Abstract
NAD+ is an essential metabolite participating in cellular biochemical processes and signaling. The regulation and interconnection among multiple NAD+ biosynthesis pathways are incompletely understood. Yeast (Saccharomyces cerevisiae) cells lacking the N-terminal (Nt) protein acetyltransferase complex NatB exhibit an approximate 50% reduction in NAD+ levels and aberrant metabolism of NAD+ precursors, changes that are associated with a decrease in nicotinamide mononucleotide adenylyltransferase (Nmnat) protein levels. Here, we show that this decrease in NAD+ and Nmnat protein levels is specifically due to the absence of Nt-acetylation of Nmnat (Nma1 and Nma2) proteins and not of other NatB substrates. Nt-acetylation critically regulates protein degradation by the N-end rule pathways, suggesting that the absence of Nt-acetylation may alter Nmnat protein stability. Interestingly, the rate of protein turnover (t½) of non-Nt-acetylated Nmnats did not significantly differ from those of Nt-acetylated Nmnats. Accordingly, deletion or depletion of the N-end rule pathway ubiquitin E3 ligases in NatB mutants did not restore NAD+ levels. Next, we examined whether the status of Nt-acetylation would affect the translation of Nmnats, finding that the absence of Nt-acetylation does not significantly alter the polysome formation rate on Nmnat mRNAs. However, we observed that NatB mutants have significantly reduced Nmnat protein maturation. Our findings indicate that the reduced Nmnat levels in NatB mutants are mainly due to inefficient protein maturation. Nmnat activities are essential for all NAD+ biosynthesis routes, and understanding the regulation of Nmnat protein homeostasis may improve our understanding of the molecular basis and regulation of NAD+ metabolism.
Keywords: NAD biosynthesis, cell metabolism, metabolic regulation, yeast genetics, yeast metabolism, cell signaling, NAD homeostasis, NatB, nicotinamide mononucleotide adenylyltransferase (Nmnat), protein acetylation
Introduction
NAD+ and its reduced form NADH are primary redox carriers involved in various cellular biochemical reactions. NAD+ also serves as a co-substrate in protein modifications, such as protein deacetylation (mediated by the sirtuins, Sir2 family proteins) and ADP-ribosylation (mediated by the poly(ADP-ribose)polymerase). These modified proteins contribute to the regulation of chromatin structure, DNA repair, circadian rhythm, metabolic responses, and life span (1–5). Essentially, cells must balance NAD+ biosynthesis with its consumption to maintain optimal cellular function. Moreover, abnormalities in NAD+ metabolism have been associated with several metabolic disorders and diseases, including diabetes, cancers, and neuron degeneration (6–9). Understanding the regulation of NAD+ homeostasis may help elucidate the mechanisms of these disorders.
NAD+ biosynthesis in yeast is maintained by three major pathways: de novo biosynthesis, nicotinic acid (NA)2 and nicotinamide (NAM) salvage (10, 11), and nicotinamide riboside (NR) salvage (Fig. 1A) (12). NAD+ biosynthesis by the de novo pathway begins at tryptophan and requires Bna proteins to synthesize nicotinic acid mononucleotide (NaMN). This pathway is the most resource-intensive of the three and is heavily regulated (13–15). NaMN is also produced by the NA/NAM salvage branch, which begins with precursors like NA and NAM. Under NA abundant conditions, NA/NAM salvage is the preferred NAD+ biosynthesis route, and BNA genes are silenced by the NAD+-dependent deacetylase Hst1 (13, 14). NaMN produced from both branches is converted to nicotinic acid adenine dinucleotide by nicotinamide mononucleotide adenylyltransferases (Nmnats) Nma1 and Nma2 (16–19), followed by the amidation to NAD+ by Qns1 (20, 21). In the NR salvage pathway, NR is phosphorylated by NR kinase Nrk1 to produce nicotinamide mononucleotide (NMN) (12). Nma1, Nma2, or Pof1 transfer the AMP moiety of ATP to NMN to produce NAD+ (16–19, 22). It has been shown that Nma1 and Nma2 have dual-substrate specificity toward both NMN and NaMN (16–18), whereas Pof1 activity is specific for NMN (22). NR can also enter the NAM salvage branch when converted to NAM by nucleosidases Urh1 and Pnp1 (23). Moreover, yeast cells also release and re-uptake small NAD+ precursors such as NA, NAM, QA, and NR (Fig. 1A) (14, 22, 24). Specific transporters Tna1 (for NA and QA) (25, 26) and Nrt1 (for NR) (27) are responsible for the uptake of NAD+ precursors, whereas the mechanisms of precursor release remain unclear. The three branches of NAD+ biosynthesis are thus coordinated together and provide the cell with NAD+ tuned to the cellular requirements and environmental conditions.
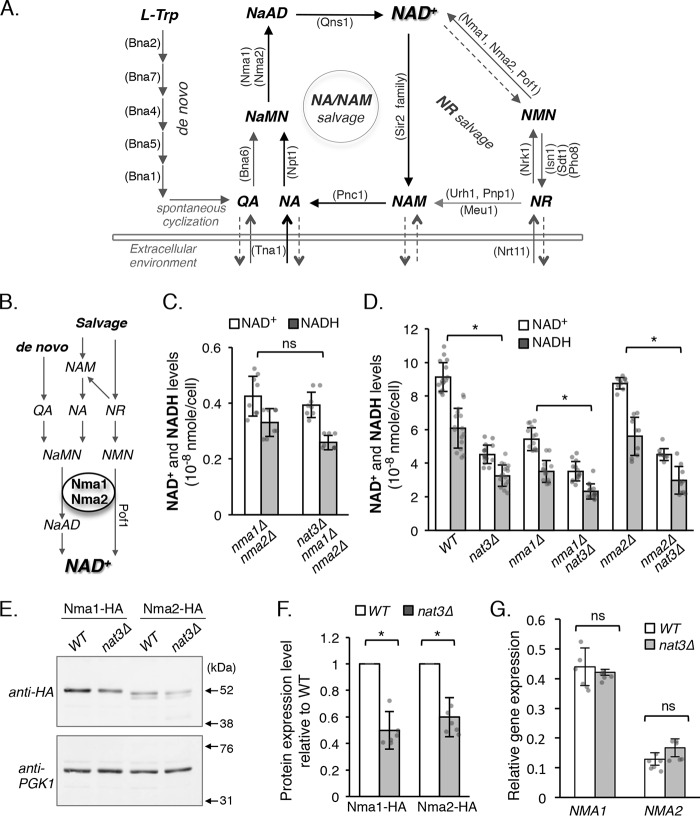
Figure 1.
NAD+ deficiency of the NatB mutant is primarily due to Nma1 and Nma2 defects. A, simplified model of NAD+ biosynthesis pathways in S. cerevisiae. NAD+ can be synthesized de novo from tryptophan (l-Trp) and by salvaging NAD+ intermediates through the NA/NAM and NR cycles. Yeast cells also release and re-uptake small NAD+ precursors; however, the mechanisms are not completely understood. Abbreviations of protein names are shown in parentheses. B, schematic diagram showing that all three NAD+ biosynthesis pathways require Nma1 and Nma2 activities. A third Nmnat Pof1 is part of the NR salvage pathway exclusively. C, nma1Δnma2Δ and nma1Δnma2Δnat3Δ mutants have similar levels of NAD+(H). NR was supplemented to the cell culture at 10 μm. Cells lacking NMA1 and NMA2 can grow in the presence of NR using Pof1. The graph is based on data of two independent experiments. Error bars represent data from four biological replicates per strain each with two technical replicates (total n = 8 per strain). Individual data points are shown as dots in the bar graph. D, NAD+(H) levels of nma1Δ and nma2Δ were compared combining nma1Δ or nma2Δ with nat3Δ deletions. Both nma1Δnat3Δ and nma2Δnat3Δ had lower levels of NAD+ compared with the single Nmnat deletion. The graph is based on data of six independent experiments. Error bars represent data from eight (WT and nat3Δ), seven (nma1Δ and nma1Δnat3Δ), or five (nma2Δ and nma2Δnat3Δ) biological replicates per strain each with two technical replicates (total n = 16, 14, or 10 per strain). E, Western blot analysis of Nma1 and Nma2 tagged with HA in combination with PGK1 control in both WT and nat3Δ cells. Arrows mark the positions of the molecular mass markers. F, quantitative analysis of E showing Nma1 and Nma2 protein depletions due to nat3Δ deletion. Nma1–HA and Nma2–HA are normalized to PGK1, and WT is set to 1. The graph is based on data of three independent experiments. Error bars represent data from six biological replicates per strain (total n = 6 per strain). G, gene expression qPCR analysis of NMA1 and NMA2 mRNA in WT and nat3Δ cells. Data shown are representative of at least three independent experiments. The graph is based on data from two independent experiments. Error bars represent data from two biological replicates per strain each with three technical replicates (total n = 6 per strain). The error bars denote standard deviations. The p values are calculated using Student's t test (*, p < 0.05; ns, not significant).
The Nmnat proteins Nma1 and Nma2 are the only enzymes that are responsible for generating NAD+ in all three biosynthesis pathways (Fig. 1B). Therefore, Nma1 and Nma2 may be critical for the co-regulation of de novo, NA/NAM salvage, and NR salvage pathways. In fact, we have previously shown that overexpression of NMA1 caused a significant increase in the total NAD+ content of the cell (24). It was also shown in a recent study that Nma1 is the only NAD+ biosynthesis enzyme that upon overexpression can increase the NAD+ content in the cell and modulates NAD+ to correlate with the concentrations of ATP (15). Overall, these studies suggest Nmnats are rate-limiting factors in NAD+ biosynthesis pathways. Previously, we identified the NatB complex, composed of Nat3 (catalytic subunit) and Mdm20 (regulatory subunit), as a potential regulator of NAD+ biosynthesis, possibly through N-terminal (Nt-) acetylation of Nma1 and Nma2 (24). The levels of Nma1 and Nma2 proteins were reduced in NatB deletion mutants compared with the wildtype (WT). Nt-acetylation is primarily a co-translational protein modification carried out by Nt-acetyltransferases (28). This protein modification may affect protein stability, complex formation, and subcellular localization (28). NatB carries out Nt-acetylation of ∼15% of encoded yeast proteins, which are Met-retained peptides with Asp, Asn, Glu, or Gln at position 2 (29). NatB-mediated Nt-acetylation is efficient, and nearly all proteins that code as NatB substrates are indeed NatB substrates, which is not true for other Nt-acetyltransferases. Interestingly, yeast NatB substrates are predominantly near 100% as found in the acetylated state (28–30). In line with these observations, we showed that Nma1 and Nma2 peptides identified from WT cells were 100% acetylated, whereas Nma1 or Nma2 peptides identified from NatB mutants were 95–100% nonacetylated (24). Therefore, Nt-acetylation appears to be critical for maintaining proper Nmnat protein levels and thus NAD+ biosynthesis. To date, a direct link between NatB-mediated Nt-acetylation of Nmnats and NAD+ biosynthesis has not been established because NatB also has many other substrates in the cell.
It also remains unclear why NatB mutants have lower Nmnat protein levels. It is possible non-Nt-acetylated Nmnats are more susceptible to degradation. N-terminal regions of proteins contain specific degrons, termed N-degrons (31). These degrons are residue-specific and are described by the N-end rule pathways. The Arg/N-end rule pathway is dedicated to peptides with non-Nt-acetylated N-terminal Arg-, Lys-, His-, Leu-, Trp-, Phe-, Ile-, Tyr-, or Met-retained peptides followed by a bulk hydrophobic residue at position 2. Ubr1 is the E3 ligase that targets these substrates for ubiquitination and subsequent degradation by the proteasome. The Ac/N-end rule pathway is typically specific for Nt-acetylated substrates, which are targeted by the E3 ligases Doa10 or Not4. Without Nt-acetylation, Ac/N-end rule proteins may become unrecognizable to their E3 ligases and thus long-lived (32). Whereas Nma1 and Nma2 do not fulfill the requirements for degradation by the Arg/N-end rule pathway, it is worth noting that examples of Ac/N-substrates becoming Arg/N-substrates in the absence of Nt-acetylation have been observed (33, 34). In addition to the possibility of being degraded by the N-end rule pathways, the status of Nt-acetylation may alter the translation efficiency of Nmnats.
In this study, we aim to examine whether NatB regulates NAD+ biosynthesis directly through Nt-acetylation of Nma1 and Nma2 and why the absence of Nt-acetylation causes the reduction of Nmnat protein levels in NatB mutants. Our studies may help us understand the roles of Nt-acetylation on Nmnats and provide a molecular basis for the regulation of NAD+ homeostasis factors.
Results
Nma1 and Nma2 play a primary role in NatB deletion associated NAD+ deficiency
We previously reported altered NAD+ homeostasis in NatB deletion mutants, nat3Δ and mdm20Δ (24). These NatB deletion mutants have low levels of NAD+ with an increased flux of NAD+ precursors through the vacuolar NR salvage branch, which promotes the formation and release of NAM and NA. This defect was associated with a decrease in Nma1 and Nma2 protein levels, both of which are NatB substrates (24) and catalyze a reaction shared by all three pathways (Fig. 1B). Nearly 15% of all yeast proteins are Nt-acetylated by the NatB complex. To confirm that the low NAD+ phenotype in NatB deletion mutants is primarily due to Nma1 and Nma2 defects, we compared NAD+ levels of the nma1Δnma2Δ and nma1Δnma2Δnat3Δ mutants (Fig. 1C). If the low NAD+ levels in nat3Δ cells are due to additional defects, we would expect to see further decreases in NAD+ and NADH levels in the nma1Δnma2Δnat3Δ mutant. Under normal growth conditions, the nma1Δnma2Δ mutants are inviable unless NR is supplemented to the growth media. It was shown that Pof1, a third Nmnat that is not a NatB substrate, can convert NMN to NAD+ in the NR salvage branch (22). Therefore, adding NR to growth media supports the growth of the nma1Δnma2Δ mutants through the activity of Pof1 (Fig. 1A). In line with our expectations, there is no significant difference in NAD+ and NADH levels between the nma1Δnma2Δ and nma1Δnma2Δnat3Δ strains grown in NR-supplemented media. Furthermore, to examine whether NatB activity is important for both Nma1 and Nma2 functions, we paired the nat3Δ deletion with either nma1Δ or nma2Δ single deletions. As shown in Fig. 1D, both the nma1Δnat3Δ and nma2Δnat3Δ double mutants showed a further decrease in NAD+ and NADH levels compared with the respective single Nmnat deletions alone (Fig. 1D). The levels of decrease in nma1Δnat3Δ and nma2Δnat3Δ cells were ∼30 and ∼50%, respectively. Because Nma1 is the major Nmnat, it is anticipated that the absence of Nma1 Nt-acetylation (nat3Δnma2Δ versus nma2Δ cells) has a larger impact on the NAD+ level compared with the absence of Nma2 Nt-acetylation (nat3Δnma1Δ versus nma1Δ cells). These results indicate that the low NAD+ defects in NatB mutants act primarily through both Nma1 and Nma2.
Next, we determined whether Nma1 and Nma2 protein levels were decreased to a similar extent in the NatB deletion nat3Δ mutant when a different control is employed. In previous work we used actin, whose expression appeared stable in NatB mutants, as an internal control (24). However, because actin is also a NatB substrate (35), we chose Pgk1 as the internal control for protein turnover studies in this work. Pgk1 has already been used in many other protein degradation studies for the same purpose (36–40). Consistent with previous findings (24), Nma1 and Nma2 protein levels were significantly reduced in nat3Δ cells (Fig. 1E) to about 60% of the WT levels (Fig. 1F). A smaller band was observed right below the Nma2–HA band, which might be an Nma2-specific background band or a variant of Nma2. Although Nt-acetylation is primarily a co-translational protein modification, we also wanted to confirm that the low protein levels of Nma1 and Nma2 were not due to reduced transcription. As shown in Fig. 1G, no significant difference in the level of mRNAs (determined by RT-qPCR) was found when comparing the nat3Δ mutant to WT. Together, these results suggest that Nt-acetylation is required for maintaining NAD+ by modulating the Nma1 and Nma2 protein levels.
NatB promotes NAD+ biosynthesis directly through Nt-acetylation of Nma1 and Nma2
We previously found overexpression of Nma1 can increase the levels of NAD+ in both WT and nat3Δ cells (24). This result is supported by the finding that overexpression of Nma1 can bypass an ATP-dependent regulatory mechanism (15). However, this may also indicate that the low NAD+ defect in NatB mutants is not directly due to lack of Nt-acetylation of Nmnats but may rather stem from lower intracellular concentrations of ATP or other unforeseen consequences because NatB acetylates many other proteins. To test whether the Nt-acetylation modification is indeed important for Nma1- and Nma2-mediated NAD+ biosynthesis, we replaced the endogenous Nma1 and Nma2 with mutant versions expected to be NatA substrates instead of NatB substrates (Fig. 2A). NatA substrates are not Met-retained like NatB substrates, but they first undergo processing by removal of the initiator methionine by methionine aminopeptidases, and the new N terminus is then acetylated by NatA (Fig. 2A, left panel) (28). Additionally, potential NatB substrates (as predicted by their second amino acids) are almost always acetylated by NatB. This is not true for all potential NatA substrates. Based on reported proteomics data, we chose to insert Ser between the first two amino acids, Met and Asp, in Nma1 and Nma2 (Nma1/2MD → Nma1/2MSD) (Fig. 2A, right panel) because this peptide sequence is almost always acetylated by NatA (28, 30).
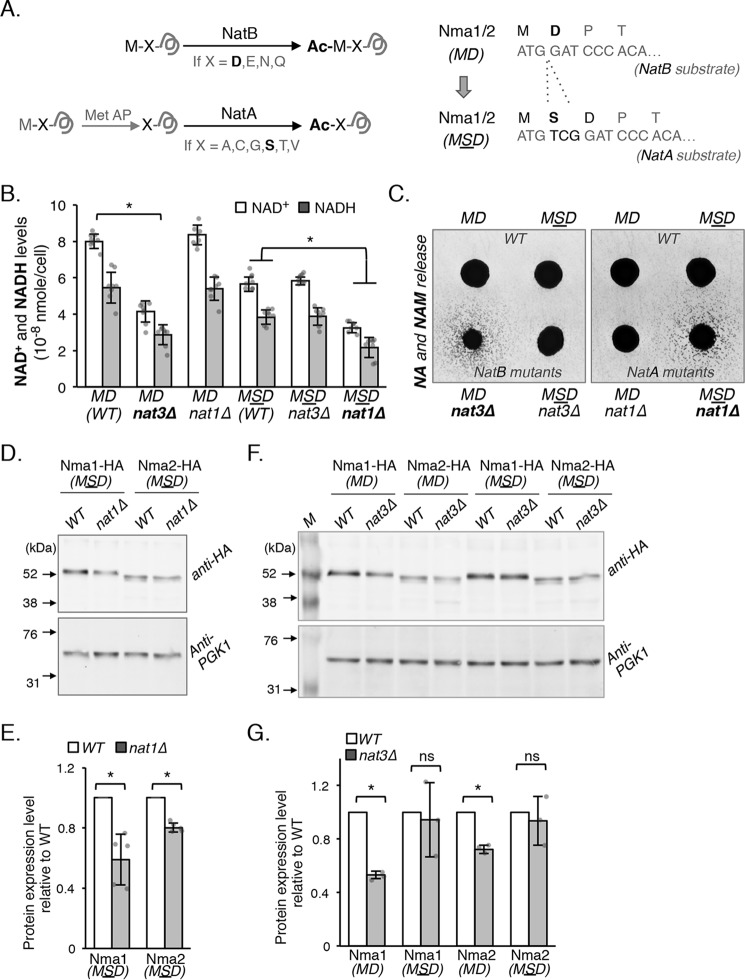
Figure 2.
NAD+ metabolism defect of the NatB mutant is directly linked to absence of Nt-acetylation and associated reduction of Nma1 and Nma2 protein levels. A, schematic showing amino acid sequences that result in Nt-acetylation by NatB or NatA (left panel). NatB acetylates Met-retained peptides when the second amino acid is Asp (D), Asn (N), Glu (E), and Gln (Q). NatA acetylates substrates where the second amino acid is Ala (A), Cys (C), Gly (G), Ser (S), Thr (T), or Val (V). NatA substrates are first processed by methionine aminopeptidases, which remove the N-terminal methionine. To make Nma1 and Nma2 become NatA substrates, Ser was inserted as the second amino acid (right panel), which converts Nma1/2MD to Nma1/2MSD. B, NAD+(H) levels of nma1Δnma2Δ cells harboring NatB-specific (MD) and NatA-specific (MSD) Nma1 and Nma2 combined with NatB-specific (nat3Δ) or NatA-specific (nat1Δ) deletions. Changing Nma1 and Nma2 to NatA substrates makes NAD+(H) levels sensitive to nat1Δ, whereas nat3Δ no longer decreases NAD+(H). The graph is based on data from three independent experiments. Error bars represent data from four biological replicates per strain each with two technical replicates (total n = 8 per strain). C, cell-based NA/NAM cross-feeding assay showing the levels of NA/NAM release. NA/NAM-dependent recipient cells (bna6Δnrk1Δnrt1Δ) were plated as a lawn on niacin-free SD. Next, nma1Δnma2Δ cells harboring NatB-specific (MD) or NatA-specific (MSD) Nma1 and Nma2 combined with nat3Δ or nat1Δ were spotted on top of the lawn as feeder cells. Growth of the recipient cells indicates the level of NA/NAM released by each specific feeder strain. D, levels of NatA-specific Nma1-HAMSD and Nma2–HAMSD are reduced in nat1Δ (a NatA mutant) cells determined by Western blot analysis. E, quantification of Western-blotting protein studies shown in D. NatA-specific Nma1-HAMSD and Nma2–HAMSD are normalized to PGK1, and WT is set to 1. The graph is based on data from five independent experiments. Error bars represent data from five biological replicates per strain (total n = 5 per strain). F, levels of NatA-specific Nma1MSD and Nma2MSD are not significantly changed in nat3Δ cells (a NatB mutant). Western blot analysis of NatB-specific (MD) and NatA-specific (MSD) Nma1-HA and Nma2–HA in WT and nat3Δ cells is shown. G, quantification of protein studies shown in F. Protein levels are normalized to PGK1, and WT is set to 1. The graph is based on data from three independent experiments. Error bars represent data from three biological replicates per strain (total n = 3 per strain). The error bars denote standard deviations. The p values are calculated using Student's t test (*, p < 0.05; ns, not significant).
The MD (WT) strain carries Nma1 and Nma2 in their native form as NatB substrates, which is made by integrating pRG207–NMA1MD (WT) and pRG205–NMA2MD (WT) into nma1Δnma2Δ cells. The MSD (WT) strain carries modified Nma1 and Nma2, which are now NatA substrates, and is made by integrating pRG207–NMA1MSD and pRG205–NMA2MSD into nma1Δnma2Δ cells. These strains were then paired with either nat3Δ (NatB mutant) or nat1Δ (NatA mutant) deletions for NAD+ measurements (Fig. 2B). As expected, the NAD+ levels of MD strains carrying NMA1MD and NMA2MD (NatB substrates) were decreased when combined with nat3Δ but not nat1Δ, indicating Nma1MD and Nma2MD were still sensitive to deletions of NAT3 but not NAT1. However, NAD+ levels of MSD strains carrying NMA1MSD and NMA2MSD (NatA substrates) were sensitive to deletions of NAT1 but not NAT3 (Fig. 2B). These results suggest the low NAD+ phenotype of NatB deletion mutants is indeed due to the absence of Nt-acetylation on Nma1 and Nma2. Notably, the MSD (WT) strain has a lower basal NAD+ level compared with the MD (WT) strain, which suggests that acetylation of Nma1MSD and Nma2MSD by NatA may not be optimal. Because it has been shown that the level of acetylation of a specific NatA substrate may vary (28, 30), we determined the percentage of Nt-acetylation of Nma1MSD and Nma2MSD in both MSD (WT) and NatA mutant cells by MS. Our results showed that all N-terminal peptides of Nma1MSD and Nma2MSD identified from WT cells were Nt-acetylated. Conversely, all Nma1MSD and Nma2MSD peptides identified from nat1Δ cells were not Nt-acetylated (Fig. S1). These results indicate that the low basal NAD+ level in the MSD (WT) strain is not due to inefficient Nt-acetylation of Nmnats.
Next, we examined whether observed NAD+ reduction in the nat1Δ mutant carrying NMA1MSD and NMA2MSD was due to a blockage in NA/NAM salvage. If so, this mutant is expected to show increased release of NA and NAM as reported previously for the nat3Δ mutant (24). A cross-feeding reporter assay was employed to determine the level of NA and NAM released by cells. In this system, cell growth of the NA- and NAM- dependent mutant bna6Δnpt1Δnrk1Δ (recipient cells) was used as a readout of the amount of NA and NAM released by the feeder cells (mutants of interests) (24). As expected, the growth of recipient cells was only observed when the nat3Δ mutant harboring NMA1MD and NMA2MD (Fig. 2C, left panel) and the nat1Δ mutant harboring NMA1MSD and NMA2MSD (Fig. 2C, right panel) were used as the feeders. Moreover, the nat3Δ mutant harboring NMA1MSD and NMA2MSD no longer releases more NA and NAM when compared with nat3Δ harboring NMA1MD and NMA2MD (Fig. 2C, left panel). Therefore, it appears that NAD+ biosynthesis mediated by Nma1 and Nma2 requires Nt-acetylation by NatB for optimal product formation, and the lack of this modification reduces the NAD+ content of the cell resulting in the release of NAD+ precursors like NAM and NA into the environment.
To confirm whether the protein levels of Nma1MSD and Nma2MSD were indeed depleted in the nat1Δ mutant, whole-cell lysates were generated from cells expressing C-terminally HA-tagged Nma1MSD and NMA2MSD followed by Western blot analysis. As shown in Fig. 2, D and E, Nma1MSD proteins were depleted in nat1Δ cells to about 60% of the control MSD (WT) strain, which is in agreement with the results observed in the nat3Δ cells where Nma1 is still a NatB substrate (Fig. 1, E and F). Interestingly, Nma2MSD proteins showed a smaller decrease in nat1Δ cells to about 80% of the control MSD (WT) cells (Fig. 2E). This decrease was less significant compared with the ∼40% decrease observed in nat3Δ cells (Fig. 1, E and F), suggesting that the absence of Nt-acetylation had a lesser effect on Nma2MSD levels. This difference was not reflected in the NAD+ levels (Figs. 1D and 2B) because Nma1 is the major Nmnat, and deleting Nma2 does not decrease NAD+ levels unless Nma1 is depleted (Fig. 1D). Importantly, Nma1MSD and Nma2MSD protein levels were not significantly depleted in nat3Δ mutants (Fig. 2, F and G). These results support a positive correlation between Nmnat protein levels and NAD+ levels. In addition, Nt-acetylation appears to play a direct role in maintaining the level of Nmnat proteins and NAD+ biosynthesis.
Degradation by the N-end rule pathways is not the main cause of reduced Nma1 and Nma2 protein levels and NAD+ levels in NatB mutants
Next, we examined whether Nma1 and Nma2 proteins are less stable and degraded at a faster rate in NatB mutants. To test whether the N-end rule pathways play an important role in the reduction of Nma1 and Nma2 protein levels, E3 ligases associated with the Arg-N and Ac/N-end rule pathways were deleted or depleted in combination with deletion of NAT3. If any of these E3 ligases were important for rapid ubiquitination and subsequent degradation of the nonacetylated Nma1 and Nma2 proteins, we expected to see NAD+ levels restored. As shown in Fig. 3A, deletions of E3 ligases DOA10 (Ac-N) or UBR1 (Arg-N) did not restore NAD+ levels. Next, because double deletions of NOT4 (Ac-N) and NAT3 were synthetically lethal, we employed an auxin-inducible degron (AID) system (41, 42) to transiently deplete Not4 in the nat3Δ mutant (Fig. 3B). In this system, the minimum AID sequence (41, 42) was inserted into the chromosome region corresponding to the C terminus of Not4, generating NOT4–AID* strain. Next, a plasmid carrying a specific E3 ubiquitin ligase OsTIR1–9myc was introduced to the NOT4–AID* strain. Upon addition, auxin (indole-3-acetic acid, IAA) binds to toll-interleukin receptor (TIR) and promotes the interaction of TIR and the AID degron of Not4 resulting in Not4 degradation. We first validated that Not4 was indeed depleted under our experimental conditions within 2 h upon auxin induction by Western blot analysis (Fig. 3B, upper right panel). However, NAD+ levels were not restored in nat3Δ mutants by Not4 depletion (Fig. 3B). Therefore, it appears that Ubr1, Doa10, and Not4 may not play a major role in the rapid degradation of nonacetylated Nma1 and Nma2. Interestingly, a small but not significant increase of NAD+ was observed when Not4 was depleted in WT cells (Fig. 3B) suggesting the Ac/N-end rule pathway may play a role in maintaining proper protein turnover of Nt-acetylated Nma1 and Nma2.
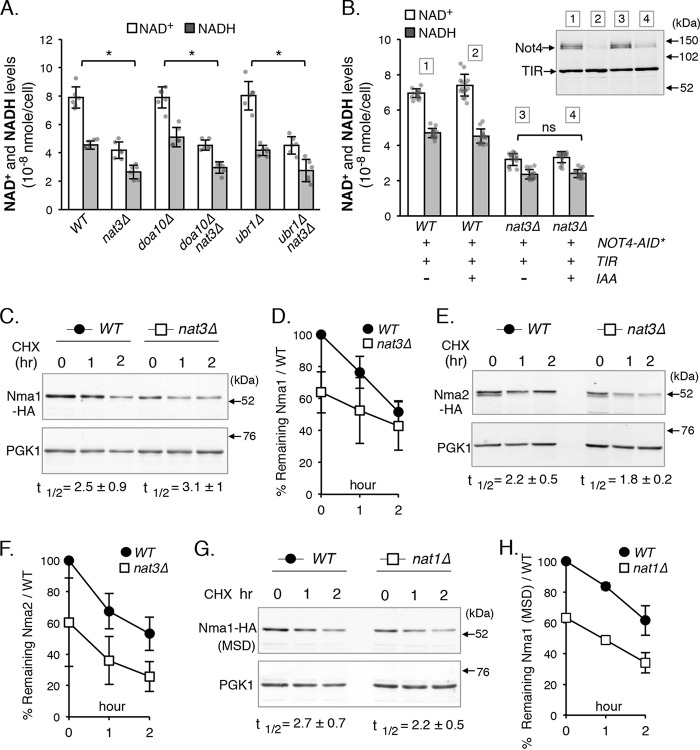
Figure 3.
Post-translational degradation by the N-end rule pathways is not the main cause of reduced Nma1 and Nma2 proteins and NAD+ levels in NatB mutants. A, deletions of E3 ligases of the Ac/N-end rule pathway (doa10Δ) and Arg/N-end rule pathway (ubr1Δ) do not restore NAD+(H) levels in nat3Δ cells. The graph is based on data from two independent experiments. Error bars represent data from three biological replicates per strain each with two technical replicates (total n = 6 per strain). B, depletion of Not4 (an Ac/N-end rule E3 ligase) does not restore NAD+(H) levels in nat3Δ cells (left panel). Not4 depletion was induced in cells harboring Not4–AID*–myc9 and TIR by addition of auxin (IAA) to 1 mm for 2 h (upper right panel). The graph is based on data from four independent experiments. Error bars represent data from eight biological replicates per strain each with two technical replicates (total n = 16 per strain). C, CHX chase studies of Nma1-HA in WT versus nat3Δ cells over 2 h determined by Western blotting. CHX was added to the cell culture to a final concentration of 0.4 mg/ml. Half-life (t½) displayed is based on three independent experiments. D, quantification of protein studies shown in C. Protein levels are normalized to PGK1, and WT at 0 h is set to 100%. E, cycloheximide chase studies of Nma2–HA in WT versus nat3Δ cells over 2 h determined by Western blotting. Half-life displayed is based on three independent experiments. F, quantification of protein studies shown in E. Proteins levels are normalized to PGK1, and WT at 0 h is set to 100%. G, cycloheximide chase studies of NatA-specific Nma1-HAMSD. Half-life (t½) displayed is based on three independent experiments. H, quantification of protein studies shown in G. Protein levels are normalized to PGK1, and WT at 0 h is set to 100%. D, F, and H, graphs are based on data from three independent experiments. Error bars represent data from three biological replicates per strain (total n = 3 per strain). The error bars denote standard deviations. The p values are calculated using Student's t test (*, p < 0.05; ns, not significant).
Although no known E3 ligase of the two N-end rule pathways has a significant role in the rapid degradation of nonacetylated Nma1 and Nma2, we could not rule out the possibility of an unknown E3 ligase. To test the possibility that nonacetylated Nma1 and Nma2 were being targeted for rapid degradation compared with their acetylated counterparts, we employed cycloheximide chases followed by Western blot analysis to determine the half-lives (t½) of the proteins. A small but insignificant change in the rate of degradation was observed when comparing acetylated Nma1 and Nma2 with their nonacetylated counterparts (Fig. 3, C–F), which does not account for an approximate 50% reduction in Nma1 and Nma2 in NatB deletion mutants (Fig. 1, E and F). Additionally, we determined the half-lives of nonacetylated and acetylated Nma1MSD in WT and nat1Δ cells (Fig. 3, G and H), and the results were in agreement with those shown in Fig. 3, C–F. Therefore, it appears that the low levels of Nma1 and Nma2 proteins in NatB mutants are not due to more rapid post-translational protein degradation of the non-Nt-acetylated Nma1 and Nma2 by the N-end rule pathways.
Nt-acetylation is important for Nmnat protein maturation
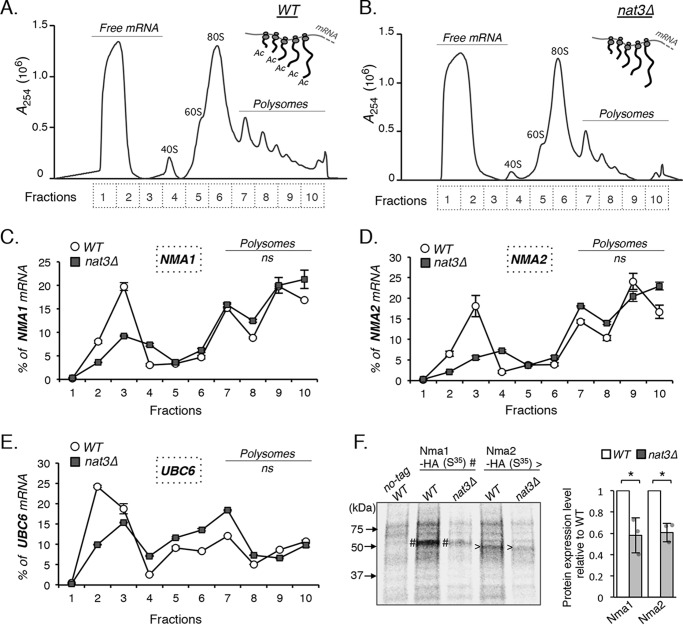
Next, we examined whether the NatB complex facilitates Nmnat translation, and therefore, the levels of Nmnats are reduced in the absence of NatB. In fact, Nt-acetyltransferase complexes have been shown to associate with both nontranslating and translating ribosome subunits (43). To address this question, we determined the rate of translation of NMA1 and NMA2 mRNAs by polysome profile analysis as described (44–46). Cell lysates generated from WT and NatB mutant cells were fractionated on a 10–50% sucrose gradient, which gave rise to the polysome profile shown in Fig. 4, A (WT) and B (nat3Δ). Sucrose gradient solutions were collected in 10 equal volume fractions: fraction 1 corresponds to the top of the gradient, and fraction 10 corresponds to the bottom of the gradient. Fractions 7–10 correspond to translating mRNAs associated with heavy polysomes. The amount of gene-specific mRNA in each fraction was calculated to obtain the proportion (%) of the target mRNA found in each fraction as described (45, 47). As a result, relative abundance and distribution of specific mRNA in 10 fractions were shown for NMA1 (Fig. 4C), NMA2 (Fig. 4D), and a non-NatB substrate control UBC6 (Fig. 4E). Overall, the percentages of NMA1 and NMA2 mRNA associated with the polysome fractions 7–10 were not significantly altered in the NatB mutant. Statistical analysis of the entire curve using the area under the curve method (48, 49) also indicated that there was no significant difference between the WT and the nat3Δ curves. These results suggest that the Nt-acetylation status does not appear to affect the rate of Nmnat translation as determined by polysome formation. However, Nt-acetylation may affect other aspects of translation.
Figure 4.
Nt-acetylation is important for Nmnat protein maturation. A–D show results of polysome fractionation and studies of NMA1 and NMA2 mRNA-associated polysome fractions in WT and NatB mutant (nat3Δ) cells. A, polysome profile of WT cells. Optical density profile (A254) of WT cell lysate in 10–50% sucrose gradient is shown. Sucrose gradient solutions are collected in 10 equal volume fractions. Fraction 1 corresponds to the top of the gradient, and fraction 10 corresponds to the bottom of the gradient. Translated mRNAs are associated with the heavy polysome fractions (fractions 7–10). 40S, small ribosomal subunit; 60S, large ribosomal subunit; 80S, ribosome (monosome). B, polysome profile of nat3Δ cells. Optical density profile (A254) of nat3Δ cell lysate in 10–50% sucrose gradient is shown. Results shown in C and D indicate that the status of Nt-acetylation does not alter the rate of translation initiation of Nmnats because NMA1 and NMA2 mRNA associated polysome fractions are not significantly different between WT and nat3Δ cells. C–E, mRNAs coding for NMA1 and NMA2 are actively translated. UBC6 is a control for non-NatB substrate. The x axis indicates the number of fractions collected from the sucrose gradient. The y axis shows % of gene-specific mRNA present in each fraction. The sum of gene-specific mRNA in all 10 fractions is set at 100%. Statistical analysis of the entire curve using “area under the curve” method (48, 49) indicates that there is no significant difference between the WT and the nat3Δ curves. The graphs are representative of the trend observed across three independent experiments (three biological replicates). Error bars represent data from three technical replicates for each strain in an experiment. F, pulse–chase studies to determine the levels of nascent Nma1 and Nma2 protein synthesis. WT (no-tag) and cells carrying HA-tagged Nma1 and Nma2 were pulsed with [35S]methionine/cysteine followed by a chase with excess nonradioactive amino acids to label nascent Nmna1 and Nma2. Proteins immunoprecipitated by an anti-HA Ab were analyzed on SDS-PAGE (left panel). The image shown is representative of the trend observed across three independent experiments. # marks Nma1–HA bands, and > marks Nma2 bands. Nma1 and Nma2 protein levels are normalized to WT (with WT set to 1) (right panel). The cell numbers (A600 unit) and total protein levels (determined by relative 35S radioactivity) are matched for all strains in the study, and these factors are included in the quantification. Error bars represent data from three biological replicates for each strain in three independent experiments. The error bars denote standard deviations. The p values are calculated using Student's t test (*, p < 0.05; ns, not significant).
Another possibility is that Nt-acetylation may be required for optimal Nmnat protein maturation. To test this, we determined the levels of nascent Nma1 and Nma2 protein synthesis by pulse–chase studies as described (50–52) in both WT and NatB mutant cells. The parental control strain (nontagged) and cells expressing HA-tagged Nma1 and Nma2 were pulsed with [35S]methionine/cysteine followed by a chase with nonlabeled methionine/cysteine. Nma1–HA and Nma2–HA proteins were then immunoprecipitated and analyzed by SDS-PAGE. As shown in Fig. 4F, a significant reduction of nascent Nma1 and Nma2 protein levels was observed in the nat3Δ mutants. Notably, although the cell numbers and protein levels were matched for all strains in the assays, the nat3Δ mutants appeared to have lower background signals. This suggests that NatB-mediated Nt-acetylation may also affect the maturation of other proteins. Collectively, our results indicate that absence of Nt-acetylation decreases the rate of Nmnat protein maturation, which contributes to the low Nmnat levels in the NatB mutants.
Discussion
In this study, we showed that Nt-acetylation by NatB is important for maintaining the levels of Nma1 and Nma2 proteins and therefore NAD+ biosynthesis (Fig. 1). We also established a direct link between NatB and Nmnats in the regulation of NAD+ metabolism (Fig. 2). NatB Nt-acetylates ∼15% of cellular proteins; hence, the observed NAD+ defects might be indirectly due to unknown off-target effects. Therefore, we asked whether we could alter NAD+ levels by modulating Nt-acetylation of Nma1 and Nma2 by replacing the endogenous Nma1 and Nma2 with mutant versions expected to be NatA substrates instead of NatB substrates. As expected, the NAD+ levels of strains carrying NatA-specific Nma1 and Nma2 (MSD) became sensitive to deletions of NAT1 but not NAT3 (Fig. 2B). These results indicate that Nt-acetylation is indeed important for Nma1- and Nma2-mediated NAD+ biosynthesis. Notably, NatA-specific Nmnat strains (MSD, WT) have lower basal levels of NAD+ compared with the NatB-specific Nmnat strains (MD, WT) (Fig. 2B). This suggests the level of Nmnat Nt-acetylation may not be optimal in the MSD WT strains. However, because the levels of Nmnats are similar in both WT (MSD and MD) strains (Fig. 2B), we did not anticipate a reduced percentage of Nmnat Nt-acetylation in the MSD WT strains. To address this, we determined the levels of Nt-acetylation in Nmnats from the MSD strains and confirmed that all identified Nmnat peptides were indeed Nt-acetylated in a NatA-dependent manner (Fig. S1). Therefore, NatA-specific Nmnats may have lower basal activities.
We also addressed why might NatB mutants have reduced levels of Nmnats. We previously showed that Nma1 and Nma2 proteins in NatB mutant cells were almost 100% non-Nt-acetylated (24), suggesting non-Nt-acetylated Nmnat proteins may be more susceptible for protein degradation. Because the N-end rule pathways are closely associated with the degradation of Nt-acetylated peptides, and Nmnat protein levels are correlated with NAD+ levels, we examined whether deleting specific E3 ligases in the N-end rule pathways would restore NAD+ levels. Our results suggest that post-translational degradation by the N-end rule pathways is not a major cause of Nmnat protein reduction in NatB mutants and that absence of Nt-acetylation does not significantly affect the half-lives of Nmnats under our assay conditions (Fig. 3, C–H). The half-lives of WT Nma1 (∼2.2 h) and Nma2 (∼2.5 h) obtained in our studies were within the range of reported half-lives for Nma1 and Nma2 (between ∼1 and ∼10 h) (53–55). Different growth and physiological conditions, experimental methods, and calculations may account for some of these differences. For instance, in a study that employed cycloheximide treatment and tagged proteins, an endogenous control was not included, and the protein dilution by cell division was not considered (53). In a different study that employed a SILAC MS approach, protein dilution by cell division was considered (54), which appeared to circumvent some pitfalls of the cycloheximide approach. However, most proteome-wide studies have been unable to agree on protein half-lives (53–56). Admittedly, the cycloheximide method we used in this study has its limitations. For instance, cycloheximide is a global translation inhibitor and prevents translation of the degradation systems that also turn over. Therefore, these degradation systems may deplete and alter the rate of degradation of their target proteins. In addition, blocking translation elicits a stress response that may not reflect the degradation process under normal growth conditions. Finally, this method relies on tagging proteins, which can alter protein stability. However, even if the protein half-lives we reported for HA-tagged Nmnats are different from the nontagged versions (54), our main conclusion that the rates of Nmnat degradation are not significantly different between the WT and the nat3Δ mutant is largely valid. Additionally, we did include an endogenous reference (Pgk1) (36–40) in our studies to account for protein dilution by cell division.
Our results show that the status of Nt-acetylation likely affects Nmnat translation at the level of protein maturation (Fig. 4F). Although our studies indicate that absence of Nt-acetylation does not significantly affect polysome formation on NMA1 and NMA2 mRNAs (Fig. 4, C and D), it is possible that our experimental conditions are not sensitive enough to reveal small differences. Interestingly, variations in fractions 1–3 were observed across different sets of experiments, which suggested mRNA recruitment to ribosomes might be affected by the presence of NatB. Although our statistical analysis indicates that there is no significant difference between the WT and the nat3Δ ribosome fractions, it remains possible that Nt-acetylation can affect other aspects of translation initiation.
Nt-acetylation may promote Nmnat protein maturation by a few possible ways. Interestingly, an intrinsically disordered region has been predicted at the N terminus of Nma1 and Nma2 (57). Intrinsically disordered regions have been suggested to be important for mediating interactions with co-translational chaperones, including Hsp70 (58, 59). Nt-acetylation may promote interactions of Nmnats and their chaperones, which facilitates proper folding and maturation. In addition, Nt-acetylation may play a role in Nmnat complex formation. It has been reported that Nma1 and Nma2 likely function as tetramers (16–18). Although it was suggested that Nma1 likely functions as a homotetramer (16), Nma1 and Nma2 also co-immunoprecipitate suggesting other stoichiometry may exist (24, 60). Furthermore, Nt-acetylation is important for protein–protein interactions and can facilitate up to a 1000-fold increase in affinity (61, 62), which may be important for mediating co-translational complex formation and folding. It is therefore possible that NatB deletion mutants have low Nma1 and Nma2 protein levels because co-translational complex formation is partially mediated through Nt-acetylation, and in the absence of Nt-acetylation, the association of these nascent peptides with their partners (including chaperones) is decreased, resulting in lower protein maturation. Supporting this possibility, studies in bacteria, yeast, and mammalian cells suggest many multisubunit complexes are built during translation, where proteins undergoing translation already begin interacting with their partners (63–65). Preventing co-translational binding and folding may result in degradation of the translating partners, suggesting a competition between protein folding and complex formation with degradation. Nt-acetylation of Nma1 and Nma2 may be important for mediating transient interactions with chaperones or for complex formation within the Nmnats. Nascent Nmnat peptides that fail to reach maturation are likely degraded co-translationally. It has been shown that all newly-synthesized polypeptides are monitored by a ribosome-based quality control system during translation (66–69). As a result, significant portions of nascent polypeptides are subject to co-translational degradation (68, 70–72). However, detailed mechanisms of this protein quality control system are not completely understood.
There are three different Nmnat proteins in humans: Nmnat1, Nmnat2, and Nmnat3 (73). Sequence conservation between human and yeast Nmnats ranges from 39 to 47% for Nma1 and 36 to 42% for Nma2. Yeast Nmnats have an ∼150-amino acid N-terminal extension not seen with human Nmnats (73). However, even with the large difference in sequences, Nmnats from yeast and human have two well-conserved functions. The first function is that they wield dual NAMN- and NMN-adenylyltransferase activities. Second, both yeast and human Nmnats have a chaperone role in mediating protein folding (74–76). Based on the protein sequences, Nmnat1 is the only human Nmnat that is likely a potential NatB substrate, but this has not been confirmed. In conclusion, our studies help us understand the mechanisms of NatB-mediated regulation of NAD+ biosynthesis by modulating Nt-acetylation of Nmnats. It would be informative to further investigate whether and how Nt-acetylation of Nmnat affects its complex formation and function. These studies may also provide insight into the regulation of NAD+ homeostasis and the molecular basis of disorders associated with aberrant NAD+ metabolism.
Experimental procedures
Yeast strains, growth media, and plasmids
Yeast strain BY4742 MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 acquired from Open Biosystems (77) was used for this study. Standard media such as synthetic complete (SC) media, synthetic minimal media (SD), and yeast extract/peptone/dextrose (YPD)-rich media were made as described (78). NA/NAM-free SD and NA/NAM-free SC were made by using niacin-free yeast nitrogen base (Sunrise Science Products). SC was used for most experiments unless noted otherwise. Gene deletions were generated by replacing the coding regions of WT genes with gene-specific PCR products generated using either the pAG32–hphMX4 (hygromycin resistance) or reusable loxP–kanr–loxP cassettes (79, 80). Multiple deletions were carried out by removing the kanr marker using a galactose-inducible Cre recombinase (79). NMA1 and NMA2 were tagged by the HA tag directly in the genome using the pFA6a–3HA–HIS3MX system as described previously (81, 82). Plasmids carrying NMA1 and NMA2 were generated by ligating NMA1 (−803 to +1496) and NMA2 (−522 to +1702) PCR fragments, which include the promoter and terminator regions, into pRG205 and pRG207 (cut with KpnI and HindIII enzymes), and the plasmids were linearized with AscI and integrated into the genome as described (83). Using these parent plasmids, mutation and insertion variants of NMA1 and NMA2, NMA1–MSD (M1D2 → M1S2D3, Ser inserted between the first Met and the second Asp), NMA2–MSD (M1D2 → M1S2D3), and NMA1-int HA-tag (K30S31 → K30-YPYDVPDYA-S40; HA tag added between K30 and S31) were generated using the Q5® site-directed mutagenesis kit (New England Biolabs). Strains carrying Not4–AID*–9myc were generated by integration using PCR product amplified from the pKan–AID*–9myc plasmid (42). Plasmid pRG207–pADH1–OsTIR1–9MYC was made by ligation of the pADH1–OsTIR1–9MYC fragment from pNHK53 into pRG207 (41). This allows integration of pRG207–pADH1–osTIR1–9MYC to the LYS2 locus because our strain does not have the URA3 locus required for pNHK53 integration. The pKan–AID*–9myc and pNHK53 plasmids were obtained from the National BioResource Project–Yeast. The pRG205 and pRG207 plasmids were obtained from Addgene. All plasmid constructs were verified by DNA sequencing.
NAD+ and NADH measurements
NAD+(H) was measured by an enzymatic cycling assay as described previously (84, 85). In brief, cells were grown to early-mid log phase (A600 of ∼1), and 1 A600 unit (1 A600 unit = 1 × 107 cells/ml) of cells was collected in duplicate. Acid extraction was performed in one tube to obtain NAD+, and alkali extraction was performed in the other tube to obtain NADH for 40 min at 60 °C. Amplification of NAD+ or NADH in the form of malate was carried out using 3 or 4 μl of neutralized acid or alkali-extracted lysate in 100 μl of cycling reaction for 1 h at room temperature, and the reaction was terminated by heating at 100 °C for 5 min. Once cooled, malate produced from the cycling reaction was converted to oxaloacetate and then to aspartate and α-ketoglutarate by the addition of 1 ml of malate indicator reagent for 20 min at room temperature. The reaction produced a corresponding amount of NADH as readout, which was measured fluorometrically with excitation at 365 nm and emission monitored at 460 nm. Standard curves for determining NAD+ and NADH concentrations were obtained as follows: NAD+ and NADH were added into the acid and alkali buffer to a final concentration of 0, 2.5, 5, and 7.5 μm, which were then treated with the same procedure along with other samples. The fluorometer was calibrated each time before use with 0, 5, 10, 20, 30, and 40 μm NADH to ensure that the detection was within a linear range.
RNA extraction, cDNA synthesis, and qPCR
Cells were grown in SC to early–mid-log phase (A600 of ∼1), and 40 A600 units of cells were collected. Total RNA was isolated using GeneJET RNA purification kit (Thermo Fisher Scientific), and cDNA was synthesized using QuantiTect Reverse Transcription kit (Qiagen) according to the manufacturer's instructions. For each qPCR, 100 ng of cDNA and 500 nm of each primer were used. qPCR was run on the LightCycler 480 using LightCycler 480 SYBR Green I Master Mix (Roche Applied Science), as described previously (22). The target mRNA transcripts were normalized to TAF10 transcript levels (86).
Cycloheximide chase and Western blots
For cycloheximide chase studies, cells were grown in SC for 5–6 h to early–mid-log phase (A600 of ∼1) before either harvesting (0-h time point) or treating with 0.4 mg/ml cycloheximide (CHX) followed by harvesting at 1 and 2 h later. This step was skipped if measuring basal protein levels. Cell lysates for Western blotting were made as described (34). Briefly, 1 A600 unit of cells was pelleted, and the supernatant was removed. Treatment with 800 μl of 2 m lithium acetate was carried out on ice for 5 min, followed by pelleting and the removal of supernatant. 800 μl of 0.4 m NaOH was then added for 5 min on ice, followed by pelleting and removal of supernatant. Cells were lysed by boiling the pellet in 80 μl of HU buffer (8 m urea, 0.2 m Tris-HCl, pH 6.8, 5% SDS, 1 mm EDTA, 0.1 m DTT, 0.005% bromphenol blue) with EDTA-free cOmplete mini protease inhibitor mixture (Roche Applied Science) at 70 °C for 10 min. Lysates were clarified by centrifugation for 10 min and stored or loaded into a 10% SDS-PAGE. Equal volume (∼20 μl) of lysate was loaded into each lane. Following electrophoresis, protein was transferred to low fluorescence polyvinylidene difluoride (GE Healthcare). Blocking was carried out using OneBlockTM Western-FL Blocking Buffer (20–314), followed by blotting using a combination of anti-HA rabbit (Cell Signaling 3724S), anti-c-Myc rabbit (Novus NBP2-52636), and anti-PGK1 mouse (Invitrogen 459250). Secondary antibodies of goat anti-rabbit IgG Alexa Fluor Plus 647 (A32733) and goat anti-mouse IgG Alexa Fluor 555 (A32727) were used to visualize the protein using an Amersham Biosciences Imager 600RGB imaging system (GE Healthcare). Protein intensity was calculated using the Amersham Biosciences Imager 600 software.
NA/NAM cross-feeding spot assays
The bna6Δnrk1Δnrt1Δ mutant (which could utilize NA and NAM) cells were used as “recipient cells,” and yeast strains of interest were used as “feeder cells.” First, recipient cells were plated onto NA/NAM-free SD as a lawn (∼104 cells/cm2), as described previously (24). Next, 2 μl of each feeder cell strain (∼104 cells, cell suspension was made in sterile water at A600 of 1) was spotted onto the lawn of recipient cells. Plates were then incubated at 30 °C for 3 days. The extent of the recipient cell growth indicates the levels of NA/NAM released by feeder cells.
Polysome profiling and isolation of mRNA
Polysome profiling was carried out using early–mid-log phase cells grown in 100 ml of SC or YPD as described (44–46) with modifications. Once cells reached approximately A600 of 1, 0.2 mg/ml cycloheximide was added to the culture and allowed to grow for 10 min. Cells were collected by centrifugation and washed once in polysome extraction buffer (20 mm Tris-HCl, pH 7.5, 50 mm KCl, 10 mm MgCl2, 5 mm DTT, and 0.2 mg/ml cycloheximide). Cell pellets were flash-frozen in liquid nitrogen and stored at −80 °C overnight. 10–50% sucrose gradients were made as follows: ∼2.2 ml of 10, 20, 30, 40, and 50% sucrose solutions (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 0.1 mg/ml cycloheximide, 1 mm DTT, and 5 units/ml RiboLock RNase Inhibitor (EO0381)) were overlaid with freezing of each layer at −80 °C before the next layer was added. Once all layers were added, the gradients were kept indefinitely at −80 °C. 24 h prior to use, gradients were thawed at 4 °C to become linear. The next day, pre-chilled 500 μl of glass beads and 400 μl of polysome extraction buffer with the addition of 1× EDTA-free cOmplete Mini protease inhibitor mixture (Roche Applied Science) and 1 unit/μl RiboLock RNase inhibitor were added to the cell pellet. Bead beating was carried out continuously at 3000 rpm for 15 min at 4 °C. The supernatant was transferred to a new tube and clarified for 20 min at 21,130 × g at 4 °C. About 400 μl of cell lysate was layered onto the gradients and centrifuged at 45,000 rpm in a SW41Ti swinging bucket rotor for 2.5 h. Sucrose gradients were collected using a Brandel Density Gradient Fractionation System into 10 fractions (∼1 ml for each fraction). RNAs in each fraction were concentrated by addition of 600 μl of TRIzol and 240 μl of chloroform to 600 μl from each fraction. Samples were mixed by vortexing, and aqueous and organic layers were separated by centrifugation at 21,310 × g for 15 min at 4 °C. 500 μl of the upper layer was transferred to a new tube containing 1 ml of isopropyl alcohol and 2 μl of 15 mg/ml GlycoBlue. RNA was precipitated overnight at −20 °C and pelleted by centrifugation at 21,310 × g for 15 min at 4 °C. The supernatant was removed, and the pellet was washed once with 1 ml of ice-cold 70% ethanol followed by pelleting and removal of the supernatant. The pellet was air-dried for 5–10 min at room temperature and dissolved in 12 μl of nuclease-free water. cDNA synthesis and qPCR were carried out as described above with the following modifications. For each fraction, 1 μg of mRNA was used for cDNA synthesis in every 20-μl reaction. 2 μl of the resultant cDNA (∼100 ng) was used for each qPCR. The amount of gene-specific mRNA in each fraction was calculated as described (45, 47). In brief, relative levels of gene-specific cDNA in each fraction were first normalized to fraction 1 and then normalized to the total amount (the sum of 10 fractions) to obtain the proportion (%) of the target gene found in each fraction. As a result, relative abundance and distribution of specific mRNA in all 10 fractions were shown for each gene.
Pulse–chase analysis for studying Nma1 and Nma2 protein synthesis and maturation
[35S]Methionine/cysteine protein labeling and pulse-chase studies were carried as described (50–52) with modifications. WT (nontagged) control cells and cells expressing HA-tagged Nma1 and Nma2 were grown in 20 ml of SC to approximately A600 of 1. Cells were collected by centrifugation, and cell pellets were washed three times in a wash medium (SD with only His, Lys, Leu, and Ura). The cells were then resuspended in 0.3 ml of the wash medium and transferred to a tube containing [35S]methionine/cysteine-labeling mix (PerkinElmer Life Sciences Express label) at a concentration of 25 μCi/A600 of cells. This was followed by incubation in a 30 °C water bath for 7 min with occasional tapping. The cells were then pelleted, resuspended in 1.8 ml of chase medium (SD with 10 mm cysteine and 10 mm methionine), and placed on ice. 0.6 ml of the cells was added into a new tube containing 102 μl of extraction buffer (1.85 m NaOH and 7.4% β-mercaptoethanol) and incubated on ice for 10 min. Then 42 μl of 100% TCA was added followed by a 15-min incubation on ice. The extracts were centrifuged at 14,000 rpm at 4 °C for 5 min. The pellets were washed with 1 ml of ice-cold acetone and centrifuged as before at 4 °C. The pellets were further dried using a SpeedVac centrifuge and resuspended in 100 μl of resuspension solution containing 2.5% SDS, 5 mm EDTA, and protease inhibitor mixture (Pierce). The samples were then heated at 95 °C for 10 min, and the supernatants were collected after spinning in a microcentrifuge at room temperature for 5 min. 2 μl of supernatant from each sample was used to determine the incorporation of 35S into the proteins by a scintillation counter (Beckman Coulter). Equal amounts of incorporated radioactivity was added to a tube containing anti-HA magnetic beads (Pierce) in 1 ml of immunoprecipitation (IP) buffer (25 mm Tris-HCl, 100 mm NaCl, 3 mm of EDTA, 5% glycerol, 1% NP-40, and protease inhibitor mixture) and incubated overnight at 4 °C in a rotator. The beads were separated on a magnetic stand and washed two times with IP buffer containing 150 mm NaCl and washed one time with IP buffer containing 200 mm NaCl. The washed beads were then resuspended in 20 μl of 2× sample buffer (65.8 mm Tris-HCl, pH 6.8, 26.3% (w/v) glycerol, 2.1% SDS, and 0.01% bromphenol blue) and boiled at >90 °C for 5 min. The beads were separated by centrifugation, and the liquid samples were analyzed by SDS-PAGE. The gel was then dried on a gel dryer (Bio-Rad) for 2 h and exposed to a phosphorimaging plate (Amersham Biosciences) for ∼24 h. The images were visualized and quantitated using a phosphorimager (Typhoon GE FLA 9500).
LC-MS/MS analysis of N-terminal acetylation of Nma1 and Nma2
Nma1 and Nma2 proteins in the MSD (WT) and MSD NatA mutant (nat1Δ) cells were immunoprecipitated using an anti-HA Ab, as described previously (24). Chymotrypsin-digested peptides were analyzed by LC-MS/MS on a Thermo Fisher Scientific Q Exactive Plus Orbitrap mass spectrometer in conjunction with Proxeon Easy-nLC II HPLC (Thermo Fisher Scientific) and Proxeon nanospray source. The digested peptides were loaded onto a 100-μm × 25-mm Magic C18 100 Å 5U reverse-phase trap where they were desalted on line before being separated using a 75-μm × 150-mm Magic C18 200 Å 3U reverse-phase column. Peptides were eluted using a 60-min gradient with a flow rate of 300 nl/min. An MS survey scan was obtained for the m/z range 350–1600; MS/MS spectra were acquired using a top 15 method, where the top 15 ions in the MS spectra were subjected to high-energy collisional dissociation. An isolation mass window of 1.6 m/z was for the precursor ion selection, and normalized collision energy of 27% was used for fragmentation. A 20-s duration was used for the dynamic exclusion.
LC-MS/MS data analysis
Tandem mass spectra were extracted and charge states deconvoluted by Proteome Discoverer (Thermo Fisher Scientific). All MS/MS samples were analyzed using X! Tandem (GPM, SCR_015645, version TORNADO (2013.02.01.1)). X! Tandem was set up to search the 20181003 Uniprot Saccharomyces cerevisiae database (12,320 entries), the cRAP database of common laboratory contaminants (RRID: SCR_018187, 114 entries), plus an equal number of reverse protein sequences assuming the digestion enzyme chymotrypsin. X! Tandem was searched with a fragment ion mass tolerance of 20 ppm and a parent ion tolerance of 20 ppm. Deamidation of asparagine and glutamine, oxidation of methionine and tryptophan, sulfone of methionine, tryptophan oxidation to formylkynurenine of tryptophan, and acetylation of the N terminus were specified in X! Tandem as variable modifications. Scaffold (Scaffold_4.8.4, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 86.0% probability to achieve an FDR less than 1.0% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 9.0% probability to achieve an FDR of less than 1.0% and contained at least two identified peptides. Protein probabilities were assigned by the ProteinProphetTM algorithm (87). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. The total number of identified Nma1 and Nma1-specific peptide sequences was determined by Scaffold software (Fig. S1A). Skyline software (88) was used to calculate the peak area of the acetylated N-terminal peptides using both the precursor and the most intense product ions specified by a spectral library generated from the search results above (Fig. S1, B and C).
Data availability
All data are contained within the article and supporting materials.
Author contributions
T. Croft, P. V., and S.-J. L. conceptualization; T. Croft, P. V., C. J. T. R., B. G., T. Cater, M. R. S., B. P., and S.-J. L. data curation; T. Croft, P. V., C. J. T. R., M. R. S., B. P., and S.-J. L. formal analysis; T. Croft, P. V., C. J. T. R., B. G., T. Cater, M. R. S., B. P., and S.-J. L. validation; T. Croft, P. V., C. J. T. R., B. G., T. Cater, and S.-J. L. investigation; T. Croft, P. V., C. J. T. R., B. G., M. R. S., B. P., and S.-J. L. visualization; T. Croft, P. V., C. J. T. R., B. G., M. R. S., B. P., and S.-J. L. methodology; T. Croft, P. V., M. R. S., B. P., and S.-J. L. writing-original draft; T. Croft, P. V., C. J. T. R., B. G., T. Cater, M. R. S., B. P., and S.-J. L. writing-review and editing; P. V., C. J. T. R., and S.-J. L. supervision; S.-J. L. funding acquisition; S.-J. L. project administration.
Supplementary Material
Acknowledgments
We thank Dr. C. Fraser for assistance with polysome profile studies and Dr. J. Roth and Dr. S. Collins for suggestions and discussions.
This work was supported by National Institutes of Health Grant GM102297 from NIGMS. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1.
- NA
- nicotinic acid
- NAM
- nicotinamide
- QA
- quinolinic acid
- NR
- nicotinamide riboside
- NaMN
- nicotinic acid mononucleotide
- NMN
- nicotinamide mononucleotide
- CHX
- cycloheximide
- qPCR
- quantitative PCR
- Nt
- N-terminal
- IAA
- indole-3-acetic acid
- Ab
- antibody
- Nmnat
- nicotinamide mononucleotide adenylyltransferase
- AID
- auxin-inducible degron
- FDR
- false discovery rate
- TIR
- toll-interleukin receptor.
References
- 1. Kato M., and Lin S. J.. 2014) Regulation of NAD+ metabolism, signaling and compartmentalization in the yeast Saccharomyces cerevisiae. DNA Repair 23, 49–58 10.1016/j.dnarep.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nikiforov A., Kulikova V., and Ziegler M.. 2015) The human NAD metabolome: Functions, metabolism and compartmentalization. Crit. Rev. Biochem. Mol. Biol. 50, 284–297 10.3109/10409238.2015.1028612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chini C. C. S., Tarragó M. G., and Chini E. N.. 2017) NAD and the aging process: role in life, death and everything in between. Mol. Cell. Endocrinol. 455, 62–74 10.1016/j.mce.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Imai S., and Guarente L.. 2014) NAD and sirtuins in aging and disease. Trends Cell Biol. 24, 464–471 10.1016/j.tcb.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshino J., Baur J. A., and Imai S. I.. 2018) NAD+ intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab. 27, 513–528 10.1016/j.cmet.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hosseini L., Vafaee M. S., Mahmoudi J., and Badalzadeh R.. 2019) Nicotinamide adenine dinucleotide emerges as a therapeutic target in aging and ischemic conditions. Biogerontology 20, 381–395 10.1007/s10522-019-09805-6 [DOI] [PubMed] [Google Scholar]
- 7. Okabe K., Yaku K., Tobe K., and Nakagawa T.. 2019) Implications of altered NAD metabolism in metabolic disorders. J. Biomed. Sci. 26, 34 10.1186/s12929-019-0527-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pehar M., Harlan B. A., Killoy K. M., and Vargas M. R.. 2018) Nicotinamide adenine dinucleotide metabolism and neurodegeneration. Antioxid. Redox Signal. 28, 1652–1668 10.1089/ars.2017.7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Demarest T. G., Babbar M., Okur M. N., Dan X., Croteau D. L., Fakouri N. B., Mattson M. P., and Bohr V. A.. 2019) NAD+ metabolism in aging and cancer. Annu. Rev. Cancer Biol. 3, 105–130 10.1146/annurev-cancerbio-030518-055905 [DOI] [Google Scholar]
- 10. Preiss J., and Handler P.. 1958) Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J. Biol. Chem. 233, 488–492 [PubMed] [Google Scholar]
- 11. Preiss J., and Handler P.. 1958) Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J. Biol. Chem. 233, 493–500 [PubMed] [Google Scholar]
- 12. Bieganowski P., and Brenner C.. 2004) Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117, 495–502 10.1016/S0092-8674(04)00416-7 [DOI] [PubMed] [Google Scholar]
- 13. Bedalov A., Hirao M., Posakony J., Nelson M., and Simon J. A.. 2003) NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 7044–7054 10.1128/MCB.23.19.7044-7054.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. James Theoga Raj C., Croft T., Venkatakrishnan P., Groth B., Dhugga G., Cater T., and Lin S. J.. 2019) The copper-sensing transcription factor Mac1, the histone deacetylase Hst1, and nicotinic acid regulate de novo NAD+ biosynthesis in budding yeast. J. Biol. Chem. 294, 5562–5575 10.1074/jbc.RA118.006987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinson B., Ceschin J., Saint-Marc C., and Daignan-Fornier B.. 2019) Dual control of NAD+ synthesis by purine metabolites in yeast. eLife 8, e43808 10.7554/eLife.43808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emanuelli M., Carnevali F., Lorenzi M., Raffaelli N., Amici A., Ruggieri S., and Magni G.. 1999) Identification and characterization of YLR328W, the Saccharomyces cerevisiae structural gene encoding NMN adenylyltransferase. Expression and characterization of the recombinant enzyme. FEBS Lett. 455, 13–17 10.1016/S0014-5793(99)00852-2 [DOI] [PubMed] [Google Scholar]
- 17. Emanuelli M., Amici A., Carnevali F., Pierella F., Raffaelli N., and Magni G.. 2003) Identification and characterization of a second NMN adenylyltransferase gene in Saccharomyces cerevisiae. Protein Expr. Purif. 27, 357–364 10.1016/S1046-5928(02)00645-9 [DOI] [PubMed] [Google Scholar]
- 18. Natalini P., Ruggieri S., Raffaelli N., and Magni G.. 1986) Nicotinamide mononucleotide adenylyltransferase. Molecular and enzymatic properties of the homogeneous enzyme from baker's yeast. Biochemistry 25, 3725–3729 10.1021/bi00360a037 [DOI] [PubMed] [Google Scholar]
- 19. Kornberg A. 1950) Reversible enzymatic synthesis of diphosphopyridine nucleotide and inorganic pyrophosphate. J. Biol. Chem. 182, 779–793 [PubMed] [Google Scholar]
- 20. Bieganowski P., Pace H. C., and Brenner C.. 2003) Eukaryotic NAD+ synthetase Qns1 contains an essential, obligate intramolecular thiol glutamine amidotransferase domain related to nitrilase. J. Biol. Chem. 278, 33049–33055 10.1074/jbc.M302257200 [DOI] [PubMed] [Google Scholar]
- 21. Yu C. K., and Dietrich L. S.. 1972) Purification and properties of yeast nicotinamide adenine dinucleotide synthetase. J. Biol. Chem. 247, 4794–4802 [PubMed] [Google Scholar]
- 22. Kato M., and Lin S. J.. 2014) YCL047C/POF1 is a novel nicotinamide mononucleotide adenylyltransferase (NMNAT) in Saccharomyces cerevisiae. J. Biol. Chem. 289, 15577–15587 10.1074/jbc.M114.558643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Belenky P., Racette F. G., Bogan K. L., McClure J. M., Smith J. S., and Brenner C.. 2007) Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129, 473–484 10.1016/j.cell.2007.03.024 [DOI] [PubMed] [Google Scholar]
- 24. Croft T., James Theoga Raj C., Salemi M., Phinney B. S., and Lin S.-J.. 2018) A functional link between NAD+ homeostasis and N-terminal protein acetylation in Saccharomyces cerevisiae. J. Biol. Chem. 293, 2927–2938 10.1074/jbc.M117.807214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Llorente B., and Dujon B.. 2000) Transcriptional regulation of the Saccharomyces cerevisiae DAL5 gene family and identification of the high affinity nicotinic acid permease TNA1 (YGR260w). FEBS Lett. 475, 237–241 10.1016/S0014-5793(00)01698-7 [DOI] [PubMed] [Google Scholar]
- 26. Ohashi K., Kawai S., and Murata K.. 2013) Secretion of quinolinic acid, an intermediate in the kynurenine pathway, for utilization in NAD+ biosynthesis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 12, 648–653 10.1128/EC.00339-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Belenky P. A., Moga T. G., and Brenner C.. 2008) Saccharomyces cerevisiae YOR071C encodes the high affinity nicotinamide riboside transporter Nrt1. J. Biol. Chem. 283, 8075–8079 10.1074/jbc.C800021200 [DOI] [PubMed] [Google Scholar]
- 28. Aksnes H., Ree R., and Arnesen T.. 2019) Co-translational, post-translational, and non-catalytic roles of N-terminal acetyltransferases. Mol. Cell 73, 1097–1114 10.1016/j.molcel.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Damme P., Lasa M., Polevoda B., Gazquez C., Elosegui-Artola A., Kim D. S., De Juan-Pardo E., Demeyer K., Hole K., Larrea E., Timmerman E., Prieto J., Arnesen T., Sherman F., Gevaert K., and Aldabe R.. 2012) N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc. Natl. Acad. Sci. U.S.A. 109, 12449–12454 10.1073/pnas.1210303109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Varland S., Aksnes H., Kryuchkov F., Impens F., Van Haver D., Jonckheere V., Ziegler M., Gevaert K., Van Damme P., and Arnesen T.. 2018) N-terminal acetylation levels are maintained during acetyl-CoA deficiency in Saccharomyces cerevisiae. Mol. Cell. Proteomics 17, 2309–2323 10.1074/mcp.RA118.000982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varshavsky A. 2019) N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. U.S.A. 116, 358–366 10.1073/pnas.1816596116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hwang C.-S., Shemorry A., and Varshavsky A.. 2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 10.1126/science.1183147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim H.-K., Kim R.-R., Oh J.-H., Cho H., Varshavsky A., and Hwang C.-S.. 2014) The N-terminal methionine of cellular proteins as a degradation signal. Cell 156, 158–169 10.1016/j.cell.2013.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oh J.-H., Hyun J.-Y., and Varshavsky A.. 2017) Control of Hsp90 chaperone and its clients by N-terminal acetylation and the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 114, E4370–E4379 10.1073/pnas.1705898114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Polevoda B., Cardillo T. S., Doyle T. C., Bedi G. S., and Sherman F.. 2003) Nat3p and Mdm20p are required for function of yeast NatB Nα-terminal acetyltransferase and of actin and tropomyosin. J. Biol. Chem. 278, 30686–30697 10.1074/jbc.M304690200 [DOI] [PubMed] [Google Scholar]
- 36. Buchanan B. W., Lloyd M. E., Engle S. M., and Rubenstein E. M.. 2016) Cycloheximide chase analysis of protein degradation in Saccharomyces cerevisiae. J. Vis. Exp. 2016 10.3791/53975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buchanan B. W., Mehrtash A. B., Broshar C. L., Runnebohm A. M., Snow B. J., Scanameo L. N., Hochstrasser M., and Rubenstein E. M.. 2019) Endoplasmic reticulum stress differentially inhibits endoplasmic reticulum and inner nuclear membrane protein quality control degradation pathways. J. Biol. Chem. 294, 19814–19830 10.1074/jbc.RA119.010295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koch B. A., Jin H., Tomko R. J. Jr, and Yu H. G.. 2019) The anaphase-promoting complex regulates the degradation of the inner nuclear membrane protein Mps3. J. Cell Biol. 218, 839–854 10.1083/jcb.201808024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neal S., Jaeger P. A., Duttke S. H., Benner C., K Glass C., Ideker T., and Hampton R. Y.. 2018) The Dfm1 Derlin is required for ERAD retrotranslocation of integral membrane proteins. Mol. Cell 69, 306–320.e4 10.1016/j.molcel.2017.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Habeck G., Ebner F. A., Shimada-Kreft H., and Kreft S. G.. 2015) The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J. Cell Biol. 209, 261–273 10.1083/jcb.201408088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- 42. Morawska M., and Ulrich H. D.. 2013) An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30, 341–351 10.1002/yea.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Polevoda B., Brown S., Cardillo T. S., Rigby S., and Sherman F.. 2008) Yeast Nα-terminal acetyltransferases are associated with ribosomes. J. Cell. Biochem. 103, 492–508 10.1002/jcb.21418 [DOI] [PubMed] [Google Scholar]
- 44. Panasenko O. O., and Collart M. A.. 2012) Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase. Mol. Microbiol. 83, 640–653 10.1111/j.1365-2958.2011.07957.x [DOI] [PubMed] [Google Scholar]
- 45. Chassé H., Boulben S., Costache V., Cormier P., and Morales J.. 2017) Analysis of translation using polysome profiling. Nucleic Acids Res. 45, e15 10.1093/nar/gkw907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zeidan Q., He F., Zhang F., Zhang H., Jacobson A., and Hinnebusch A. G.. 2018) Conserved mRNA-granule component Scd6 targets Dhh1 to repress translation initiation and activates Dcp2-mediated mRNA decay in vivo. PLoS Genet. 14, e1007806 10.1371/journal.pgen.1007806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pringle E. S., McCormick C., and Cheng Z.. 2019) Polysome profiling analysis of mRNA and associated proteins engaged in translation. Curr. Protoc. Mol. Biol. 125, e79 10.1002/cpmb.79 [DOI] [PubMed] [Google Scholar]
- 48. Fekedulegn D. B., Andrew M. E., Burchfiel C. M., Violanti J. M., Hartley T. A., Charles L. E., and Miller D. B.. 2007) Area under the curve and other summary indicators of repeated waking cortisol measurements. Psychosom. Med. 69, 651–659 10.1097/PSY.0b013e31814c405c [DOI] [PubMed] [Google Scholar]
- 49. Pruessner J. C., Kirschbaum C., Meinlschmid G., and Hellhammer D. H.. 2003) Two formulas for computation of the area under the curve represent measures of total hormone concentration versus time-dependent change. Psychoneuroendocrinology 28, 916–931 10.1016/S0306-4530(02)00108-7 [DOI] [PubMed] [Google Scholar]
- 50. Fritzsche S., and Springer S.. 2014) Pulse-chase analysis for studying protein synthesis and maturation. Curr. Protoc. Protein Sci. 78, 30.3.1–30.3.23 10.1002/0471140864.ps3003s78 [DOI] [PubMed] [Google Scholar]
- 51. Simon E., and Kornitzer D.. 2014) Pulse-chase analysis to measure protein degradation. Methods Enzymol. 536, 65–75 10.1016/B978-0-12-420070-8.00006-4 [DOI] [PubMed] [Google Scholar]
- 52. Graham T. R. 2000) Metabolic labeling and immunoprecipitation of yeast proteins. Curr. Protoc. Cell Biol. 2001 Chapter 7, Unit 7.6 10.1002/0471143030.cb0706s06 [DOI] [PubMed] [Google Scholar]
- 53. Belle A., Tanay A., Bitincka L., Shamir R., and O'Shea E. K.. 2006) Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009 10.1073/pnas.0605420103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Christiano R., Nagaraj N., Fröhlich F., and Walther T. C.. 2014) Global proteome turnover analyses of the yeasts S. cerevisiae and S. pombe. Cell Rep. 9, 1959–1965 10.1016/j.celrep.2014.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martin-Perez M., and Villén J.. 2017) Determinants and regulation of protein turnover in yeast. Cell Syst. 5, 283–294.e5 10.1016/j.cels.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martin-Perez M., and Villén J.. 2015) Feasibility of protein turnover studies in prototroph Saccharomyces cerevisiae strains. Anal. Chem. 87, 4008–4014 10.1021/acs.analchem.5b00264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mészáros B., Erdos G., and Dosztányi Z.. 2018) IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 10.1093/nar/gky384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Willmund F., del Alamo M., Pechmann S., Chen T., Albanèse V., Dammer E. B., Peng J., and Frydman J.. 2013) The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152, 196–209 10.1016/j.cell.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Truman A. W., Kristjansdottir K., Wolfgeher D., Ricco N., Mayampurath A., Volchenboum S. L., Clotet J., and Kron S. J.. 2015) The quantitative changes in the yeast Hsp70 and Hsp90 interactomes upon DNA damage. Data Brief. 2, 12–15 10.1016/j.dib.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 10.1038/nature04670 [DOI] [PubMed] [Google Scholar]
- 61. Johnsson N., Marriott G., and Weber K.. 1988) p36, the major cytoplasmic substrate of src tyrosine protein kinase, binds to its p11 regulatory subunit via a short amino-terminal amphiphatic helix. EMBO J. 7, 2435–2442 10.1002/j.1460-2075.1988.tb03089.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Becker T., Weber K., and Johnsson N.. 1990) Protein-protein recognition via short amphiphilic helices; a mutational analysis of the binding site of annexin II for p11. EMBO J. 9, 4207–4213 10.1002/j.1460-2075.1990.tb07868.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kamenova I., Mukherjee P., Conic S., Mueller F., El-Saafin F., Bardot P., Garnier J.-M., Dembele D., Capponi S., Timmers H. T. M., Vincent S. D., and Tora L.. 2019) Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun. 10, 1740 10.1038/s41467-019-09749-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shiber A., Döring K., Friedrich U., Klann K., Merker D., Zedan M., Tippmann F., Kramer G., and Bukau B.. 2018) Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272 10.1038/s41586-018-0462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shieh Y.-W., Minguez P., Bork P., Auburger J. J., Guilbride D. L., Kramer G., and Bukau B.. 2015) Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science 350, 678–680 10.1126/science.aac8171 [DOI] [PubMed] [Google Scholar]
- 66. Kramer G., Boehringer D., Ban N., and Bukau B.. 2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 10.1038/nsmb.1614 [DOI] [PubMed] [Google Scholar]
- 67. Pechmann S., Willmund F., and Frydman J.. 2013) The ribosome as a hub for protein quality control. Mol. Cell 49, 411–421 10.1016/j.molcel.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ha S. W., Ju D., Hao W., and Xie Y.. 2016) Rapidly translated polypeptides are preferred substrates for cotranslational protein degradation. J. Biol. Chem. 291, 9827–9834 10.1074/jbc.M116.716175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Duttler S., Pechmann S., and Frydman J.. 2013) Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393 10.1016/j.molcel.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Qian S.-B., Princiotta M. F., Bennink J. R., and Yewdell J. W.. 2006) Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J. Biol. Chem. 281, 392–400 10.1074/jbc.M509126200 [DOI] [PubMed] [Google Scholar]
- 71. Schubert U., Antón L. C., Gibbs J., Norbury C. C., Yewdell J. W., and Bennink J. R.. 2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 10.1038/35008096 [DOI] [PubMed] [Google Scholar]
- 72. Turner G. C., and Varshavsky A.. 2000) Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117–2120 10.1126/science.289.5487.2117 [DOI] [PubMed] [Google Scholar]
- 73. Lau C., Niere M., and Ziegler M.. 2009) The NMN/NaMN adenylyltransferase (NMNAT) protein family. Front. Biosci. 14, 410–431 10.2741/3252 [DOI] [PubMed] [Google Scholar]
- 74. Ocampo A., Liu J., and Barrientos A.. 2013) NAD+ salvage pathway proteins suppress proteotoxicity in yeast models of neurodegeneration by promoting the clearance of misfolded/oligomerized proteins. Hum. Mol. Genet. 22, 1699–1708 10.1093/hmg/ddt016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ali Y. O., Allen H. M., Yu L., Li-Kroeger D., Bakhshizadehmahmoudi D., Hatcher A., McCabe C., Xu J., Bjorklund N., Taglialatela G., Bennett D. A., De Jager P. L., Shulman J. M., Bellen H. J., and Lu H. C.. 2016) NMNAT2: HSP90 complex mediates proteostasis in proteinopathies. PLoS Biol. 14, e1002472 10.1371/journal.pbio.1002472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhai R. G., Zhang F., Hiesinger P. R., Cao Y., Haueter C. M., and Bellen H. J.. 2008) NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration. Nature 452, 887–891 10.1038/nature06721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., and Boeke J. D.. 1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 78. Burke D., Dawson D., and Sterns T.. 2000) Methods in Yeast Genetics. pp. 171–174, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 79. Güldener U., Heck S., Fielder T., Beinhauer J., and Hegemann J. H.. 1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524 10.1093/nar/24.13.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goldstein A. L., and McCusker J. H.. 1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 81. Longtine M. S., McKenzie A. 3rd., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R.. 1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 82. Sheff M. A., and Thorn K. S.. 2004) Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- 83. Gnügge R., Liphardt T., and Rudolf F.. 2016) A shuttle vector series for precise genetic engineering of Saccharomyces cerevisiae. Yeast 33, 83–98 10.1002/yea.3144 [DOI] [PubMed] [Google Scholar]
- 84. Easlon E., Tsang F., Dilova I., Wang C., Lu S. P., Skinner C., and Lin S. J.. 2007) The dihydrolipoamide acetyltransferase is a novel metabolic longevity factor and is required for calorie restriction-mediated life span extension. J. Biol. Chem. 282, 6161–6171 10.1074/jbc.M607661200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lin S. S., Manchester J. K., and Gordon J. I.. 2001) Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 276, 36000–36007 10.1074/jbc.M103509200 [DOI] [PubMed] [Google Scholar]
- 86. Teste M.-A., Duquenne M., François J., and Parrou J.-L.. 2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 10, 99 10.1186/1471-2199-10-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nesvizhskii A. I., Keller A., Kolker E., and Aebersold R.. 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 88. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J.. 2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting materials.