Abstract
Under internal or external insults such as aging and oxidative stresses, cells are induced into a senescent state and stop cellular division permanently. As senescent cells (SnCs) accumulate, the regeneration capacity of biological tissue would be compromised, which has been found to be associated with a plethora of age-related disorders. Therefore, isolating SnCs becomes necessary. To address the lack of effective surface markers for SnCs isolation, a label-free microfluidic device was proposed in this paper, in which a spiral microchannel was deployed to isolate SnCs based on their size differences. We adopted a well-received cellular senescence model by exerting excessive oxidative stress to murine mesenchymal stem cells. This model was then validated through a series of SnCs characterizations including size measurement, p16INK4a expression level, senescence-associated beta-galactosidase, and doubling time. The senescence chip demonstrated an efficiency of 75% and viability over 85% at a flow rate of 5 ml/min. The average cell size from the inner outlet was 5 μm larger than that from the outer outlet. The isolated cells had a sixfold higher p16INK4a expression level. Overall, the chip had an area under curve of 0.719 in the receiver operating characteristic analysis, showing decent performance in sorting SnCs. By having the ability to perform size-based sorting at a high flow rate, such a microfluidic device can provide high-throughput and label-free isolation of SnCs. To further improve the isolation performance, the device can be modified to introduce additional physical biomarkers of SnCs such as stiffness. This device poses a good potential in purification for cytotherapy or estimation of biological age.
I. INTRODUCTION
Cellular senescence was first identified in 1965 as the stable cellular state of proliferation arrest regardless of sufficient supply of nutrients and space.1 In recent years, several biochemical and biophysical hallmarks of senescent cells (SnCs) have been discovered. Demaria and his group gave a comprehensive summary and discussion on state-of-the-art understanding of the cellular senescence hallmarks.2 Among different hallmarks, an essential group is the senescence-associated secretory phenotype (SASP).3,4 In general, SASP refers to a group of secretomes including growth factors, proteases, chemokines, and cytokines secreted by SnCs.5 The accumulation of SASP influences the neighboring environment, affecting tissue functions. A variety of age-related diseases such as Alzheimer's diseases and osteoarthritis are found to be associated with the senescence process.6 In a recent study, osteoarthritis-like lesions were triggered in the knee joints of mice after transplantation of SnCs into the synovial joint.7 The transplanted SnCs exhibited a 20-fold higher production of Interleukin 26, a key component of SASP, over non-SnCs. As a result of SASP secretion, SnCs could have possibly provoked inflammation leading to osteoarthritis. In addition, SnCs are also not favored when it comes to mesenchymal stem cell (MSC) therapy due to decreasing stemness.8 The loss in stemness eventually leads to a degeneration of differential potential, resulting in poor therapeutic efficiency. It has been shown that removing senescent MSCs contributes to chondrogenic potential when using MSCs for cartilage regeneration.9 In view of these undesired impacts of SnCs, being able to isolate them will be greatly beneficial in treating diseases and improving therapeutic efficiency.
The intrinsic features of SnCs make them difficult to be separated. First, SnCs account for a rather small portion in both tissues and biofluids. For example, it was rare to find more than two senescent dermal fibroblasts in 1 mm2 of the dermis.10 Besides, SnCs usually exist in a heterogeneous population together with other non-SnCs. Therefore, selecting a proper biomarker is essential to effectively and efficiently isolate SnCs. However, the gold standard in cell sorting, fluorescence-activated cell sorting (FACS), is not suitable for isolating SnCs. This is because of the unavailability of a single biomarker to selectively target at SnCs. Instead, multiple biomarkers have to be labeled in order to identify SnCs.11 As suggested, a combination of several biomarkers including senescence-associated beta-galactosidase (SA-βgal), Lamin B1, high mobility group box 1 protein, and p16INK4a (p16) may be necessary for detecting SnCs. Employing multiple biomarkers for labeling not only raises the cost but also increases the complexity of the system. Considering the aforementioned limitations of label-based cell sorting, label-free sorting based on biophysical markers may provide a better solution in isolating SnCs.
When cells become senescent, the mTOR pathway is initiated, which is a senescence-stimuli response pathway regulating cell size.12,13 In consequence, cells enlarge their size and become hypertrophy.14 This opens up a door for developing a cost-effective and simpler label-free cell sorting method based on the size difference between normal cells and SnCs. The feasibility of size-based SnCs sorting has been shown in some recently published work. A microfluidic filter was fabricated to capture large senescent fibroblasts when flushing the sample perpendicular to the filter surface.15 This dead-end flow filtration was effective but low in efficiency. The filter could be easily saturated when SnCs started to block all the gaps. To overcome such drawbacks, another microfluidic chip integrated the design of dead-end flow and cross-end flow filtration, where an array of pillars were placed at a certain angle inclined to the fluidic flow.16 Although this design demonstrated a higher efficiency, it essentially relies on filtration for isolation which inevitably causes clogging. In contrast, spiral microfluidic devices utilizing inertial forces to sort cells of different sizes can effectively address the above-mentioned issues. In the curved channel, a particle undergoes two opposing forces and becomes focused at a certain equilibrium position.17 As the values of both forces are size-dependent, cells with different sizes are driven to different equilibrium positions and get sorted accordingly. In the past few years, accumulating studies have demonstrated the feasibility of such a design by its success in isolating circulating tumor cells from blood and MSCs from bone marrow.18,19 Given the unmet need for an effective system to investigate cellular senescence, this work employed spiral microfluidics to conduct size-based cell sorting on SnCs. To achieve that, a cellular model for senescence induction was first established and verified. Subsequently, the microfluidic device was fabricated and calibrated for isolation using fluorescent beads at particular sizes. As a proof of concept, the induced SnCs were used to evaluate the performance of the microfluidic device. The device demonstrated a fairly good performance in isolating senescent MSCs and has a great potential in many biomedical applications.
II. METHODS
A. Device design and fabrication
The design of the device was first developed via SolidWorks (Dassault Systèmes, USA). Specifically, the spiral microfluidics had a width of 600 μm and a height of 130 μm. At the bifurcation, the width of the inner outlet and the outer outlet were 100 μm and 500 μm, respectively. The design was then fabricated into an aluminum mold through micro-milling. The microfluidic device was fabricated via molding and casting on the mold following previous work.20 Briefly, mixed polydimethylsiloxane (PDMS) (monomer:crosslinker = 10:1, Sylgard 184, Dow Corning Inc., USA) was first cast onto the mold and baked in an oven. Subsequently, cured PDMS was peeled from the mold and holes were punched for the inlets and outlets. Finally, the PDMS was bonded onto a glass slide after plasma treating both surfaces.
B. In vitro cellular senescence induction
Cellular senescence was induced by either replication or oxidative stress. For both approaches, murine MSCs (mMSCs) were first cultured in Minimum Essential Medium Eagle—Alpha Modification (α-MEM) (GibcoTM) with 10% fetal bovine serum (GibcoTM) and 1% penicillin/streptomycin (GibcoTM) in 25-cm2 flask at 37 °C with 5% CO2. In the replicative senescence group, mMSCs were passaged to a new flask at around 95% confluency. Approximately 20% of those mMSCs were seeded to the new flask. mMSCs were continuously passaged from passage 13 up to passage 39. In this work, cells with a passage number of less than 20 were defined as early passage. Cells at passage 20–29 were defined as middle passage. All the cells with a passage number higher than 30 were defined as late passage. In the oxidative stress-induced senescence group, H2O2 was selected as the oxidizing agent to raise oxidative stress. At around 80% confluency, H2O2 was added into the media to induce cellular senescence on Day 0. Cells were exposed to media containing 100 μM H2O2 for three days without any replacement of the media. As for control, cells were cultured with normal α-MEM media. On Day 3, cells were collected. In the oxidative stress-induced senescence group, cells at early, middle, and late passages were all chosen and exposed to the oxidative stress.
C. Experimental setup and calibration
The senescence chip was set up as shown in Fig. 1(a). In the calibration of the isolation efficiency, two sizes of fluorescent microspheres (15 μm, excitation 400 nm, emission 450 nm; 20 μm, excitation 570 nm, emission 610 nm; Tianjin BaseLine, China) were used to simulate the behavior of normal cells and SnCs in the microchannel under different flow rates. Microspheres were first dispersed in phosphate-buffered saline (PBS) and loaded into a disposable 10 ml syringe (Terumo). An infusion pump (SPLab01, Baoding Shenchen Precision Pump) was used to initiate and maintain the flow. To visualize the orbit of microparticles with different sizes under different flow rates, images of the bifurcation were captured with a fluorescent microscope (Leica TCS SPE Confocal Microscope).
FIG. 1.
(a) The experimental setup of the microfluidic device for SnCs isolation. (b) Real-size image of the device. (c) Simulation illustration of the principle of the size-based SnCs isolation at the flow rate of 5 ml/min.
D. Device operation
Sample cells were first dispersed in serum-free media and loaded into a disposable 10 ml syringe (Terumo). To reduce non-specific cell-to-cell or cell-to-channel binding, 0.1% v/v Pluronic 188 poloxamer was added into the sample solution. The microfluidic chip was placed under a fluorescent microscope and observed through a camera (Canon EOS600D). The induced senescent mMSCs were used to conduct isolation. Based on the results of calibration, the flow rates were further fine-tuned in an attempt to figure out optimal conditions for cell isolation. To quantitatively evaluate the performance of the size-based cell isolation, the efficiency was defined as
| (1) |
where cells with size larger than 20 μm were considered as “large cells.”
E. Sample characterization
1. Cell size measurement
The cell size was measured via an image-based analysis method. Images of the suspended cells on the hemocytometer were captured with bright field microscopy and analyzed with ImageJ (NIH, USA), where the diameters of the cells were measured manually.
2. Cell doubling time
Cell doubling time was calculated by counting the cell numbers using a hemocytometer at different time points. Sample cells were cultured on six-well plates with equal seeding density on Day 0. After 24 h of incubation, first counting was conducted. Subsequent cell counting was done every 24 h until Day 5. The cell doubling time was calculated with the equation,
where was the time difference, Nt was the final cell number, and Ni was the initial cell number.
3. SA-βgal staining
SA-βgal was used to verify whether cells were in senescence. The sample cells were stained with a commercial staining kit following the manufacturer's protocol (SA-βgal Staining Kit, Cell Signalling Technology #9860). Cells were first washed and fixed at room temperature. The fixed cells were then stained at 37 °C without the supply of CO2 overnight. Stained cells were observed under the bright field of a microscope.
4. Quantitative polymerase chain reaction (qPCR)
qPCR of p16 was conducted to measure the expression of p16 at the RNA level. The RNA was first extracted using Total RNA Kit I (Omega Bio-Tek). After RNA extraction, reverse transcription was conducted using RevertAid First Strand cDNA Synthesis kit (ThermoScientific). Subsequently, cDNA of p16 (5′-GAACTCTTTCGGTCGTACCC-3′ and 5′-CGAATCTGCACCGTAGTTGA-3′) and β-actin (5′-TGAGAGGGAAATCGTGCGTG-3′ and 5′-TGCTTGCTGATCCACATCTGC-3′) was amplified through qPCR. The expression level of a certain RNA was reflected as the cycle number for amplification. β-actin was chosen as the reference to conduct relative quantification on p16 expression level.
5. Cell viability
The cell viability was measured by trypan blue staining. 10 μl of cells were taken from the sample and mixed with the same volume of trypan blue staining solution. The viability was then calculated by counting the cell numbers of trypan blue positive vs trypan blue negative using a hemocytometer.
6. Numerical simulation
Numerical simulation was conducted to study the flow velocity and pressure distribution at the optimal separation flow rated. The device design was imported to COMSOL Multiphysics (COMSOL Inc.). Fluid properties were set to be laminar flow and water was selected as the liquid as cells were suspended in culture media during the isolation. The flow rate used for simulation was 5 ml/min based on both microsphere calibration and cell trails.
F. Data analysis
Receiver operating characteristic (ROC) analysis was conducted with SPSS (IBM, USA) to evaluate the performance of the chip under optimal conditions (flow rate: 5 ml/min). The test outcome was collected from the size measurement of the two outlets. The condition was that all the cells at the inner outlet were senescent and all the cells at the outer outlet were non-senescent. The corresponding area under the curve (AUC) was calculated by the software. An AUC value lower than 0.6 was classified as a failure, whereas a value higher than 0.7 was defined to be fairly good. An excellent performance was expected to have a value higher than 0.8.
III. RESULTS
A. Cellular senescence induction
Both replicative senescence group and oxidative stress-induced senescence group underwent a series of assays including SA-βgal staining, size measurement, qPCR on p16, and cell doubling time to verify whether senescence had been successfully induced. In SA-βgal staining, no obvious positive signals were spotted in cells at early, middle, and late passage without H2O2 treatment [Fig. 2(a) i–iii]. Although mMSCs at early passage lacked SA-βgal positive signals after exposure to H2O2 [Fig. 2(a) iv], positive SA-βgal signals were found in mMSCs at the middle and late passages (Figs. 2(a) v and 2(a) vi). Besides, the mMSCs at middle and late passage showed a significant morphology change under oxidative stress, becoming more irregular in shape and occupying a larger spreading area. The results from qPCR on p16 demonstrated a similar trend with SA-βgal staining [Fig. 2(b)]. There was no significant difference in the p16 expression level among cells at different passage numbers without exposure to oxidative stress. By introducing H2O2 into culture media, p16 expression levels increased in mMSCs at the middle and late passage for 2.5 and 2.8 folds, respectively. As for mMSCs at the early passage, the expression of p16 was diminished. In terms of cell size, there was no significant difference among cells at different passage numbers without oxidative stress. However, all three groups exhibited different degrees of enlargement in size after being exposed to oxidative stress [Fig. 2(c)]. The results on cell doubling time show that mMSCs at the late passage proliferated around three times as slow as the cells at the early and middle passage [Fig. 2(d)]. After exposure to the excessive oxidative stress, all of these cells slowed down their proliferation rates with the cells at the late passage having the most statistically significant reduction.
FIG. 2.
(a) SA-βgal staining of mMSCs at different passage numbers with or without H2O2 treatment. (i–iii) mMSCs without H2O2 treatment; (iv–vi) mMSCs with H2O2 treatment. (b) Fold changes in p16 expression level from cells at different passage numbers with or without H2O2 treatment. (c) Average sizes of cells at different passage numbers with or without H2O2 treatment. (d) Doubling time of cells at different passage numbers with or without H2O2 treatment. *p < 0.05, **p < 0.01, and ***p > 0.1.
B. Calibration of spiral microfluidic chip
Figure 3 showed the different focusing behavior of particles of different sizes. At the flow rate of 1 ml/min, both particles were not focused and flowed freely across the channel [Fig. 3(a)]. Starting from 2 ml/min, both particles began to focus and flowed into the inner inlet [Figs. 3(b)–3(d)]. At 5 ml/min, small particles (15 μm) started to be dragged outward and tended to focus into the outer outlet [Fig. 3(e)]. Meanwhile, large particles (20 μm) remained close to the inner wall of the channel and continuously flowed into the inner outlet [Fig. 3(e)].
FIG. 3.
Fluorescent images on different sizes of microspheres under different flow rates. Flow rate: (a) 1 ml/min, (b) 2 ml/min, (c) 3 ml/min, (d) 4 ml/min, and (e) 5 ml/min. Blue microspheres: 15 μm; red microspheres: 20 μm.
C. Isolation of SnCs
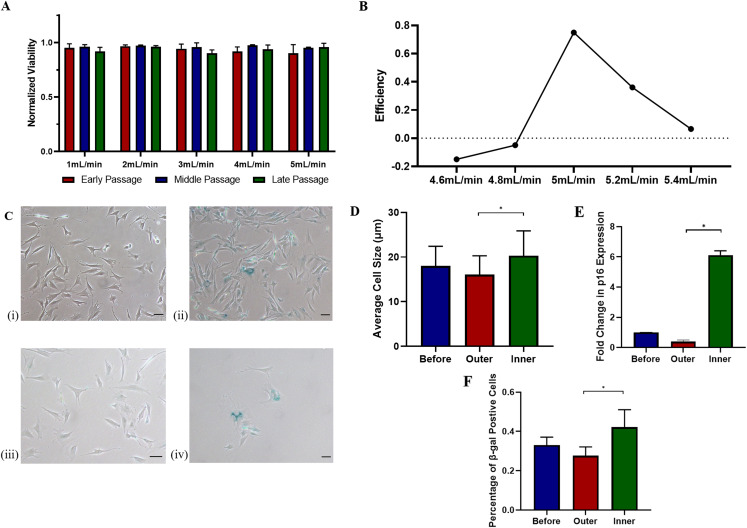
The simulation of flow velocity demonstrated that a secondary dean flow was induced inside the spiral channel compared to a homogeneous flow profile in the inlet [Fig. 1(c)]. The large and senescent cells were pushed close to the inner wall of the channel as a result of the balance between the dean drag force and the inertial lift force. The pressure was highest in the inlet and gradually reduced along the channel. To ensure that flow rates used in calibration were compatible with cells, viability was measured immediately after the operation. All flow rates were proved to be compatible with cells at different passage numbers as the cell viability remained higher than 85% after isolation [Fig. 4(a)]. The flow rate was further tuned around 5 ml/min to determine an optimal flow rate. As shown in Fig. 4(b), the efficiency reached a maximum of 0.75 at a flow rate of 5 ml/min. Once the optimal flow rate was selected, the device's performance on isolating induced senescent mMSCs was evaluated via SA-βgal staining, qPCR on p16, size measurement, and ROC analysis. After the successful induction of senescence, induced senescent mMSCs collected at the inner outlet remained senescent after isolation [Fig. 4(c)]. Cells collected at the inner outlet were larger than those at the outer outlet [Fig. 4(d)]. It is also consistent with the results from qPCR, where the p16 expression level of the inner outlet was almost six times as much as that of the outer outlet [Fig. 4(e)]. Meanwhile, the p16 expression level of the outer outlet was lower than that of the inlet, suggesting fewer SnCs were collected from the outer outlet. A similar trend was observed when quantitatively analyzing the SA-βgal staining [Fig. 4(f)]. On top of that, ROC analysis provided a quantitative evaluation of the performance of isolation (Fig. 5). As the ROC curve lies above the random guess curve [Fig. 5(a)], it manifests that the chip was effective in isolating senescent mMSCs. The calculated AUC was 0.719 [Fig. 5(b)], which was considered to be fairly good.
FIG. 4.
(a) Normalized viability of H2O2-treated mMSCs from different passage numbers collected at the outlet under different flow rates (n = 3). (b) The efficiency of the spiral microfluidics under different flow rates. (c) SA-βgal staining on mMSCs (i) Normal mMSCs without senescence induction. (ii) Induced senescent mMSCs before isolation. (iii) Induced senescent mMSCs collected from the outer outlet after isolation. (iv) Induced senescent mMSCs collected from the inner outlet after isolation. (d) Average sizes of the induced senescent mMSCs collected before and after isolation at the optimal flow rate. (e) qPCR on p16 expression from induced senescent mMSCs collected before and after isolation at the optimal flow rate. (f) Percentage of SA-βgal positive cells collected before and after isolation at the optimal flow rate. Scale bar: 50 μm. *p < 0.01.
FIG. 5.
ROC analysis of the performance of SnCs isolation. (a) ROC curve for the performance of isolation (flow rate: 5 ml/min; blue line: test outcome; red line: random guess). (b) Statistic analysis on the AUC.
IV. DISCUSSION
To validate the induction of cellular senescence, four different assays were conducted including proliferation rate measurement, size measurement, SA-βgal staining, and qPCR on p16. Cellular senescence was first identified as the termination in proliferation.1 Hence, the decrease in the proliferation rate is the most straightforward indicator. Measuring cell size indicates the degree of hypertrophy which, as aforementioned, is a biophysical marker of cellular senescence. As for SA-βgal, it is a lysosomal enzyme used as the indicator for the increased lysosomal content in SnCs. The positive SA-βgal signals are normally detected only in SnCs but absent in pre-senescent and quiescent cells.21,22 p16 is a tumor suppressor protein that inhibits the progression from the G1 phase to the S phase in a cell cycle. The increasing expression of this marker along the development of senescence has been discovered both in vitro and in vivo models.23,24 Both SA-βgal and p16 are commonly used markers for cellular senescence at the biomolecular level.25
Based on the above four assays, oxidative stress-induced senescence seems to be more effective than replicative senescence. Among mMSCs at the early, middle, and late passage, none of them exhibited the phenotype of cellular senescence [Figs. 2(a)i–iii]. Although a decrease in the proliferation rate was observed for late passage mMSCs in the experiment [Fig. 2(d)], these cells may not have eventually reached the point of termination of proliferation. Compared to Hayflick limit at around 40–60 doubling for fibroblasts,26 there could be several times of doubling before this lineage of mMSCs eventually became senescent. Moreover, the exact passage number when cells become senescent could vary among different lineages of the same type of cells. Taking human MSCs (hMSCs) as an example, over 80% of the hMSCs at Passage 10 became SA-βgal positive in the study of Wagner et al, whereas hMSCs at Passage 10 in the work of Yin et al. were still considered as the early passage.9,27 In consideration of these findings, replicative senescence may not be an effective method to induce senescence with consistent outcomes.
In the oxidative stress-induced senescence group, both mMSCs at middle and late passage showed positive signals after SA-βgal staining [Fig. 2(a) v–vi], had a higher expression of p16 [Fig. 2(b)], became larger [Fig. 2(c)], and had a prolonged doubling time [Fig. 2(d)], exhibiting hallmarks of senescence. However, except for the increase in cell size and doubling time, the other two hallmarks were not observed in cells at early passage after exposure to H2O2 [Figs. 2(b) and 2(c)]. When confronted with the same level of oxidative stress, cells at the early passage seemed to have better resilience to the stress. The term cell resilience is defined as the capability of cells to remain viable and protect themselves against external stress.28 The attenuation in cell resilience is believed to happen prior to senescence and deteriorate further with the aging process.29,30 For cells at middle and late passages, even though senescence was not detected without H2O2 treatment, their resilience against oxidative stress had reduced. As a result, cells became senescent after H2O2 treatment. Hence, cells at the middle or late passage were more suitable for oxidative stress-induced senescence. This is also consistent with the previous study where old hMSCs demonstrated a reduced tolerance to oxidative stress.31 In order to bring senescence to cells at the early passage, a more severe oxidative stress may be necessary to break their robust resilience.
The calibration of the device with fluorescent microspheres proved the feasibility of isolating SnCs (larger than 20 μm) from non-SnCs (around 15 μm) via the spiral microfluidic device (Fig. 3). The size of the fluorescent beads was intentionally selected based on the size measurement in Fig. 2(c). The average cell size before H2O2 induction was around 15 μm, whereas the size after successful induction was around 20 μm. With beads separation at the flow rate of 5 ml/min [Fig. 3(e)], the optimal flow rate for size-based cell sorting was studied within the range from 4.6 to 5.4 ml/min using the induced SnCs with the mentioned induction method. Prior to the investigation on efficiency, the viability test confirmed the safe use of flow rates within such a range [Fig. 4(a)]. In the numerical simulation, even though the pressure was relatively high at the inlet, it quickly dropped along the channel. In other words, the cells were exposed to the high-stress environment for only seconds [Fig. 1(c)], whereas it usually takes more than 10 min for the shear stress to have detrimental effects on MSCs.32,33 In Fig. 4(b), negative efficiency values were gained at a flow rate lower than 5 ml/min. This implied that some large cells inevitably flowed to the outlet, while many small cells still flowed to the inner outlet. It is consistent with Fig. 3(d), where many small beads flowed to the inner outlet. The device reached a maximum efficiency of 75% at a flow rate of 5 ml/min. The efficiency dropped down with a further increased flow rate. This is because more and more large cells started to flow to the outer outlet. To ensure that the large cells collected from the inner outlets were SnCs, SA-βgal staining and qPCR on p16 expression were conducted. In consistence with efficiency, the SnC concentration increased in the population collected from the inner outlet and decreased in the outer outlet [Fig. 4(f)]. Notably, the p16 expression of cells from the inner outlet was almost 6 times higher than that of the outer outlet in Fig. 4(e) even though the number of SA-βgal positive cells was 1.5 times larger in Fig. 4(f). As the expression level of p16 associates with the degree of senescence,34 it is highly likely that our device is capable of isolating severely senescent mMSCs. Moreover, an AUC value of 0.719 based on ROC analysis indicated a fairly good performance in isolation (Fig. 5). Although the AUC value did not go as high as over 0.8 for excellent performance, the proposed microfluidic platform is still considered to be effective in isolating SnCs for proof-of-concept trails. Since this size-based sorting approach via spiral microfluidics waived the process of cell labeling, the complexity and cost of operation were significantly reduced. Meanwhile, the flow rate used for isolation was sufficiently high for high-throughput isolation on SnCs without sacrificing the viability.
Although the spiral microfluidic device was proved to be effective in isolating senescent mMSCs, some limitations have to be solved to achieve excellent performance. First, the sensitivity of the device can be further improved. In Fig. 3(e), some of the small particles still flowed into the inner outlet at the optimal flow rate. This is probably because of the fluctuation of flow during the experiments and particle–particle interactions inside the flow. As a result, when the majority of the small cells were focused toward the outer outlet, there were still some small cells focused toward the inner outlet. This could be solved by either changing the cross section of the channel to a trapezoidal shape or introducing a sheath flow at the inlet of the chip.35,36 Second, the cell size could be a weak biomarker for isolation. Comparing Figs. 2(c), the cell size at early passage increased after H2O2 induction. However, neither qPCR nor SA-βgal staining confirmed the onset of senescence. In other words, SnCs were larger, but not all the larger cells were senescent. In an attempt to improve the specificity of this chip, another biomarker such as stiffness could be considered for secondary isolation after size-based sorting. The mechanical deformability of cytoplasm was reported to decrease with the progression of aging.37 Several microfluidic devices have already been successfully developed to conduct stiffness-based label-free cell sorting.38–40 Applying this biophysical feature of cellular senescence as a secondary isolation marker would not only preserve the strengths of label-free cell sorting but also improve the specificity of the device.
The isolation of senescent MSCs with the microfluidic device could have a wide range of applications. For example, the device could be used for eliminating SnCs from a heterogeneous population of cells. The senescent MSCs are undesired for regenerative medicine due to their low stemness.41 By removing SnCs with this device, we could improve the quality of the cell source for cytotherapy, thus increasing the therapeutic efficiency. The device can be further modified for quick estimations of biological ages. At the molecular scale, around 20% of the cellular senescence genes overlap with longevity-associated genes.42 In other words, biological ages can be derived by counting the number of senescent MSCs isolated from the biofluids. Therefore, the high-throughput and label-free approach offered by this device provides a simple and effective solution to these biomedical applications.
V. CONCLUSIONS
In summary, a spiral microfluidic device for high-throughput and label-free SnCs isolation was fabricated and tested in this work. Prior to using the device, the optimal senescence induction model was first established. Employing the loss in the cell resilience, exposing cells at the middle or late passage to H2O2 yielded the best performance in senescence induction. In subsequent tests with induced SnCs, the chip demonstrated efficiency of 75% for size-based sorting. After isolation, the p16 expression level increased six times in the concentrated population as compared to the original population. Over 85% of the cells remained viable after isolation. Although the device developed in this study is effective in isolating senescence cells, the efficiency can be further improved by modifying the device design. We believe that in its current version or with minor improvement in performance, this chip can be readily used for fundamental cellular senescence studies regarding chronic diseases, biological age estimation, and cell-based therapies.
ACKNOWLEDGMENTS
This study was supported by the Research Grants Council of Hong Kong Early Career Scheme (No. PolyU 251008/18M), the PROCORE-France/Hong Kong Joint Research Scheme (No. F-PolyU504/18), the Institute for Entrepreneurship at the Hong Kong Polytechnic University (PolyU) under Award No. POC-19(01)-004. K.J. acknowledges support from the MBI for the Graduate Scholarship. The authors would like to acknowledge Professor Mo Yang and Dr. Xin Zhao at PolyU for providing their research facilities, Mr. Sina Kheiri at the University of Toronto for his assistance in numerical simulation, and Dr. Clara Hung from University Research Facility in Life Sciences at PolyU for taking the fluorescent images in the device calibration. The authors declare no competing interests.
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Hayflick L., Exp. Cell Res. 37(3), 614–636 (1965). 10.1016/0014-4827(65)90211-9 [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Segura A., Nehme J., and Demaria M., Trends Cell Biol. 28(6), 436–453 (2018). 10.1016/j.tcb.2018.02.001 [DOI] [PubMed] [Google Scholar]
- 3.Farr J. N., Xu M., Weivoda M. M., Monroe D. G., Fraser D. G., Onken J. L., Negley B. A., Sfeir J. G., Ogrodnik M. B., and Hachfeld C. M., Nat. Med. 23(9), 1072 (2017). 10.1038/nm.4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campisi J., Andersen J. K., Kapahi P., and Melov S., “Cellular senescence: A link between cancer and age-related degenerative disease?,” in Seminars in Cancer Biology, Dec. 1, 2011 (Academic Press; ), Vol. 21, No. 6, pp. 354–359. 10.1016/j.semcancer.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freund A., Orjalo A. V., Desprez P.-Y., and Campisi J., Trends Mol. Med. 16(5), 238–246 (2010). 10.1016/j.molmed.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muñoz-Espín D. and Serrano M., Nat. Rev. Mol. Cell Biol. 15(7), 482 (2014). 10.1038/nrm3823 [DOI] [PubMed] [Google Scholar]
- 7.Xu M., Bradley E. W., Weivoda M. M., Hwang S. M., Pirtskhalava T., Decklever T., Curran G. L., Ogrodnik M., Jurk D., Johnson K. O., Lowe V., Tchkonia T., Westendorf J. J., and Kirkland J. L., J. Gerontol. Ser. A 72(6), 780–785 (2017). 10.1093/gerona/glw154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choumerianou D. M., Martimianaki G., Stiakaki E., Kalmanti L., Kalmanti M., and Dimitriou H., Cytotherapy 12(7), 881–887 (2010). 10.3109/14653249.2010.501790 [DOI] [PubMed] [Google Scholar]
- 9.Yin L., Wu Y., Yang Z., Tee C. A., Denslin V., Lai Z., Lim C. T., Lee E. H., and Han J., Lab Chip 18(6), 878–889 (2018). 10.1039/C7LC01005B [DOI] [PubMed] [Google Scholar]
- 10.Waaijer M. E., Parish W. E., Strongitharm B. H., van Heemst D., Slagboom P. E., de Craen A. J., Sedivy J. M., Westendorp R. G., Gunn D. A., and Maier A. B., Aging Cell 11(4), 722–725 (2012). 10.1111/j.1474-9726.2012.00837.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiley C. D., Flynn J. M., Morrissey C., Lebofsky R., Shuga J., Dong X., Unger M. A., Vijg J., Melov S., and Campisi J., Aging Cell 16(5), 1043–1050 (2017). 10.1111/acel.12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd A. C., Cell 154(6), 1194–1205 (2013). 10.1016/j.cell.2013.08.053 [DOI] [PubMed] [Google Scholar]
- 13.Blagosklonny M. V., Aging (Albany NY) 4(3), 159–165 (2012). 10.18632/aging.100443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blagosklonny M. V., J. Cell. Physiol. 209(3), 592–597 (2006). 10.1002/jcp.20750 [DOI] [PubMed] [Google Scholar]
- 15.Kim M. S., Jo S., Park J. T., Shin H. Y., Kim S. S., Gurel O., and Park S. C., Anal. Chem. 87(19), 9584–9588 (2015). 10.1021/acs.analchem.5b00445 [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., Mao P., Snijders A. M., and Wang D., Aging Cell, 17(2), e12722 (2018). 10.1111/acel.12722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuntaegowdanahalli S. S., Bhagat A. A. S., Kumar G., and Papautsky I., Lab Chip 9(20), 2973–2980 (2009). 10.1039/b908271a [DOI] [PubMed] [Google Scholar]
- 18.Khoo B. L., Grenci G., Lim Y. B., and Lee S. C., Han J. and Lim C. T., Nat. Protocols 13(1), 34 (2018). 10.1038/nprot.2017.125 [DOI] [PubMed] [Google Scholar]
- 19.Lee L. M., Rosano J. M., Wang Y., Klarmann G. J., Garson C. J., Prabhakarpandian B., Pant K., Alvarez L. M., and Lai E., Anal. Methods 10(7), 713–721 (2018). 10.1039/C7AY02500A [DOI] [Google Scholar]
- 20.Warkiani M. E., Guan G., Luan K. B., Lee W. C., Bhagat A. A. S., Chaudhuri P. K., Tan D. S.-W., Lim W. T., Lee S. C., and Chen P. C., Lab Chip 14(1), 128–137 (2014). 10.1039/C3LC50617G [DOI] [PubMed] [Google Scholar]
- 21.Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., LINskENs M., Rubelj I., and Pereira-Smith O., Proc. Natl. Acad. Sci. U.S.A. 92(20), 9363–9367 (1995). 10.1073/pnas.92.20.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho S. and Hwang E. S., Mol. Cells 33(6), 597–604 (2012). 10.1007/s10059-012-0042-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burd C. E., Sorrentino J. A., Clark K. S., Darr D. B., Krishnamurthy J., Deal A. M., Bardeesy N., Castrillon D. H., Beach D. H., and Sharpless N. E., Cell 152(1–2), 340–351 (2013). 10.1016/j.cell.2012.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorrentino J. A., Krishnamurthy J., Tilley S., Alb J. G., Burd C. E., and Sharpless N. E., J. Clin. Invest. 124(1), 169–173 (2014). 10.1172/JCI70960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He S. H. and Sharpless N. E., Cell 169(6), 1000–1011 (2017). 10.1016/j.cell.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayflick L. and Moorhead P. S., Exp. Cell Res. 25(3), 585–621 (1961). 10.1016/0014-4827(61)90192-6 [DOI] [PubMed] [Google Scholar]
- 27.Wagner W., Horn P., Castoldi M., Diehlmann A., Bork S., Saffrich R., Benes V., Blake J., Pfister S., and Eckstein V., PLoS One 3(5), e2213 (2008). 10.1371/journal.pone.0002213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finch C. E., Morgan T. E., Longo V. D., and De Magalhaes J. P., Aging cell 9(4), 519–526 (2010). 10.1111/j.1474-9726.2010.00578.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkland J. L., Stout M. B., and Sierra F., J. Gerontol. Ser. A: Biomed. Sci. Med. Sci. 71(11), 1407–1414 (2016). 10.1093/gerona/glw086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkland J. L. and Tchkonia T., EBioMedicine 21, 21–28 (2017). 10.1016/j.ebiom.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandl A., Meyer M., Bechmann V., Nerlich M., and Angele P., Exp. Cell Res. 317(11), 1541–1547 (2011). 10.1016/j.yexcr.2011.02.015 [DOI] [PubMed] [Google Scholar]
- 32.Lv Y. G., Hao X. Y., Sha Y. Q., and Yang L., Biotechnol. Lett. 36(12), 2559–2569 (2014). 10.1007/s10529-014-1625-z [DOI] [PubMed] [Google Scholar]
- 33.Chen W. T., Hsu W. T., Yen M. H., Changou C. A., Han C. L., Chen Y. J., Cheng J. Y., Chang T. H., Lee O. K. S., and Ho J. H. C., Biomaterials 190, 1–10 (2019). 10.1016/j.biomaterials.2018.10.026 [DOI] [PubMed] [Google Scholar]
- 34.Xiao Y. Y., Zhang Y. Y., and Xiao F., Drug Chem. Toxicol. 43(2), 213–218 (2020). 10.1080/01480545.2018.1551407 [DOI] [PubMed] [Google Scholar]
- 35.Guan G., Wu L., Bhagat A. A., Li Z., Chen P. C., Chao S., Ong C. J., and Han J., Sci. Rep. 3, 1475 (2013). 10.1038/srep01475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhagat A. A. S., Hou H. W., Li L. D., Lim C. T., and Han J. Y., Lab Chip 11(11), 1870–1878 (2011). 10.1039/c0lc00633e [DOI] [PubMed] [Google Scholar]
- 37.Phillip J. M., Wu P.-H., Gilkes D. M., Williams W., McGovern S., Daya J., Chen J., Aifuwa I., Lee J. S., and Fan R., Nat. Biomed. Eng. 1(7), 0093 (2017). 10.1038/s41551-017-0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G. H., Mao W. B., Byler R., Patel K., Henegar C., Alexeev A., and Sulchek T., PLoS One 8(10), e68910 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Islam M., Brink H., Blanche S., DiPrete C., Bongiorno T., Stone N., Liu A., Philip A., Wang G. H., Lam W., Alexeev A., Waller E. K., and Sulchek T., Sci. Rep. 7, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Islam M., Mezencev R., McFarland B., Brink H., Campbell B., Tasadduq B., Waller E. K., Lam W., Alexeev A., and Sulchek T., Cell Death Dis. 9, 1–12 (2018). 10.1038/s41419-018-0266-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z., Luo X., Zhao X., Yang M., and Wen C., J. Orthop. Transl. 17, 55–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tacutu R., Budovsky A., Yanai H., and Fraifeld V. E., Aging (Albany NY) 3(12), 1178 (2011). 10.18632/aging.100413 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.