Abstract
Sepsis due to antimicrobial resistant pathogens is a major health problem worldwide. The inability to rapidly detect and thus treat bacteria with appropriate agents in the early stages of infections leads to excess morbidity, mortality, and healthcare costs. Here we report a rapid diagnostic platform that integrates a novel one-step blood droplet digital PCR assay and a high throughput 3D particle counter system with potential to perform bacterial identification and antibiotic susceptibility profiling directly from whole blood specimens, without requiring culture and sample processing steps. Using CTX-M-9 family ESBLs as a model system, we demonstrated that our technology can simultaneously achieve unprecedented high sensitivity (10 CFU/ml) and rapid sample-to-answer assay time (one hour). In head-to-head studies, by contrast, real time PCR and BioRad ddPCR only exhibited a limit of detection of 1,000 CFU/ml and 50–100 CFU/ml, respectively. In a blinded test inoculating clinical isolates into whole blood, we demonstrated 100% sensitivity and specificity in identifying pathogens carrying a particular resistance gene. We further demonstrated that our technology can be broadly applicable for targeted detection of a wide range of antibiotic resistant genes found in both Gram-positive (vanA, nuc, and mecA) and Gram-negative bacteria, including ESBLs (blaCTX-M-1 and blaCTX-M-2 families) and CREs (blaOXA-48 and blaKPC), as well as bacterial speciation (E. coli and Klebsiella spp.) and pan-bacterial detection, without requiring blood culture or sample processing. Our rapid diagnostic technology holds great potential in directing early, appropriate therapy and improved antibiotic stewardship in combating bloodstream infections and antibiotic resistance.
One Sentence Summary:
We report a rapid diagnostic platform that integrates novel one-step blood droplet PCR assay and a high throughput droplet counting system to perform bacterial identification and antibiotic susceptibility profiling directly from whole blood specimens.
Introduction
Sepsis due to bloodstream infections (BSI) affects 30 million people worldwide, leading to an estimated 6 million deaths every year(1). Sepsis is also an expensive condition managed in hospitals, costing more than $20 billion a year in the U.S. alone(2). This high mortality rate is due in part to the inability to accurately diagnose underlying infections in the first hours of illness when treatment is most effective(3). Approximately 30–50% of patients receive ineffective initial antibiotic therapy because physicians must treat immediately using first-line antibiotics without a precise diagnosis(3–6). Furthermore, patient survival decreases by 10% for each hour of delay in administering the correct antibiotics(4–6). Further exacerbating these health concerns, antibiotic-resistant bacteria like methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), Gram-negative extended spectrum β-lactamase (ESBL)–producing Enterobacteriaceae, and carbapenem-resistant Enterobacteriaceae (CRE), are on the rise(7,8). According to the Centers for Disease Control and Prevention (CDC), more than 2 million people are infected annually with antibiotic-resistant bacteria, with >23,000 deaths in the United States alone(7). The continuous rise of antibiotic-resistant Gram-negative rods has fuelled concerns about the lack of antibiotics to treat these pathogens(9,10). This is especially true for CREs, which produce carbapenemases that hydrolyze beta-lactam antibiotics, including carbapenems that are considered the “drug of last resort” to treat serious Gram-negative infections. As a result, mortality rates in the setting of BSI due to CREs are over 50%(7,8,11). Furthermore, aggressive bacterial infections associated with antibiotic resistance are often managed within intensive care units (ICUs) with high associated costs, imposing significant healthcare, economic, and social burdens(2).
The development and use of rapid tests for bacterial detection and antibiotic susceptibility testing (AST) has been identified as one of the priorities to combat antibiotic resistance(7,8,12–14). A critical barrier to managing sepsis and antibiotic resistance is the lack of rapid diagnostics, resulting in either the use of unnecessarily broad first-line antibiotics, or a long delay in administering the appropriate antibiotic(s)(3,7,15). The present gold standard, bacterial culture coupled with antibiotic susceptibility testing, requires several days. Recent phenotypic assays including imaging- (e.g., Accelerate Pheno™(16)) and microfluidic culture-(17) or morphology-based methods still operate on post-culture specimens(18). Several molecular and cellular detection methods have recently been reported including polymerase chain reaction (PCR) (e.g., FilmArray BCID(19), GeneXpert®(20)), nanoparticle-based assays (e.g., Nanosphere’s Verigene(21), T2 Biosystems T2MR), microfluidic-based capture and enrichment(22–25), electrochemical sensors(26–28), synthetic biology methods (e.g., programmable toehold switches(29), CRISPR(30), phages(31)), sequencing methods (e.g., Oxford Nanopore MinION(32)), and detection based on host responses(33–35). Many of these assays that target bacterial identification and drug resistance genes can reduce assay time to hours but are often not sensitive enough to detect bacteria at low concentrations (<1 to 100 Colony-Forming Unit (CFU) /ml; as is commonly found in adult BSIs(36,37)), therefore still requiring a culture-enrichment step(19,21,38,39). Furthermore, existing methods typically require expensive equipment and lengthy, complex sample processing (e.g., cell lysis, nucleic acid extraction) for target purification and enrichment, which results in significant loss of low number targets and inconsistency between tests(40).
To address these challenges in BSIs and antibiotic resistance, we present a rapid diagnostic platform that integrates a novel one-step, culture- and processing-free, blood droplet digital PCR assay and a high throughput 3D particle counter system for droplet analysis (“Integrated Comprehensive Droplet Digital Detection” or “IC3D”, hereinafter collectively referred to as “IC3D blood ddPCR”). In our work-flow, as illustrated in Figure 1, unprocessed whole blood specimens are mixed with reagents for blood inhibitor-resistant PCR within a microfluidic device, which are rapidly encapsulated into millions of individual picoliter-sized droplets. Following the digital PCR reaction, droplets containing the target bacterium become fluorescent, which can be detected and quantified by a high throughput 3D particle counter. Our technology can simultaneously achieve unprecedented high sensitivity, short assay time and robustness required to detect low-abundance bacteria in BSIs, owing to the following innovative features: 1) the one-step, blood inhibitor-resistant PCR eliminates any sample processing steps and therefore reduces target loss, inter-assay variability and assay time; 2) running the blood PCR in a digital fashion greatly improves precision and sensitivity compared to conventional PCR systems. The compartmentalization of single target bacteria in droplet “microreactors” significantly increases the effective target concentration following PCR amplification and at the same time reduces interference from background, permitting absolute, digital (“1 or 0”) quantification of targets with single-cell sensitivity without the need for internal references; and 3) our high-throughput 3D particle counter can robustly and accurately detect single fluorescent particles from a large volume (ml) within several minutes, which significantly shortens the assay time compared to conventional 1D or 2D particle counting systems and allows us to accommodate a larger sample input, thereby increasing the chance to detect rare targets. Previously, we have demonstrated the utility of this platform using DNAzyme chemistry for sensitive detection of E. coli in whole blood(41) and isothermal exponential amplification (EXPAR) chemistry for the detection of low-abundance miRNA in plasma(42). While these methods underline the ability of the IC3D system to incorporate a range of detection techniques, significant advancements have been made in this current study to enable innovative, whole blood digital PCR, which is much more versatile and modular, allowing future assays to be efficiently developed for a broad range of pathogen targets including antibiotic resistance.
Figure 1.

IC3D Workflow for rapid bacterial ID/AST. a) raw blood sample and PCR reagents are mixed and then encapsulated in picoliter-sized droplets. b) PCR produces a fluorescence signal in the droplets that contain bacterial targets. c-d) Droplets are collected in a cuvette and analyzed using our high throughput 3D particle counter.
Using blaCTX-M-9 family ESBLs as a model system, we demonstrate our technology can detect antibiotic resistant bacteria in a one-step process directly from inoculated whole blood specimens with a sensitivity of 10 CFU/ml and sample-to-answer time < 1 hour. We further demonstrated our technology can be broadly applicable to detect a wide range of antibiotic resistant genes in both Gram-positive and Gram-negative organisms, to bacterial speciation, and to pan-bacterial detection, without requiring blood culture or sample processing.
Results
One-step blood PCR chemistry in bulk
To achieve the necessary sensitivity and specificity, many existing tests require time-consuming sample processing steps to either enrich analyte concentrations or eliminate the inhibitory-effects of complex matrices like whole blood. In this paper, we set out to develop a one-step, inhibitor-resistant PCR chemistry to enable rapid, sensitive detection of bacteria straight from whole blood without the need for any sample processing steps. A primary problem with PCR-based diagnostic tests of blood samples is the low-sensitivity or false-negative reactions, due to PCR inhibitors (e.g. hemoglobin) found in blood. Therefore, DNA purification methods are typically employed prior to PCR to remove inhibitors from blood samples, which not only increases the assay complexity and assay time but can also lead to loss of the targets during purification steps, further decreasing the detection sensitivity.
With recent advances in the development of inhibitor-resistant Taq mutants and PCR enhancer cocktails (PEC) for improving enzymatic performance(43,44), we decided to leverage these innovative findings and incorporate them into our one-step blood PCR assay in our IC3D blood ddPCR platform. The design of the primer and probe chemistry developed for one-step blood PCR for each unique bacterial target involved iterative optimization and testing involving gel electrophoresis to validate primer specificity, qPCR to evaluate inhibitor-resistant PCR using first engineered targets and then clinical isolates in bulk, and ultimately transition to blood droplet digital PCR (see Supplemental Materials and Methods and presented in detail below). Final sequences for the different bacterial genetic panels used in the studies can be found in Table S1. Optimized chemistry for one-step blood PCR was able to achieve target amplification in the presence of 20% whole blood with minimal non-specific amplification of negative controls with qPCR using representative engineered targets and clinical isolates (Figure S1 and 2, respectively).
Blood droplet generation and blood droplet digital PCR
In order to efficiently process large volumes of blood specimens, a microfluidic flow-focusing 4-nozzle design was implemented to achieve high-throughput droplet generation (1 ml sample / 20 minutes) with exceptional droplet uniformity (Figure 2). Due to the potential for complex matrices like whole blood to clog microfluidic devices and cause chip failure, a series of micro-posts with variable spacing were integrated into the sample and oil inlets to isolate large debris from the downstream droplet generation nozzles (Figure 2.b).
Figure 2.
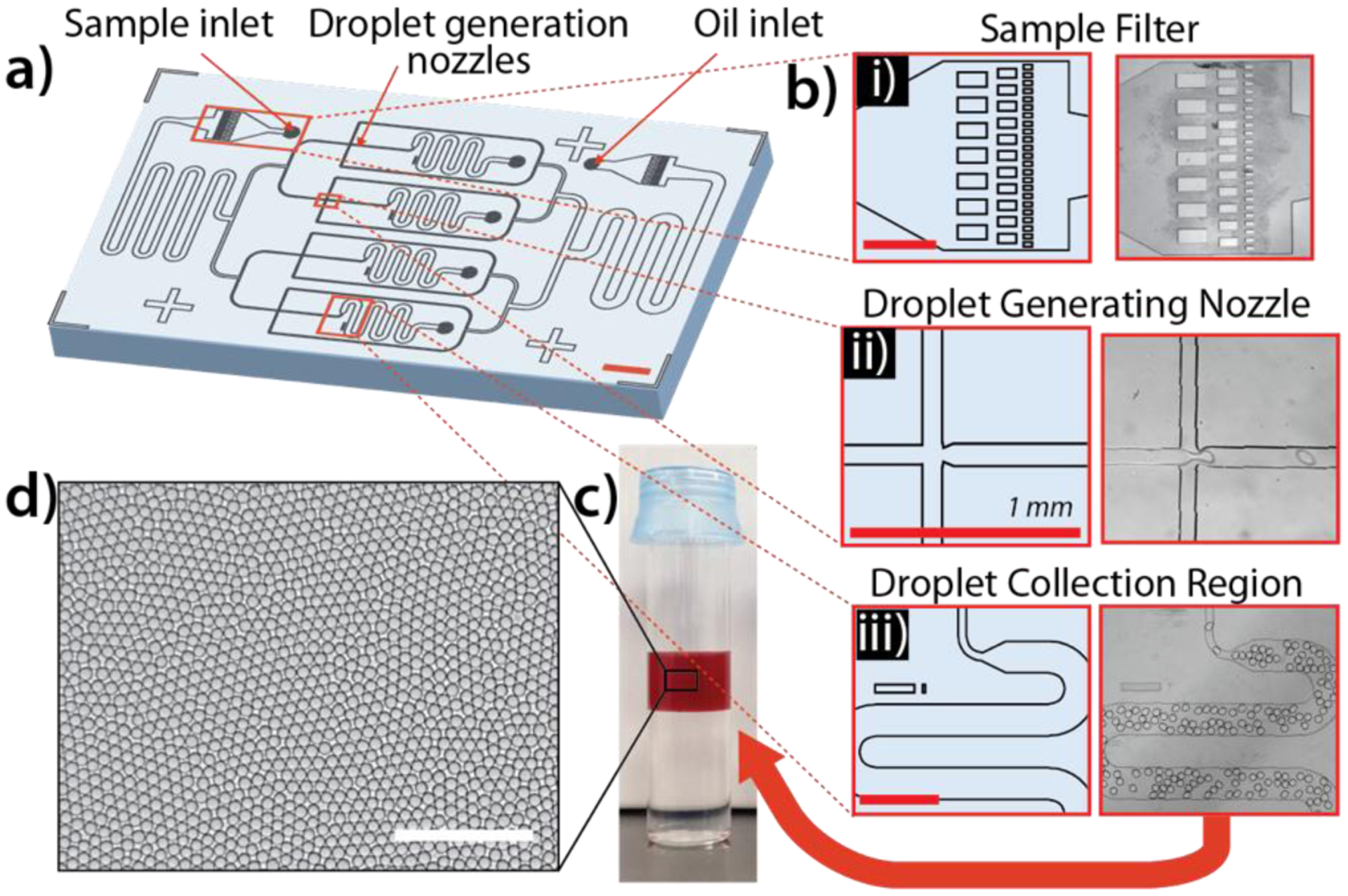
High throughput droplet generation chip design and characterization. a) Schematic of PDMS microfluidic droplet generator employing flow-focusing principle across 4 parallel nozzles. b) Key design features are identified including i) micropillar sample filter, ii) droplet generation nozzle, and iii) droplet collection chamber. (scale bar = 1 mm) c) Photograph of droplets containing 10% whole blood in cuvette before 3D scanning. Droplet layer (approximately 1 ml total volume) floats on top of a layer of HFE-2000 oil due to lower density. d) 2D monolayer of droplets for brightfield microscopy to assess droplet uniformity (average droplet diameter = 85 μm, %CV = <5%) (Scale bar = 500μm).
Existing digital PCR assays use purified nucleic acid samples, therefore requiring sample processing and significantly longer assay time. To our knowledge, we are the first to demonstrate robust, inhibitor-resistant blood digital PCR in droplets due to the difficulties in maintaining uniform, thermally-stable droplets with complex matrices. In order to maximize droplet yield and prevent droplet coalescence, several strategies were evaluated including modifications to the microfluidic droplet generator, the droplet generation process in either the continuous or dispersed phase, and during the thermocycling process. Careful evaluation of each of these modifications uncovered several key parameters. First, while addition of Tween-20 was not found to significantly affect droplet thermal stability (Figure 3.a), we determined that a higher concentration of PFPE surfactant in the oil layer of PCR tubes during thermocycling resulted in more uniform droplet sizes. For example, droplets generated using 2% PFPE surfactant oil but transferred to PCR tubes containing 5% PFPE surfactant oil were more stable (percent coefficient of variation (%CV) < 10%) than when the same 2% PFPE surfactant oil was used in PCR tubes (%CV > 14%) (Figure 3.b). Additionally, the concentration of the detergent NP-40 in the dispersed phase was found to play a critical role in droplet thermal stability, with an optimal concentration of 0.2% (stable droplet size distribution from 5.0% pre-PCR to 4.6% CV post-PCR). Widespread droplet coalescence was observed (%CV = 28%) with 0% NP-40, and droplet splitting was observed with ≥ 0.4% NP-40 (size variation increase from 4.9% pre-PCR to 8.3% CV post-PCR) (Figure 3.c). Finally, the addition of a layer of mineral oil on top of the droplet layer within PCR tubes was found to improve droplet uniformity (3.8% pre-PCR, 4.5% CV post-PCR) and yield, both microscopically by the evaluation of droplet diameter variation and macroscopically by observing the lack of large merged droplets (Figure 3.d). Ultimately, we identified the optimal conditions for robust blood droplet PCR to be: 0.2% NP-40 in the dispersed phase and 2% PFPE surfactant oil in the continuous phase for droplet generation, followed by thermocycling droplets in 5% PFPE surfactant oil with a top layer of mineral oil to prevent evaporation (Figure 3.e).
Figure 3.
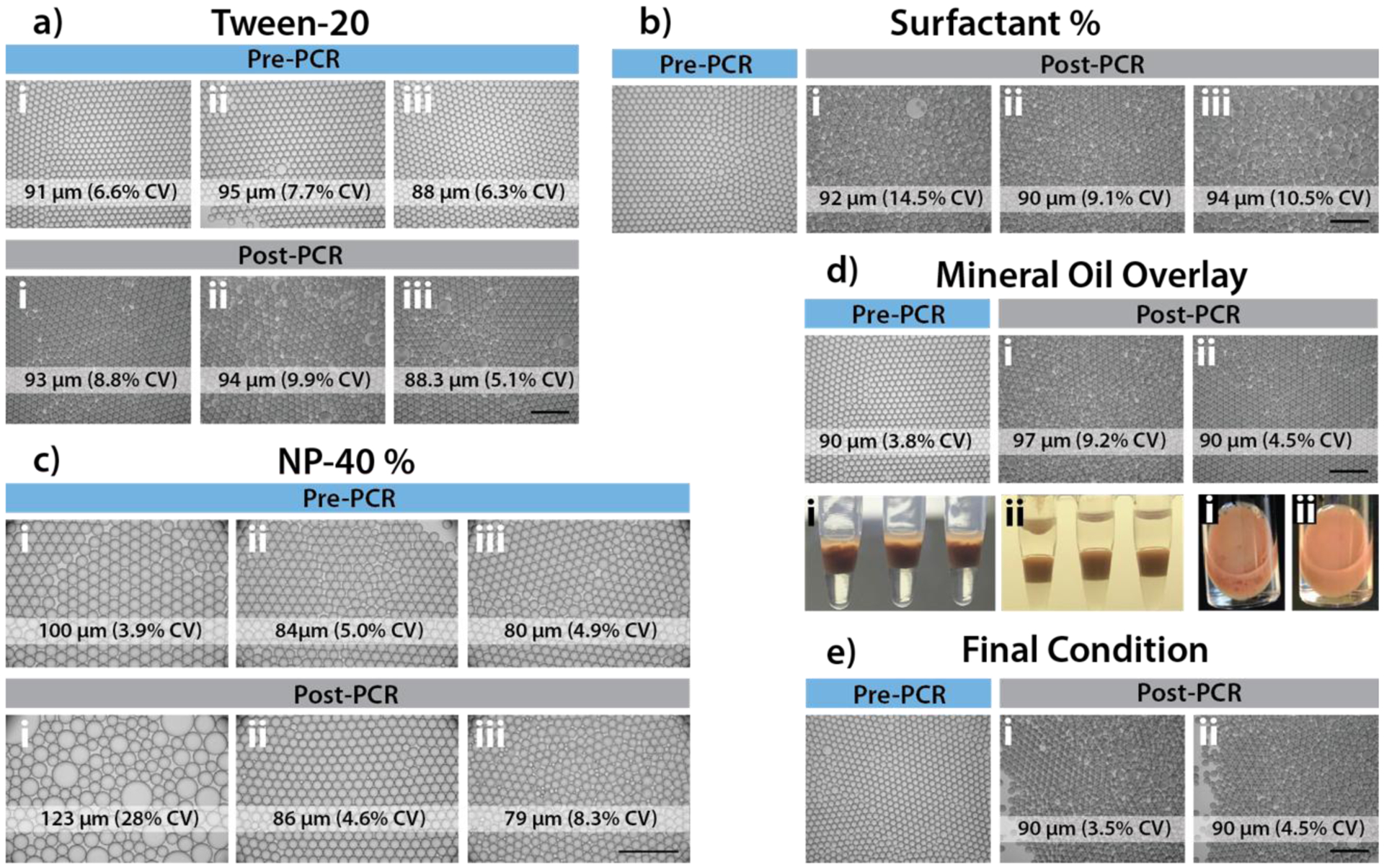
Blood droplet thermal stability optimization. a) Addition of Tween-20 to PCR mixture prior to encapsulation (i: 0%; ii: 0.01%; iii: 0.2%). Addition of Tween-20 was not found to significantly affect droplet thermal stability. b) Optimization of surfactant concentration for oil in PCR tubes. Emulsions of 2% surfactant with PCR mixtures containing 10% whole blood were loaded into PCR tubes containing preloaded various percentage of surfactant in HFE7500 (i: 2%; ii: 5%, iii: 10%). Droplets before and after PCR thermocycling were imaged (BF, 4x) and quantified for droplet CV. 5% surfactant oil in PCR tubes was found to be optimal. c) Optimization of NP-40 concentration in PCR buffer mixture prior to encapsulation (i: 0%; ii: 0.2%; iii: 0.4%). Final concentration of 0.2% NP-40 was found to result in improved droplet thermal stability. d) Addition of mineral oil overlay during 40 cycles of thermocycling (i: without mineral oil, ii: with 70 μl mineral oil overlay). Mineral oil overlay was found to significantly improve droplet thermal stability and yield. e) Final condition combining optimal parameters found in a-d. A portion of 10% whole blood droplets created with 2% PFPE surfactant oil were continuously mixed with 5% (w/w) surfactant in HFE7500 oil for 30 mins, before loading into PCR tubes. For PCR thermocycling, 70 μl of droplets (without mixing condition (i) or with mixing (ii)) were transferred into PCR tubes preloaded with 70 μl of mineral oil and 30 μl of 5% (w/w) surfactant in HFE7500 oil. The addition of a mixing step was not found to significantly improve droplet uniformity or thermal stability.
In addition to the challenges in producing uniform, thermally-stable droplets, one-step blood PCR presents many unique challenges for diagnostic instruments, both in terms of robust target amplification and sensitive optical detection. Prior to the studies featured in this paper, we optimized several additional parameters to overcome the inhibitory effects and optical interference of this complex matrix. In order to offset the high baseline fluorescence signal commonly attributed to blood autofluorescence, the far-red-emitting fluorophore, Quasar 670 (LGC Biosearch Technologies), was shown to achieve the highest signal-to-noise ratio (SNR) of selected fluorophores with different emission spectra (SNR of 5.5 compared to SNR < 2.5 for Alexa-Fluor 488 and CAL Fluor Gold 540, Figure S3), and was selected for the majority of TaqMan probes designed for the following studies. Additionally, by testing different droplet sizes, we determined that larger droplets (on the order of 100 μm in diameter) resulted in more robust detection with lower baseline fluorescent signals than smaller droplets under the same conditions (Figure S4), and therefore utilized droplets with an average diameter of approximately 85 μm (%CV < 5%) in the following studies.
To overcome PCR inhibition in the presence of whole blood, several additional parameters were optimized to achieve robust signal amplification for fluorescence detection on our 3D particle counter instrument. First, bacterial lysis within droplets was evaluated using several different strategies including heat treatment, enzymatic lysis, sonication, and UV exposure, resulting in the selection of a 5-minute incubation at 95°C, which was sufficient to efficiently lyse both Gram negative and Gram positive (though less efficiently) bacteria strains in this study (Supplemental Methods, Figure S5). Primer and probe concentrations were varied in order to determine the optimal conditions to achieve maximum fluorescence signal while maintaining a low background signal for negative droplets, resulting in the selection of concentrations of 1 μM for primers and 500 nM for probes (Supplemental Methods). By experimenting with the concentration of whole blood in the final PCR samples, 10% whole blood was selected for the present studies as a conservative approach to achieve robust detection across a broad panel of molecular targets. Though target amplification was possible in 20% and 30% whole blood, the increased concentration of whole blood led to additional complications with stable droplet production in some samples (Figure S6.a, b). Concentrations of bovine serum albumin (BSA), a known PCR amplification facilitator often applied to overcome inhibition(45), were tested from 0–20 mg/ml resulting in a significant boost in signal amplification at 5 mg/ml followed by a gradual plateau for increasing concentrations (Figure S6.c). Additionally, we have also optimized other parameters including MgCl2 concentration and PCR cycle number, which are presented in the Supplemental Methods section and Figure S6.d–f. Final conditions determined through these optimization studies were utilized in the main studies presented here, resulting in robust, specific PCR amplification within positive droplets containing target bacteria with no amplification in negative droplets using an engineered E. coli strain containing the synthetic blaCTX-M-9 target gene as a model (Figure 4) (fluorescent droplet images for additional genetic targets including pan-bacteria (16s rDNA), ESBLs (blaCTX-M-1, blaCTX-M-2), CREs (blaKPC, blaOXA-48, blaNDM, blaVIM), VREs (vanA), MRSA (mecA, nuc), and species-specific genes (E. coli: uidA) are shown in Figure S7). An extensive literature search determined that these targets represent a large fraction of the most prevalent clinical cases of ESBLs and CREs(46).
Figure 4.
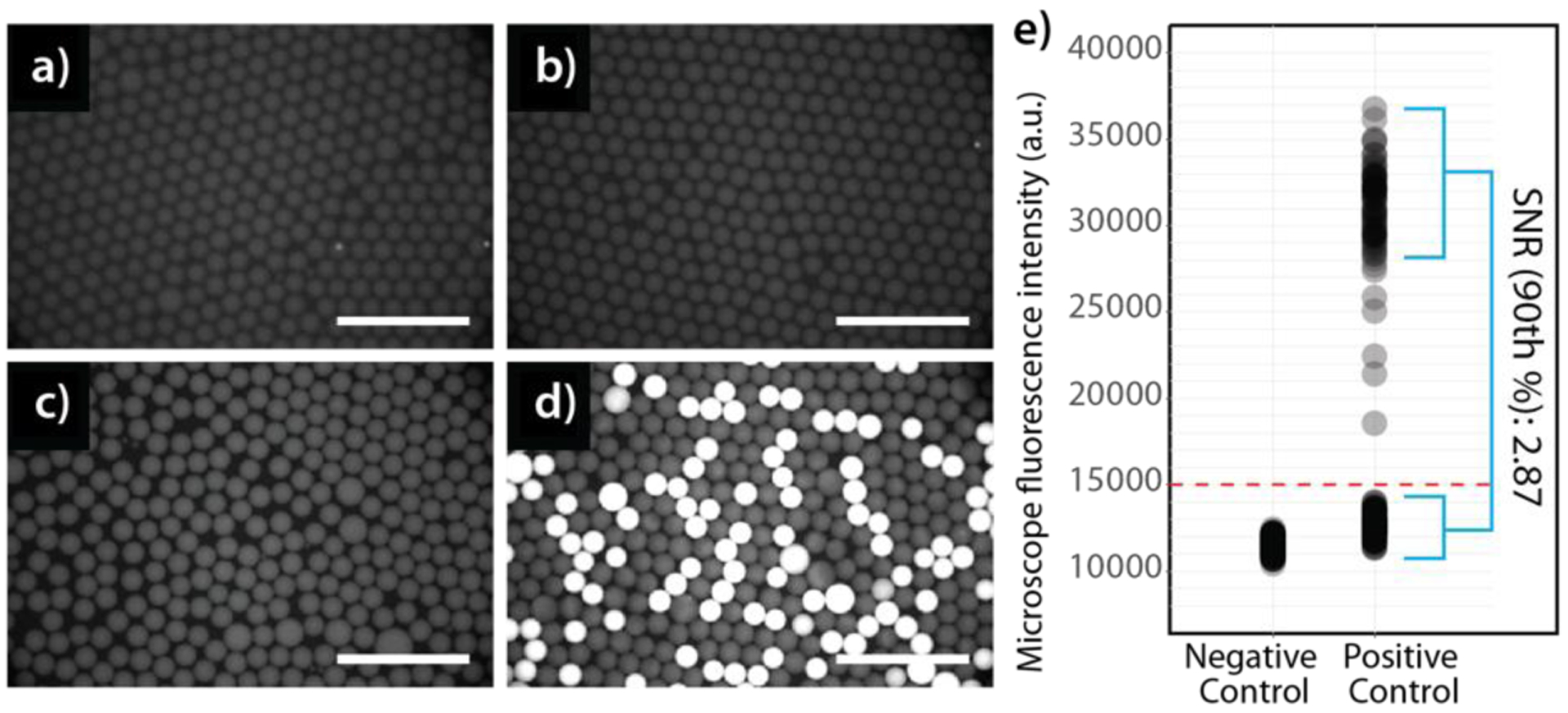
Microscope images of negative (a,b) and positive (c,d) droplet samples encapsulating an engineered E. coli strain containing the synthetic blaCTX-M-9 target gene before (a,c) and after (b,d) thermocycling. Cy5 fluorescence images were recorded to measure droplet SNR for different replicates. (Same intensity range for all panels, scaled to visualize trace fluorescent signal in negative control droplets. Scalebar = 500 μm). e) Quantitative intensity measurements for droplets in panels B (negative control) and D (positive control), where overall SNR averages 2.87 (defined as 90th % positive droplet intensity divided by average negative droplet intensity), (n = 473 individual droplets). The optimal conditions used in this experiment were 10% whole blood, 40 PCR cycles, 2:1 primer-to-probe ratio (1μM primer, 500 nM probe), 5mg/ml BSA, 0.2% NP-40, and a 70 μl mineral oil overlay over droplets during thermocycling.
Single-digit bacteria detection in whole blood using the IC3D blood ddPCR system
The IC3D blood ddPCR system leverages the principles of digital PCR by partitioning a blood sample into millions of picoliter-sized droplets, significantly increasing the concentration of released or amplified molecular targets in droplets containing bacteria (dramatically increasing the target-to-background ratio) and minimizing interference from inhibitors and nonspecific noise. Unlike conventional 1D or 2D digital droplet PCR systems, the IC3D technology is optimized for high-throughput analysis of large sample volumes and can therefore accommodate significantly more reaction partitions than existing systems (e.g. BioRad ddPCR), enabling much higher detection sensitivity(41,42,47).
In the following studies, blood samples spiked with bacteria at pre-defined concentrations were mixed with the appropriate PCR reagents (depending on the target molecular panel), resulting in a final dilution of 10% whole blood. These samples were then encapsulated into millions of picoliter-sized droplets (1 ml PCR sample = ~ 3M droplets) using a custom high-throughput microfluidic droplet generation device (Figure 1.a), thermocycled using a conventional PCR instrument (Figure 1.b), and transferred to a large-volume cuvette (1 cm diameter) for rapid fluorescence scanning using our 3D particle counter (Figure 1.c).
While actuating the cuvette to create a spiral trace with fast rotation (200 rpm) and slower vertical motion (5 mm/s), the fixed, horizontally-oriented, confocal microscope focuses excitation light generated by diode lasers within the cuvette through an objective. Emission light from the sample is collected by the same objective, transmitted through a set of dichroic filters, focused by a lens through a pinhole and collected by a photodetector, which records the temporal fluorescence signal (Figure 5.a). A shape-fitting algorithm is then applied to the fluorescence signal data with a fixed width and variable amplitude that specifically converts fluctuations in the signal data corresponding to positive, florescent droplets into absolute counts over time (Figure 1.d, Figure 5.b).
Figure 5.
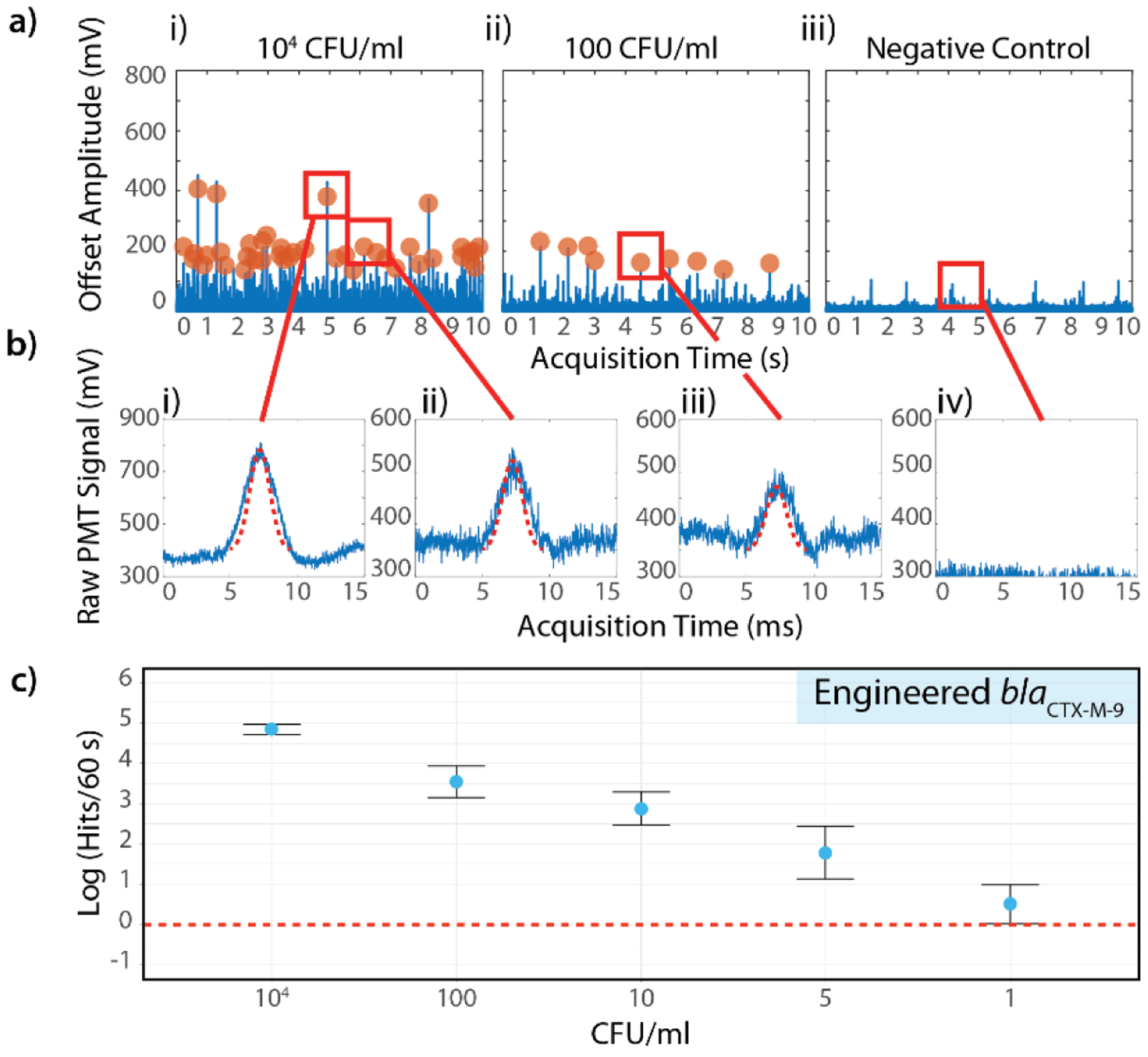
Single-digit sensitivity using the IC3D blood ddPCR system. a) Representative fluorescence time trace data from (i) high concentration sample (104 CFU/ml), (ii) low concentration sample (100 CFU/ml) and (iii) negative control. E. coli strain containing the synthetic blaCTX-M-9 target gene were used as a model target. b) Representative peaks identified by the shape fitting algorithm based on variable peak amplitude, fixed width, and fixed statistical significance. i) High amplitude and ii) lower amplitude peaks both fit acceptance criteria (104 CFU/ml sample), iii) low amplitude peak identified in low concentration sample (100 CFU/ml), iv) false positive rate = 0% for negative control. c) Demonstration of IC3D sensitivity in detecting low concentrations of bacteria spiked into whole blood. Y-axis = log (events per 60s), error bars = relative error (for symmetrical display on log-scale), defined as ±0.434*stDev/y) across 3 independent sample replicates. CFU/ml (x axis) represents the number of spiked bacteria in the final measurement volume.
Using an engineered E. coli strain containing the synthetic blaCTX-M-9 target gene (approximately 5 – 15 copies/bacterium), a carefully-designed study was performed to determine the analytical performance of our IC3D blood ddPCR system in detecting low concentrations of bacteria in unprocessed whole blood (Table S2). Briefly, 6 concentrations were prepared in triplicate for 2 distinct biological preparations on 2 different days including ultra-low bacteria concentrations (1, 5, and 10 CFU/ml in the final measurement volume) prepared using a microinjector (see Materials and Methods, Figure S8). Higher concentrations of 104 and 100 CFU/ml were prepared via serial dilution from a high concentration stock solution of the engineered E. coli with concentration confirmed by optical density and standard CFU plating.
Overall, the ultra-low 1 CFU/ml and 5 CFU/ml samples were each detected in 3 of the corresponding 6 total replicates (50%) (Figure 5.c). Likewise, the 10 CFU/ml sample was detected in 5/6 replicates (83%) and the 100 and 104 CFU/ml samples were detected in 100% of their corresponding replicates with a false positive rate of 0% (Table S3). The aggregate response of all 6 replicates demonstrated a strong linear relationship between the sample bacteria concentration and the number of IC3D hits with average numbers of hits ranging from 1.5 (1 CFU/ml) to 126 (104 CFU/ml) for a 60 second scan. Note that because the sample is not discarded during the detection process, the IC3D system can perform statistical over-sampling by repeatedly interrogating the same sample volume, which increases the probability of detecting ultra-rare events. To express this hit rate in absolute concentrations, reference to a calibration curve with known particle concentrations can be performed.
For comparison of IC3D and conventional real-time PCR assays for quantitative detection of bacteria, PCR mixtures were mixed with serial dilutions of the same bacterial stock (Supplemental Methods). In four replications per PCR reaction, no fluorescence was observed for the negative samples, but successful detection was observed in samples with final bacterial concentrations ≥ 1,000 CFU/ml (Table S4, Figure S9). However, no or little fluorescence was detected for samples with final bacterial concentrations of 50, 100, and 500 CFU/ml. Hence, stable amplification was only observed for 1,000 CFU/ml or higher, indicating the lowest limit of detection by conventional real-time PCR for the same engineered target is 1,000 CFU/ml, compared to 1–10 CFU/ml on the IC3D system. This data again demonstrates droplet digital detection format can greatly improve PCR sensitivity.
We further compared the performance of our one-step blood PCR assay to a commercially available droplet digital PCR platform (BioRad) that counts droplets one at a time in a 1D channel. Because the BioRad ddPCR system is not amenable to directly analyze targets in blood specimens, we first had to extract DNA from a sample of bacteria spiked into whole blood utilizing the manufacturer recommended protocol. Due to limitations of accommodating more than 66ng of gDNA per assay, the limit of detection on the conventional BioRad 1D droplet scanning system was between 50 – 100 CFU/ml with a sample-to-answer turnaround of 6 hours including the sample processing steps (Figure S11, Table S7). In addition to being laborious and undesirable, sample processing also poses risks of losing rare targets from the original sample and leads to increased measurement variation. We further summarized and compared several other representative 1D and 2D droplet counting systems (Table S7). Unfortunately, none of them have the required sensitivity, assay time and simplicity, and therefore, are not suitable for BSI bacterial ID/AST applications.
Specific detection of clinical isolates using the IC3D blood ddPCR system
Following the single-target sensitivity experiments, the specificity of the IC3D blood ddPCR system was assessed in a single-blinded study to detect the presence of blaCTX-M-14 (under the CTX-M-9 family) with 10 clinical isolates of unknown identities. In brief, each clinical isolate and an engineered positive control were mixed with a PCR mixture containing 10% EDTA-treated whole blood at 104 CFU/ml with probes targeting the beta-lactamase CTX-M-9 family gene and encapsulated into droplets.
IC3D blood ddPCR analysis identified four blaCTX-M-14 positive samples appropriately differentiated from six blaCTX-M-14 negative samples with different beta-lactamase genes (Figure 6), including one or more of the following: blaACT-5, blaCMY-2, blaCTX-M-2, blaCTX-M-15, blaDHA-1, blaOXA-1, blaOXA-9, blaOXA-10, blaSHV-1, blaSHV-11, blaSHV-83, blaTEM-1A, blaTEM-1B, blaCMY-80, blaCMY-94, blaKPC-3, blaKPC-4, blaNDM-1 (Table S5). Despite the small number of samples, we demonstrated 100% sensitivity and specificity in this blinded study for the detection of blaCTX-M-14. In addition, quantitative measures demonstrated strong agreement for detecting the isolates at a concentration of 104 CFU/ml (avg hits/60s = 417 ± 92 for positive samples, all negative samples = 0 hits).
Figure 6.
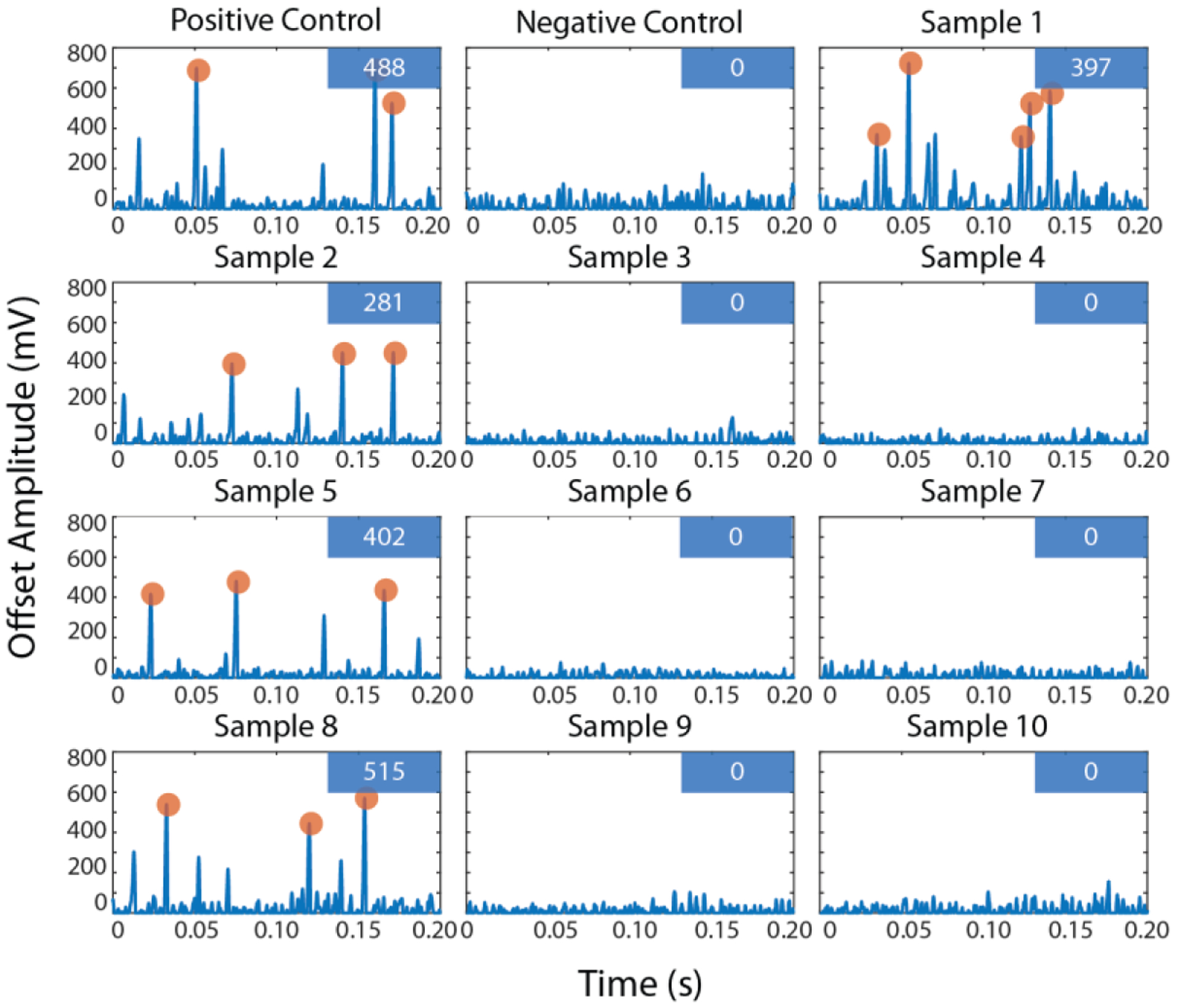
Clinical isolate specificity demonstration with IC3D blood ddPCR raw fluorescent time trace. Each box represents a 200 ms sample of data from a particular clinical sample (see Table S5). The numbers displayed in the box in the upper right corner of each panel are the number of confirmed hits in a 60s scan. (X-axis = acquisition time in seconds, y-axis = offset signal amplitude in mV).
Expansive detection of resistance and speciation markers with the IC3D blood ddPCR system
While the analytical performance of the IC3D system was evaluated with a specific genetic target (blaCTX-M-9) as a model, the capabilities of this technology as a rapid diagnostic platform for managing sepsis and antibiotic resistance are not limited to a particular target. By developing one-step PCR chemistries targeting a wide selection of molecular markers, the IC3D blood ddPCR system has potential to provide more comprehensive information depending on the specific clinical scenario. For example, in some settings, a rapid diagnostic test to quickly rule-in or rule-out a bacterial infection using a pan-bacteria molecular marker could help streamline triage efforts before administering additional diagnostic tests. Additionally, in order to make appropriate antibiotic treatment recommendations, it may be necessary to rapidly obtain more comprehensive information such as Gram-positive vs Gram-negative determination and/or species identification in addition to the status of different antimicrobial resistance markers.
As a demonstration of the IC3D blood ddPCR technology’s ability to cover an extensive range of different molecular targets for the diagnosis and management of BSIs, a series of experiments were performed using clinical isolates spiked into whole blood and interrogated for pan-bacteria (16s rDNA), bacteria speciation (E. coli (uidA) and Klebsiella (khe)), and antibiotic resistance molecular markers including both Gram-positive (nuc and mec A for MRSA and vanA for VRE) and Gram-negative organisms (blaCTX-M-1, blaCTX-M-2, blaOXA-48, blaKPC in both ESBL and CRE classes) (Figure 7). Except for MRSA which used a simultaneous multiplex detection of two different molecular markers (nuc and mecA genes), all probes were labelled with the far-red dye, Quasar 670. Due to differences in probe length and tertiary structure which affect the relative proximity of the fluorophore and quencher, different baseline fluorescence signals and peak amplitudes were observed for different targets across this panel. While this could create issues for particle counting instruments that employ a simple fixed threshold to define positive and negative events, the use of a variable-amplitude shape-fitting algorithm enables the IC3D instrument to adapt to differences in the absolute signal, resulting in higher specificity and more robust detection for a variety of targets. By applying this shape-fitting algorithm, samples containing clinical isolates were confirmed positive by the identification of signal fluctuations matching stringent shape-fitting criteria consistent with positive, fluorescent target-containing droplets. Additionally, the simultaneous detection of two different genetic targets (nuc-FAM and mecA-Quasar 670) for the identification of MRSA demonstrates the ability of the IC3D blood ddPCR system to perform multiplex assays for detecting targets co-localized within the same droplet (and therefore the same single bacterium) or different droplets.
Figure 7.
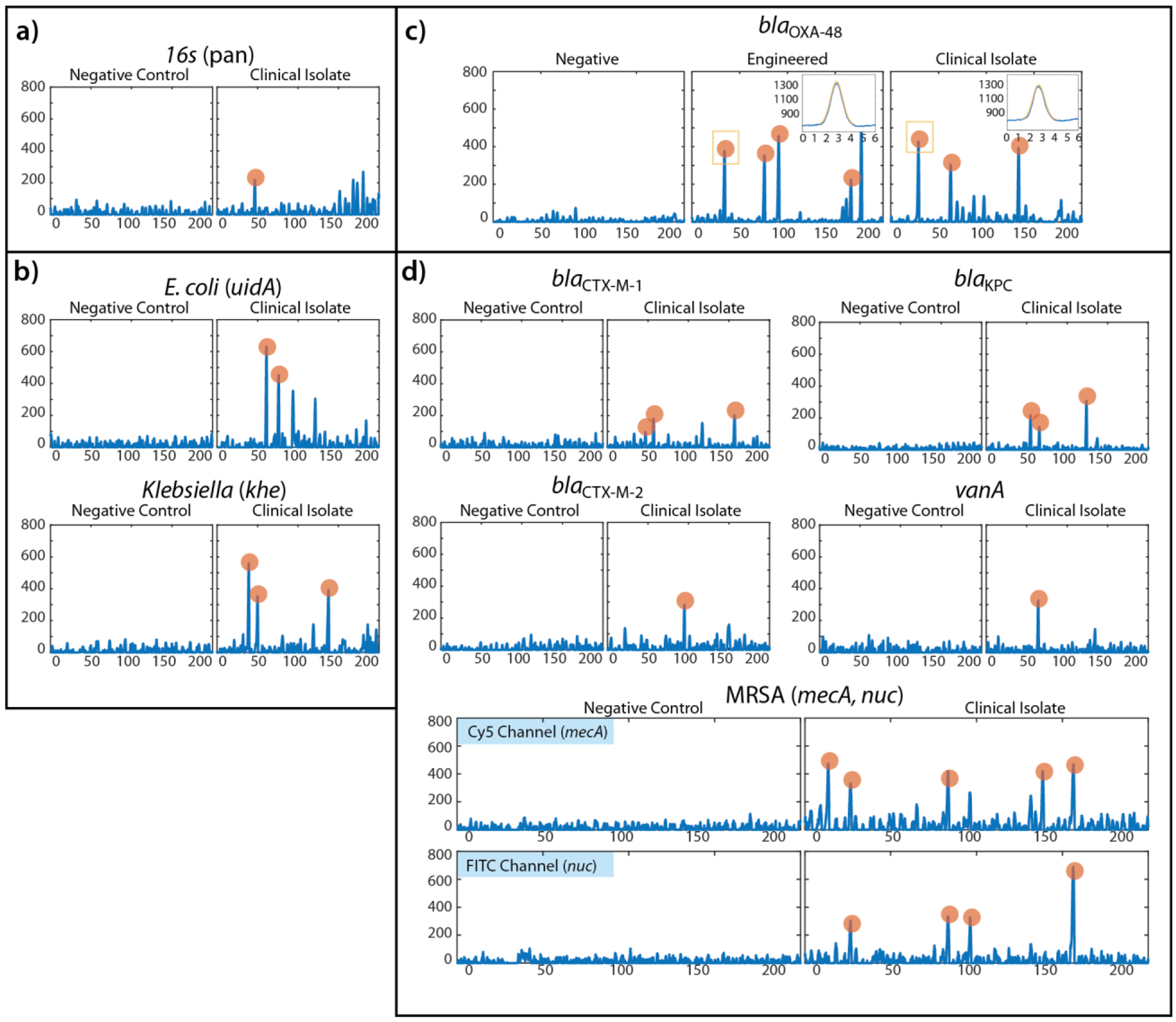
Demonstration of wide applicability of IC3D blood ddPCR technology for the rapid diagnosis and mangement of BSI and antibiotic resistance. a) Pan-bacteria detection with 16s gene panel. b) Species-identification panel including E. coli- (uidA) and Klebsiella- (khe) specific genetic markers. c) Representative example of blaOXA-48 as a model using internal negative control and positive control (engineered bacteria) along with clinical isolate. Inset panels for engineered control and clinical isolates demonstrate representative peaks identified with shape-fitting algorithm (X-axis = 6ms of data at 64kHz, Y-axis = raw signal intensity in mV). d) Antibiotic resistance genetic panel including blaCTX-M-1, blaCTX-M-2, vanA, and blaKPC, and MRSA (nuc, mecA). Each panel is a representative portion of the raw fluorescence signal recorded by the particle counter instrument. X-axis = 200ms of data at 64kHz, Y-axis = offset signal amplitude in mV). Orange highlighted peaks denote signal fluctuations that match the stringent shape-fitting criteria consistent with positive, fluorescent target-containing droplets.
Demonstration of sample-to-answer results in 1 hour with the IC3D blood ddPCR system
Finally, as a demonstration of rapid sample-to-answer turnaround, an experiment was conducted with a modified protocol to increase droplet generation throughput and shorten the time required for signal amplification. Since the IC3D blood ddPCR workflow involves high-throughput droplet generation and rapid 3D scanning, the clear bottleneck remains the time required to achieve complete PCR amplification. In this experiment, the droplet generation flow rate was increased from 50 μl/min (1 ml/ 20 min) to 100 μl/min (1 ml/ 10 min) without any observable compromise in droplet stability. Additionally, the PCR protocol was modified from the standard 40 cycles of 30s denaturation, 30s annealing/extension to 35 cycles of 10s denaturation/ annealing/ extension, reducing the average time to completion from 100 minutes to 42 minutes (Supplemental Methods). By comparing the fluorescence intensity from a positive control run with the standard-length PCR protocol and the rapid-PCR protocol, a minor loss (<5% signal loss; average SNR = 1.5 (standard PCR protocol), SNR = 1.45 (rapid-PCR protocol)) in fluorescence intensity was attributed to the compressed PCR protocol (Figure S10, Table S6). However, due to the sensitivity of the 3D particle counter, positive droplets were detected at approximately the same rate (avg hits/60s = 488 (standard PCR protocol), = 515 (rapid-PCR protocol)) with both protocols, demonstrating that the IC3D blood ddPCR system has the potential to detect antibiotic resistant bacteria from a blood sample in less than one hour.
Discussion
Rapid diagnostic tests can be exceedingly valuable in directing early therapy, improving clinical outcome, and enabling better antibiotic stewardship in BSI management. Yet, comprehensive tests directly applied to whole blood are currently lacking. Indeed, recent studies have shown that reduced time for bacterial ID/AST is correlated with improved mortality and reduced healthcare cost(48–50). Through innovative integration of a novel one-step, blood digital PCR and a large volume, high-throughput particle counting system, we demonstrated targeted bacterial detection and AST directly from blood samples without requiring culture or sample processing. To our knowledge, we are the first to demonstrate robust blood digital PCR, which can be broadly applied to numerous diagnostic settings as it eliminates the need for sample processing steps and improves assay precision, sensitivity and inter-assay reproducibility. In addition, since our IC3D employs a volumetric detection methodology (rather than “flow-based”), it can interrogate a large volume of droplets at a rate of approximately 470 μl/min (see Table S7 for analysis). This high throughput significantly shortens the assay time compared to conventional 1D or 2D particle counting systems for digital PCR and allows us to accommodate a larger sample input, thereby further increasing the chance to detect rare targets. We have performed additional comparisons with promising droplet counting technologies that claim throughputs of 250 kHz(51) – 1 MHz(52), though these have only been evaluated in ideal cases with fluorescent dye and buffer and not yet in a true assay format (Table S7). Indeed, the unique features of our IC3D blood ddPCR allow us to simultaneously achieve unprecedented high sensitivity (10 CFU/ml) with rapid sample-to-answer turnaround (one hour) that no currently available technologies can deliver, which are essential for BSI bacterial ID/AST applications.
Our targeted molecular diagnostic for bacterial detection and AST profiling is intended to serve as a rapid “rule-in” rather than “rule-out” function in the clinical workflow to aid early treatment decisions(53). Given the complexity of BSI pathogens and antibiotic resistance and that molecular assays do not always correlate to phenotypic resistance profiles especially for ESBLs and CREs, we envision our targeted assay will be used in conjunction with, rather than replacing, culture-based phenotypic methods and other molecular assays including sequencing that can survey broader genetic profiles that would be more suited for broad species identification and detection of unknown pathogens or resistance markers and polymicrobial bacteremia. Another limitation of this method is the increased cost due to the large volume of samples and reagents (e.g. polymerases) we employ in our assay, which could be reduced through mass production in the future. With our highly sensitive system, false positives can be an issue, especially in the case of pan-bacteria PCR detection, due to circulating bacterial genetic material or contamination from reagents and work environments which requires careful screening of vendors and using methods that remove or suppress such contamination. Currently, our prototype involves several manual transfer steps that have potential to lose rare targets in concentrations of <10 CFUs/ml; future efforts to integrate and automate this process will eliminate these steps to further improve assay sensitivity and robustness. We are currently performing a clinical study to assess the feasibility of fitting the IC3D blood ddPCR system into current clinical workflows of BSI management as well as its potential clinical value.
Materials and Methods
Sample Preparation
As a model system, an engineered E. coli strain containing a low copy number plasmid (approx. 5 – 15 copies/bacterium) pBR322 blaCTX-M-9 kanr with a synthetic blaCTX-M-9 target gene was used to evaluate the analytical performance of the IC3D blood ddPCR system (see Supplemental Methods). To assess the analytical detection sensitivity, a total of six bacterial concentrations were prepared in triplicate including a negative control; concentrations of 104 and 100 CFU/ml were prepared by serial dilution while the concentrations of 1, 5, and 10 CFU/ml were prepared using a microinjector (see below). To assess the target specificity within a particular family of genes and across multiple antibiotic resistance and species-specific genes, clinical isolates and those obtained from the CDC and FDA Antimicrobial Resistance Bank were subcultured twice onto 5% sheep blood agar plates. Serial dilutions were prepared from overnight cultures by touching the top of three well isolated colonies which were emulsified in sterile saline, adjusted to a 0.5 McFarland standard. Serial dilutions of this standardized suspension were made in saline and plated to verify the colony count and to check for purity.
De-identified and healthy donor blood samples used in this study were obtained with informed consent from donors and approval from the Institutional Review Board (IRB 2012–9023) via the Blood Donor Program at the UCI Institute for Clinical and Translational Science (ICTS). One day prior to each experiment, a PCR master mix containing EDTA-treated whole blood at a dilution of 10% for droplet generation was prepared with all components added, except bacteria (or PBS for negative control samples). A PCR master mix for droplet generation was prepared containing 10% whole blood (EDTA-treated), inhibitor-resistant Taq (VitaNavi Technologies), PCR buffer (1x), PEC buffer (1x), dNTP (0.2 mM), BSA (5 mg/ml), forward and reverse primers (1 μM each), and target-specific probe (0.5 μM). The PCR master mix was subjected to filtration with a 10-micron filter and stored at −20 °C. On the day of each experiment, final samples were prepared by spiking bacteria (or PBS for negative control samples) into the corresponding thawed PCR master mix. Droplets were prepared for each sample using separate microfluidic devices to avoid carry-over contamination according to methods described below.
Low CFU Sample Preparation via Microinjection
Low CFU samples (1, 5, and 10 CFU/ml) were prepared using a microinjector for superior precision and repeatability compared to serial dilution. Samples were prepared on the day preceding each experiment at the UC Irvine California Institute of Regenerative Medicine (CIRM) IVF workstation (TransferMan® NK2 micromanipulator, CellTram Vario microinjector, Eppendorf). Individual bacteria were aspirated from a stock suspension of 106 CFU/ml in LB media with a 4 μm inner diameter capillary (Eppendorf ICSI TransferTips) and dispensed into 50 μL aliquots of PBS to achieve the desired target concentration. Immediately after the desired target concentration was confirmed, samples were stored on ice until all remaining samples were prepared. All samples were then stored overnight at 4 °C to prevent bacterial growth (see Supplemental Methods, Figure S8).
Droplet Generation
Samples were emulsified with the use of 3M™ Novec™ 7500 Engineered Fluid (fluorinated oil) containing 2% PFPE surfactant (prepared by Velox Biosystems with our proprietary formulation). Samples were loaded into syringes on top of pure HFE-7500 fluorocarbon oil for volume displacement in the needle and tubing. A second syringe was used to load fluorinated oil with 2% PFPE surfactant. Both syringes were vertically mounted onto syringe pumps (Harvard Apparatus, PhD Ultra™) and connected to the sample and oil inlets of individual microfluidic devices via needles and tubing. Optical microscopes (Celestron® LCD Digital Microscope) were used to monitor droplet generation. Samples for IC3D analysis were prepared with custom designed and fabricated PDMS droplet generation chips (Figure 2.a). Each chip contained 4 parallel flow-focusing droplet-generation nozzles with a single aqueous inlet and oil inlet. Droplets were collected in sterile collection tubes in 2% PFPE surfactant oil to preserve the integrity of the droplet size and shape.
Blood droplet generation and blood droplet digital PCR
Prior to thermocycling, droplet samples were transferred to thin-walled PCR tubes, each containing 30 μl Novec™ 7500 oil with 5% PFPE surfactant and 70 μl droplets, overlaid with 60 μl of mineral oil. For droplet PCR, the following PCR protocol was used: (1) 5 min at 95 °C (lysis and initial denaturation), (2) 10 sec at 95 °C (denaturation), (3) 30 sec at 50 °C, (4) Repeat Steps 2 & 3 for 40 cycles. Following thermocycling, fluorescence droplet images were acquired for negative and positive control samples (see below). Droplet samples for IC3D scanning were manually transferred from individual PCR tubes into thoroughly cleaned, borosilicate glass cuvettes loaded with 1.5 ml of 3M™ Novec™ 7500 Engineered Fluid (fluorinated oil) containing 2% surfactant. For PCR (droplet PCR and real time PCR), the primer and probe sequences can be found in Table S1.
Microscope Evaluation
For the negative and positive control samples (Pre/Post-thermocycling), approximately 10 μl of droplets were dispensed onto a disposable hemocytometer (Invitrogen™ Countess Cell Counting Chamber Slides) for droplet size analysis. Three unique fields were imaged using both a brightfield and CY5 filter set at 4x and 10x magnification (Nikon Eclipse TE-1000) (Figure 3). A custom GUI programmed in Matlab™ (MathWorks®, R2017a) was used to identify droplets in the recorded images and measure droplet diameters and fluorescence intensity. Droplet generation consistency was assessed by measuring the coefficient of variation of droplet diameters (%CV = [StDev/Average]*100). Droplets generated in this study averaged approximately 85 μm in diameter with a %CV < 5%.
IC3D droplet scanning and analysis
The droplet scanning instrument used in this study consisted of a low-cost, bench-top, horizontal-geometry confocal microscope (Figure 1.c). In this embodiment, a visible-wavelength excitation laser beam (λ = 647 nm with ~5 mW typical output power) was focused by an under-filled low numerical aperture 20x objective (MV-20x, Newport) inside a cylindrical cuvette (Abbott) containing the droplet sample. Slow (5 mm/s) vertical translation and fast (200 rpm) rotation of the cuvette transported the target fluorescent droplets across the Gaussian-shaped excitation volume. The emitted fluorescence signal was collected by the same objective, transmitted through the dichroic filters and a 1mm pinhole and collimated onto the sensitive area of a photomultiplier tube (H9305–04 PMT, Hamamatsu) for the time-trace acquisition (acquisition time = 60 seconds). To improve sampling, two additional 60 second recordings were obtained for each sample after thoroughly re-mixing the droplets using a large orifice 1000 μl pipette.
To achieve high specificity while counting the number of detection events (hits), fluorescence time trace data was fit to a pre-determined Gaussian profile of fixed standard deviation (peak width, based on droplet size) and variable amplitude. Hits were only counted if the chi-square value of the shape-fit was statistically significant at the desired level (e.g. Χ2 < 0.05) and the peak amplitude was above a user-defined minimum threshold.
Supplementary Material
Acknowledgments:
We thank Neto Sosa, Dr. Christopher Heylman, Dr. Dong-Ku Kang, Dr. Monsur Ali, Dr. Egest Pone, Dr. Hongzhang (Simon) He, and Dr. Xuning (Emily) Guo for their assistance on the project; Dr. Luis Mota-Bravo and Dr. Saahir Khan for useful discussions; and Dr. Beniamino Barbieri at ISS Inc. for input on instrumentation.
Funding: This study is supported by NIH/NIAID (1 R01 AI117061), UCI Applied Innovation’s Proof of Product (POP) Grants. T. V. is supported by NSF GRFP (DGE-1839285). J. Z. is supported by NIH TL1 training grant (TR001415). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing interests: Weian Zhao is the founder of Velox Biosystems Inc that develops in vitro diagnostics.
Data and materials availability: Primer and probe DNA sequences for all genes presented in this study are provided in Table S1. All additional data that supports the findings of this study are available from the corresponding author [WZ] on reasonable request.
References and Notes:
- 1.World Health Organization (WHO). Fact sheet: Sepsis [Internet]. [cited 2019 Mar 29]. Available from: https://www.who.int/news-room/fact-sheets/detail/sepsis
- 2.The Alliance for the, Prudent Use of Antibiotics. The cost of antibiotic resistance to U.S. families and the health care system. 2010.
- 3.Li Y, Yang X, Zhao W. Emerging Microtechnologies and Automated Systems for Rapid Bacterial Identification and Antibiotic Susceptibility Testing. SLAS Technol. 2017. Dec;22(6):585–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006. June;34(6):1589–96. [DOI] [PubMed] [Google Scholar]
- 5.Kang C-I, Kim S-H, Park WB, Lee K-D, Kim H-B, Kim E-C, et al. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother. 2005. February;49(2):760–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luna CM, Aruj P, Niederman MS, Garzon J, Violi D, Prignoni A, et al. Appropriateness and delay to initiate therapy in ventilator-associated pneumonia. Eur Respir J. 2006. January;27(1):158–64. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States, 2013. 2013.
- 8.World Health Organization (WHO). Antimicrobial resistance. Global report on surveillance. 2014.
- 9.Boucher HW. Challenges in anti-infective development in the era of bad bugs, no drugs: a regulatory perspective using the example of bloodstream infection as an indication. Clin Infect Dis. 2010. January 1;50 Suppl 1:S4–9. [DOI] [PubMed] [Google Scholar]
- 10.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, et al. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008. January 15;46(2):155–64. [DOI] [PubMed] [Google Scholar]
- 11.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013. Jan;34(1):1–14. [DOI] [PubMed] [Google Scholar]
- 12.White House, Washington DC: The National Action Plan for Combating Antibiotic-Resistant Bacteria. 2015. [Google Scholar]
- 13.Pluddemann Annette, Onakpoya Igho, Harrison Sian, Shinkins Bethany, Tompson Alice, Davis Ruth, et al. Position Paper on Anti-Microbial Resistance Diagnostics – Centre for Evidence-Based Medicine, University of Oxford June 2015; 2015. [Google Scholar]
- 14.van Belkum A, Bachmann TT, Lüdke G, Lisby JG, Kahlmeter G, Mohess A, et al. Developmental roadmap for antimicrobial susceptibility testing systems. Nature Reviews Microbiology. 2019. January 1;17(1):51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley SO. New Technologies for Rapid Bacterial Identification and Antibiotic Resistance Profiling. SLAS Technol. 2017. Apr;22(2):113–21. [DOI] [PubMed] [Google Scholar]
- 16.Neely LA, Audeh M, Phung NA, Min M, Suchocki A, Plourde D, et al. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci Transl Med. 2013. April 24;5(182):182ra54. [DOI] [PubMed] [Google Scholar]
- 17.Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip. 2008. Aug;8(8):1265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi J, Yoo J, Lee M, Kim E-G, Lee JS, Lee S, et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci Transl Med. 2014. December 17;6(267):267ra174. [DOI] [PubMed] [Google Scholar]
- 19.Altun O, Almuhayawi M, Ullberg M, Ozenci V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol. 2013. December;51(12):4130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marlowe EM, Novak-Weekley SM, Cumpio J, Sharp SE, Momeny MA, Babst A, et al. Evaluation of the Cepheid Xpert MTB/RIF assay for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J Clin Microbiol. 2011. April;49(4):1621–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, et al. Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J Clin Microbiol. 2013. July;51(7):2072–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Issadore D, Chung HJ, Chung J, Budin G, Weissleder R, Lee H. μHall Chip for Sensitive Detection of Bacteria. Advanced Healthcare Materials. 2013. Sep 1;2(9):1224–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou HW, Bhattacharyya RP, Hung DT, Han J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip. 2015. May 21;15(10):2297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohlsson P, Evander M, Petersson K, Mellhammar L, Lehmusvuori A, Karhunen U, et al. Integrated Acoustic Separation, Enrichment, and Microchip Polymerase Chain Reaction Detection of Bacteria from Blood for Rapid Sepsis Diagnostics. Anal Chem. 2016. October 4;88(19):9403–11. [DOI] [PubMed] [Google Scholar]
- 25.Park KS, Huang C-H, Lee K, Yoo Y-E, Castro CM, Weissleder R, et al. Rapid identification of health care-associated infections with an integrated fluorescence anisotropy system. Sci Adv. 2016. May;2(5):e1600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sage AT, Besant JD, Lam B, Sargent EH, Kelley SO. Ultrasensitive electrochemical biomolecular detection using nanostructured microelectrodes. Acc Chem Res. 2014. August 19;47(8):2417–25. [DOI] [PubMed] [Google Scholar]
- 27.Nemr CR, Smith SJ, Liu W, Mepham AH, Mohamadi RM, Labib M, et al. Nanoparticle-Mediated Capture and Electrochemical Detection of Methicillin-Resistant Staphylococcus aureus. Anal Chem. 2019. February 19;91(4):2847–53. [DOI] [PubMed] [Google Scholar]
- 28.Liao JC, Mastali M, Gau V, Suchard MA, Moller AK, Bruckner DA, et al. Use of electrochemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens. J Clin Microbiol. 2006. February;44(2):561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014. November 6;159(4):925–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018. April 27;360(6387):439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerging infectious diseases. 2012. Sep;18(9):1503–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao MD, Ganesamoorthy D, Elliott AG, Zhang H, Cooper MA, Coin LJM. Streaming algorithms for identification of pathogens and antibiotic resistance potential from real-time MinION(TM) sequencing. Gigascience. 2016. Jul 26;5(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015. May 13;7(287):287ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crawford K, DeWitt A, Brierre S, Caffery T, Jagneaux T, Thomas C, et al. Rapid Biophysical Analysis of Host Immune Cell Variations Associated with Sepsis. Am J Respir Crit Care Med. 2018. Jul 15;198(2):280–2. [DOI] [PubMed] [Google Scholar]
- 35.Hassan U, Ghonge T, Reddy BJ, Patel M, Rappleye M, Taneja I, et al. A point-of-care microfluidic biochip for quantification of CD64 expression from whole blood for sepsis stratification. Nat Commun. 2017. Jul 3;8:15949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Currie B Impact of molecular diagnostics on infection control. Infect Dis Spec Edn; 2011;14(5). [Google Scholar]
- 37.Lee A, Mirrett S, Reller LB, Weinstein MP. Detection of bloodstream infections in adults: how many blood cultures are needed? J Clin Microbiol. 2007. Nov;45(11):3546–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida A, Nagashima S, Ansai T, Tachibana M, Kato H, Watari H, et al. Loop-mediated isothermal amplification method for rapid detection of the periodontopathic bacteria Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. J Clin Microbiol. 2005. May;43(5):2418–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Nordmann P. Evaluation of a DNA microarray for the rapid detection of extended-spectrum beta-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM). J Antimicrob Chemother. 2012. Aug;67(8):1865–9. [DOI] [PubMed] [Google Scholar]
- 40.Muthukumar A, Zitterkopf NL, Payne D. Molecular Tools for the Detection and Characterization of Bacterial Infections: A Review. Laboratory Medicine. 2008. Jul 1;39(7):430–6. [Google Scholar]
- 41.Kang D-K, Ali MM, Zhang K, Huang SS, Peterson E, Digman MA, et al. Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection. Nat Commun. 2014. November 13;5:5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K, Kang D-K, Ali MM, Liu L, Labanieh L, Lu M, et al. Digital quantification of miRNA directly in plasma using integrated comprehensive droplet digital detection. Lab Chip. 2015. Nov 7;15(21):4217–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Kermekchiev MB, Barnes WM. Direct DNA amplification from crude clinical samples using a PCR enhancer cocktail and novel mutants of Taq. The Journal of molecular diagnostics : JMD. 2010. March;12(2):152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kermekchiev MB, Kirilova LI, Vail EE, Barnes WM. Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic acids research. 2009. April;37(5):e40–e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Applied and environmental microbiology. 1996. March;62(3):1102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010. March;54(3):969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ou C-Y, Vu T, Grunwald JT, Toledano M, Zimak J, Toosky M, et al. An ultrasensitive test for profiling circulating tumor DNA using integrated comprehensive droplet digital detection. Lab Chip. 2019;19:993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doern GV, Vautour R, Gaudet M, Levy B. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J Clin Microbiol. 1994. July;32(7):1757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beekmann SE, Diekema DJ, Chapin KC, Doern GV. Effects of rapid detection of bloodstream infections on length of hospitalization and hospital charges. J Clin Microbiol. 2003. July;41(7):3119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endimiani A, Jacobs MR. The Changing Role of the Clinical Microbiology Laboratory in Defining Resistance in Gram-negatives. Infect Dis Clin North Am. 2016. June;30(2):323–45. [DOI] [PubMed] [Google Scholar]
- 51.Kim M, Pan M, Gai Y, Pang S, Han C, Yang C, et al. Optofluidic ultrahigh-throughput detection of fluorescent drops. Lab Chip. 2015;15(6):1417–23. [DOI] [PubMed] [Google Scholar]
- 52.Yelleswarapu VR, Jeong H-H, Yadavali S, Issadore D. Ultra-high throughput detection (1 million droplets per second) of fluorescent droplets using a cell phone camera and time domain encoded optofluidics. Lab Chip. 2017;17(6):1083–94. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Yang X, Zhao W. Emerging Microtechnologies and Automated Systems for Rapid Bacterial Identification and Antibiotic Susceptibility Testing. SLAS TECHNOLOGY: Translating Life Sciences Innovation. 2017. Aug 29;22(6):585–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pekin D, Skhiri Y, Baret J-C, Le Corre D, Mazutis L, Salem CB, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011. Jul 7;11(13):2156–66. [DOI] [PubMed] [Google Scholar]
- 55.Laboratories BioRad, Inc. ddPCR Supermix for Probes: “Recommendations for Optimal Results.” [Google Scholar]
- 56.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(1):257–66. [DOI] [PubMed] [Google Scholar]
- 57.Silkie SS, Tolcher MP, Nelson KL. Reagent decontamination to eliminate false-positives in Escherichia coli qPCR. J Microbiol Methods. 2008. March;72(3):275–82. [DOI] [PubMed] [Google Scholar]
- 58.Hartman LJ, Selby EB, Whitehouse CA, Coyne SR, Jaissle JG, Twenhafel NA, et al. Rapid real-time PCR assays for detection of Klebsiella pneumoniae with the rmpA or magA genes associated with the hypermucoviscosity phenotype: screening of nonhuman primates. J Mol Diagn. 2009. Sep;11(5):464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim SW, Abate AR. Ultrahigh-throughput sorting of microfluidic drops with flow cytometry. Lab Chip. 2013. Dec 7;13(23):4563–72. [DOI] [PubMed] [Google Scholar]
- 60.Terekhov SS, Smirnov IV, Stepanova AV, Bobik TV, Mokrushina YA, Ponomarenko NA, et al. Microfluidic droplet platform for ultrahigh-throughput single-cell screening of biodiversity. Proc Natl Acad Sci U S A. 2017. March 7;114(10):2550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


