNs in Z4. A, B, Immunofluorescence for OMP (green) and STMN1 (magenta) on hemisections of OE in control and OMP-Cyp mice, is shown. Dashed line represents the Z1–Z2 border. Note the relative decrease of OMP in Z4 (marked by yellow lines) in OMP-Cyp mice (B) compared with controls (A). C–L, Analyses of OE, metaplastic patches in Z4 (Z4 patch), and RE of OMP-Cyp mice are shown. Where relevant, the shape of one Z4 patch cell is outlined by a dotted yellow line. C, D, Hematoxylin and eosin (H&E) staining shows the large volume of patch cells. E, F, Shows that patch cells express SOX2 (magenta) but not CYP26B1 (green). G, Double-immunofluorescence analysis of the OE/RE border (vertical line) shows that both SUS cells in OE and RE cells express SOX2 (magenta) and that both HBCs in OE and basal cells in RE express Krt5 (green). H, Analyses for OMP (yellow) in OE and FoxJ1 (magenta) demarcate the border between OE and RE cells with FoxJ1+ nuclei (arrowheads). I, J, Shows analyses for cilia markers Ac-Tub (green) and AC3 (magenta). Patch cells do not stain for AC3 or Ac-Tub, while cilia of OSNs in OE are positive for both (note that green plus magenta shows as white). K, L, SUS cells and patch cells both have CD36 (green) positive microvilli, while SUS cells but not Z4 patch cells express RALDH1 (magenta). Dashed lines in C–L indicate basal lamina. Scale bars: A, 300 µm; C–L, 12.5 µm.
Figure 6.
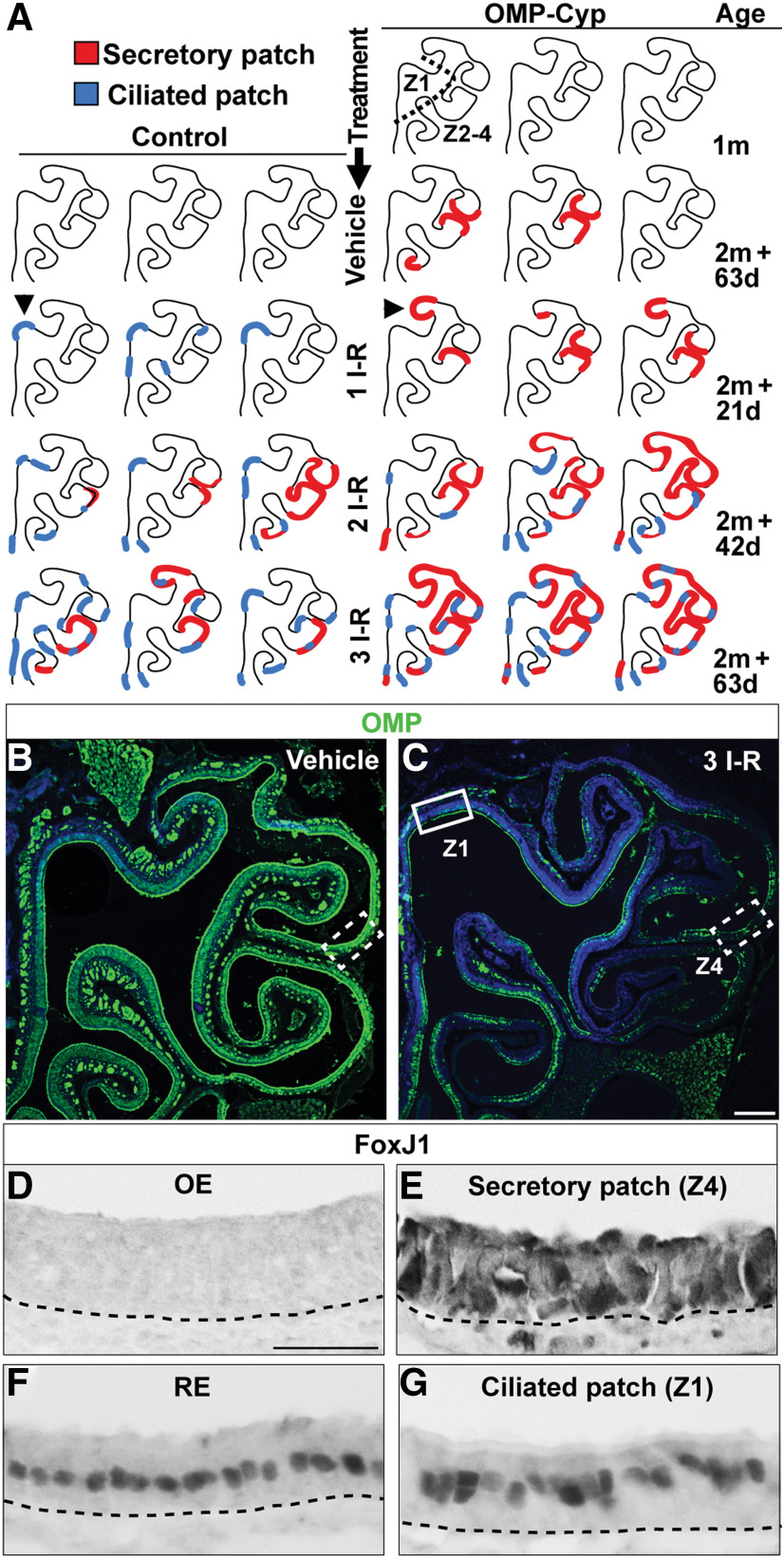
Two types of metaplastic patches appear after methimazole injury or high-level transgenic expression of CYP26B1. A, Schematic illustrations of OE hemisections representing three OMP-Cyp mice at 1 month of age (1m). Also illustrated are OE hemisections of control and OMP-Cyp mice after one, two, and three I–R cycles according to the schedule outlined in Figure 4O. Red lines indicate patches with metaplastic secretory respiratory cells (secretory patch), whereas blue lines indicate patches with metaplastic ciliated respiratory cells (ciliated patch). Arrowheads indicate examples of OE regions without flanking RE where isolated patches within otherwise normal OE histologically were identified. B, C, Distribution of OMP+ OSNs (green) in control mice after vehicle (B) and after three I–R cycles (C) is shown. Nuclei are in blue. Indicated Z1 region (solid box) and Z4 region (dashed lined boxes) correspond to the regions analyzed in D, E, and G. D–G, Show FoxJ1 immunostaining in Z4 (D), a patch of secretory respiratory metaplasia in Z4 (E), RE (F), and a patch of ciliated respiratory metaplasia in Z1 patch (G). Scale bars: C, 300; D, 12.5 µm.
RA-dependent OE regeneration shows zonal differences
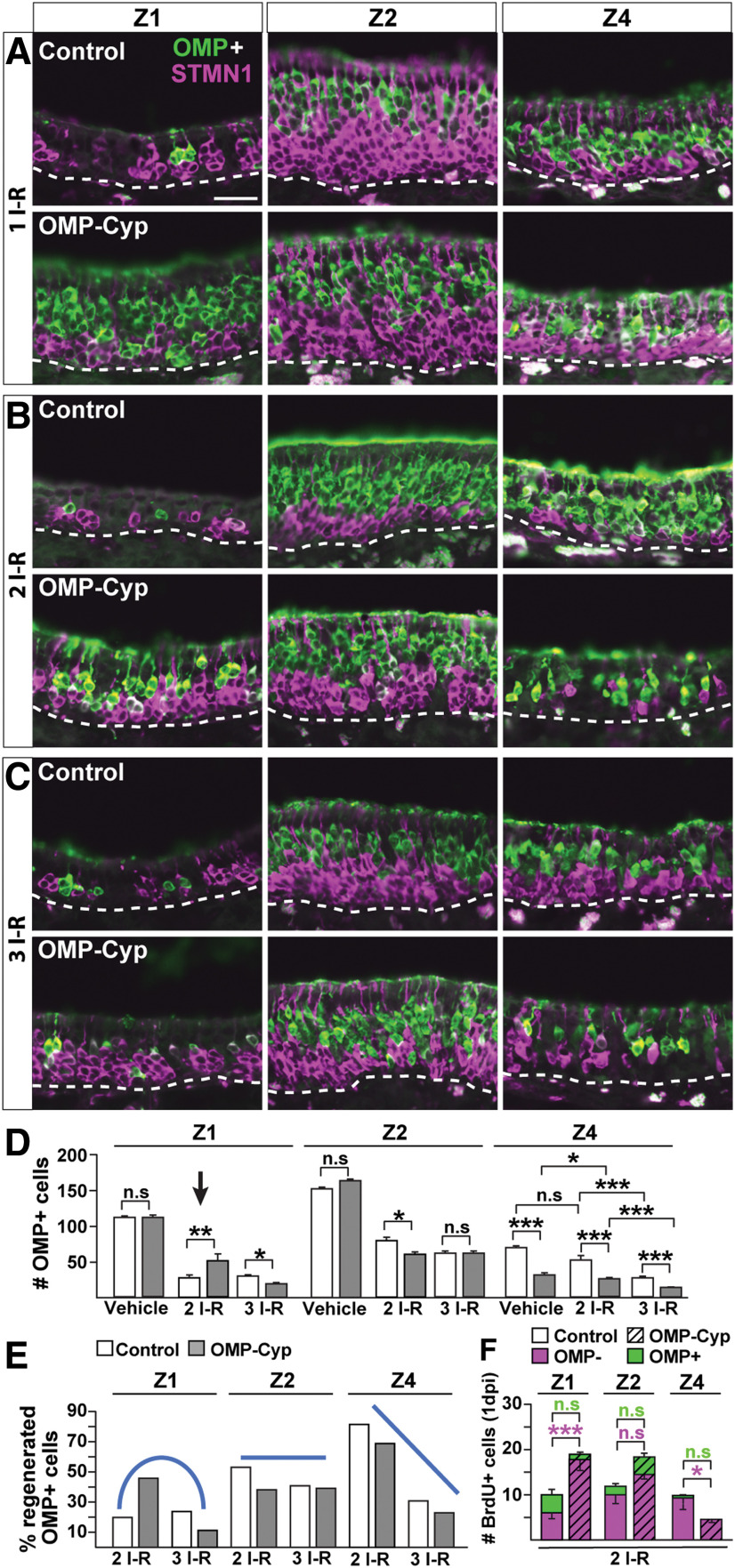
As Z4 normally showed the highest proliferation, fastest transit time, and fastest cell turnover, one plausible explanation for the metaplasia was that Z4 inherently might be most vulnerable to exhaustion of the self-renewal capacity of HBCs. We hypothesized that several episodes of methimazole-induced OE injury in control mice would cause repeated HBC activation and phenocopy the Z4 phenotype in OMP-Cyp mice. Activated HBCs regenerate the entire OE in <30 d after intraperitoneal injection of methimazole (Bergman et al., 2002). To experimentally challenge the renewal capacity of OE, 2-month-old mice were injected with methimazole or vehicle every third week. Mice maximally received three methimazole doses, according to schedules in Figure 4O. Double OMP/STMN1 immunofluorescence analysis 21 d after one, two, or three methimazole injury–regeneration (I–R) cycles showed an increased ratio of immature to mature OSNs in all zones of control and OMP-Cyp mice (compare Fig. 2D,E, 5A–C). Surprisingly, after up to two I–R cycles, there were more immature STMN1+ and mature OMP+ OSNs in Z1 of OMP-Cyp mice than in controls (Fig. 5A,B, compare control and OMP-Cyp). Quantification after two I–R cycles showed that the number of OMP+ cells in Z1 in OMP-Cyp mice was twice as high as that of controls (Fig. 5D, arrow). This suggested that a reduction in RA bioavailability actually improved OSN regeneration in Z1, in contrast to the situation in Z4 where regeneration was impaired (Fig. 5A,B). However, the positive effect of CYP26B1+ OSNs on neurogenesis in Z1 could not sustainably counterbalance the negative effect of repeated injuries. Accordingly, there was an abrupt change from increased to decreased recovery of Z1 OSNs between the second and third I–R cycle in OMP-Cyp mice (Fig. 5B,C). In Z1, OMP+ OSNs were increased 40–50% after two cycles, followed by a 40–50% decrease after three cycles (Fig. 5D). Interestingly, the regenerative capacity of Z2 was quite resistant to repeated injury both in control and OMP-Cyp mice (Fig. 5A–D).
Figure 5.
RA-dependent OE regeneration shows zonal differences. A–C, Double immunofluorescence for OMP (green) and STMN1 (magenta) in Z1, Z2, and Z4 of OE in littermate control and OMP-Cyp mice 21 d after methimazole-induced one (A), two (B), and three (C) I–R cycles. Scale bar, A, 25 µm. D, Quantification of OMP+ cells in Z1, Z2, and Z4 of littermate controls (open bars) and OMP-Cyp mice (gray bars) after vehicle, two I–R, and three I–R cycles is shown. E, Percentage of regenerated OMP+ cells after two I–R and three I–R cycles. The percentage was calculated relative to the number of OMP+ OSNs per 300 μm OE length in vehicle-treated control and OMP-Cyp mice, respectively. Blue lines indicate the trends of regenerative capacity in Z1, Z2, and Z4 in OMP-Cyp mice compared with control mice following two I–R and three I–R cycles, respectively. F, Quantification of the number of BrdU+/OMP− (magenta) and BrdU+/OMP+ (green) double-positive cells in OMP-Cyp mice (striped bars) and littermate controls (open bars) after two I–R cycles at 1 dpi of BrdU. Graphs show result from N = 3 mice and n = 8 hemisections per mouse. Error bars represent the mean ± SEM. Two-tailed Student's t test: n.s. nonsignificant, *p < 0.05, **p < 0.01, ***p < 0.001.
In Z4 of OMP-Cyp mice, there was already only 42% OMP+ cells per unit length compared with control mice before the first methimazole injection (Fig. 5D). The regenerative response of Z4 was different from that of Z1 and Z2. After two and three I–R cycles, the relative numbers of Z4 OSNs were further reduced by ∼30% and ∼60%, respectively, compared with control mice (Fig. 5D). Based on these results, we categorized zonal differences in OE regenerative capacity when RA is low as “increased then decreased” for Z1, “indifferent” for Z2, and “decreased” for Z4 (Fig. 5E). To relate these zonal differences in the effect of high CYP26B1 on neurogenesis to differences in proliferation, we quantified the progeny of acutely BrdU-labeled OMP− cells at 3 weeks after the second injury–repair cycle (Fig. 4O). This experiment showed that the effect of increased CYP26B1 on OSN-regenerated numbers (Fig. 5D) was mirrored by the effect on progenitor cell proliferation, which also varied in an “increased,” “indifferent,” and “decreased” pattern for Z1, Z2, and Z4, respectively (Fig. 5F).
Inverse effects of CYP26B1 on two different types of respiratory metaplasia
Fascinatingly, the secretory respiratory cell metaplasia that appeared “spontaneously” in Z4 in OMP-Cyp mice between 1 and 4 months of age (Figs. 3, 4) also appeared in Z4 of 4-month-old control mice following more than two I–R cycles [Fig. 6A–D (marked red in A)]. The surface area of metaplastic patches after two I–R cycles in controls was similar to that of vehicle-treated 4-month-old OMP-Cyp mice (Fig. 6A). Moreover, increased CYP26B1 aggravated the expansion of the patches with each I–R cycle. These results suggested that reduced RA bioavailability and repeated injuries negatively affected Z4 regeneration in a similar way.
Close-up examination of OE after a single I–R cycle revealed an additional type of metaplasia in Z1 of controls, but not in OMP-Cyp mice [Fig. 6A–C,F,G (marked blue in A)]. The cell type in these “Z1 patches” had apically located FoxJ1+ nuclei (Fig. 6G) and Ac-Tub+/AC3− cilia (Fig. 7A). In similarity to respiratory cells, the patch cells in Z1 also expressed Keratin 19 (Krt19; Fig. 7B,D). The cell morphology and antibody staining pattern in the Z1 type of patch were thus identical to ciliated respiratory cells, but, in contrast to RE, this type of metaplasia did not harbor Reg3γ+ or FcγBP+ respiratory cells (Figs. 4L, 7C,E). In OE of control mice, ciliated respiratory metaplasia initially appeared in Z1, but then in Z2-4 after repeated injury (Fig. 6A, blue). Both ciliated and secretory epithelial patches appeared at the OE–RE boundary and as isolated patches within otherwise normal OE. The isolated patches were most readily distinguished by histology in OE regions that are devoid of flanking RE, such as the septum and dorsal roof of the nasal cavity (Fig. 6A, arrowheads). Moreover, both types of metaplastic patches consisted either solely of metaplastic cells or of a mixture of metaplastic and OE cells (Fig. 7, compare F′, G).
Figure 7.
Metaplastic cells in Z1 are ciliated respiratory cells. A, Ac-Tub (green) and AC3 (magenta) analyses of a Z1 patch showing that Ac-Tub cilia (arrowheads) staining of the metaplastic cells does not overlap with OSN-specific AC3+ cilia staining. B, D, Krt19 and Krt5 immunofluorescence shows that both Z1 patch cells (B) and RE cells (D) express Krt19. C, E, Analysis for Reg3γ (green) and RALDH1 (magenta) shows that the Z1 patch cell type is RALDH1+/Reg3γ− (C), while both RALDH1+ and Reg3γ+ cells are present in RE (E). F, RALDH1+ ciliated respiratory cells (magenta) and OMP+ OSNs (green) along the dorsal nasal recess (i.e., Z1) of control mice after three I–R cycles. F´, Close-up of a patch with RALDH1+ respiratory cells that lacks OMP+ OSNs. G, A patch that has a mixture of RALDH1+ respiratory cells and OMP+ OSNs. Scale bars: A–E, G, 12.5 µm; F, 200 µm. Nuclei are in blue.
Interestingly, in OMP-Cyp mice the appearance of ciliated respiratory metaplasia was prevented in Z1 following up to two I–R cycles. That the transgenic increase in CYP26B1 inhibited metaplasia in Z1 was in line with the stimulatory effect of CYP26B1 on proliferation and neurogenesis, also for up to two I–R cycles (Fig. 4A–F). Furthermore, in similarity to cells in the RE, and in contrast to the RALDH− SUS cells in Z1, the respiratory cells appearing in Z1 were RALDH1+ (Figs. 1B,C, 7C–F). Together, repeated I–R cycles led to intriguing inverse zone-specific niche changes in RA metabolism in such a way that RALDH− SUS cells in Z1 were replaced by one type of respiratory cell that was RALDH+, while RALDH+ SUS cells in Z4 were replaced by another type of respiratory cell that instead was RALDH−.
CYP26B1-induced HBC activation resembles injury-induced HBC activation
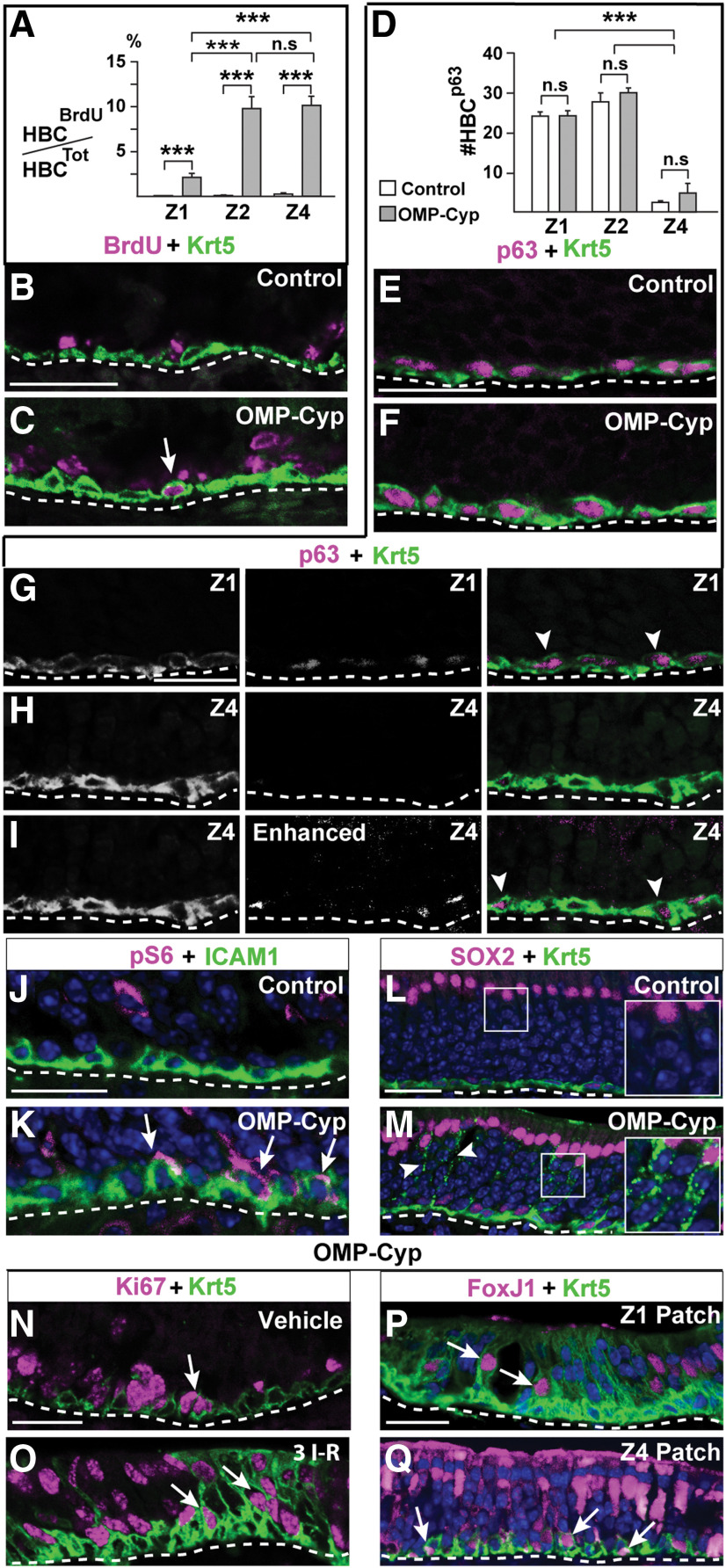
To address whether reduced RA bioavailability activated dormant HBCs to enter the cell cycle, the number of cells double positive for BrdU and the HBC marker Krt5 was determined. As expected the proportions of BrdU+/Krt5+ double-positive cells in Z1, Z2, and Z4 of control mice at 1 dpi of BrdU were very low: 0%, 0.2%, and 0.5%, respectively (Fig. 8A,B). In OMP-Cyp littermates on the other hand, BrdU+/Krt5+ cells in Z1, Z2, and Z4 were 2.5%, 10%, and 10%, respectively (Fig. 8A,C). This was interesting since it showed that increased CYP26B1 in OSNs activated dormant HBCs in the niche to proliferate, and the effect was most pronounced in Z2-4.
Figure 8.
Increased CYP26B1 in OSNs activates HBCs. A, Quantification of the percentage of BrdU+ HBCs in Z1, Z2, and Z4 of in 5-month-old control (open bars) and OMP-Cyp (gray bars) mice. B, C, Double immunofluorescence for BrdU (magenta) and Krt5 (green). Arrow in C indicates a double-positive HBC. D, Quantification of the number of p63+ HBCs in Z1, Z2, and Z4 of OE. E, F, Krt5 and p63 (magenta) in Z1 of control and OMP-Cyp mice. G, H, Krt5 (green) and p63 (magenta) double-positive cells in Z1 (G, arrowheads) and Krt5+ cells with p63 immunofluorescence below detection threshold in Z4 (H) of OE. I, Enhanced exposure of the p63 signal in H showing that p63 immunofluorescence is present in Z4 HBCs (arrowhead), but with a significantly lower intensity compared with Z1 HBCs. J, K, pS6 (magenta) and ICAM1 (green) analyses of control (J) and OMP-Cyp (K) mice. Arrows in K show the presence of double-positive HBCs. Note that overlap in white is highly localized as ICAM and pS6 are in different cellular compartments. L, M, SOX2 (magenta) and Krt5 (green) analysis showing that SUS cells with Krt5+ puncta (arrowheads) are present in OMP-Cyp (M), but not in control mice (L). Magnified inserts have been resampled to increase resolution. N, O, Ki67 (magenta) and Krt5 (green) in OMP-Cyp mice treated either with vehicle (K) or after three I–R cycles. Arrows indicate double-positive HBCs. P, Q, FoxJ1 (magenta) and Krt5 (green) fluorescence in a Z1 (P) and a Z4 (Q) patch after three I–R cycles in OMP-Cyp mice. Arrows indicate double-positive HBCs. Scale bars, 25 μm. n.s. nonsignificant, ***p < 0.001.
It is known that the loss of p63 activates HBCs in a gene dosage-dependent manner (Schnittke et al., 2015). Interestingly, examination of p63 in 4-month-old mice showed that Z4 had fewer HBCs that expressed p63 to a high level, compared with Z1 and Z2, in control as well as in OMP-Cyp mice (Fig. 8D–I). Thus, a high fraction of HBCs with p63 virtually below detection threshold coincided with sensitivity for exhaustion of HBCs in Z4 and the appearance of secretory respiratory metaplasia. The number of p63+ HBCs did not differ significantly between control and OMP-Cyp mice (Fig. 8D). This indicated that CYP26B1 did not activate HBCs via p63 downregulation.
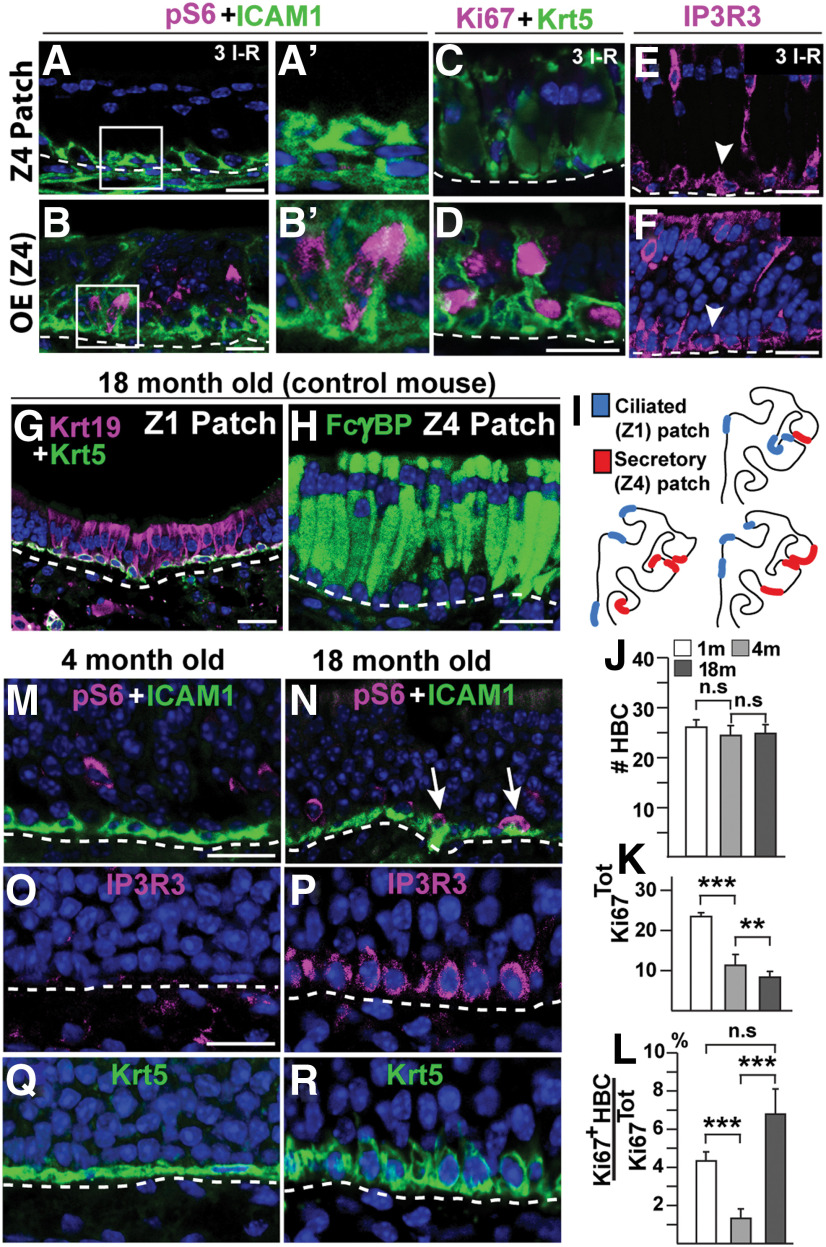
A characteristic of injury-induced HBC activation is a transient increase in mTORC1 signaling, which can be assessed by phosphorylation of ribosome S6 (pS6; Gadye et al., 2017; Haller et al., 2017). Since anti-pS6 and anti-Krt5 antibodies were raised in the same species, we used ICAM1 as an HBC marker. In contrast to Krt5, ICAM1 selectively stain the basal HBC membrane surface juxtaposed to basal lamina (Carter et al., 2004). Immunofluorescence demonstrated double-positive pS6+/ICAM1+ HBCs in OMP-Cyp mice, but not in control mice (Fig. 8J,K).
Signs of HBC transdifferentiation
Close-up examination of OMP-Cyp OE showed that the cytoplasm of SOX2+ SUS cells contained Krt5+ puncta that were not found in controls (Fig. 8L,M). Krt5 could persist for some time during the differentiation of an HBC into a Krt5-negative cell (Xie et al., 2013; Chen et al., 2017; Child et al., 2018). Moreover, recent results showed that Krt5+ HBCs can differentiate directly to SUS cells without cell division (Fletcher et al., 2017). Consequently, the Krt5+ SUS cells were likely newly generated as a result of the increased HBC activation and OSN turnover in OMP-Cyp mice. In line with such reasoning, a high percentage (∼20%) of Krt5+ cells was not confined to the basal lamina at an early time point (6 d) after the start of the third I–R cycle (Fig. 8N,O). A fraction (∼13%) of these “nonbasal” Krt5+ cells were not dormant as these were positive for Ki67, which labels cells that are no longer in G0 (Fig. 8O). The relatively high percentage of Ki67+/Krt5+cells leaving the basal lamina during acute methimazole-induced regeneration, compared with when CYP26B1 is chronically high in OMP-Cyp mice, likely was a consequence of methimazole inducing a synchronous wave of activation of HBCs 6 d earlier. Moreover, at this early time point of methimazole-induced regeneration, Krt5 could be detected in quite a few ciliated respiratory patch cells as well as in some Z4 secretory respiratory patch cells (Fig. 8P,Q). Conceivably, some Krt5+ HBCs might just have differentiated to ciliated and secretory respiratory cells in a manner similar to the direct transdifferentiation, as has been described for SUS cells (Fletcher et al., 2017).
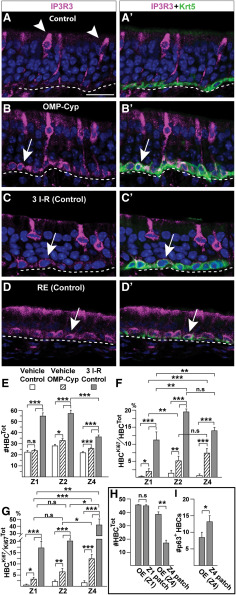
IP3R3 marks a state of activation of HBCs and RE basal cells
To analyze all OE niche cells, we included IP3R3, which marks MVCs (Hegg et al., 2010; Jia et al., 2013). Surprisingly, in addition to MVCs, HBCs in uninjured OMP-Cyp mice were IP3R3+ while HBCs in control mice were negative, as expected (Fig. 9A,B). Moreover, HBCs in control mice expressed IP3R3 6 d after methimazole treatment (Fig. 9C). Unlike the few scattered HBCs positive for pS6 and BrdU, IP3R3 was in most if not all HBCs throughout OE. This uniform induction of IP3R3 expression in HBCs correlated with a change from a flat to a rounded cellular morphology (Fig. 9, compare A′,B′,C′, Krt5 immunofluorescence). Intriguingly, Krt5+ basal cells in the RE of untreated control mice expressed IP3R3 constitutively to a level that was comparable to that of HBCs in OMP-Cyp mice and in injured controls (Fig. 8D). Thus, following activation, HBCs resembled RE basal cells both with regard to high IP3R3 expression and the potential to produce some RE cell types.
Figure 9.
Activating stimuli induce all HBCs to express IP3R3 and change morphology. A–D′, Double-immunofluorescence analyses for Krt5 (green) and IP3R3 (magenta). A, A′, IP3R3 in apically located MVCs (arrowheads) in controls. Note the lack of IP3R3 in Krt5+ HBCs with a flat cellular morphology at basal lamina. B, B′, IP3R3 and a rounded morphology characterize Krt5+ HBCs in OMP-Cyp mice (arrow). C, C′, IP3R3 is turned on in HBCs and HBCs also change to a rounded morphology in control mice after methimazole injury (6 d after three I–R cycles). D, D′, Krt5+ basal cells in RE of control mice constitutively express IP3R3 (arrow). E–I, Quantification of cell numbers in Z1, Z2, Z4, and metaplastic patches in vehicle-treated controls (open bars), OMP-Cyp mice (striped bars), and control mice 6 d after three I–R cycles (gray bars). E, The number of Krt5+ HBCs. F, G, The percentage of Ki67+ HBCs (HBCKi67) of the total number of HBCs (HBCTot; F), and the percentage of Ki67+ HBCs (HBCKi67) of the total number Ki67+ cells (Ki67Tot; G). H, The number of Krt5+ HBCs in Z1 patch, Z4 patch, and adjacent OE. I, The number of p63+ HBCs in Z4 patches and adjacent Z4 OE. Graphs show N = 3 mice and n = 8 hemisections per mouse. Error bars represent the mean ± SEM. Two-tailed Student's t test: n.s. nonsignificant, *p < 0.05, **p < 0.01, ***p < 0.001. Scale bar, A, 12.5 µm.
Direct contribution of HBCs to regeneration is more common in Z4 than in Z1 and Z2
To compare the relative impact of chronically low RA to that of three I–R cycles, while also taking zonal differences in HBC numbers and their activation state into account, we quantified the number of Krt5+ HBCs (HBCTot) and Ki67+/Krt5+ nonquiescent HBCs (HBCKi67). In OMP-Cyp mice, the HBCTot number was as in controls for Z1, while HBCTot was increased ∼15% for Z2 and Z4 (Fig. 9E). Six days after the last methimazole injury in control mice, HBCTot was increased ∼30–50% in all zones (Fig. 9E). These results indicated that HBC self-renewal in Z1 was less influenced by CYP26B1 than by injury, while both conditions increased HBC self-renewal in Z2-4. As expected, the fractions of nonquiescent HBCs (HBCKi67/HBCTot) were very low in control mice (Z1, 0.4%; Z2, 1.2%; and Z4, 0.7%; Fig. 9F). Following three I–R cycles, the nonquiescent HBCs in Z1, Z2, and Z4 increased to 11.6%, 19.7%, and 14.3%, respectively. The corresponding numbers in OMP-Cyp mice were 2.0%, 5.6%, and 7.3%, respectively (Fig. 9F). Thus, nonquiescent HBCs were consistently lowest in Z1 and similar between Z2 and Z4. To get an estimate of HBCs in the cell cycle (HBCKi67) relative to the total number of cells in the cell cycle (mainly GBCs at these time points), we determined the HBCKi67/CellKi67Tot ratios for Z1, Z2, and Z4 (Fig. 9G). This result indicated that the contribution of HBCs to the total pool of cycling cells, in response to CYP26B1 transgenic expression or repeated injury was greater in Z4 compared with Z1 and Z2. Thus, Z4 generated fewer of the GBCs that also could contribute to injury-induced regeneration.
Analyses of the two types of metaplasia showed that the number of Krt5+ cells was reduced in secretory, but not in ciliated respiratory metaplastic patches, compared with the number of Krt5+ HBCs in OE flanking these patches (Fig. 9H). Moreover, Krt5+ cells remaining in the secretory respiratory metaplastic patches were quiescent (i.e., they expressed a relatively high level of p63; Fig. 9I), were pS6− and Ki67− compared with adjacent Z4 OE (Fig. 10A–D). However, the HBCs in secretory patches showed high levels of IP3R3 (Fig. 10E,F).
Figure 10.
Quiescent HBCs in Z4 patches and HBC activation in old mice. A–F, Immunofluorescence analyses of control mice 6 d after three I–R cycles. A–B′, pS6 (magenta) and ICAM1 (green) analysis of a Z4 patch (A, A′) and adjacent Z4 OE (B, B′). Boxed regions in A and B are magnified and resampled in A′ and B′. C, D, Lack of Ki67 (magenta) in Krt5+ (green) HBCs in a Z4 patch (C) while Ki67+/Krt5+ double-positive HBCs are found in adjacent Z4 OE (D). E, F, IP3R3+ HBCs (arrowheads) in both a Z4 patch and Z4 OE after three I–R cycles. G, Krt19 (magenta) in metaplastic ciliated respiratory cells in a Z1 patch of an 18-month-old control mouse. H, Immunofluorescence for FcγBP (green) in metaplastic secretory cells in a Z4 patch of an 18-month-old control mouse. I, Schematic illustration of the distribution of patches in OE hemisections of three different 18-month-old control mice is shown. J–L, Quantification of cell numbers in OE of 1- (open bars), 4- (gray bars), and 18-month-old (black bars) control mice is shown. J, The total number of Krt5+ HBCs. K, L, The total number of Ki67+ cells (K) and the percentage of Ki67+ HBCs of the total number of Ki67+ cells (Ki67Tot; L). M, N, An increase in the number of ICAM1+ HBCs (green) that are also positive for pS6 (magenta, arrows) in 4-month-old compared with 18-month-old controls. O–R, Induction of IP3R3 (magenta) in HBCs and change from a flat to a rounded cellular morphology of HBCs in control mice between 4 months (O, Q) and 18 months (P, R) of age. Graphs show data from N = 3 mice and n = 8 hemisections per mouse. Error bars represent the mean ± SEM. Two-tailed Student's t test: n.s. nonsignificant, **p < 0.01, ***p < 0.001. Scale bars, 25 µm.
Aging of OE is akin to repeated injury of OE
The similarities between the injury-induced changes in OE identified in this study and the aging-associated changes noted in the literature (Naessen, 1971; Nakashima et al., 1984; Loo et al., 1996; Genter and Ali, 1998; Kondo et al., 2009) motivated analyses of OE in old mice for the expression of ciliated and secretory respiratory metaplastic cell markers Krt19 and FcγBP, respectively. Interestingly, age-related patches in 18-month-old normal mice indeed resembled patches after more than one I–R cycle in 4-month-old control mice with respect to immunoreactivity, morphology, and zonal distribution (Fig. 10G–I). Quantification of HBCs and nonquiescent OE cells (Ki67Tot) was done at 1, 4, and 18 months of age (Fig. 10K). In accordance with previous studies, the total number of nonquiescent OE cells decreased between 4 and 18 months while the number HBCs remained the same (Brann and Firestein, 2014). Under normal homeostatic conditions, it was estimated that as few as 2.4% of S-phase cells are HBCs, while the remaining 97.6% are GBCs (Huard and Schwob, 1995). Interestingly, our experiments showed that the ratio of nonquiescent HBCs to the total number of nonquiescent cells (i.e., Ki67+ HBC/Ki67Tot) increased between 4 and 18 months (Fig. 10L). Thus, aging caused a decline in GBC proliferation, which appeared to correlate with an increase in the activation of HBCs. Indeed, immunofluorescence showed that pS6+/ICAM1+ double-positive HBCs were detected in 18-month-old mice, but not in 4-month-old mice (Fig. 10M,N). Moreover, similar to the activation of HBCs by CYP26B1 and injury, IP3R3 was induced at 18 months of age in HBCs throughout OE, which again correlated with a change from a flat to a rounded HBC morphology (Fig. 10O–R). These results in summary indicated that HBCs were released from dormancy with age and that there was a strong correlation between increased activation of HBCs and the appearance of age-related metaplasia of the OE.
Discussion
We herein show that SUS cells in Z2-4 begin to express RALDH1 after birth. Another characteristic of OE in adult mice is odor/air-induced Z1high-Z4low CYP26B1 in OSNs (Login et al., 2015a,b). Moreover, proliferation in OE is stimulated by increased CYP26B1 expression in OSNs, while it is inhibited by nasal administration of 13-cis-RA or a CYP26B1 inhibitor, liarozole. These results indicate that the activities of CYP26B1 and RALDH1 are important regulators of the adult stem cell niche.
Previous studies of the OE niche in adult mice have led to seemingly contradictory results regarding the question of whether HBCs are activated or not during normal tissue homeostasis (Leung et al., 2007; Iwai et al., 2008; Schwob et al., 2017). We find that HBCs, in addition to being activated by injury, are activated by increased CYP26B1 in OSNs and by old age. Thus, our finding that HBCs are activated specifically in old mice indicates a role for this stem cell also during normal life. The activation correlates with induced IP3R3 in HBCs and that the effect of CYP26B1 on neurogenesis in OE differs along the dorsomedial (Z1)–ventrolateral (Z4) axis. HBCs located in Z4 express low levels of p63 and decrease in number when activated continuously. Fewer HBCs in Z4 correlates with respiratory metaplasia consisting of a novel cell type, which we identify as a secretory respiratory cell type that expresses FcγBP. In Z1, CYP26B1 stimulates neurogenesis and inhibits a distinct type of injury-induced and age-related metaplasia with a ciliated respiratory cell type.
Recent results show that HBCs are not activated even when the GBC population is experimentally exhausted by continuous high cell turnover as a result of OMP promoter-driven diphtheria toxin-induced death of OSNs (Child et al., 2018). In contrast, we find that HBCs are activated by OMP promoter-driven CYP26B1 expression, which also results in increased cell turnover and OSN death (Login et al., 2015b). A straightforward interpretation of our result is that HBCs are non-cell-autonomously activated by a CYP26B1-dependent reduction of RA bioavailability. With similar reasoning, injury that causes death of RA-producing RALDH1+ SUS cells should also lead to reduced RA, which in turn would signal activation of HBCs postinjury. HBCs are indeed activated when SUS cells are genetically ablated (Herrick et al., 2017). By inference, it was suggested that SUS cells, via the Notch ligand Jagged1, normally serve to maintain Notch+ HBCs in a quiescent state via upregulation of p63. We do not find p63 levels to be altered in HBCs by increased CYP26B1. Thus, CYP26B1 activates HBCs independently or downstream of p63.
We show that CYP26B1, injury, or old age results in the induction of IP3R3 in HBCs. Interestingly, IP3R3-deficient mice have fewer HBCs and impaired recovery following toxin-induced OE damage but not following bulbectomy, which selectively kills OSNs (Jia et al., 2013; Jia and Hegg, 2015). The function of IP3R3 in OE regeneration has been attributed to IP3R3+ MVCs. However, in another study it was shown that methimazole-induced OE regeneration was not effected by selective MVC ablation (Weng et al., 2016). These seemingly contradictory results regarding IP3R3 can be explained by our finding that IP3R3 can function cell autonomously in HBCs when induced by CYP26B1, injury, or old age.
In contrast to the scattered distribution of nonquiescent HBCs is the induced IP3R3 in virtually all HBCs. We find that the uniform induction of IP3R3 correlates with flat HBCs taking on a rounded morphology. IP3R3 signaling may therefore play a more lasting role during an activation process. Induction of IP3R3 likely is associated with the injury-induced and sequential states of HBC activation recently defined by Gadye et al. (2017). By single-cell RNA sequencing, it was shown that HBC activation includes transient and heterogeneous states during which multiple cell fates are specified, including renewed stem cells and committed differentiating progenitors. During this early window of HBC activation, genes with function in barrier formation and proliferation are induced. The change to a rounded morphology is compatible with the preparation for cell division, and IP3R3 regulates Ca2+ transfer from the endoplasmic reticulum to mitochondria, which promotes mitochondrial respiration required for self-renewal and differentiation (Filadi et al., 2018). One possible function of IP3R3 could be to “prime” HBCs to respond to IP3-dependent signaling activated during inflammation and/or tissue repair. In this regard, it is interesting that EGF receptor signaling, which uses IP3 as one second messenger, is associated with an early HBC activation state, as defined by Gadye et al. (2017) and as shown by Getchell et al. (2000), which induces HBC proliferation when transgenically activated in mice.
Extensive activation of HBCs in ventrolateral OE by continuous high CYP26B1 expression, repeated injury, or old age gives rise to metaplasia with a previously unspecified RE cell type. This RE cell type is associated with extracellular globules and produces FcγBP, which is a large secreted mucin-like glycoprotein implicated in mucus barrier function (Harada et al., 1997; Kobayashi et al., 2002; Johansson et al., 2009). Mucus accumulation in airways is a protective reaction to inflammation and injury. FcγBP+ cells first appear in highly proliferative Z4. Thus, a history of many cell divisions and replicative senescence may underlie the appearance of metaplastic FcγBP+ cells. The finding that few and dormant HBCs remain in the metaplastic patches supports this hypothesis. However, we find that HBC increased proliferation equally in Z2 and Z4. Thus, many HBC cell divisions per se do not fully explain why FcγBP+ metaplasia first appears in Z4. The results herein suggest that rapid differentiation, high cell turnover, and minimum lineage amplification via dividing GBCs distinguishes Z4 from Z2. Moreover, in Z4 we find a relatively low level of p63, which cell-autonomously represses HBC differentiation (Fletcher et al., 2011). That the Z4 stem cell niche first becomes exhausted may thus be due to the combination of a high HBC proliferation and low p63 that biases activated Z4 HBCs toward differentiating rather than symmetric renewing divisions.
In contrast to the response in Z4, a distinct type of age-related metaplasia with ciliated respiratory cells already appears in Z1 after one I–R cycle in normal mice. The regenerative capacity of Z1 HBCs is thus lower than in other zones. This response is consistent with previous results from examinations of histologic sections of injury-induced, as well as age-related, lesions showing that ciliated respiratory-like cells first appear dorsomedially (Loo et al., 1996; Genter and Ali, 1998; Kondo et al., 2009). Genetic cell linage tracing experiments have shown that ciliated respiratory cells in OE are derived from HBC cells (Xie et al., 2013; Child et al., 2018). We show that the ciliated respiratory cell type is FoxJ1+ and that these metaplastic patches do not harbor other respiratory cell types.
A striking observation is that neurogenesis and regeneration proceed more efficiently in Z1 at subphysiological RA levels, which is inverse to the situation in Z4. Specifically, in Z1, but not in Z4, increased CYP26B1 in OSNs increases neurogenesis in OSNs and inhibits the metaplastic response following initial I–R cycles. We find that fewer Z1 HBCs are activated to proliferate in response to injury and show a low contribution to the total number of proliferating cells. Instead, neurogenesis and regeneration of Z1 appear to rely more on an RA-sensitive phase that comprises amplification of OSN numbers through GBCs rather than HBCs.
Our results suggest that cell types derived from HBCs (i.e., RALDH1+ SUS cells and CYP26B1+ OSNs) largely regulate RA bioavailability in the adult stem cell niche. RA-regulated homeostasis in OE is thus akin to that of some other stem cell niches for which feedback signals from differentiated progeny to their parent stem cells are key homeostatic niche regulators (Hsu and Fuchs, 2012). Epithelial metaplasia is generally considered as an adaptive process resulting from the transformation of one type of epithelium into another type to mitigate the effects of chronic injury (Herfs et al., 2009). An interesting question then becomes how a metaplastic state is maintained. We show that RALDH− SUS cells in Z1 are replaced with RALDH+-ciliated RE cells, while the scenario in Z4 is the reverse, with a change from RALDH+ SUS to RALDH− secretory RE cells. These cell type replacements, and thus switch in local RA production, may serve to maintain metaplasia and thereby ensure a barrier when the OE stem cell niche is exhausted. We here have found that Krt5+ RE basal cells also express IP3R3, although in a constitutive manner. The state of HBCs after activation thus bears resemblance to that of RE stem cells with regard to IP3R3 expression and the capacity to generate RE cell types. Thus, the consequences to OSNs of, for example, repeated toxic exposures, infectious insults, or chronic inflammatory rhinitis may over time interfere with inter-stem cell niche regulation. Disturbed inter-stem cell niche regulation may lead to reprogramming of HBCs toward respiratory cell fates or changed lineage segregation of Krt5+ basal cell subpopulations, the latter of which is shown to occur for different RE lineages following injury (Pardo-Saganta et al., 2015).
Our study demonstrates the role of the neural activity-regulated gene CYP26B1 as an important spatial stem cell regulator in a neuroepithelium where changes in RA influence age-related tissue pathology. Combined, our data suggest that RA serves to delay olfactory stem cell aging. RA homeostasis in the adult human brain is regulated by CYP26B1 (Stoney et al., 2016). Whether CYP26B1+ neurons regulate dormant stem/progenitor cells after CNS injury or at old age by mechanisms similar to those suggested in the present study is an interesting question for future investigations.
Footnotes
The authors declare no competing financial interests.
This work was supported by the Swedish Brain Foundation (project no. FO20180195). We thank Dr. Sara Wilson for critical reading of the manuscript.
References
- Alenius M, Bohm S (1997) Identification of a novel neural cell adhesion molecule-related gene with a potential role in selective axonal projection. J Biol Chem 272:26083–26086. 10.1074/jbc.272.42.26083 [DOI] [PubMed] [Google Scholar]
- Asson-Batres MA, Zeng MS, Savchenko V, Aderoju A, McKanna J (2003) Vitamin A deficiency leads to increased cell proliferation in olfactory epithelium of mature rats. J Neurobiol 54:539–554. 10.1002/neu.10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman U, Ostergren A, Gustafson AL, Brittebo B (2002) Differential effects of olfactory toxicants on olfactory regeneration. Arch Toxicol 76:104–112. 10.1007/s00204-002-0321-2 [DOI] [PubMed] [Google Scholar]
- Brann JH, Firestein SJ (2014) A lifetime of neurogenesis in the olfactory system. Front Neurosci 8:182. 10.3389/fnins.2014.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittebo EB. (1995) Metabolism-dependent toxicity of methimazole in the olfactory nasal mucosa. Pharmacol Toxicol 76:76–79. 10.1111/j.1600-0773.1995.tb00107.x [DOI] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N, Buettner F, Sommerkamp P, Klimmeck D, Ladel L, Thalheimer FB, Pastor-Flores D, Roma LP, Renders S, Zeisberger P, Przybylla A, Schönberger K, Scognamiglio R, Altamura S, Florian CM, Fawaz M, Vonficht D, Tesio M, Collier P, Pavlinic D, et al. (2017) Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell 169:807–823.e19. 10.1016/j.cell.2017.04.018 [DOI] [PubMed] [Google Scholar]
- Caggiano M, Kauer JS, Hunter DD (1994) Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication-incompetent retrovirus. Neuron 13:339–352. 10.1016/0896-6273(94)90351-4 [DOI] [PubMed] [Google Scholar]
- Carter LA, MacDonald JL, Roskams AJ (2004) Olfactory horizontal basal cells demonstrate a conserved multipotent progenitor phenotype. J Neurosci 24:5670–5683. 10.1523/JNEUROSCI.0330-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Reed RR, Lane AP (2017) Acute inflammation regulates neuroregeneration through the NF-κB pathway in olfactory epithelium. Proc Natl Acad Sci U S A 114:8089–8094. 10.1073/pnas.1620664114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child KM, Herrick DB, Schwob JE, Holbrook EH, Jang W (2018) The neuroregenerative capacity of olfactory stem cells is not limitless: implications for aging. J Neurosci 38:6806–6824. 10.1523/JNEUROSCI.3261-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. (2008) Retinoic acid synthesis and signaling during early organogenesis. Cell 134:921–931. 10.1016/j.cell.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CW, Bowles J, Koopman P (2014) Control of mammalian germ cell entry into meiosis. Mol Cell Endocrinol 382:488–497. 10.1016/j.mce.2013.09.026 [DOI] [PubMed] [Google Scholar]
- Filadi R, Leal NS, Schreiner B, Rossi A, Dentoni G, Pinho CM, Wiehager B, Cieri D, Cali T, Pizzo P, Ankarcrona M (2018) TOM70 sustains cell bioenergetics by promoting IP3R3-mediated ER to mitochondria Ca(2+) transfer. Curr Biol 28:369–382.e6. 10.1016/j.cub.2017.12.047 [DOI] [PubMed] [Google Scholar]
- Fletcher RB, Prasol MS, Estrada J, Baudhuin A, Vranizan K, Choi YG, Ngai J (2011) p63 regulates olfactory stem cell self-renewal and differentiation. Neuron 72:748–759. 10.1016/j.neuron.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, Cole MB, Flores Q, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J (2017) Deconstructing olfactory stem cell trajectories at single-cell resolution. Cell Stem Cell 20:817–830.e8. 10.1016/j.stem.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadye L, Das D, Sanchez MA, Street K, Baudhuin A, Wagner A, Cole MB, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J, Fletcher RB (2017) Injury activates transient olfactory stem cell states with diverse lineage capacities. Cell Stem Cell 21:775–790.e9. 10.1016/j.stem.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genter MB, Ali SF (1998) Age-related susceptibility to 3,3′-iminodipropionitrile-induced olfactory mucosal damage. Neurobiol Aging 19:569–574. 10.1016/S0197-4580(98)00104-3 [DOI] [PubMed] [Google Scholar]
- Getchell TV, Narla RK, Little S, Hyde JF, Getchell ML (2000) Horizontal basal cell proliferation in the olfactory epithelium of transforming growth factor-alpha transgenic mice. Cell Tissue Res 299:185–192. 10.1007/s004419900149 [DOI] [PubMed] [Google Scholar]
- Goodman T, Crandall JE, Nanescu SE, Quadro L, Shearer K, Ross A, McCaffery P (2012) Patterning of retinoic acid signaling and cell proliferation in the hippocampus. Hippocampus 22:2171–2183. 10.1002/hipo.22037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziadei PP, Graziadei GA (1979) Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol 8:1–18. 10.1007/BF01206454 [DOI] [PubMed] [Google Scholar]
- Grubb BR, Rogers TD, Kulaga HM, Burns KA, Wonsetler RL, Reed RR, Ostrowski LE (2007) Olfactory epithelia exhibit progressive functional and morphological defects in CF mice. Am J Physiol Cell Physiol 293:C574–583. 10.1152/ajpcell.00106.2007 [DOI] [PubMed] [Google Scholar]
- Gudas LJ, Wagner JA (2011) Retinoids regulate stem cell differentiation. J Cell Physiol 226:322–330. 10.1002/jcp.22417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussing F, Bohm S (2004) NQO1 activity in the main and the accessory olfactory systems correlates with the zonal topography of projection maps. Eur J Neurosci 19:2511–2518. 10.1111/j.0953-816X.2004.03331.x [DOI] [PubMed] [Google Scholar]
- Haller S, Kapuria S, Riley RR, O'Leary MN, Schreiber KH, Andersen JK, Melov S, Que J, Rando TA, Rock J, Kennedy BK, Rodgers JT, Jasper H (2017) mTORC1 activation during repeated regeneration impairs somatic stem cell maintenance. Cell Stem Cell 21:806–818.e5. 10.1016/j.stem.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Iijima S, Kobayashi K, Yoshida T, Brown WR, Hibi T, Oshima A, Morikawa M (1997) Human IgGFc binding protein (FcgammaBP) in colonic epithelial cells exhibits mucin-like structure. J Biol Chem 272:15232–15241. 10.1074/jbc.272.24.15232 [DOI] [PubMed] [Google Scholar]
- Hegg CC, Jia C, Chick WS, Restrepo D, Hansen A (2010) Microvillous cells expressing IP3 receptor type 3 in the olfactory epithelium of mice. Eur J Neurosci 32:1632–1645. 10.1111/j.1460-9568.2010.07449.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke J, Erhardt W (2004) Speziesspezifische anästesie; Nager. In: Anästesie und analgesie beim klein- und heimtier sowie bei vögeln, reptilien, amphibien und fischen (Erhardt W, Henke J, Haberstroth J, eds), pp 642–658. Stuttgart, Germany: Schattauer. [Google Scholar]
- Herfs M, Hubert P, Delvenne P (2009) Epithelial metaplasia: adult stem cell reprogramming and (pre)neoplastic transformation mediated by inflammation? Trends Mol Med 15:245–253. 10.1016/j.molmed.2009.04.002 [DOI] [PubMed] [Google Scholar]
- Herrick DB, Lin B, Peterson J, Schnittke N, Schwob JE (2017) Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proc Natl Acad Sci USA 114:E5589–E5598. 10.1073/pnas.1701333114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Fuchs E (2012) A family business: stem cell progeny join the niche to regulate homeostasis. Nat Rev Mol Cell Biol 13:103–114. 10.1038/nrm3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard JM, Schwob JE (1995) Cell cycle of globose basal cells in rat olfactory epithelium. Developmental dynamics: an official publication of the. Dev Dyn 203:17–26. 10.1002/aja.1002030103 [DOI] [PubMed] [Google Scholar]
- Isoherranen N, Zhong G (2019) Biochemical and physiological importance of the CYP26 retinoic acid hydroxylases. Pharmacol Ther 204:107400. 10.1016/j.pharmthera.2019.107400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai N, Zhou Z, Roop DR, Behringer RR (2008) Horizontal basal cells are multipotent progenitors in normal and injured adult olfactory epithelium. Stem Cells 26:1298–1306. 10.1634/stemcells.2007-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S, Lie DC, DeCicco KL, Shi Y, DeLuca LM, Gage FH, Evans RM (2006) Retinoic acid is required early during adult neurogenesis in the dentate gyrus. Proc Natl Acad Sci U S A 103:3902–3907. 10.1073/pnas.0511294103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W, Youngentob SL, Schwob JE (2003) Globose basal cells are required for reconstitution of olfactory epithelium after methyl bromide lesion. J Comp Neurol 460:123–140. 10.1002/cne.10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C, Hegg CC (2015) Effect of IP3R3 and NPY on age-related declines in olfactory stem cell proliferation. Neurobiol Aging 36:1045–1056. 10.1016/j.neurobiolaging.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C, Hayoz S, Hutch CR, Iqbal TR, Pooley AE, Hegg CC (2013) An IP3R3- and NPY-expressing microvillous cell mediates tissue homeostasis and regeneration in the mouse olfactory epithelium. PLoS One 8:e58668. 10.1371/journal.pone.0058668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Thomsson KA, Hansson GC (2009) Proteomic analyses of the two mucus layers of the colon barrier reveal that their main component, the Muc2 mucin, is strongly bound to the Fcgbp protein. J Proteome Res 8:3549–3557. 10.1021/pr9002504 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Ogata H, Morikawa M, Iijima S, Harada N, Yoshida T, Brown WR, Inoue N, Hamada Y, Ishii H, Watanabe M, Hibi T (2002) Distribution and partial characterisation of IgG Fc binding protein in various mucin producing cells and body fluids. Gut 51:169–176. 10.1136/gut.51.2.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Watanabe K, Sakamoto T, Suzukawa K, Nibu K, Kaga K, Yamasoba T (2009) Distribution and severity of spontaneous lesions in the neuroepithelium and Bowman's glands in mouse olfactory mucosa: age-related progression. Cell Tissue Res 335:489–503. 10.1007/s00441-008-0739-9 [DOI] [PubMed] [Google Scholar]
- Lee S, Eguchi A, Tsuzuki S, Matsumura S, Inoue K, Iwanaga T, Masuda D, Yamashita S, Fushiki T (2015) Expression of CD36 by olfactory receptor cells and its abundance on the epithelial surface in mice. PLoS One 10:e0133412. 10.1371/journal.pone.0133412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR (2007) Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci 10:720–726. 10.1038/nn1882 [DOI] [PubMed] [Google Scholar]
- Login H, Butowt R, Bohm S (2015a) Activity-dependent and graded BACE1 expression in the olfactory epithelium is mediated by the retinoic acid metabolizing enzyme CYP26B1. Brain Struct Funct 220:2143–2157. 10.1007/s00429-014-0783-z [DOI] [PubMed] [Google Scholar]
- Login H, Haglin S, Berghard A, Bohm S (2015b) The stimulus-dependent gradient of Cyp26B1+ olfactory sensory neurons is necessary for the functional integrity of the olfactory sensory map. J Neurosci 35:13807–13818. 10.1523/JNEUROSCI.2247-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo AT, Youngentob SL, Kent PF, Schwob JE (1996) The aging olfactory epithelium: neurogenesis, response to damage, and odorant-induced activity. Int J Dev Neurosci 14:881–900. 10.1016/s0736-5748(96)00046-9 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Duronio RJ (2002) Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol 14:692–699. 10.1016/s0955-0674(02)00387-3 [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R (1996) Visualizing an olfactory sensory map. Cell 87:675–686. 10.1016/s0092-8674(00)81387-2 [DOI] [PubMed] [Google Scholar]
- Naessen R. (1971) An enquiry on the morphological characteristics and possible changes with age in the olfactory region of man. Acta Otolaryngol 71:49–62. 10.3109/00016487109125332 [DOI] [PubMed] [Google Scholar]
- Nakashima T, Kimmelman CP, Snow JB Jr (1984) Structure of human fetal and adult olfactory neuroepithelium. Arch Otolaryngol 110:641–646. 10.1001/archotol.1984.00800360013003 [DOI] [PubMed] [Google Scholar]
- Niederreither K, McCaffery P, Dräger UC, Chambon P, Dollé P (1997) Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev 62:67–78. 10.1016/s0925-4773(96)00653-3 [DOI] [PubMed] [Google Scholar]
- Norlin EM, Alenius M, Gussing F, Hägglund M, Vedin V, Bohm S (2001) Evidence for gradients of gene expression correlating with zonal topography of the olfactory sensory map. Mol Cell Neurosci 18:283–295. 10.1006/mcne.2001.1019 [DOI] [PubMed] [Google Scholar]
- Öztokatli H, Hörnberg M, Berghard A, Bohm S (2012) Retinoic acid receptor and CNGA2 channel signaling are part of a regulatory feedback loop controlling axonal convergence and survival of olfactory sensory neurons. FASEB J 26:617–627. 10.1096/fj.11-192450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard A, Schnittke N, Romano RA, Sinha S, Schwob JE (2011) DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. J Neurosci 31:8748–8759. 10.1523/JNEUROSCI.0681-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A, Law BM, Tata PR, Villoria J, Saez B, Mou H, Zhao R, Rajagopal J (2015) Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell 16:184–197. 10.1016/j.stem.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschaki M, Cammas L, Muta Y, Matsuoka Y, Mak SS, Rataj-Baniowska M, Fraulob V, Dolle P, Ladher RK (2013) Retinoic acid regulates olfactory progenitor cell fate and differentiation. Neural Dev 8:13. 10.1186/1749-8104-8-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso CE, Jang W, Dräger UC, Schwob JE (2012) Differential expression of components of the retinoic acid signaling pathway in the adult mouse olfactory epithelium. J Comp Neurol 520:3707–3726. 10.1002/cne.23124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R, Brix A, Harkema J, Herbert R, Kittel B, Lewis D, March T, Nagano K, Pino M, Rittinghausen S, Rosenbruch M, Tellier P, Wohrmann T (2009) Proliferative and nonproliferative lesions of the rat and mouse respiratory tract. Toxicol Pathol 37:5S–73S. 10.1177/0192623309353423 [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB (1993) A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73:597–609. 10.1016/0092-8674(93)90145-g [DOI] [PubMed] [Google Scholar]
- Rock JR, Randell SH, Hogan BL (2010) Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech 3:545–556. 10.1242/dmm.006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammeta N, McClintock TS (2010) Chemical stress induces the unfolded protein response in olfactory sensory neurons. J Comp Neurol 518:1825–1836. 10.1002/cne.22305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittke N, Herrick DB, Lin B, Peterson J, Coleman JH, Packard AI, Jang W, Schwob JE (2015) Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proc Natl Acad Sci U S A 112:E5068–E5077. 10.1073/pnas.1512272112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, Hewitt Coleman J (2017) Stem and progenitor cells of the mammalian olfactory epithelium: taking poietic license. J Comp Neurol 525:1034–1054. 10.1002/cne.24105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoney PN, Fragoso YD, Saeed RB, Ashton A, Goodman T, Simons C, Gomaa MS, Sementilli A, Sementilli L, Ross AW, Morgan PJ, McCaffery PJ (2016) Expression of the retinoic acid catabolic enzyme CYP26B1 in the human brain to maintain signaling homeostasis. Brain Struct Funct 221:3315–3326. 10.1007/s00429-015-1102-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzukawa K, Kondo K, Kanaya K, Sakamoto T, Watanabe K, Ushio M, Kaga K, Yamasoba T (2011) Age-related changes of the regeneration mode in the mouse peripheral olfactory system following olfactotoxic drug methimazole-induced damage. J Comp Neurol 519:2154–2174. 10.1002/cne.22611 [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R (1993) Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74:309–318. 10.1016/0092-8674(93)90422-m [DOI] [PubMed] [Google Scholar]
- Vedin V, Slotnick B, Berghard A (2004) Zonal ablation of the olfactory sensory neuroepithelium of the mouse: effects on odorant detection. Eur J Neurosci 20:1858–1864. 10.1111/j.1460-9568.2004.03634.x [DOI] [PubMed] [Google Scholar]
- Vedin V, Molander M, Bohm S, Berghard A (2009) Regional differences in olfactory epithelial homeostasis in the adult mouse. J Comp Neurol 513:375–384. 10.1002/cne.21973 [DOI] [PubMed] [Google Scholar]
- Wang TW, Zhang H, Parent JM (2005) Retinoic acid regulates postnatal neurogenesis in the murine subventricular zone-olfactory bulb pathway. Development 132:2721–2732. 10.1242/dev.01867 [DOI] [PubMed] [Google Scholar]
- Weng PL, Vinjamuri M, Ovitt CE (2016) Ascl3 transcription factor marks a distinct progenitor lineage for non-neuronal support cells in the olfactory epithelium. Sci Rep 6:38199. 10.1038/srep38199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Fang C, Schnittke N, Schwob JE, Ding X (2013) Mechanisms of permanent loss of olfactory receptor neurons induced by the herbicide 2,6-dichlorobenzonitrile: effects on stem cells and noninvolvement of acute induction of the inflammatory cytokine IL-6. Toxicol Appl Pharmacol 272:598–607. 10.1016/j.taap.2013.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapiec B, Mombaerts P (2020) The zonal organization of odorant receptor gene choice in the main olfactory epithelium of the mouse. Cell Rep 30:4220–4234.e5. 10.1016/j.celrep.2020.02.110 [DOI] [PubMed] [Google Scholar]