Retinoic acid synthesis is not essential for meiosis in fetal ovaries, challenging the dogma about the meiosis-inducing factors.
Abstract
In mammals, the timing of meiosis entry is regulated by signals from the gonadal environment. All-trans retinoic acid (ATRA) signaling is considered the key pathway that promotes Stra8 (stimulated by retinoic acid 8) expression and, in turn, meiosis entry. This model, however, is debated because it is based on analyzing the effects of exogenous ATRA on ex vivo gonadal cultures, which not accurately reflects the role of endogenous ATRA. Aldh1a1 and Aldh1a2, two retinaldehyde dehydrogenases synthesizing ATRA, are expressed in the mouse ovaries when meiosis initiates. Contrary to the present view, here, we demonstrate that ATRA-responsive cells are scarce in the ovary. Using three distinct gene deletion models for Aldh1a1;Aldh1a2;Aldh1a3, we show that Stra8 expression is independent of ATRA production by ALDH1A proteins and that germ cells progress through meiosis. Together, these data demonstrate that ATRA signaling is dispensable for instructing meiosis initiation in female germ cells.
INTRODUCTION
Germ cells exhibit the unique capacity to generate haploid gametes, eventually giving rise to an embryo after fertilization. In mice, primordial germ cells (PGCs) are specified in the epiblast around 6.25 days post-coitum (dpc) and colonize the gonads at around 10.5 dpc (1). At this stage, the gonads start differentiating as testes in XY embryos, or as ovaries in XX embryos (2). In parallel, germ cells loose pluripotency, becoming either prospermatogonia in testes or oogonia in ovaries, both of which further progress into meiotic divisions (3). However, the timing of the initiation of meiosis is sexually dimorphic (4), starting around 8 days postpartum in the testis versus 13.5 dpc in the ovary.
The nature of the signal(s) instructing oogonia to transition from mitosis to meiosis is still debated. Notably, Stra8 (stimulated by retinoic acid 8) is the only gatekeeper currently described that engages the meiotic program. This is evidenced by the failure of premeiotic DNA replication in female Stra8-deficient gonads (5). As Stra8 was originally identified as an all-trans retinoic acid (ATRA)–responsive gene in P19 embryonic carcinoma cells (6), ATRA signaling has been proposed as a primary meiosis-instructing factor. This concept is supported by the up-regulation of meiotic markers including Stra8 in embryonic ovaries cultured ex vivo in the presence of ATRA and ATRA receptor (RAR) agonists, or the down-regulation of Stra8 using pan-RAR antagonists (7, 8). Nevertheless, these findings have been brought into question by experiments that demonstrated Stra8 expression and meiosis initiation in embryonic ovaries lacking two of the three ATRA-synthesizing enzymes (ALDH1A2 and ALDH1A3, encoded by the Aldh1a2 and Aldh1a3 genes) (9). Along the same lines, genetic ablation of Aldh1a1 alone does not impair meiosis, although it reduces Stra8 expression and delays meiosis initiation (10). In both mouse models, the remaining Aldh1a isotype(s) that is (are) remaining may, however, be sufficient to produce ATRA and, as a result, induce meiosis.
To clarify the contribution of endogenous ATRA in vivo, we have generated mice deficient for all three Aldh1a isotypes either in the somatic cells of the embryonic ovary or ubiquitously. Using this approach, we have robustly decreased ATRA signaling during PGC colonization of the developing gonad. Detailed analysis of the mutant phenotypes revealed that ALDH1A1, ALDH1A2, and ALDH1A3 are dispensable for meiotic initiation in oogonia.
RESULTS
Expression of ALDH1A1 and ALDH1A2 is restricted to the somatic cells of the ovary
It has been shown that two potential sources of ATRA coexist in the female urogenital ridges. First, the mesonephros, a transient organ adjacent to the gonad, exhibits both Aldh1a2 mRNA expression at 10.5 and 12.5 dpc and a strong ATRA responsiveness according to the Tg(RARE-Hspa1b/lacZ)12Jrt transgenic reporter (7, 11). Second, ALDH1A1 and ALDH1A2 expression has been detected in the somatic cells within the developing ovary at 12.5 and 13.5 dpc (10, 12). These findings suggest the existence of both external and endogenous sources of ATRA in the female gonad at the onset of meiosis initiation.
Single-cell RNA sequencing and immune-localization analyses (Fig. 1, A and B) revealed that Aldh1a1 expression became readily detectable in the supporting cells of the ovary at the time of meiosis entry (~13.5 dpc), as previously reported (10, 12). Aldh1a2 exhibited robust expression in the somatic progenitor cells of the gonad and in the mesonephros from 10.5 to 13.5 dpc. At 13.5 dpc, Aldh1a2 was also highly expressed in the supporting cells. Aldh1a3 mRNA was nearly absent in the ovary as evidenced by transcriptomic analysis (Fig. 1A). All these observations indicate that the somatic cells of the ovary, but not the germ cells, are able to synthesize endogenous ATRA as early as 10.5 dpc, i.e., 3 days before meiosis entry.
Fig. 1. ATRA signaling is active in the developing ovary.
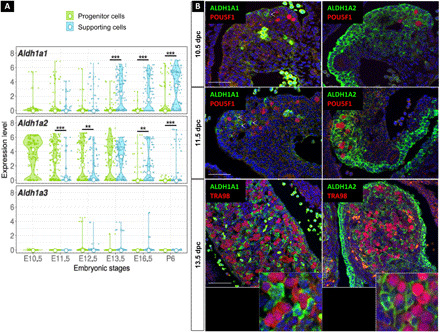
(A) Violin plots showing the expression of Aldh1a1, Aldh1a2, and Aldh1a3 in the progenitor and supporting cells between 10.5 to 16.5 dpc and postnatal day 6 (E10.5 to E16.5 and P6) determined by single-cell RNA sequencing analysis of Sf1-positive somatic cells from female gonads. Expression values are log-transformed reads per kilobase of transcript per million mapped reads (RPKM); small points represent expression in individual cells, and the white point is the median of expression. Statistical analyses were performed using the Wilcoxon-Mann-Whitney test, and P value was adjusted for false discovery rate. (B) Immunodetection of ALDH1A1 or ALDH1A2 (green) and POU5F1 or TRA98 (germ cells, red) in 10.5, 11.5, and 13.5 dpc ovaries. DAPI (blue), nuclei. Scale bars (white), 50 μm. Arrowheads highlight examples of ALDH1A-positive cells. **P <0.01 and ***P <0.001 (adjusted p-values, Wilcoxon-Mann-Whitney test).
Germ cells respond to ATRA in the developing ovary
Using an ATRA-reporter mouse model carrying the Tg(RARE-Hspa1b/lacZ)12Jrt transgene (11), the mesonephros exhibits a strong response to endogenous ATRA, whereas the ATRA responsiveness is relatively weak in gonads (7, 10), suggesting that only very few cells are responsive to endogenous ATRA in the ovaries. To investigate the nature and fate of these ATRA-responsive cells, we used a novel transgenic reporter mouse line called Tg(RARE-Hspa1b-cre/ERT2), in which RA response elements (RAREs) coupled to the Hspa1b minimal promoter drive expression of the tamoxifen (TAM)–inducible CreERT2 recombinase, thus allowing cell lineage tracing of ATRA-responsive cells. In this model, the expression of the membrane-tagged green fluorescent protein (GFP) from the Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo reporter (13) is activated in cells that are ATRA responsive at the time of TAM administration. These cells then become permanently labeled by GFP (fig. S1A). As expected from previous studies (11, 14), the periocular mesenchyme and the developing heart, two tissues that are sensitive to endogenous ATRA, harbored GFP-positive cells ~24 hours after TAM administration (fig. S1B). We then asked whether germ cells in vivo were sensitive to endogenous ATRA. To this aim, TAM was administered to the pregnant females and the gonads from the embryos were collected ~24 hours later for GFP labeling analyses (Fig. 2A). Although a small number of somatic (POU5F1-negative) cells were responsive to endogenous ATRA at 9.5 dpc (evidenced as GFP-positive cells at 10.5 dpc), only a few germ cells were ATRA responsive between 12.5 and 13.5 dpc (because they were GFP positive at 13.5 dpc) (Fig. 2A). Confocal microscopy on whole-mount organs further revealed that the vast majority of germ cells were not ATRA responsive between 13.5 and 14.5 dpc because they were GFP negative at 14.5 dpc (Fig. 2B), although there were a few exceptions (arrowheads in Fig. 2B). Thus, less than 5% of germ cells were ATRA responsive as previously reported (7). We next dissected ovaries at 13.5 dpc and cultured them for 24 hours in the presence of 4-hydroxy-TAM (4-OH-TAM) at either a physiological dose of ATRA (1 nM) (9) or a pharmacological dose of ATRA (100 nM). Dimethyl sulfoxide (DMSO) was used as a negative control (Fig. 2C). Immunostaining analysis using anti-GFP and anti-TRA98 (a germ cell marker) antibodies demonstrated that germ cells from the ex vivo transgenic ovaries lacked GFP-positive germ cells in DMSO-treated samples but were able to respond to a very low concentration of exogenous ATRA (many GFP-positive germ cells in 1 nM ATRA–treated samples). As expected, cells of the mesonephros also responded to ATRA treatment (7). Together, these results indicate that the vast majority of germ cells failed to activate ATRA-responsive genes in vivo, ultimately questioning the requirement of ATRA for meiosis entry.
Fig. 2. Most of the germ cells do not respond to ATRA before 13.5 dpc.
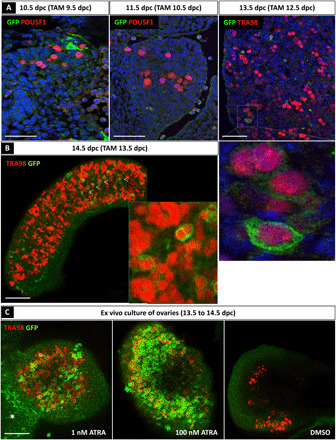
(A) Immunodetection of ATRA-responsive cells (GFP positive, green) and germ cells (POU5F1- or TRA98-positive cells, red) in 10.5, 11.5, and 13.5 dpc Tg(RARE-Hspa1b-cre/ERT2) ovarian sections after TAM induction at 9.5, 10.5, and 12.5 dpc, respectively. DAPI (blue), nuclei. Scale bars (white), 50 μm. (B) Immunodetection of ATRA-responsive cells (GFP, green) and TRA98 (germ cells, red) in 14.5 dpc Tg(RARE-Hspa1b-cre/ERT2) whole ovaries after TAM induction at 13.5 dpc. Scale bars (white), 50 μm. White arrowheads, GFP-positive germ cells. (C) Immunodetection of TRA98 (germ cells) (red) and GFP (ATRA-responsive cells) (green) in the presence of either DMSO (control) or 1 or 100 nM ATRA in cultured Tg(RARE-Hspa1b-cre/ERT2) ovaries from 13.5 to 14.5 dpc. Asterisk, GFP-positive cells within the mesonephros.
Generation of three mouse models deficient for Aldh1a1;Aldh1a2;Aldh1a3 in the embryonic ovaries
To functionally test the contribution of ATRA signaling to meiosis entry, we performed conditional deletion of all three ATRA-producing enzymes (Aldh1a1;Aldh1a2;Aldh1a3, hereafter referred to as Aldh1a1-3) using three distinct Cre driver lines (fig. S2, A and B). First, we used the Tg(Nr5a1-cre)2Klp transgenic line (hereafter named Sf1-cre) to induce deletion in SF1-positive somatic cells in the gonads from 11.5 dpc (15). Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) analysis in Sf1-Cre;Aldh1a1-3flox/flox embryos at 13.5 dpc demonstrated the efficient loss of mRNA expression for all three Aldh1a1-3 genes (Fig. 3A). Second, to induce an earlier deletion than the Sf1-Cre, we used the Wt1tm2(cre/ERT2)Wtp strain (16) (hereafter referred to Wt1-CreERT2) that expresses a TAM-inducible CreERT2 recombinase in most, if not all, somatic cells of the gonads at 10.5 dpc (17). TAM administration at 10.5 and 11.5 dpc triggered a strong reduction in Aldh1a1, Aldh1a2, and Aldh1a3 mRNA levels at 13.5 dpc (Fig. 3B). The highly efficient ablation of ALDH1A1 and ALDH1A2 was further confirmed by immunodetection at 13.5 dpc (Fig. 3D). Next, we used the Tg(CAG-cre/ERT2)#Rlb line (18) (hereafter referred to as CAGG-CreER), which ubiquitously expresses a TAM-inducible CreERT2 to delete Aldh1a1-3 in all cell types. In TAM-treated CAGG-CreER;Aldh1a1-3flox/flox ovaries, Aldh1a1, Aldh1a2, and Aldh1a3 mRNA levels were significantly reduced in the gonads, as previously reported (19). This was accompanied by a significant decrease of ALDH1A1 and ALDH1A2 protein levels at 13.5 dpc, resulting in severe heart malformations (Fig. 3E and fig. S2C). To summarize, all three mouse models demonstrated efficient elimination of Aldh1a1, Aldh1a2, and Aldh1a3 expression in the embryonic ovaries.
Fig. 3. Generation of three mouse models deficient for Aldh1a1;Aldh1a2;Aldh1a3 in the embryonic ovaries.
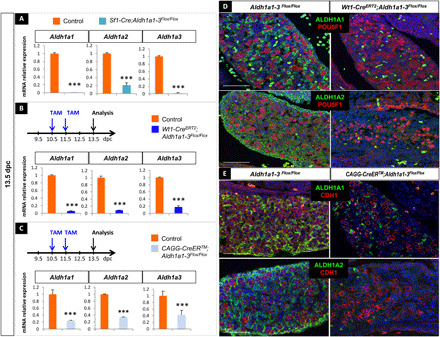
(A) RT-qPCR analysis of Aldh1a1, Aldh1a2, and Aldh1a3 expression in 13.5 dpc control (orange) and Sf1-Cre;Aldh1a1-3flox/flox (blue) gonads. Student’s t test, unpaired. Bars represent mean + SEM; n = 10 individual gonads. ***P < 0.001. (B) Top: Protocol of induction of Aldh1a1-3 deletion (10.5 dpc onward). Bottom: RT-qPCR analysis of Aldh1a1, Aldh1a2, and Aldh1a3 expression in 13.5 dpc control (orange) and Wt1-CreERT2;Aldh1a1-3flox/flox (blue) gonads. Student’s t test, unpaired. Bars represent mean + SEM; n = 10 individual gonads. ***P < 0.001. (C) Top: Protocol of induction of Aldh1a1-3 deletion (10.5 dpc onward). Bottom: RT-qPCR analysis of Aldh1a1, Aldh1a2, and Aldh1a3 expression in 13.5 dpc control (orange) and CAGG-CreER;Aldh1a1-3 flox/flox (blue) gonads. However, the deletion was less efficient than using the Wt1-CreERT2 or Sf1-Cre transgenes. Student’s t test, unpaired. Bars represent mean + SEM; n = 15 individual gonads. ***P < 0.001. (D) Immunodetection of ALDH1A1 or ALDH1A2 (green) and POU5F1 (germ cells, red) in 13.5 dpc control and Wt1-CreERT2;Aldh1a1-3flox/flox ovaries. DAPI (blue), nuclei. Scale bars (white), 50 μm. (E) Immunodetection of ALDH1A1 or ALDH1A2 (green) and CDH1 (germ cells, red) in 13.5 dpc control and CGAG-CreERT2;Aldh1a1-3 flox/flox ovaries. DAPI (blue), nuclei. Scale bars (white), 50 μm.
To assess the impact of Aldh1a1-3 deletion on ATRA synthesis, we performed metabolomic investigations and evaluated endogenous production of ATRA in mesonephroi, control testes, control ovaries, or Wt1-CreERT2;Aldh1a1-3flox/flox ovaries from 13.5 dpc embryos (fig. S3A). RA can be isomerized in ATRA, the most abundant form, in 9-cis-RA (9cRA), which is almost undetectable in vivo, and in 13-cis-RA (13cRA), which is mostly present in the serum (20). RA isomers have different affinities for nuclear receptors and therefore exert different biological activities: ATRA has the highest affinity for RA receptors (RAR) and 9cRA can bind RAR/RXR. In contrast, 13cRA has a 100-fold lower affinity to RARs than the two others (21). Using single-ion mass spectrometry, we were able to detect all three RA isomers in vivo, even 9cRA isomer, which shows the high sensitivity of this method, and to quantify the relative abundance of ATRA isomer (fig. S3A). The ATRA level was abundant in mesonephroi as previously described (7) but was strongly decreased in Wt1-CreERT2;Aldh1a1-3flox/flox ovaries compared to control ovaries. In addition, this level was similar to the level detected in testes (fig. S3, B and C).
To determine whether this level of ATRA was sufficient to trigger a biological activity in the mutant ovaries, we designed an experimental ATRA-reporter assay by transfecting Chinese hamster ovary (CHO) cells with a plasmid containing the ATRA-responsive hsp68 mouse promoter (11) controlling the expression of the acGFP1 gene encoding the Aequorea coerulescens GFP. We cultured the transfected CHO cells in the absence or presence of increasing ATRA concentrations, or in the presence of cellular suspensions from either mesonephroi, control, or Wt1-CreERT2;Aldh1a1-3flox/flox ovaries dissected from 13.5 dpc embryos (Fig. 4A). Although GFP expression (quantified by Western blot) was stimulated by commercial ATRA from 1 nM onward, mesonephroi, or control ovaries, the GFP level of expression induced by mutant ovaries barely reached the residual level of GFP measured in cultures with medium depleted in RA, confirming that the genetic deletion of Aldh1a1-3 was efficient enough to inhibit ATRA synthesis (Fig. 4B).
Fig. 4. Remaining ATRA levels present in ovaries after genetic deletion of Aldh1a1, Aldh1a2, and Aldh1a3 genes are not sufficient to trigger ATRA biological activity.
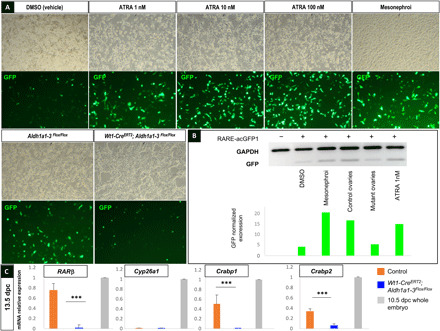
(A) CHO cells transfected with the ATRA-reporter construct and incubated with either DMSO (vehicle), increasing concentrations of ATRA from 1 to 100 nM, cellular suspensions of mesonephroi, Aldh1a1-3 flox/flox, or Wt1-CreERT2;Aldh1a1-3flox/flox ovaries dissected from 13.5 dpc embryos (top, brightfield picture; bottom, GFP detection). (B) Immunodetection by Western blot of GFP and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) in the same CHO cells transfected (+) or not (−) with the ATRA-reporter construct (RARE-acGFP1 plasmid) after protein extraction (upper), and corresponding quantification of GFP band intensity after normalization with GAPDH expression (lower). (C) RT-qPCR analysis of RARβ, Cyp26a1, Crabp1, and Crabp2 expression in 13.5 dpc control (orange), Wt1-CreERT2;Aldh1a1-3flox/flox (blue) gonads, or whole 10.5 dpc embryos (positive control, gray). Student’s t test, unpaired. Bars represent mean + SEM; n = 10 individual gonads. ***P < 0.001.
We next analyzed the expression of RARβ, Cyp26a1, and Crabp1-2, four ATRA-target genes used to monitor the inhibition of RA signaling (9) in control and Wt1-CreERT2;Aldh1a1-3flox/flox ovaries at 13.5 dpc. Although RARβ, Crabp1, and Crabp2 mRNA levels were significantly down-regulated in the Wt1-CreERT2;Aldh1a1-3flox/flox ovaries compared to the controls, we did not detect any Cyp26a1 expression, neither in the control nor in the mutant ovaries (Fig. 4C), demonstrating that RA signaling was impaired in Wt1-CreERT2;Aldh1a1-3flox/flox ovaries. Together, these results indicate that the very low level of ATRA remaining in Wt1-CreERT2;Aldh1a1-3flox/flox ovaries was not sufficient to promote the expression of a RARE reporter and of universal target genes.
Genetic deletion of all Aldh1a isotypes in the embryonic ovaries does not impair PGC proliferation and differentiation
Because ATRA signaling has been reported to regulate germ cell proliferation in the developing ovary (22), we next examined the proliferation of POU5F1- or DDX4-positive germ cells at 11.5 and 12.5 dpc in control and Wt1-CreERT2;Aldh1a1-3flox/flox ovaries treated by TAM at 9.5 and 10.5 dpc. A 3-hour pulse of 5-bromo-2′-deoxyuridine (BrdU) incorporation permanently labeled the cells transitioning through the S phase of the cell cycle. Quantification of the number of BrdU-positive germ cells indicated that despite the significant loss of Aldh1a1, Aldh1a2, and Aldh1a3 expression upon TAM treatment (Fig. 5A), proliferation was not affected in Wt1-CreERT2;Aldh1a1-3flox/flox gonads (Fig. 5B). Moreover, the pluripotency markers Pou5f1 (also known as Oct3/4), Sox2, and Dazl, driving oogonia differentiation (23), were expressed at similar levels in control and Aldh1a1-3–deficient gonads at 11.5 and 12.5 dpc (fig. S4). Together, these results indicate that the genetic ablation of ALDH1A1, ALDH1A2, and ALDH1A3 does not hinder the proliferation or differentiation of PGCs.
Fig. 5. Genetic ablation of all three Aldh1a1, Aldh1a2, and Aldh1a3 genes does not impair germ cell proliferation in vivo.
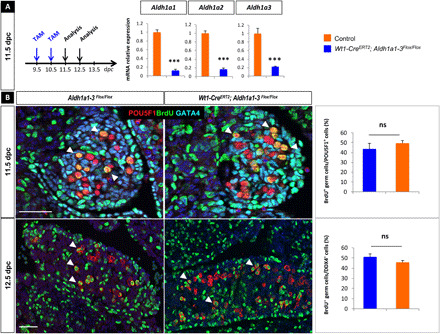
(A) Protocol of induction of Aldh1a1-3 deletion (9.5 dpc onward). RT-qPCR analysis of Aldh1a1, Aldh1a2, and Aldh1a3 expression in 11.5 dpc control (orange) and Wt1-CreERT2;Aldh1a1-3flox/flox (blue) ovaries. Student’s t test, unpaired. Bars represent mean + SEM; n = 10 individual gonads. ***P < 0.001. (B) Immunodetection of BrdU (proliferating cells, green), POU5F1 (PGCs, red), and GATA4 (gonadal somatic cells, cyan) at 11.5 dpc in control (left) and Wt1-CreERT2;Aldh1a1-3flox/flox (right) ovaries. Histograms: Percentage of BrdU-positive (proliferating) versus POU5F1-positive (total) germ cells in control (orange) and Wt1-CreERT2;Aldh1a1-3flox/flox (mutant, blue) ovaries at 11.5 dpc. Student’s t test, unpaired. Bars represent mean + SEM; n = 10 sections of each genotype (four to seven ovaries per genotype). ns, not significant. Bottom: Immunodetection of BrdU (proliferating cells, green) and DDX4 (germ cells, red) at 12.5 dpc in control (left) and Wt1-CreERT2;Aldh1a1-3flox/flox (right) ovaries. Histograms: Percentage of BrdU-positive (proliferating) versus DDX4-positive (total) germ cells in control (orange) and Wt1-CreERT2;Aldh1a1-3flox/flox (mutant, blue) ovaries at 12.5 dpc. Student’s t test, unpaired. Bars represent mean + SEM; n = 10 sections of each genotype (four to seven ovaries per genotype). Arrowheads highlight examples of BrdU-positive germ cells.
Germ cells enter meiosis in ovaries lacking all Aldh1a isotypes
We next assessed whether germ cells initiated meiosis after genetic deletion of Aldh1a1-3 using the different mouse models described above. In Sf1-Cre;Aldh1a1-3flox/flox embryos, Stra8 expression was reduced by ~20% at 13.5 dpc (fig. S5A). Stra8 expression was also reduced at 13.5 dpc in ovaries of Wt1-CreERT2;Aldh1a1-3flox/flox embryos treated by TAM at 9.5 and 10.5 dpc (fig. S5B). Nevertheless, under these conditions, TAM treatment induced frequent embryonic lethality. Accordingly, TAM was administrated at 10.5 and 11.5 dpc in the following experiments, leading to an almost identical decrease of Aldh1a1 and Aldh1a2 mRNA expression (fig. S6). Upon this TAM treatment, germ cells in Wt1-CreERT2;Aldh1a1-3flox/flox ovaries expressed Stra8 at 13.5 dpc, although its mRNA levels were reduced by ~25% when compared to controls (Fig. 6, A and C). At 14.5 dpc, this deficit was partly compensated in the Wt1-CreERT2;Aldh1a1-3flox/flox ovaries, suggesting a delay in the onset of Stra8 expression in the absence of ALDH1A isotypes (Fig. 6B). Notably, the reduction in Stra8 expression did not impair meiosis entry, as evidenced by the normal levels of Rec8 mRNA, a gene that encodes a component of the cohesin complex accumulating during the meiotic S phase (Fig. 6B), and of Spo11 mRNA, which is expressed during the leptotene stage of meiotic prophase I (Fig. 6A). Together, these results demonstrate that genetic deletion of the three Aldh1a1-3 genes does not impair meiosis entry.
Fig. 6. Meiosis progresses in the absence of endogenous ATRA synthesis in vivo.
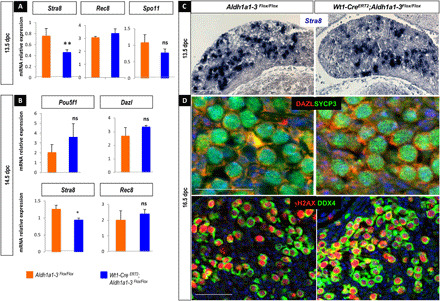
(A) RT-qPCR analysis of Stra8, Rec8, and Spo11 expression in 13.5 dpc control (orange) and Wt1-CreERT2;Aldh1a1-3flox/flox (blue) ovaries. Student’s t test, unpaired. Bars represent mean + SEM; n = 10 individual ovaries. **P < 0.01. (B) RT-qPCR analysis of Pou5f1, Dazl, Stra8, and Rec8 expression in 14.5 dpc control (orange) and Wt1-CreERT2;Aldh1a1-3flox/flox (blue) ovaries. Student’s t test, unpaired. Bars represent mean + SEM; n = 10 individual gonads. *P < 0.05. (C) In situ hybridization using Stra8 riboprobe at 13.5 dpc in control (left) and Wt1-CreERT2;Aldh1a1-3flox/flox (right) ovaries. Scale bars (white), 50 μm. (D) Immunodetection of DAZL (red) and SYCP3 (green) in 16.5 dpc control and Wt1-CreERT2;Aldh1a1-3flox/flox ovaries. Bottom: Immunodetection of phospho–histone γH2AX (red) and DDX4 (green) in 16.5 dpc control and mutant ovaries. DAPI (blue), nuclei. Scale bars (white), 50 μm.
We next examined whether meiosis further progressed despite Aldh1a1-3 ablation by looking at specific chromosomal features of meiosis. During meiotic progression, homologous chromosomes pair and the synaptonemal complex promotes chromosome recombination. Immunodetection of the synaptonemal complex protein 3 (SYCP3), which appears in leptotene stage and becomes enriched in the zygotene stage, revealed similar thread-like synaptonemal complex structures in TAM-treated control and Wt1-CreERT2;Aldh1a1-3flox/flox ovaries (Fig. 6D), further demonstrating that germ cells entered meiosis in the absence of all ALDH1A isotypes. Quantification of the germ cells positive for phospho-γH2AX, which is associated with DNA double-strand breaks occurring during the leptotene stage, confirmed these observations (Fig. 6D and fig. S7). Hence, these results demonstrate that female germ cells progress to meiotic prophase even when Aldh1a1-3 are deleted from the ovary from 10.5 dpc onward.
In TAM-treated CAGG-CreER;Aldh1a1-3flox/flox ovaries, the mRNA levels of meiotic markers such as Mei1, Dmc1, Rec8, Syce1, and Spo11 were not significantly changed as evidenced by RT-qPCR and in situ hybridization experiments (fig. S8, A and B). Accordingly, phospho-γH2AX expression was comparable between TAM-treated control and CAGG-CreER;Aldh1a1-3flox/flox ovaries at 16.5 dpc (fig. S8C). Together, these results indicate that genetic ablation of Aldh1a1-3 does not prevent meiosis entry in the developing mouse ovary.
DISCUSSION
Evidence that PGCs do not enter meiosis because of an intrinsic autonomous property (i.e., cell division timing) but are rather instructed by a signaling molecule produced by somatic ovarian cells led to a search for a “meiosis-inducing substance” (MIS) (24). Although ATRA has been suggested to be one such MIS, its role in this biological process has been questioned, giving rise to conflicting reports (7–9). In all three distinct models of Aldh1a1-3 genetic deletion we have used, we observed only a minor decrease in Stra8 expression, and this reduction was not robust enough to prevent meiosis initiation and germ cells from progressing into meiotic prophase I. Thus, we conclude that ATRA signaling does not initiate but contributes to meiosis by making Stra8 expression on time. Accordingly, ectopic expression of Cyp26a1, which has the most efficient catalytic activity on ATRA degradation, does not affect meiosis entry or Stra8 expression, although Stra8 mRNA levels were reduced by ~30%, i.e., in the same range than the mRNA reduction observed in the present study (25). This indicates that ATRA does regulate Stra8 transcription maintenance rather than Stra8 initiation of transcription. In agreement with our findings, the study from Vernet et al. (this issue) describes a similar delay in expression of Stra8 and a normal progression into meiosis in fetal ovaries of mice lacking the RARs RARα, RARβ, and RARγ. Because the fetal lethality in our different mice models prevented us to study the postnatal ovary, the fate of the ATRA-responsive female germ cells in vivo requires further investigations.
When identified, Stra8 was not classified in the class of immediate and early, ATRA-responsive gene, as indicated by the kinetics of its mRNA accumulation in P19 pluripotent carcinoma cells treated with ATRA (6). It was shown that ATRA regulates the phosphorylation status of the STRA8 protein in P19 cells (6), indicating that ATRA might also control STRA8 posttranslational modifications. The functional relevance of ATRA-induced phosphorylation in STRA8 activity or stability has not been characterized. Nevertheless, our results indicate that Aldh1a1-3 are not required for Stra8 expression in germ cells, suggesting that either ATRA does not phosphorylate STRA8 in vivo or STRA8 phosphorylation has no impact on its activity in germ cells. Together, our results, those from Vernet et al. and Bellutti et al. (25), indicate that ATRA signaling is dispensable for meiosis entry.
In PGC-like cells (PGCLCs) derived from embryonic stem cells, Stra8 expression is induced without ATRA treatment (26), further highlighting that ATRA is not instrumental for meiosis initiation in this system and that other signals are responsible for initiation of Stra8 expression. In vitro generation of female PGCLCs expressing Stra8, SYCP3, and other meiotic markers requires bone morphogenetic proteins (BMPs), which activate transforming growth factor β (TGFβ)/SMAD signaling. In these experiments, ATRA increased the number of meiotic germ cells, indicating that ATRA signaling served to enhance a preexisting situation (26).
Meiosis initiation is timed by epigenetic factors such as polycomb repressive complex PRC1 that promotes structural modifications of chromatin and consequently times the expression of Stra8 (27). Recent data show that deficiency in vitamin C during gestation induces incomplete DNA demethylation of key germline genes and thus delays meiosis initiation in the embryos (28), indicating that molecules from the maternal nutrition participate in regulating meiotic gene expression in the germ cells of the progeny. In addition, in the absence of Rspo1, an activator of WNT/β-catenin signaling in the female fetal gonad, a proportion of PGCs neither expressed Stra8 nor entered meiosis (29), suggesting a control of Stra8 expression by WNT/β-catenin. The Msx genes, which encode homeodomain transcription factors, are direct targets of WNT/β-catenin signaling in murine embryonic stem cells (30), and in mutant embryos lacking both Msx1 and Msx2, germ cells failed to initiate meiosis (31). In gonadal cultures, BMP4 stimulates the expression of Msx1 and Msx2 that, in turn, directly regulate Stra8 expression (29), suggesting that TGFβ signaling is also involved in Stra8 regulation. However, in mouse germ cells, the central transducer Smad4 is dispensable for Stra8 expression, although it is required to up-regulate other key genes involved in meiosis (30). Last, the DMRT1 transcription factor also contributes to the switch from mitosis to meiosis by directly regulating Stra8 expression in female germ cells (32). Together, these studies indicate that convergent pathways other than ATRA signaling collaborate to regulate Stra8 expression and the transition from mitosis to meiosis cycle.
Early studies based on the observation that female germ cells cultured together with fetal testes were prevented from initiating meiosis (33, 34) led to the concept of a secreted masculinizing meiosis preventing substance (MPS) in the male gonad (35). The P450 enzyme CYP26B1, expressed in the supporting cells of the fetal testis but not the ovary, has been proposed to be MPS. Analysis of Cyp26b1-null mutant mice demonstrated that CYP26B1 activity prevented germ cells from entering into meiosis in male mice (36). Whereas CYP26B1 is able to degrade ATRA (37), the data from Kumar et al. and Bellutti et al. (25) suggest that CYP26B1 metabolizes a substrate other than ATRA to prevent meiosis initiation (9). So far, knowledge about the metabolic ligands of CYP26B1 is poor, and the nature and the molecular identity of CYP26B1 substrate(s) in the fetal testis remain unknown.
Identifying the molecules controlling the fundamental decision of germ cell to enter meiosis and defining whether they have MPS or MIS functions represents a major challenge for the reproductive medicine community in the upcoming years. The main clinical consequences of defects in germ cell development are infertility and increased susceptibility to germ cell tumors. Therefore, understanding how germ cells change their gene expression profiles in response to somatic signals will provide knowledge on the etiology of human genetic diseases.
MATERIALS AND METHODS
Experimental design
The aim of this study was to investigate the role of endogenous ATRA signaling by using an in vivo genetic approach of deletion of Aldh1a1, Aldh1a2, and Aldh1a3 genes (i.e., mice lacking all sources of ATRA in vivo) to challenge the admitted concept of ATRA being the MIS. The number of samples was determined on the basis of experimental approach, availability, and feasibility required to obtain definitive results. No data were excluded from the analyses. The numbers of replicates are specified in Materials and Methods. The researchers were not blinded during data collection or analysis.
Mouse strains and genotyping
The experiments described here were carried out in compliance with the relevant institutional and European animal welfare laws, guidelines, and policies. All the experiments were approved by the French Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche (APAFIS # 3771-2016012110545580v13). All mice were kept on a 129/Sv-C57BL/6J mixed background. Mouse lines were obtained from the Jackson Laboratory. The Wt1tm2(cre/ERT2)Wtp, Tg(Nr5a1-cre)2Klp, Tg(CAG-cre/ERT2)#Rlb, Aldh1a1flox/flox, Aldh1a2flox/flox, Aldh1a3flox/flox, and Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo mice were described previously and genotyped as reported (13, 15, 16, 38, 39, 40). Genotyping was performed using DNA extracted from tail tips or ear biopsies of mice. To activate the CreERT recombinase in embryos, TAM (catalog no. T5648, Sigma-Aldrich) was directly diluted in corn oil to a concentration of 40 mg/ml, and two TAM treatments (200 mg/kg body weight) were administrated to pregnant females by force-feeding at 9.5 and 10.5 dpc or 10.5 and 11.5 dpc. This resulted in embryos in which Aldh1a1-3 were deleted upon TAM induction when they were carrying the creERT transgene in contrast with their control littermates. For proliferation assays, BrdU (catalog no. B5002, Sigma-Aldrich) was diluted to a concentration of 10 mg/ml in sterile H2O and administrated to the pregnant females at a final concentration of 10 μg/ml by intraperitoneal injection, and pregnant females and their embryos were sacrificed after 3 hours.
Single-cell sequencing
Single-cell RNA sequencing was performed as described in (41). Briefly, somatic cells of developing mouse female gonads were purified by fluorescence-activated cell sorting (FACS) using the Tg(Nr5a1-GFP) at six stages of development (10.5, 11.5, 12.5, 13.5, and 16.5 dpc and postnatal day 6). Single-cell isolation, reverse transcription, and complementary DNA (cDNA) amplification were performed using the Fluidigm C1 Auto Prep system, and single-cell sequencing library was prepared with Illumina Nextera XT following Fluidigm protocol. Library was multiplexed and sequenced on an Illumina HiSeq2000 platform with 100–base pair (bp) paired-end reads at an average depth of 10 million reads per single cell. Obtained reads were mapped on the mouse reference genome (GRCm38.p3), and gene expression was quantified in RPKMs (reads per kilobase of exon per million reads mapped). Cells were clustered using principal components analysis and hierarchical clustering, and cell types were identified according to the expression level of marker genes and gene ontology enrichment tests. Significance of the difference in expression level between cell types was assessed in R version 3.6.0 using Wilcoxon-Mann-Whitney tests, and P values were adjusted for false discovery rate using Bonferroni.
qPCR analyses
Individual gonads without mesonephroi were dissected in phosphate-buffered saline (PBS) from 11.5, 12.5, 13.5, and 14.5 dpc embryos. RNA was extracted using the Qiagen RNeasy Kit and reverse-transcribed using the RNA RT-PCR Kit (Stratagene). Primers and probes were designed by the Roche Assay Design Center (https://www.rocheappliedscience.com/sis/rtpcr/upl/adc.jsp). All real-time PCR assays were carried out using the LightCycler FastStart DNA Master Kit (Roche) according to the manufacturer’s instructions. qPCR was performed on cDNA from one gonad and compared to a standard curve. They were repeated at least twice. Relative expression levels of each sample were quantified in the same run and normalized by measuring Sdha expression (which represents the total gonadal cells). For Aldh1a1, Aldh1a2, and Aldh1a3 real-time PCR, the following specific primers were used: 5′-actttcccaccattgagtgc-3′ and 5′-caccatggatgcttcagaga-3′ (Aldh1a1), 5′-catggtatcctccgcaatg-3′ and 5′-gcgcatttaaggcattgtaac-3′ (Aldh1a2), and 5′-tctgggaatggcagagaact-3′ and 5′-ttgatggtgacggttttcac-3′ (Aldh1a3). For each sample, relative expression levels were quantified and normalized. For each genotype (n = 10), the mean of these 10 absolute expression levels (i.e., normalized) was calculated and then divided by the mean of the 10 absolute expression levels of the control samples considered as the reference (=1 when divided by itself), leading to the fold of change.
Statistical analysis
For each genotype, the mean of the normalized expression levels was calculated, and graphs show mean fold change + SEM. All the data were analyzed by Student’s t test using Microsoft Excel. Asterisks highlight the pertinent comparisons and indicate levels of significance: *P < 0.05, **P < 0.01, and ***P < 0.001. Data are shown as mean + SEM.
Mass spectrometry
For metabolomic analysis, 12 mesonephroi or 12 gonads of each genotype (Aldh1a1-3flox/flox ovaries, Aldh1a1-3flox/flox testes, Wt1-CreERT2;Aldh1a1-3flox/flox ovaries) from 13.5 dpc littermate embryos were dissected and subjected to methanol-chloroform extraction. The methanol phase was collected for mass spectrometry, whereas the interphase was used for quantification of protein concentration. Single ion monitoring analysis of RA was performed using a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Compounds were loaded onto a Phenomenex Synergi 4 μm Hydro-RP 80A 250 × 2 mm. RA isomers were separated with a 25-min gradient at a flow rate of 0.350 ml/min (mobile phase A water and 0.1% formic acid, mobile phase B acetonitrile and 0.1% formic acid gradient, 30% A for 5 min and 90% B in 15-min return initial condition for 5 min). The column was then thoroughly cleaned with 10 blank runs using a 2-μl injection of 30% MeCN, 30% isopropanol, and 0.1% formic acid after each run. The mass spectrometry method was done in positive electrospray ionization mode, which increases the sensitivity for RA isomers. We used a targeted SIM (Selected Ion Monitoring) scan on the M+H: 301.216. This led to an improved signal-to-noise ratio and to lower detection limits. The method consisted of full scans and targeted SIM scan. Full scans were acquired with AGC (Automatic Gain Control) target value of 1 × 106, resolution of 70,000 full width at half maximum (FWHM) at 200 mass/charge ratio (m/z), and maximum ion injection time of 100 ms. The target was monitored with a 4-min window, AGC target value of 1 × 105, resolution of 280,000 FWHM at 200 m/z, and maximum ion injection time of 500 ms.
RA levels (all isomers) were quantified by normalizing the peak area given by the mass spectrometer by the protein concentration of each sample. ATRA-specific peak area was measured using ImageJ software and normalized with the protein concentration.
Detection of ATRA activity
CHO cells were cultured in Dulbecco’s minimum essential medium (DMEM):F12 medium (1:1) (Gibco) supplemented with recombinant human epidermal growth factor (EGF) (10 ng/ml; catalog no. PHG0314, Gibco), ITS (insulin-transferrin-selenium; 100×; catalog no. 41400045, Gibco), and 100× nonessential amino acids (catalog no. 1140068, Gibco) in the absence of serum. Cells (15,000 per well) were seeded in a 24-well plate 24 hours before transfection. Then, CHO cells were transfected with 800 ng of plasmid described hereafter, using Lipofectamine 2000 reagent (catalog no. 11668019, Invitrogen) according to the manufacturer’s procedure. The ATRA-reporter construct was designed as follows: A DNA fragment encompassing the RA-responsive hsp68 mouse promoter as previously published (10), the rabbit β-globin intron, the coding sequence of the acGFP1 gene (Aequorea coerulescens GFP), two STOP codons, and the SV40 late polyadenylation signal was synthesized by Sigma-Aldrich and cloned by the manufacturer in the kanamycin-resistant plasmid pUC57. Two copies in tandem of the 5′HS4 insulator were then added by cloning in appropriate restriction sites at the 3′ end of the polyadenylation signal. The sequence of the whole construct is available upon request. Twenty-four hours after transfection, CHO cells were incubated with either DMSO (vehicle), commercial ATRA (catalog no. R2625, Sigma-Aldrich) from 1 to 100 nM in DMSO, or cellular suspensions from mesonephroi, Aldh1a1-3flox/flox, or Wt1-CreERT2;Aldh1a1-3flox/flox ovaries from 13.5 dpc littermate embryos freshly dissected (n = 6 mesonephroi or gonads per experiment, experiment performed in two replicates). Twenty-four hours later, GFP endogenous signal was visualized with an Axio Imager Z1 microscope (Zeiss) coupled with an AxioCam MRm camera (Zeiss) and images were processed with AxioVision LE and ImageJ.
Protein extracts and immunoblotting
Cells were solubilized in 50-ml lysis buffer containing 50 mM tris-HCl (pH 7.4), 200 mM NaCl, 1 mM EDTA, 0.2% NP-40, protease inhibitor (cOmplete; catalog no. 4693116001, Sigma-Aldrich), and phosphatase inhibitor cocktails (PhosSTOP; catalog no. 4906845001, Sigma-Aldrich). Proteins (25 mg per lane) were resolved on SDS polyacrylamide gel electrophoresis and transferred onto Immobilon-P membrane (Millipore). The membrane was saturated with PBS supplemented with 5% milk and 0.1% Tween 20 and incubated with the following antibodies: GFP (1:1000; catalog no. ab6673, Abcam) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:5000; sc32233, Santa Cruz Biotechnology). Western blot chemiluminescence detection was performed using a Fusion FX7 Spectra imager (Vilber). Band intensity was quantified and normalized using ImageJ software.
In situ hybridization
Samples were dissected and fixed overnight with 4% (w/v) paraformaldehyde and then processed for paraffin embedding. Microtome sections of 7-μm thickness were processed for in situ hybridization. Stra8 digoxigenin-labeled riboprobe was synthesized, and in situ hybridization analyses were performed as described in (29). Imaging was performed on an MZ9.5 microscope (Leica) coupled with a DHC490 camera (Leica) and Leica application suite V3.3.0 software and processed with Adobe Photoshop. For each genotype, n = 3 to 5 embryos.
Immunological analyses
Samples were fixed overnight with 4% (w/v) paraformaldehyde and then processed either for paraffin embedding or directly for whole-mount immunostaining. Microtome sections of 5-μm thickness were processed for immunostaining. Immunofluorescence analyses were performed as described (29). The following dilutions of primary antibodies were used: ALDH1A1 (1:50; catalog no. 52492, Abcam), ALDH1A2 (1:500; catalog no. HPA010022, Sigma-Aldrich), CDH1 (1:100; catalog no. 610182, BD Transduction Laboratories), DAZL (1:200; catalog no. GTX89448, GeneTex), DDX4 (1:200; catalog no. 13840, Abcam), GATA4 (1:200; catalog no. 1237, Santa Cruz Biotechnology), GFP (1:750; catalog no. TP401, Torrey Pines Bio Labs), POU5F1 (1:250; catalog no. 611202, BD Transduction Laboratories), phospho-γH2AX (1:300; catalog no. 16193, Millipore), SOX2 (1:200; catalog no. 97959, Abcam), SYCP3 (1:200; catalog no. 15093, Abcam), TRA98 (1:150; catalog no. 82527, Abcam), and FUT4 (SSEA1) (1:200; catalog no. 21702, Santa Cruz Biotechnology). Slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) diluted in the mounting medium at 10 μg/ml (Vectashield, Vector Laboratories) to detect nuclei. Imaging was performed with a motorized Axio Imager Z1 microscope (Zeiss) coupled with an AxioCam MRm camera (Zeiss), and images were processed with AxioVision LE and ImageJ. ImageJ software was used for quantification of proliferating germ cells versus total germ cells. Whole-organ imaging was performed on an LSM 780 NLO inverted Axio Observer Z1 confocal microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) using a Plan Apo 10× dry NA (numerical aperture) 0.45 objective. In monophoton mode, images were acquired using an argon laser (488 nm) and DPSS (green and yellow diode-pumped solid state) (561 nm). The microscope z-drive was used for z acquisitions, and an automated xy stage was used for multiposition recording acquisitions (Märzhäuser, Wetzlar, Germany). For each genotype, n = 3 to 5 embryos.
Immunodetection and quantification of proliferating germ cells
Paraffin sections from each genotype were processed for immunohistological experiments with POU5F1, DDX4, or GATA4 antibody. Then, proliferation analysis was performed on the same sections by BrdU labeling, and detection was performed using an appropriate kit (catalog no. 11 296 736 001, Roche). Total germ cells and proliferating cells were quantified on the entire section using ImageJ software. For each picture, the number of BrdU-positive germ cells (proliferating) and the number of either OCT4- or DDX4-positive cells (total) were counted. Then, the percentage of BrdU-positive versus OCT4- or DDX4-positive germ cells was determined. For each genotype (n = 4 to 7; 15 pictures per genotype), the mean and mean + SEM of these percentages were calculated and reported on a graph after statistical analysis (for details, see the “Statistical analysis” section).
Supplementary Material
Acknowledgments
We thank L. Turchi, F. Massa, S. Lachambre, and S. Rekima for their help. Funding: This work was supported by grants from the Agence Nationale de la Recherche (ANR-13-BSV2-0017-02 ARGONADS, ANR-11-LABX-0028-01, and ANR-18-CE14-0012 SexSpecs). M.L.R. was supported by a fellowship from La Ligue Nationale Contre le Cancer. A.S. was supported by a grant from La Ligue Nationale Contre le Cancer (Equipe Labellisée Ligue Nationale Contre le Cancer). The microscopy was done in the Prism facility, “Plateforme Prism,” and the histological experiments were done at the “Experimental Histopathology Platform,” IBV-CNRS UMR 7277-INSERM U1091-UNS. Author contributions: A.-A.C., M.-C.C., and N.B.G. designed the study, analyzed the data, and wrote the paper. A.-A.C., M.L.R., I.S., G.J., J.-M.G., and F.D.S. performed the experiments and analyzed the data. E.P., S.N., and A.S. discussed the results and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/21/eaaz1261/DC1
REFERENCES AND NOTES
- 1.Mikedis M. M., Downs K. M., Mouse primordial germ cells: A reappraisal. Int. Rev. Cell Mol. Biol. 309, 1–57 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Nef S., Stévant I., Greenfield A., Characterizing the bipotential mammalian gonad. Curr. Top. Dev. Biol. 134, 167–194 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Kocer A., Reichmann J., Best D., Adams I. R., Germ cell sex determination in mammals. Mol. Hum. Reprod. 15, 205–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams I. R., McLaren A., Sexually dimorphic development of mouse primordial germ cells: Switching from oogenesis to spermatogenesis. Development 129, 1155–1164 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Baltus A. E., Menke D. B., Hu Y. C., Goodheart M. L., Carpenter A. E., de Rooij D. G., Page D. C., In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat. Genet. 38, 1430–1434 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Oulad-Abdelghani M., Bouillet P., Décimo D., Gansmuller A., Heyberger S., Dollé P., Bronner S., Lutz Y., Chambon P., Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J. Cell Biol. 135, 469–477 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowles J., Knight D., Smith C., Wilhelm D., Richman J., Mamiya S., Yashiro K., Chawengsaksophak K., Wilson M. J., Rossant J., Hamada H., Koopman P., Retinoid signaling determines germ cell fate in mice. Science 312, 596–600 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Koubova J., Menke D. B., Zhou Q., Capel B., Griswold M. D., Page D. C., Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. U.S.A. 103, 2474–2479 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Chatzi C., Brade T., Cunningham T. J., Zhao X., Duester G., Sex-specific timing of meiotic initiation is regulated by Cyp26b1 independent of retinoic acid signalling. Nat. Commun. 2, 151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowles J., Feng C. W., Miles K., Ineson J., Spiller C., Koopman P., ALDH1A1 provides a source of meiosis-inducing retinoic acid in mouse fetal ovaries. Nat. Commun. 7, 10845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossant J., Zirngibl R., Cado D., Shago M., Giguere V., Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 5, 1333–1344 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Teletin M., Vernet N., Ghyselinck N. B., Mark M., Roles of retinoic acid in germ cell differentiation. Curr. Top. Dev. Biol. 125, 191–225 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L., A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Molotkov A., Molotkova N., Duester G., Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development 133, 1901–1910 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bingham N. C., Verma-Kurvari S., Parada L. F., Parker K. L., Development of a steroidogenic factor 1/Cre transgenic mouse line. Genesis 44, 419–424 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Zhou B., Ma Q., Rajagopal S., Wu S. M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K. R., Pu W. T., Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109–113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong J. F., Pritchard-Jones K., Bickmore W. A., Hastie N. D., Bard J. B., The expression of the Wilms’ tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 40, 85–97 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Hayashi S., McMahon A. P., Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244, 305–318 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Minkina A., Lindeman R. E., Gearhart M. D., Chassot A. A., Chaboissier M. C., Ghyselinck N. B., Bardwell V. J., Zarkower D., Retinoic acid signaling is dispensable for somatic development and function in the mammalian ovary. Dev. Biol. 424, 208–220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. A. Kane, J. L. Napoli, Quantification of endogenous retinoids, in Retinoids, H. Sun, G. H. Travis, Eds. (Humana Press, 2010), vol. 652, pp. 1–54. [DOI] [PMC free article] [PubMed]
- 21.Allenby G., Janocha R., Kazmer S., Speck J., Grippo J. F., Levin A. A., Binding of 9-cis-retinoic acid and all-trans-retinoic acid to retinoic acid receptors α, β, and γ. Retinoic acid receptor γ binds all-trans-retinoic acid preferentially over 9-cis-retinoic acid. J. Biol. Chem. 269, 16689–16695 (1994). [PubMed] [Google Scholar]
- 22.Morita Y., Tilly J. L., Segregation of retinoic acid effects on fetal ovarian germ cell mitosis versus apoptosis by requirement for new macromolecular synthesis. Endocrinology 140, 2696–2703 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Gill M. E., Hu Y. C., Lin Y., Page D. C., Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc. Natl. Acad. Sci. U.S.A. 108, 7443–7448 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans E. P., Ford C. E., Lyon M. F., Direct evidence of the capacity of the XY germ cell in the mouse to become an oocyte. Nature 267, 430–431 (1977). [DOI] [PubMed] [Google Scholar]
- 25.Bellutti L., Abby E., Tourpin S., Messiaen S., Moison D., Trautmann E., Guerquin M.-J., Rouiller-Fabre V., Habert R., Livera G., Divergent roles of CYP26B1 and endogenous retinoic acid in mouse fetal gonads. Biomolecules 9, E536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyauchi H., Ohta H., Nagaoka S., Nakaki F., Sasaki K., Hayashi K., Yabuta Y., Nakamura T., Yamamoto T., Saitou M., Bone morphogenetic protein and retinoic acid synergistically specify female germ-cell fate in mice. EMBO J. 36, 3100–3119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokobayashi S., Liang C. Y., Kohler H., Nestorov P., Liu Z., Vidal M., van Lohuizen M., Roloff T. C., Peters A. H., PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 495, 236–240 (2013). [DOI] [PubMed] [Google Scholar]
- 28.DiTroia S. P., Percharde M., Guerquin M.-J., Wall E., Collignon E., Ebata K. T., Mesh K., Mahesula S., Agathocleous M., Laird D. J., Livera G., Ramalho-Santos M., Maternal vitamin C regulates reprogramming of DNA methylation and germline development. Nature 573, 271–275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chassot A. A., Gregoire E. P., Lavery R., Taketo M. M., de Rooij D. G., Adams I. R., Chaboissier M. C., RSPO1/β-catenin signaling pathway regulates oogonia differentiation and entry into meiosis in the mouse fetal ovary. PLOS ONE 6, e25641 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussein S. M., Duff E. K., Sirard C., Smad4 and β-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J. Biol. Chem. 278, 48805–48814 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Le Bouffant R., Souquet B., Duval N., Duquenne C., Herve R., Frydman N., Robert B., Habert R., Livera G., Msx1 and Msx2 promote meiosis initiation. Development 138, 5393–5402 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Krentz A. D., Murphy M. W., Sarver A. L., Griswold M. D., Bardwell V. J., Zarkower D., DMRT1 promotes oogenesis by transcriptional activation of Stra8 in the mammalian fetal ovary. Dev. Biol. 356, 63–70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaren A., Meiosis and differentiation of mouse germ cells. Symp. Soc. Exp. Biol. 38, 7–23 (1984). [PubMed] [Google Scholar]
- 34.Byskov A. G., Saxén L., Induction of meiosis in fetal mouse testis in vitro. Dev. Biol. 52, 193–200 (1976). [DOI] [PubMed] [Google Scholar]
- 35.Best D., Sahlender D. A., Walther N., Peden A. A., Adams I. R., Sdmg1 is a conserved transmembrane protein associated with germ cell sex determination and germline-soma interactions in mice. Development 135, 1415–1425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacLean G., Li H., Metzger D., Chambon P., Petkovich M., Apoptotic extinction of germ cells in testes of Cyp26b1 knockout mice. Endocrinology 148, 4560–4567 (2007). [DOI] [PubMed] [Google Scholar]
- 37.MacLean G., Abu-Abed S., Dollé P., Tahayato A., Chambon P., Petkovich M., Cloning of a novel retinoic-acid metabolizing cytochrome P450, Cyp26B1, and comparative expression analysis with Cyp26A1 during early murine development. Mech. Dev. 107, 195–201 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Dupé V., Matt N., Garnier J. M., Chambon P., Mark M., Ghyselinck N. B., A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc. Natl. Acad. Sci. U.S.A. 100, 14036–14041 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matt N., Dupé V., Garnier J. M., Dennefeld C., Chambon P., Mark M., Ghyselinck N. B., Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development 132, 4789–4800 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Vermot J., Garnier J. M., Dierich A., Niederreither K., Harvey R. P., Chambon P., Dollé P., Conditional (loxP-flanked) allele for the gene encoding the retinoic acid-synthesizing enzyme retinaldehyde dehydrogenase 2 (RALDH2). Genesis 44, 155–158 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Stévant I., Kühne F., Greenfield A., Chaboissier M. C., Dermitzakis E. T., Nef S., Dissecting cell lineage specification and sex fate determination in gonadal somatic cells using single-cell transcriptomics. Cell Rep. 26, 3272–3283.e3 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/21/eaaz1261/DC1


