Abstract
Severe congenital neutropenia (SCN) is characterized by a near absence of neutrophils, rendering individuals with this disorder vulnerable to recurrent life-threatening infections. The majority of SCN cases arise because of germline mutations in the gene elastase, neutrophil-expressed (ELANE) encoding the neutrophil granule serine protease neutrophil elastase. Treatment with a high dose of granulocyte colony-stimulating factor increases neutrophil production and reduces infection risk. How ELANE mutations produce SCN remains unknown. The currently proposed mechanism is that ELANE mutations promote protein misfolding, resulting in endoplasmic reticulum stress and activation of the unfolded protein response (UPR), triggering death of neutrophil precursors and resulting in neutropenia. Here we studied the ELANE mutation p.G185R, often associated with greater clinical severity (e.g. decreased responsiveness to granulocyte colony-stimulating factor and increased leukemogenesis). Using an inducible expression system, we observed that this ELANE mutation diminishes enzymatic activity and granulocytic differentiation without significantly affecting cell proliferation, cell death, or UPR induction in murine myeloblast 32D and human promyelocytic NB4 cells. Impaired differentiation was associated with decreased expression of genes encoding critical hematopoietic transcription factors (Gfi1, Cebpd, Cebpe, and Spi1), cell surface proteins (Csf3r and Gr1), and neutrophil granule proteins (Mpo and Elane). Together, these findings challenge the currently prevailing model that SCN results from mutant ELANE, which triggers endoplasmic reticulum stress, UPR, and apoptosis.
Keywords: neutrophil, enzyme mutation, unfolded protein response (UPR), differentiation, hematopoiesis, innate immunity, ELANE, granulocyte colony-stimulating factor, immune deficiency, neutrophil elastase, severe congenital neutropenia
Introduction
Severe congenital neutropenia (SCN)3 is a condition of extremely low absolute numbers of circulating neutrophils (<500/μl), which results in recurrent life-threatening infections (1). Accumulation of granulocyte precursors (e.g. promyelocytes) is found in the bone marrow of affected individuals. Chronic administration of high-dose granulocyte colony-stimulating factor (GCSF) increases the number of neutrophils and prevents infection. GCSF (also known as CSF3) is an important cytokine promoting survival, proliferation, and differentiation of neutrophils and their precursors (2), as evidenced by severe neutropenia in mice with genetic ablation of Csf3 or its cognate receptor Csf3r. One major concern is that a subset of patients with SCN acquires a mutation in CSF3R and develops myeloid malignancy. The leading cause of SCN are autosomally dominant mutations in ELANE, which encodes neutrophil elastase. The mechanism by which mutations in ELANE result in SCN remains elusive, and greater understanding of its role in the pathogenesis of SCN may identify the mechanism of action for GCSF and prevent development of myeloid malignancies.
Previous studies have suggested that mutations in ELANE result in improper folding, promoting endoplasmic reticulum (ER) stress), activation of the unfolded protein response (UPR) (3–5), and/or subcellular mislocalization (6, 7), which may lead to cell death or cell survival with an altered transcriptional profile and protein synthesis (8). By the promyelocyte stage of granulopoiesis, ELANE has become the most abundant protein, achieving millimolar concentration in neutrophils (9). Thus, ER stress could be sufficiently potent to cause cell death. Apoptosis induced by mutant neutrophil elastase expression can be responsible for reduced neutrophil counts (6, 10, 11). An alternate hypothesis is that mutated ELANE produces neutropenia through a block in differentiation. The block in terminal granulocytic differentiation may be attributed to aberrant transcription factor activity (12).
Progress toward understanding the pathogenesis of SCN has been hampered by lack of an animal model and technical challenges associated with patient-derived induced pluripotent stem cells. Here we used an inducible expression system of WT and mutant ELANE to investigate its biochemical and cellular properties. An inducible expression system avoids the problem of bias of selection of only surviving cells that express mutant ELANE. Furthermore, we used murine myeloblast 32D and human promyelocytic NB4 cells, which model granulocytic differentiation. We report here that mutant ELANE impaired granulocytic differentiation without eliciting ER stress/UPR or apoptotic response.
Results
Mutant ELANE exhibited reduced activity and altered subcellular localization
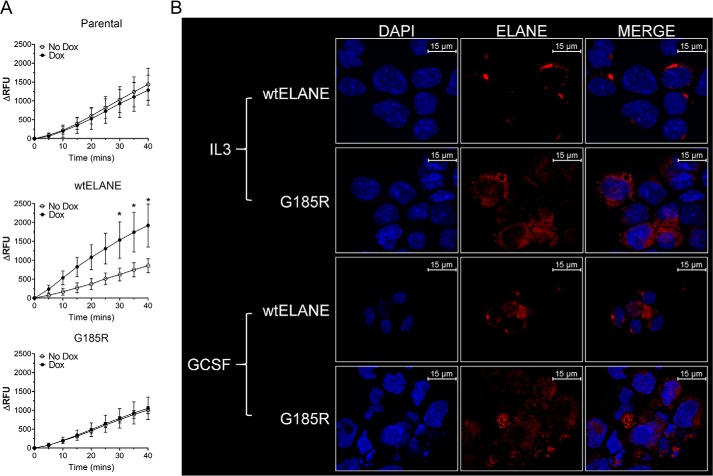
To investigate the effect of ELANE mutations on myeloid cell growth, we deployed a doxycycline-inducible model of ELANE expression in 32D and NB4 cells. In this model, we induced expression of WT ELANE or ELANE G185R. The G185R mutation is one of the more clinically deleterious ones causing SCN, requiring a greater GCSF dose, and is associated with a higher risk of transformation to myeloid malignancy (16). Induction of ELANE expression occurred in a dose-dependent manner, with maximal expression observed at 1 μg/ml in 32D and NB4 cells (Fig. 1, A and B). A dose of 1 μg/ml doxycycline was used for all subsequent experiments. Induction of ELANE using 1 μg/ml doxycycline was comparable with the amount of ELANE present in human neutrophils (Fig. S1). We next evaluated the enzymatic activity of mutant ELANE. Mutant ELANE demonstrated loss of enzymatic activity (Fig. 2A). Immunofluorescence studies indicated diffuse localization of mutant ELANE compared with granular expression of WT ELANE upon GCSF and IL3 treatment (Fig. 2B). We observed that mutant ELANE G185R is enzymatically inactive and that its localization is different compared with WT ELANE.
Figure 1.
Inducible expression of WT and G185R ELANE. A and B, Western blots and quantitation of blots as mean relative intensity ± S.D. (n = 2), showing induction of human neutrophil elastase (NE) (huELANE, WT and G185R) upon doxycycline (Dox) treatment of 32D cells (A) and NB4 cells (B) at the indicated concentrations. Mean relative intensity was calculated relative to actin. Statistical significance, calculated using two-way ANOVA and Bonferroni multiple comparisons post-test, showed no significant differences between induction of WT or G185R ELANE.
Figure 2.
Enzymatic activity and localization of WT and G185R ELANE. A, neutrophil elastase activity profile of ELANE (WT and G185R)-expressing 32D cells over the indicated time period using fluorometric quantification. 32D parental cells were used as controls. Data from two independent experiments performed in triplicates are represented as mean ± S.D. Statistical significance was computed using two-way ANOVA followed by multiple comparisons Bonferroni post-test (*, p < 0.05). B, immunofluorescence staining of 32D cells with Alexa Fluor 594–conjugated anti-ELANE antibodies and 4′,6-diamidino-2-phenylindole (DAPI) upon indicated treatment for 48 h. RFU, relative fluorescence units.
Inducible expression of mutant ELANE did not affect proliferation, viability, or apoptosis
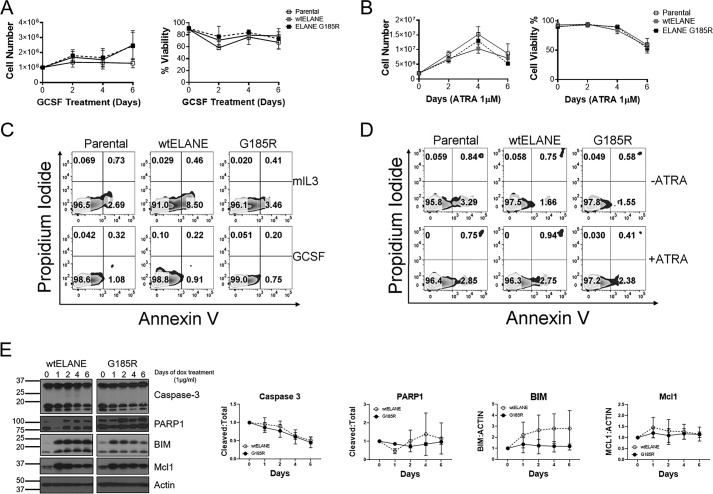
To determine the effect of mutant ELANE expression on proliferation, 32D cells were treated with 100 ng/ml GCSF with or without doxycycline. Cell counting was performed using trypan blue to exclude counting of dead cells. Doxycycline treatment of 32D cells did not affect cell growth, survival, or differentiation (Fig. S2, A–D). Similar observations regarding cell growth and viability were noted upon doxycycline treatment of NB4 cells (Fig. S3, A–C). Treatment with GCSF or ATRA for 6 days revealed no effect of mutant ELANE expression on cell proliferation or viability in 32D or NB4 cells, respectively (Fig. 3, A and B). Cell cycle analysis in 32D cells further showed no differences in cell cycle initiation and progression in 32D cells upon expression of WT or mutant ELANE (Fig. S4), supporting the observations of no effect on cell growth. The expression of mutant ELANE did not elicit cell death or apoptosis, as determined using Annexin V and propidium iodide staining by flow cytometry in 32D or NB4 cells (Fig. 3, C and D). The findings were supported by Western blot analysis for intracellular anti-apoptotic (Mcl1) and pro-apoptotic (BIM, caspase-3, and PARP1) mediators that did not demonstrate any differences between WT ELANE and mutant ELANE G185R induction in 32D cells (Fig. 3E). Together, these data demonstrate that inducible expression of mutant ELANE does not inhibit proliferation or promote cell death.
Figure 3.
Effect of WT and G185R ELANE on proliferation, viability, and apoptosis. A, trypan blue exclusion–based cell count and viability analysis of 32D cells expressing WT ELANE and G185R upon GCSF treatment (100 ng/ml). B, NB4 cells upon ELANE (WT and G185R) induction with ATRA (1 μm) treatment. Data are presented as mean ± S.D. (n = 3) C, Annexin V–PI–based apoptosis analysis of ELANE (WT and G185R)–expressing 32D cells with IL3 (2 ng/ml) and GCSF (100 ng/ml) treatment for 3 days. This assay is representative of three independent experiments. D, flow cytometry–based apoptosis analysis of NB4 cells with or without ATRA (1 μm) treatment for 3 days, representative of three independent experiments. E, immunoblotting was performed to determine the effect of ELANE (WT and G185R) expression on the indicated pro- and anti-apoptotic markers. Quantitation was performed using ImageJ. Data are represented as mean relative intensity normalized to day 0 ± S.D.; n = 3. Relative intensity for cleaved caspase-3 and PARP1 was determined with respect to total caspase-3 (uncleaved and cleaved) and total PARP1 (cleaved and uncleaved), respectively. Relative levels of BIM and Mcl1 were determined with respect to actin. Statistical significance, calculated using two-way ANOVA and Bonferroni multiple comparisons post-test, showed no significant differences between induction of WT or G185R ELANE.
Mutant ELANE failed to trigger ER stress/UPR
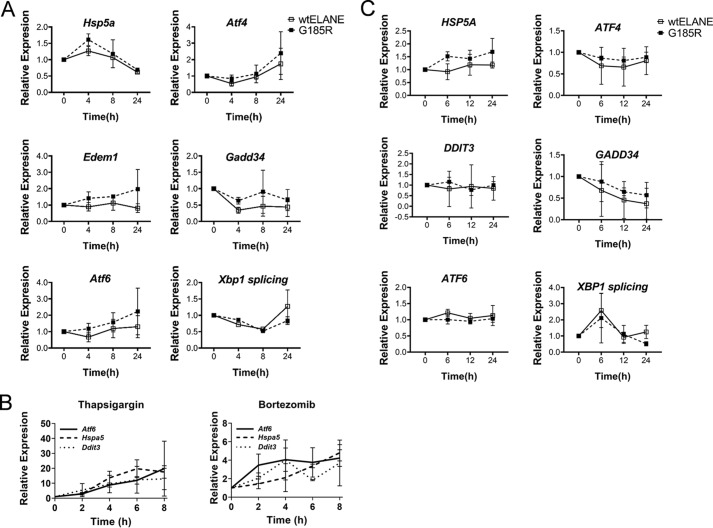
Induction of the UPR has been proposed in several studies to explain the pathogenesis of SCN associated with mutant ELANE (4, 5, 17, 18). Interestingly, the mouse Elane G193X knock-in strain requires concomitant administration of bortezomib, which inhibits proteasomal activity and produces ER stress to induce UPR to generate neutropenia (18). Using our inducible system, we measured time-dependent changes in UPR markers. Doxycycline was added to 32D cells growing in murine IL3 to induce ELANE expression, and cells were harvested at 0, 4, 8, and 24 h for mRNA extraction and qPCR analysis. There was no statistically significant up-regulation of the UPR markers Hspa5, Atf4, Atf6, Gadd34, Edem1, and Xbp1 splicing following induction of mutant ELANE versus WT ELANE (Fig. 4A). Thapsigargin, an inducer of UPR, and bortezomib were used as positive controls (Fig. 4B). There was no significant increase in the UPR markers ATF4, ATF6, HSPA5, DDIT3, GADD34, and XBP1 splicing upon induction of mutant ELANE in NB4 cells (Fig. 4C). Together, these data demonstrate that mutant ELANE does not mediate ER stress/UPR.
Figure 4.
Induction of G185R ELANE did not enhance UPR markers. A, 32D cells expressing WT and G185R ELANE were analyzed by qPCR for expression of the indicated UPR markers over a period of 24 h. Data are presented as mean ± S.D. (n = 3). B, qPCR to determine expression of the UPR genes Atf6, Hspa5, and Ddit3 in 32D parent cells treated with thapsigargin or bortezomib for 0, 2, 4, 6, and 8 h. Data are presented as mean ± S.D. (n = 3). C, NB4 cells expressing WT and G185R ELANE were analyzed by qPCR for expression of the indicated UPR markers (n = 3, mean ± S.D.). Statistical significance, computed using two-way ANOVA followed by multiple comparisons post-test using the Bonferroni method, showed no significant differences between WT and G185R ELANE–expressing cells. Xbp1 or XBP1 splicing is defined as ratio of Xbp1s/Xbp1 gene expression and XBP1s/XBP1 gene expression for murine and human cells, respectively.
Mutant ELANE impaired granulocytic differentiation
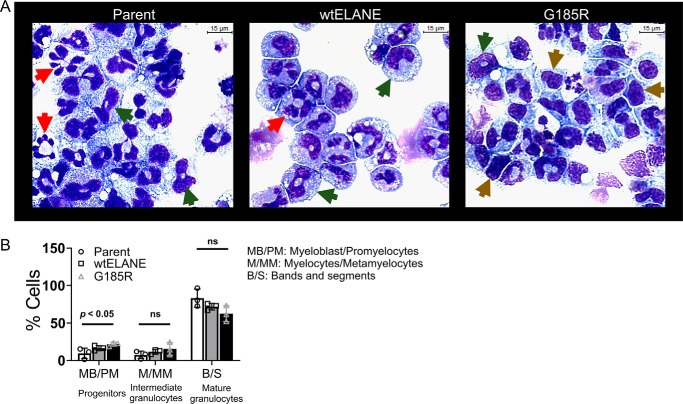
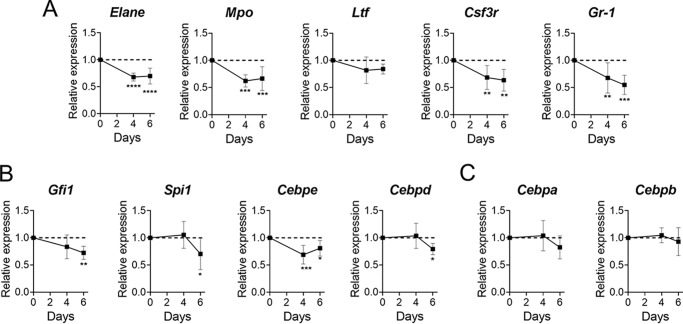
We next determined the effect of mutant ELANE expression on GCSF-induced differentiation in 32D cells. The cells were treated with 100 ng/ml GCSF with or without 1 μg/ml doxycycline. At day 6, cells were harvested, cytospun, and stained using Wright–Giemsa–type staining to determine differentiation status based on morphology. We observed that cells expressing mutant ELANE were more frequently undifferentiated (Fig. 5A). Differential counts were performed on 200 cells and grouped into three stages representing progenitors (myeloblasts and promyelocytes), intermediate granulocytes (myelocytes and metamyelocytes), and mature granulocytes (bands and segmented) (19). Blinded differential cell counting showed a statistically significant increase in progenitor cells when induced with doxycycline to express mutant ELANE (Fig. 5B). We next assessed gene expression of granulopoietic transcription factors (Cebpa, Cebpb, Cebpd, Cebpe, Gfi1, and Spi1) and other markers of granulocyte differentiation (Mpo, Elane, Ltf, Csf3r, and Gr1) using qPCR. Normalizing the expression of these markers to that of WT ELANE, we observed that cells expressing mutant ELANE had, in multiple genes, 50% reduced expression (Fig. 6, A and B). However, Cebpa and Cebpb did not show mutant ELANE–dependent suppression of gene expression (Fig. 6C). This are similar to a report of SCN patient neutrophils, which, even after GCSF-mediated differentiation, lack antimicrobial activity because of defects in granule protein expression and reduced myeloperoxidase (MPO) and ELANE (6, 17, 20). The doxycycline-inducible system for 32D WT ELANE and ELANE G185R cells showed a similar efficacy of expression (Fig. S5)
Figure 5.
G185R ELANE impaired morphological differentiation. A, Wright–Giemsa–based morphological analysis of cytospin-immobilized 32D parental cells and ones expressing ELANE (WT and G185R). For quantitation, cells were classified into three different stages of differentiation: brown arrows, myeloblasts/promyelocytes; green arrows, myelocytes/metamyelocytes; red arrows, bands/segmented. B, graphical description of differential cells counts of 32D parental and 32D cells expressing WT ELANE and G185R. Differential counts are shown as bar charts. Data are represented as mean ± S.D. (n = 3). Statistical significance was computed using Student's t test (ns, nonsignificant).
Figure 6.
G185R ELANE decreased granulocytic differentiation inducers and markers. A–C, the differentiation markers Elane, Mpo, Ltf, Csf3r, and Gr-1 (A); the differentiation transcription factors Gfi1, Spi1, Cebpd, and Cebpe (B); and Cebpa and Cebpe (C) in 32D cells expressing ELANE (WT and G185R) treated with 100 ng/ml GCSF and 1 μg/ml doxycycline for 6 days. The expression levels for each day in cells expressing mutant ELANE are normalized to expression on the respective day in cells expressing WT ELANE. The data are represented as mean ± S.D. of n = 4. Statistical significance was computed using two-way ANOVA followed by multiple comparisons post-test using the Bonferroni method (*, p < 0.05; **, p < 0.01; ***, p < .001; ****, p < 0.0001).
Discussion
Granulocytic development is a prodigious process (1011 cells/day) divided into multiple stages and tightly regulated by a small set of ligand–receptor reactions and transcription factors (21). SCN is characterized by profound deficiency of neutrophils with production arrest at the promyelocyte stage. This stage of neutrophil production is marked by formation of primary granules consisting of MPO, neutrophil elastase, cathepsin G, and proteinase 3. The association of SCN and promyelocytic arrest in individuals with monoallelic mutations of ELANE demonstrates that granulopoiesis is central to SCN pathogenesis (22). Although Elane−/− and Elane G193X knock-in mouse strains do not display neutropenia (at least in the absence of further protease inhibition in the latter model), impaired granulopoiesis occurs in human patient–derived induced pluripotent stem cells (iPSCs), and ectopic expression of WT ELANE rescues neutrophil production (4).
The mechanism by which ELANE mutations cause SCN remains poorly understood. A prevailing explanation is that, as one of the most abundant proteins in the promyelocyte, mutant ELANE triggers ER stress and cell death (4, 5, 17). This is supported by the observation that a single dose of bortezomib elicits neutropenia in the mouse ELANE G193X knock-in strain (17). The fact that a common side effect of therapeutic bortezomib is neutropenia, even when normally administered in patients with WT ELANE, exposes the limitations of this mouse model. Alternative explanations, which are not as well supported by experimental data, include that mislocalization of ELANE causes cell death, that the protease activity of ELANE degrades a critical factor for differentiation and survival, and that mutant ELANE blocks differentiation without causing cell death.
Experimental data are limited by the paucity of experimental models. Use of transient expression in cell lines, as performed here, has significant limitations. Nevertheless, as noted, the mouse model is also imperfect. Patient-derived iPSCs require precise culture conditions. Myeloid and nonhematopoietic cell lines relied on stable transfectants, which have the shortcoming of studying those cells that only survive expression of the mutation.
Here we deployed human and murine myeloid cell lines with an inducible expression system of human ELANE. We found that mutated ELANE did not affect survival, viability, or proliferation. Mutated ELANE impaired granulocytic differentiation at the promyelocyte stage. Furthermore, mutated ELANE demonstrated absent enzymatic activity and did not elicit a strong UPR. This is consistent with previous studies where ELANE G185R did not induce the UPR (6, 10). In addition, we also observed diffused expression of ELANE G185R compared with localized expression of WT ELANE, which corroborates other reports of mutant ELANE mislocalization (4, 6, 7). However, our findings do not allude to invariable observation of apoptosis because of mutant ELANE expression, as reported previously (4, 6, 7, 10, 17). We observed mutant ELANE–dependent impaired granulocytic differentiation in 32D cells. This is attributed to suppression of gene expression of transcription factors involved in granulocytic differentiation. Mutant ELANE, by its mislocalization, may perturb transcriptional regulation of granulopoiesis. One possible mechanism for impaired differentiation is defective GCSF/GCSF receptor signaling. WT neutrophil elastase has been shown to degrade GCSF and its cognate receptor (23). However, enzymatic activity was abrogated. Other aberrant functions could impair terminal granulocytic differentiation. Because patients with deletion of the ELANE locus do not display neutropenia (24), mutant ELANE acting as a scaffolding protein could sequester a critical mediator of differentiation.
SCN is a life-threatening disease most frequently caused by dominantly acting mutations in ELANE, which encodes a neutrophil serine protease. Current dogma holds that the neutropenia results from excess deaths because of mutations that cause misfolding and an enhanced UPR. Our study, with an inducible expression system for mutant ELANE, suggests a new explanation of impaired differentiation with only modest UPR effects.
Materials and methods
Cell lines and culture conditions
The factor-dependent murine myeloblast cell line 32D cl3 and the human promyelocytic leukemic NB4 cell line harboring t(15;17) were used. 32D cells were grown in Iscove's modified Dulbecco's medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (Gemini Bio-Products, West Sacramento, CA), 1% PenStrep (Thermo Fisher Scientific), 2 ng/ml murine IL3 (Peprotech, Rocky Hill, NJ), and 1× Glutamax (Thermo Fisher Scientific). NB4 were cultured in RPMI (Corning Inc, Corning, NY) supplemented with 10% FBS, 1% PenStrep and 1X Glutamax. All cell lines were grown at 37 °C and 5% CO2. Doxycycline-inducible expression of ELANE was performed using the pInducer20 lentiviral vector system (13), and ELANE was introduced into 32D and NB4 cells by lentiviral transduction. Lentiviral particles were made as described elsewhere (14). The pInducer 21 containing either WT or mutant ELANE G185R were cloned employing Gateway cloning. The original source of the ELANE inserts was the pCS2+ vector, which was generated as described previously (15). To induce expression of ELANE, the medium was further supplemented with 1 μg/ml doxycycline hyclate (Sigma-Aldrich, St. Louis, MO) unless otherwise specified. The medium was replenished with fresh doxycycline every 48 h. 32D cells were differentiated using 100 ng/ml GCSF (Amgen Inc., Thousand Oaks, CA). All-trans retinoic acid (ATRA, Sigma-Aldrich) treatment of NB4 cells was achieved by adding ATRA (in 0.1% (v/v) DMSO) at a final concentration of 1 μm. Cell counts and viability were assessed using trypan blue exclusion. Cells were stained with 0.4% trypan blue (Thermo Fisher Scientific) and counted manually using a hemocytometer or the Luna II automated cell counter (Logos Biosystems, Annandale, VA). Cells stained with trypan blue were counted as dead, and those that excluded the dye were counted as live. Live cell counts were used to report cell number, and the percentage of live cells over total cells was used to report viability. To determine granulocytic differentiation using changes in cellular morphology, cells were subjected to cytocentrifugation at a speed of 500 rpm for 5 min. Samples were subsequently fixed and stained using the PROTOCOL Hema 3 staining kit (Fisher Scientific, Hampton, NH), which is comparable with Wright–Giemsa staining. Slides were analyzed and pictures were obtained using an Axion microscope (Zeiss, White Plains, NY).
Western blotting
Cells were lysed using radioimmune precipitation assay buffer supplemented with 1× protease inhibitor mixture, 1 mm PMSF, and 1 mm sodium orthovanadate (all from EMD Millipore, Burlington, MA). The lysates were quantified for protein concentration using a BCA assay (Thermo Fisher). Finally, the proteins were denatured with 1× Laemmli buffer (Bio-Rad) containing β-mercaptoethanol. Lysates were then electrophoresed on a polyacrylamide mini-gel and transferred onto nitrocellulose membranes (Bio-Rad). Membranes were subsequently blocked with Tris-buffered saline (Fisher Scientific) and 0.1% Tween 20 (Sigma-Aldrich) containing 5% nonfat dry milk (Bio-Rad). Primary antibodies (Table S1A) were applied in Tris-buffered saline and Tween 20 (TBST) with 5% nonfat dry milk/TBST overnight at 4 °C, followed by incubation with a horseradish peroxidase–conjugated secondary antibody (Rockland Immunochemicals, Gilbertsville, PA) dissolved in TBST solution with 5% nonfat dry milk for 1 h at room temperature and addition of ECLTM Start Western blot detection reagent (GE Healthcare). The immunoblots were visualized on photographic film (Agfa Healthcare) developed using SRX-101a Developer (Konica Medical Corp., Wayne, NJ). Band intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
Apoptosis analysis
Flow cytometry was used to determine apoptosis using Annexin V–propidium iodide (PI) staining. Treated cells were stained using Alexa Fluor 488 Annexin V and propidium iodide according to the manufacturer's protocol (Dead Cell Apoptosis Kit, Thermo Fisher Scientific). Data for all flow cytometry experiments were acquired using a BD LSRFortessa cytometer (BD Biosciences). The data were analyzed using FlowJo v10.5.3 (FlowJo LLC, Ashland, OR). Overnight treatment with 1 μm camptothecin (Sigma-Aldrich) was used as a positive control.
Cell cycle analysis
After overnight serum and cytokine starvation to induce G1 arrest, IL3- and FBS-containing medium was freshly added and then, at the indicated time points, cells were collected and resuspended in PBS (Fisher Scientific), and ice-cold 70% ethanol (Fisher Scientific) was added dropwise while mildly vortexing. After removing ethanol, cells were washed with 0.3% Tween 20 (Fisher Scientific) in PBS and then resuspended in DNA staining buffer (50 μg/ml PBS/PI and 100 μg/ml RNase A). After 15-min incubation at 37 °C, the PI signal was acquired on a BD LSRFortessa flow cytometer. Diploid (G1/G0) and tetraploid (G2/M) cells were quantified by applying the Dean–Jett–Fox model using FlowJo v10.5.3 (FlowJo LLC).
RNA isolation and qPCR-based gene expression analysis
Total RNA was isolated from the harvested cells using TRIzol reagent (Thermo Fisher) according to the manufacturer's instructions. Reverse transcription was performed using the iScript II cDNA Synthesis Kit (Bio-Rad), which involves a propriety blend of oligo(dT)s and random hexamers. Complementary DNA was used for real-time quantitative PCR (qPCR) to determine gene expression using custom-designed primers (Table S2). qPCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems, Foster City, CA) on a StepOne Plus system (Thermo Fisher). Gene expression analysis was performed using the ΔΔCT method, with Actin/ACTIN as the reference gene and time 0 as reference sample to determine normalized expression.
Neutrophil elastase activity
Fluorometric analysis of neutrophil elastase activity was performed using a kit (Sigma-Aldrich). Briefly, 32D cells with inducible expression of WT and G185R ELANE were treated with or without doxycycline for 48 h. This was followed by treatment of the cell pellets according to the manufacturer's instructions. The readings were taken on 96-well black optical plates (Thermo Fisher) using a Gen5 plate reader (BioTek Instruments, St. Winooski, VT) for enzyme kinetics over a period of 40 min with 5-min intervals. Purified neutrophil elastase activity and its inhibition with the specific inhibitor sivelestat (Sigma-Aldrich) served as positive and negative controls, respectively.
Immunofluorescence
Commercially purchased human neutrophils (Astarte Biologics, Bothell, WA) and 32D cells with inducible expression of WT and G185R ELANE were harvested following 48-h treatment with GCSF or IL3. A concentration of 500,000 cells/ml was collected for cytospin and subsequently fixed with 4% formaldehyde in PBS, permeabilized with 0.5% Triton X-100 in Dulbecco's Phosphate Buffered Saline, and blocked with 3% BSA using the Image-iT Fixation/Permeabilization Kit (Invitrogen) according to the manufacturer's instructions. Following three washes in 1× PBS, the cells were incubated with Alexa Fluor 594–conjugated ELANE (E9C9L) XP rabbit mAb (Cell Signaling Technology, Danvers, MA) diluted 1:100 in 3% BSA for 1 h at 37 °C. The cells were washed three times with 1× PBS, and nuclei were counterstained with 1 μg/ml 4′,6-diamidino-2-phenylindole for 30 min at room temperature. Slides were then mounted and imaged with a Leica SP8 confocal microscope (Leica Microsystems, Buffalo Grove, IL).
Statistical analysis
Appropriate statistical analyses (as indicated in the figure legends) were performed using GraphPad Prism 8.0 software (GraphPad, La Jolla, CA).
Data availability
All data are contained in the manuscript.
Author contributions
B. G., H. M. M., B. W., and R. K. data curation; B. G., H. M. M., B. W., R. K., and S. J. C. formal analysis; B. G. validation; B. G., H. M. M., R. K., and S. J. C. investigation; B. G., H. M. M., M. S. H., and S. J. C. methodology; H. M. M. and S. J. C. conceptualization; H. M. M., M. S. H., and S. J. C. resources; H. M. M. and S. J. C. supervision; H. M. M. funding acquisition; B. G., H. M. M., B. W., M. S. H., and S. J. C. writing-original draft; S. J. C. project administration.
Supplementary Material
This work was supported by supported by National Institutes of Health Grants R01HL128173-05 (to S. J. C.) and R01HL130472-04 (to M. S. H.), an Alex's Lemonade Stand Foundation innovation award (to S. J. C.), a Leukemia Research Foundation young investigator award (to H. M.), and a VeloSano catalyst award (to B. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5 and Tables S1 and S2.
- SCN
- severe congenital neutropenia
- GCSF
- granulocyte colony-stimulating factor
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- qPCR
- quantitative PCR
- ATRA
- all-trans retinoic acid
- ANOVA
- analysis of variance
- PI
- propidium iodide.
References
- 1. Skokowa J., Dale D. C., Touw I. P., Zeidler C., and Welte K.. 2017) Severe congenital neutropenias. Nat. Rev. Dis. Primers 3, 17032 10.1038/nrdp.2017.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Panopoulos A. D., and Watowich S. S.. 2008) Granulocyte colony-stimulating factor: molecular mechanisms of action during steady state and “emergency” hematopoiesis. Cytokine 42, 277–288 10.1016/j.cyto.2008.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xia J., and Link D. C.. 2008) Severe congenital neutropenia and the unfolded protein response. Curr. Opin. Hematol. 15, 1–7 10.1097/MOH.0b013e3282f13cd2 [DOI] [PubMed] [Google Scholar]
- 4. Nayak R. C., Trump L. R., Aronow B. J., Myers K., Mehta P., Kalfa T., Wellendorf A. M., Valencia C. A., Paddison P. J., Horwitz M. S., Grimes H. L., Lutzko C., and Cancelas J. A.. 2015) Pathogenesis of ELANE-mutant severe neutropenia revealed by induced pluripotent stem cells. J. Clin. Invest. 125, 3103–3116 10.1172/JCI80924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nustede R., Klimiankou M., Klimenkova O., Kuznetsova I., Zeidler C., Welte K., and Skokowa J.. 2016) ELANE mutant-specific activation of different UPR pathways in congenital neutropenia. Br. J. Haematol. 172, 219–227 10.1111/bjh.13823 [DOI] [PubMed] [Google Scholar]
- 6. Massullo P., Druhan L. J., Bunnell B. A., Hunter M. G., Robinson J. M., Marsh C. B., and Avalos B. R.. 2005) Aberrant subcellular targeting of the G185R neutrophil elastase mutant associated with severe congenital neutropenia induces premature apoptosis of differentiating promyelocytes. Blood 105, 3397–3404 10.1182/blood-2004-07-2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Köllner I., Sodeik B., Schreek S., Heyn H., von Neuhoff N., Germeshausen M., Zeidler C., Krüger M., Schlegelberger B., Welte K., and Beger C.. 2006) Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood 108, 493–500 10.1182/blood-2005-11-4689 [DOI] [PubMed] [Google Scholar]
- 8. Hetz C. 2013) The biological meaning of the UPR. Nat. Rev. Mol. Cell Biol. 14, 404 10.1038/nrm3606 [DOI] [PubMed] [Google Scholar]
- 9. Fouret P., du Bois R. M., Bernaudin J. F., Takahashi H., Ferrans V. J., and Crystal R. G.. 1989) Expression of the neutrophil elastase gene during human bone marrow cell differentiation. J. Exp. Med. 169, 833–845 10.1084/jem.169.3.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Makaryan V., Kelley M. L., Fletcher B., Bolyard A. A., Aprikyan A. A., and Dale D. C.. 2017) Elastase inhibitors as potential therapies for ELANE-associated neutropenia. J. Leukoc. Biol. 102, 1143–1151 10.1189/jlb.5A1016-445R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qiu Y., Zhang Y., Hu N., and Dong F.. 2017) A truncated granulocyte colony-stimulating factor receptor (G-CSFR) inhibits apoptosis induced by neutrophil elastase G185R mutant: implication for understanding CSF3R gene mutations in severe congenital neutropenia. J. Biol. Chem. 292, 3496–3505 10.1074/jbc.M116.755157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horwitz M. S., Duan Z., Korkmaz B., Lee H. H., Mealiffe M. E., and Salipante S. J.. 2007) Neutrophil elastase in cyclic and severe congenital neutropenia. Blood 109, 1817–1824 10.1182/blood-2006-08-019166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meerbrey K. L., Hu G., Kessler J. D., Roarty K., Li M. Z., Fang J. E., Herschkowitz J. I., Burrows A. E., Ciccia A., Sun T., Schmitt E. M., Bernardi R. J., Fu X., Bland C. S., Cooper T. A., et al. (2011) The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 3665–3670 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mehta H. M., Futami M., Glaubach T., Lee D. W., Andolina J. R., Yang Q., Whichard Z., Quinn M., Lu H. F., Kao W. M., Przychodzen B., Sarkar C. A., Minella A., Maciejewski J. P., and Corey S. J.. 2014) Alternatively spliced, truncated GCSF receptor promotes leukemogenic properties and sensitivity to JAK inhibition. Leukemia 28, 1041–1051 10.1038/leu.2013.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li F. Q., and Horwitz M.. 2001) Characterization of mutant neutrophil elastase in severe congenital neutropenia. J. Biol. Chem. 276, 14230–14241 10.1074/jbc.M010279200 [DOI] [PubMed] [Google Scholar]
- 16. Bellanné-Chantelot C., Clauin S., Leblanc T., Cassinat B., Rodrigues-Lima F., Beaufils S., Vaury C., Barkaoui M., Fenneteau O., Maier-Redelsperger M., Chomienne C., and Donadieu J.. 2004) Mutations in the ELA2 gene correlate with more severe expression of neutropenia: a study of 81 patients from the French Neutropenia Register. Blood 103, 4119–4125 10.1182/blood-2003-10-3518 [DOI] [PubMed] [Google Scholar]
- 17. Grenda D. S., Murakami M., Ghatak J., Xia J., Boxer L. A., Dale D., Dinauer M. C., and Link D. C.. 2007) Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood 110, 4179–4187 10.1182/blood-2006-11-057299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nanua S., Murakami M., Xia J., Grenda D. S., Woloszynek J., Strand M., and Link D. C.. 2011) Activation of the unfolded protein response is associated with impaired granulopoiesis in transgenic mice expressing mutant Elane. Blood 117, 3539–3547 10.1182/blood-2010-10-311704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou T., Kinney M. C., Scott L. M., Zinkel S. S., and Rebel V. I.. 2015) Revisiting the case for genetically engineered mouse models in human myelodysplastic syndrome research. Blood 126, 1057–1068 10.1182/blood-2015-01-624239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Donini M., Fontana S., Savoldi G., Vermi W., Tassone L., Gentili F., Zenaro E., Ferrari D., Notarangelo L. D., Porta F., Facchetti F., Notarangelo L. D., Dusi S., and Badolato R.. 2007) G-CSF treatment of severe congenital neutropenia reverses neutropenia but does not correct the underlying functional deficiency of the neutrophil in defending against microorganisms. Blood 109, 4716–4723 10.1182/blood-2006-09-045427 [DOI] [PubMed] [Google Scholar]
- 21. Fiedler K., and Brunner C.. 2012) The role of transcription factors in the guidance of granulopoiesis. Am. J. Blood Res. 2, 57–65 [PMC free article] [PubMed] [Google Scholar]
- 22. Dale D. C., Person R. E., Bolyard A. A., Aprikyan A. G., Bos C., Bonilla M. A., Boxer L. A., Kannourakis G., Zeidler C., Welte K., Benson K. F., and Horwitz M.. 2000) Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood 96, 2317–2322 10.1182/blood.V96.7.2317 [DOI] [PubMed] [Google Scholar]
- 23. Hunter M. G., Druhan L. J., Massullo P. R., and Avalos B. R.. 2003) Proteolytic cleavage of granulocyte colony-stimulating factor and its receptor by neutrophil elastase induces growth inhibition and decreased cell surface expression of the granulocyte colony-stimulating factor receptor. Am. J. Hematol. 74, 149–155 10.1002/ajh.10434 [DOI] [PubMed] [Google Scholar]
- 24. Horwitz M. S., Laurino M. Y., and Keel S. B.. 2019) Normal peripheral blood neutrophil numbers accompanying ELANE whole gene deletion mutation. Blood Adv. 3, 2470–2473 10.1182/bloodadvances.2019000498 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the manuscript.