Abstract
Blast-related mild traumatic brain injury (mTBI) is considered the “signature” injury of the wars in Iraq and Afghanistan. Identifying biomarkers that could aid in diagnosis and assessment of chronic mTBI are urgently needed, as little progress has been made toward identifying blood-based biomarkers of repetitive mTBI in the chronic state. Addressing this knowledge gap is especially important in the population of military veterans who are receiving assessment and care often years after their last exposure. Circulating microRNAs (miRNAs), especially those encapsulated in extracellular vesicles (EVs), have gained interest as a source of biomarkers for neurological conditions. To identify biomarkers for chronic mTBI, we used next generation sequencing (NGS) to analyze miRNAs in plasma and plasma-derived EVs from 27 Iraq and Afghanistan war veterans with blast-related chronic mTBI, 11 deployed veteran non-TBI controls, and 31 civilian controls. We identified 32 miRNAs in plasma and 45 miRNAs in EVs that significantly changed in the chronic mTBI cohort compared with control groups. These miRNAs were predominantly associated with pathways involved in neuronal function, vascular remodeling, blood–brain barrier integrity, and neuroinflammation. In addition, the plasma proteome was analyzed and showed that the concentrations of C-reactive protein (CRP) and membrane metalloendopeptidase (MME) were elevated in chronic mTBI samples. These plasma miRNAs and proteins could potentially be used as biomarkers and provide insights into the molecular processes associated with the long-term health outcomes associated with blast-related chronic mTBI.
Keywords: biomarkers, EVs, miRNAs, mTBI, veterans
Introduction
Military service members of the wars in Afghanistan and Iraq are at high risk for exposure to explosive blast resulting in traumatic brain injury (TBI). With >370,000 United States military service members diagnosed since 2000,1 blast induced mild TBI (mTBI) is one of the most prevalent combat-related injuries occurring among deployed service members and has been termed the “signature injury” of these wars. mTBI (blast and/or impact) imparts a spectrum of immediate-phase primary biomechanical and delayed secondary insults to the brain. Immediate “primary” injuries are mainly the result of mechanical trauma. In this regard, blast induced injuries are especially complex; in addition to possible blast-associated impact injuries (e.g., being hit with shrapnel or the head being propelled into other objects), high explosives generate intense high-velocity blast overpressures (BOPs) that are capable of inflicting significant biomechanical injuries to the brain, as well as other organs, as they propagate through the body.2 These initial blast-related insults also trigger delayed “secondary” pathogenic, as well as neuroprotective, biological responses that evolve over hours to years after the initial injury. These insults engage a variety of cellular and molecular systems that regulate neurovascular, neuroimmune, white matter/axonal, neuronal, and metabolic processes.3–10 Most civilians who experience single impact mTBI recover within 90 days; ∼ 15% develop long-term chronic post- concussive symptoms; however, the percentage is much higher in combat veterans because of repetitive mTBI.
The current diagnosis of chronic mTBI is largely based on retrospective or recall of acute symptoms following the event, as well as behavioral and cognitive assessments. Therefore, molecular markers that allow a more accurate diagnosis for identifying and managing individuals with mTBI, particularly in the chronic state, would be very useful. Although there has been progress in identifying biomarkers for the acute phase of mTBI,11,12 there remains a substantial gap in chronic mTBI.
MicroRNAs (miRNAs) are a class of small regulatory RNAs that attenuate the cellular proteome through the destabilization of messenger RNA (mRNA) or inhibition of mRNA translation.13 The discovery of extracellular RNAs, especially miRNAs, suggests that some of the cell-free circulating RNAs may function as paracrine or endocrine signaling molecules between cells. These cell-free RNAs are either encapsulated in various extracellular vesicles (EVs), including microvesicles, exosomes, and apoptotic bodies, or complexed with RNA binding proteins or lipoproteins outside of EVs.14 These protein-associated miRNAs are often released passively by cells, and may not involve active cell–cell signaling.15 In addition to signaling between cells, the molecular content in EVs has emerged as a rich resource for biomarkers for various health conditions, including neurodegenerative diseases, such as post-traumatic stress disorder (PTSD), Alzheimer's disease, and Parkinson's disease.16–24 To identify the impact of chronic mTBI on the spectrum of plasma miRNAs and proteins, different measurement approaches, including a mass spectrometry based global protein profiling, a targeted measurement on a set of neurological function related proteins, and small RNA, were employed. Using well-characterized cohorts5,8,25–29 including: (1) veterans with blast-related chronic mTBI; (2) veterans deployed to Iraq or Afghanistan, but with no lifetime history of TBI (Deployed Controls [DC]); and (3) age-matched civilian controls with no history of TBI, the concentration changes of several proteins and miRNAs in plasma samples from chronic mTBI patients were identified. Many of the miRNAs identified were found to target genes and biological processes associated with neurobiological and neuroinflammatory responses, suggesting their function as potential regulators of pathways and processes associated with brain injury.
Methods
Human subjects
All human studies were approved by the VA Puget Sound Health Care System Human Subjects Committee. All participants provided written informed consent before study. The study conformed to institutional regulatory guidelines and principles of human subject protection in the Declaration of Helsinki. Veterans were determined to have chronic mTBI by results of physical and neurological examinations, behavioral assessments including the PTSD Checklist-Military version (PCL-M) Patient Health Questionnaire (PHQ)-9 for depression symptom assessment, and the Alcohol Use Disorders Identification Test Consumption items (AUDIT-C).30,31 Two separate control groups, veterans deployed to Iraq or Afghanistan, but with no lifetime history of TBI (DC); and age-matched civilian controls (Community Controls [CC]) with no history of TBI, were included. Lifetime history of both blast-related and impact-related TBI was obtained using a semistructured interview with two expert clinicians as described.5 All participants were male. Females were eligible for study inclusion, but no female with blast-related mTBI was enrolled. For this reason, only males were included in the control groups. Where appropriate, inclusion criteria for all participants included documented hazardous duty in Iraq and/or Afghanistan with the United States Armed Forces during Operation Iraqi Freedom (OIF) and/or Operation Enduring Freedom (OEF). For inclusion, veterans with mTBI had at least one blast exposure with acute symptoms that met VA/Department of Defense (DoD)/American Congress of Rehabilitation Medicine criteria for mTBI. For all study participants (veterans and civilian controls), exclusion criteria included moderate-severe TBI, seizure disorder, insulin-dependent diabetes, current Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) diagnosis of alcohol abuse or other substance abuse, schizophrenia or other psychotic disorders, bipolar disorder, or dementia, or taking medications likely to affect cognitive function. See Tables S1 and S2 for full details on the participants of this study.
Statistical analysis of subject characteristics
Apolipoprotein E (APOE4) positivity was determined by previously described polymerase chain reaction (PCR) conditions,32 and the HhaI restriction digest method.33 Categorical data (APOE4 positivity, race, and Hispanic ethnicity) were analyzed by χ2 tests. Between-group differences in age were determined by one-way analysis of variance (ANOVA) tests. PCL-M score, PHQ-9, Pittsburgh Sleep Quality Index (PSQI), and AUDIT-C measures were evaluated using between subject t tests where all reported p values denote two-tailed critical value outcomes. Statistical analyses were conducted using SPSS software (IBM, Armonk, NY).
Plasma preparation and EV isolation
Blood samples were collected via an intravenous catheter placed in an antecubital vein into polypropylene tubes containing sodium ethylenediaminetetraacetic acid (EDTA). Blood cells were removed by low speed centrifugation at 1000g for 10 min at 4°C. The supernatant was centrifuged again at 2000g for 15 min at 4°C to remove cell debris. Following centrifugation, the supernatant (plasma) was transferred into a clean polypropylene tube and stored at -80°C. Blood processing occurred within 1 h of collection. Before RNA isolation, the plasma samples were spun at 10,000g at 4°C for 10 min to remove cell debris and platelets. EVs were enriched from plasma using the Total Exosome Isolation Kit (Thermo Fisher, Waltham, MA) according to the manufacturer's instructions.
miRNA isolation and library construction
RNA was isolated from plasma and plasma derived EVs using the miRNeasy kit (QIAGEN, Germantown, MD) according to the manufacturer's instructions. The RNA was eluted in nuclease-free water followed by quantity and quality assessment using a Bioanalyzer (Agilent Technologies, Santa Clara, CA). To profile miRNA in plasma and EVs, the NEBNext Small RNA Library Prep kit (New England Biolabs, Ipswich, MA) was used and the library concentrations were assessed by NEBNext Library Quant Kit (New England Biolabs). The libraries were pooled (2 nM final concentration) and then run on a NextSeq500 sequencer (Illumina, San Diego, CA).
Data analysis
Sequence files were processed and analyzed using an in-house small RNA analysis pipeline—sRNAnalyzer,34 Briefly, the adapters were trimmed, and low complexity (such as homopolymer and simple repeat sequences), low quality and short reads (< 15 nucleotides) were removed. The processed reads were then mapped against various sequence databases. For miRNA, miRBase (www.mirbase.org) v21 was used. The miRNA mapping results (based on no mismatch allowed) were normalized using read count per million of processed read and log2 transformed. The miRTarbase (mirtarbase.mbc.nctu.edu.tw/) was used to obtain validated mRNA targets by at least three different experimental techniques to identify biological processes that may be regulated by specific miRNAs. Gene enrichment analysis was performed with Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/). After mapping against the miRNA database, remaining unmapped reads from samples were run through mirdeep235 to identify putative novel miRNAs (Fig. S1A).
Quantitative Reverse Transcription (qRT)-PCR verification
TaqMan Advanced miRNA assays (Thermo Fisher, Waltham, MA) were used to confirm the miRNA concentration changes. The level of miR-16-5p was used as a normalizer, because it was identified as a miRNA with low concentration variation (low coefficient of variance across samples) based on the miRNA mapping results. Relative miRNA concentrations are presented by ΔΔCt values (Ct reference – Ct target).36
Plasma proteomics
The impact of chronic mTBI on blood proteome was analyzed by isobaric tag for relative and absolute quantitation (iTRAQ). For iTRAQ analysis, plasma samples were pooled and two pools from each group with a total of six samples were analyzed (see Table S3, Fig. S2). The labeling and sample preparation methods follow manufacturer's protocol. Briefly, the top 14 high abundant blood proteins were removed with a human Mars14 LC column (4.6 × 100 mm, Agilent, Santa Clara, CA). The flowthrough of each sample was collected and concentrated, and the protein concentrations were determined by bicinchoninic acid (BCA) assay (Pierce, Rockford, IL). The proteins were digested with trypsin following a 2,2,2-trifluoroethanol (TFE)-denaturation and Oasis C18 cartridges (Waters, Milford, MA) desalting before iTRAQ labeling. Approximately 60 μg of tryptic peptides from each pooled sample were labeled with one of the eight isobaric tags (AB SCIEX, Framingham, MA) on N-termini and lysine residues. Samples were combined and fractionated into 96 fractions by a high-pH reverse-phase high-performance liquid chromatography (HPLC) fractionation before being combined into 12 fractions and analyzed on a Q Exactive™ Plus Hybrid Quadrupole-Orbitrap mass spectrometer. Acquired data were processed in Trans Proteomic Pipeline TPP (V4.8) including PeptideProphet and ProteinProphet. Protein identifications were filtered with a threshold of false discovery rate (FDR) <0.01 at protein level. Libra, an in-house script, was used to extract ratios of isobaric tags in MS/MS spectra. Only proteins with more than two independent peptides identified and quantified in all channels were included.37,38 Ratios for each protein were reported as significantly different if they passed a fold change criterion of 50% or more. Besides iTRAQ- based global profiling, the Proximity Extension Assay (PEA) (Olink Proteomics, Uppsala, Sweden) was used to measure the concentrations of 92 neurological function related proteins (Table S4) from plasma samples, as per the manufacturer's instructions.
Results
Study participant characteristics
The study analyzed plasma samples from a cohort with a total of 69 participants composed of 27 United States military OEF/OIF veterans with blast-related mTBI, 11 OEF/OIF DC veterans with no lifetime history of TBI, and 31 civilian CC who were statistically comparable in terms of age at time of blood draw, race, and the APOE4 genotype (Table S1). The chronic mTBI group experienced an average of 14 blast-related mTBIs (median = 6) and a mean of two loss of consciousnesses (LOCs) lasting <30 min that occurred between 1.6 and 7.7 years prior to the plasma sample collections used in this study (Table S1). As expected of this population, the chronic mTBI group had significantly greater comorbid PTSD symptoms, more depressive symptoms, and poorer sleep quality, but no significant increase in alcohol use compared with controls39–41 (Table S1).
Small-RNA sequencing
To explore whether there was a change in the profile of circulating miRNAs in chronic mTBI, we characterized the cell-free small RNA using next generation sequencing (NGS) from both the whole plasma and EVs purified from the same plasma sample. From whole plasma, we obtained ∼7,900,000 reads on average across all samples, of which ∼5,000,000 reads passed the quality assessment (“processed” reads) (Table 1). Approximately 1,000,000 processed reads mapped to various known human sequences. Among them, ∼830,000 reads mapped to miRNAs. We observed 466 miRNAs (with at least one mapped read) on average in each sample, and 287 of them had ≥10 mapped reads (Table 1). From the corresponding EV samples, we obtained ∼9,000,000 reads on average, of which ∼6,000,000 were processed reads (Table 1). Among them, ∼269,000 processed reads on average mapped to miRNAs. In each sample, we observed 334 different miRNAs on average; of these 211 had ≥10 mapped reads (Table 1).
Table 1.
General Sequencing Statistics
| Sample type | Plasma |
|
EV |
|
||||
|---|---|---|---|---|---|---|---|---|
| CC | DC | mTBI | Average | CC | DC | mTBI | Average | |
| Raw reads | 4,333,418 | 8,573,008 | 10,749,056 | 7,885,161 | 11,313,785 | 8,927,924 | 6,537,316 | 8,926,342 |
| Processed reads | 2,857,023 | 5,562,843 | 6,783,140 | 5,067,669 | 7,823,015 | 6,072,330 | 4,570,299 | 6,155,215 |
| All human reads | 503,358 | 955,787 | 1,870,701 | 1,109,949 | 637,665 | 523,810 | 287,044 | 482,840 |
| miRNA reads | 365,298 | 688,534 | 1,427,134 | 826,989 | 365,053 | 292,205 | 149,627 | 268,962 |
| piRNA reads | 7,388 | 11,614 | 20,645 | 13,216 | 13,482 | 10,333 | 7,968 | 10,594 |
| snoRNA reads | 5,755 | 8,285 | 17,489 | 10,510 | 11,806 | 9,484 | 7,626 | 9,638 |
| miRNA with ≥1 mapped read | 468 | 469 | 462 | 466 | 475 | 283 | 243 | 334 |
| miRNA with ≥10 mapped reads | 240 | 321 | 301 | 287 | 250 | 208 | 174 | 211 |
General statistics of breakdown of sequencing results for each category, included raw sequencing reads, processed reads that passed initial quality control, reads mapping to the human genome, and reads mapping to various ncRNAs (miRNA, piRNA, and snoRNA). For miRNA, the number for miRNA with >1 and 10 mapped reads are given for each category.
EV, extracellular vesicle; CC, community controls; DC, deployed controls; mTBI, mild traumatic brain injury; miRNA, microRNA; piRNA, piwi-interacting RNA; snoRNA, small nucleolar RNA.
Circulating miRNAs associated with chronic mTBI
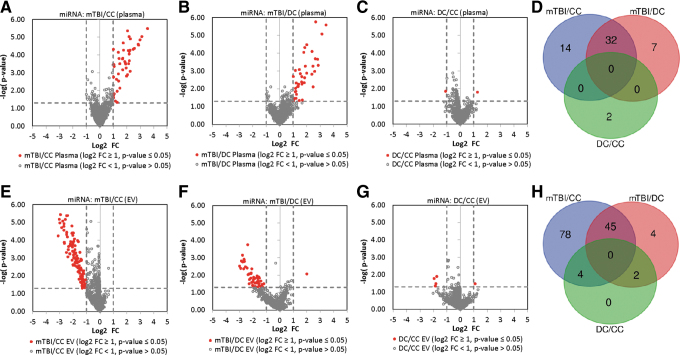
To identify circulating miRNAs associated with chronic mTBI, we compared the spectrum of miRNA from whole plasma and EV between chronic mTBIs and two separate control groups: CC and DC. The affected miRNAs were determined based on a log twofold concentration change (log2 FC) of ≥ ±1 (at least twofold concentration change) with p < 0.05 (unpaired t test). From plasma, 46 miRNAs exhibited significant concentration differences between chronic mTBIs and the CC group (Fig. 1A) and 39 miRNAs between chronic mTBIs and DC samples (Fig. 1B). Between the two, 32 affected miRNAs were in common and all showed an increased concentration in chronic mTBI samples (Fig. 1D, Table 2).
FIG. 1.
Plasma and extracellular vesicle (EV) microRNAs (miRNAs) that show differential concentration changes between traumatic brain injured (TBI) and control study participants. (A–C) Volcano plots showing plasma miRNA log2 fold change (log2FC) versus p value between (A) chronic mild TBI (mTBI) subjects and community controls (CC), (B) chronic mTBI subjects and deployed veterans with no history of TBI (DC), and (C) DC and CC. (D) Overlap between miRNAs that show concentration differences in A–C. (E–F) Volcano plots showing plasma EV miRNA log2FC versus p value between (E) chronic mTBI subjects and CC, (F) chronic mTBI subjects and DC, and (G) DC and CC. (H) Overlap between miRNAs that show concentration differences in E–F. For volcano plots, gray unfilled circles represent miRNAs with a p value >0.05, and red circles represent miRNAs with a p value <0.05, and a log2FC > ±1 (fold change >2). Color image is available online.
Table 2.
Overlapping miRNAs Showing Differences between mTBI and Controls in Plasma and EVs
| |
Plasma mTBI vs. CC |
Plasma mTBI vs. DC |
EV mTBI vs. CC |
EV mTBI vs. DC |
||||
|---|---|---|---|---|---|---|---|---|
| miRNA | Log2 fold change | p value | Log2 fold change | p value | Log2 fold change | p value | Log2 fold change | p value |
| let-7b-5p | – | – | – | – | −1.500 | 0.007255 | −1.550 | 0.029722 |
| let-7d-3p | – | – | – | – | −3.110 | 0.000057 | −3.010 | 0.003029 |
| let-7f-1-5p | 1.370 | 0.000069 | 1.540 | 0.001410 | – | – | – | – |
| let-7f-2-5p | – | – | – | – | −1.560 | 0.015669 | −1.970 | 0.017791 |
| miR-103a-1-3p | 1.690 | 0.000165 | 2.010 | 0.000033 | – | – | – | – |
| miR-103b-1-5p | 2.260 | 0.000814 | 2.480 | 0.007663 | – | – | – | – |
| miR-106a-5p | 2.140 | 0.000302 | 1.820 | 0.004509 | – | – | – | – |
| miR-106b-5p | 1.710 | 0.000151 | 1.790 | 0.000089 | – | – | – | – |
| miR-10a-5p | – | – | – | – | −1.850 | 0.001792 | −2.090 | 0.004312 |
| miR-1246-5p | 1.860 | 0.000235 | 1.250 | 0.020410 | – | – | – | – |
| miR-126-3p | – | – | – | – | −1.520 | 0.005143 | −1.690 | 0.010909 |
| miR-128-1-3p | – | – | – | – | −2.420 | 0.000065 | −2.150 | 0.007397 |
| miR-1307-3p | – | – | – | – | −1.880 | 0.000973 | −1.620 | 0.022368 |
| miR-132-5p | 3.540 | 0.000003 | 2.860 | 0.000472 | – | – | – | – |
| miR-139-5p | – | – | – | – | −2.490 | 0.000004 | −2.180 | 0.014812 |
| miR-140-3p | – | – | – | – | −1.540 | 0.002687 | −1.640 | 0.015714 |
| miR-142-3p | 1.700 | 0.000678 | 1.400 | 0.005392 | – | – | – | – |
| miR-143-3p | – | – | – | – | −2.680 | 0.000012 | −2.390 | 0.000177 |
| miR-144-3p | 2.250 | 0.000053 | 2.400 | 0.000078 | – | – | – | – |
| miR-144-5p | 2.880 | 0.000027 | 3.140 | 0.000008 | −2.000 | 0.002375 | −2.270 | 0.008368 |
| miR-146a-5p | – | – | – | – | −1.920 | 0.000464 | −1.540 | 0.038702 |
| miR-146b-5p | – | – | – | – | −2.380 | 0.000863 | −2.170 | 0.014685 |
| miR-148a-5p | 1.480 | 0.000307 | 1.340 | 0.007504 | – | – | – | – |
| miR-148b-3p | – | – | – | – | −1.510 | 0.015277 | −1.950 | 0.013167 |
| miR-151a-3p | – | – | – | – | −2.640 | 0.000149 | −2.370 | 0.009147 |
| miR-15a-5p | 2.050 | 0.001272 | 2.640 | 0.000201 | – | – | – | – |
| miR-15b-5p | 2.190 | 0.000060 | 2.390 | 0.001502 | – | – | – | – |
| miR-16-1-5p | 1.740 | 0.000021 | 1.860 | 0.000047 | – | – | – | – |
| miR-16-2-3p | – | – | – | – | −2.980 | 0.000011 | −2.790 | 0.000715 |
| miR-17-5p | 1.430 | 0.000155 | 1.910 | 0.000531 | – | – | – | – |
| miR-181a-1-5p | – | – | – | – | −1.770 | 0.001842 | −1.910 | 0.015319 |
| miR-182-5p | 1.760 | 0.000053 | 1.170 | 0.009230 | – | – | – | – |
| miR-183-5p | 2.060 | 0.000004 | 1.380 | 0.006866 | – | – | – | – |
| miR-184-3p | 2.570 | 0.000042 | 1.680 | 0.041781 | −2.760 | 0.000008 | −1.850 | 0.015228 |
| miR-185-3p | – | – | – | – | −2.090 | 0.000016 | −1.430 | 0.042859 |
| miR-18a-5p | 1.490 | 0.007164 | 2.000 | 0.001294 | – | – | – | – |
| miR-192-5p | – | – | – | – | −3.020 | 0.000007 | −2.920 | 0.004058 |
| miR-199a-1-3p | – | – | – | – | −2.850 | 0.000020 | −2.650 | 0.001332 |
| miR-19a-3p | 1.010 | 0.018209 | 1.150 | 0.014389 | – | – | – | – |
| miR-203a-3p | – | – | – | – | −1.140 | 0.017134 | −1.310 | 0.033100 |
| miR-20a-5p | 2.270 | 0.000012 | 2.680 | 0.000002 | – | – | – | – |
| miR-20b-5p | 2.120 | 0.000056 | 1.850 | 0.002363 | – | – | – | – |
| miR-21-5p | – | – | – | – | −1.320 | 0.049644 | −1.670 | 0.046573 |
| miR-222-3p | – | – | – | – | −2.150 | 0.000200 | −2.560 | 0.003303 |
| miR-223-3p | 1.230 | 0.002305 | 1.460 | 0.000817 | – | – | – | – |
| miR-223-5p | – | – | – | – | −2.080 | 0.001320 | −1.860 | 0.028278 |
| miR-2355-3p | 2.180 | 0.000086 | 1.650 | 0.005462 | – | – | – | – |
| miR-26a-1-5p | – | – | – | – | −2.350 | 0.000335 | −2.670 | 0.001216 |
| miR-27a-3p | – | – | – | – | −2.580 | 0.000007 | −2.100 | 0.025788 |
| miR-27b-3p | – | – | – | – | −2.480 | 0.000047 | −2.870 | 0.001527 |
| miR-30a-5p | – | – | – | – | −2.490 | 0.000488 | −2.440 | 0.003852 |
| miR-30d-5p | – | – | – | – | −2.660 | 0.000129 | −2.770 | 0.001729 |
| miR-30e-5p | – | – | – | – | −1.780 | 0.001842 | −1.700 | 0.012906 |
| miR-3158-1-3p | – | – | – | – | −1.350 | 0.021697 | −2.030 | 0.037187 |
| miR-32-5p | 2.180 | 0.000009 | 2.470 | 0.000528 | – | – | – | – |
| miR-340-5p | – | – | – | – | −3.470 | 0.000000 | −2.690 | 0.004273 |
| miR-342-5p | – | – | – | – | −1.980 | 0.000039 | −1.420 | 0.021622 |
| miR-3613-5p | 1.010 | 0.024744 | 1.370 | 0.007090 | – | – | – | – |
| miR-369-5p | – | – | – | – | −1.970 | 0.000000 | −1.850 | 0.044017 |
| miR-374b-5p | 1.070 | 0.014987 | 1.640 | 0.002041 | – | – | – | – |
| miR-379-5p | – | – | – | – | −1.720 | 0.001542 | −1.990 | 0.043900 |
| miR-411-5p | 2.480 | 0.000047 | 2.810 | 0.000216 | – | – | – | – |
| miR-423-3p | – | – | – | – | −2.250 | 0.000563 | −2.150 | 0.008162 |
| miR-423-5p | – | – | – | – | −1.780 | 0.001526 | −1.670 | 0.028619 |
| miR-4433b-3p | – | – | – | – | −1.690 | 0.001924 | −2.010 | 0.023562 |
| miR-4440-3p | 2.200 | 0.000207 | 1.450 | 0.044562 | – | – | – | – |
| miR-483-5p | – | – | – | – | −2.940 | 0.000004 | −2.290 | 0.030866 |
| miR-484-5p | 1.020 | 0.001705 | 1.160 | 0.000907 | – | – | – | – |
| miR-486-1-5p | – | – | – | – | −1.160 | 0.034682 | −1.520 | 0.042751 |
| miR-490-3p | 3.120 | 0.000009 | 2.590 | 0.001038 | – | – | – | – |
| miR-584-5p | – | – | – | – | −2.680 | 0.000027 | −2.640 | 0.001609 |
| miR-664a-5p | – | – | – | – | −2.210 | 0.000165 | −2.270 | 0.017894 |
| miR-7-5p | – | – | – | – | −1.390 | 0.004122 | −1.740 | 0.027532 |
| miR-7974-3p | – | – | – | – | −1.160 | 0.017294 | −1.190 | 0.030883 |
| miR-96-5p | 2.990 | 0.000014 | 3.440 | 0.000003 | – | – | – | – |
The affected miRNAs were determined based on a log 2-fold concentration change (log2 FC) of ≥ ±1 (a concentration change ≥ ± 2-fold) with p < 0.05 (unpaired t test). miRNAs that show concentration difference between chronic mTBI/CC and mTBI/DC are shown here.
CC, community controls; DC, deployed controls; EV, EV, extracellular vesicle; miRNA, microRNA;. mTBI, mild traumatic brain injury
From EV, 127 EV-miRNAs showed significant concentration differences between the chronic mTBI and CC groups (Fig. 1E), and 51 EV-miRNAs exhibited significant concentration differences between the chronic mTBI and DC groups (Fig. 1F). The majority of these chronic mTBI-affected miRNAs identified in the DC group (45 out of 51) overlapped with affected miRNAs from the chronic mTBI/CC comparison (Table 2, Fig. 1H). Even though most of the 127 affected miRNAs in the chronic mTBI and CC comparison were not identified as affected miRNAs in the DC group, the direction of concentration changes of the 127 miRNAs were the same but did not meet the selection criteria for affected miRNA (Fig. 1E and F). In contrast to whole plasma, all EV-affected miRNAs showed decreased concentrations in chronic mTBI veterans compared with controls. Between the two controls, the DC and CC groups, we identified two miRNAs in whole plasma and six miRNAs in EV that showed statistically significant concentration differences (Fig. 1 C,D and G,H). These findings suggest that the DC and CC samples have very similar circulating miRNA profiles, as was expected.
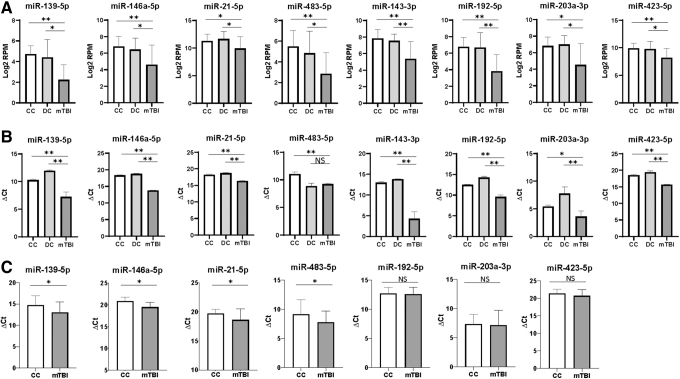
To verify the chronic mTBI affected miRNAs, 8 affected miRNAs in EVs (miR-139-5p, miR-143-3p, miR-146a-5p, miR-192-5p, miR-203a-3p, miR-21-5p, miR-423-5p, and miR-483-5p) were selected based on concentration differences between chronic mTBI veterans and controls, and their relevance to TBI, cerebrovascular disease, or neuroinflammation based on prior reports (Fig. 2A, Table 2).42–44 We performed qRT-PCR verification and were able to confirm the concentration changes in chronic mTBI samples, with the exception of miR-483-5p, in which we could only verify a change between chronic mTBI and CC (Fig. 2B). The concentration changes of some of these EV miRNAs (miR-139-5p, miR-146a-5p, miR-21-5p, and miR-483-5p) were further verified using qRT-PCR on a separate validation cohort of 40 individuals (20 chronic mTBI, and 20 CC –Table S2), showing that the concentrations of these EV miRNAs were significantly lower in chronic mTBI than in controls (Fig. 2C).
FIG. 2.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) verification of extracellular vesicle (EV) microRNAs (miRNAs) that show differential concentration changes between traumatic brain injury (TBI) and control subjects. (A) Concentration levels of eight EV miRNAs selected for follow-up for qRT-PCR verification. (B) qRT-PCR verification of eight EV miRNAs in the same set of samples. (C) qRT-PCR verification of EV miRNAs in a separate cohort of 40 individuals (20 mild TBI [mTBI] and 20 community controls [CC]). Results are shown as log2 fold change of ΔCt values (max cycles – [Ct reference – Ct target]) for qRT-PCR or log2 fold change of reads per million (RPM) for small RNA sequencing (sRNAseq). miR-16-5p was used as a reference standard for qRT-PCR. Statistically significant results are designated by an asterisks (*p < 0.05, **p < 0.01). mTBI, dark gray bars, CC, white bars, DC (deployed controls), light gray bars.
Novel circulating miRNAs associated with chronic mTBI
In addition to the known miRNAs, several putative novel (not previously identified) miRNA candidates were identified from NGS data (Fig. S1A). Using a cutoff of having at least five mapped reads in >50% of the samples, 54 putative miRNAs in whole plasma, and 36 in EVs were identified. Of these, 21 were unique to plasma, 3 were unique to EVs, and the remaining 33 were in common between whole plasma and EVs (Fig. S1B). Several of these novel miRNA candidates showed significant concentration changes between chronic mTBI and either CC or DC (log2 FC ≥ ±1, p value <0.05) in EVs, but none in whole plasma (Table S5). There were five chronic mTBI affected novel miRNAs in EVs (one elevated and four decreased) when compared with CC (Fig. S1C), and four affected novel miRNAs in EVs (one elevated and three decreased) when compared with DC (Fig. S1D). Three of these affected novel EV miRNAs are in common between chronic mTBI/CC and chronic mTBI/DC comparisons (Fig. S1E).
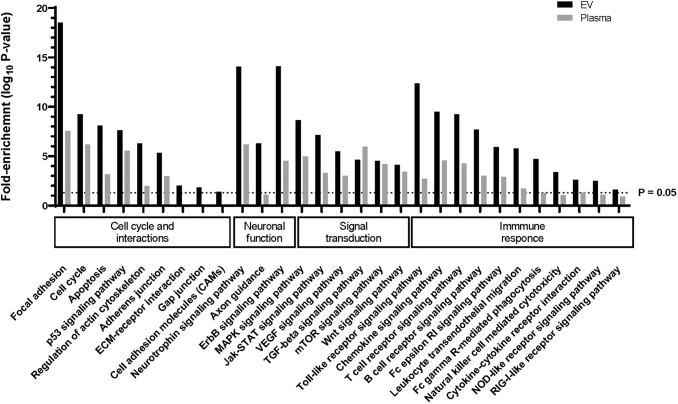
To identify biological processes associated with chronic mTBI affected circulating miRNAs, we used experimentally validated targets based on overlapping chronic mTBI affected miRNAs between the mTBI/CC and mTBI/DC groups in whole plasma (32 miRNAs) and in EVs (45 miRNAs). The majority of the affected pathways are associated with signal transduction and the immune system, such as T and B cell signaling, leukocyte migration, vascular endothelial growth factor (VEGF), ErbB, and mitogen-activated protein kinase (MAPK) signaling pathways (Fig. 3).
FIG. 3.
Enriched biological pathways and processes associated with traumatic brain injury (TBI). Selected pathway analysis showing fold enrichment (based on Database for Annotation, Visualization and Integrated Discovery [DAVID] functional analysis) of Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathways/processes associated with plasma (gray bars) and extracellular vesicle (EV) (black bars) microRNA (miRNA) targets. The enrichment factor is calculated as –log10(false discovery rate [FDR] corrected p value), with >1.3 enrichment factor considered statistically significant (FDR corrected p value <0.05 – dashed line).
Blood proteins associated with chronic mTBI
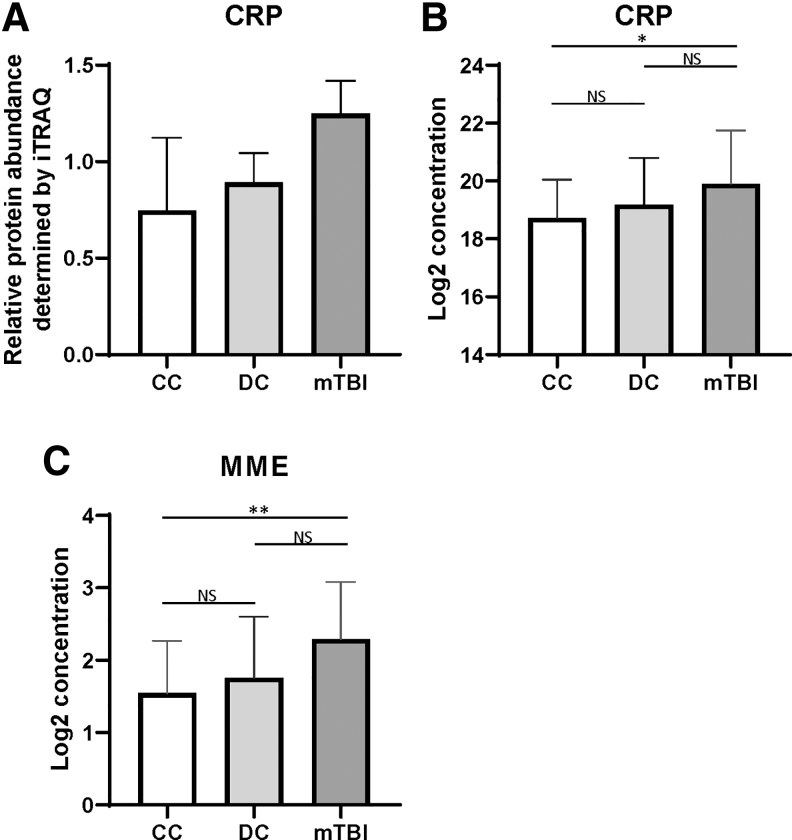
In addition to circulating RNAs, the blood proteins associated with chronic mTBI were analyzed with two different approaches. Using iTRAQ, 345 plasma proteins were identified and quantified among chronic mTBI, CC, and DC samples. Several proteins including ubiquitin-conjugating enzyme E2 D2-like protein (UBE2DNL), endoplasmic reticulum aminopeptidase 20 (ERAP2), golgin subfamily A member 4 (GOLGA4), proprotein convertase subtilisin/kexin type 4 9 PCSK4) and C-reactive protein (CRP) showed common concentration changes between the chronic mTBI and either the CC or DC group (Table S6). Based on the abundance and availability of an enzyme-linked immunosorbent assay (ELISA) kit, we measured the CRP level of individual samples and the results were in agreement with the iTRAQ finding (Fig. 4A, B). Second, to complement the global profiling approach, we measured the concentration changes of 92 neurological function-related proteins (Table S4) using proximity extension assay (PEA) which revealed one protein, membrane metalloendopeptidase (MME), showing a significant concentration increase in chronic mTBI samples compared with both CC and DC samples (≥ ± 1.5-fold concentration change with p < 0.05 using an unpaired t test) (Fig. 5C). MME is a neutral endopeptidase shown to inactivate several classes of peptide hormones.45,46
FIG. 4.
Circulating plasma levels of C-reactive protein (CRP) and metalloendopeptidase (MME) are associated with traumatic brain injury (TBI). (A) Relative protein abundance for CRP levels measured by isobaric tag for relative and absolute quantitation (iTRAQ) for community controls (CC), deployed controls (DC), and mild TBI (mTBI) subject pools (average of two pools each). (B) log2 transformed CRP levels measured by enzyme-linked immunosorbent assay (ELISA) for individual CC, DC, and mTBI individuals. (C) log2 transformed MME levels (in normalized protein eXpression [NPX] units) measured by Proximity Extension Assay (PEA) for individual CC, DC, and mTBI.
FIG. 5.
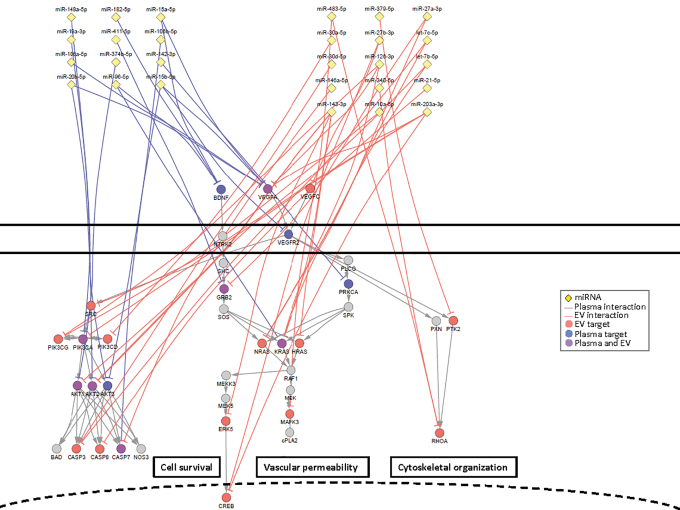
Regulation of vascular endothelial growth factor (VEGF) and neurotrophin signaling pathways by microRNAs (miRNAs) associated with mild traumatic brain injury (mTBI). Network diagram showing miRNAs associated with TBI from plasma or extracellular vesicles (EVs) targeting mRNAs of the VEGF and neurotrophin signaling pathway, including downstream signaling effectors that regulate key biological processes associated with these pathways. miRNAs (yellow diamonds) are shown regulating plasma targets (blue edges and nodes), EV targets (red edges and nodes), or targets identified in both plasma and EV (purple edges and nodes). Color image is available online.
Discussion
There is a lack of study and molecular biomarkers for chronic mTBI. Using different molecular profiling approaches to characterize the impact of chronic mTBI on the spectrum of plasma proteome and circulating miRNA from a well-characterized cohort including veteran participants with chronic mTBI and two different groups of healthy controls: one being CC and the other being DC, mall RNA sequencing was used to characterize the changes of miRNA profiles in whole plasma, and plasma-derived EVs in chronic mTBI veterans. The overall sequencing reads obtained between EV and whole plasma were similar, but the sequence reads mapped to miRNAs were much lower in EV samples (∼ 30% of the corresponding whole plasma). This finding supports the conclusion that most of the cell-free miRNAs in circulation are not encapsulated in EVs, as described previously.15 Even though most of the circulating miRNAs are not in EVs, various studies have demonstrated the functionality of EVs and suggested their involvement in cell–cell communication.16 Therefore, characterizing the molecular content, such as miRNAs, in EVs may lead to some functional insights associated with pathophysiological conditions.
From the small RNA sequencing results, 32 miRNAs that showed significant concentration changes in the plasma of veterans with blast-related chronic mTBI compared with both non-TBI DCs and CCs were identified. Of these miRNAs, three have been previously linked to TBI (miR-20a-5p, miR-142-3p, and miR-148a-5p). The concentration of miR-142-3p was found to be elevated in the plasma of TBI subjects,47 whereas miR-20a-5p was elevated in serum.48 Using a murine model of blast-induced TBI, Sajja and coworkers found that the plasma level of miR-148a-5p, was elevated.49
In the EVs, 45 miRNAs that showed significant concentration changes in chronic mTBI veterans were identified. Several of these chronic mTBI-affected EV-encapsulated miRNAs were verified by qRT-PCR in the original discovery sample set and then in a separate validation cohort of chronic mTBI and control individuals. To our knowledge, this is the first report to investigate the effect of chronic mTBI on EV-encapsulated miRNAs. Three of the chronic mTBI-affected EV miRNAs (miR-21-5p, miR-30d-5p, and miR-423-3p) have been described as TBI-associated circulating miRNAs (serum/plasma) in prior studies.24,26,31 In addition, three other miRNAs (let-7b-5p, let-7d-3p, and miR-223-3p) have been observed in plasma from a blast-induced murine TBI model.50 Interestingly, of the chronic mTBI affected miRNAs, only one miRNA, miR-144-5p, was identified in both plasma and EVs, but it showed opposite direction of concentration change (increased concentration in whole plasma, but decreased concentration in EVs [Table 2]). One possible hypothesis is that some miRNAs from affected cells are no longer being loaded into EVs or that the EV releasing processes are impaired in chronic mTBI-affected cells, and that instead, these miRNAs enter circulation complexed with RNA-binding proteins and/or high-density lipoprotein (HDL)/low-density lipoprotein (LDL) through other mechanisms. This would result in concentration differences between EV-encapsulated and non-EV-associated miRNAs in circulation.
In addition, 54 novel miRNA candidates in whole plasma and 36 novel miRNAs in EVs were identified from small RNA sequencing results. Among them, 21 novel miRNAs were unique to plasma, 3 were unique to EVs, and the remaining 33 were in common. Several of these miRNA candidates showed concentration differences between chronic mTBI and control groups in EVs. Although these results are promising, as they exemplify one of the advantages of sequencing-based small RNA analysis, these candidate miRNAs need to be further validated with different sample types.
From gene targets of chronic mTBI-affected miRNAs, we were able to identify several enriched biological processes. Some of these processes are associated with neuronal function (e.g., axon guidance, focal adhesion, adherence junctions, and neurotrophin and gonadotropin-releasing hormone [GnRH] signaling pathways); immune response (leukocyte transendothelial migration, and chemokine, mammalian target of rapamycin [mTOR], Toll-like, T-cell/B-cell receptor, nucleotide-binding oligomerization domain-like [NOD-like], and retinoic acid-inducible gene-I-like [RIG-1-like] signaling pathways); or cellular regulation and signal transduction that promote as vascular remodeling (VEGF signaling pathways, apoptosis, and cell cycle) (Fig. 3). These miRNA-regulated pathways represent critical processes associated with different pathologies associated with chronic mTBI. For example, brain-derived neurotrophic factor (BDNF), which plays an important role in regulating neural regeneration and synaptic efficiency through the neurotrophin signaling pathway, is dysregulated in TBI and associated disorders such as PTSD (Fig. 5).51 The concentration of three miRNAs, miR-182-5p, miR-96-5p, and miR-15a-5p, which were elevated in the plasma of veterans with chronic mTBI, are potential regulators of BDNF.52,53 Further, a direct regulatory role between elevated miR-182-5p and decreased BDNF in circulation has been proposed in individuals with depression, which is one of the major comorbidities among people with TBI (Fig. 5).52,54 One of the critical effectors of BDNF/neurotrophin signaling pathway is extracellular receptor kinase (Erk5)/MAPK7, which facilitates the survival of neurons in the hippocampus through activation of the transcription factor, cyclic adenosine monophosphate (cAMP) response element-binding protein (CREB).55,56 Moreover, disruption of Erk5 and subsequent CREB activation is a hallmark for TBI.57 miR-143-3p is a well-characterized direct regulator of Erk5,58–60 and miR-203a-3p and miR-27b-3p can directly suppress CREB.61,62 This suggests that the decreased concentrations of miR-143-3p, miR-203a-3p, and miR27b-3p in EVs of chronic mTBI may signal through CREB to facilitate responses after injury (Fig. 5). Leukocyte transendothelial migration allows for the activation and recruitment of leukocytes to migrate to the site of injury.63,64 This process depends on signaling through interactions between the C-X-C chemokine receptor type 4 (CXCR4) receptor on the surface of leukocytes and its ligand C-X-C motif chemokine 12 (CXCL12) on the endothelium. CXCR4 has been previously associated with TBI pathology and plays a critical role in regulating the adult neural stem cell population.65–67 miR-146a-5p, a chronic mTBI-affected miRNA in EV, is a well-characterized regulator of CXCR4 68–70 and can also regulate the level of CXCL12,71
A growing body of reports places fresh emphasis on the significance of vascular factors and the blood–brain barrier as systems that are disturbed by blast-induced mTBI.7,8,72–78 In addition, several reports indicate that the VEGF level in blood is elevated after blast-induced mTBI,79–81 including in serum from blast-exposed mice.82 Several chronic mTBI-affected miRNAs in whole plasma and EVs interact with genes involved in the VEGF pathway, as well as the downstream effectors that regulate cell survival, cytoskeletal organization, and vascular permeability (Fig. 5).
There are more biological pathways with stronger significances from chronic mTBI-affected miRNAs in EVs, compared with those from whole plasma (Fig. 3). Of these pathways, several are only significantly enriched (-log10[p value] >1.3) in the EV-encapsulated miRNAs. For example, ubiquitin mediated proteolysis, natural killer (NK) cell-mediated cytotoxicity, axon guidance, and NOD-like and RIG-1-Like signaling pathways are only significantly enriched in EVs associated miRNAs compared with whole plasma; whereas ECM receptor interactions, cell adhesion, gap junction, and the GnRH signaling pathway are significantly enriched in EVs, but show no enrichment in whole plasma. As discussed, these pathways and processes are crucial in maintaining neuronal function, inflammation, and vascular remodeling, and may imply EV's involvement in these activities.
In addition to circulating miRNA, the impact of chronic mTBI on plasma proteome has also been examined by two complementary approaches; iTRAQ for global profiling and PEA for targeted analysis on a set of proteins related to neurological function. The targeted approach allowed us to focus on a subset of the functionally relevant proteome that may be present at lower levels in circulation, whereas a global profiling approach such as iTRAQ is most likely to detect and measure proteins with higher concentrations.83 CRP, an inflammatory response protein, was elevated in mTBI veterans based on the iTRAQ measurement and subsequently validated with ELISA, as has been previously reported in TBI.84 Through the use of PEAs, the MME protein showed elevated levels in chronic mTBI plasma samples compared with controls. Interestingly, MME has been shown to be associated with the clearance of amyloid β (Aβ) plaques that are sometimes found in the brains of TBI subjects, similar to those found in the brains of individuals with Alzheimer's disease.85 Further, increased MME staining has been observed in damaged axons in the brains of long-term TBI survivors, and MME levels negatively correlate with Aβ burden, suggesting a protective role of MME in TBI.86 This is the first report that increased circulating levels of MME are associated with chronic mTBI.
One note on interpreting the findings from this study is the usually unavoidable comorbidity of PTSD in Iraq and Afghanistan veterans with mTBI caused by explosive ordnance in the combat theater. It is well established that PTSD is the major comorbidity for mTBI among veterans of the wars in Iraq and Afghanistan.87,88 Even though it would be ideal to identify markers exclusively for either chronic mTBI or PTSD, 75% of the mTBI participants involved in this study met DSM-IV criteria for PTSD, and this percentage is even greater when one includes subsyndromal PTSD. This comorbidity is unavoidable, given that repetitive mTBI caused by exploding ordnance is also a terrifying, life-threatening event that is the etiology for PTSD.89,90 Importantly, there is also a growing appreciation that regardless of whether PTSD-related symptoms arise from physiological (e.g., blast-related sheer stresses to white matter and cellular structures) or psychological stressors, both mTBI and PTSD are likely mediated by a constellation of interrelated pathophysiological processes involving inflammation,91,92 neuroendocrine disturbances,93,94 and catecholamine and other neurotransmitter abnormalities.95–97
Conclusion
In summary, circulating miRNAs that showed concentration changes in whole plasma and EVs collected from veterans with chronic blast-related mTBI when compared with control groups were identified. Computational analyses demonstrated that some of these miRNAs are associated with TBI-related genes and pathways reported in literature. The increase of CRP and MME protein levels in plasma from chronic mTBI veterans was also identified. One important aspect of the chronic mTBI-affected proteins and miRNAs identified is whether these miRNAs and proteins are also associated with acute TBI. Studies from rodent models have shown time-dependent molecular profile changes in blood, suggesting that levels of these circulating markers may change over time from the initial injury, and the recovery rate may also affect the levels of these circulating markers.98 Further studies in animal models and longitudinal samples from independent cohorts are needed to evaluate the prognostic utility of circulating markers identified in chronic mTBI samples.
Supplementary Material
Acknowledgments
We acknowledge technical assistance from Molly Tang and Li Gray.
Funding Information
This work is supported by research contracts from Department of Defeuse (DoD) W81XWH-16-1-0301 and W911NF-17-2-0086 (Dr. Wang) and Defense Threat Reduction Agency (DTRA) HDTRA1-13-C-0055 (Dr. Wang), and National Institutes of Health (NIH) grants U01HL126496-02, R56HL133887, U01CA213330, and R01DA040395 (Dr. Wang) as well as by Department of Veterans Affairs Rehabilitation Research and Development Service Merit Review Grant #B77421 (Dr. Peskind), Department of Veterans Affairs Office of Research and Development Medical Research Service grant RDIS# 0005 (Dr. Cook), Department of Veterans Affairs Biomedical Laboratory Research and Development Service (BLR&D) grant IK2 BX003258 (Dr. Schindler), R01AG046619 (Dr. Banks), T32AG052354 (Dr. Logsdon), and VA Puget Sound Health Care System Seed Grant (Dr. Meabon). We also thank Brain Injury Law of Seattle for their financial contributions.
Author Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- 1. Defense and Veterans Brain Injury Center (2016). DoD Worldwide Numbers for TBI. DVBIC Available from: http://dvbic.dcoe.mil/dod-worldwide-numbers-tbi (Last accessed April20, 2018)
- 2. Cernak I., and Noble-Haeusslein L.J. (2010). Traumatic brain injury: an overview of pathobiology with emphasis on military populations. J. Cereb. Blood Flow Metab. 30, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cernak I., Ignjatović D., Andelić G., and Savić J. (1991). Metabolic changes as part of the general response of the body to the effect of blast waves [in Serbian]. Vojnosanit. Pregl. 48, 515–522 [PubMed] [Google Scholar]
- 4. Mac Donald C., Johnson A., Cooper D., Malone T., Sorrell J., Shimony J., Parsons M., Snyder A., Raichle M., Fang R., Flaherty S., Russell M., and Brody D.L. (2013). Cerebellar white matter abnormalities following primary blast injury in US military personnel. PloS One 8, e55823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petrie E.C., Cross D.J., Yarnykh V.L., Richards T., Martin N.M., Pagulayan K., Hoff D., Hart K., Mayer C., Tarabochia M., Raskind M.A., Minoshima S., and Peskind E.R. (2014). Neuroimaging, behavioral, and psychological sequelae of repetitive combined blast/impact mild traumatic brain injury in Iraq and Afghanistan war veterans. J. Neurotrauma 31, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stocker R.P.J., Cieply M.A., Paul B., Khan H., Henry L., Kontos A.P., and Germain A. (2014). Combat-related blast exposure and traumatic brain injury influence brain glucose metabolism during REM sleep in military veterans. NeuroImage 99, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elder G.A., Gama Sosa M.A., De Gasperi R., Stone J.R., Dickstein D.L., Haghighi F., Hof P.R., and Ahlers S.T. (2015). Vascular and inflammatory factors in the pathophysiology of blast-induced brain injury. Front. Neurol. 6, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meabon J.S., Huber B.R., Cross D.J., Richards T.L., Minoshima S., Pagulayan K.F., Li G., Meeker K.D., Kraemer B.C., Petrie E.C., Raskind M.A., Peskind E.R., and Cook D.G. (2016). Repetitive blast exposure in mice and combat veterans causes persistent cerebellar dysfunction. Sci. Transl. Med. 8, 321ra6. [DOI] [PubMed] [Google Scholar]
- 9. Agoston D., Arun P., Bellgowan P., Broglio S., Cantu R., Cook D., da Silva U.O., Dickstein D., Elder G., Fudge E., Gandy S., Gill J., Glenn J.F., Gupta R.K., Hinds S., Hoffman S., Lattimore T., Lin A., Lu K.P., Maroon J., Okonkwo D., Perl D., Robinson M., Rosen C., and Smith D. (2017). Military blast injury and chronic neurodegeneration: research presentations from the 2015 International State-of-the-Science Meeting. J. Neurotrauma 34, S6–S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simon D.W., McGeachy M.J., Bayır H., Clark R.S.B., Loane D.J., and Kochanek P.M. (2017). The far-reaching scope of neuroinflammation after traumatic brain injury. Nat. Rev. Neurol. 13, 171–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Papa L., Edwards D., and Ramia M. (2015). Exploring serum biomarkers for mild traumatic brain injury, in: F.H. Kobeissy (ed.). Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects , pp. 299–306. CRC Press/Taylor & Francis: Boca Raton FL; [PubMed] [Google Scholar]
- 12. Fiandaca M.S., Mapstone M., Mahmoodi A., Gross T., Macciardi F., Cheema A.K., Merchant-Borna K., Bazarian J., and Federoff H.J. (2018). Plasma metabolomic biomarkers accurately classify acute mild traumatic brain injury from controls. PloS One 13, e0195318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Djuranovic S., Nahvi A., and Green R. (2012). miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang J., Li S., Li L., Li M., Guo C., Yao J., and Mi S. (2015). Exosome and exosomal microrna: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L., Tait J.F., and Tewari M. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U. S. A. 108, 5003–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghai V., and Wang K. (2016). Recent progress toward the use of circulating microRNAs as clinical biomarkers. Arch. Toxicol. 90, 2959–2978 [DOI] [PubMed] [Google Scholar]
- 17. Lee M.Y., Baxter D., Scherler K., Kim T.-K., Wu X., Abu-Amara D., Flory J., Yehuda R., Marmar C., Jett M., Lee I., Wang K., and Hood L. (2019). Distinct Profiles of cell-free microRNAs in plasma of veterans with post-traumatic stress disorder. J. Clin. Med. 8, 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dean K.R., Hammamieh R., Mellon S.H., Abu-Amara D., Flory J.D., Guffanti G., Wang K., Daigle B.J., Gautam A., Lee I., Yang R., Almli L.M., Bersani F.S., Chakraborty N., Donohue D., Kerley K., Kim T.-K., Laska E., Lee M.Y., Lindqvist D., Lori A., Lu L., Misganaw B., Muhie S., Newman J., Price N.D., Qin S., Reus V.I., Siegel C., Somvanshi P.R., Thakur G.S., Zhou Y., Hood L., Ressler K.J., Wolkowitz O.M., Yehuda R., Jett M., Doyle F.J., and Marmar C. (2019). Multi-omic biomarker identification and validation for diagnosing warzone-related post-traumatic stress disorder. Mol. Psychiatry [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun E., and Shi Y. (2015). MicroRNAs: small molecules with big roles in neurodevelopment and diseases. Exp. Neurol. 268, 46–53 [DOI] [PubMed] [Google Scholar]
- 20. Kumar P., Dezso Z., MacKenzie C., Oestreicher J., Agoulnik S., Byrne M., Bernier F., Yanagimachi M., Aoshima K., and Oda Y. (2013). Circulating miRNA biomarkers for Alzheimer's disease. PloS One 8, e69807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Margis R., Margis R., and Rieder C.R.M. (2011). Identification of blood microRNAs associated to Parkinsońs disease. J. Biotechnol. 152, 96–101 [DOI] [PubMed] [Google Scholar]
- 22. Cardo L.F., Coto E., de Mena L., Ribacoba R., Moris G., Menéndez M., and Alvarez V. (2013). Profile of microRNAs in the plasma of Parkinson's disease patients and healthy controls. J. Neurol. 260, 1420–1422 [DOI] [PubMed] [Google Scholar]
- 23. Tetta C., Ghigo E., Silengo L., Deregibus M.C., and Camussi G. (2013). Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine 44, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Basso M., and Bonetto V. (2016). Extracellular vesicles and a novel form of communication in the brain. Front. Neurosci. 10, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peskind E.R., Petrie E.C., Cross D.J., Pagulayan K., McCraw K., Hoff D., Hart K., Yu C.-E., Raskind M.A., Cook D.G., and Minoshima S. (2011). Cerebrocerebellar hypometabolism associated with repetitive blast exposure mild traumatic brain injury in 12 Iraq war Veterans with persistent post-concussive symptoms. NeuroImage 54, Suppl. 1, S76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pagulayan K.F., Petrie E.C., Cook D.G., Hendrickson R.C., Rau H., Reilly M., Mayer C., Meabon J.S., Raskind M.A., Peskind E.R., and Kleinhans N. (2018). Effect of blast-related mTBI on the working memory system: a resting state fMRI study. Brain Imaging Behav. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pagulayan K.F., Rau H., Madathil R., Werhane M., Millard S.P., Petrie E.C., Parmenter B., Peterson S., Sorg S., Hendrickson R., Mayer C., Meabon J.S., Huber B.R., Raskind M., Cook D.G., and Peskind E.R. (2018). Retrospective and prospective memory among OEF/OIF/OND Veterans with a self-reported history of blast-related mTBI. J. Int. Neuropsychol. Soc. 24, 324–334 [DOI] [PubMed] [Google Scholar]
- 28. Schindler A.G., Meabon J.S., Pagulayan K.F., Hendrickson R.C., Meeker K.D., Cline M., Li G., Sikkema C., Wilkinson C.W., Perl D.P., Raskind M.R., Peskind E.R., Clark J.J., and Cook D.G. (2017). Blast-related disinhibition and risk seeking in mice and combat Veterans: Potential role for dysfunctional phasic dopamine release. Neurobiol. Dis. 106, 23–34 [DOI] [PubMed] [Google Scholar]
- 29. Hendrickson R.C., Raskind M.A., Millard S.P., Sikkema C., Terry G.E., Pagulayan K.F., Li G., and Peskind E.R. (2018). Evidence for altered brain reactivity to norepinephrine in Veterans with a history of traumatic stress. Neurobiol. Stress 8, 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Forbes D., Creamer M., and Biddle D. (2001). The validity of the PTSD checklist as a measure of symptomatic change in combat-related PTSD. Behav. Res. Ther. 39, 977–986 [DOI] [PubMed] [Google Scholar]
- 31. Bush K., Kivlahan D.R., McDonell M.B., Fihn S.D., and Bradley K.A. (1998). The AUDIT alcohol consumption questions (AUDIT-C): an effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP). Alcohol Use Disorders Identification Test. Arch. Intern. Med. 158, 1789–1795 [DOI] [PubMed] [Google Scholar]
- 32. Emi M., Wu L.L., Robertson M.A., Myers R.L., Hegele R.A., Williams R.R., White R., and Lalouel J.M. (1988). Genotyping and sequence analysis of apolipoprotein E isoforms. Genomics 3, 373–379 [DOI] [PubMed] [Google Scholar]
- 33. Hixson J.E., and Vernier D.T. (1990). Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J. Lipid Res. 31, 545–548 [PubMed] [Google Scholar]
- 34. Wu X., Kim T.-K., Baxter D., Scherler K., Gordon A., Fong O., Etheridge A., Galas D.J., and Wang K. (2017). sRNAnalyzer-a flexible and customizable small RNA sequencing data analysis pipeline. Nucleic Acids Res. 45, 12,140–12, 151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Friedländer M.R., Mackowiak S.D., Li N., Chen W., and Rajewsky N. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Henningsen K., Palmfeldt J., Christiansen S., Baiges I., Bak S., Jensen O.N., Gregersen N., and Wiborg O. (2012). Candidate hippocampal biomarkers of susceptibility and resilience to stress in a rat model of depression. Mol. Cell. Proteomics MCP 11, M111..016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sleddering M.A., Markvoort A.J., Dharuri H.K., Jeyakar S., Snel M., Juhasz P., Lynch M., Hines W., Li X., Jazet I.M., Adourian A., Hilbers P.A.J., Smit J.W.A., and Dijk K.W.V. (2014). Proteomic analysis in type 2 diabetes patients before and after a very low calorie diet reveals potential disease state and intervention specific biomarkers. PloS One 9, e112835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peskind E.R., Petrie E.C., Cross D.J., Pagulayan K., McCraw K., Hoff D., Hart K., Yu C.-E., Raskind M.A., Cook D.G., and Minoshima S. (2011). Cerebrocerebellar hypometabolism associated with repetitive blast exposure mild traumatic brain injury in 12 Iraq war Veterans with persistent post-concussive symptoms. NeuroImage 54, Suppl. 1, S76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petrie E.C., Cross D.J., Yarnykh V.L., Richards T., Martin N.M., Pagulayan K., Hoff D., Hart K., Mayer C., Tarabochia M., Raskind M.A., Minoshima S., and Peskind E.R. (2014). Neuroimaging, behavioral, and psychological sequelae of repetitive combined blast/impact mild traumatic brain injury in Iraq and Afghanistan war veterans. J. Neurotrauma 31, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ryan-Gonzalez C., Kimbrel N.A., Meyer E.C., Gordon E.M., DeBeer B.B., Gulliver S.B., Elliott T.R., and Morissette S.B. (2019). Differences in post-traumatic stress disorder symptoms among post-9/11 veterans with blast- and non-blast mild traumatic brain injury. J. Neurotrauma 36, 1584–1590 [DOI] [PubMed] [Google Scholar]
- 42. Di Pietro V., Ragusa M., Davies D., Su Z., Hazeldine J., Lazzarino G., Hill L.J., Crombie N., Foster M., Purrello M., Logan A., and Belli A. (2017). MicroRNAs as novel biomarkers for the diagnosis and prognosis of mild and severe traumatic brain injury. J. Neurotrauma 34, 1948–1956 [DOI] [PubMed] [Google Scholar]
- 43. Wu D., Cerutti C., Lopez-Ramirez M.A., Pryce G., King-Robson J., Simpson J.E., van der Pol S.M., Hirst M.C., de Vries H.E., Sharrack B., Baker D., Male D.K., Michael G.J., and Romero I.A. (2015). Brain endothelial miR-146a negatively modulates T-cell adhesion through repressing multiple targets to inhibit NF-κB activation. J. Cereb. Blood Flow Metab. 35, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Müller A.H., Povlsen G.K., Bang-Berthelsen C.H., Kruse L.S., Nielsen J., Warfvinge K., and Edvinsson L. (2015). Regulation of microRNAs miR-30a and miR-143 in cerebral vasculature after experimental subarachnoid hemorrhage in rats. BMC Genomics 16, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rice G.I., Thomas D.A., Grant P.J., Turner A.J., and Hooper N.M. (2004). Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism. Biochem. J. 383, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vanneste Y., Michel A., Dimaline R., Najdovski T., and Deschodt-Lanckman M. (1988). Hydrolysis of alpha-human atrial natriuretic peptide in vitro by human kidney membranes and purified endopeptidase-24.11. Evidence for a novel cleavage site. Biochem. J. 254, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mitra B., Rau T.F., Surendran N., Brennan J.H., Thaveenthiran P., Sorich E., Fitzgerald M.C., Rosenfeld J.V., and Patel S.A. (2017). Plasma micro-RNA biomarkers for diagnosis and prognosis after traumatic brain injury: a pilot study. J. Clin. Neurosci. 38, 37–42 [DOI] [PubMed] [Google Scholar]
- 48. Bhomia M., Balakathiresan N.S., Wang K.K., Papa L., and Maheshwari R.K. (2016). A panel of serum MiRNA biomarkers for the diagnosis of severe to mild traumatic brain injury in humans. Sci. Rep. 6, 28148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sajja V.S.S.S., Jablonska A., Haughey N., Bulte J.W.M., Stevens R.D., Long J.B., Walczak P., and Janowski M. (2018). Sphingolipids and microRNA changes in blood following blast traumatic brain injury: an exploratory study. J. Neurotrauma 35, 353–361 [DOI] [PubMed] [Google Scholar]
- 50. Martinez B., and Peplow P.V. (2017). MicroRNAs as diagnostic markers and therapeutic targets for traumatic brain injury. Neural Regen. Res. 12, 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaplan G.B., Vasterling J.J., and Vedak P.C. (2010). Brain-derived neurotrophic factor in traumatic brain injury, post-traumatic stress disorder, and their comorbid conditions: role in pathogenesis and treatment. Behav. Pharmacol. 21, 427–437 [DOI] [PubMed] [Google Scholar]
- 52. Li Y., Li S., Yan J., Wang D., Yin R., Zhao L., Zhu Y., and Zhu X. (2016). miR-182 (microRNA-182) suppression in the hippocampus evokes antidepressant-like effects in rats. Prog. Neuropsychopharmacol. Biol. Psychiatry 65, 96–103 [DOI] [PubMed] [Google Scholar]
- 53. Long J., Jiang C., Liu B., Fang S., and Kuang M. (2016). MicroRNA-15a-5p suppresses cancer proliferation and division in human hepatocellular carcinoma by targeting BDNF. Tumour Biol. 37, 5821–5828 [DOI] [PubMed] [Google Scholar]
- 54. Li Y.-J., Xu M., Gao Z.-H., Wang Y.-Q., Yue Z., Zhang Y.-X., Li X.-X., Zhang C., Xie S.-Y., and Wang P.-Y. (2013). Alterations of serum levels of BDNF-related miRNAs in patients with depression. PloS One 8, e63648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Obara Y., and Nakahata N. (2010). The signaling pathway leading to extracellular signal-regulated kinase 5 (ERK5) activation via G-proteins and ERK5-dependent neurotrophic effects. Mol. Pharmacol. 77, 10–16 [DOI] [PubMed] [Google Scholar]
- 56. Finegan K.G., Wang X., Lee E.-J., Robinson A.C., and Tournier C. (2009). Regulation of neuronal survival by the extracellular signal-regulated protein kinase 5. Cell Death Differ. 16, 674–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Atkins C.M., Falo M.C., Alonso O.F., Bramlett H.M., and Dietrich W.D. (2009). Deficits in ERK and CREB activation in the hippocampus after traumatic brain injury. Neurosci. Lett. 459, 52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Esau C., Kang X., Peralta E., Hanson E., Marcusson E.G., Ravichandran L.V., Sun Y., Koo S., Perera R.J., Jain R., Dean N.M., Freier S.M., Bennett C.F., Lollo B., and Griffey R. (2004). MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 279, 52,361–52,365 [DOI] [PubMed] [Google Scholar]
- 59. Akao Y., Nakagawa Y., Iio A., and Naoe T. (2009). Role of microRNA-143 in Fas-mediated apoptosis in human T-cell leukemia Jurkat cells. Leuk. Res. 33, 1530–1538 [DOI] [PubMed] [Google Scholar]
- 60. Clapé C., Fritz V., Henriquet C., Apparailly F., Fernandez P.L., Iborra F., Avancès C., Villalba M., Culine S., and Fajas L. (2009). miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PloS One 4, e7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu J., Zou Z., Nie P., Kou X., Wu B., Wang S., Song Z., and He J. (2016). Downregulation of microRNA-27b-3p enhances tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1 expression. Cell Death Dis. 7, e2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wong K.-Y., Liang R., So C.-C., Jin D.-Y., Costello J.F., and Chim C.-S. (2011). Epigenetic silencing of MIR203 in multiple myeloma. Br. J. Haematol. 154, 569–578 [DOI] [PubMed] [Google Scholar]
- 63. Carlos T.M., Clark R.S., Franicola-Higgins D., Schiding J.K., and Kochanek P.M. (1997). Expression of endothelial adhesion molecules and recruitment of neutrophils after traumatic brain injury in rats. J. Leukoc. Biol. 61, 279–285 [DOI] [PubMed] [Google Scholar]
- 64. Schwarzmaier S.M., Kim S.-W., Trabold R., and Plesnila N. (2010). Temporal profile of thrombogenesis in the cerebral microcirculation after traumatic brain injury in mice. J. Neurotrauma 27, 121–130 [DOI] [PubMed] [Google Scholar]
- 65. Itoh T., Satou T., Ishida H., Nishida S., Tsubaki M., Hashimoto S., and Ito H. (2009). The relationship between SDF-1alpha/CXCR4 and neural stem cells appearing in damaged area after traumatic brain injury in rats. Neurol. Res. 31, 90–102 [DOI] [PubMed] [Google Scholar]
- 66. Nichols J.E., Niles J.A., DeWitt D., Prough D., Parsley M., Vega S., Cantu A., Lee E., and Cortiella J. (2013). Neurogenic and neuro-protective potential of a novel subpopulation of peripheral blood-derived CD133+ ABCG2+CXCR4+ mesenchymal stem cells: development of autologous cell-based therapeutics for traumatic brain injury. Stem Cell Res. Ther. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gyoneva S., and Ransohoff R.M. (2015). Inflammatory reaction after traumatic brain injury: Therapeutic potential of targeting cell-cell communication by chemokines. Trends Pharmacol. Sci. 36, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Alfano D., Gorrasi A., Li Santi A., Ricci P., Montuori N., Selleri C., and Ragno P. (2015). Urokinase receptor and CXCR4 are regulated by common microRNAs in leukaemia cells. J. Cell. Mol. Med. 19, 2262–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jurkin J., Schichl Y.M., Koeffel R., Bauer T., Richter S., Konradi S., Gesslbauer B., and Strobl H. (2010). miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J. Immunol. 184, 4955–4965 [DOI] [PubMed] [Google Scholar]
- 70. Labbaye C., Spinello I., Quaranta M.T., Pelosi E., Pasquini L., Petrucci E., Biffoni M., Nuzzolo E.R., Billi M., Foà R., Brunetti E., Grignani F., Testa U., and Peschle C. (2008). A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat. Cell Biol. 10, 788–801 [DOI] [PubMed] [Google Scholar]
- 71. Hsieh J.-Y., Huang T.-S., Cheng S.-M., Lin W.-S., Tsai T.-N., Lee O.K., and Wang H.-W. (2013). miR-146a-5p circuitry uncouples cell proliferation and migration, but not differentiation, in human mesenchymal stem cells. Nucleic Acids Res. 41, 9753–9763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rodriguez U.A., Zeng Y., Deyo D., Parsley M.A., Hawkins B.E., Prough D.S., and DeWitt D.S. (2018). Effects of mild blast traumatic brain injury on cerebral vascular, histopathological, and behavioral outcomes in rats. J. Neurotrauma 35, 375–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kuriakose M., Rao K.V.R., Younger D., and Chandra N. (2018). Temporal and spatial effects of blast overpressure on blood-brain barrier permeability in traumatic brain injury. Sci. Rep. 8, 8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Logsdon A.F., Meabon J.S., Cline M.M., Bullock K.M., Raskind M.A., Peskind E.R., Banks W.A., and Cook D.G. (2018). Blast exposure elicits blood-brain barrier disruption and repair mediated by tight junction integrity and nitric oxide dependent processes. Sci. Rep. 8, 11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huber B.R., Meabon J.S., Hoffer Z.S., Zhang J., Hoekstra J.G., Pagulayan K.F., McMillan P.J., Mayer C.L., Banks W.A., Kraemer B.C., Raskind M.A., McGavern D.B., Peskind E.R., and Cook D.G. (2016). Blast exposure causes dynamic microglial/macrophage responses and microdomains of brain microvessel dysfunction. Neuroscience 319, 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Readnower R.D., Chavko M., Adeeb S., Conroy M.D., Pauly J.R., McCarron R.M., and Sullivan P.G. (2010). Increase in blood-brain barrier permeability, oxidative stress, and activated microglia in a rat model of blast-induced traumatic brain injury. J. Neurosci. Res. 88, 3530–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Elder G.A., Gama Sosa M.A., De Gasperi R., Stone J.R., Dickstein D.L., Haghighi F., Hof P.R., and Ahlers S.T. (2015). Vascular and inflammatory factors in the pathophysiology of blast-induced brain injury. Front. Neurol. 6, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gama Sosa M.A., De Gasperi R., Perez Garcia G.S., Perez G.M., Searcy C., Vargas D., Spencer A., Janssen P.L., Tschiffely A.E., McCarron R.M., Ache B., Manoharan R., Janssen W.G., Tappan S.J., Hanson R.W., Gandy S., Hof P.R., Ahlers S.T., and Elder G.A. (2019). Low-level blast exposure disrupts gliovascular and neurovascular connections and induces a chronic vascular pathology in rat brain. Acta Neuropathol. Commun. 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hubbard W.B., Greenberg S., Norris C., Eck J., Lavik E., and VandeVord P. (2017). Distinguishing the unique neuropathological profile of blast polytrauma. Oxid. Med. Cell. Longev. 2017, 5175249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sajja V.S.S.S., Tenn C., McLaws L.J., and Vandevord P.J. (2012). A temporal evaluation of cytokines in rats after blast exposure. Biomed. Sci. Instrum. 48, 374–379 [PubMed] [Google Scholar]
- 81. Kovesdi E., Gyorgy A.B., Kwon S.-K.C., Wingo D.L., Kamnaksh A., Long J.B., Kasper C.E., and Agoston D.V. (2011). The effect of enriched environment on the outcome of traumatic brain injury; a behavioral, proteomics, and histological study. Front. Neurosci. 5, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ahmed F., Plantman S., Cernak I., and Agoston D.V. (2015). The temporal pattern of changes in serum biomarker levels reveals complex and dynamically changing pathologies after exposure to a single low-intensity blast in mice. Front. Neurol. 6, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Aggarwal K., Choe L.H., and Lee K.H. (2006). Shotgun proteomics using the iTRAQ isobaric tags. Brief. Funct. Genomic. Proteomic. 5, 112–120 [DOI] [PubMed] [Google Scholar]
- 84. Su S.-H., Xu W., Li M., Zhang L., Wu Y.-F., Yu F., and Hai J. (2014). Elevated C-reactive protein levels may be a predictor of persistent unfavourable symptoms in patients with mild traumatic brain injury: a preliminary study. Brain. Behav. Immun. 38, 111–117 [DOI] [PubMed] [Google Scholar]
- 85. Johnson V.E., Stewart W., and Smith D.H. (2010). Traumatic brain injury and amyloid-β pathology: a link to Alzheimer's disease? Nat. Rev. Neurosci. 11, 361–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chen X.-H., Johnson V.E., Uryu K., Trojanowski J.Q., and Smit D.H. (2009). A lack of amyloid β plaques despite persistent accumulation of amyloid β in axons of long-term survivors of traumatic brain injury. Brain Pathol. 19, 214–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yurgil K.A., Barkauskas D.A., Vasterling J.J., Nievergelt C.M., Larson G.E., Schork N.J., Litz B.T., Nash W.P., Baker D.G., and Marine Resiliency Study Team (2014). Association between traumatic brain injury and risk of posttraumatic stress disorder in active-duty Marines. JAMA Psychiatry 71, 149–157 [DOI] [PubMed] [Google Scholar]
- 88. Kennedy J.E., Jaffee M.S., Leskin G.A., Stokes J.W., Leal F.O., and Fitzpatrick P.J. (2007). Posttraumatic stress disorder and posttraumatic stress disorder-like symptoms and mild traumatic brain injury. J. Rehabil. Res. Dev. 44, 895–920 [DOI] [PubMed] [Google Scholar]
- 89. Kaplan G.B., Leite-Morris K.A., Wang L., Rumbika K.K., Heinrichs S.C., Zeng X., Wu L., Arena D.T., and Teng Y.D. (2018). Pathophysiological bases of comorbidity: traumatic brain injury and post-traumatic stress disorder. J. Neurotrauma 35, 210–225 [DOI] [PubMed] [Google Scholar]
- 90. Hendrickson R.C., Schindler A.G., and Pagulayan K.F. (2018). Untangling PTSD and TBI: challenges and strategies in clinical care and research. Curr. Neurol. Neurosci. Rep. 18, 106. [DOI] [PubMed] [Google Scholar]
- 91. Deslauriers J., Powell S., and Risbrough V.B. (2017). Immune signaling mechanisms of PTSD risk and symptom development: insights from animal models. Curr. Opin. Behav. Sci. 14, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Simon D.W., McGeachy M.J., Bayır H., Clark R.S.B., Loane D.J., and Kochanek P.M. (2017). The far-reaching scope of neuroinflammation after traumatic brain injury. Nat. Rev. Neurol. 13, 171–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Molaie A.M., and Maguire J. (2018). Neuroendocrine abnormalities following traumatic brain injury: an important contributor to neuropsychiatric sequelae. Front. Endocrinol. 9, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tanriverdi F., and Kelestimur F. (2015). Neuroendocrine disturbances after brain damage: an important and often undiagnosed disorder. J. Clin. Med. 4, 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Schindler A.G., Meabon J.S., Pagulayan K.F., Hendrickson R.C., Meeker K.D., Cline M., Li G., Sikkema C., Wilkinson C.W., Perl D.P., Raskind M.R., Peskind E.R., Clark J.J., and Cook D.G. (2017). Blast-related disinhibition and risk seeking in mice and combat Veterans: Potential role for dysfunctional phasic dopamine release. Neurobiol. Dis. 106, 23–34 [DOI] [PubMed] [Google Scholar]
- 96. Charney D.S., Deutch A.Y., Krystal J.H., Southwick S.M., and Davis M. (1993). Psychobiologic mechanisms of posttraumatic stress disorder. Arch. Gen. Psychiatry 50, 295–305 [DOI] [PubMed] [Google Scholar]
- 97. McAllister T.W. (2011). Neurobiological consequences of traumatic brain injury. Dialogues Clin. Neurosci. 13, 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Thelin E.P., Just D., Frostell A., Häggmark-Månberg A., Risling M., Svensson M., Nilsson P., and Bellander B.-M. (2018). Protein profiling in serum after traumatic brain injury in rats reveals potential injury markers. Behav. Brain Res. 340, 71–80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.