Abstract
Objective
The present study aimed to investigate the correlation of protein phosphatase Mg2+/Mn2+ dependent 1D (PPM1D) with the risk stratification, treatment response, and survival profile in acute myeloid leukemia (AML) patients.
Methods
Totally 221 de novo AML patients and 50 healthy donors were enrolled. The bone marrow samples were collected before treatment from AML patients and acquired after enrollment from healthy donors. And bone marrow mononuclear cells were separated for detecting the mRNA/protein expressions of PPM1D by reverse transcription‐quantitative polymerase chain reaction and Western blot. Complete remission (CR) was assessed after induction treatment, and event‐free survival (EFS) and overall survival (OS) were calculated in AML patients.
Results
PPM1D mRNA (P < .001)/protein (P < .001) relative expressions were increased in AML patients compared with healthy donors, and receiver operating characteristic curve presented that PPM1D mRNA (AUC: 0.728, 95% CI: 0.651‐0.806)/protein (AUC: 0.782, 95% CI: 0.707‐0.857) relative expressions could differentiate AML patients from healthy donors. In AML patients, PPM1D mRNA (P < .001)/protein (P < .001) high relative expressions were correlated with poor‐risk stratification. As for its association with prognosis, PPM1D mRNA (P < .001)/protein (P = .010) relative expressions were elevated in CR patients compared with non‐CR patients. Patients with PPM1D mRNA (P < .001 for EFS; P = .004 for OS)/protein (P < .001 for EFS; P = .006 for OS) high relative expressions exhibited reduced EFS and OS compared with those with low expressions.
Conclusion
PPM1D high expression correlates with poor‐risk stratification and might serve as a potential biomarker for worse prognosis in AML patients, suggesting its potential to guide AML management.
Keywords: acute myeloid leukemia, complete remission, protein phosphatase Mg2+/Mn2+ dependent 1D, risk stratification, survival profile
1. INTRODUCTION
Acute myeloid leukemia (AML) is the most prevalent adult acute leukemia, which is characterized by abnormal proliferation and the accumulation of immature myeloid precursor cells in the bone marrow, peripheral blood, and even some tissues, contributing to the destruction of the hematopoietic system.1 Nowadays, the management of AML has experienced great improvements, consisting of chemotherapy, hematopoietic stem cell transplantation, molecularly targeted therapy, transfusion support, etc2, 3 However, the event‐free survival (EFS) and overall survival (OS) are still unsatisfied.4, 5 Therefore, it is essential to discover potential biomarkers which could predict prognosis and guide AML management effectively in AML patients.
Protein phosphatase Mg2+/Mn2+ dependent 1D (PPM1D) is a major serine/threonine phosphatase of the protein phosphatase 2C (PP2C) family, and PP2C family members serve important roles in regulating cell stress response pathways.6 PPM1D is identified to regulate p38 MARK/p53 pathway and functions as oncogene in various types of human solid malignancies.7, 8 Regarding hematologic malignancies, there is evidence that PPM1D expression is inhibited by a potent cancer chemotherapeutic agent for acute promyelocytic leukemia (APL), which activates p38 MARK/p53 signaling and promotes APL cell apoptosis.9 Additionally, PPM1D is reported to contribute to tumorigenesis through inducing the transformation of leukemic cells in adult T‐cell leukemia/lymphoma (ATLL), and the inhibition of PPM1D is revealed to mediate neutrophil differentiation in human APL, implying that PPM1D is also involved in the initiation and development of hematologic malignancies.10, 11 As for in AML, PPM1D mutant strongly outcompetes the wild‐type PPM1D and correlates with increased drug resistance in the treatment.12 According to the previous studies, we hypothesized that PPM1D might be of value in predicting AML risk as well as prognosis in AML patients. Therefore, we conducted this study to investigate PPM1D expression in AML patients compared to healthy donors and explore the correlation of PPM1D with the risk stratification, treatment response, and survival profile in AML patients.
2. MATERIALS AND METHODS
2.1. Participants
Between January 2016 and June 2019, 221 de novo AML patients and 50 healthy donors were consecutively recruited. The inclusion criteria of AML patients were as follows: (a) newly diagnosed as primary AML based on morphology, cytochemistry, immunophenotyping, cytogenetics and molecular genetics, according to the criteria of 2008 WHO classification13; (b) age above 18 years; (c) no history of systematic treatments (eg, chemotherapy, radiotherapy, or stem cell transplantation); and (d) could be followed up regularly. The exclusion reasons of AML patients were as follows: (a) M3 in French‐American‐Britain (FAB) classification (acute promyelocytic leukemia); (b) complicated with other malignant myeloid diseases (eg, polycythemia vera or primary thrombocytosis) or malignancies; (c) human immunodeficiency virus (HIV) positive; and (d) pregnant or lactating woman. For the healthy bone marrow donors, their health conditions were confirmed before donation. This study was approved by the Ethics Committee of our hospital, and all participants signed informed consents.
2.2. Data collection
For the AML patients, the demographic characteristics including age and gender were collected on the enrollment, while the clinical characteristics were acquired from blood and bone marrow examinations, including French‐American‐Britain (FAB) classification, cytogenetics abnormalities (such as normal karyotype (NK), complex karyotype (CK), inv(16) or t(16;16), t(8;21), +8, −7 or 7q‐, t(9;11), 11q23, t(9;22), inv(3) or t(3;3), −5 or 5q‐, t(6;9), and so on), monosomal karyotype (MK), molecular genetics mutation (such as internal tandem duplications in the FMS‐like tyrosine kinase 3 (FLT3‐ITD) mutation, isolated biallelic CCAAT/enhancer‐binding protein α (CEBPA) mutation, and nucleophosmin 1 (NPMI) mutation), and white blood cell (WBC) level.
2.3. Risk assessment
The risk stratification was assessed based on cytogenetics and molecular abnormalities, and AML patients were classified as favorable‐risk stratification (cytogenetics: inv(16) or t(16;16), or t(8;21), t(15;17); molecular abnormalities: normal cytogenetics, NPM1 mutation in the absence of FLT3‐ITD, or isolated biallelic CEBPA mutation); intermediate‐risk stratification (cytogenetics: normal cytogenetics, +8 alone, t(9;11), other non‐defined; molecular abnormalities: t(8;21), inv(16), t(16;16): with c‐KIT mutation), and poor‐risk stratification (cytogenetics: ≥3 clonal chromosomal abnormalities, MK, −5, 5q‐, −7, 7q‐, 11q23‐non t(9;11) inv(3), t(3;3), t(6;9), t(9;22); molecular abnormalities: normal cytogenetics, with FLT3‐ITD mutation), according to NCCN guideline of AML.14
2.4. Sample collection
Bone marrow samples of AML patients were collected before initiation of treatment, and bone marrow samples of healthy donors were acquired when examining their eligibility for bone marrow transplantation. After bone marrow samples collection, the bone marrow mononuclear cells (BMMCs) were separated by density gradient centrifugation. Then, the expression of PPM1D mRNA in mononuclear cells was detected by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR), and the expression of PPM1D protein in mononuclear cells was measured by Western blot.
2.5. RT‐qPCR
Total RNA was extracted from mononuclear cells using TRIzol reagent (Invitrogen) and then reversely transcribed to cDNA using ReverTra Ace qPCR RT Kit (Toyobo). After that, qPCR was performed using KOD SYBR qPCR Mix (Toyobo) to quantify PPM1D expression. The procedures of amplification were carried out as follows: first, 3 minutes at 95 degrees centigrade, 40 cycles of PCR then followed by standard conditions with 15 seconds denaturation at 95 degrees centigrade, next elongation for 1 minute at 61 degrees centigrade. And the result was calculated using 2−ΔΔCt method with GAPDH as an internal reference. The primers were listed as follows: PPM1D forward primer: CAATTGGCCTTGTGCCTACT, reverse primer: TCTTTCGCTGTGAGGTTGTG; GAPDH, forward primer: GAGTCCACTGGCGTCTTCAC, reverse primer: ATCTTGAGGCTGTTGTCATACTTCT.
2.6. Western blot
Total protein was extracted with RIPA buffer (Sigma‐Aldrich). The protein concentration in each sample was then measured using the Bicinchoninic Acid Kit (Sigma‐Aldrich). 20 μg protein was loaded to NUPAGETM Bis‐Tris 4%‐8% protein gels (Thermo Scientific) and presented with electrophoresis, followed by transferring onto polyvinylidene fluoride membrane (Millipore, USA). After blocking with BSA (Sigma‐Aldrich, Louis, MO, USA), the membranes were incubated with the primary antibodies overnight at 4°C. Then, the membranes were incubated with the secondary antibody for 90 minutes at 37°C. PierceTM Fast Western Blot Kit, ECL Substrate (Thermo Scientific) was used to illuminized the bands, and Gel Imager (Thermo Scientific) was used to visualize the result. The antibodies used in this study were as follows:
Primary antibodies: Mouse Anti‐PPM1D/WIP1 antibody (dilution, 1:1000, Abcam), Mouse Anti‐GAPDH antibody (dilution, 1:5000, Abcam); secondary antibody: Goat Anti‐Mouse IgG H&L (HRP) (dilution, 1:10 000, Abcam).
2.7. Treatment and follow‐up
After induction therapy, the remission status was assessed for all patients, and based on the remission status, patients were classified as complete remission (CR) group and non‐CR group. Besides, intensive follow‐up was conducted for all patients, and the last follow‐up date was June 31, 2019. During follow‐up, induction therapy failure, relapse from CR, or death were recorded. Event‐free survival (EFS) was defined as the duration from the date of initiation of treatment to the date of induction therapy failure, or relapse from CR or death, and patients not known to have any of these events were censored on the date they were last examined.2 Overall survival (OS) was defined as the duration from the date of initiation of treatment to the date of death, and patients not known to have died at last follow‐up were censored on the date they were last known to be alive.2
2.7.1. Grouping
According to the median values of PPM1D mRNA relative expression in AML patients, all AML patients were divided into patients with PPM1D mRNA high expression and those with PPM1D mRNA low expression. And according to the median values of PPM1D protein relative intensity in AML patients, all AML patients were further divided into patients with PPM1D protein high intensity and those with PPM1D protein low intensity.
2.8. Statistical analysis
All statistical analyses were performed using SPSS 22.0 (IBM), and all figures were plotted using GraphPad Prism 7.00 (GraphPad Software). Continuous variables were displayed as mean ± standard deviation (SD) or median and interquartile range (IQR). Categorical variables were summarized as frequency (percentage). Comparisons of PPM1D mRNA/protein expression between two groups were determined by Wilcoxon rank‐sum test, while comparisons of PPM1D mRNA/protein expression among three groups were analyzed by Kruskal‐Wallis H test. Receiver operating characteristic (ROC) curves and the areas under the curve (AUC) with 95% confidence intervals (CI) were used to assess the ability of PPM1D mRNA/protein in discriminating AML and healthy donors. Kaplan‐Meier curves were used to display EFS and OS, and the difference of EFS and OS between PPM1D high expression group and PPM1D low expression group (classified by the median values of PPM1D mRNA/protein relative expression/intensity) was determined by log‐rank test. P value < .05 was considered significant.
3. RESULTS
3.1. Clinical characteristics of AML patients
There were total of 221 AML patients enrolled in our present study, and their mean age was 52.1 ± 14.9 years (Table 1). There were 85 (38.5%) females and 136 (61.5%) males among all patients. As for FAB classification, there were 79 (35.7%), 65 (29.4%), 66 (29.9%) and 11 (5.0%) patients in M2, M4, M5 and M6 respectively. And regarding risk stratification, the number of patients with favorable‐risk, intermediate‐risk and poor‐risk were 58 (26.3%), 88 (39.8%), and 75 (33.9%) respectively. As for the induction therapy regimens, the number of patients who received daunorubicin + cytarabine, idarubicin + cytarabine, and anthracenedione mitoxantrone + cytarabine was 96 (43.4%), 85 (38.5%), and 40 (18.1%), respectively. Information of other clinical characteristics was listed in Table 1.
Table 1.
Clinical characteristics of AML patients
| Items | AML patients (N = 221) |
|---|---|
| Age (years), mean ± SD | 52.1 ± 14.9 |
| Gender, No. (%) | |
| Female | 85 (38.5) |
| Male | 136 (61.5) |
| FAB classification, No. (%) | |
| M2 | 79 (35.7) |
| M4 | 65 (29.4) |
| M5 | 66 (29.9) |
| M6 | 11 (5.0) |
| Cytogenetics, No. (%) | |
| NK | 113 (51.1) |
| CK | 25 (11.3) |
| inv(16) or t(16;16) | 17 (7.7) |
| t(8;21) | 10 (4.5) |
| +8 | 7 (3.2) |
| −7 or 7q‐ | 7 (3.2) |
| t(9;11) | 7 (3.2) |
| 11q23 | 6 (2.7) |
| t(9;22) | 4 (1.8) |
| inv(3) or t(3;3) | 2 (0.9) |
| −5 or 5q‐ | 1 (0.5) |
| t(6;9) | 1 (0.5) |
| Others (non‐defined) | 21 (9.5) |
| MK, No. (%) | 19 (8.6) |
| Molecular genetics mutation, No. (%) | |
| FLT3‐ITD mutation | 48 (21.7) |
| Isolated biallelic CEBPA mutation | 22 (10.0) |
| NPMI mutation | 78 (35.3) |
| Risk stratification, No. (%) | |
| Favorable‐risk | 58 (26.3) |
| Intermediate‐risk | 88 (39.8) |
| Poor‐risk | 75 (33.9) |
| WBC (×109/L), median (IQR) | 17.0 (8.5‐29.2) |
| Induction therapy regimens, No. (%) | |
| Daunorubicin + cytarabine | 96 (43.4) |
| Idarubicin + cytarabine | 85 (38.5) |
| Anthracenedione mitoxantrone + cytarabine | 40 (18.1) |
Abbreviations: AML, acute myeloid leukemia; CEBPA, CCAAT/enhancer‐binding protein α; CK, complex karyotype; FAB classification, French‐American‐Britain classification; FLT3‐ITD, internal tandem duplications in the FMS‐like tyrosine kinase 3; IQR, interquartile range; MK, monosomal karyotype; NK, normal karyotype; NPM1, nucleophosmin 1; SD, standard deviation; WBC, white blood cell.
3.2. Correlation of PPM1D with AML risk
Protein phosphatase Mg2+/Mn2+ dependent 1D mRNA relative expression was increased in AML patients (1.640 [1.221‐2.939]) compared with health donors (1.025 [0.649‐1.649]) (P < .001) (Figure 1A). And PPM1D mRNA relative expression was of acceptable value in differentiating AML patients from health donors (AUC: 0.728, 95% CI: 0.651‐0.806) (Figure 1B). Furthermore, PPM1D protein relative intensity was also elevated in AML patients (0.584 [0.417‐0.887]) compared with health donors (0.303 [0.212‐0.482]) (P < .001) (Figure 1C,D). PPM1D protein relative intensity was also of good value in differentiating AML patients from health donors (AUC: 0.782, 95% CI: 0.707‐0.857) (Figure 1E).
Figure 1.
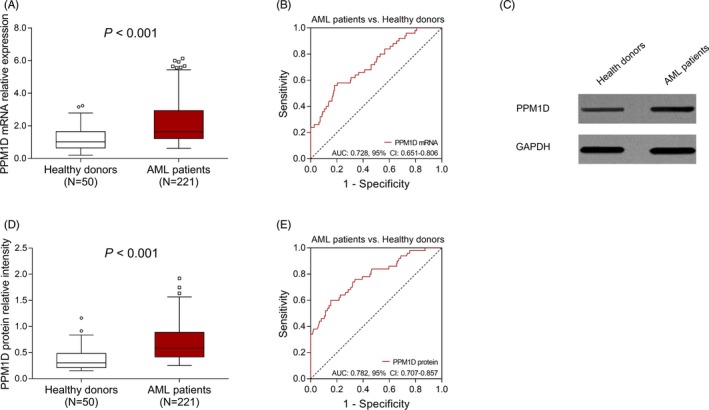
PPM1D expression between AML patients and health donors. The comparison of PPM1D mRNA relative expression between AML patients and healthy donors (A). The performance of PPM1D mRNA relative expression in distinguishing AML patients from healthy donors (B). Representative Western blot images exhibiting the PPM1D protein relative expression in AML patients and healthy donors (C). The comparison of PPM1D protein relative intensity between AML patients and healthy donors (D). The performance of PPM1D protein relative intensity in distinguishing AML patients from healthy donors (E). Comparisons of PPM1D mRNA/protein expression between two groups were determined by Wilcoxon rank‐sum test. ROC curves and the AUC with 95% CI were used to assess the ability of PPM1D mRNA/protein in discriminating AML and healthy donors. P value < .05 was considered significant. AML, acute myeloid leukemia; mRNA, messenger RNA; AUC, area under the curve; CI, confidence interval; PPM1D, protein phosphatase Mg2+/Mn2+ dependent 1D; ROC, receiver operating characteristic
3.3. Correlation of PPM1D with risk stratification in AML patients
PPM1D mRNA relative expression was the highest in patients with poor‐risk (2.219 [1.432‐3.386]), followed by patients with intermediate‐risk (1.639 [1.156‐2.388]) and then patients with favorable‐risk (1.391 [0.962‐2.194]) (P < .001) (Figure 2A). And PPM1D protein relative intensity was also the highest in patients with poor‐risk (0.713 [0.488‐0.976]), followed by patients with intermediate‐risk (0.580 [0.366‐0.864]) and then patients with favorable‐risk (0.478 [0.374‐0.648]) (P < .001) (Figure 2B,C).
Figure 2.
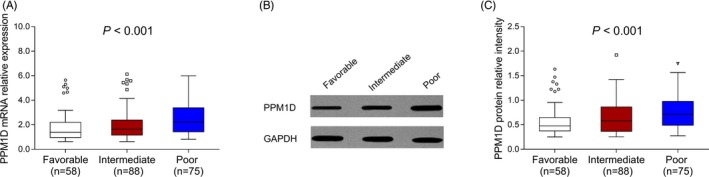
PPM1D expression among AML patients with favorable‐risk, intermediate‐risk, and poor‐risk. The comparison of PPM1D mRNA relative expression among AML patients with favorable‐risk stratification, intermediate‐risk stratification, and poor‐risk stratification (A). Representative Western blot images presenting the PPM1D protein relative expression among AML patients with favorable‐risk stratification, intermediate‐risk stratification, and poor‐risk stratification (B). The comparison of PPM1D protein relative intensity among AML patients with favorable‐risk stratification, intermediate‐risk stratification, and poor‐risk stratification (C). Comparisons of PPM1D mRNA/protein relative expression among three groups were analyzed by Kruskal‐Wallis H test. P value < .05 was considered significant. AML, acute myeloid leukemia; mRNA, messenger RNA; PPM1D, protein phosphatase Mg2+/Mn2+ dependent 1D
3.4. Correlation of PPM1D with mutation in AML patients
Protein phosphatase Mg2+/Mn2+ dependent 1D protein relative intensity was positively correlated with FLT3‐ITD mutation (P = .029) (Figure S1A), while there was no association between PPM1D protein relative intensity with CEBPA mutation (P = .328) (Figure S1B) or NPMI mutation (P = .843) (Figure S1C). Similarly, PPM1D mRNA relative expression (P = .013) was positively correlated with FLT3‐ITD mutation (Figure S1C), while there was no association between PPM1D mRNA relative expression with CEBPA mutation (P = .725) (Figure S1E) or NPMI mutation (P = .979) (Figure S1F).
3.5. Correlation of PPM1D with EFS in AML patients
All AML patients were divided into CR group (n = 174) and non‐CR group (n = 47) based on the induction remission status. As to the clinical characteristics of CR and non‐CR patients, we observed that the mean age of CR patients was decreased compared with non‐CR patients (P = .020) (Table S1). Regarding molecular genetics mutation, the number of CR patients with isolated biallelic CEBPA mutation was decreased compared with non‐CR patients (P = .027), while the number of CR patients with NPMI mutation was increased compared with non‐CR patients (P = .009). As for the risk stratification, CR patients trended to have favorable‐risk stratification compared with non‐CR patients, but without statistical significance (P = .083). However, there was no difference of gender (P = .483), FAB classification (P = .727), cytogenetics (P = .411), MK (P = .137), FLT3‐ITD mutation (P = .266), WBC (P = .353), or induction therapy regimens (P = .322) between CR patients and non‐CR patients. More detailed clinical characteristics of CR patients and non‐CR patients were listed in Table S1.
And we further analyzed the correlation of PPM1D with CR in AML patients and found that PPM1D mRNA relative expression was increased in non‐CR group (2.518 [1.416‐3.983]) compared with CR group (1.522 [1.172‐2.449]) (P < .001) (Figure 3A). And PPM1D protein relative intensity was also elevated in non‐CR group (0.731 [0.454‐1.185]) compared with CR group (0.562 [0.411‐0.782]) (P = .010) (Figure 3B,C).
Figure 3.
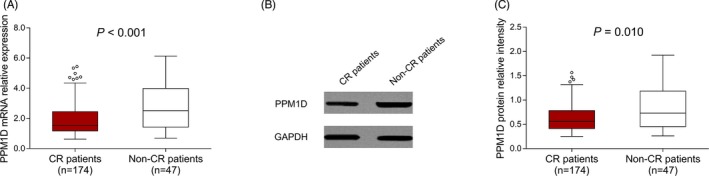
PPM1D expression between CR patients and non‐CR patients. The comparison of PPM1D mRNA relative expression between CR patients and non‐CR donors (A). Representative Western blot images presenting the PPM1D protein relative expression between CR patients and non‐CR patients (B). The comparison of PPM1D protein relative intensity between CR patients and non‐CR patients (C). Comparisons of PPM1D mRNA/protein expression between two groups were determined by Wilcoxon rank‐sum test. P value < .05 was considered significant. AML, acute myeloid leukemia; CR, complete remission; mRNA, messenger RNA; PPM1D, protein phosphatase Mg2+/Mn2+ dependent 1D
3.6. Correlation of PPM1D with EFS in AML patients
All AML patients were classified into patients with PPM1D mRNA/protein high expression and PPM1D mRNA/protein low expression according to the median values of PPM1D mRNA/protein relative expression at baseline, and EFS was reduced in patients with PPM1D mRNA high expression compared with patients with PPM1D mRNA low expression (P < .001) (Figure 4A). EFS was also shorter in patients with PPM1D protein high expression compared with patients with PPM1D protein low expression (P < .001) (Figure 4B).
Figure 4.
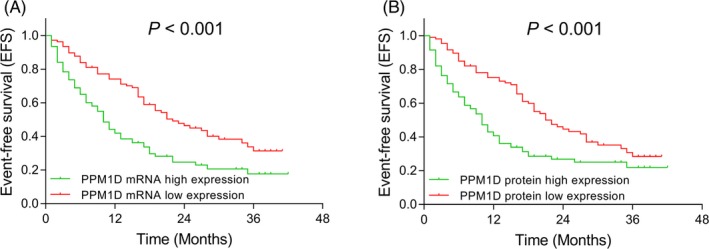
Comparison of EFS between AML patients with PPM1D high and low expressions. The comparison of EFS between AML patients with PPM1D mRNA high expression and PPM1D mRNA low expression (A). The comparison of EFS between AML patients with PPM1D protein high expression and PPM1D protein low expression (B). Kaplan‐Meier curves were used to display EFS, and the difference of EFS between PPM1D high expression group and PPM1D low expression group was determined by log‐rank test. P value < .05 was considered significant. AML, acute myeloid leukemia; EFS, event‐free survival; mRNA, messenger RNA; PPM1D, protein phosphatase Mg2+/Mn2+ dependent 1D
3.7. Correlation of PPM1D with OS in AML patients
OS was reduced in patients with PPM1D mRNA high expression compared with patients with PPM1D mRNA low expression (P = .004) (Figure 5A). And OS was also reduced in patients with PPM1D protein high expression compared with PPM1D protein low expression (P = .006) (Figure 5B).
Figure 5.
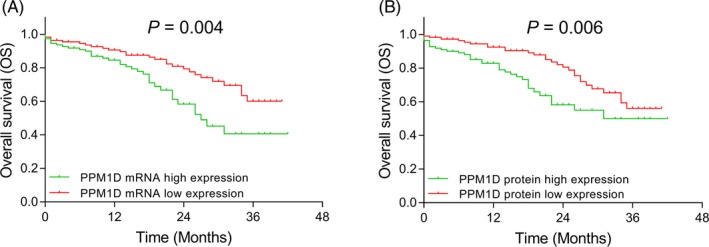
Comparison of OS between AML patients with PPM1D high and low expressions. The comparison of OS between AML patients with PPM1D mRNA high expression and PPM1D mRNA low expression (A). The comparison of OS between AML patients with PPM1D protein high expression and PPM1D protein low expression (B). Kaplan‐Meier curves were used to display OS, and the difference of OS between PPM1D high expression group and PPM1D low expression group was determined by log‐rank test. P value < .05 was considered significant. AML, acute myeloid leukemia; mRNA, messenger RNA; OS, overall survival; PPM1D, protein phosphatase Mg2+/Mn2+ dependent 1D
4. DISCUSSION
In the present study, we found that (a) PPM1D was of acceptable value in predicting AML risk and its high expression was associated with poor‐risk stratification in AML patients. (b) PPM1D high expression was associated with worse CR, EFS, and OS in AML patients.
Protein phosphatase Mg2+/Mn2+ dependent 1D is reported to be a growth‐promoting phosphatase via exerting negative control on several tumor suppressor pathways and functions as an oncogene in various solid tumors.6, 7, 8, 15, 16 For example, clinical experiments indicate that PPM1D is highly expressed in non–small‐cell lung cancer tissues compared with normal lung tissues, and PPM1D overexpression is correlated with advanced tumor features (increased tumor size and lower histological differentiation) in NSCLC patients.8 Another study reveals that PPM1D is overexpressed in hepatocellular carcinoma, and PPM1D overexpression promotes cell viability and invasion via inhibition of p38MARK/p35/p16 signaling pathway. And the latter signaling pathway is known as an inactivation signaling of both solid tumors and hematological malignancies, such as APL.7, 9, 17 Additionally, existing evidences suggests that PPM1D induces transformation virus‐infected T cells into leukemic cells, which contributes to the development of ATLL.10 And in another type of leukemia, APL, PPM1D inhibition is observed to induce neutrophil differentiation in human APL cell line HL‐60, suggesting that targeting PPM1D inhibits the APL progression.11 Although the previous studies indicate that PPM1D play an important role in some solid tumors as well as hematologic malignancies, the role of PPM1D in AML has not been explored yet. Therefore, we performed the present study to explore the correlation of PPM1D with AML risk, and AML clinical features. We found that PPM1D was of acceptable value in predicting AML risk and its high expression was associated with poor‐risk stratification in AML patients. The possible reasons might include that (a) increased expression of PPM1D might enhance its downstream oncogenic target genes (such as MMP‐9, VEGF‐C), inducing the transformation of AML cells, which further contributed to the initiation of AML in AML patients. Therefore, PPM1D was associated with higher risk of AML (b). Additionally, upregulation of PPM1D might inactivate the tumor suppressor signaling pathway (Chk2/p53 signaling), leading to the inhibiting effect on AML apoptosis but the promoting effects on cytogenetic abnormality and AML‐related gene mutations, and thus, AML patients with increased PPM1D expression had poor‐risk stratification. Interestingly, we also observed that PPM1D protein relative intensity was positively correlated with FLT3‐ITD mutation, which could validate our explanation.
Regarding the correlation of PPM1D with prognosis, some recent studies report the positive association of PPM1D with high chemotherapy resistance and undesirable survival profile in several solid tumors.9, 16, 18, 19, 20, 21, 22 For example, one study indicates that downregulation of PPM1D activates Chk1 and p53, which further increases the ovarian cancer cell sensibility to cisplatin treatment.20 Another study reveals that in breast cancer treatment, decreased expression of PPM1D improves the effect of doxorubicin‐induced apoptosis via activating p53‐mediated signaling pathway in MCF‐7 breast cancer cell line.21 In addition, the predictive role of PPM1D on prognosis has been reported by several researches in solid tumors.16, 22, 23 For example, in colorectal cancer, patients with high levels of PPM1D show worse five‐year OS and recurrence‐free survival compared with those with low levels of PPM1D.16 Another study in esophageal squamous cell carcinoma (ESCC) exemplifies that PPM1D expression is elevated in metastatic ESCC patients compared with those without metastasis, and PPM1D high expression is considered to be an independent prognostic factor in ESCC patients.22 Based on these previous studies, PPM1D is of potential in predicting worse treatment response as well as survival profile in patients with solid tumors; however, the correlation of PPM1D expression with treatment response and survival profile in AML patients remained unknown. In our present study, we found that PPM1D high expression was associated with worse CR, EFS, and OS in AML patients. The possible reasons might include that: (a) According to the previous results, PPM1D high expression was associated with poor‐risk stratification, which indirectly led to unfavorable prognosis via affecting cytogenetics and molecular abnormalities. (b) PPM1D high expression inactivated its downstream anti‐tumor signaling pathways (Chk2/p53 signaling and p38/p53 signaling), contributing to the decreased chemotherapy sensitivity; therefore, AML patients with PPM1D high expression reported decreased CR, EFS, and OS.
There still existed some limitations in our study: (a) The sample size of the healthy donors was relatively small, which might lead to relatively low statistical significance. (b) Although previous study indicated that PPM1D might affect cell activities via regulating its downstream anti‐tumor genes in solid tumors, the underlying mechanism of PPM1D in AML still needs further cellular experiments for exploration. (c) Considering that our study was single‐centered, which might lead to regional selective bias, therefore patients from more centers were needed for validation.
In conclusion, PPM1D high expression correlates with poor‐risk stratification, worse CR, and unfavorable survival profile in AML patients, suggesting its potential to guide AML management.
Supporting information
Yu M, Hu J, He D, et al. Potentiality of Protein phosphatase Mg2+/Mn2+ dependent 1D as a biomarker for predicting prognosis in acute myeloid leukemia patients. J Clin Lab Anal. 2020;34:e23171 10.1002/jcla.23171
REFERENCES
- 1. Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136‐1152. [DOI] [PubMed] [Google Scholar]
- 2. Dohner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453‐474. [DOI] [PubMed] [Google Scholar]
- 3. Piemontese S, Ciceri F, Labopin M, et al. A survey on unmanipulated haploidentical hematopoietic stem cell transplantation in adults with acute leukemia. Leukemia. 2015;29(5):1069‐1075. [DOI] [PubMed] [Google Scholar]
- 4. Ramos NR, Mo CC, Karp JE, et al. Current approaches in the treatment of relapsed and refractory acute myeloid leukemia. J Clin Med. 2015;4(4):665‐695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tallman MS, Wang ES, Altman JK, et al. Acute myeloid leukemia, version 3.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2019;17(6):721‐749. [DOI] [PubMed] [Google Scholar]
- 6. Fiscella M, Zhang H, Fan S, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53‐dependent manner. Proc Natl Acad Sci U S A. 1997;94(12):6048‐6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu B, Guo BM, Kang J, et al. PPM1D exerts its oncogenic properties in human pancreatic cancer through multiple mechanisms. Apoptosis. 2016;21(3):365‐378. [DOI] [PubMed] [Google Scholar]
- 8. Yang S, Dong S, Qu X, et al. Clinical significance of Wip1 overexpression and its association with the p38MAPK/p53/p16 pathway in NSCLC. Mol Med Rep. 2017;15(2):719‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoda A, Toyoshima K, Watanabe Y, et al. Arsenic trioxide augments Chk2/p53‐mediated apoptosis by inhibiting oncogenic Wip1 phosphatase. J Biol Chem. 2008;283(27):18969‐18979. [DOI] [PubMed] [Google Scholar]
- 10. Zane L, Yasunaga J, Mitagami Y, et al. Wip1 and p53 contribute to HTLV‐1 Tax‐induced tumorigenesis. Retrovirology. 2012;9:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kamada R, Kudoh F, Yoshimura F, et al. Inhibition of Ser/Thr phosphatase PPM1D induces neutrophil differentiation in HL‐60 cells. J Biochem. 2017;162(4):303‐308. [DOI] [PubMed] [Google Scholar]
- 12. Hsu JI, Dayaram T, Tovy A, et al. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell. 2018;23(5): 700–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swerdlow SHCE, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, Vol 2, 4th ed. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 14. NCCN:Acute Myeloid Leukemia Version 1. 2015; https://www.nccn.org/
- 15. Buss MC, Remke M, Lee J, et al. The WIP1 oncogene promotes progression and invasion of aggressive medulloblastoma variants. Oncogene. 2015;34(9):1126‐1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li ZT, Zhang L, Gao XZ, et al. Expression and significance of the Wip1 proto‐oncogene in colorectal cancer. Asian Pac J Cancer Prev. 2013;14(3):1975‐1979. [DOI] [PubMed] [Google Scholar]
- 17. Xu Z, Cao C, Xia H, et al. Protein phosphatase magnesium‐dependent 1delta is a novel tumor marker and target in hepatocellular carcinoma. Front Med. 2016;10(1):52‐60. [DOI] [PubMed] [Google Scholar]
- 18. Wang P, Ye JA, Hou CX, et al. Combination of lentivirus‐mediated silencing of PPM1D and temozolomide chemotherapy eradicates malignant glioma through cell apoptosis and cell cycle arrest. Oncol Rep. 2016;36(5):2544‐2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oghabi Bakhshaiesh T, Majidzadeh AK, Esmaeili R. Wip1: a candidate phosphatase for cancer diagnosis and treatment. DNA Repair (Amst). 2017;54:63‐66. [DOI] [PubMed] [Google Scholar]
- 20. Ali AY, Abedini MR, Tsang BK. The oncogenic phosphatase PPM1D confers cisplatin resistance in ovarian carcinoma cells by attenuating checkpoint kinase 1 and p53 activation. Oncogene. 2012;31(17):2175‐2186. [DOI] [PubMed] [Google Scholar]
- 21. Kong W, Jiang X, Mercer WE. Downregulation of Wip‐1 phosphatase expression in MCF‐7 breast cancer cells enhances doxorubicin‐induced apoptosis through p53‐mediated transcriptional activation of Bax. Cancer Biol Ther. 2009;8(6):555‐563. [DOI] [PubMed] [Google Scholar]
- 22. Li K, Liu Y, Xu S, et al. PPM1D functions as oncogene and is associated with poor prognosis in esophageal squamous cell carcinoma. Pathol Oncol Res. 2018; 10.1007/s12253-018-0518-1 [DOI] [PubMed] [Google Scholar]
- 23. Kadam PD, Chuan HH. Erratum to: rectocutaneous fistula with transmigration of the suture: a rare delayed complication of vault fixation with the sacrospinous ligament. Int Urogynecol J. 2016;27(3):505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


