Abstract
The hereditary aspect of obesity is a major focus of modern medical genetics. The genetic background is known to determine a higher-than-average prevalence of obesity in certain regions, like Oceania. There is evidence that dysfunction of brown adipose tissue (BAT) may be a risk factor for obesity and type 2 diabetes (T2D). A significant number of studies in the field focus on the UCP family. The Ucp genes code for electron transport carriers. UCP1 (thermogenin) is the most abundant protein of the UCP superfamily and is expressed in BAT, contributing to its capability of generating heat. Single nucleotide polymorphisms (SNPs) of Ucp1–Ucp3 were recently associated with risk of cardiometabolic diseases. This review covers the main Ucp SNPs A–3826G, A–1766G, A–112C, Met229Leu, Ala64Thr (Ucp1), Ala55Val, G–866A (Ucp2), and C–55 T (Ucp3), which may be associated with the development of obesity, disturbance in lipid metabolism, T2D, and cardiovascular diseases.
Background
In recent years, there has been a growing interest in the genetics of obesity as one of the major risk factors for cardiovascular diseases and type 2 diabetes (T2D). The prevalence of obesity is increasing worldwide, and being overweight often becomes a chronic, relapsing disease.
Genetic predisposition to obesity can play important role in its etiology together with environmental influences. This idea was confirmed in studies involving twins (Herskind et al. 1996) and adopted children (Stunkard et al. 1986). A high heritability of body mass index (BMI) was shown in 1233 Danish adults, females were found to have a greater heritability than males, and the BMI heritability in males appeared to increase with age (Herskind et al. 1996). A large study including 540 adoptees and their biological and adoptive parents found a significant relationship between the weight class (thin, median weight, overweight, and obese) of the adoptees and the BMI of their biologic parents, but no relationship was observed with the weight class of the adoptive parents (Stunkard et al. 1986). According to published studies, heritability explains most of the variation (61–80%) in BMI, a primary measure of adiposity (Nan et al. 2012), reflecting a high genetic contribution to the etiology of overweight and obesity. This seems promising for those who begin to study the genetic aspect of obesity. Based on the special role that brown adipose tissue (BAT) plays in systemic metabolism, the Ucp genes could be of importance. Five homologous genes of the Ucp family have been described in humans.
All of the UCP proteins are located on the inner membrane of mitochondria and act to reduce the proton gradient to a greater or lesser degree and thereby to decrease the mitochondrial electrochemical potential (Cline et al. 2001; Gong et al. 1997; Krauss et al. 2002; Mao et al. 1999; Paulik et al. 1998; Sanchis et al. 1998). Their expression is tissue specific: UCP4 and UCP5 play their role in cells of the central nervous system (Ramsden et al. 2012); UCP3 is present in skeletal muscle cells (Boss et al. 1997), BAT (Vidal-Puig et al. 1997), and cardiomyocytes (Razeghi et al. 2002); and UCP2 is found in considerable amounts in white adipose tissue, skeletal muscle, the heart, the lung, the spleen, the thymus, cells of the immune system, and vascular cells and is expressed in lower amounts in the brain, liver, and kidney (Fisler and Warden 2006; Pierelli et al. 2017). UCP1, or thermogenin, is specific to BAT and determines its main function, which is heat generation. Thermogenesis in BAT is essential for the majority of mammals and especially for true hibernators (marmots, hamsters, and bats), non-hibernating small rodents (rats and mice) (Cannon and Nedergaard 2004) and rabbits (Hardman et al. 1969); certain large mammals such as horses (Ertelt et al. 2014), bears (Nedergaard and Cannon 1990) and cetaceans (Hashimoto et al. 2015); and primates (Génin et al. 2003), including humans (Ricquier 2016). BAT is vital in human newborns because their thermoregulation is not developed enough to allow efficient cold adaptation. BAT activation is a promising approach to controlling obesity and, therefore, diabetes and cardiovascular diseases and has attracted substantial interest of researchers after important milestones were achieved. First, the presence of brown fat cells in a human adult body was proved (Heaton 1972). Second, animal experiments showed that BAT is capable of eliminating excess calories consumed with excess food (Rothwell and Stock 1997). UCP1, being the ultimate marker of BAT, is of interest for the genetics of obesity and the focus of this review.
Brown adipose tissue and UCP1: general characteristics
Apart from muscles, the liver, the pancreas and the brain, BAT has emerged as an important metabolic organ that is evolutionarily designed to generate heat and thus protect mammals from hypothermia (Cannon and Nedergaard 2004). BAT and its putative role in hibernation were first described in 1551 by the Swiss naturalist Konrad Gessner, who found BAT in the interscapular area of marmots (Cannon and Nedergaard 2008). Different functions were assigned to BAT since that time. In 1670, BAT was incorrectly characterized as part of the thymus. Later, in 1817, BAT was considered to be a ductless gland with hemopoietic properties. Since 1863, BAT was classified as a form of adipose tissue that functions as a nutrient pool. In 1902, the endocrine function of BAT was addressed again (Rasmussen 1923). The thermogenic function of BAT was demonstrated in the 1960s by in vivo studies in rodents, and then brown fat depots were similarly found in human newborns (Ricquier 2016). Brown fat contributes to nonshivering thermogenesis (Lichtenbelt 2012) in the infant’s body and prevents hypothermia until the body is capable of independent thermoregulation. The blood flowing through brown fat depots gets warmed up and then increases the temperature of vital organs: the brain, the heart, the lung, the kidney, and even skeletal muscles (Oppert et al. 1994; Ricquier 2011). Heat is not generated by contractile activity in brown adipocytes, which act mostly by increasing the metabolic rate and oxygen consumption of the body. The content of brown fat decreases significantly as the body ages, but there is evidence that small amounts of BAT still remain in adults (Cannon and Nedergaard 2004). The presence of BAT in adults was confirmed in many studies (Heaton 1972; Huttunen et al. 1981). Although the BAT contribution was thought to decrease with age, it was proved about a decade ago that BAT found in human adults is functionally active (Cypess et al. 2009; van Marken Lichtenbelt et al. 2009; Virtanen et al. 2009). This finding renewed interest to BAT as an important metabolic organ in humans.
BAT differs from white adipose tissue in performing other functions and having a different morphological structure and anatomical localization. Brown adipocytes are smaller, and their cytoplasm contains a large number of small lipid droplets, while white fat cells contain one large lipid droplet (Cannon and Nedergaard 2004). Brown fat cells have many mitochondria, which ensure their high metabolic activity. The innervation and vascularization of BAT are several times higher than in white adipose tissue, and BAT is therefore capable of responding faster to a decrease in ambient temperature and delivering heat to vital organs (Saely et al. 2011). While white adipose tissue is distributed almost throughout the body, BAT is located in the supraclavicular, cervical, axillary, and mediastinal regions (Lee et al. 2013; Saely et al. 2011; Sampath et al. 2016).
Today, fluorine-18-fluorodeoxyglucose (18F-FDG) positron emission tomography (PET) is the main tool for visualizing BAT in humans (Sampath et al. 2016). BAT, which actively generates heat, accumulates the tracer more intensely than surrounding tissues as a result of a higher glucose uptake. PET studies were conducted in various populations, and metabolically active BAT was found in 1.2–7.2% of adult subjects (Au-Yong et al. 2009; Zhang et al. 2014b). However, the studies were performed in different climatic conditions and the subjects differed in age and BMI, which is a ratio of body weight to height squared (kg/m2). People with active BAT were younger and had lower BMI values (Yoneshiro et al. 2011; Zhang et al. 2014b). In addition, the probability to detect BAT activation is higher in subjects with previous exposure to low air temperatures (Sampath et al. 2016). It is interesting to note that active BAT is more frequent in women than in men (Cypess et al. 2009). Thus, functionally active BAT may contribute to maintaining the normal body weight in some individuals, and exposure to cold triggers a physiological mechanism that induces nonshivering thermogenesis (van Marken Lichtenbelt et al. 2009).
The therapeutic potential of BAT is promising. Despite its low content in the body, BAT can exert a significant metabolic effect. In adults, BAT contributes 1–5% to overall energy metabolism of the body and defects in its function might lead to an excess weight gain of 1–2 kg per year (Urhammer et al. 1997). Scientific interest in BAT is reasonable due to the possibility to develope treatment for metabolic disorders. In fact, it was noted that, white adipose tissue can transdifferentiate into BAT during chronic cold exposure and that BAT loses its capability of thermogenesis and transdifferentiates into white adipose tissue during a prolonged positive energy balance (Barbatelli et al. 2010; Cinti et al. 1997; Rosenwald et al. 2013). Constant sympathetic stimulation was shown to result in the formation of thermogenin-expressing loci in white adipose tissue (Cinti et al. 1997). This process is called browning (Bartelt and Heeren 2014). Beige adipocytes, which are also known as brown-like-in-white cells, could be found within white adipose tissue. Beige adipocytes possess multiple lipid droplets and a high number of mitochondria, thus morphologically resembling brown adipocytes (Phillips 2019). Since beige cells express Ucp1 similarly to BAT cells, they are capable of thermogenesis (Harms and Seale 2013; Rui 2017). Brown-like loci were first identified in adult mice and were found to emerge in white adipose tissue of the parametrium in response to cold acclimation (Young et al. 1984). Beige adipocytes predominantly originate from the Myf5 (myogenic factor 5)-negative lineage, like white adipocytes, while brown adipocytes arise from Myf + precursors (Phillips 2019). Recent studies in the field revealed that the exercise-induced myokine irisin can trigger the formation of beige adipocytes within white fat pads (Seale et al. 2007). A beige adipocyte-specific transcriptional pattern (Ucp1, Ppargc1α, PRDM16, TMEM26, and CD137) emerges in response to irisin expression, and protein levels of UCP1 increase in subcutaneous white adipose tissue (Li et al. 2019). Such transformation of white adipose tissue might be used to treat metabolic disorders. The possibility of Ucp1 activation in white adipose tissue, including the use of irisin, is considered as one of the strategies to combat obesity and thus to fight against T2D and cardiovascular disorders. Two cell-specific transcriptional coactivators, PPARγ coactivator 1α (PGC-1α, encoded by Ppargc1a) and PR domain-containing 16 (PRDM16), were described as major molecular determinants of the brown adipose phenotype (Kajimura et al. 2009). Generation of Ppargc1a-null mice revealed that PGC-1α is essential for expression of Ucp1, but not for brown fat cell differentiation (Lin et al. 2004; Uldry et al. 2006). Cell-based studies showed that, upon forskolin treatment (which imitates noradrenergic stimulation), PRDM16 could be recruited to the enhancer of Ucp1 to induce its expression in undifferentiated murine embryonic fibroblasts. PRDM16 was identified as a universal brown fat cell-fate determination factor (Seale et al. 2007) that promotes differentiation of Myf5-positive myogenic precursor cells into brown adipocytes while banning myogenic differentiation (Seale et al. 2008). PRDM16 was additionally shown to induce acute expression of BAT-specific genes such as Ucp1 and Ppargc1a, when ectopically expressed in murine embryonic fibroblasts (Seale et al. 2007) and to play a role in BAT-specific gene expression in beige fat cells (Cohen et al. 2014; Seale et al. 2011). The molecular mechanism of transcriptional activation by PRDM16 is still unclear (Iida et al. 2015). Recent studies showed that early B-cell factors (EBF) are required for chronic cold-induced BAT recruitment. Both EBF1 and EBF2 cooperate with estrogen-related receptor α and PGC-1 α to promote Ucp1 transcription (Angueira et al. 2020).
Uncoupling protein 1
UCP1 is a transmembrane protein carrier with a molecular weight of 32 kDa and, like all members of the UCP family, is located in the inner membrane of mitochondria (Hoang et al. 2013). UCP1 allows protons from the intermembrane space to penetrate back into the mitochondrial matrix, bypassing the oxidative phosphorylation of ADP. As a result, ATP synthesis is reduced and free cell respiration is intensified to release energy in the form of heat. Despite the existence of several homologues, UCP1 is the only member of the family that performs a true thermogenic function (Ricquier 2011). UCP1 constitutes about 10% of the brown adipocyte mitochondrial membrane protein, in contrast to UCP2 и UCP3, the content of which does not exceed 0.1% (Kozak and Anunciado-Koza 2009).
UCP1 was found in BAT in the 1970s and was sequenced later. It was believed earlier that Ucp1 is only expressed in newborns, but both the UCP1 protein and its mRNA were found in adults more recently (Bouillaud et al. 1988; Lean et al. 1986). Brown fat cells most intensely express Ucp1 in the human body, but the Ucp1 mRNA becomes detectable in subcutaneous white fat in response to certain stimuli, such as exercise training (Norheim et al. 2014; Otero-Díaz et al. 2018) and cold exposure (Barbatelli et al. 2010; Finlin et al. 2018; Kern et al. 2014), and is found in skeletal muscle of mice (Gorski et al. 2018).
Thermogenesis in BAT is directly controlled by the endings of sympathetic nerve fibers, which are abundant in the tissue. Norepinephrine released by sympathetic nerve fibers activates brown adipose hormone-sensitive lipase, which hydrolyzes intracellular triglycerides through a cascade of reactions. UCP1 is activated by the resulting free fatty acids, and the phosphorylation-uncoupling process intensifies (Fedorenko et al. 2013).
Free radicals are generated when electrons are transferred to oxygen molecules during oxidative phosphorylation. However, these toxic compounds are balanced with present antioxidants in normal conditions. When this balance is disrupted, oxidative stress occurs to induce a number of pathophysiological conditions, including T2D (Brondani et al. 2012). By reducing the membrane potential, UCP1 and its homologues reduce the generation of free radicals and thus protect the cell against oxidative stress (Brand et al. 2004).
Despite the fact that most members of the UCP family are capable of reducing the proton gradient, the true thermogenic function is only performed by UCP1. Thus, UCP1 is one of the main focuses of research into the molecular biological processes associated with energy metabolism and its disorders. A significant part of Ucp1 studies aimed at detecting its single nucleotide polymorphisms (SNPs) and analysing their phenotypic manifestations.
Here we review the data on the most significant SNPs of the Ucp family genes and their association with cardiometabolic diseases (CMDs) and CMD risk factors.
Role of Ucp1 polymorphisms in cardiometabolic diseases
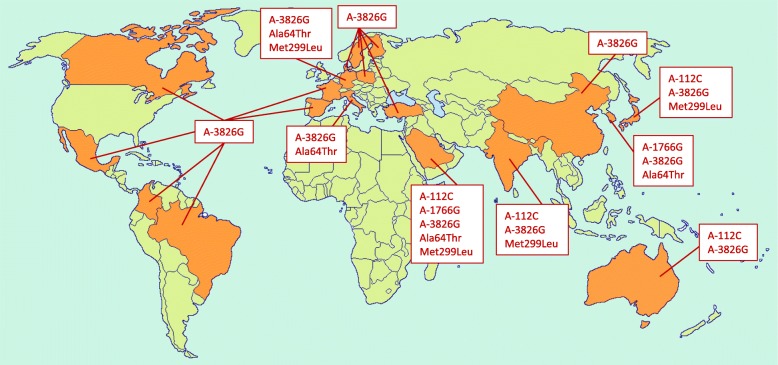
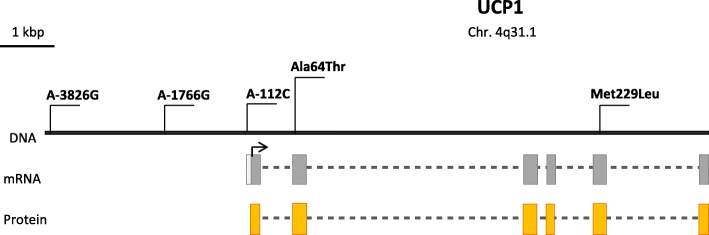
CMDs are chronic diseases that develop latently throughout life and progress to an advanced stage by the time symptoms appear. CMDs are lifestyle dependent and include a set of risk factors for the development of chronic pathological conditions with adverse effects on the cardiovascular function, such as atherosclerosis, arterial thrombosis, and metabolic disorders leading to T2D and related complications (Flouris et al. 2017; Scottish Intercollegiate Guidelines Network (SIGN) 2017). Many CMDs have a multifactorial origin; i.e., both genetic and environmental factors are involved in their development. Numerous studies showed that an increased BMI, insulin secretion, and increased blood pressure may be due to a genetic predisposition. Mutations in the Ucp genes can increase the risk of obesity and associated metabolic disorders (Brondani et al. 2014a). The Ucp1 gene was assigned to the long arm of chromosome 4 in q31.1. Ucp1 is split into six exons, spans about 13 kb, and contains a 9-kb transcribed region (Cassard et al. 1990). A large number of studies (Table 1) described SNPs in the noncoding region of the Ucp1 gene, such as A–3826G (rs1800592), A–1766G (rs3811791), and A–112C (rs10011540), as well as in its coding part, which includes Ala64Thr (rs45539933) and Met229Leu (rs2270565) (Fig. 1). However, the influence of Ucp1 genetic variants was studied poorly, if at all, in Russia and Eastern Europe, where the CMD prevalence rates remain high (Flouris et al. 2017) (Fig. 1).
Table 1.
Studies on the association between the Ucp1 polymorphisms and CMDs or CMD risk factors in different populations
| SNP(s) | Population and participants | Association | Allele/genotype frequencies | Reference | |
|---|---|---|---|---|---|
| +/− | Condition | ||||
| A–3826G | Australian: overweight/obese F = 526 | + | ↑ BMI (P = 0.02), ↑ insulin (P = 0.03, not adjusted), and ↑ fasting glucose concentrations (P = 0.01, adjusted for BMI) | G allele: 0.23 | (Heilbronn et al. 2000) |
| A–3826G | Brazil: T2D patients N = 981 and controls N = 534 | – | T2D (P > 0.05) |
AA: 49.9%, AG: 37.7%, GG: 12.4%, G allele: 0.313 in diabetics; AA: 49.3%, AG: 39.5%, GG: 11.2%, G allele: 0.310 in controls |
(de Souza et al. 2013) |
| A–3826G | Canadian: parents N = 123 and offsprings N = 138 from 64 families | – | Body fat, RMR, and absolute changes in body fat over a 12-year period | G allele: 0.28 | (Oppert et al. 1994) |
| + | ↑ body fat gain over time (P < 0.05) | ||||
| A–3826G |
Chinese: T2D patients N = 792, including DR group: PDR N = 220 and NPDR N = 228; DNR group: N = 334 |
+ | ↑ risk of PDR (additive model OR = 1.72, 95% CI: 1.06–2.79, P = 0.03) |
AA: 20.7%, AG: 49.3%, GG: 30.0%, G allele: 54.6% in PDR group; AA: 28.1%, AG: 48.2%, GG: 23.7%, G allele: 47.8% in DNR group |
(Zhang et al. 2015) |
| + | PDR (OR = 1.32, 95% CI: 1.03–1.68, P = 0.03) | ||||
| – | DR and DNR or NPDR and DNR | ||||
| A–3826G | Chinese: hypertensive subjects M = 573 and F = 589; normotensive subjects M = 373 and F = 672 | – | EH |
AA: 25.12%, AG: 50.85%, GG: 24.06%, G allele: 49.47% in normotensives; AA: 25.22%, AG: 52.01%, GG: 22.77%, G allele: 48.78% in hypertensives |
(Sun et al. 2018) |
| A–3826G | Chinese: T2D patients N = 3107, including DR = 662 | – | DR (OR = 1.001, P = 0.993) | AA (n): 2578, AC (n): 469, CC (n): 14 | (Jin et al. 2020) |
| A–1766G | – | DR (OR = 0.949, P = 0.488) | |||
| A–112C | + | DR (OR = 1.368, P = 0.004) | |||
| A–1766G | Chinese: T2D patients N = 929, nondiabetic controls N = 1044 | + | ↑ risk of T2D (OR = 1.42, P = 0.042), ↑ level of TG (β = 0.048, P = 0.034) under recessive model |
AA: 53.8%, AG: 37.3%, GG: 8.9%, G allele: 27.6% in T2D patients; AA: 53%, AG: 40.6%, GG: 6.4%, G allele: 26.7% in controls |
(Dong et al. 2020) |
| A–3826G | Colombian: T2D cases M = 190 and F = 355; controls M = 126 and F = 323 | + | A allele and T2D (OR = 0.78; 95% CI: 0.63–0.97; P = 0.02) | A allele: 0.54 in T2D cases and 0.60 in controls | (Franco-Hincapié et al. 2014) |
| A–3826G | Danes:young healthy subjects M = 177 and F = 176 | – | BMI, fat mass, or weight gain during childhood and adolescence | G allele: 25.3% (95% CI: 22.2–28.4%) | (Urhammer et al. 1997) |
| Ala64Thr | Danes: males with juvenile obesity N = 156; controls N = 205: lean controls N = 79 | – | Obesity and weight gain during childhood or adolescence, or insulin sensitivity | 64Thr allele: 8.2% in juvenile obese subjects, 8.8% in controls, and 8.2% in lean controls | |
| Met229Leu | – | 229Leu allele: 8.2% in obese subjects, 8.1% in controls, and 5.6% in lean controls | |||
| A–3826G | Finnish: T2D patients M = 38 and F = 32; controls M = 55 and F = 68 | – | T2D, body weight or BMI in diabetics or controls | G allele: 38.6% in diabetics, 34.1% in controls | (Sivenius et al. 2000) |
| A–3826G | Finnish: obese premenopausal women N = 77 | – | Weight gain after a VLCD | G allele: 0.19 | (Fogelholm et al. 1998) |
| A–3826G | French: overweight patients N = 163 | + | ↓ weight loss after low calorie diet (P < 0.05) | G allele: 0.27 | (Fumeron et al. 1996) |
| A–3826G | German: T2D patients N = 517 | – |
Neuropathy (P = 0.79), retinopathy (P = 0.48), and nephropathy (P = 0.93) |
AA: 49.9%, AG: 45.6%, GG: 4.5%, G allele: 0.27 |
(Rudofsky et al. 2007) |
| Ala64Thr | German children and adolescents: obese subjects N = 293, F = 53%; lean subjects N = 134, F = 46% | – | Early-onset obesity | 64Thr allele: 8.2% in obese, 4.1% in lean individuals | (Hamann et al. 1998) |
| Met229Leu | – | 229Leu allele: 10.4% in obese, 12.0% in lean individuals | |||
| A–3826G | Indian: M = 49 and F = 47 | + |
GG genotype and BMI (P < 0.01), SBP (P < 0.01) and DBP (P < 0.05) in females |
GG:13.5%, AG: 46.5%, AA: 39.9% | (Dhall et al. 2012) |
| A–3826G | Indian: T2D subjects M = 353 and F = 457; normal glucose-tolerant subjects M = 374 and F = 616 | – | T2D |
AA: 36%, AG: 46%, GG: 18% G allele: 0.41 in T2D subjects. AA: 40%, AG: 45%, GG: 15% G allele: 0.38 in normal glucose-tolerant subjects |
(Vimaleswaran et al. 2009) |
| A–112C | – |
AA: 63%, AC: 33%, CC: 4%, C allele: 0.21 in T2D subjects. AA: 62%, AC: 34%, CC: 4%, C allele: 0.21 in normal glucose- tolerant subjects |
|||
| Met299Leu | – | MetMet: 76%, MetLeu: 23%, LeuLeu: 1%, 299Leu allele: 0.12 in T2D subjects; MetMet: 73%, MetLeu: 26%, LeuLeu: 1%, 299Leu allele: 0.14 in normal glucose-tolerant subjects | |||
| A–3826G | Italian: severely obese nondiabetic individuals M = 40 and F = 72 | + | IR in morbid obesity |
AA: 25.9%, AG + GG: 74.1% in total obese population; AG + GG: 88% in IR (+) and 63% in IR (−) (OR = 4.3, 95% CI: 1.6–11.7; P = 0.003) |
(Bracale et al. 2012) |
| A–3826G | Italian: T2D individuals M = 56.6%; controls M = 41.2% | – | T2D (OR = 0.85, 95% CI: 0.65–1.11; P = 0.221) | G allele: 0.24 | (Montesanto et al. 2018) |
| + | Risk of nephropathy (OR = 0.57, 95% CI: 0.33–0.98; P = 0.031) | ||||
| – | Ischemic heart disease and stroke (OR = 1.10, 95% CI: 0.74–1.64; P = 0.643) | ||||
| Ala64Thr | – |
T2D (OR = 0.99, 95% CI: 0.61–1.6; P = 0.969) |
64Thr allele: 0.063 | ||
| + | Risk of retinopathy (OR = 0.31, 95% CI: 0.12–0.82; P = 0.010) | ||||
| – | Ischemic heart disease and stroke (OR = 1.08, 95% CI: 0.52–2.26; P = 0.837) | ||||
| A–3826G | Japanese: individuals sampled during cold N = 1080 and hot season N = 979 | + | VFA during cold season (P = 0.0197) | G allele: 0.51 | (Nakayama et al. 2013) |
| A–3826G | Japanese: subjects without a history of cardiovascular disease M = 231 and F = 347 | + | GG genotype and HT in males (OR = 2.32, 95% CI: 1.08–4.99) and older subjects (OR = 1.89, 95% CI: 1.00–3.57) |
AA: 24.0%, AG: 50.0%, GG: 26.0%, G allele: 0.51 |
(Kotani et al. 2007) |
| A–112C | Japanese: T2D patients M = 180 and F = 140, controls M = 145 and F = 105 | + | T2D (P = 0.017) | С allele: 6.2% in healthy controls, 10.2% in T2D patients | (Mori et al. 2001) |
| Met299Leu | + | T2D (P = 0.038) | 229Leu allele: 7.2% in healthy controls, 10.8% in T2D patients | ||
| A–3826G | – | T2D | G allele: 53.6% in healthy controls, 49.7% in T2D patients | ||
| A–112C | Japanese: T2D cases M = 55 and F = 38 | + | ↑ FIRI (P = 0.0085), HOMA-IR (P = 0.0089), and HLC (P = 0.012) | AA: 88.2%, AC: 10.7%, CC:1.1% | (Fukuyama et al. 2006) |
| A–3826G | – | T2D | AA: 32.3%, AG: 48.4%, GG: 19.3% | ||
| A–3826G | Japanese: obese F = 113, healthy non-obese F = 76 | + | GG genotype and resistance to weight loss among obese (P < 0.05) | G allele: 0.46 in obese and 0.45 in healthy non-obese | (Kogure et al. 1998) |
| A–3826G |
Japanese: healthy postmenopausal group F = 182 and premenopausal group F = 99 |
+ | ↑ body weight (P = 0.048) in premenopausal women and changes in serum TG (P = 0.049) and HDL (P = 0.020) levels in postmenopausal women |
AA: 23.7%, AG: 53.2%, GG: 23.1%, G allele: 0.50 |
(Matsushita et al. 2003) |
| A–3826G | Japanese: healthy boys N = 22 | + |
GG genotype and ↓ TEM to fat intake (P < 0.05) |
AA+AG (n): 13, GG (n): 9 | (Nagai et al. 2003) |
| A–3826G | Japanese: healthy children N = 19 | + | GG genotype and ↓ cold-induced thermogenesis (P < 0.05) | AA+AG (n): 14, GG (n): 5 | (Nagai et al. 2007) |
| A–3826G | Japanese: M = 251 | + | AG genotype and BMI (P = 0.016) | AA: 24.3%, AG: 48.2%, GG: 27.5% | (Nakano et al. 2006) |
| A–3826G | Korean: obese patients M = 44 and F = 146 | + | AG/GG genotypes and ↑ DBP (P = 0.023) and LDL-C (P = 0.011); GG genotype and ↓ HDL-C (P < 0.05) and ↑ atherogenic index |
AA: 22.1%, AG: 53.7%, GG: 24.2%, G allele: 0.51 |
(Oh et al. 2004) |
| A–3826G | Korean: F = 387 | + | [−3826G/−1766G] Ht and ↑ body fat percent (P = 0.045) | NA | (Kim et al. 2005) |
| A–1766G | + | AG/GG genotypes and abdominal subcutaneous fat (P = 0.015), abdominal visceral fat (P = 0.013), ↑ WHR (P = 0.008), body fat mass (P = 0.023), and percent body fat (P = 0.014) | AA:57.4%, AG: 37.7%, GG: 4.9%, G allele: 0.238 | ||
|
A–3826G A–1766G Ala64Thr |
Korean: overweight F = 453 | + |
[−3826G/ –1766A/64Thr] Ht and ↓ abdominal fat tissue area (P = 0.02), fat tissue area at thigh (P = 0.008), body fat mass (P = 0.002), and WHR (P = 0.01) [−3826G/ –1766A/64Ala] Ht and reduction of WHR and body fat mass by VLCD (P = 0.05–0.006) |
NA | (Shin et al. 2005) |
| A–3826G | Mexican adolescents: normal weight N = 159 and overweight/obese N = 111 | + | ↑ percentage of fat (P = 0.002) and muscle weight (P = 0.019) in a recessive model | AA: 16.2%, AG: 55.9%, GG: 27.9% in obese; AA: 17%, AG: 50.9%, GG: 32.1% in controls | (Sámano et al. 2018) |
| A–3826G | Mixed ethnicity population: obese patients N = 150, F = 80% | + |
↓ values of weight, body fat and free fat mass (P < 0.05); GG genotype and ↓ tendency of T2D |
AA: 41.3%, AG: 45.3%, GG: 13.4%, G allele: 0.36 |
(Nicoletti et al. 2016) |
| A–3826G | Polish: overweight/obese individuals M = 38 and F = 80 | – | BMI | AA: 51.38%, AG: 33.94%, GG:14.68%; G allele: 30.5%. | (Kieć-Wilk et al. 2002) |
| + |
↑ fasting levels of TG (P < 0.04) and ↓ HDL-C (P = 0.004) |
||||
| A–3826G | Polish: embers of obese Caucasian families M = 38 and F = 84 | – | Susceptibility to obesity and glucose tolerance parameters | NA | (Malczewska-Malec et al. 2004) |
| A–112C | Saudi Arabian: obese patients M = 138 and F = 199; healthy volunteers M = 76 and F = 79 | – | Obesity | A allele/C allele (n/n): 627/47 in obese patients, 283/27 in controls | (Chathoth et al. 2018) |
| A–1766G | + | A allele and moderate obesity (OR = 2.89, 95% CI 1.33–6.25; P = 0.007) | A allele/G allele (n/n): 624/50 in obese patients, 298/12 in controls | ||
| A–3826G | + | Obesity after adjusting age, sex, and T2D (OR = 1.52, 95% CI: 1.10–2.08; P = 0.009) | A allele/G allele (n/n): 443/231 in obese patients, 227/83 in controls | ||
| A–3826G | Spanish: obese individuals N = 159, normal weight, N = 154 | + |
↑ BMI (P = 0.03), ↑ percentage of body fat (P < 0.04), ↑ SBP (P = 0.009), ↑ DBP (P = 0.02) in obese group |
G allele: 0.21 in healthy, 0.19 in obese individuals (P = 0.574) | (Forga et al. 2003) |
| A–3826G | Spanish: obese M = 38 and F = 55; non-obese M = 122 and F = 117 | + | Obesity in women (P = 0.019) | G allele: 0.3 in obese (0.28 in males and 0.31 in females), 0.24 in non-obese (0.32 in males and 0.17 in females) (P = 0.008) | (Ramis et al. 2004) |
| A–3826G | Swedish: obese subjects M = 310 and F = 364, non-obese subjects M = 54 and F = 257 | – | Obesity-related phenotypes and weight gain | G allele: 0.25 in obese, 0.24 in non-obese subjects | (Gagnon et al. 1998) |
| A–3826G | Turkish:obese M = 83 and F = 63; lean individuals M = 77 and F = 17 | + | ↑cholesterol levels | G allele: 0.30 in obese, 0.31 in lean individuals | (Proenza et al. 2000) |
| A–3826G | Turkish children and adolescents: obese N = 268 and non-obese = 185 | + | GG denotype and obesity (OR = 2.02, 95% CI 1.17–3.47; P = 0.010),↑ TG levels in obese subjects (P = 0.048), ↓ HDL-C (P = 0.017) |
AA: 46.3%, AG: 33.2%, GG: 20.5%, G allele: 37.1% in obese; AA: 46.5%, AG: 42.2%, GG: 11.4%, G allele: 32.4% in controls |
(Gul et al. 2017) |
| Ala64Thr | White subjects: obese N = 93 and non-obese N = 69 | – | BMI or obesity | 64Thr allele: 12.0% | (Herrmann et al. 2003) |
| + | WHR (P = 0.003) after adjustment for gender, age, BMI, and T2D | ||||
Abbreviations: F Females, M Males, BMI Body mass index, RMR Resting metabolic rate, DR Diabetic retinopathy, PDR Prolifirative diabetic retinopathy, NPDR Non-prolifirative diabetic retinopathy, DNR Diabetes without retinopathy, EH Essential hypertension, T2D Type 2 diabetes, VLCD Very low calorie diet, IR Insulin resistance, VFA Visceral fat accumulation, HT Hypertension, FIRI Fasting insulin resistance index, HOMA-IR Homoeostasis model assessment of insulin resistance, HLC Hepatic lipid content, HDL-C High density lipoprotein cholesterol, TEM Thermic effect of a meal, Ht Haplotype, SBP Systolic blood pressure, DPB Diastolic blood pressure, LDL-C Low density lipoprotein cholesterol, WHR Waist-to-hip ratio, TG Trygliceride, NA Not available
Fig. 1.
World map showing the investigation of the association of Ucp1 polymorphisms with CMDs or CMD risk factors
A–3826G polymorphism of the Ucp1 gene
The A/G SNP at position − 3826 bp (rs1800592) of the 5′ noncoding region of the gene (Fig. 2) was the focus of initial publicationsin the field. The SNP was found by Canadian scientists in 1994 in a study of the genetic characteristics and family histories of patients with elevated BMIs. The study was the first to associate the G allele with a higher BMI, the risk of T2D, and lipid metabolism disorders (Oppert et al. 1994).
Fig. 2.
Schematic structure of the Ucp1 gene and its best-studied polymorphisms
It was later established that the SNP is in the regulatory region of the gene and that obese G allele carriers have reduced Ucp1 mRNA expression (Esterbauer et al. 1998), suggesting functional significance for the SNP. An enhancer sequence with five hexameric binding sites for nuclear retinoic acid receptors—RARα, RARβ, retinoid X receptors (RXR) and peroxisome proliferator-activated receptors (PPARγ)—was found in close proximity to the A–3826G SNP (del Mar Gonzalez-Barroso et al. 2002). Polymorphisms of this region are likely to disrupt the transcription factor binding sites, thus reducing Ucp1 expression of to some extent.
A statistically significant correlation between A–3826G and phenotypic characteristics was observed in numerous studies of the BMI, body fat content, and weight loss or gain intensity, the AA genotype being associated with the lowest risk in all cases (Brondani et al. 2012; Forga et al. 2003). However, the contribution of the mutant allele to the pathogenesis of diseases was not confirmed in some studies of A–3826G in patients with metabolic disorders and cardiovascular diseases from different populations (Gagnon et al. 1998; Kieć-Wilk et al. 2002; Schäffler et al. 1999; Urhammer et al. 1997).
Japanese researchers showed that a decrease in body weight in obese G allele carriers was lower than in A allele carriers, with similar low-calorie diet and exercise regimen (4.3 ± 2.6 vs. 7.4 ± 4.2 kg; p < 0.05) (Kogure et al. 1998). Similar results were obtained by scientists from France: the GG genotype determined a resistance to low calorie diet (P < 0.05) (Fumeron et al. 1996), and Finland (Fogelholm et al. 1998), but the results did not reach statistical significance. In addition, G allele carriers with the genomic existence of the Trp64Arg substitution in the beta 3-adrenergic receptor gene were more susceptible to weight gain than A allele carriers in the Finnish population (Sivenius et al. 2000).
The G allele was associated with an increased BMI (P = 0.02) and a higher blood glucose concentration in the Australian female population (P = 0.01, adjusted for BMI), and its higher frequency was observed in women with T2D (P = 0.06, adjusted for BMI) (Heilbronn et al. 2000). In the work by Spanish authors, the G allele was more common in men, but the association with overweight was revealed only in women: the G allele frequency in obese female patients was higher than in non-obese female subjects (0.31 vs. 0.17, P = 0.008) (Ramis et al. 2004). The association of the GG genotype with obesity and a higher blood pressure was also seen among Indian females but not among males (Dhall et al. 2012).
In contrast, in Japan, the nucleotide substitution was not associated with hypertension in women, while susceptibility to the disease in the male population was determined by the GG genotype togetger with the age and increased BMI (Kotani et al. 2007). In addition, male G allele carriers had higher BMIs as compared with the male carries of the AA genotype. However, the highest BMI values were observed in individuals with the AG genotype among young Japanese males (P = 0.016) (Nakano et al. 2006). In the Italian population, the G allele variant was associated with the development of insulin resistance in obese patients (Bracale et al. 2012), which increases the risk for diabetes. In northwestern Colombia, the A allele negatively correlated with T2D and, according to the authors, could act as a protective factor in the population studied (Franco-Hincapié et al. 2014). In Saudi Arabia, a statistically significant association was found for the minor G allele and an increased BMI (Chathoth et al. 2018). In Mexican adolescents, the –3826G variant was associated with higher percentage of fat (P = 0.002) and muscle mass (P = 0.019) in the body, but no differences in allele frequencies was found between corpulent and lean students (Sámano et al. 2018). Opposite results were obtained by Brazilian researchers. In a sample of patients with BMI ≥ 35 kg/m2, the G allele was associated with lower weight and body fat content and GG genotype carriers were less likely to be diagnosed with T2D as compared with carriers of the wild-type A allele (Nicoletti et al. 2016).
Proenza et al. observed increases in total cholesterol levels associated with a high BMI and low high density lipoprotein cholesterol (HDL-C) in GG carriers that among 271 patients of Turkish origin (Proenza et al. 2000). The G allele was more frequent in patients with triglyceridemia as compared with subjects without it (42.9 vs. 34.4%, P = 0.048) among obese children and adolescents (Gul et al. 2017). In a study of obese families from southern Poland, the GG genotype was similarly associated with decreased HDL-C levels and higher levels of triglycerides and free fatty acids as compared with the AG and AA genotypes (Kieć-Wilk et al. 2002). Similar results were obtained for HDL-C levels in a group of Japanese postmenopausal women (P = 0.020) (Matsushita et al. 2003) and obese patients from the Korean population. In addition, Korean G allele carriers showed an increased diastolic blood pressure and higher low-density liprotein cholesterol (LDL-C) levels (Oh et al. 2004). In studies conducted in East Asians, the atherogenic index, which reflects the LDL-C/HDL-C ratio in the blood, was higher in GG genotype carriers. A high LDL-C content and an elevated atherogenic index increase the risk of atherosclerosis and cardiovascular disease (Jia et al. 2010).
The A–3826G SNP is associated with the development of microvascular complications in diabetes mellitus. In the Chinese population, the G allele was associated with an increased risk of proliferative diabetic retinopathy in patients with T2D (Zhang et al. 2015). However, no association of the SNP with microvascular complications of diabetes was found in a study involving a European sample (Rudofsky et al. 2007). On the contrary, the G allele was recently reported to act as a protective factor against the development of retinopathy in residents of central Italy suffering from T2D (Montesanto et al. 2018).
No significant association with overweight or diabetes was found in studies of the A–3826G polymorphism in the Danish (Urhammer et al. 1997), Swedish (Gagnon et al. 1998; Mottagui-Tabar et al. 2007), German (Schäffler et al. 1999), and Polish (Kieć-Wilk et al. 2002; Malczewska-Malec et al. 2004) populations.
Mutations in the Ucp1 gene can impare the main BAT function, that is, nonshivering thermogenesis. Healthy children with the GG genotype showed a lowere capacity of the thermic effect of a high fat meal (P < 0.05) (Nagai et al. 2003) and lower capacity for thermogenesis after exposure to cold (Nagai et al. 2007) as compared with AA and AG genotype carriers in the Japanese population. Accumulation of visceral fat, which is one of the main risk factors for cardiovascular diseases, may also be associated with a decrease in BAT activity. Nakayama et al. studied associations of the A–3826G polymorphism with visceral fat accumulation during cold and hot seasons. An association of the G allele with increased visceral fat (P = 0.0198) was only observed in the cold months of the year (Nakayama et al. 2013). Such observations confirm the fact that BAT becomes metabolically more active in adults after cold exposure and is detected in winter more frequently than in summer in humans (Saito et al. 2009). The A/G substitution is in the regulatory region of the gene and may decrease Ucp1 transcription, thus impairing nonshivering thermogenesis. G allele carriers are consequently more prone to visceral obesity in winter. Nonshivering thermogenesis merely contributes to energy expenditure during the hot season, and no association between the mutation and the amount of fat in the abdominal cavity was therefore detected in summer (Nakayama et al. 2013).
Inconsistent results of Ucp1 A–3826G studies may partly be explained by the differences in climatic conditions in which they were carried out. As mentioned above, there is evidence that active BAT is more frequently observed in cold seasons in humans (van Marken Lichtenbelt et al. 2009; Saito et al. 2009), yet the season was not taken into consideration in most of the association studies of Ucp1 polymorphisms. There is no doubt that weather and climate play a major role in human health. A worldwide population analysis showed increased rates of coronary events during increased cold periods in warm climates (Barnett et al. 2005). Populations of the coldest regions (Northern Sweden, North Karelia, and Kuopio) demonstrated little changes in coronary event rates with changes in temperatures (Barnett et al. 2005). This trend was supported by findings from a study in Siberia, where mortality from ischaemic heart disease did not rise with the decreasing temperature. In contrast, death rates rose as temperature fell below 18 °C in western Europe. This could be explained in part by genetic and lifetime adaptations to cold (Donaldson et al. 1998).
Ethnic differences in the allele frequencies of the polymorphism are also important to consider. A population analysis showed that the A allele is more frequent at high latitudes and that its frequency inversely correlates with the level of solar radiation. Of 52 populations studied, the lowest frequency of the A allele was observed in Africa (Hancock et al. 2011). Thus, environmental factors (climate, geographical location, dietary habbits, and daily activity) could modify the association between the Ucp1 polymorphisms and CMDs and contribute to the differences in association (Flouris et al. 2017). Moreover, the substitutions may alter the genotypic response to environmental influences by activating different molecular factors and pathways, thus modifying the risk for the disease (Slavich and Cole 2013).
A variation in haplotypes containing the A–3826G polymorphism and other SNPs of the Ucp genes can also affect the study results. Shin et al. studied three haplotypes containing the –3826G allelic variant in a group of Korean obese women. Both positive and negative correlations with obesity were observed with different haplotypes (Shin et al. 2005). It is interesting to note that the –3826G allele contained in different haplotypes with Ucp1, Ucp2 and Ucp3 polymorphisms reduced the risk of hypertension, while the A allele increased the probability of disease development in a Chinese population (Sun et al. 2018).
A–1766G polymorphism of the Ucp1 gene
The substitution of A for G at nucleotide position − 1766 (rs3811791) is another polymorphism in the upstream region of the Ucp1 gene (Fig. 2). The SNP was first discovered by Korean researchers in 2004. The SNP is upstream of the promoter region of the gene and is about 2 kb closer to the transcription start site than the A–3826G SNP described earlier (Kim et al. 2005). The –1766G allelic variant was associated with several obesity parameters, such as the waist–hip ratio (WHR) (P = 0.008), body fat mass (P = 0.023), body fat percentage (P = 0.014), and abdominal fat amount in the same study. The interaction of the A–3826G and A–1766G polymorphisms was additionally considered in the Korean population. Patients with the haplotype [−3826G/−1766G] had the highest body fat percentage, while the haplotype [−3826G/−1766A] was associated with lower body fat (Kim et al. 2005). The data from the study were consistent with the results obtained by Shin et al. The –1766G allele showed a significant association with high values of the WHR, fat mass, body fat percentage, and abdominal fat amount in the Korean population (Shin et al. 2005). In Saudi Arabia, the association of the A–1766G substitution with obesity was detected after dividing the sample by BMI into subgroups with grade II and III obesity. A significant increase in –1766G allele frequency was observed in the subgroup with grade II obesity (BMI = 35–39.9) as compared with a control cohort [OR = 2.89, 95% CI 1.33–6.25; P = 0.007] (Chathoth et al. 2018). In a recent study conducted in China, the polymorphism was associated with a significantly higher risk of T2D (adjusted OR = 1.42, 95% CI: 1.01–1.99, P = 0.042) and higher TG levels (β = 0.048, P = 0.034) under the recessive model (Dong et al. 2020).
A–112C polymorphism of the Ucp1 gene
Phenotypic manifestations were much less studied for the A–112C polymorphism (rs10011540) (Table 1). It is known that the A–112C polymorphism affects the consensus motif of the (T(G/A)TTT(T/G)(G/T)) insulin response element (IRS). The mutant С allele reduced the activity of the Ucp1 gene promoter in in vitro experiments, confirming the involvement of the IRS in regulating transcription (Mori et al. 2001). Insulin was shown to induce Ucp1 expression (Teruel et al. 1998; Valverde et al. 2003). It is therefore possible to assume that the A–112C substitution weakens IRS affinity for transcription factors, thus reducing the activity of the Ucp1 promoter.
There are data on the association of the A–112C polymorphism with insulin resistance parameters in T2D patients. The C allele was more frequent in patients with T2D than in a control group in the Japanese population (P = 0.017) (Mori et al. 2001). Another group of Japanese researchers showed that, among T2D patients, carriers of the –112C allelic variant had higher levels of fasting insulin resistance index (FIRI) (P = 0.0085), homeostasis model assessment (HOMA) index (P = 0.0089), and hepatic lipid content (HLC) (P = 0.012). In addition, the –112C allele was in linkage disequilibrium with the –3826G variant. However, no statistically significant association was detected between –112C and BMI or visceral fat content (Fukuyama et al. 2006).
In the Indian population, CC genotype carriers had a significantly higher systolic blood pressure as compared with carriers of the AA genotype (126 ± 20 mmHg vs. 119 ± 17 mmHg, P = 0.05). The A–112C polymorphism was not associated with T2D in the study, but acted as part of the haplotype [−3826A/−112C/229Met] to increase the risk of T2D in Indians (Vimaleswaran et al. 2009). In a Chinese population, the –112C allelic variant was identified as a risk factor for diabetic retinopathy (OR = 1.368 P = 0.004) (Jin et al. 2020).
Polymorphisms of the coding region of the Ucp1 gene
A number of polymorphisms in exons of the gene can also be attributed to risk factors for cardiovascular diseases and metabolic disorders (Table 1). The SNP rs45539933 is a substitution of G for A at nucleotide position + 1068 and results in a substitution of Ala for Thr at amino acid position 64 (Fig. 2). The SNP was detected in exon 2 of the Ucp1 gene by Hamann et al. The frequency of the mutant allele in a group of obese adults and children was two times higher than in a control group in the German population (8.2 vs. 4.1%, nominal P = 0.029, corrected P = 0.058) (Hamann et al. 1998). The Ala64Thr polymorphism was associated with the WHR in Caucasians. Carriers of the 64Thr allele had higher waist circumference values and, consequently, higher WHRs (P = 0.003) (Herrmann et al. 2003). On the contrary, the 64Thr allele was associated with lower fatness, lower WHR, and lower fat mass in a study involving Korean female patients (Shin et al. 2005). Carriage of the 64Thr allele reduced the risk of retinopathy in T2D patients from the Italian population (Montesanto et al. 2018).
It is difficult to provide a clear explanation for the discrepancies observed in the results at the moment. Ethnic differences in SNP allele frequencies are necessary to take into account when studying different populations; e.g., the 64Thr allele in Koreans was less frequent than in Caucasians. Gender differences between samples are also of importance; e.g., only women were included in the study by Korean authors. Because obesity and diabetes are multifactorial in nature, the diet can distort the results of studies on the association between polymorphisms and obesity (Luan et al. 2001). Different dietary patterns of Caucasians and Asians might contribute to the inconsistency of research results.
Hamann et.al. detected three other point mutations in the coding region of the Ucp1 gene: Arg40Trp (rs573078239) in exon 1 and Lys257Arg (rs1431343082) and Met229Leu (rs2270565) in exon 5, but none of them showed an association with the development of obesity (Hamann et al. 1998). The Ala64Thr and Met229Leu substitutions did not correlate with BMI in Caucasians of Danish ancestry. The mutant alleles 64Thr and 229Leu are found in the amino acid sequences of UCP1 in rodents (hamsters and rats) and rabbits, whose BAT plays a more significant metabolic role than in humans, and are evolutionarily conserved (Urhammer et al. 1997). The fact may indicate a functional significance of the respective protein sites. Further studies showed that the Met229Leu substitution was more frequent in T2D patients than in healthy people from the Japanese population. In addition, Met229Leu was in linkage disequilibrium with the A–112C substitution (Mori et al. 2001). However, the association of the polymorphism with the disease was not confirmed in T2D patients from India. Yet the frequency of the haplotype [−3826A/−112C/229Met] was significantly higher in the T2D patients (Vimaleswaran et al. 2009).
The effects of polymorphisms of the other Ucp family genes on metabolism
The Ucp1 homologues, such as Ucp2 and Ucp3 also deserve attention among candidate genes whose polymorphisms may be associated with obesity and risk factors for cardiovascular diseases. Ucp2 and Ucp3 are in the q13 region of chromosome 11 and are 60% similar to Ucp1. The genes coding for UCP2 and UCP3 are involved in regulating free fatty acid metabolism and prevent the formation of reactive oxygen species, protecting cells from oxidative stress (Bondareva et al. 2018).
Obesity and T2D are often accompanied by the formation of reactive oxygen species in mitochondria and the accumulation of lipids in cardiomyocytes, skeletal muscle cells, and hepatocytes. Animal studies showed that lipids accumulated in cardiomyocytes exert cytotoxic effects and can lead to heart failure. An adaptive mechanism based on increased expression of Ucp2 and Ucp3 may be activated in some tissues, including the myocardium, to prevent oxidative stress and lipid toxicity. Being in excess, UCP2 and UCP3 neutralize the harmful effects of reactive oxygen species and reduce the accumulation of long-chain fatty acids, thus preventing cell death (Cadenas 2018; Ruiz-Ramírez et al. 2016).
UCP2 was shown to exert an anti-atherogenic effect on vessel walls (Blanc et al. 2003), thus improving tolerance to coronary heart disease (Cheurfa et al. 2008; McLeod et al. 2005), and to protect cardiomyocytes from death due to oxidative stress (Teshima et al. 2003). Most studies of the Ucp2 and Ucp3 polymorphisms focus on the SNPs that are in the Ucp2 upstream region (G–866A (rs659366)), Ucp2 exon 4 (Ala55Val (rs660339)), and the Ucp3 promoter region (C–55 T (rs1800849)). These polymorphisms may be associated with T2D, obesity, fat metabolism, and mRNA expression (Wang et al. 2004).
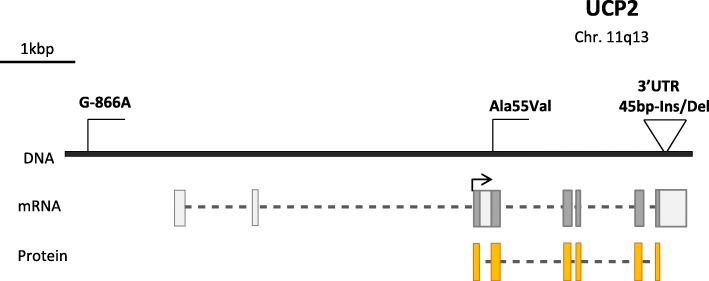
G–866A polymorphism of the Ucp2 gene
The G–866A polymorphism is upstream of the promoter of the Ucp2 gene (Fig. 3) and affects the multifunctional cis-regulatory element that is responsible for the tissue-specific binding of various nuclear proteins (Esterbauer et al. 2001). The location of the polymorphism determines its influence on Ucp2 expression. Studies showed that the minor A allele of the G–866A polymorphism increases Ucp2 expression in vivo and enhances gene transcription in experiments with cell lines. A study of the G–866A substitution in obese and normal-weight patients showed that the A allele reduces the risk of obesity in middle-aged humans (Esterbauer et al. 2001). At the same time, such an association was not detected in children and adolescents (Le Fur et al. 2004; Oguzkan-Balci et al. 2013).
Fig. 3.
Schematic structure of the Ucp2 gene and its best-studied polymorphisms
Increased Ucp2 expression leads to a reduced ATP synthesis, which is among the factors that decrease insulin secretion and increase the risk of T2D. For example, healthy –866A allele carriers showed a decreased response to insulin when administered with glucose (D’Adamo et al. 2004; Krempler et al. 2002; Sasahara et al. 2004). A glucose tolerance test showed a decreased level of insulin secretion in –866A allele carriers with T2D (Gomathi et al. 2019), and insulin therapy was required for them more often and prescribed at earlier stages of the disease as compared with –866G allele carriers (D’Adamo et al. 2004). The –866A allele was associated with T2D in women aged 40 to 65 years (OR = 2.18, P < 0.05) in a study by Russian authors (Lapik et al. 2015). However, the opposite was demonstrated in several studies; e.g., the –866A allele was associated with reduced risk of T2D in the Italian population (Bulotta et al. 2005) and a decreased insulin sensitivity was detected in the GG genotype carriers (P = 0.05) in the Danish population (Andersen et al. 2013). The –866A allele was associated with a decrease in the conductivity of the peripheral nervous system in patients with T2D (Hiroshi et al. 2006) and was found to reduce the risk of diabetic neuropathy in type 1 diabetes (Rudofsky et al. 2006).
The G allele of the G–866A polymorphism is associated with reduced mRNA expression (Esterbauer et al. 2001; Sasahara et al. 2004), high BMI values, increased body fat mass (Yoon et al. 2007), high risk of obesity, and decreased risk of T2D (Krempler et al. 2002). Increased levels of triglycerides, total cholesterol, and LDL-C in the blood in –866G allele carriers were more frequent than in subjects carrying the –866A allele (Reis et al. 2004). On the other hand, the –866A allele was associated with obesity (P = 0.003) and hyperinsulinemia in the Indian population (Srivastava et al. 2010). The data are consistent with the results of a study by Wang et al., who showed that AA genotype carriers had decreased Ucp2 mRNA levels relative to GG carriers (Wang et al. 2004). A study in Saudi Arabia showed that subjects with the AA and AG genotypes of the G–866A polymorphism were more prone to have nonalcoholic fatty liver disease (Mohseni et al. 2017).
Currently available data on the contribution of the G–866A polymorphism to the development of obesity and T2D are rather contradictory. However, the results of independent meta-analyses are generally consistent with each other. The –866A allele was shown to reduce the risk of obesity in European populations, but this association was not observed in Asians (Brondani et al. 2014b; Liu et al. 2013; Zhang et al. 2014a). Meta-analyses of studies of T2D did not reveal any association between the G–866A polymorphism and the disease among either Europeans and Asians (Qin et al. 2013; de Souza et al. 2013; Xu et al. 2011).
Because Ucp2 is widely expressed in various tissue types, association of its polymorphisms are not limited to obesity and diabetes. As already mentioned, UCP2 reduces the production of reactive oxygen species, which are one of the main sources of oxidative stress and, as a consequence, may cause cell damage and death. Oxidative stress plays an important role in the pathogenesis of atherosclerosis and coronary heart disease. Studies associated the G–688A genetic substitution with cardiovascular pathologies. The –866A allelic variant was associated with reduced risk of coronary artery disease in French men with T2D (Cheurfa et al. 2008). AA genotype carriers who survived ischemic stroke showed a higher functional status as compared with GG and GA genotype carriers (Díaz-Maroto Cicuéndez et al. 2017). At the same time, the A allele increased the risk of developing coronary artery disease in the Mexican population (Gamboa et al. 2018) and in males of the European population (Dhamrait et al. 2004).
Ala55Val polymorphism in exon 4 and Ins/Del 45 bp in the 3’UTR of the Ucp2 gene
The Ala55Val polymorphism is in Ucp2 exon 4, where a C/T nucleotide substitution leads to a substitution of alanine for valine at position 55 of the UCP2 amino acid sequence (Fig. 3). The results of studies of this polymorphism do not allow a clear conclusion about its association with obesity, T2D, and related metabolic disorders. A number of studies showed that Val/Val genotype carriers are at higher risk of T2D and obesity and have a higher BMI as compared with Ala/Val or Ala/Ala genotype carriers (Surniyantoro et al. 2018; Xiu et al. 2004; Yu et al. 2005). However, other studies found no association between the Ala55Val polymorphism and BMI, obesity, metabolic syndrome, T2D, and insulin secretion (Rosmond et al. 2002; Surniyantoro et al. 2018).
According to the results of meta-analyses (Brondani et al. 2014a; de Souza et al. 2011; Xu et al. 2011; Zhang et al. 2014a), the 55Val allele was more often associated with diseases in Asian populations.
The Ins/Del 45 bp polymorphism is in the 3’UTR of Ucp2 exon 8 and is 158 bp downstream of the stop codon (Fig. 3). The insertion was shown to disturb the stability of the Ucp2 mRNA in vitro (Esterbauer et al. 2001); i.e., the polymorphism is possibly of functional significance. However, this effect was not observed in vivo (Walder et al. 1998). The Ins allele frequency in obese patients was found to be significantly higher than in subjects with normal body weights. Ins allele carriers had higher BMI values, which increased the risk of obesity when the genome contained the Ins allele of Ucp2 (Lee et al. 2008; Marti et al. 2004; Yanovski et al. 2000). The Ins/Ins genotype was associated with higher metabolic rates (P = 0.038), higher energy consumption, and lower BMI values in the Indian population (Walder et al. 1998). Del/Del genotype carriers were more efficient in reducing weight as compared with Ins/Del and Ins/Ins genotype carriers in the Japanese population (Mutombo et al. 2012). On the contrary, a weight loss program was more effective for Ins/Del and Ins/Ins genotype carriers in Caucasians of the Greek population (Papazoglou et al. 2012). Other studies found neither positive nor negative association between the Ins/Del polymorphism and changes in weight and BMI (Berentzen et al. 2005; Dalgaard et al. 1999; Maestrini et al. 2003; Papazoglou et al. 2012; Yiew et al. 2010).
The question of the biological significance of the Ins/Del polymorphism remains open today. It is of interest to study the effect of the insertion on the functionality of UCP2.
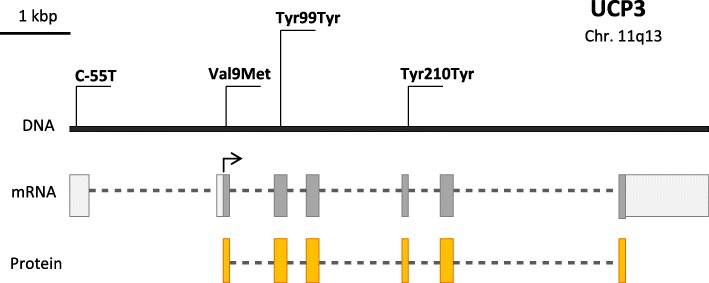
Ucp3 gene polymorphisms
The most detailed data are available for the C/T nucleotide substitution at position − 55 (Fig. 4), which is in the Ucp3 promoter region and is close to the presumed TATA box and 4 bp downstream of the presumed PPAR response element (Acín et al. 1999). The SNP location suggests an effect on transcription of the Ucp3 gene.
Fig. 4.
Schematic structure of the Ucp3 gene and its best-studied polymorphisms
Schrauwen et al. associated the − 55 T allele with increased expression of the Ucp3 mRNA in skeletal muscles (Schrauwen et al. 1999b). Ucp3 mRNA expression in muscle cells positively correlated with the resting metabolic rate (Schrauwen et al. 1999a). In Europeans and the Japanese, − 55 T allele carriers had lower BMIs (Halsall et al. 2001; Hamada et al. 2008; Lapice et al. 2014; Liu et al. 2005) and higher HDL-C levels (Hamada et al. 2008). The polymorphism inversely correlated with waist circumference in Mexican adolescents (Muñoz et al. 2017). However, opposite results were obtained in other studies. The BMI was higher in carriers of the − 55 T allele in Scandinavians (Lindholm et al. 2004). TT carriers showed higher BMIs in both obese patients and healthy controls in a study by French authors (Otabe et al. 2000). The results of a meta-analysis did not confirm the association between the polymorphism and the risk of obesity in Europeans or Asians (Qian et al. 2013). A study of the Chinese population similarly found no association between the polymorphism and obesity (Cai et al. 2017).
Contradictory results are similarly available for the association between the C–55 T substitution and the risk of T2D in different populations. The risk of T2D in − 55 T allele carriers was lower than in CC genotype carriers in the Brazilian (Schnor et al. 2017) and French (Meirhaeghe et al. 2000) populations, while no significant association with prediabetes was detected in the Chinese population (Su et al. 2018). Data from independent meta-analyses agree that the association of the C–55 T polymorphism with T2D is observed only in Asians (de Souza et al. 2013; Xu et al. 2011).
Less is known for the polymorphisms Val9Met (GTG/ATG) of exon 2, Tyr99Tyr (TAT/TAC) of exon 3, and Tyr210Tyr (TAC/TAT) of exon 5 of the Ucp3 gene (Fig. 4). The TT genotype of the Tyr99Tyr polymorphism was weakly associated with diabetes, but not with obesity, in the French population (Otabe et al. 1999). A nearly statistically significant association of Tyr99Tyr with BMI and body fat mass was revealed by Lanouette et al. (Lanouette et al. 2002). The mutant alleles of the Tyr99Tyr and Tyr210Tyr polymorphisms were associated with high caloric intake and lean mass, and Tyr99Tyr was found to be in linkage disequilibrium with the C–55 T substitution (Damcott et al. 2004).
A population-based study of the Ucp3 C–55 T and Ucp2 Ins/Del polymorphisms gave rise to a hypothesis about the role of the UCP family proteins in the regulation of angiotensin-converting enzyme (ACE). ACE is a component of the renin-angiotensin system, and acts to convert inactive angiotensin I into the active form angiotensin II, which causes constriction of blood vessels and thus increases the blood pressure. ACE is a target for a class of modern drugs that reduce the blood pressure by inhibiting the ACE molecule and thus blocking the formation of angiotensin II. A study conducted in patients with diabetes from Scandinavia and healthy UK residents revealed a link between the allelic variants of Ucp3 –55C and Ucp2 Ins and a higher level of ACE activity in the blood plasma. At the same time, a sixfold decrease in the activity of the Ucp2 mRNA as a result of RNA interference was found to cause a parallel increase in ACE gene expression. On a more global scale, this hypothesis may lead to a revision of views on the ratio of extracellular and intracellular mechanisms of metabolic regulation. The applied medical value of such a mechanism may also be great. According to the authors of the article, these facts may provide direction for new research investigating the interface between local cellular metabolism and endocrine metabolic regulation (Dhamrait et al. 2016).
Conclusions
A growing number of studies address the polymorphic variations in the Ucp gene family, and a major part of them focus on several polymorphisms of the Ucp1 gene. The data sets concerning their association with CMD risk are largely mosaic and contradictory. An association of the polymorphisms with CMDs was not found in some European populations. However, most studies showed an associations with certain metabolic disorders. The discrepancy can be explained by small samples, ethnic differences, different climatic conditions in which the research was conducted, and differences in age and lifestyle. Because Ucp mutations are associated with disease and current research portrays conflicting findings, we need more thoughtfuly and carefully verify the studies in this field. In recent years, polymorphisms of the Ucp2 and Ucp3 genes have been increasingly included in the genetic factors to be analyzed. At the same time, the biological role of their genetic variations has not been fully understood or has been described based on only few in vitro studies. The lack of research on the biological role of genetic variations in the Ucp family largely influences the prognostic power in CMDs. In addition, data on the regulation of Ucp gene expression, signaling pathways and bioactive molecules involved in regulating the Ucp genes, and the role of polymorphisms in regulating the gene functions have just begun to accumulate. The greatest effect on gene expression is known for the polymorphisms of the regulatory gene regions, such as A–3826G in the 5′ noncoding region of Ucp1, G–866A in Ucp2, and C–55 T in the promoter of Ucp3. Many studies have associated these polymorphisms with the level of Ucp1–Ucp3 gene expression, increased BMI, susceptibility to T2D, higher blood lipid levels, and elevated blood pressure. In some cases, the polymorphisms show an association with a disease only when considered together as components of a haplotype. Despite the great interest in the genes of the Ucp family, such studies have not been conducted at all in a number of Eastern European countries. Further research on the polymorphisms of the Ucp genes in representative samples of different ethnic groups and the accumulation of data on the biological role of genetic variations could have possible applications, such as treatment of metabolic syndrome.
Acknowledgments
We thank Dr. Andreas Flouris of the University of Thessaly for shared vision and discussion of the manuscript.
Authors’ contributions
Anna E. Pravednikova, Sergey Y. Shevchenko, Svetlana N. Larina, Zaur M. Kachaev, Alexander D. Egorov, Yulii V. Shidlovskii, − manuscript preparation, writing the initial draft, Victor V. Kerchev, Manana R. Skhirtladze – data presentation, Yulii V. Shidlovskii – acquisition of the financial support for the paper. The author(s) read and approved the final manuscript.
Funding
This work was supported by a grant from the Russian Foundation for Basic Research (project no.18–34-20065).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Anna E. Pravednikova, Email: pravednikova.anya@yandex.ru
Sergey Y. Shevchenko, Email: simurg87@list.ru
Victor V. Kerchev, Email: vi_dok@citydom.ru
Manana R. Skhirtladze, Email: lagerind.igb.ras@gmail.com
Svetlana N. Larina, Email: snlarina07@yandex.ru
Zaur M. Kachaev, Email: k-z-m@mail.ru
Alexander D. Egorov, Email: egorov.alek@gmail.com
Yulii V. Shidlovskii, Email: yul.biogen@gmail.com
References
- Acín A, Rodriguez M, Rique H, Canet E, Boutin JA, Galizzi JP. Cloning and characterization of the 5′ flanking region of the human uncoupling protein 3 (UCP3) gene. Biochem Biophys Res Commun. 1999;258(2):278–283. doi: 10.1006/bbrc.1999.0530. [DOI] [PubMed] [Google Scholar]
- Andersen G, Dalgaard LT, Justesen JM, Anthonsen S, Nielsen T, Thørner LW, et al. The frequent UCP2-866G>A polymorphism protects against insulin resistance and is associated with obesity: a study of obesity and related metabolic traits among 17 636 Danes. Int J Obes. 2013;37(2):175–181. doi: 10.1038/ijo.2012.22. [DOI] [PubMed] [Google Scholar]
- Angueira AR, Shapira SN, Ishibashi J, Lazar MA, Lim H-W, Seale P. Early B cell factor activity controls developmental and adaptive thermogenic gene programming in adipocytes. Cell Rep. 2020;30(9):2869–2878. doi: 10.1016/j.celrep.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au-Yong ITH, Thorn N, Ganatra R, Perkins AC, Symonds ME. Brown adipose tissue and seasonal variation in humans. Diabetes. 2009;58(11):2583–2587. doi: 10.2337/db09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbatelli G, Murano I, Madsen L, Hao Q, Jimenez M, Kristiansen K, et al. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am J Physiol - Endocrinol Metab. 2010;298(6):E1244–E1253. doi: 10.1152/ajpendo.00600.2009. [DOI] [PubMed] [Google Scholar]
- Barnett AG, Dobson AJ, McElduff P, Salomaa V, Kuulasmaa K, Sans S. Cold periods and coronary events: an analysis of populations worldwide. J Epidemiol Community Health. 2005;59(7):551–557. doi: 10.1136/jech.2004.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelt A, Heeren J. Adipose tissue browning and metabolic health. Nat Rev Endocrinol. 2014;10(1):24–36. doi: 10.1038/nrendo.2013.204. [DOI] [PubMed] [Google Scholar]
- Berentzen T, Dalgaard LT, Petersen L, Pedersen O, Sørensen TIA. Interactions between physical activity and variants of the genes encoding uncoupling proteins −2 and −3 in relation to body weight changes during a 10-y follow-up. Int J Obes. 2005;29(1):93–99. doi: 10.1038/sj.ijo.0802841. [DOI] [PubMed] [Google Scholar]
- Blanc J, Alves-Guerra MC, Esposito B, Rousset S, Gourdy P, Ricquier D, et al. Protective role of uncoupling protein 2 in atherosclerosis. Circulation. 2003;107(3):388–390. doi: 10.1161/01.cir.0000051722.66074.60. [DOI] [PubMed] [Google Scholar]
- Bondareva EA, Parfenteva OI, Kozlov AV, Zhuravleva US, Kosyakova EV, Karelina EE, et al. The Ala/Val polymorphism of the UCP2 gene is reciprocally associated with aerobic and anaerobic performance in athletes. Hum Physiol. 2018;44(6):673–678. [Google Scholar]
- Boss O, Samec S, Paoloni-Giacobino A, Rossier C, Dulloo A, Seydoux J, et al. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408(1):39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- Bouillaud F, Villarroya F, Hentz E, Raimbault S, Cassard A-M, Ricquier D. Detection of brown adipose tissue uncoupling protein mRNA in adult patients by a human genomic probe. Clin Sci. 1988;75(1):21–27. doi: 10.1042/cs0750021. [DOI] [PubMed] [Google Scholar]
- Bracale R, Labruna G, Finelli C, Daniele A, Sacchetti L, Oriani G, et al. The absence of polymorphisms in ADRB3, UCP1, PPARγ, and ADIPOQ genes protects morbid obese patients toward insulin resistance. J Endocrinol Investig. 2012;35(1):2–4. doi: 10.1007/BF03345413. [DOI] [PubMed] [Google Scholar]
- Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, et al. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic Biol Med. 2004;37(6):755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- Brondani LA, Assmann TS, de Souza BM, Bouças AP, Canani LH, Crispim D. Meta-analysis reveals the association of common variants in the uncoupling protein (UCP) 1-3 genes with body mass index variability. PLoS One. 2014;9(5):e96411. doi: 10.1371/journal.pone.0096411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondani LA, de Souza BM, Assmann TS, Bouças AP, Bauer AC, Canani LH, et al. Association of the UCP polymorphisms with susceptibility to obesity: case-control study and meta-analysis. Mol Biol Rep. 2014;41(8):5053–5067. doi: 10.1007/s11033-014-3371-7. [DOI] [PubMed] [Google Scholar]
- Brondani LDA, Assmann TS, Duarte GCK, Gross JL, Canani LH, Crispim D. The role of the uncoupling protein 1 (UCP1) on the development of obesity and type 2 diabetes mellitus. Arq Bras Endocrinol Metabol. 2012;56(4):215–225. doi: 10.1590/s0004-27302012000400001. [DOI] [PubMed] [Google Scholar]
- Bulotta A, Ludovico O, Coco A, Di Paola R, Quattrone A, Carella M, et al. The common -866G/A polymorphism in the promoter region of the UCP-2 gene is associated with reduced risk of type 2 diabetes in Caucasians from Italy. J Clin Endocrinol Metab. 2005;90(2):1176–1180. doi: 10.1210/jc.2004-1072. [DOI] [PubMed] [Google Scholar]
- Cadenas S. Mitochondrial uncoupling, ROS generation and cardioprotection. BBA - Bioenerg. 2018;1859(9):940–950. doi: 10.1016/j.bbabio.2018.05.019. [DOI] [PubMed] [Google Scholar]
- Cai W, Mao L, Shi Y, Chen J, Wang L, Cai W. Association between peroxisome proliferator-activated receptor, UCP3 and lipoprotein lipase gene polymorphisms and obesity in Chinese adolescents. Obes Res Clin Pract. 2017;11(1):27–33. doi: 10.1016/j.orcp.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Neither fat nor flesh. Nature. 2008;454(7207):947–948. doi: 10.1038/454947a. [DOI] [PubMed] [Google Scholar]
- Cassard A-M, Bouillaud F, Mattei M-G, Hentz E, Raimbault S, Thomas M, et al. Human uncoupling protein gene: structure, comparison with rat gene, and assignment to the long arm of chromosome 4. J Cell Biochem. 1990;43(3):255–264. doi: 10.1002/jcb.240430306. [DOI] [PubMed] [Google Scholar]
- Chathoth S, Ismail MH, Vatte C, Cyrus C, Al AZ, Ahmed KA, et al. Association of uncoupling protein 1 (UCP1) gene polymorphism with obesity: a case-control study. BMC Med Genet. 2018;19(1):1–11. doi: 10.1186/s12881-018-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheurfa N, Dubois-Laforgue D, Ferrarezi DAF, Reis AF, Brenner GM, Bouche C, et al. The common-866G> a variant in the promoter of UCP2 is associated with decreased risk of coronary artery disease in type 2 diabetic men. Diabetes. 2008;57(4):1063–1068. doi: 10.2337/db07-1292. [DOI] [PubMed] [Google Scholar]
- Cinti S, Frederich RC, Zingaretti MC, De Matteis R, Flier JS, Lowell BB. Immunohistochemical localization of leptin and uncoupling protein in white and brown adipose tissue. Endocrinology. 1997;138(2):797–804. doi: 10.1210/endo.138.2.4908. [DOI] [PubMed] [Google Scholar]
- Cline GW, Vidal-Puig AJ, Dufour S, Cadman KS, Lowell BB, Shulman GI. In vivo effects of uncoupling protein-3 gene disruption on mitochondrial energy metabolism. J Biol Chem. 2001;276(23):20240–20244. doi: 10.1074/jbc.M102540200. [DOI] [PubMed] [Google Scholar]
- Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, et al. Dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156(1–2):304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Goldfine AB, Kuo FC, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1526. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Adamo M, Perego L, Cardellini M, Marini MA, Frontoni S, Andreozzi F, et al. The -866A/A genotype in the promoter of the human uncoupling protein 2 gene is associated with insulin resistance and increased risk of type 2 diabetes. Diabetes. 2004;53(7):1905–1910. doi: 10.2337/diabetes.53.7.1905. [DOI] [PubMed] [Google Scholar]
- Dalgaard LT, Sørensen TIA, Andersen T, Hansen T, Pedersen O. An untranslated insertion variant in the uncoupling protein 2 gene is not related to body mass index and changes in body weight during a 26-year follow-up in Danish Caucasian men. Diabetologia. 1999;42(12):1413–1416. doi: 10.1007/s001250051312. [DOI] [PubMed] [Google Scholar]
- Damcott CM, Feingold E, Moffett SP, Barmada MM, Marshall JA, Hamman RF, et al. Genetic variation in uncoupling protein 3 is associated with dietary intake and body composition in females. Metabolism. 2004;53(4):458–464. doi: 10.1016/j.metabol.2003.11.019. [DOI] [PubMed] [Google Scholar]
- de Souza BM, Assmann TS, Kliemann LM, Gross JL, Canani LH, Crispim D. The role of uncoupling protein 2 (UCP2) on the development of type 2 diabetes mellitus and its chronic complications. Arq Bras Endocrinol Metabol. 2011;55(4):239–248. doi: 10.1590/s0004-27302011000400001. [DOI] [PubMed] [Google Scholar]
- de Souza BM, Brondani LA, Bouças AP, Sortica DA, Kramer CK, Canani LH, et al. Associations between UCP1 -3826A/G, UCP2 -866G/A, Ala55Val and Ins/Del, and UCP3 -55C/T polymorphisms and susceptibility to type 2 diabetes mellitus: case-control study and meta-analysis. PLoS One. 2013;8(1):1–11. doi: 10.1371/journal.pone.0054259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mar Gonzalez-Barroso M, Pecqueur C, Gelly C, Sanchis D, Alves-Guerra M-C, Bouillaud F, et al. Transcriptional activation of the human UCP1 gene in a rodent cell line. J Biol Chem. 2002;275(41):31722–31732. doi: 10.1074/jbc.M001678200. [DOI] [PubMed] [Google Scholar]
- Dhall M, Chaturvedi MM, Rai U, Kapoor S. Sex-dependent effects of the UCP1 -3826 A/G polymorphism on obesity and blood pressue. Ethn Dis. 2012;22(2):181–184. [PubMed] [Google Scholar]
- Dhamrait SS, Maubaret C, Pedersen-Bjergaard U, Brull DJ, Gohlke P, Payne JR, et al. Mitochondrial uncoupling proteins regulate angiotensin-converting enzyme expression: crosstalk between cellular and endocrine metabolic regulators suggested by RNA interference and genetic studies. BioEssays. 2016;38(Suppl 1):S107–S118. doi: 10.1002/bies.201670909. [DOI] [PubMed] [Google Scholar]
- Dhamrait SS, Stephens JW, Cooper JA, Acharya J, Mani AR, Moore K, et al. Cardiovascular risk in healthy men and markers of oxidative stress in diabetic men are associated with common variation in the gene for uncoupling protein 2. Eur Heart J. 2004;25(6):468–475. doi: 10.1016/j.ehj.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Díaz-Maroto Cicuéndez I, Fernández-Díaz E, García-García J, Jordán J, Fernández-Cadenas I, Montaner J, et al. The UCP2-866G/a polymorphism could be considered as a genetic marker of different functional prognosis in ischemic stroke after recanalization. NeuroMolecular Med Springer US. 2017;19(4):571–578. doi: 10.1007/s12017-017-8470-x. [DOI] [PubMed] [Google Scholar]
- Donaldson GC, Ermakov SP, Komarov YM, McDonald CP, Keatinge WR. Cold related mortalities and protection against cold in Yakutsk, eastern Siberia: observation and interview study. Br Med J. 1998;317(7164):978–982. doi: 10.1136/bmj.317.7164.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Lv Y, Xie L, Yang R, Chen L, Zhang L, et al. Association of UCP1 polymorphisms with type 2 diabetes mellitus and their interaction with physical activity and sedentary behavior. Gene. 2020;739:144497. doi: 10.1016/j.gene.2020.144497. [DOI] [PubMed] [Google Scholar]
- Ertelt A, Barton A-K, Schmitz RR, Gehlen H. Metabolic syndrome: is equine disease comparable to what we know in humans? Endocr Connect. 2014;3(3):R81–R93. doi: 10.1530/EC-14-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterbauer H, Oberkofler H, Liu YM, Breban D, Hell E, Krempler F, et al. Uncoupling protein-1 mRNA expression in obese human subjects: the role of sequence variations at the uncoupling protein-1 gene locus. J Lipid Res. 1998;39(4):834–844. [PubMed] [Google Scholar]
- Esterbauer H, Schneitler C, Oberkofler H, Ebenbichler C, Paulweber B, Sandhofer F, et al. A common polymorphism in the promoter of UCP2 is associated with decreased risk of obesity in middle-aged humans. Nat Genet. 2001;28(2):178–183. doi: 10.1038/88911. [DOI] [PubMed] [Google Scholar]
- Fedorenko A, Lishko PV, Yuriy K. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell. 2013;71(2):233–236. doi: 10.1016/j.cell.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin BS, Memetimin H, Confides AL, Kasza I, Zhu B, Vekaria HJ, et al. Human adipose beiging in response to cold and mirabegron. JCI insight. 2018;3(15):1–15. doi: 10.1172/jci.insight.121510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisler JS, Warden CH. Uncoupling proteins, dietary fat and the metabolic syndrome. Nutr Metab. 2006;3:38. doi: 10.1186/1743-7075-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouris AD, Shidlovskii YV, Shaposhnikov AV, Yepiskoposyan L, Nadolnik L, Karabon L, et al. Role of UCP1 gene variants in interethnic differences in the development of cardio-metabolic diseases. Front Genet. 2017;8:7. doi: 10.3389/fgene.2017.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogelholm M, Valve R, Kukkonen-Harjula K, Nenonen A, Hakkarainen V, Laakso M, et al. Additive effects of the mutations in the beta3-adrenergic receptor and uncoupling protein-1 genes on weight loss and weight maintenance in Finnish women. J Clin Endocrinol Metab. 1998;83(12):4246–4250. doi: 10.1210/jcem.83.12.5339. [DOI] [PubMed] [Google Scholar]
- Forga L, Corbalán M, Marti A, Fuentes C, Martínez-González MA, Martínez A. Influence of the polymorphism −3826 A → G in the UCP1 gene on the components of metabolic syndrome. An Sist Sanit Navar. 2003;26(2):231–236. doi: 10.23938/ASSN.0449. [DOI] [PubMed] [Google Scholar]
- Franco-Hincapié L, Duque CE, Parra MV, Gallego N, Villegas A, Ruiz-Linares A, et al. Association between polymorphism in uncoupling proteins and type 2 diabetes in a northwestern Colombian population. Biomédica. 2014;29(1):108–118. [PubMed] [Google Scholar]
- Fukuyama K, Ohara T, Hirota Y, Maeda K, Kuno S, Zenibayashi M, et al. Association of the -112A > C polymorphism of the uncoupling protein 1 gene with insulin resistance in Japanese individuals with type 2 diabetes. Biochem Biophys Res Commun. 2006;339:1212–1216. doi: 10.1016/j.bbrc.2005.11.140. [DOI] [PubMed] [Google Scholar]
- Fumeron F, Durack-Bown I, Betoulle D, Cassard-Doulcier A, Tuzet S, Bouillaud F, et al. Polymorphisms of uncoupling protein (UCP) and beta 3 adrenoreceptor genes in obese people submitted to a low calorie diet. Int J Obes Relat Metab Disord. 1996;20(12):1051–1054. [PubMed] [Google Scholar]
- Gagnon J, Lago F, Chagnon YC, Pérusse L, Näslund I, Lissner L, et al. DNA polymorphism in the uncoupling protein 1 (UCP1) gene has no effect on obesity related phenotypes in the Swedish obese subjects cohorts. Int J Obes. 1998;22(6):500–505. doi: 10.1038/sj.ijo.0800613. [DOI] [PubMed] [Google Scholar]
- Gamboa R, Huesca-Gómez C, López-Pérez V, Posadas-Sánchez R, Cardoso-Saldaña G, Medina-Urrutia A, et al. The UCP2-866G/A, Ala55Val and UCP3-55C/T polymorphisms are associated with premature coronary artery disease and cardiovascular risk factors in Mexican population. Genet Mol Biol. 2018;41(2):371–378. doi: 10.1590/1678-4685-GMB-2017-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Génin F, Nibbelink M, Galand M, Perret M, Ambid L. Brown fat and nonshivering thermogenesis in the gray mouse lemur (Microcebus murinus) Am J Physiol - Regul Integr Comp Physiol. 2003;284(3):811–818. doi: 10.1152/ajpregu.00525.2002. [DOI] [PubMed] [Google Scholar]
- Gomathi P, Samarth AP, Raj NBAJ, Sasikumar S, Murugan PS, Nallaperumal S, et al. The -866G/A polymorphism in the promoter of the UCP2 gene is associated with risk for type 2 diabetes and with decreased insulin levels. Gene. 2019;701:125–130. doi: 10.1016/j.gene.2019.03.041. [DOI] [PubMed] [Google Scholar]
- Gong DW, He Y, Karas M, Reitman M. Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, β3-adrenergic agonists, and leptin. J Biol Chem. 1997;272(39):24129–24132. doi: 10.1074/jbc.272.39.24129. [DOI] [PubMed] [Google Scholar]
- Gorski T, Mathes S, Krützfeldt J. Uncoupling protein 1 expression in adipocytes derived from skeletal muscle fibro/adipogenic progenitors is under genetic and hormonal control. J Cachexia Sarcopenia Muscle. 2018;9(2):384–399. doi: 10.1002/jcsm.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul A, Ateş Ö, Özer S, Kasap T, Ensari E, Demir O, et al. Role of the polymorphisms of uncoupling protein genes in childhood obesity and their association with obesity-related disturbances. Genet Test Mol Biomarkers. 2017;21(9):531–538. doi: 10.1089/gtmb.2017.0068. [DOI] [PubMed] [Google Scholar]
- Halsall DJ, Luan J, Saker P, Huxtable S, Farooqi IS, Keogh J, et al. Uncoupling protein 3 genetic variants in human obesity: the c-55t promoter polymorphism is negatively correlated with body mass index in a UK Caucasian population. Int J Obes Relat Metab Disord. 2001;25(4):472–477. doi: 10.1038/sj.ijo.0801584. [DOI] [PubMed] [Google Scholar]
- Hamada T, Kotani K, Fujiwara S, Sano Y, Domichi M, Tsuzaki K, et al. The common −55 C/T polymorphism in the promoter region of the uncoupling protein 3 gene reduces prevalence of obesity and elevates serum high-density lipoprotein cholesterol levels in the general Japanese population. Metabolism. 2008;57(3):410–415. doi: 10.1016/j.metabol.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Hamann A, Tafel J, Busing B, Munzberg H, Hinney A, Mayer H, et al. Analysis of the uncoupling protein-1 (UCP1) gene in obese and lean subjects: identification of four amino acid variants. Int J Obes Relat Metab Disord. 1998;22(9):939–941. doi: 10.1038/sj.ijo.0800725. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Clark VJ, Qian Y, Di Rienzo A. Population genetic analysis of the uncoupling proteins supports a role for UCP3 in human cold resistance. Mol Biol Evol. 2011;28(1):601–614. doi: 10.1093/molbev/msq228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman MJ, Hey EN, Hull D. Fat metabolism and heat production in young rabbits. J Physiol. 1969;205(1):51–59. doi: 10.1113/jphysiol.1969.sp008950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19(10):1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- Hashimoto O, Ohtsuki H, Kakizaki T, Amou K, Sato R, Doi S, et al. Brown adipose tissue in cetacean blubber. PLoS One. 2015;10(2):1–14. doi: 10.1371/journal.pone.0116734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton JM. The distribution of brown adipose tissue in the human. J Anat. 1972;112(Pt 1):35–39. [PMC free article] [PubMed] [Google Scholar]
- Heilbronn LK, Kind KL, Pancewicz E, Morris AM, Noakes M, Clifton PM. Association of ± 3826 G variant in uncoupling protein-1 with increased BMI in overweight Australian women. Diabetologia. 2000;43(2):242–244. doi: 10.1007/s001250050036. [DOI] [PubMed] [Google Scholar]
- Herrmann S-M, Wang J-G, Staessen JA, Kertmen E, Schmidt-Petersen K, Zidek W, et al. Uncoupling protein 1 and 3 polymorphisms are associated with waist-to-hip ratio. J Mol Med (Berl) 2003;81(5):327–332. doi: 10.1007/s00109-003-0431-1. [DOI] [PubMed] [Google Scholar]
- Herskind AM, McGue M, Sorensen TI, Harvald B. Sex and age specific assessment of genetic and environmental influences on body mass index in twins. Int J Obes Relat Metab Disord. 1996;20(2):106–113. [PubMed] [Google Scholar]
- Hiroshi M, Hideyyuki S, Kenichi O. Uncoupling protein 2 promoter polymorphism -866G/A affects peripheral nerve dysfunction in Japanese type 2 diabetic patients. Diabetes Care. 2006;29(4):888–894. doi: 10.2337/diacare.29.04.06.dc05-1984. [DOI] [PubMed] [Google Scholar]
- Hoang T, Smith MD, Jelokhani-Niaraki M. Expression, folding, and proton transport activity of human uncoupling protein-1 (ucp1) in lipid membranes. J Biol Chem. 2013;288(51):36244–36258. doi: 10.1074/jbc.M113.509935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen P, Hirvonen J, Kinnula V. The occurrence of brown adipose tissue in outdoor workers. Eur J Appl Physiol Occup Physiol. 1981;46(4):339–345. doi: 10.1007/BF00422121. [DOI] [PubMed] [Google Scholar]
- Iida S, Chen W, Nakadai T, Ohkuma Y, Roeder RG. PRDM16 enhances nuclear receptordependent transcription of the brown fat-specific Ucp1 gene through interactions with mediator subunit MED1. Genes Dev. 2015;29(3):308–321. doi: 10.1101/gad.252809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JJ, Tian YB, Cao ZH, Tao LL, Zhang X, Gao SZ, et al. The polymorphisms of UCP1 genes associated with fat metabolism, obesity and diabetes. Mol Biol Rep. 2010;37(3):1513–1522. doi: 10.1007/s11033-009-9550-2. [DOI] [PubMed] [Google Scholar]
- Jin P, Li Z, Xu X, He J, Chen J, Xu X, et al. Analysis of association between common variants of uncoupling proteins genes and diabetic retinopathy in a Chinese population. BMC Med Genet. 2020;21(1):1–12. doi: 10.1186/s12881-020-0956-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, et al. Initiation of myoblast/brown fat switch through a PRDM16-C/EBP-b transcriptional complex. Nature. 2009;460(7259):1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern PA, Finlin BS, Zhu B, Rasouli N, McGehee RE, Westgate PM, et al. The effects of temperature and seasons on subcutaneous white adipose tissue in humans: evidence for thermogenic gene induction. J Clin Endocrinol Metab. 2014;99(12):E2772–E2779. doi: 10.1210/jc.2014-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieć-Wilk B, Wybrańska I, Malczewska-Malec M, Leszczyńska-Gołabek L, Partyka L, Niedbał S, et al. Correlation of the −3826A >G polymorphism in the promoter of the uncoupling protein 1 gene with obesity and metabolic disorders in obese families from southern Poland. J Physiol Pharmacol. 2002;53(3):477–490. [PubMed] [Google Scholar]
- Kim KS, Cho D-Y, Kim YJ, Choi SM, Kim JY, Shin SU, et al. The finding of new genetic polymorphism of UCP-1 A-1766G and its effects on body fat accumulation. Biochim Biophys Acta. 2005;1741(1–2):149–155. doi: 10.1016/j.bbadis.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Kogure A, Yoshida T, Sakane N, Umekawa T, Takakura Y, Kondo M. Synergic effect of polymorphisms in uncoupling protein 1 and β3- adrenergic receptor genes on weight loss in obese Japanese. Diabetologia. 1998;41(11):1399. doi: 10.1007/s001250051084. [DOI] [PubMed] [Google Scholar]
- Kotani K, Sakane N, Tsuzaki K, Shimohiro H, Tabata M, Kurozawa Y. The uncoupling protein-1 gene −3826A/G polymorphism and hypertension in Japanese subjects. Clin Chem Lab Med. 2007;45(9):1186–1189. doi: 10.1515/CCLM.2007.253. [DOI] [PubMed] [Google Scholar]
- Kozak LP, Anunciado-Koza R. UCP1: its involvement and utility in obesity. Int J Obes. 2009;32(Suppl 7):32–38. doi: 10.1038/ijo.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Zhang CY, Lowell BB. A significant portion of mitochondrial proton leak in intact thymocytes depends on expression of UCP2. Proc Natl Acad Sci U S A. 2002;99(1):118–122. doi: 10.1073/pnas.012410699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempler F, Esterbauer H, Weitgasser R, Ebenbichler C, Patsch JR, Miller K, et al. A functional polymorphism in the promoter of UCP2 enhances obesity risk but reduces type 2 diabetes risk in obese middle-aged humans. Diabetes. 2002;51(11):3331–3335. doi: 10.2337/diabetes.51.11.3331. [DOI] [PubMed] [Google Scholar]
- Lanouette C-M, Chagnon YC, Rice T, Pérusse L, Muzzin P, Giacobino J-P, et al. Uncoupling protein 3 gene is associated with body composition changes with training in HERITAGE study. J Appl Physiol. 2002;92(3):1111–1118. doi: 10.1152/japplphysiol.00726.2001. [DOI] [PubMed] [Google Scholar]
- Lapice E, Monticelli A, Cocozza S, Pinelli M, Giacco A, Rivellese AA, et al. The energy intake modulates the association of the -55CT polymorphism of UCP3 with body weight in type 2 diabetic patients. Int J Obes. 2014;38(6):873–877. doi: 10.1038/ijo.2013.174. [DOI] [PubMed] [Google Scholar]
- Lapik IA, Sharafetdinov KK, Plotnikova OA, Sorokina EY, Sentsova TB, Baturin AK. Personalized therapy in patients with type 2 diabetes based molecular genetic studies. Russ Med J. 2015;23(29):32–34. [Google Scholar]
- Le Fur S, Le Stunff C, Dos Santos C, Bougnères P. The common - 866 G/A polymorphism in the promoter of uncoupling protein 2 is associated with increased carbohydrate and decreased lipid oxidation in juvenile obesity. Diabetes. 2004;53(1):235–239. doi: 10.2337/diabetes.53.1.235. [DOI] [PubMed] [Google Scholar]
- Lean MEJ, James WPT, Jennings G, Trayhurn P. Brown adipose tissue uncoupling protein content in human infants, children and adults. Clin Sci. 1986;71(3):291–297. doi: 10.1042/cs0710291. [DOI] [PubMed] [Google Scholar]
- Lee P, Swarbrick MM, Ho KKY. Brown adipose tissue in adult humans: a metabolic renaissance. Endocr Rev. 2013;34(3):413–438. doi: 10.1210/er.2012-1081. [DOI] [PubMed] [Google Scholar]
- Lee YH, Kim W, Yu BC, Park BL, Kim LH, Shin HD. Association of the ins/del polymorphisms of uncoupling protein 2 (UCP2) with BMI in a Korean population. Biochem Biophys Res Commun. 2008;371(4):767–771. doi: 10.1016/j.bbrc.2008.04.144. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang Y, Wang F, Donelan W, Zona MC, Li S, et al. Effects of irisin on the differentiation and browning of human visceral white adipocytes. Am J Transl Res. 2019;11(12):7410–21. [PMC free article] [PubMed]
- Lichtenbelt WVM. Brown adipose tissue and the regulation of nonshivering thermogenesis. Curr Opin Clin Nutr Metab Care. 2012;15(6):547–552. doi: 10.1097/MCO.0b013e3283599184. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119(1):121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lindholm E, Klannemark M, Agardh E, Groop L, Agardh CD. Putative role of polymorphisms in UCP1-3 genes for diabetic nephropathy. J Diabetes Complicat. 2004;18(2):103–107. doi: 10.1016/S1056-8727(03)00019-9. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhao X, Kang S, Zhang D. An association between -866G/A polymorphism in the promoter of UCP2 and obesity: a meta-analysis. Gene. 2013;514(1):41–47. doi: 10.1016/j.gene.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Liu Y-J, Liu P-Y, Long J, Lu Y, Elze L, Recker RR, et al. Linkage and association analyses of the UCP3 gene with obesity phenotypes in Caucasian families. Physiol Genomics. 2005;22(2):197–203. doi: 10.1152/physiolgenomics.00031.2005. [DOI] [PubMed] [Google Scholar]
- Luan J, Browne PO, Harding AH, Halsall DJ, O’Rahilly S, Chatterjee VKK, et al. Evidence for gene-nutrient interaction at the PPARγ locus. Diabetes. 2001;50(3):686–689. doi: 10.2337/diabetes.50.3.686. [DOI] [PubMed] [Google Scholar]
- Maestrini S, Podesta F, Di Blasio AM, Savia G, Brunani A, Tagliaferri A, et al. Lack of association between UCP2 gene polymorphisms and obesity phenotype in Italian Caucasians. J Endocrinol Investig. 2003;26(10):985–990. doi: 10.1007/BF03348196. [DOI] [PubMed] [Google Scholar]
- Malczewska-Malec M, Wybranska I, Leszczynska-Golabek I, Partyka L, Hartwich J. Analysis of candidate genes in polish families with obesity. Clin Chem Lab Med. 2004;42(5):487–493. doi: 10.1515/CCLM.2004.083. [DOI] [PubMed] [Google Scholar]
- Mao W, Yu XX, Zhong A, Li W, Brush J, Sherwood SW, et al. UCP4, a novel brain-specific mitochondrial protein that reduces membrane potential in mammalian cells. FEBS Lett. 1999;443(3):326–330. doi: 10.1016/s0014-5793(98)01713-x. [DOI] [PubMed] [Google Scholar]
- Marti A, Corbalán MS, Forga LL, Martinez-González MA, Martinez JA. Higher obesity risk associated with the exon-8 insertion of the UCP2 gene in a Spanish case-control study. Nutrition. 2004;20(6):498–501. doi: 10.1016/j.nut.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Matsushita H, Kurabayashi T, Tomita M, Kato N, Tanaka K. Effects of uncoupling protein 1 and β3-adrenergic receptor gene polymorphisms on body size and serum lipid concentrations in Japanese women. Maturitas. 2003;45(1):39–45. doi: 10.1016/s0378-5122(03)00088-4. [DOI] [PubMed] [Google Scholar]
- McLeod CJ, Aziz A, Hoyt RF, McCoy JP, Sack MN. Uncoupling proteins 2 and 3 function in concert to augment tolerance to cardiac ischemia. J Biol Chem. 2005;280(39):33470–33476. doi: 10.1074/jbc.M505258200. [DOI] [PubMed] [Google Scholar]
- Meirhaeghe A, Amouyel P, Helbecque N, Cottel D, Otabe S, Froguel P, et al. An uncoupling protein 3 gene polymorphism associated with a lower risk of developing type II diabetes and with atherogenic lipid profile in a French cohort. Diabetologia. 2000;43(11):1424–1428. doi: 10.1007/s001250051549. [DOI] [PubMed] [Google Scholar]
- Mohseni F, Farajnia S, Farhangi MA, Khoshbaten M, Jafarabadi M-A. Association of UCP2 –866G>A polymorphism with nonalcoholic fatty liver disease in patients from north-west of Iran. Lab Med. 2017;48(1):65–72. doi: 10.1093/labmed/lmw052. [DOI] [PubMed] [Google Scholar]
- Montesanto A, Bonfigli AR, Crocco P, Garagnani P, De M, Boemi M, et al. Genes associated with type 2 diabetes and vascular complications. Aging (Albany NY) 2018;10(2):178–196. doi: 10.18632/aging.101375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Okazawa H, Iwamoto K, Maeda E, Hashiramoto M, Kasuga M. A polymorphism in the 5′ untranslated region and a met 229->Leu variant in exon 5 of the human UCP1 gene are associated with susceptibility to type II diabetes mellitus. Diabetologia. 2001;44(3):373–376. doi: 10.1007/s001250051629. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Hoffstedt J, Brookes AJ, Jiao H, Arner P, Dahlman I. Association of ADRB1 and UCP3 gene polymorphisms with insulin sensitivity but not obesity. Horm Res. 2007;69(1):31–36. doi: 10.1159/000111793. [DOI] [PubMed] [Google Scholar]
- Muñoz AM, Velásquez CM, Agudelo GM, Uscátegui RM, Estrada A, Patiño FA, et al. Examining for an association between candidate gene polymorphisms in the metabolic syndrome components on excess weight and adiposity measures in youth: a cross-sectional study. Genes Nutr. 2017;12(1):1–11. doi: 10.1186/s12263-017-0567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutombo PB, Yamasaki M, Shiwaku K. UCP2 I/D modulated change in BMI during a lifestyle modification intervention study in Japanese subjects. Genet Test Mol Biomarkers. 2012;17(1):16–20. doi: 10.1089/gtmb.2012.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai N, Sakane N, Fujishita A, Fujiwara R, Kimura T, Kotani K, et al. The −3826 A → G variant of the uncoupling protein-1 gene diminishes thermogenesis during acute cold exposure in healthy children. Obes Res Clin Pract. 2007;1(2):99–107. doi: 10.1016/j.orcp.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Nagai N, Sakane N, Ueno LM, Hamada T, Moritani T. The −3826 A→G variant of the uncoupling protein-1 gene diminishes postprandial thermogenesis after a high fat meal in healthy boys. J Clin Endocrinol Metab. 2003;88(12):5661–5667. doi: 10.1210/jc.2003-030672. [DOI] [PubMed] [Google Scholar]
- Nakano T, Shinka T, Masako S, Yoichi S. A/G heterozygote of the A-3826G polymorphism in the UCP-1 gene has higher BMI than A/A and G/G homozygote in young Japanese males. J Med Investig. 2006;53:87–92. doi: 10.2152/jmi.53.218. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Miyashita H, Yanagisawa Y, Iwamoto S. Seasonal effects of UCP1 gene polymorphism on visceral fat accumulation in Japanese adults. PLoS One. 2013;8(9):1–6. doi: 10.1371/journal.pone.0074720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan Cassandra, Guo Boliang, Warner Claire, Fowler Tom, Barrett Timothy, Boomsma Dorret, Nelson Tracy, Whitfield Keith, Beunen Gaston, Thomis Martine, Maes Hermine Hendrik, Derom Catherine, Ordoñana Juan, Deeks Jonathan, Zeegers Maurice. Heritability of body mass index in pre-adolescence, young adulthood and late adulthood. European Journal of Epidemiology. 2012;27(4):247–253. doi: 10.1007/s10654-012-9678-6. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Cannon B. Mammalian hibernation. Philos Trans R Soc B. 1990;326(1237):669–686. doi: 10.1098/rstb.1990.0038. [DOI] [PubMed] [Google Scholar]
- Nicoletti CF, de Oliveira APRP, Brochado MJF, de Oliveira BP, Pinhel MAS, Marchini JS, et al. UCP1 -3826 A>G polymorphism affects weight, fat mass, and risk of type 2 diabetes mellitus in grade III obese patients. Nutrition. 2016;32:83–87. doi: 10.1016/j.nut.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Norheim F, Langleite TM, Hjorth M, Holen T, Kielland A, Stadheim HK, et al. The effects of acute and chronic exercise on PGC-1α, irisin and browning of subcutaneous adipose tissue in humans. FEBS J. 2014;281(3):739–749. doi: 10.1111/febs.12619. [DOI] [PubMed] [Google Scholar]
- Oguzkan-Balci S, Col-Araz N, Nacak M, Araz M, Sabanci H, Balat A, et al. Mitochondrial uncoupling protein 2 (UCP2) gene polymorphisms are associated with childhood obesity and related metabolic disorders. J Pediatr Endocrinol Metab. 2013;26(3–4):277–283. doi: 10.1515/jpem-2012-0267. [DOI] [PubMed] [Google Scholar]
- Oh HH, Kim KS, Choi SM, Yang HS, Yoon Y. The effects of uncoupling protein-1 genotype on lipoprotein cholesterol level in Korean obese subjects. Metabolism. 2004;53(8):1054–1059. doi: 10.1016/j.metabol.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Oppert J, Vohl M, Chagnon M, Dionne F, Cassard-Doulcier A, Ricquier D, et al. DNA polymorphism in the uncoupling protein (UCP) gene and human-body fat. Int J Obes. 1994;18(8):526–531. [PubMed] [Google Scholar]
- Otabe S, Clement K, Dina C, Pelloux V, Guy-Grand B, Froguel P, et al. A genetic variation in the 5′ flanking region of the UCP3 gene is associated with body mass index in humans in interaction with physical activity. Diabetologia. 2000;43(2):245–249. doi: 10.1007/s001250050037. [DOI] [PubMed] [Google Scholar]
- Otabe S, Clement K, Lepretre F, Pelloux V, Leibel R, Chung W, et al. Mutation screening and association studies of the human UCP 3 gene in normoglycemic and NIDDM morbidly obese patients. Diabetes. 1999;48:46–48. [PubMed] [Google Scholar]
- Otero-Díaz B, Rodríguez-Flores M, Sánchez-Muñoz V, Monraz-Preciado F, Ordoñez-Ortega S, Becerril-Elias V, et al. Exercise induces white adipose tissue browning across the weight Spectrum in humans. Front Physiol. 2018;9:1781. [DOI] [PMC free article] [PubMed]
- Papazoglou D, Papathanasiou P, Papanas N, Papatheodorou K, Chatziangeli E, Nikitidis I, et al. Uncoupling protein-2 45-base pair insertion/deletion polymorphism: is there an association with severe obesity and weight loss in morbidly obese subjects? Metab Syndr Relat Disord. 2012;10(4):307–311. doi: 10.1089/met.2012.0003. [DOI] [PubMed] [Google Scholar]
- Paulik MA, Buckholz RG, Lancaster ME, Dallas WS, Hull-Ryde EA, Weiel JE, et al. Development of infrared imaging to measure thermogenesis in cell culture: thermogenic effects of uncoupling protein-2, troglitazone, and β- adrenoceptor agonists. Pharm Res. 1998;15(6):944–9. [DOI] [PubMed]
- Phillips Kevin J. Beige Fat, Adaptive Thermogenesis, and Its Regulation by Exercise and Thyroid Hormone. Biology. 2019;8(3):57. doi: 10.3390/biology8030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierelli G, Stanzione R, Forte M, Migliarino S, Perelli M, Volpe M, et al. Uncoupling protein 2: a key player and a potential therapeutic target in vascular diseases. Oxid Med Cell Longev Hindawi. 2017;2017:1–11. doi: 10.1155/2017/7348372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proenza AM, Poissonnet CM, Ozata M, Ozen S, Guran S, Palou A, et al. Association of sets of alleles of genes encoding β3-adrenoreceptor, uncoupling protein 1 and lipoprotein lipase with increased risk of metabolic complications in obesity. Int J Obes. 2000;24(1):93–100. doi: 10.1038/sj.ijo.0801091. [DOI] [PubMed] [Google Scholar]
- Qian L, Xu K, Xu X, Gu R, Liu X, Shan S, et al. UCP2 -866G/A, Ala55Val and UCP3 -55C/T polymorphisms in association with obesity susceptibility - a meta-analysis study. PLoS One. 2013;8(4):e58939. doi: 10.1371/journal.pone.0058939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin LJ, Wen J, Qu YL, Huang QY. Lack of association of functional UCP2 -866G/A and Ala55Val polymorphisms and type 2 diabetes in the Chinese population based on a case-control study and a meta-analysis. Genet Mol Res. 2013;12(3):3324–3334. doi: 10.4238/2013.September.3.9. [DOI] [PubMed] [Google Scholar]
- Ramis JM, González-Sánchez JL, Proenza AM, Martínez-Larrad MT, Fernández-Pérez C, Palou A, et al. The Arg64 allele of the β 3 -adrenoceptor gene but not the -3826G allele of the uncoupling protein 1 gene is associated with increased leptin levels in the Spanish population. Metabolism. 2004;53(11):1411–1416. doi: 10.1016/j.metabol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Ramsden DB, Ho PWL, Ho JWM, Liu HF, So DHF, Tse HM, et al. Human neuronal uncoupling proteins 4 and 5 (UCP4 and UCP5): structural properties, regulation, and physiological role in protection against oxidative stress and mitochondrial dysfunction. Brain Behav. 2012;2(4):468–478. doi: 10.1002/brb3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AT. The so-called hibernating gland. J Morphol. 1923;38(1):147–205. [Google Scholar]
- Razeghi P, Young ME, Ying J, Depre C, Uray IP, Kolesar J, et al. Downregulation of metabolic gene expression in failing human heart before and after mechanical unloading. Cardiology. 2002;97(4):203–209. doi: 10.1159/000063122. [DOI] [PubMed] [Google Scholar]
- Reis AF, Dubois-Laforgue D, Bellanné-Chantelot C, Timsit J, Velho G. A polymorphism in the promoter of UCP2 gene modulates lipid levels in patients with type 2 diabetes. Mol Genet Metab. 2004;82(4):339–344. doi: 10.1016/j.ymgme.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Ricquier D. Uncoupling protein 1 of brown adipocytes, the only uncoupler: a historical perspective. Front Endocrin. 2011;2:1–7. doi: 10.3389/fendo.2011.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricquier D. UCP1, the mitochondrial uncoupling protein of brown adipocyte: a personal contribution and a historical perspective. Biochimie. 2016;134:3–8. doi: 10.1016/j.biochi.2016.10.018. [DOI] [PubMed] [Google Scholar]
- Rosenwald M, Perdikari A, Rülicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol. 2013;15(6):659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- Rosmond R, Bouchard C, Björntorp P. Lack of association between the uncoupling protein-2 Ala55Val gene polymorphism and phenotypic features of the metabolic syndrome. Biochim Biophys Acta - Mol Basis Dis. 2002;1588(2):103–105. doi: 10.1016/s0925-4439(02)00152-7. [DOI] [PubMed] [Google Scholar]
- Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced obesity. Obes Res. 1997;5(6):1–7. doi: 10.1002/j.1550-8528.1997.tb00591.x. [DOI] [PubMed] [Google Scholar]
- Rudofsky G, Schrödter A, Voron’ko OE, Schlotterer A, Humpert PM, Tafel J, et al. Promoter polymorphisms of UCP1, UCP2, and UCP3 are not associated with diabetic microvascular complications in type 2 diabetes. Horm Metab Res. 2007;39(4):306–309. doi: 10.1055/s-2007-973816. [DOI] [PubMed] [Google Scholar]
- Rudofsky G, Schroedter A, Schlotterer A, Voron’ko OE, Schlimme M, Tafel J, et al. Functional polymorphisms of UCP2 and UCP3 are associated with a reduced prevalence of diabetic neuropathy in patients with type 1 diabetes. Diabetes Care. 2006;29(1):89–94. doi: 10.2337/diacare.29.01.06.dc05-0757. [DOI] [PubMed] [Google Scholar]
- Rui L. Brown and beige adipose tissues in health and disease. Compr Physiol. 2017;7(4):1281–1306. doi: 10.1002/cphy.c170001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ramírez A, López-Acosta O, Barrios-Maya MA, El-Hafidi M. Cell death and heart failure in obesity: role of uncoupling proteins. Oxidative Med Cell Longev. 2016;2016:1–11. doi: 10.1155/2016/9340654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saely CH, Geiger K, Drexel H. Brown versus white adipose tissue: a mini-review. Gerontology. 2011;58(1):15–23. doi: 10.1159/000321319. [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58(7):1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sámano R, Huesca-gómez C, López-marure R. Association between UCP polymorphisms and adipokines with obesity in Mexican adolescents. J Pediatr Endocrinol Metab. 2018;1:1–8. doi: 10.1515/jpem-2017-0262. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Srinath CS, Breedella MA, Cypess AM, Torriani M. Imaging of brown adipose tissue: state of the art. Radiology. 2016;280(1):4–19. doi: 10.1148/radiol.2016150390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis D, Fleury C, Chomiki N, Goubern M, Huang Q, Neverova M, et al. BMCP1, a novel mitochondrial carrier with high expression in the central nervous system of humans and rodents, and respiration uncoupling activity in recombinant yeast. J Biol Chem. 1998;273(51):34611–34615. doi: 10.1074/jbc.273.51.34611. [DOI] [PubMed] [Google Scholar]
- Sasahara M, Nishi M, Kawashima H, Ueda K, Sakagashira S, Furuta H, et al. Uncoupling protein 2 promoter polymorphism - 866G/A affects ots expression in β-cells and modulates clinical profiles of Japanese type 2 diabetic patients. Diabetes. 2004;53(2):482–485. doi: 10.2337/diabetes.53.2.482. [DOI] [PubMed] [Google Scholar]
- Schäffler A, Palitzsch K-D, Watzlawek E, Drobnik W, Schwer H, Schölmerich J, et al. Frequency and significance of the A → G (−3826) polymorphism in the promoter of the gene for uncoupling protein-I with regard to metabolic parameters and adipocyte transcription factor binding in a large population-based Caucasian cohort. Eur J Clin Investig. 1999;29(9):770–779. doi: 10.1046/j.1365-2362.1999.00529.x. [DOI] [PubMed] [Google Scholar]
- Schnor NPP, Verlengia R, Novais PFS, Crisp AH, Leite CV d S, Rasera-Junior I, et al. Association of 5-HT2C (rs3813929) and UCP3 (rs1800849) gene polymorphisms with type 2 diabetes in obese women candidates for bariatric surgery. Arch Endocrinol Metab. 2017;61(4):326–331. doi: 10.1590/2359-3997000000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen P, Xia J, Bogardus C, Pratley RE, Ravussin E. Skeletal muscle uncoupling protein 3 expression is a determinant of energy expenditure in Pima Indians. Diabetes. 1999;48(September):146–149. doi: 10.2337/diabetes.48.1.146. [DOI] [PubMed] [Google Scholar]
- Schrauwen P, Xia J, Walder K, Snitker S, Ravussin E. A novel polymorphism in the proximal UCP3 promoter region: effect on skeletal muscle UCP3 mRNA expression and obesity in male nondiabetic Pima Indians. Int J Obes. 1999;23(12):1242–1245. doi: 10.1038/sj.ijo.0801057. [DOI] [PubMed] [Google Scholar]
- Scottish Intercollegiate Guidelines Network (SIGN). Risk estimation and the prevention of cardiovascular disease. Available: https://www.sign.ac.uk/sign-149-risk-estimation-and-the-prevention-of-cardiovascular-disease.html.
- Seale Patrick, Bjork Bryan, Yang Wenli, Kajimura Shingo, Chin Sherry, Kuang Shihuan, Scimè Anthony, Devarakonda Srikripa, Conroe Heather M., Erdjument-Bromage Hediye, Tempst Paul, Rudnicki Michael A., Beier David R., Spiegelman Bruce M. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121(1):53–56. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6(1):38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HD, Kil SK, Min HC, Yoon Y. The effects of UCP-1 polymorphisms on obesity phenotypes among Korean female subjects. Biochem Biophys Res Commun. 2005;335(2):624–630. doi: 10.1016/j.bbrc.2005.07.096. [DOI] [PubMed] [Google Scholar]
- Sivenius K, Valve R, Lindi V, Niskanen L, Laakso M, Uusitupa M. Synergistic effect of polymorphisms in uncoupling protein 1 and β3-adrenergic receptor genes on long-term body weight change in Finnish type 2 diabetic and non-diabetic control subjects. Int J Obes. 2000;24(4):514–519. doi: 10.1038/sj.ijo.0801194. [DOI] [PubMed] [Google Scholar]
- Slavich GM, Cole SW. The emerging field of human social genomics. Clin Psychol Sci. 2013;1(3):331–348. doi: 10.1177/2167702613478594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava N, Prakash J, Lakhan R, Agarwal CG, Pant DC, Mittal B. A common polymorphism in the promoter of UCP2 is associated with obesity and hyperinsulenemia in northern Indians. Mol Cell Biochem. 2010;337(1–2):293–298. doi: 10.1007/s11010-009-0311-2. [DOI] [PubMed] [Google Scholar]
- Stunkard AJ, Sørensen TIA, Hanis C, Teasdale TW, Chakraborty R, Schull WJ, et al. An adoption study of human obesity. N Engl J Med. 1986;314(4):193–198. doi: 10.1056/NEJM198601233140401. [DOI] [PubMed] [Google Scholar]
- Su M, Chen X, Chen Y, Wang C, Li S, Ying X, et al. UCP2 and UCP3 variants and gene-environment interaction associated with prediabetes and T2DM in a rural population: a case control study in China. BMC Med Genet. 2018;19:1–9. doi: 10.1186/s12881-018-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Zhang JT, Xie XR, Li T, Li XY, Wang NN, et al. Association of uncoupling protein gene polymorphisms with essential hypertension in a northeastern Han Chinese population. J Hum Hypertens. 2018;33:524–530. doi: 10.1038/s41371-018-0141-3. [DOI] [PubMed] [Google Scholar]
- Surniyantoro HNE, Sadewa AH, Hastuti P. Uncoupling protein 2 ( UCP2 ) as genetic risk factor for obesity in Indonesia is different in gender stratification. Kobe J Med Sci. 2018;64(2):64–72. [PMC free article] [PubMed] [Google Scholar]
- Teruel T, Valverde AM, Navarro P, Benito M, Lorenzo M. Inhibition of PI 3-kinase and RAS blocks IGF-I and insulin-induced uncoupling protein 1 gene expression in brown adipocytes. J Cell Physiol. 1998;176(1):99–109. doi: 10.1002/(SICI)1097-4652(199807)176:1<99::AID-JCP12>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Teshima Y, Akao M, Jones SP, Marbán E. Uncoupling protein-2 overexpression inhibits mitochondrial death pathway in cardiomyocytes. Circ Res. 2003;93(3):192–200. doi: 10.1161/01.RES.0000085581.60197.4D. [DOI] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3(5):333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Urhammer SA, Fridberg M, Sørensen TI, Echwald SM, Andersen T, Tybjaerg-Hansen A, et al. Studies of genetic variability of the uncoupling protein 1 gene in Caucasian subjects with juvenile-onset obesity. J Clin Endocrinol Metab. 1997;82(12):4069–4074. doi: 10.1210/jcem.82.12.4414. [DOI] [PubMed] [Google Scholar]
- Valverde AM, Arribas M, Mur C, Navarro P, Pons S, Cassard-Doulcier AM, et al. Insulin-induced up-regulated uncoupling protein-1 expression is mediated by insulin receptor substrate 1 through the phosphatidylinositol 3-kinase/Akt signaling pathway in fetal brown adipocytes. J Biol Chem. 2003;278(12):10221–10231. doi: 10.1074/jbc.M209363200. [DOI] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JMAFL, Kemerink GJ, Bouvy ND, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Vidal-Puig A, Solanes G, Grujic D, Flier JS, Lowell BB. UCP3: an uncoupling protein homologue expressed preferentially and abundantly in skeletal muscle and brown adipose tissue. Biochem Biophys Res Commun. 1997;235(1):79–82. doi: 10.1006/bbrc.1997.6740. [DOI] [PubMed] [Google Scholar]
- Vimaleswaran KS, Radha V, Ghosh S, Majumder PP, Rao MRS, Mohan V. A haplotype at the UCP1 gene locus contributes to genetic risk for type 2 diabetes in Asian Indians (CURES-72) Metab Syndr Relat Disord. 2009;8(1):63–68. doi: 10.1089/met.2009.0039. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Walder K, Norman RA, Hanson RL, Schrauwen P, Neverova M, Jenkinson CP, et al. Association between uncoupling protein polymorphisms (UCP2-UCP3) and energy metabolism/obesity in Pima Indians. Hum Mol Genet. 1998;7(9):1431–1435. doi: 10.1093/hmg/7.9.1431. [DOI] [PubMed] [Google Scholar]
- Wang H, Chu WS, Lu T, Hasstedt SJ, Kern PA, Elbein SC. Uncoupling protein-2 polymorphisms in type 2 diabetes, obesity, and insulin secretion. Am J Physiol Metab. 2004;286(1):E1–E7. doi: 10.1152/ajpendo.00231.2003. [DOI] [PubMed] [Google Scholar]
- Xiu LL, Weng JP, Sui Y, Wang J, Yan JH, Huang ZM. Common variants in beta 3-adrenergic-receptor and uncoupling protein-2 genes are associated with type 2 diabetes and obesity. Zhonghua Yi Xue Za Zhi. 2004;84(5):375–379. [PubMed] [Google Scholar]
- Xu K, Zhang M, Cui D, Fu Y, Qian L, Gu R, et al. UCP2 -866G/A and Ala55Val, and UCP3 -55C/T polymorphisms in association with type 2 diabetes susceptibility: a meta-analysis study. Diabetologia. 2011;54(9):2315–2324. doi: 10.1007/s00125-011-2245-y. [DOI] [PubMed] [Google Scholar]
- Yanovski JA, Diament AL, Sovik KN, Nguyen TT, Li H, Sebring NG, et al. Associations between uncoupling protein 2, body composition, and resting energy expenditure in lean and obese African American, white, and Asian children. Am J Clin Nutr. 2000;71(6):1405–1412. doi: 10.1093/ajcn/71.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiew SK, Khor LY, Tan ML, Pang CL, Chai VY, Kanachamy SS, et al. No association between peroxisome proliferator-activated receptor and uncoupling protein gene polymorphisms and obesity in Malaysian university students. Obes Res Clin Pract. 2010;4(4):e325–e331. doi: 10.1016/j.orcp.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Yoneshiro T, Aita S, Matsushita M, Okamatsu-ogura Y, Kameya T, Kawai Y, et al. Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obesity Nature Publishing Group. 2011;19(9):1755–1760. doi: 10.1038/oby.2011.125. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Park BL, Cha MH, Kim KS, Cheong HS, Choi YH, et al. Effects of genetic polymorphisms of UCP2 and UCP3 on very low calorie diet-induced body fat reduction in Korean female subjects. Biochem Biophys Res Commun. 2007;359(3):451–456. doi: 10.1016/j.bbrc.2007.05.110. [DOI] [PubMed] [Google Scholar]
- Young P, Arch JRS, Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Lett. 1984;167(1):10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
- Yu X, Jacobs DR, Schreiner PJ, Gross MD, Steffes MW, Fornage M. The uncoupling protein 2 Ala55Val polymorphism is associated with diabetes mellitus: the CARDIA study. Clin Chem. 2005;51(8):1451–1456. doi: 10.1373/clinchem.2004.044859. [DOI] [PubMed] [Google Scholar]
- Zhang M, Wang M, Zhao ZT. Uncoupling protein 2 gene polymorphisms in association with overweight and obesity susceptibility: a meta-analysis. Meta Gene. 2014;2:143–159. doi: 10.1016/j.mgene.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Meng N, Lv Z, Li H, Qu Y. The gene polymorphisms of UCP1 but not PPAR γ and TCF7L2 are associated with diabetic retinopathy in Chinese type 2 diabetes mellitus cases. Acta Ophthalmol. 2015;93(3):e223–e229. doi: 10.1111/aos.12542. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Cypess AM, Miao Q, Ye H, Liew CW, Zhang Q, et al. The prevalence and predictors of active brown adipose tissue in Chinese adults. Eur J Endocrinol. 2014;170(3):359–366. doi: 10.1530/EJE-13-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.