Significance
Although numerous studies demonstrate an impact of protein motions on function, the spatial distribution of protein flexibility or rigidity that is optimal for efficient enzyme catalysis is often ambiguous. Here we present a temperature-dependent hydrogen/deuterium exchange mass spectrometry (HDX-MS) study of wild type and mutant forms of human catechol O-methyltransferase (COMT). The results show that optimal catalysis is linked to the maintenance of protein flexibility within a large network of helices that extend from behind the methyl-bearing sulfur of AdoMet to the entire region surrounding the extended conformation of bound cofactor.
Keywords: hydrogen deuterium exchange, enzyme mechanism, methyl transfer, protein flexibility
Abstract
Human catechol O-methyltransferase (COMT) has emerged as a model for understanding enzyme-catalyzed methyl transfer from S-adenosylmethionine (AdoMet) to small-molecule catecholate acceptors. Mutation of a single residue (tyrosine 68) behind the methyl-bearing sulfonium of AdoMet was previously shown to impair COMT activity by interfering with methyl donor–acceptor compaction within the activated ground state of the wild type enzyme [J. Zhang, H. J. Kulik, T. J. Martinez, J. P. Klinman, Proc. Natl. Acad. Sci. U.S.A. 112, 7954–7959 (2015)]. This predicts the involvement of spatially defined protein dynamical effects that further tune the donor/acceptor distance and geometry as well as the electrostatics of the reactants. Here, we present a hydrogen/deuterium exchange (HDX)-mass spectrometric study of wild type and mutant COMT, comparing temperature dependences of HDX against corresponding kinetic and cofactor binding parameters. The data show that the impaired Tyr68Ala mutant displays similar breaks in Arrhenius plots of both kinetic and HDX properties that are absent in the wild type enzyme. The spatial resolution of HDX below a break point of 15–20 °C indicates changes in flexibility across ∼40% of the protein structure that is confined primarily to the periphery of the AdoMet binding site. Above 20 °C, Tyr68Ala behaves more like WT in HDX, but its rate and enthalpic barrier remain significantly altered. The impairment of catalysis by Tyr68Ala can be understood in the context of a mutationally induced alteration in protein motions that becomes manifest along and perpendicular to the primary group transfer coordinate.
The impact of protein motions on enzyme function is most obvious when large conformational changes occur that can be readily detectable by X-ray crystallography (1–3). However, a growing body of evidence indicates that subtle and transient protein conformational substates are necessary for many chemical transformations catalyzed by enzymes (4–13). Such dynamical interconversions are often rapid (picoseconds to microseconds) (14), and, due to their short lifetimes, exceedingly difficult to measure experimentally and to link to active site catalysis. Experimental tools to address these processes include the combined application of site-specific mutagenesis with biophysical measurements of fluorescence lifetimes and Stokes shifts (15), elevation of pressure (16), T-jump FRET (17), and rapid, time-dependent X-ray crystallography (18). Alternatively, hydrogen/deuterium exchange mass spectrometry (HDX-MS) can be used for the spatial resolution of protein motions (19–24). We have previously demonstrated that performing HDX as a function of temperature can uncover the regions within a protein that dominate the thermal activation of active site chemistry. In the case of C-H activating enzymes such as the thermophilic alcohol dehydrogenase (ht-ADH) (25–27) and soybean lipoxygenase (SLO) (28, 29), where quantum mechanical tunneling dominates the chemical coordinate, in-depth kinetic and biophysical probes have revealed a model for catalysis in which the thermal activation barrier to reaction comes from the protein scaffold via well-resolved networks that connect solvent-exposed surfaces to the enzyme active site (26–31).
In order to extend the HDX approach to broader classes of enzyme reactions, we turned to the widespread family of methyltransferases, using catechol O-methyltransferase (COMT) as a prototype (32). COMT catalyzes methyl transfer from S-adenosyl methionine (AdoMet) to the ring oxygens of catechols (such as dopamine), and belongs to a large superfamily of AdoMet-dependent enzymes that act on a wide range of methyl acceptors (small-molecule metabolites, proteins, RNA, and DNA) (32). Primary kinetic isotope effects (KIEs) measured using [methyl-14C]AdoMet demonstrated rate-limiting methyl transfer and ruled out any significant contribution of quantum mechanical tunneling in the physiological temperature range (33, 34). This makes COMT an excellent system in which to interrogate the role of protein dynamical networks in enzyme reactions that occur via “more traditional” reaction coordinates (35). In an earlier comparison of wild type (WT) COMT to a series of mutations at residue Tyr68 located near the AdoMet cofactor binding site, we observed an unexpected linear correlation between the second-order rate constant, kcat/Km(dopamine), and the secondary KIE, measured using [methyl-3H]AdoMet (34). The secondary KIE increased as kcat/Km decreased upon mutation, signifying loss of compaction of the transition state upon mutational impairment of activity. These findings are consistent with analyses of ternary complexes between COMT, AdoMet, and catechol-based inhibitors by X-ray crystallography (36) and QM-MM analyses (34, 37) showing stable energetically minimized ground state structures with short methyl donor–acceptor distances in the wild type enzyme. Together, these observations led to a model involving a methyl donor–acceptor distance that is compacted in the activated ground state of WT COMT and becomes elongated in the catalytically impaired Tyr68 mutants (33, 34, 37).
To interrogate internal protein motions in COMT that could be related to catalytic efficiency, we previously employed fluorescence spectroscopy to monitor a single tryptophan residue (Trp143) located near the AdoMet adenine and ca. 7 Å away from Tyr68 (33). Measurements of nanosecond Stokes shifts showed moderate differences between Tyr68Ala (Y68A) and WT apo-enzymes (72 cm−1 in Y68A vs. 114 cm−1 in WT) , which became more pronounced in binary complexes with AdoMet (148 cm−1 in Y68A vs. 752 cm−1 in WT). These results suggested that mutation of Tyr68 constrains the active site environment in a manner that reduces its ability to adapt to and stabilize the Trp143 excited-state dipole, providing evidence that the Y68A mutation alters the dynamics of COMT. However, the findings were limited to only a small region within the active site.
Here we examine flexibility/dynamics throughout the entire COMT protein by combining time-, temperature-, and mutation-dependent HDX-MS. The results reveal unanticipated differences between WT and Y68A COMT in the effects of temperature on conformational mobility. Significantly, Y68A COMT shows a discontinuity in its temperature-dependent HDX behavior that is absent in the WT enzyme and implicates a cooperative transition below 20 °C. The flexibility of the mutant protein decreases across a large fraction of its periphery, and is accompanied by an increased enthalpic barrier for kcat and kcat/Km of 9.3 and 7.9 kcal/mol, respectively. Above a kinetic breakpoint of 15–20 °C, the mutant protein recovers partial flexibility but never achieves the higher catalytic efficiency of WT. The regions in mutant COMT that undergo the conformational transition reflected by the temperature break include helices that extend in two directions along and perpendicular (orthogonal) to the primary reaction coordinate. A similar break in HDX behavior with temperature that correlates with impaired catalysis was previously seen in the native thermophilic ht-ADH, and to a greater extreme following reductions in size of second-sphere hydrophobic side chains (38, 39). The extent of parallel behaviors observed for a prototypic group (methyl) transfer reaction and numerous hydrogen tunneling reactions is compelling, expanding the evidence for a generic role of functionally evolved protein networks in the optimization of enzyme catalysis.
Results
HDX-MS provides a versatile approach to interrogate the interplay between protein structure, conformational change, and dynamics in catalysis (20, 22–24, 40–42). The analysis of the rate and extent of exchange of deuterium from solvent into the backbone amides of proteins makes it possible to assess, in a rapid manner, spatially resolved regions within proteins where changes in flexibility may play key roles in function. Few AdoMet-dependent methyltransferases have been examined by HDX-MS (43, 44), but extensive kinetic and structural investigations exist for a number of such enzymes, including COMT (45–48). This makes COMT an ideal system to construct a global dynamical map of a prototypic methyltransferase. Our studies focus on the Mg2+-bound form of COMT, performed in the absence of substrate and AdoMet cofactor in order to avoid complications which can mask effects on HDX. As discussed by Kern et al., the intrinsic flexibility in apo-forms of enzymes is expected to be reflective of and propagated into their complexes with substrate and/or cofactor (49).
Briefly, HDX measurements were made on the soluble form of human COMT (221 aa, 26 kDa) by incubation in deuterated buffer at varying times (10 s to 2 h) and temperatures (10, 20, and 37 °C), quenching of HDX at pH 2.4, and digestion into peptides with immobilized pepsin. Peptides were separated by RP-HPLC (0 °C) and peptide masses analyzed by high-resolution mass spectrometry (50). The experiment yielded 124 and 125 peptides for WT and Y68A COMT, respectively (SI Appendix, Table S1). Peptides ranged from 7 to 25 aa in length (averaging 13 aa) and covered 97% of the COMT sequence (SI Appendix, Fig. S1). HDX time courses report rates of hydrogen exchange (kobs) covering a dynamic range of ca. 10 min−1 to 0.002 min−1 (28). Under the EX-2 conditions using native proteins at neutral pH, HDX rate constants can be approximated by kobs = Kop × kint, where Kop is the equilibrium constant for the interconversion between open (i.e., exchanging) and closed (nonexchanging) states of COMT and kint is the intrinsic rate constant for exchange from the open form. Although kint increases with temperature, because most peptides were identical between WT and mutant COMT, we can assume that the temperature dependences of kint are nearly equal between the enzyme forms. Therefore, by comparing the behavior of site-specific mutants to WT enzyme, Kop can be taken as the primary determinant of mutational changes in HDX at different temperatures throughout the protein (28).
HDX-MS Analyses of WT-COMT.
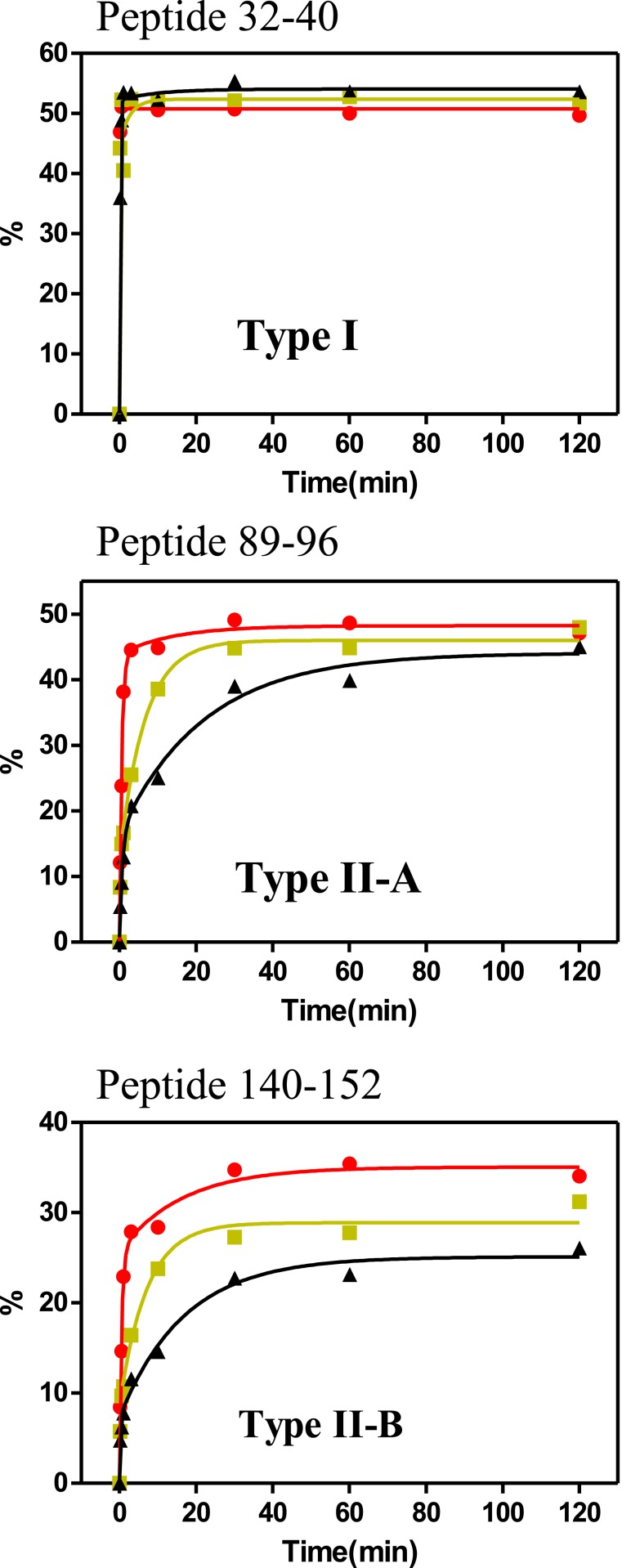
HDX time courses at different temperatures revealed two types of patterns. Approximately one third of the peptides analyzed were found to undergo rapid exchange at the upper limit of the experimental dynamic range and are named type I (Fig. 1). Type I patterns indicate little or no effects of temperature, most likely due to rates being outside the fastest time point of these measurements. The remaining peptides showed measurable increases in HDX with temperature and are named type II. Some of these approach a common plateau value at the longest time point of 2 h (named type II-A; Fig. 1), while the remainder indicate divergence at 2 h (named type II-B; Fig. 1).
Fig. 1.
Examples of different patterns of HDX responses to temperature (types I, II-A, II-B). Time courses of percent deuterium incorporation vs. time at 10 °C (black, triangle), 20 °C (yellow, square), and 37 °C (red, dots). The text includes details of these assignments, and Dataset S1 shows plots of all observed peptides.
The patterns of HDX behavior are mapped onto an X-ray structure of COMT with bound AdoMet cofactor and dinitrocatechol substrate in order to highlight the positions occupied by the methyl donor and accepting atoms (sulfonium in AdoMet and oxygen in substrate; Fig. 2). The COMT structure is comprised of eight alpha helices forming a TIM barrel-like fold. The enzyme is seen to bind to AdoMet in an extended conformation within the barrel interior, while the catechol substrate is directly coordinated to an active site metal (Mg2+) located in a more solvent-exposed position. These structural features are consistent with the kinetically ordered binding of AdoMet first, followed by Mg2+ and catechol (Scheme 1). It can be seen that, for the most part, the most rapidly exchanging regions of COMT (type I, Fig. 2, cyan; type II-A, Fig. 2, yellow) occur near the substrate and metal binding sites, defined by two long beta-strands and a helix turn at the entry point to the active site. In contrast, the regions undergoing slower exchange (type II-B, Fig. 2, raspberry) are primarily located at the periphery of the protein and surrounding the cofactor binding site.
Fig. 2.
The mapping of HDX patterns to the crystal structure of WT COMT. Shown is the structure of a ternary complex of COMT with AdoMet cofactor and dinitrocatechol substrate (PDB ID code 3BWM) (36) with the following color scheme for HDX in WT enzyme: type I, cyan; type II-A, yellow; type II-B, raspberry. Mg2+ metal is in orange. Because the reported HDX experiments represent the apo protein, we have shown the AdoMet and substrate as thin lines rather than the usual stick representation (see Fig. 4). Data for the region containing the site of mutation (gray) were not complete, but, based on two temperatures, appeared to correspond to type I behavior (see text).
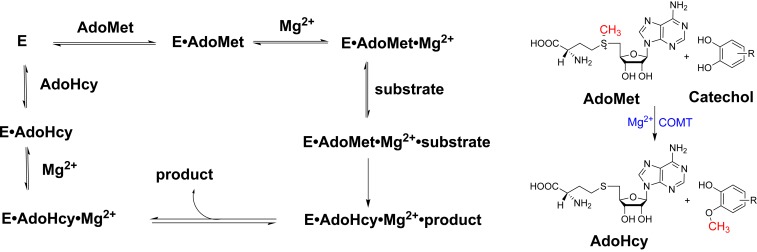
Scheme 1.
Kinetic pathway for COMT, illustrating the ordered binding of AdoMet, Mg2+, and substrate, followed by the ordered release of AdoHcy, Mg2+, and product.
HDX Analyses of Y68A COMT.
As previously described (33), Y68A in WT COMT plays a key role in controlling the position of the methyl group undergoing transfer to the substrate; specifically, the Tyr β-carbon is in van der Waals contact with the sulfonium of AdoMet (3.5 Å), while the Tyr ring hydroxyl is within hydrogen bonding distance to Glu6 (2.5 Å; SI Appendix, Fig. S2). Mutations in Tyr68 lead to a progressive reduction in the rate constants for methylation [measured by kcat/Km(dopamine)], which can be attributed to a corresponding elongation of the distance between the methyl donor and acceptor and therefore less compaction along the reaction coordinate axis (measured by the secondary KIE for [methyl-3H]AdoMet) (33). Among several mutants examined, Y68A produced the most extreme behavior, decreasing kcat/Km(dopamine) by more than three orders of magnitude and increasing the secondary KIE to a value approaching that of the reaction in solution. We therefore characterized Y68A COMT by HDX-MS in parallel with WT COMT.
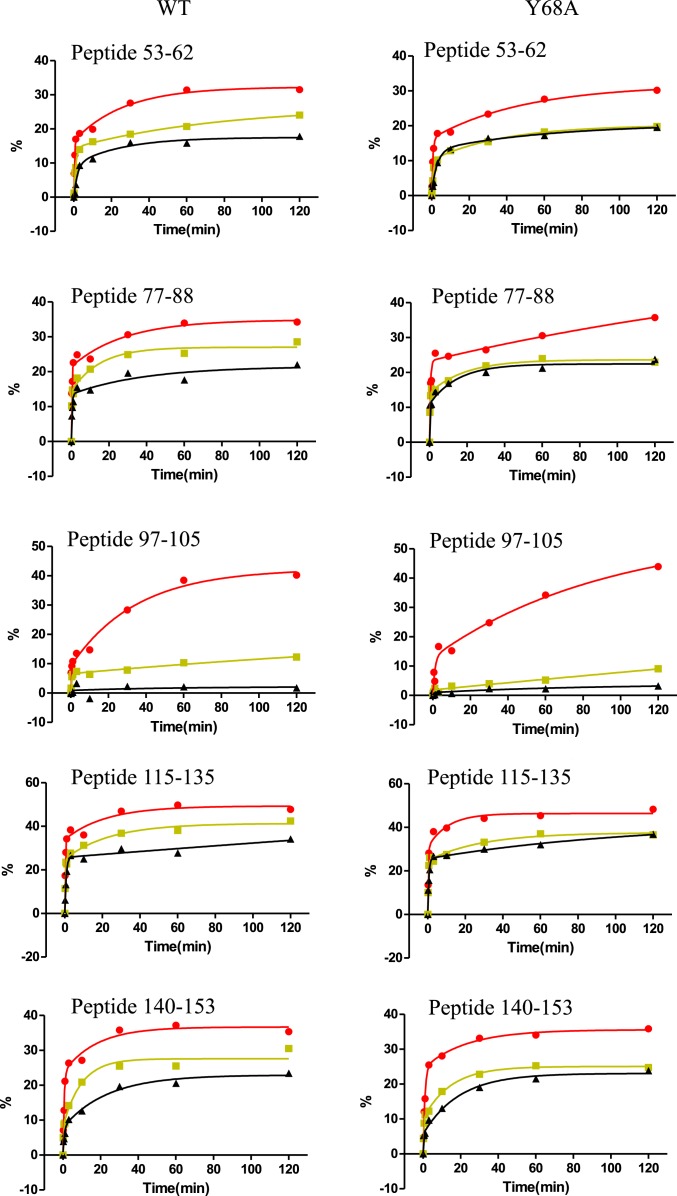
HDX-MS analysis produced nearly identical peptides from the WT and Y68A enzymes, except for those containing the Tyr-to-Ala change at position 68 (SI Appendix, Table S1). This allowed side-by-side comparisons of experimental traces across most regions in COMT (Dataset S1, Part A). Overall, differences between WT and mutant were not observable or else minimal in peptides with type I behavior. However, differences were apparent in the majority of peptides classified as type II-B (SI Appendix, Table S2). The most revealing changes occurred within four extended regions, where deuteration time courses were comparable between WT and Y68A at 37 °C and 10 °C, but markedly decreased in the Y68A mutant compared to WT at 20 °C. Representative peptides with this behavior are presented in Fig. 3. The patterns show that, whereas HDX increases continuously with temperature in the WT enzyme, HDX in the mutant at 20 °C is maintained at levels comparable to those at 10 °C, increasing in value as the temperature is raised to 37 °C. Thus, Y68A COMT displays a discontinuity, or “breakpoint,” in its temperature dependence at ∼20 °C, not seen in WT COMT.
Fig. 3.
Representative type II-B peptides showing variations in HDX between WT and Y68A COMT. Time courses of percent deuteration vs. time are shown for 37 °C (red), 20 °C (yellow), and 10 °C (black). In all cases, deuteration time courses were comparable between Y68A (Right) and WT (Left) at 37 °C and 10 °C, but markedly reduced deuteration was seen in Y68A relative to WT at 20 °C, revealing a break in the temperature dependence of HDX uptake introduced by the mutation.
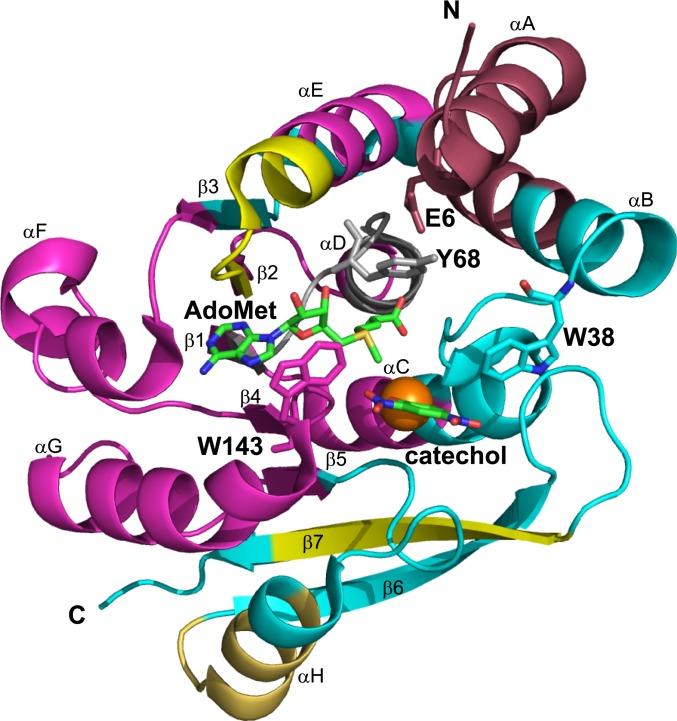
These regions are mapped onto the structure of COMT in Fig. 4 (magenta). It can be seen that the regions showing the mutation-induced temperature break in HDX primarily occur in helices αC, αE, αF, and αG, comprising the α-helical scaffold that encompasses bound AdoMet. Given similar temperature dependences of the intrinsic exchange rate, kint, for identical peptides shared between WT and mutant enzymes, the results suggest that Y68A introduces a transition in the processes that control Kop in regions surrounding the cofactor binding site. In other words, the temperature break in the mutant reports a decrease in conformational mobility that differentially exposes backbone amides to solvent at 20 °C.
Fig. 4.
The mapping of variations in HDX behavior between WT and Y68A. The regions colored magenta correspond to positions showing a break in the temperature dependence of HDX in Y68A at 20 °C. Color coding for the remainder of the protein is as in Fig. 2.
Two remaining regions of considerable interest are the loop between strand β1 and helix αD, containing the site of the Y68A mutation, and helices αA–αB at the N terminus. In the β1–αD region surrounding Tyr68, direct comparison between WT and Y68A was not possible due to differential proteolysis and lower recovery of peptides, together with the absence of data at 10 °C for many peptides (Dataset S1, Part B). In an effort to gain some insight into the backbone flexibility surrounding position 68 and its sensitivity to mutation, we focused on the HDX behavior of several peptides overlapping residues 62 to 73 at 20 and 37 °C (Dataset S1, Part B). This small stretch of COMT can be expected to be relatively free of the behavior detected for adjoining peptides extending into the N- or C-portions of the protein. The WT peptide 66–73 showed type I behavior, as did the Y68A peptides 63–70, 63–72, and 63–73, all undergoing HDX in a manner that was more or less equivalent. Thus, both WT and Y68A COMT were dominated by rapid and largely temperature-independent exchange properties in this region, precluding further analysis.
Glu6 in helix αA is located within hydrogen bonding distance of the ring hydroxyl of Tyr68 (SI Appendix, Fig. S2). The loss of this H-bonding interaction in Y68A may have been expected to open up the region 1–28 to faster HDX. However, after inspection of seven peptides within positions 1 to 33 at all three temperatures (Dataset S1, Part A), the only discernible difference was a small increase in the 10 °C traces, changing the pattern of exchange in Y68A closer to type II-A, instead of the type II-B behavior seen in WT. This provides a valuable control, indicating that the loss of a hydrogen bond to Glu6 in Y68A COMT has only minor impact, and that the effect of the mutation must arise from changes in other types of interactions.
Comparative Temperature Dependencies of kcat, kcat/Km, and Kd Values for AdoMet and AdoHcy.
In order to compare the temperature break in HDX for Y68A with catalytic behavior, we also pursued the temperature dependence of kinetic and binding parameters for WT and Y68A. The kinetic parameters kcat and kcat/Km between 5 and 41 °C are summarized in SI Appendix, Tables S3 and S4, with the relevant Arrhenius relationships presented in Fig. 5 and the resulting activation energies in Table 1. We note that the evaluation of kcat with Y68A below 20 °C required long extrapolations to saturating substrate due to a precipitous rise in Km(catechol) to almost 50 mM; we did not attempt kinetic analyses beyond an upper limit of 20 mM catechol, given the potential for substrate inhibition as well as possible damaging redox side reactions. Therefore, kcat values below 20 °C were derived from fitting data collected at substrate concentrations of ca. Km/2.
Fig. 5.
Temperature dependences for kinetic parameters of WT and Y68A COMT. Shown are Arrhenius plots of lnkcat vs. 1/T for (A) WT and (B) Y68A COMT and lnkcat/Km vs. 1/T for (C) WT and (D) Y68A. Whereas WT shows a single straight line at all temperatures, Y68A reveals a breakpoint in slope at ca. 15 to 20 °C. Data are summarized in SI Appendix, Tables S3 and S4, and tests of fitting are shown in SI Appendix, Fig. S3.
Table 1.
Different trends in the temperature dependences of kcat and kcat/Km for WT and Y68A COMT, highlighting the breakpoint in Ea with temperature with Y68A
| COMT | kcat, s−1 | kcat/Km, M−1, s−1 | ||||
| Ea(bb) | Ea(ab) | ΔEa (Ea(bb)-Ea(ab)) | Ea(bb) | Ea(ab) | ΔEa (Ea(bb)-Ea(ab)) | |
| WT | ← 13.1(0.5) → | ← 18.8(0.5)→ | ||||
| Y68A | 27.3( 0.6) | 18.0(0.9) | 9.3(1.4) | 28.7(0.5) | 20.7(0.8) | 7.9(1.3) |
| ΔEa(Y68A-WT) | 14.2(1.1) | 4.9(1.4) | 9.8(0.8) | 1.9(1.3) | ||
The Ea and ΔEa values are all kcal/mol. Ea(ab), activation energies above the breakpoint; Ea(bb), activation energies below the breakpoint.
Analysis of kcat vs. 1/T for WT COMT showed a straight line (Fig. 5A). In contrast, data for Y68A COMT showed a noticeable discontinuity above and below the temperature range of 15 to 20 °C (Fig. 5B). These differences were apparent in the best fits generated after multiple trials of fitting the WT and Y68A data to straight or discontinuous lines with varying breakpoints (SI Appendix, Fig. S3). In pursuing an analysis of the temperature dependence of kcat/Km using extrapolated values for kcat and Km, we noted the large propagated errors below 20 °C for Y68A. We therefore pursued a direct analysis of kcat/Km using the linear portions of v vs. S plots. Plotting the resulting values once again shows a breakpoint of ca. 15 to 20 °C for Y68A but not WT (SI Appendix, Table S4 and Fig. 5 C and D).
As summarized in Table 1, the kinetic data indicate an increased energy of activation in the Y68A mutant, in proceeding from temperatures above the breakpoint to below the breakpoint, with similar effects on kcat and kcat/Km (ΔEa ∼ 8 to 9 kcal/mol; Table 1). The similarities between ΔEa for the first- and second-order kinetic parameters may be fortuitous, given the contribution of both substrate binding and catalysis to kcat/Km. As a result of this break in temperature dependence, the energy of activation increased from WT to the Y68A mutant in a manner that was larger below the temperature break (14 and 10 kcal/mol for kcat and kcat/Km, respectively) and smaller above the temperature break (5 and 2 kcal/mol for kcat and kcat/Km, respectively).
We further examined the impact of Y68A on the binding of AdoMet and its demethylated product AdoHcy. The mutation increased Kd for both reactant and product by ca. 30 fold, but in an apparently temperature-independent manner in both instances (Fig. 6 and SI Appendix, Tables S5 and S6). The observation of temperature independence for the binding of ligands to protein has already been noted for a different methyltransferase (please see refs. 51 and 52). Increased Kd with mutation has been ascribed to many diverse effects contributing to the strength of ligand binding, becoming especially complex in the case of large extended cofactors such as AdoMet, and can lead to a compensation of ΔH° by TΔS° (please see ref. 53). The absence of a temperature break for cofactor binding is especially apparent for Y68A, and this property is considered in greater detail in the Discussion.
Fig. 6.
Dissociation constants (Kd) for AdoMet (reactant) and AdoHcy (product) with COMT at varying temperatures. Plots of Kd vs. temperature are shown for WT and Y68A enzymes as indicated. Data are summarized in SI Appendix, Tables S5 and S6; the data points in brackets for WT COMT are considered outliers, as described in SI Appendix, Table S6.
Confirmation of the Rate-Determining Step.
Previous measurements of the primary 14C and secondary 3H KIEs on kcat/Km provided strong evidence for a rate-limiting methyl transfer reaction in the case of both WT and Y68A (33, 34). A similar conclusion was reported based on primary 13C kinetic isotope effect on kcat for the COMT-catalyzed methyl transfer from AdoMet to an alternate substrate, 3,4-dihydroxyacetophenone (54). In order to confirm that cofactor release does not affect the kcat values for WT and Y68A in the present studies, a final experiment was undertaken involving stopped-flow fluorescence spectroscopy to estimate koff for the product cofactor AdoHcy. Intrinsic fluorescence quenching was examined at various concentrations of AdoHcy (SI Appendix, Fig. S4) at a single temperature (5 °C), leading to measured values of kon and calculated values of koff derived from Kd (Table 2). A study of temperature dependence was not possible in this case due to the very rapid rate of binding of AdoHcy to WT even at the lowest temperature studied. The values of koff for both WT (264 ± 78 min−1) and Y68A (8 ± 2 min−1) are estimated to be ca. 100-fold faster than their respective kcat values (54), corroborating the rate-limiting methyl transfer step in this reaction.
Table 2.
AdoHcy kon and koff rates for WT and Y68A COMT at 5 °C
| Rate | WT | Y68A |
| kon (M−1s−1)* | 8.4 ± 2.1 × 105 | 925 ± 226 |
| koff (min−1)† | 264 ± 78 | 8 ± 2 |
| kcat (min−1) | 2.47 ± 0.03 | (0.09 ± 0.02)‡ |
The kon is derived from the slope of the fitting of kobs of stopped-flow fluorescence quenching vs. [AdoHcy].
The koff = Kd × kon.
The value of kcat for Y68A is approximate; see Discussion.
Discussion
Impacts of Y68A on Protein Flexibility.
From the HDX studies (Figs. 3 and 4), a reduction in protein flexibility (type II-B pattern) is observed at 20 °C within a large portion of the helical scaffold of the Rossman fold that surrounds AdoMet and positions it for reaction. A separate region of the protein that contains residues within the portal for substrate binding is far more flexible (type I) and fails to show any detectable change upon mutation. Earlier studies of the impact of Y68A and Y68F on Stokes shifts at Trp143 had indicated a remote rigidification of protein in the mutants that disrupted the ability of protein to accommodate the excited state dipole of Trp143 in WT in apo-enzyme and, even more so, in the AdoMet complex. Of note, the time-dependent Stokes shifts in COMT were collected at 37 °C, a temperature where the Y68A variant appears to recover WT HDX behavior (Fig. 3). Clearly, the increase in rate and extent of exchange in HDX for Y68A above 20 °C is complex, leading to a more flexible/exchangeable conformational landscape that, however, remains distinctive from that of WT.
Impacts of Y68A on the Temperature Dependence of Catalysis and Cofactor Binding.
As shown in earlier structural and kinetic investigations of COMT, the catalyzed reaction occurs via an ordered binding of cofactor AdoMet, followed by the addition of metal ion Mg2+ and then substrate catechol (dopamine in the present work) (55) (please see Scheme 1). The chemical step involves an irreversible SN2-type methyl transfer mechanism, followed by dissociation of the methylated catechol, then Mg2+, and finally S-adenosyl-L-homocysteine (Scheme 1). Although enzyme reactions can be rate-limited by substrate/product release, there is no evidence for such a feature in COMT, both from published kinetic isotope effect measurements (33, 34, 54) and the evidence (Table 2) showing that loss of product AdoHcy is rapid, even in the low-temperature regime (5 °C) where Y68A flexibility is most compromised (Fig. 4, magenta). Thus, the available data support the view that methyl transfer is rate-determining under conditions of both kcat and kcat/Km in the temperature range of 5 to 41 °C.
Very significantly, from the measurements of the temperature dependence of rate constants described herein (Fig. 5), a temperature break for Y68A can be seen to be present and to mirror the pattern observed for HDX (Fig. 3). We interpret these effects on catalysis for Y68A as arising from the cooperative induction of a more rigid conformational landscape below 20 °C that is poorly positioned for catalysis, requiring an input of additional thermal activation to access the reactive protein substates characteristic of WT. Extending the combined study of Y68A and temperature to the binding of AdoMet and its product AdoHcy, we further found changes in the magnitude of the cofactor Kd but no evidence for a break in behavior with temperature for Y68A (Fig. 6). These experimental binding properties are fully compatible with the model derived from kinetic measurements: whereas the generation of transiently achieved and improperly aligned protein substates in Y68A below 20 °C is expected to impair the chemical steps of catalysis, there is no reason to expect a priori a similar impact on the equilibrium properties of cofactor binding.
One final observation of importance is the retention of a difference in the energy barrier on kcat for WT vs. Y68A above the breakpoint (Table 1), a feature that is fully compatible with the previously reported changes in measured rate constants and secondary kinetic isotope effects at 37 °C (33, 34). The onset of increased flexibility that is seen from the HDX profiles of Y68A above 20 °C, though a necessary component of improved catalysis, is insufficient to produce the high turnover rates of the WT enzyme. As described later, we attempt to rationalize all of the impacts of Y68A within a structure-linked dynamical picture of catalysis in COMT.
Parsing Protein Motions in COMT into “Action Along the Reaction Coordinate vs. Perpendicular to the Reaction Coordinate.”
Increasingly, our interrogation of the nature of enzyme catalysis has moved beyond the active site toward the participation of large portions of the protein in achieving optimal catalytic activity (e.g., refs. 56 and 57). While some investigators limit their discussion of protein motions to conformational changes or loop closures around bound substrates (e.g., ref. 57 and 58), we have been pursuing the role of spatially resolved protein networks that can facilitate a thermal transfer from solvent interfaces to the active site, activating the dominant ground state E–S complexes for conversion from E–S to E–P. In contrast to high-temperature gas-phase kinetics, where transition state theory can be formulated in the context of a direct transfer of thermal energy into the bonds of the reactant, catalysis in condensed phase near room temperature presents different theoretical and experimental challenges. For instance, in enzyme catalysis, the available thermal energy is recognized to be small (kT, ca. 200 cm−1), producing, instead, a partitioning of thermal excitation into multiple low-energy protein and protein solvent breathing modes (4–6, 9, 11, 13, 14, 59, 60). One of the major challenges has been to link such motions to the enhanced rate at which bonds are broken/formed within the enzyme active site. In this context, the HDX-MS methodology has proven to be of great versatility, enabling the identification of thermally activated “solvent to active site networks of protein flexibility” with demonstrable properties that mirror those of catalysis (25, 28)
Starting with the early studies of C–H activation by the tetrameric extremophilic alcohol dehydrogenases (ADHs) (25), a correlation between global protein flexibility and active site optimization has become apparent. Specifically, through the application of site-specific mutagenesis, time-dependent fluorescence spectroscopic methods, and HDX-MS, two distinct long-range dynamical networks have been identified that connect solvent exposed regions of protein to the substrate binding domain (26); additionally, enthalpic barriers from kinetic measurements of Stokes shifts and T-jump FRET are seen to be identical to catalysis (15, 17, 27). In recent studies of a very different monomeric, single-substrate enzyme (soybean lipoxygenase), HDX-MS results link the thermal activation of a solvent-exposed loop to the positioning of the buried reactive C–H bond of substrate 15 to 30 Å away (28), once again with an enthalpic barrier for catalysis that is mirrored in Stokes shifts of a solvent-exposed chromophore (29). The goal of the present work was to extend these developed methodologies to other types of reaction, specifically the well-studied methyl transfer catalyzed by COMT (33, 34, 37).
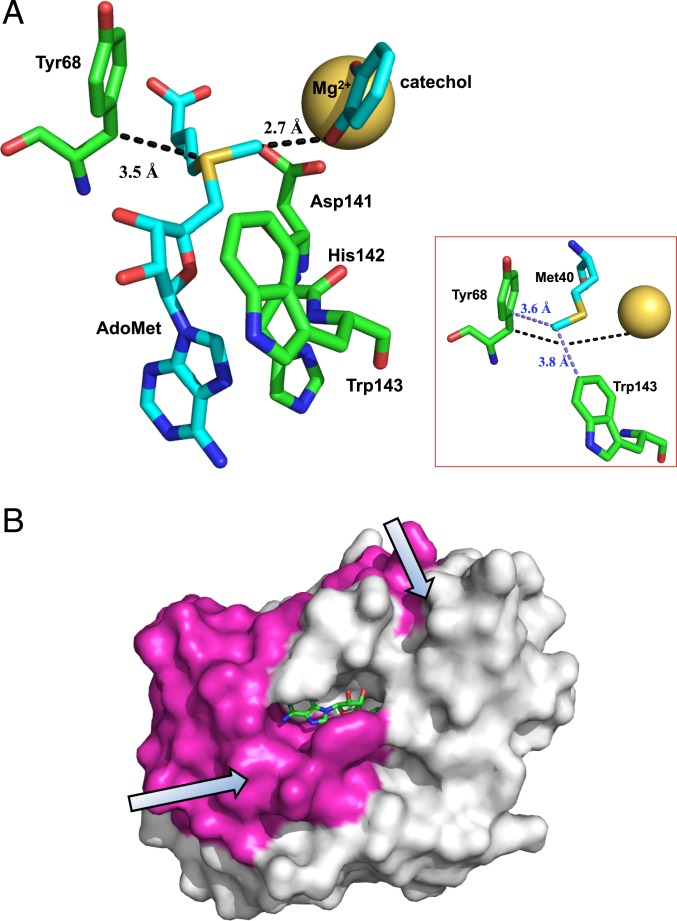
From the aggregate studies now available for COMT, we present a model for COMT that integrates the HDX and kinetic findings presented herein with the growing body of literature on this enzyme (17, 34, 61, 62). As shown in Fig. 7A, a minimal representation of the active site of COMT will include bound cofactor (AdoMet) and catecholate substrate complexed to the active site Mg2+. The positioning of Tyr-68 in Fig. 7A shows the van der Waals interaction of its beta-carbon with the sulfonium ion of cofactor, at a distance of 3.5 Å. The most prominent kinetic data regarding Tyr68, prior to this study, were focused on the insertion of a series of side chains of smaller size into position 68, yielding a linear correlation in both the rates of turnover and the size of the secondary kinetic isotope effect (34). These findings led to the proposed role of active side compaction in WT enzyme that was later supported by extended QM/MM computations (34, 37). The latter were able to reproduce the very short methyl-to-oxyanion transfer distance (2.7 Å) characterized in X-ray structures of COMT-AdoMet-nitrocatecholate complexes, and to rationalize the mutational impacts on the kinetic isotope effects as arising from increased distance between the methyl donor and acceptor atoms. These impacts are represented as “along the reaction coordinate” by the dashed black line in Fig. 7A, and can be seen to be propagated into the HDX behavior of helix αE that resides behind αD (Fig. 4).
Fig. 7.
A model for the COMT reaction integrating the HDX and kinetic findings. (A) The active site of COMT, illustrating the proposed duality of roles for Tyr68 in controlling the methyl donor–acceptor distance along the reaction coordinate (dashed black line) and the sampling of multiple conformational substates involving side chains orthogonal to the reaction coordinate (dashed light blue line, Inset). Structure is based on PDB ID code 3BWM (36), not showing dinitro groups on the catechol. (B) Space filling model of COMT showing the dynamical regions of the protein that extend in orthogonal directions from the protein solvent interface toward the reacting atoms in the active site.
Additional QM/MM studies have been pursued on COMT, leading to a rationalization of the experimental trends in rates arising from mutation at Tyr68, but a much poorer ability to explain the observed trends in the secondary KIEs (63, 64). The primary conclusion of the study by Wilson and Williams (64) was that effects along the reaction coordinate (involving Tyr68) are largely unimportant, and that rate effects are determined from residues orthogonal to the methyl transfer coordinate. A more recent NMR study of COMT in complex with a transition state analog (56) has detected changes in chemical shifts for a range of side chains, Asp141, His142, and Trp143, all of which are shown to reside below the axis of the methyl group transfer coordinates, illustrated in Fig. 7A. We consider it important that the changes in HDX patterns that have emerged from our comparative study of WT to Y68A COMT (Fig. 4) also mirror the region of protein that displayed changes in NMR chemical shifts in the presence of a transition state analog (56).
In an effort to uncover a structural basis for the propagation of the disruptive impact of Y68A on protein flexibility with the region orthogonal to the primary reaction coordinate, we searched for residues with potential interactions at both Tyr68 and a component of the Asp141, His142, and Trp143 triad. A role for Met40 emerged, with its methyl group positioned proximal to both C-1 of the phenyl ring of Tyr68 and C-5 of the indole ring of Trp143. As illustrated in the light blue dashed line of Fig. 7 A, Inset, this interaction cuts across the primary axis of the reaction coordinate to create an orthogonal interaction that is expected to be modulated by changes in Tyr68. We propose that, via an extraordinary economy of catalytic evolution, a single Tyr68 in COMT acts both to ensure close packing between the methyl donor and acceptor atoms and to maintain an extended region of protein flexibility in contact with bound cofactor. The latter may play key roles in transient adjustments of the angle of approach and charge distributions between the electrophilic (AdoMet) and nucleophilic (catecholate) poles of the reaction center, acting in concert with reductions in the distance between the methyl donor and acceptor atoms. These regions are illustrated in a space-filling model of COMT, showing the two primary networks (magenta in Fig. 7B) that initiate at separate protein–solvent interfaces and terminate at the confluence of Tyr68 and the methyl sulfonium of the AdoMet cofactor (Fig. 7B). These networks may inform the application of advanced computational methods to COMT that are capable of introducing enlarged QM regions into QM/MM studies while simultaneously carrying out MD simulations (please see ref. 65).
In closing, we note that the model developed for COMT is also of general interest in the context of its structural characterization as a “primitive TIM barrel” (66). As noted by many investigators, proteins within the TIM barrel superfamily comprise at least 10% of enzymes with known structures and contain five of the six identified EC classifications (67). Looking to the future, we are in the process of a formal interrogation of functionally linked HDX-MS studies among representative examples from the TIM-barrel superfamily. The goal of such studies is to uncover the extent to which proteins within a common structural scaffold have been able to undergo a synergistic evolution of protein dynamical effects to accompany emergent chemical properties.
Materials and Methods
Steady-state kinetic experiments quantified conversion of dopamine to methyldopamine measured by detection at 203 nm after separation by reversed-phase HPLC (C18). HDX experiments were performed at 10, 20, and 37 °C, each at time points in the range of 0.167 to 120 min, using immobilized pepsin proteolysis and LC/MS on a Synapt G2 HDMS mass spectrometer. Dissociation constants (Kd) for binding AdoMet or AdoHcy to COMT were measured by fluorescence quenching titration on a custom-built Fluorolog-3 spectrofluorometer (Horiba Jobin-Yvon). Stopped-flow spectrophotometry was carried out to measure the kinetic fluorescence quenching from which kon was derived. Detailed materials and methods are described in SI Appendix, Materials and Methods.
Data Availability.
All HDX time courses for WT and Y68A COMT are shown in Dataset S1 plots and the corresponding Excel spreadsheet. Raw datasets for HDX experiments can be publicly accessed on the MassIVE data repository at the University of California San Diego Center for Computational Mass Spectrometry (ID MSV000085213).
Supplementary Material
Acknowledgments
We thank Prof. Susan Marqusee at University of California, Berkeley, for the use of their stopped-flow fluorescence spectroscopy and Dr. Emily Guinn for technical assistance. This work was supported by NIH Grants to J.P.K. (GM118117-015) and to N.G.A. (S10 RR026641 and GM114594).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. G.S.A. is a guest editor invited by the Editorial Board.
Data deposition: Mass spectrometry data was deposited into the MassIVE database (ID MSV000085213).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917219117/-/DCSupplemental.
References
- 1.Mahajan S., Sanejouand Y.-H., On the relationship between low-frequency normal modes and the large-scale conformational changes of proteins. Arch. Biochem. Biophys. 567, 59–65 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Grant B. J., Gorfe A. A., McCammon J. A., Large conformational changes in proteins: Signaling and other functions. Curr. Opin. Struct. Biol. 20, 142–147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitford P. C., Miyashita O., Levy Y., Onuchic J. N., Conformational transitions of adenylate kinase: Switching by cracking. J. Mol. Biol. 366, 1661–1671 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luk L. Y., Loveridge E. J., Allemann R. K., Protein motions and dynamic effects in enzyme catalysis. Phys. Chem. Chem. Phys. 17, 30817–30827 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Callender R., Dyer R. B., The dynamical nature of enzymatic catalysis. Acc. Chem. Res. 48, 407–413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klinman J. P., Kohen A., Hydrogen tunneling links protein dynamics to enzyme catalysis. Annu. Rev. Biochem. 82, 471–496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glowacki D. R., Harvey J. N., Mulholland A. J., Protein dynamics and enzyme catalysis: The ghost in the machine? Biochem. Soc. Trans. 40, 515–521 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Zalatan J. G., Herschlag D., The far reaches of enzymology. Nat. Chem. Biol. 5, 516–520 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Schwartz S. D., Schramm V. L., Enzymatic transition states and dynamic motion in barrier crossing. Nat. Chem. Biol. 5, 551–558 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser J. S., et al. , Hidden alternative structures of proline isomerase essential for catalysis. Nature 462, 669–673 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benkovic S. J., Hammes G. G., Hammes-Schiffer S., Free-energy landscape of enzyme catalysis. Biochemistry 47, 3317–3321 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Qiu W., et al. , Dissection of complex protein dynamics in human thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 104, 5366–5371 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scrutton N. S., Basran J., Sutcliffe M. J., New insights into enzyme catalysis. Ground state tunnelling driven by protein dynamics. Eur. J. Biochem. 264, 666–671 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Henzler-Wildman K., Kern D., Dynamic personalities of proteins. Nature 450, 964–972 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Meadows C. W., Ou R., Klinman J. P., Picosecond-resolved fluorescent probes at functionally distinct tryptophans within a thermophilic alcohol dehydrogenase: Relationship of temperature-dependent changes in fluorescence to catalysis. J. Phys. Chem. B 118, 6049–6061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu S., Cattin-Ortolá J., Munos J. W., Klinman J. P., Hydrostatic pressure studies distinguish global from local protein motions in C−H activation by soybean lipoxygenase-1. Angew. Chem. Int. Ed. Engl. 55, 9361–9364 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaughn M. B., Zhang J., Spiro T. G., Dyer R. B., Klinman J. P., Activity-related microsecond dynamics revealed by temperature-jump Förster resonance energy transfer measurements on thermophilic alcohol dehydrogenase. J. Am. Chem. Soc. 140, 900–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neutze R., Moffat K., Time-resolved structural studies at synchrotrons and X-ray free electron lasers: Opportunities and challenges. Curr. Opin. Struct. Biol. 22, 651–659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oyeyemi O. A., et al. , Comparative hydrogen-deuterium exchange for a mesophilic vs thermophilic dihydrofolate reductase at 25 °C: Identification of a single active site region with enhanced flexibility in the mesophilic protein. Biochemistry 50, 8251–8260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konermann L., Pan J., Liu Y.-H., Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Oyeyemi O. A., et al. , Temperature dependence of protein motions in a thermophilic dihydrofolate reductase and its relationship to catalytic efficiency. Proc. Natl. Acad. Sci. U.S.A. 107, 10074–10079 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcsisin S. R., Engen J. R., Hydrogen exchange mass spectrometry: What is it and what can it tell us? Anal. Bioanal. Chem. 397, 967–972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wales T. E., Engen J. R., Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Englander S. W., Hydrogen exchange and mass spectrometry: A historical perspective. J. Am. Soc. Mass Spectrom. 17, 1481–1489 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Z.-X., Lee T., Resing K. A., Ahn N. G., Klinman J. P., Thermal-activated protein mobility and its correlation with catalysis in thermophilic alcohol dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 101, 9556–9561 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagel Z. D., Cun S., Klinman J. P., Identification of a long-range protein network that modulates active site dynamics in extremophilic alcohol dehydrogenases. J. Biol. Chem. 288, 14087–14097 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meadows C. W., Tsang J. E., Klinman J. P., Picosecond-resolved fluorescence studies of substrate and cofactor-binding domain mutants in a thermophilic alcohol dehydrogenase uncover an extended network of communication. J. Am. Chem. Soc. 136, 14821–14833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Offenbacher A. R., et al. , Hydrogen–deuterium exchange of lipoxygenase uncovers a relationship between distal, solvent exposed protein motions and the thermal activation barrier for catalytic proton-coupled electron tunneling. ACS Cent. Sci. 3, 570–579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaragoza J. P. T., et al. , Detecting and characterizing the kinetic activation of thermal networks in proteins: Thermal transfer from a distal, solvent-exposed loop to the active site in soybean lipoxygenase. J. Phys. Chem. B 123, 8662–8674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hekstra D. R., et al. , Electric-field-stimulated protein mechanics. Nature 540, 400–405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frauenfelder H., et al. , A unified model of protein dynamics. Proc. Natl. Acad. Sci. U.S.A. 106, 5129–5134 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Struck A. W., Thompson M. L., Wong L. S., Micklefield J., S-adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. ChemBioChem 13, 2642–2655 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., Klinman J. P., Enzymatic methyl transfer: Role of an active site residue in generating active site compaction that correlates with catalytic efficiency. J. Am. Chem. Soc. 133, 17134–17137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J., Kulik H. J., Martinez T. J., Klinman J. P., Mediation of donor-acceptor distance in an enzymatic methyl transfer reaction. Proc. Natl. Acad. Sci. U.S.A. 112, 7954–7959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfenden R., Snider M., Ridgway C., Miller B., The temperature dependence of enzyme rate enhancements. J. Am. Chem. Soc. 121, 7419–7420 (1999). [Google Scholar]
- 36.Rutherford K., Le Trong I., Stenkamp R. E., Parson W. W., Crystal structures of human 108V and 108M catechol O-methyltransferase. J. Mol. Biol. 380, 120–130 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Kulik H. J., Zhang J., Klinman J. P., Martínez T. J., How large should the QM region be in QM/MM calculations? The case of catechol O-methyltransferase. J. Phys. Chem. B 120, 11381–11394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagel Z. D., Dong M., Bahnson B. J., Klinman J. P., Impaired protein conformational landscapes as revealed in anomalous Arrhenius prefactors. Proc. Natl. Acad. Sci. U.S.A. 108, 10520–10525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagel Z. D., Meadows C. W., Dong M., Bahnson B. J., Klinman J. P., Active site hydrophobic residues impact hydrogen tunneling differently in a thermophilic alcohol dehydrogenase at optimal versus nonoptimal temperatures. Biochemistry 51, 4147–4156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Resing K. A., Hoofnagle A. N., Ahn N. G., Modeling deuterium exchange behavior of ERK2 using pepsin mapping to probe secondary structure. J. Am. Soc. Mass Spectrom. 10, 685–702 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Hoofnagle A. N., Resing K. A., Ahn N. G., Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 32, 1–25 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Garcia R. A., Pantazatos D., Villarreal F. J., Hydrogen/deuterium exchange mass spectrometry for investigating protein-ligand interactions. Assay Drug Dev. Technol. 2, 81–91 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Yu W., et al. , Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat. Commun. 3, 1288 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Zhao Y., et al. , A crystal structure of the dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Path. 11, e1004682 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Männistö P. T., Kaakkola S., Catechol-O-methyltransferase (COMT): Biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol. Rev. 51, 593–628 (1999). [PubMed] [Google Scholar]
- 46.Cheng X., Roberts R. J., AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 29, 3784–3795 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu B. T., Catechol-O-Methyltransferase (COMT)-mediated methylation metabolism of endogenous bioactive catechols and modulation by endobiotics and xenobiotics: Importance in pathophysiology and pathogenesis. Curr. Drug Metab. 3, 321–349 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Tunbridge E. M., Harrison P. J., Weinberger D. R., Catechol-o-methyltransferase, cognition, and psychosis: Val158Met and beyond. Biol. Psychiatry 60, 141–151 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Henzler-Wildman K. A., et al. , Intrinsic motions along an enzymatic reaction trajectory. Nature 450, 838–844 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Hoofnagle A. N., Resing K. A., Ahn N. G., “Practical methods for deuterium exchange/mass spectrometry” in MAP Kinase Signaling Protocols, Seger R., Ed. (Springer, 2004), pp. 283–298. [Google Scholar]
- 51.Pey A. L., Majtan T., Sanchez-Ruiz J. M., Kraus J. P., Human cystathionine β-synthase (CBS) contains two classes of binding sites for S-adenosylmethionine (SAM): Complex regulation of CBS activity and stability by SAM. Biochem. J. 449, 109–121 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Majtan T., Pey A. L., Kraus J. P., Kinetic stability of cystathionine beta-synthase can be modulated by structural analogs of S-adenosylmethionine: Potential approach to pharmacological chaperone therapy for homocystinuria. Biochimie 126, 6–13 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Nestorovich E. M., Karginov V. A., Berezhkovskii A. M., Parsegian V. A., Bezrukov S. M., Kinetics and thermodynamics of binding reactions as exemplified by anthrax toxin channel blockage with a cationic cyclodextrin derivative. Proc. Natl. Acad. Sci. U.S.A. 109, 18453–18458 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hegazi M. F., Borchardt R. T., Schowen R. L., alpha.-Deuterium and carbon-13 isotope effects for methyl transfer catalyzed by catechol O-methyltransferase. SN2-like transition state. J. Am. Chem. Soc. 101, 4359–4365 (1979). [DOI] [PubMed] [Google Scholar]
- 55.Lotta T., et al. , Kinetics of human soluble and membrane-bound catechol O-methyltransferase: A revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry 34, 4202–4210 (1995). [DOI] [PubMed] [Google Scholar]
- 56.Czarnota S., et al. , Equatorial active site compaction and electrostatic reorganization in catechol-O-methyltransferase. ACS Catal. 9, 4394–4401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richard J. P., Protein flexibility and stiffness enable efficient enzymatic catalysis. J. Am. Chem. Soc. 141, 3320–3331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malabanan M. M., Amyes T. L., Richard J. P., A role for flexible loops in enzyme catalysis. Curr. Opin. Struct. Biol. 20, 702–710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klinman J. P., Offenbacher A. R., Hu S., Origins of enzyme catalysis: Experimental findings for C–H activation, new models, and their relevance to prevailing theoretical constructs. J. Am. Chem. Soc. 139, 18409–18427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agarwal P. K., A biophysical perspective on enzyme catalysis. Biochemistry 58, 438–449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saez D. A., Zinovjev K., Tuñón I., Vöhringer-Martinez E., Catalytic reaction mechanism in native and mutant catechol-O-methyltransferase from the adaptive string method and mean reaction force analysis. J. Phys. Chem. B 122, 8861–8871 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Zhang J., Klinman J. P., Convergent mechanistic features between the structurally diverse N-and O-methyltransferases: Glycine N-methyltransferase and catechol O-methyltransferase. J. Am. Chem. Soc. 138, 9158–9165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J., Klinman J. P., CH··· O interactions are not the cause of trends in reactivity and secondary kinetic isotope effects for enzymatic SN2 methyl transfer reactions. bioRxiv:10.1101/071043 (September 1, 2016).
- 64.Wilson P. B., Williams I. H., Influence of equatorial CH⋅⋅⋅ O interactions on secondary kinetic isotope effects for methyl transfer. Angew. Chem. Int. Ed. Engl. 55, 3192–3195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Z., et al. , Revealing quantum mechanical effects in enzyme catalysis with large-scale electronic structure simulation. React. Chem. Eng. 4, 298–315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goldman A. D., Beatty J. T., Landweber L. F., The TIM barrel architecture facilitated the early evolution of protein-mediated metabolism. J. Mol. Evol. 82, 17–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wierenga R. K., The TIM-barrel fold: A versatile framework for efficient enzymes. FEBS Lett. 492, 193–198 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All HDX time courses for WT and Y68A COMT are shown in Dataset S1 plots and the corresponding Excel spreadsheet. Raw datasets for HDX experiments can be publicly accessed on the MassIVE data repository at the University of California San Diego Center for Computational Mass Spectrometry (ID MSV000085213).