Abstract
The mode of acquisition and causes for the variable clinical spectrum of coronavirus disease 2019 (COVID-19) remain unknown. We utilized a reverse genetics system to generate a GFP reporter virus to explore severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogenesis and a luciferase reporter virus to demonstrate sera collected from SARS and COVID-19 patients exhibited limited cross-CoV neutralization. High-sensitivity RNA in situ mapping revealed the highest angiotensin-converting enzyme 2 (ACE2) expression in the nose with decreasing expression throughout the lower respiratory tract, paralleled by a striking gradient of SARS-CoV-2 infection in proximal (high) versus distal (low) pulmonary epithelial cultures. COVID-19 autopsied lung studies identified focal disease and, congruent with culture data, SARS-CoV-2-infected ciliated and type 2 pneumocyte cells in airway and alveolar regions, respectively. These findings highlight the nasal susceptibility to SARS-CoV-2 with likely subsequent aspiration-mediated virus seeding to the lung in SARS-CoV-2 pathogenesis. These reagents provide a foundation for investigations into virus-host interactions in protective immunity, host susceptibility, and virus pathogenesis.
Keywords: SARS-CoV-2, COVID-19, reverse genetics, infectious clone, nasal infection, neutralization assay, primary cells, reporter virus, respiratory tropism, ACE2
Graphical Abstract
Highlights
-
•
A SARS-CoV-2 infectious cDNA clone and reporter viruses are generated
-
•
SARS-CoV-2 and SARS-CoV neutralization assays show limited cross neutralization
-
•
SARS-CoV-2 shows a gradient infectivity from the proximal to distal respiratory tract
-
•
Ciliated airway cells and AT-2 cells are primary targets for SARS-CoV-2 infection
Hou et al. present a reverse genetics system for SARS-CoV-2, which is then used to make reporter viruses to quantify the ability of patient sera and antibodies to neutralize infectious virus and to examine viral tropism along the human respiratory tract.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been identified as the causative agent of the ongoing pandemic coronavirus disease 2019 (COVID-19) (Gorbalenya et al., 2020). SARS-CoV-2 emerged in Wuhan, China in December 2019 and rapidly spread to more than 175 countries within three months (Huang et al., 2020, Zhu et al., 2020). As of May 18, 2020, about 4.7 million confirmed cases and > 316,000 deaths have been reported worldwide. The absence of approved vaccines and only a single emergency-use FDA-approved therapeutic against SARS-CoV-2 hinders pandemic control.
The genome of SARS-CoV-2 is an ∼30 kb RNA predicted to encode 16 non-structural proteins (nsp1–nsp16), four structural proteins (spike, membrane, envelope, and nucleocapsid), and eight accessory proteins (3a, 3b, 6, 7a, 7b, 8b, 9b, and 14) (Wu et al., 2020a), expressed from genome-length or sub-genomic mRNAs. The spike (S) glycoprotein mediates viral entry via binding to the human angiotensin-converting enzyme (ACE2) (Hoffmann et al., 2020, Walls et al., 2020, Yan et al., 2020), followed by proteolytic processing by transmembrane protease, serine 2 (TMPRSS2), furin, and perhaps other lung proteases, which trigger fusion of viral and cellular membranes. Spike glycoprotein is also the main target of host neutralizing antibodies (nAbs) (Hoffmann et al., 2020).
SARS-CoV-2 infection primarily targets the respiratory tract. A fraction of SARS-CoV-2 infections manifest as bilateral lower-zone pneumonias and diffuse alveolar damage (DAD) that might progress to acute respiratory distress syndrome (ARDS), especially in the aged and individuals with co-morbidities (Carsana et al., 2020, Guan et al., 2020). In comparison to symptoms of MERS-CoV and SARS-CoV 2003 infections, clinical symptoms of COVID-19 are broader and more variable (Huang et al., 2020, Pan et al., 2020a, Wu and McGoogan, 2020, Zhu et al., 2020). Differences in transmissibility and viral shedding suggest the in vivo replication sites and/or replication efficiency of SARS-CoV-2 differ significantly from SARS-CoV (Pan et al., 2020b, Wölfel et al., 2020, Zou et al., 2020).
A wealth of single-cell RNA sequencing (scRNA-seq) data have been mobilized to describe the expression of ACE2 and TMPRSS2 with emphasis on the human respiratory tract (Aguiar et al., 2020, Sajuthi et al., 2020, Sungnak et al., 2020). However, complementary techniques are needed to describe the organ-level architecture of receptor expression, improve on the sensitivity of scRNA for low-expression genes, e.g., ACE2, and to describe the function of ACE2, i.e., mediate infectivity. Accordingly, a combination of RNA in situ hybridization (RNA-ISH) techniques, a novel set of SARS-CoV-2 reporter viruses produced by reverse genetics, and primary cultures from all affected regions of the respiratory tract was assembled for our investigations.
We utilized the reverse genetics systems to test for protection and/or durability of protection afforded by convalescent serum and/or SARS-CoV-2-specific monoclonal antibodies (mAbs) and antigenicity relationships between SARS-CoV and SARS-CoV-2 after natural human infections. These tools were also utilized to contrast two non-exclusive hypotheses that might account for key aspects of SARs-CoV-2 transmission and pathogenesis: (1) transmission is mediated by airborne microparticles directly infecting the lung (Morawska and Cao, 2020, Wilson et al., 2020); or (2) the nose is the initial site of infection, followed by aspiration of the viral inoculum from the oropharynx into the lung (Dickson et al., 2016, Wölfel et al., 2020). Accordingly, we characterized the ACE2 and TMPRSS2 expression amounts in the nose and lung and in parallel the SARS-CoV-2 infection of human nasal, bronchial, bronchiolar, and alveolar epithelial cultures. These findings were compared with virus distributions and tropisms in lungs from lethal COVID-19 cases.
Results
Recombinant viruses replicate similarly to the SARS-CoV-2 clinical isolate in vitro
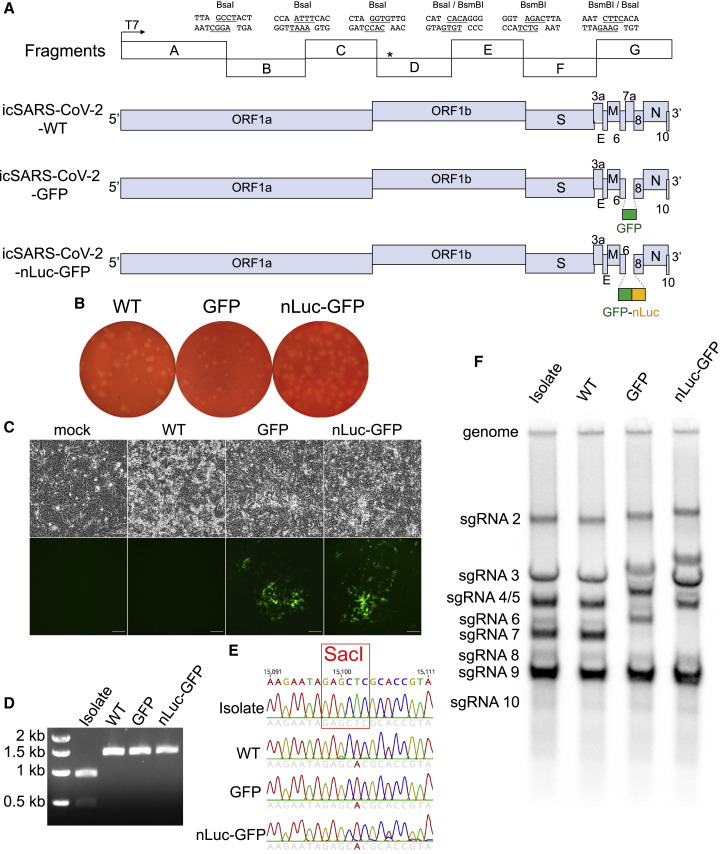
A full-length infectious complementary DNA (cDNA) clone of a US SARS-CoV-2 clinical isolate WA1 was generated by cloning seven genomic fragments separately into vector plasmids (Figure 1 A). Additionally, two reporter viruses were constructed by replacing a 276-bp region in ORF7 with a green fluorescent protein (GFP) or a GFP-fused nanoluciferase (nLuc) gene (Figure 1A). After assembly into full-length cDNA, full-length RNA was electroporated into Vero-E6 cells (Scobey et al., 2013, Yount et al., 2003). After recovering the wild-type (WT), icSARS-CoV-2-GFP, and icSARS-CoV-2-nLuc-GFP recombinant viruses, viral replication was confirmed by the presence of sub-genomic-length leader-containing RNA transcripts 20 h after electroporation (Figure S1 ). All three recombinant viruses replicated (Figure S1), generated similar plaques in Vero E6 cells, and could be passaged serially in the cell culture without exogenous trypsin (Figure 1B). We defined cytopathic effect (CPE) by cell rounding and detachment from monolayers. GFP signals were evident in cells two days after transfection with RNA transcripts from both indicator viruses (Figure 1C).
Figure 1.
Design and recovery of SARS-CoV-2 recombinant viruses
(A) Full-length cDNA clone constructs and genomes of recombinant viruses. Restriction sites, cohesive ends, and the genetic marker T15102A (∗) are indicated in the schematic diagram. GFP or GFP-fused nLuc genes were introduced into the ORF7 (replacing aas 14–104) of SARS-CoV-2 genome.
(B) Plaques of recombinant viruses.
(C) CPE and GFP signals were observed in Vero-E6 cells electroporated with sub-genomic RNA (sgRNA)-N alone (mock) or sgRNA-N mixed with full-length RNA transcripts (recombinant viruses) at two days after transfection. Scale bar, 100 μm.
(D and E) SacI digestion (D) and Sanger sequencing (E) of a 1.5-kb region covering the genetic marker in vial genomes.
(F) Northern blot analysis of genomic and sgRNAs isolated from the virus-infected cells. Abbreviations are as follows: Isolate, clinical isolate strain WA1; WT, icSARS-CoV-2-WT; GFP, icSARS-CoV-2-GFP, nLuc-GFP: icSARS-CoV-2-nLuc-GFP.
See also Figure S1.
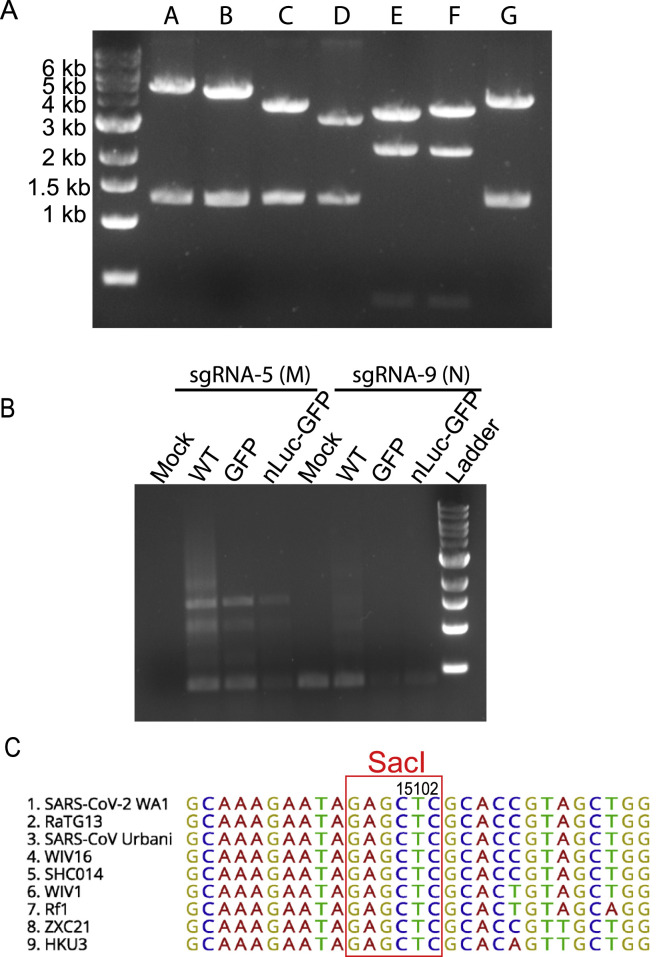
Figure S1.
Additional information for the SARS-CoV-2 infectious cDNA clone, related to Figure 1
(A) Electrophoresis of seven restriction enzyme-digested infectious cDNA clone plasmids. Plasmid A was digested with NotI and BsaI; plasmids B, C, and D were digested with BsaI; plasmids E and F were digested with BsmBI; plasmid G was digested with SalI and BsaI.
(B) Amplification SARS-CoV-2 sgRNAs using primers targeting sgRNA-5 (M) and −9 (N). Cellular RNA samples were collected from Vero-E6 cells electroporated with viral RNA transcripts at 20 h. Mock cells were electroporated with SARS-CoV-2 sgRNA-9 alone.
(C) Alignment of sequences containing the T #15102 in nsp12 gene among 9 different group 2b CoVs.
To distinguish our recombinant viruses from the circulating SARS-CoV-2 strains, we introduced a silent mutation (T15102A) into a conserved region in nsp12 to ablate an endogenous SacI site in the molecular clone (Figure S1). We confirmed the presence of this mutation in all three recombinant viruses but not in the clinical SARS-CoV-2 isolate via Sanger sequencing and PCR amplification followed by SacI digestion (Figures 1D and 1E). To evaluate viral RNA synthesis, we performed Northern blot analyses that showed that the number of sub-genomic RNA (sgRNA) bands was equivalent in the recombinant and clinical isolates, confirming the presence of eight principle sub-genomic mRNAs during infection (Figure 1F). As expected, the molecular weights of sgRNA 2 to sgRNA 7 in the two reporter viral samples were higher than those in the clinical isolate and WT samples, reflecting the insertion of the 720-bp GFP gene or the 1,233-bp nLuc-GFP gene into the 366-bp ORF7 genetic location. These data also demonstrated that ORF7 was not essential for in vitro replication of SARS-CoV-2.
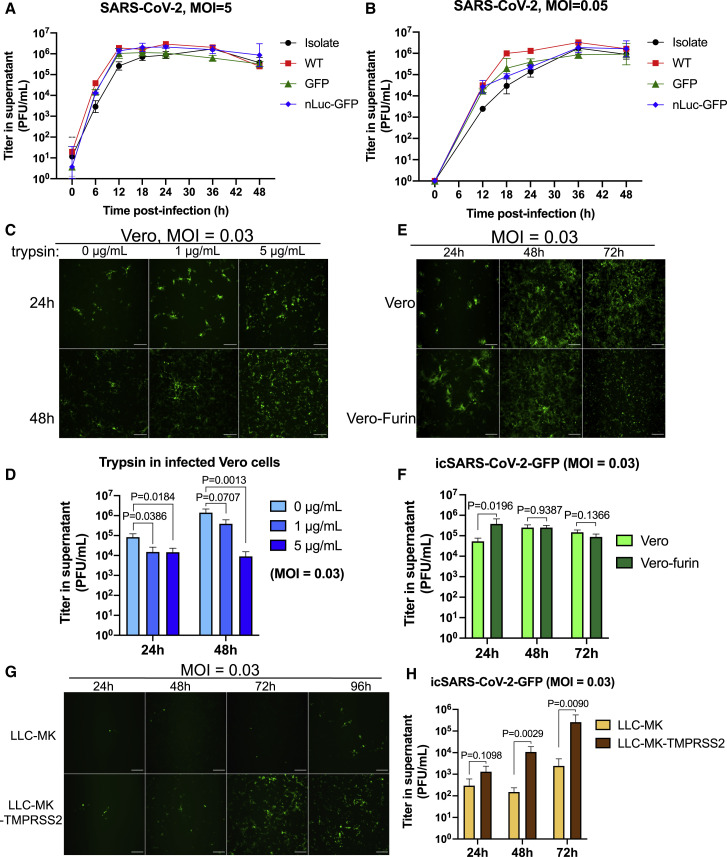
Next, we evaluated one-step (multiplicity of infection [MOI] = 5) and multi-step (MOI = 0.05) growth curves of the three recombinant viruses in Vero E6 cells in comparison to the clinical isolate WA1 strain. The titer of all SARS-CoV-2 increased and plateaued to mid-106 plaque-forming units (PFU)/mL within 12–18 h in the one-step curve and within 36–48 h in the multi-step curve (Figures 2A and 2B). In contrast to other reported indicator viruses (Thao et al., 2020), the three recombinant viruses replicated to titers equivalent to the clinical isolate.
Figure 2.
Growth curves and the role of proteases in SARS-CoV-2 replication
(A and B) One-step (A) and multi-step (B) growth curves of clinical isolate and recombinant viruses in Vero E6 cells, with MOI of 5 and 0.05, respectively.
(C and D) Fluorescent images (C) and viral titers (D) of the SARS-CoV-2-GFP replicates in Vero cells supplemented with different concentrations of trypsin.
(E and F) Fluorescent images (E) and viral titers (F) of the SARS-CoV-2-GFP replicates in normal Vero or Vero-furin cells.
(G and H) Fluorescent images (G) and viral titers (H) of the SARS-CoV-2-GFP replicates in normal LLC-MK or LLC-MK-TMPRSS2 cells.
All scale bars, 200 μm. Data are presented in mean ± SD.
See also Figure S2.
Serine proteases TMPRSS2 and Furin, but not exogenous Trypsin, enhance the replication of SARS-CoV-2
Host proteases, including cell surface and intracellular proteases, play an essential role in CoV infection by processing the S protein to trigger membrane fusion (Izaguirre, 2019, Matsuyama et al., 2010, Matsuyama et al., 2005, Menachery et al., 2020, Millet and Whittaker, 2014, Wicht et al., 2014). Therefore, we evaluated the multi-step replication (MOI = 0.03) of the icSARS-CoV-2-GFP in the presence of selected proteases via fluorescent microscopy and measurements of viral titer.
Vero cells were infected with the icSARS-CoV-2-GFP reporter virus in the presence of 0, 1, or 5 μg/mL of trypsin. Unlike some coronaviruses (CoVs) (Menachery et al., 2020, Wicht et al., 2014), trypsin did not trigger syncytium formation, and at 24 and 48 h, a slightly higher percentage of trypsin-exposed cells expressed GFP signals and CPE than did controls (Figures 2C and S2 ). Trypsin also resulted in slightly lower virus titers than controls (Figure 2D), suggesting that trypsin impairs the stability of viral particles in supernatants.
Figure S2.
Cytopathic Effect of Cells Infected with icSARS-CoV-GFP Virus, related to Figure 2
(A) Infected Vero cells supplemented with different concentrations of trypsin.
(B) Infected Vero or Vero-furin cells.
(C) Infected LLC-MK or LLC-MK-TMPRSS2 cells.
All scale bars, 200 μm.
SARS-CoV-2 S protein exhibits a novel 4 amino acid (aa) furin-cleavage site “RRAR” at the junction between S1 and S2 subunits (Andersen et al., 2020, Coutard et al., 2020). We observed increased icSARS-CoV-2-GFP expression in the furin-overexpressing versus WT cells at 24 h (Figure 2E), correlating with 1 log10 higher infectious titers than WT Vero cells at early times after infection (Figure 2F). Moreover, extensive CPE was noted in furin cells versus parental Vero cell cultures (Figure S2). In contrast, enhanced expression of TMPRSS2 in a rhesus monkey kidney epithelial cell line, LLC-MK cells, resulted in higher amounts of GFP expression and higher icSARS-CoV-2-GFP titers (Figures 2G and 2H). These data suggest that serine proteases like furin and TMPRSS2 enhance the replication efficiency and cytopathology of SARS-CoV-2 in vitro.
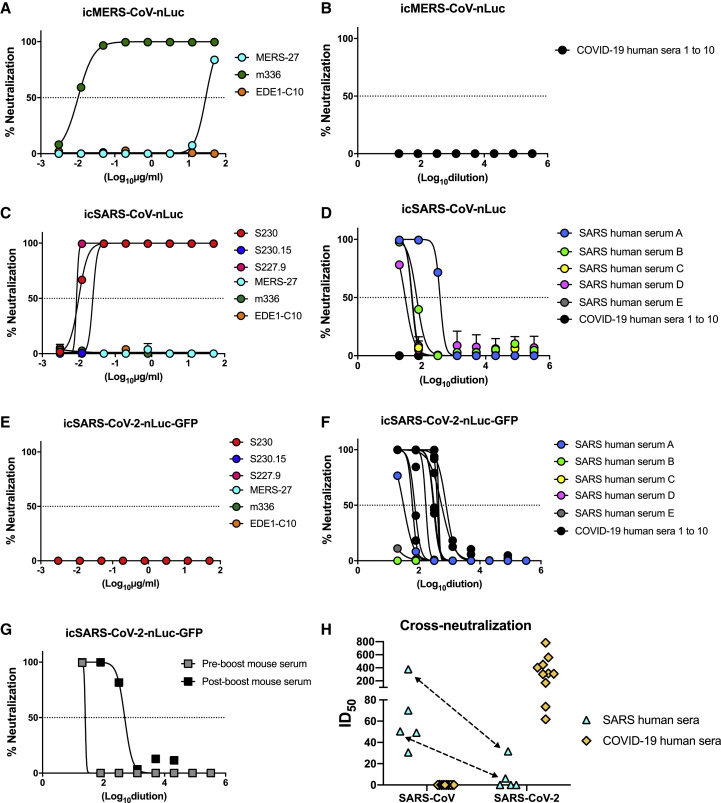
The neutralization sensitivity of SARS-CoV-2 nLuc virus to potent SARS and MERS monoclonal antibodies and polyclonal sera
Three neutralization assays were developed utilizing luciferase reporter CoVs, including SARS-CoV, MERS-CoV, and SARS-CoV-2 (Figures 3 A–3H). Previous studies have identified remarkably potent SARS and MERS nAbs that target receptor binding domains and exhibit strong neutralizing activities in vitro and in vivo (Ying et al., 2015, Yu et al., 2015, Zhu et al., 2007). We utilized three highly cross-reactive nAb against SARS-CoV (S230, S230.15, and S227.9), two nAbs against MERS-CoV (MERS-27 and m336), and one broadly cross-reactive nAb against Dengue virus (EDE1-C10). We also tested a pooled mouse serum sample collected from BALB/c mice vaccinated and boosted with a Venezuelan equine encephalitis virus viral replicon particle (VRP-SARS-COV-2-S) encoding the SARS-CoV-2 S gene. The boost was performed three weeks after vaccination, and sera were collected one week before and one week after boost.
Figure 3.
Neutralization assays using luciferase reporter coronaviruses
(A and B) mAbs (A) and COVID-19 sera (B) against icMERS-CoV-nLuc.
(C and D) mAbs (C) and SARS and COVID-19 sera (D) against icSARS-CoV-nLuc.
(E–G) mAbs (E), SARS and COVID-19 sera (F), and vaccinated mouse serum (G) against icSARS-CoV-2-nLuc-GFP.
(H) ID50 values of SARS and COVID-19 sera cross-neutralizing SARS-CoV and SARS-CoV-2. The same sera samples are indicated with arrows.
The MERS-CoV neutralizing mAbs were the following: MERS-27 and m336; the SARS-CoV neutralizing mAbs were the following: S230, S230.15, and S227.9; the Dengue virus mAb was the following: EDE1-C10. SARS patient serum samples are labeled as “A” to “E”; COVID-19 patient serum samples are labeled as “1” to “10”. Mouse serum was produced by immunized BALB/c mice with SARS-CoV-2 spike.
Both the MERS nAbs, MERS-27 and m336, neutralized the icMERS-CoV-nLuc virus but not the 2003 SARS-CoV-nLuc or 2019 SARS-CoV-2-nLuc-GFP recombinant viruses. Similarly, the three SARS nAbs, S230, S230.15, and S227.9 exhibited potent neutralization activities against icSARS-CoV-nLuc, but not icSARS-CoV-2-nLuc-GFP (Figures 3A, 3C, and 3E). As a negative control, a Dengue virus nAb EDE1-C10 did not neutralize any of the three tested CoVs. Importantly, the mouse serum sample neutralized 99.4% of the icSARS-CoV-2-nLuc-GFP virus at a 1:2 dilution after prime, and much more potent neutralization was noted after VRP-SARS-CoV-2-S boost (Figure 3G).
The S proteins of SARS-CoV and SARS-CoV-2 share 75% identity in amino acid sequences. To investigate whether SARS-CoV and SARS-CoV-2 infections elicit cross-neutralizing antibodies, we evaluated five serum samples from patients who survived the 2003 SARS-CoV Toronto outbreak and 10 serum samples from COVID-19 survivors by using nLuc neutralization assays with the two reporter CoVs. All five 2003 SARS serum samples demonstrated high neutralization titers against SARS-CoV-nLuc virus, and half-maximal inhibitory dilution (ID50) activities in the range were from 1:30.6 to 1:376.5 (Figure 3F). Surprisingly, two of these serum samples, A and E, neutralized icSARS-CoV-2-nLuc-GFP with 11.9- and 8.1-fold of decreases in ID50, respectively. In contrast, 10 COVID-19 convalescent serum samples displayed variable neutralization ID50 titers that ranged from 61.67 to 782.70 against icSARS-CoV-2-nLuc-GFP but little, if any, neutralization of 2003 icSARS-CoV-nLuc or icMERS-CoV-nLuc viruses at the lowest dilutions tested (Figures 3B, 3D, and 3H).
RNA in situ hybridization localization of the SARS-CoV-2 receptor complex in the normal human upper and lower respiratory tract
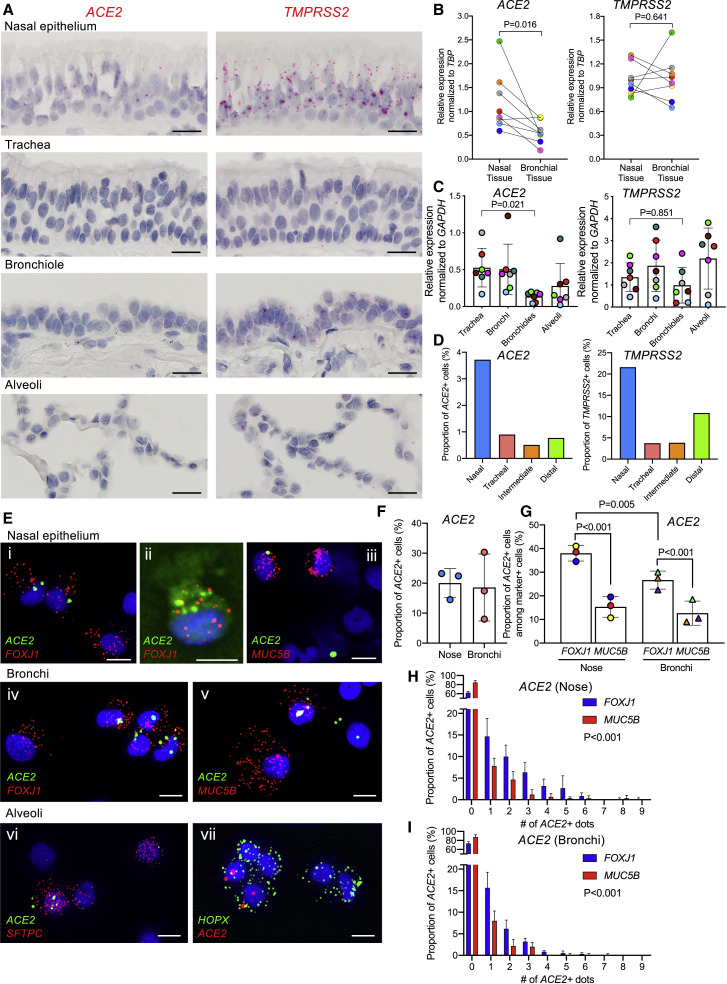
The sites of SARS-CoV-2 infection in the upper airways (nose and oropharynx) and lung (lower airways and alveoli) are under active investigation (Rockx et al., 2020). Accordingly, we characterized ACE2 and TMPRSS2 expression in these regions by using RNA-ISH (Figures 4 and S3 ). Consistent with the low amount of ACE2 expression reported from scRNA-seq data (Brann et al., 2020, Durante et al., 2020, Sajuthi et al., 2020), low amounts of ACE2 were detected in the respiratory epithelium lining the nasal cavity (Figure 4A). Scattered, low amounts of ACE2 and TMPRSS2 expression were also observed in the squamous epithelium lining oropharyngeal tonsillar tissue (Figure S3A). Notably, progressively reduced amounts of ACE2 expression were observed in the lower airway regions, culminating in minimal amounts in the alveolar region. Quantitative comparisons of nasal and bronchial airway epithelia obtained as brush samples simultaneously from the same subjects by qPCR revealed significantly higher expression of ACE2 but not TMPRSS2 in the nasal than the bronchial tissues (Figure 4B). In a separate qPCR study, there was a gradient of reduced ACE2 expression from proximal to distal intrapulmonary regions (Figure 4C). In contrast, TMPRSS2 mRNA exhibited an overall higher expression amount in all respiratory tract regions than ACE2.
Figure 4.
Intraregional ACE2 and TMPRSS2 mRNA expression in normal human airways
(A) Representative RNA-ISH images demonstrating regional distribution of ACE2 and TMPRSS2 mRNA localization (red signal) in normal human airway surface epithelium. Scale bars, 20 μm.
(B) Comparison of ACE2 and TMPRSS2 mRNA expression between matched nasal and bronchial brushed tissues obtained from seven healthy subjects.
(C) Relative expression of ACE2 and TMPRSS2 mRNA in different airway regions enriched for epithelial cells, including tracheas, bronchi, bronchiole, and alveoli, obtained from matched seven normal lungs.
(D) Frequency of ACE2+ and TMPRSS2+ cells among total cells identified in distinct anatomical airway regions in a re-analysis of scRNA-seq data (Deprez et al., 2019).
(E) RNA-ISH images depicting mRNA expression of ACE2 and cell type markers, including FOXJ1 (ciliated) (i, ii, and iv), MUC5B (secretory) (iii and v), SFTPC (alveolar type 2) (vi), and HOPX (alveolar type 1 or 2) (vii) on cytospins of nasal versus bronchial superficial epithelial and purified alveolar cells. Scale bars, 10 μm.
(F) Frequency of ACE2+ cells among nasal and bronchial preparations. A total of 1,000 cells were analyzed for ACE2 expression per donor (N = 3).
G. Frequency of ACE2+ cells among FOXJ1+ or MUC5B+ cells in nasal or bronchial preparations. A total of 200 FOXJ1+ or MUC5B+ cells were analyzed for ACE2 expression per donor (N = 3).
(H and I) Histograms depicting number of dot signals of ACE2 expression in FOXJ1 or MUC5B+ cells in nasal (H) or bronchial (I) preparations identified by scRNA-ISH. ACE2+ dot signals were counted in 200 FOXJ1 or MUC5B+ cells per donor (N = 3). Statistics for (B), (C), (F), and (G) used linear mixed-effect model with the donor as random-effect factor for comparison between groups, and pairwise comparisons of groups with more than two levels were performed using Tukey post hoc tests. (H) and (I) used generalized linear mixed-effect models with Poisson distribution to compare the difference in cell counts at varying ACE2 expression amounts between FOXJ1+ and MUC5B+ cells. Histobars and error bars represent mean ± SD. Different symbol colors indicate results from different individual donors.
See also Figure S3.
Figure S3.
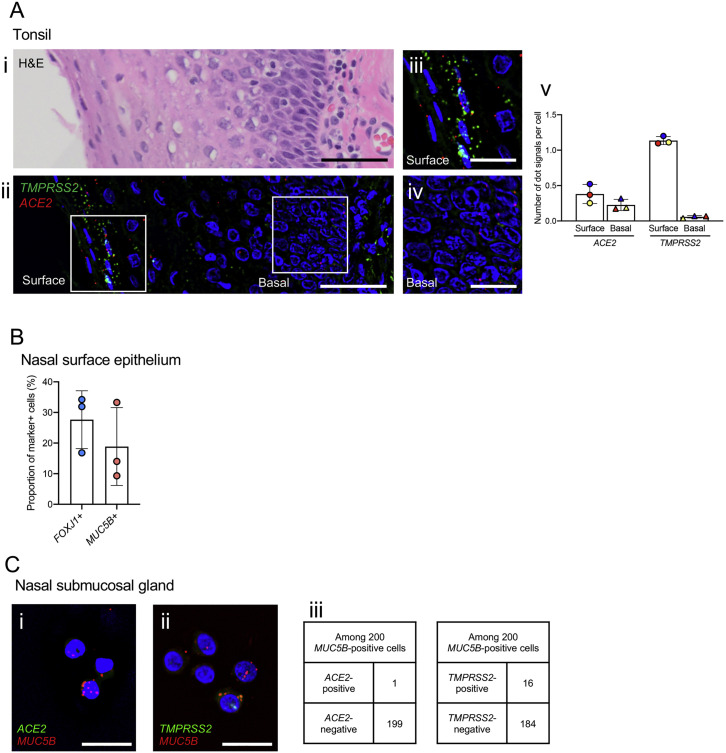
ACE2 and TMPRSS2 expression in human tonsillar epithelium and nasal surface epithelium and submucosal glands, related to Figure 4
(A) Tonsillar surface squamous epithelium stained with (i) H&E staining and (ii) dual-color-fluorescence RNA-ISH showing TMPRSS2 (green) and ACE2 (red) along with nuclear staining (blue). Scale bars, 50 um. (iii) Enlarged images of (ii) showing surface (iii) and basal (iv) expression; scale bars, 20um. Images are representative from N = 3 tonsils, N = 4-8 regions per tonsil. (v) Signal dots for ACE2 and TMPRSS2 mRNAs were counted and normalized to the number of cells in surface and basal layer of tonsillar surface epithelium as described in the STAR Methods. Each bar represents the average of N = 4-8 regions for each tonsil studied. Data are presented in mean ± SD.
(B) Frequency of FOXJ1- or MUC5B-positive cells identified by RNA-ISH among total nasal surface epithelial cells isolated. A total of 1,000 cells were analyzed for FOXJ1 or MUC5B expression per donor. N = 3.
(C) Cytospins of nasal submucosal glands cells probed by dual-color-immunofluorescent RNA-ISH. (i) shows lack of ACE2 in MUC5B-positve nasal gland cells, while (ii) depicts occasional co-expression of TMPRSS2 in a subset of MUC5B-positive cells. Scale bars, 20 μm. (iii) Frequency of detection of ACE2 or TMPRSS2 positive cells in MUC5B positive cells from nasal glands. N = 1 gland preparation, a total of 200 MUC5B positive cells were counted.
Previously reported scRNA-seq data describing ACE2 and TMPRSS2 expression in the upper and lower respiratory system have detected ACE2 in ∼5% of total cells interrogated (Deprez et al., 2019, Sajuthi et al., 2020) (Figure 4D). We recently developed a single-cell (cytospin) RNA-ISH technique that is 5–10× more sensitive at assigning cell-type-specific expression patterns than scRNA-seq (Okuda et al., 2019) (Figure 4E). This technique identified ACE2 expression in ∼20% of interrogated cells versus ∼5% by scRNA-seq (Figure 4F). These studies identified the forkhead box J1 (FOXJ1)-defined ciliated cell as the most frequent cell type in nasal scrapes (Figure S4 B) and that the percentage of ciliated cells expressing ACE2 was higher in the nose than in bronchi (Figure 4G). ACE2+- and MUC5B+-defined secretory (“club”) cells were less frequent and expressed less ACE2 than ciliated cells in each airway region (Figures 4H and 4I). Both cell types in each region exhibited considerable variability in ACE2 expression (Figures 4H and 4I). Studies of nasal submucosal glands exhibited few or no detectable ACE2+ glandular cells (Figure S4C). Finally, application of this technique to freshly excised distal lung digests revealed expression of ACE2 in a fraction of AT2 cells (Figure 4Evi). ACE2 was detected in HOPX + cells, which in humans can be AT1 or AT2 cells (Figure 4Evii) (Ota et al., 2018).
Figure S4.
Additional data of SARS-CoV and SARS-CoV-2 infected primary human cells, related to Figure 6
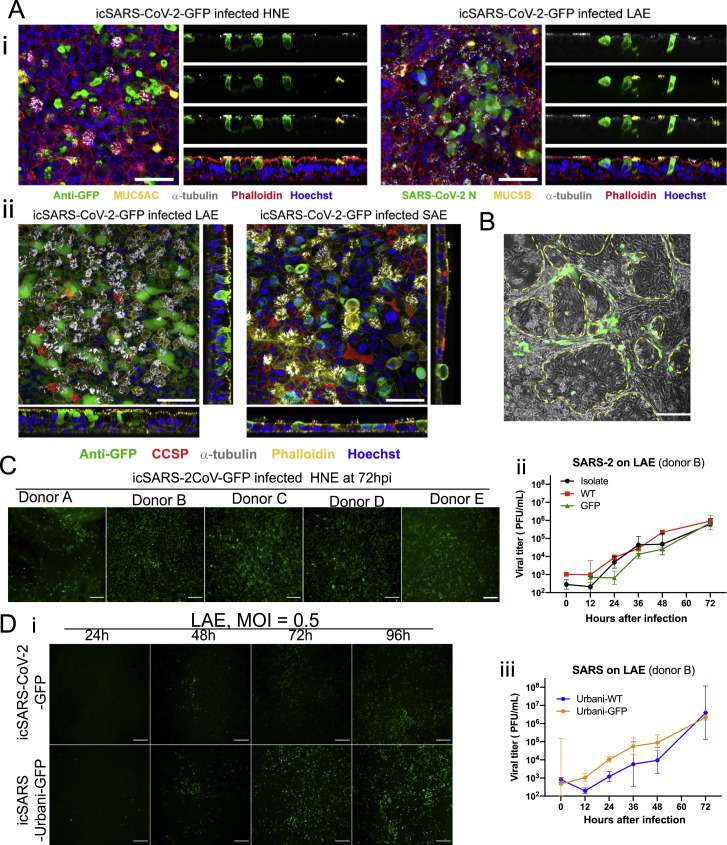
(A) Representative whole-mount extended focus views of icSARS-CoV-2-GFP-infected (i) HNE and LAE cell cultures. Red = filamentous actin (phalloidin), White = α-tubulin (multiciliated cells), Blue = nuclei (Hoechst 33342). Green = GFP (left). Green = SARS-CoV-2 Nucleocapsid (right). Yellow = MUC5AC (left). Yellow = MUC5B (right); (ii) LAE and SAE cell cultures. Yellow = filamentous actin (phalloidin), White = α-tubulin (multiciliated cells), Blue = nuclei (Hoechst 33342). Green = GFP (virus). Red = CCSP. Scale bars, 50 μm.
(B) Merged of GFP and bright field mages taken from AT1 and AT2 cells infected with icSARS-CoV-2-GFP at 48 h. The AT-1 cells are present inside the enclosed areas. Bar = 100 μm.
(C) GFP signals of icSARS2-GFP-infected HNEs collected from five different donors at 72 hpi, MOI = 3.
(D) (i) Fluorescent signals of the two viruses in LAE (ii) Growth curves of three SARS-CoV-2 viruses in LAE from the same donor. Scale bar, 200 μm. (iii) Growth curves of two SARS-Urbani viruses in LAE. Data are presented in mean ± SD. All the infections in this figure were in MOI = 0.5.
Pre-existing pulmonary disease and ACE2 and TMPRSS2 expression
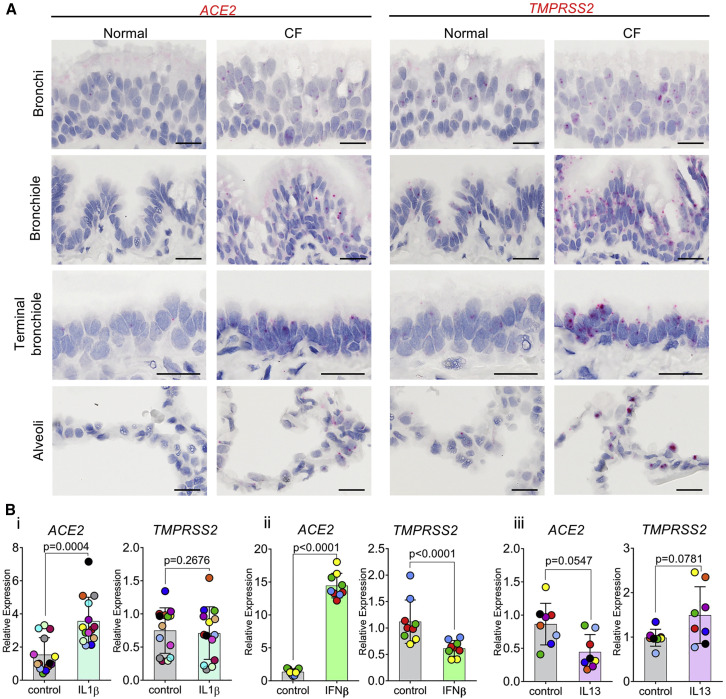
Suppurative muco-obstructive lung diseases, e.g., cystic fibrosis (CF) and non-CF bronchiectasis (NCFB), are characterized by airway mucus accumulation and neutrophilic inflammation and are reported to be at increased risk for severe SARS-CoV-2 infections (Boucher, 2019, CDC COVID-19 Response Team, 2020). To test whether dysregulation of ACE2 expression is a feature of CF, we performed RNA-ISH studies in excised CF lungs and revealed a striking upregulation of ACE2 and TMPRSS2 expression in CF airways (Figure 5 A).
Figure 5.
Inflammatory cytokines alter ACE2 and TMPRSS2 expression
(A) RNA-ISH images demonstrating regional distribution of ACE2 and TMPRSS2 mRNA localization in normal and CF human airways. Scale bars, 20 μm. Images were obtained from four different airway regions from one normal or CF subject as representative of N = 6 normal or CF subjects studied.
(B) mRNA expression of ACE2 and TMPRSS2 measured by Taqman assay after inflammatory cytokine challenge in primary human large airway epithelial cells. Shown in (i) is IL-1β (10 ng/mL, 7 days, N = 8), in (ii) is IFN-β (10 ng/mL, 3 days, N = 4 donors, 2–3 cultures per donor), and in iii is IL-13 (10 ng/mL, 7 days, N = 8). Wilcoxon matched pairs signed rank test was used for comparison between control and cytokine treatment groups. Histobars and error bars represent mean ± SD. Different symbol colors indicate results from different individual donors.
To gain insight into pathways that might contribute to dysregulation of ACE2 expression in CF lungs, we tested the effects of selected cytokines on ACE2 expression in large airway epithelial (LAE) cultures. Interleukin-1β (IL-1β), the dominant pro-mucin secretory cytokine in CF and NCFB secretions (Chen et al., 2019), upregulated ACE2, but not TMPRSS2 (Figure 5Bi). Because CF subjects experience recurrent virus-driven exacerbations, we tested the effect of interferon-β (IFN-β) on ACE2 expression. ACE2 expression was significantly increased, whereas TMPRSS2 expression decreased, by IFN-β (Figure 5Bii). In contrast, IL-13, a cytokine associated with Th2-high asthma, inhibited ACE2 expression (Figure 5Biii).
SARS-CoV-2 infectivity specific to the respiratory tract region
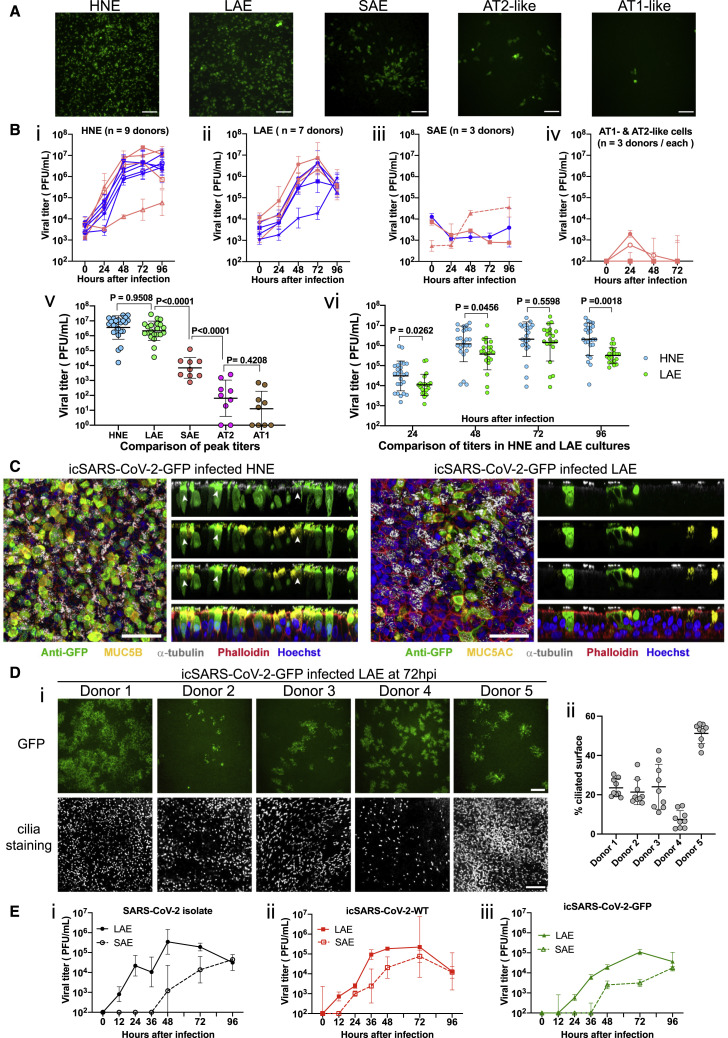
To test the relationship between ACE2 entry receptor expression and SARS-CoV-2 infection, we inoculated primary epithelial cultures from the human nasal epithelia (human nasal epithelial cells [HNE], n = 9 donors), large airway (bronchi and large airway epithelial cells (LAE), n = 7 donors), lower airway (bronchiolar and small airway epithelial cells [SAE], n = 3 donors), nasal submucosal glands (n = 2 donors), type II- and type I-like pneumocytes (AT2/AT1-like) (n = 3 donors), microvascular endothelial cells (MVE) (n = 2 donors), and fibroblasts (FBs) (n = 2 donors), and an immortalized nasal cell line (UNCNN2TS) with icSARS-CoV-2-GFP reporter virus. We observed GFP signals and detected viral titers in HNE, LAE, SAE, AT2-like, and AT1-like cell cultures (Figure 6 A). In contrast, nasal submucosal gland, UNCNN2TS, MVE, or FB cells were not susceptible, as evidenced by no GFP signals or detectable infectious titers of progeny viruses (not shown).
Figure 6.
Replication of SARS-CoV-2 in primary human respiratory cells
(A) Representative GFP signals in icSARS-CoV-2-GFP-infected HNE, LAE, SAE, AT2-like, and AT1-like cultures at 48 h. Scale bar, 80.
(B) Growth curves of icSARS-CoV-2-GFP in (i) HNE, n = 9 donors; (ii) LAE, n = 7 donors; (iii) SAE, n = 3 donors; (iv) AT1-like (empty symbols) and AT2-like (filled symbols) cells, n = 3 donors per cell type. Cells from female and male donors are labeled in pink and blue, respectively. Triplicated viral infections under MOI of 3 or 0.5 are shown in solid and dotted lines, respectively. In (v) is a comparison of the highest titers of individual culture among cell types and in (vi) is a comparison of individual titers in HNE and LAE at different time points.
(C) Representative whole-mount extended focus views of icSARS-CoV-2-GFP-infected HNE and LAE cell cultures. Color coding is as follows: red, filamentous actin (phalloidin); white, α-tubulin (multiciliated cells); green, GFP (virus); blue, nuclei (Hoechst 33342); yellow, MUC5B (left) and MUC5AC (right). An arrow represents viral-infected α-tubulin+ (ciliated) and MUC5B+ (secretory) transitional HNEs. Scale bars, 50 μm.
(D) Shown in (i) is the variability of GFP and cilia signals in icSARS-CoV-2-GFP-infected LAE cultures collected from five different donors at 72 hours after infection, scale bar, 200 μm. Shown in (ii) is the quantification of ciliated area in the LAE cultures.
(E) Growth curves of icSARS-CoV-2-GFP infected in LAE and SAE collected form the same donor. Cultures were infected with SARS-CoV-2 clinical isolate (i), WT (ii), and GFP (iii) with MOI of 0.5. Data are presented in mean ± SD.
See also Figure S4.
We measured the relative infectivity of the SARS-CoV-2 GFP virus in primary cells on the basis of the average peak titers and observed that infectivity exhibited the same pattern as the ACE2 expression amounts from the upper to lower respiratory tract (Figures 6Bi–6Biv). The icSARS-CoV-2-GFP virus replicated efficiently in the HNE and LAE, and peak viral titers were significantly higher than the titers in SAE, AT2-like, and AT1-like cultures (Figure 6Bv). Although the viral peak titers were similar, the icSARS-CoV-2-GFP infection in HNE culture resulted in significantly higher titers than LAE at 24 h, 48 h, and 96 h after infection, suggesting more robust replication in the primary nasal cells (Figure 6Bvi). Collectively, these data indicate that virus infectivity or replication efficiency varies markedly from proximal airway to alveolar respiratory regions.
We utilized whole-mount immunohistochemistry of HNE and LAE cultures to identify cell types infected by SARS-CoV-2 (Figures 6C and S4A). The ciliated cell was routinely infected and extruded. In contrast, the other major cell type facing the airway lumen, i.e., the MUC5B+ club cell, was not infected, nor was the MUC5AC+ metaplastic goblet cell. We did note a cell type co-expressing the ciliated cell marker tubulin, and MUC5B was rarely infected in the HNE, a finding consistent with infection of a secretory (club) cell transitioning to a ciliated cell phenotype.
There is debate whether AT2 and/or AT1 cells express sufficient ACE2 to mediate infection and whether AT2, AT1, or both cell types are infectable. Previous studies reported 2003 SARS-CoV infects AT2 but not AT1 pneumocytes (Mossel et al., 2008). To focus on the relative infectivity by SARS-CoV-2 for AT2 versus AT1 cells, we tested standard AT2-AT1 cell cultures and a novel cell culture approach that well preserves AT2 and AT1 cell populations over the infection or GFP expression interval. As shown in Figures 6A and S4B, AT2 cells appeared to be preferentially infected.
SARS-CoV-2 infectivity specific to the respiratory tract region
We next investigated three other aspects of SARS-CoV-2 infection of human airway epithelia. First, the variability of infectivity among HNE and LAE cultures from multiple donors was characterized. Although all nine HNE and seven LAE were infected by icSARS-CoV-2-GFP, we observed marked variability in GFP signals per culture surface area and viral growth curves. LAE cultures exhibited higher variability in susceptible cells than the HNE cultures at 72 hours after infection (Figures 6A, 6B, 6D and S4C). We quantitated ciliated cell numbers in five LAE cultures, and we noted no correlation between susceptibility and ciliated cell percentages (Figure 6Dii).
Second, to further characterize the infectivity of LAE versus SAE, we compared replication rates of three SARS-CoV-2 viruses in LAE and SAE cultures from the same donor. All three viruses replicated more slowly in SAE than LAE cells. The GFP virus replicated modestly less effectively than the clinical isolate or WT virus in the two regions (Figure 6E). This observation differs from the equivalent replication noted in the Vero-E6 cells (Figures 2A and 2B), suggesting an intact ORF7 gene contributes to SARS-CoV-2 replication, and perhaps virulence, in human tissues.
Third, we compared the replication of SARS-CoV and SARS-CoV-2 in LAE cells. SARS-Urbani WT and GFP viruses, in parallel with the three SARS-CoV-2 viruses, were administered to LAE cultures from the same donor. GFP signals were detected in LAE cultures for both viruses, but the SARS-CoV-2-GFP exhibited delayed and less-intense signals than did SARS-CoV-Urbani-GFP (Figure S4D). This phenotype is consistent with the growth curve in which a lower titer of SARS-CoV-2 was recorded at 24 h.
SARS-CoV-2 infection in COVID-19 autopsy lungs
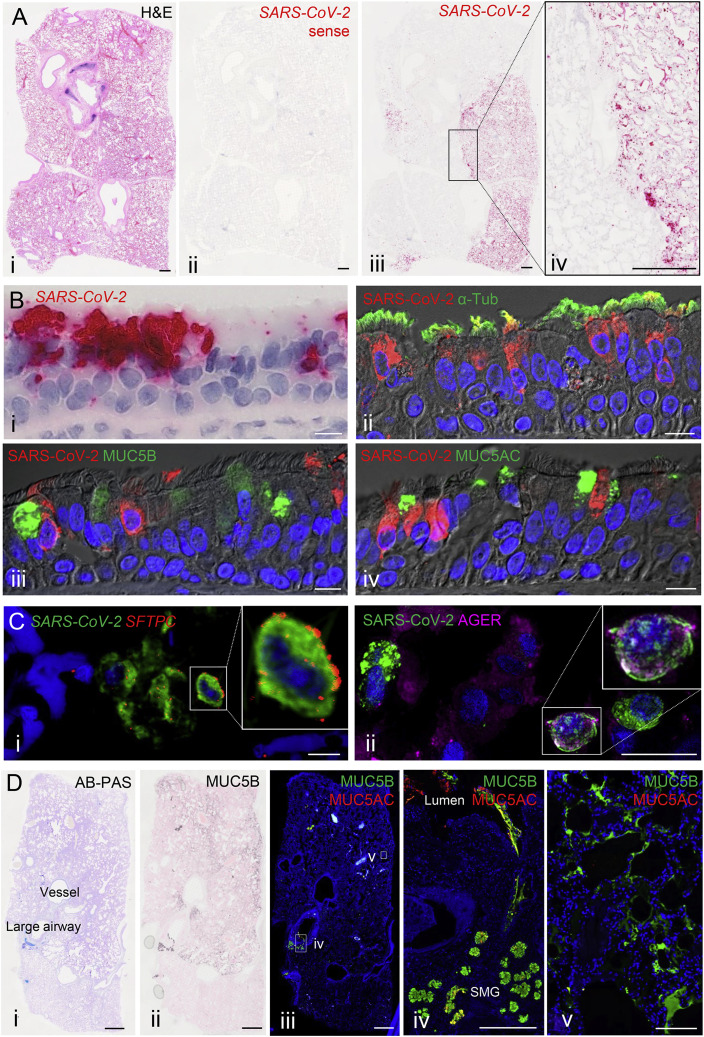
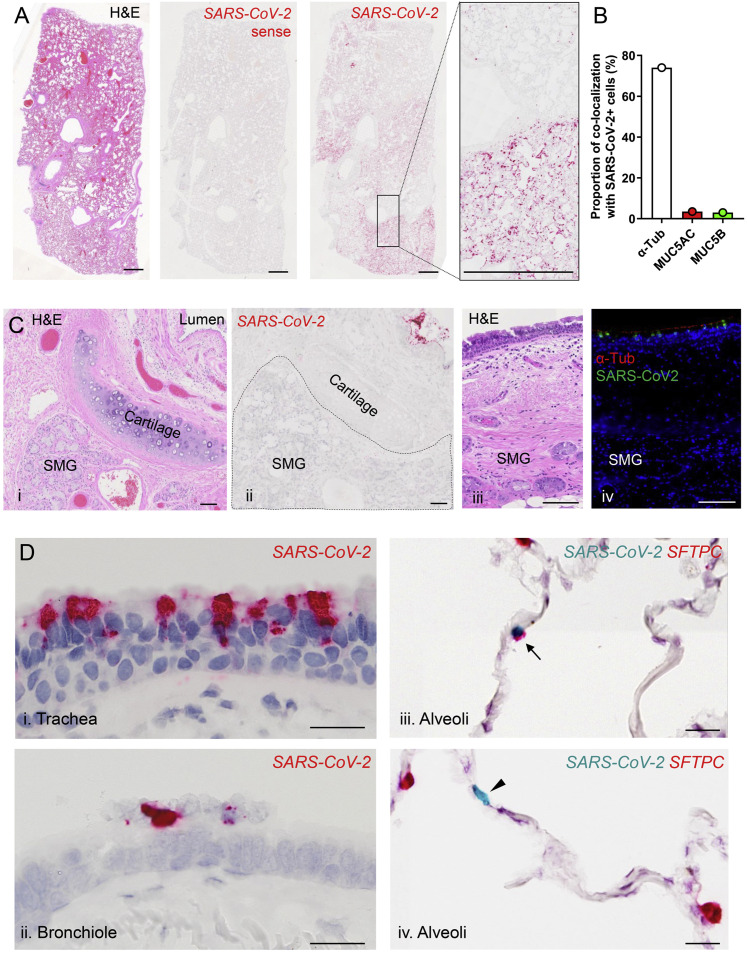
We utilized RNA-ISH and immunohistochemistry (RNA-ISH/IHC) to localize virus in four lungs from SARS-CoV-2-infected deceased subjects (Table S1). Multiple observations at different length scales were notable. First, at the macroscopic level, the infection appeared patchy, segmental, and peripheral (Figures 7 A and S5 A). These characteristics are consistent with an aspiration distribution of an infectious inoculum. Second, ciliated cells within the superficial epithelia lining proximal airway surfaces, particularly the trachea, were infected (Figures 7B and S5B). As observed in vitro, MUC5B+ club and MUC5AC+ goblet cells were not infected in vivo. Third, the submucosal glands that populate the large airway regions of the lung were not infected (Figure S5C). Fourth, alveolar cells were also infected. RNA in situ and IHC co-localization of an AT2 cell marker, SPC (SFTPC), and AT1 cell marker (AGER) with SARS-CoV-2 indicated that AT2 cells and AT1 cells (or AT2 cells that had transitioned to AT1 cells) were infected (Figures 7C and S5D).
Figure 7.
Characterization of cell types for SARS-CoV-2 infection in SARS-CoV-2 autopsy lungs
(A) Sections from an autopsy lung with SARS-CoV-2 infection were stained by hematoxylin and eosin (i) and probed for SARS-CoV-2 by RNA-ISH (ii–iv). A SARS-CoV-2 sense probe (ii) was used. Scale bars, 1 mm.
(B) The trachea from a SARS-CoV-2 autopsy was probed for SARS-CoV-2 by RNA ISH. Shown in (i) is a colorimetric detection of SARS-CoV-2 (red) showing infection of surface epithelium. Shown in (ii–iv) is the co-localization of SARS-CoV-2 (red) with cell-type-specific markers (green) determined by dual-immunofluorescent staining (ii, acetylated α-tubulin cilia marker; iii, MUC5B secretory cell marker; and iv, MUC5AC mucous (goblet)-cell marker). Scale bars, 10 μm.
(C) Co-localization of SARS-CoV-2 with alveolar cell-type-specific markers in the alveolar space from a SARS-CoV-2 autopsy. Shown in (i) is the dual color-fluorescent RNA-ISH co-localization of SARS-CoV-2 (green) with alveolar type II cell marker SFTPC (red). Shown in (ii) is the dual-immunofluorescent co-localization of SARS-CoV-2 (green) with alveolar type I cell marker AGER (magenta). Scale bars, 20 μm.
(D) Mucin expression in SARS-CoV-2 autopsy lung. Shown in (i) is the AB-PAS (blue to purple) stain for complex carbohydrate (mucin), in (ii) is MUC5B immunohistochemistry, in (iii–v) is the dual-immunofluorescent staining for MUC5B (green) and MUC5AC (red) in the large airway (iv) and the alveoli (v). Abbreviation is as follows: SM, submucosal grand. Scale bars, 2mm (i–iii) and 200 μm (iv and v).
See also Figure S5.
Figure S5.
SARS-CoV-2 infection in SARS-CoV-2 autopsy lungs, related to Figure 7
(A) Sections from of a second region of an autopsy lung with SARS-CoV-2 infection were stained by hematoxylin and eosin (H&E) (i) and probed for SARS-CoV-2 by RNA in situ hybridization (ISH) (ii, iii, and iv). Related to Figure 7A.
(B) Frequency of acetylated alpha tubulin, MUC5AC, or MUC5B colocalization with SARS-CoV-2 positive cells in the trachea from a SARS-CoV-2 autopsy. A total of 200 randomly selected SARS-CoV-2 positive cells were analyzed for each dual staining condition. Related to Figure 7B ii, iii, iv.
(C) Absence of SARS-CoV-2 infection in submucosal glands (SMG). (i-ii) H&E staining (i) and RNA-ISH (ii) for SARS-CoV-2 (red) in a large cartilaginous airway of one autopsy lung. SARS-CoV-2 is only present in the surface epithelium near the lumen, not in SMG. (iii-iv) H&E (iii) and dual-immunofluorescence staining using acetylated alpha tubulin (red) and anti-SARS-CoV-2 rabbit polyclonal antibody (green) (iv) from the trachea of a separate autopsy. Related to Figure 7B and S5Di.
(D) Regional distribution of SARS-CoV-2 RNA from trachea to alveoli identified by RNA-ISH in one SARS-2-CoV autopsy lung (in i and ii, viral staining is red; in iii, viral staining is turquoise). RNA-ISH dual color images demonstrate SARS-CoV-2 RNA and SFTPC mRNA (alveolar type 2 cell marker) localization in alveoli of a SARS-CoV-2 autopsy lung. SARS-CoV-2 (turquoise) was identified in a SFTPC (red)-positive (iii, arrow) and a SFTPC-negative cell (iv, arrowhead); Scale bars, 2mm (A); 100 μm (C); 20 μm (D).
During the routine Alcian Blue Periodic Acid Schiff (AB-PA) staining that detects mucins or mucin-like carbohydrates in SARS-CoV-2-infected autopsy lungs, we noted faint AB-PAS staining in the peripheral lung, i.e., the alveolar region in some lungs (Figures 7D). Because aberrant mucin secretion and accumulation is a feature of parenchymal diseases that can progress to fibrosis, the AB-PAS material was characterized in more detail (Figures 7Dii–7Dv). IHC studies suggested that this material in large airways was a mixture of the secreted mucins MUC5B and MUC5AC (Figure 7Div). In the alveolar parenchymal region, MUC5B alone was detected and was enriched in the peripheral subpleural area, as often observed in idiopathic pulmonary fibrosis (IPF) (Figure 7Dv) (Evans et al., 2016). Note, in none of the autopsy lungs studied was mechanical ventilation employed and the lung in graphics A and D of Figure 7 was immersion fixed. These observations, coupled to the observation that MUC5AC was not detected in the peripheral region, makes it unlikely that MUC5B selectively was mechanically spread from central to peripheral lung zones.
Discussion
We generated a SARS-CoV-2 reverse genetics system; characterized virus RNA transcription profiles; evaluated the effect of ectopically expressed proteases on virus growth; and used reporter viruses to characterize virus tropisms, ex vivo replication, and to develop a high-throughput neutralizing assay. These reagents were utilized to explore aspects of early infectivity and disease pathogenesis relevant to SARS-CoV-2 respiratory infections.
Our single-cell RNA-ISH technology extended the description of ACE2 in respiratory epithelia on the basis of scRNA-seq data (Sungnak et al., 2020). Single-cell RNA-ISH detected ∼20% of upper respiratory cells expressing ACE2 versus ∼4% for scRNA-seq (Figure 4F). Most of the RNA-ISH-detected ACE2-expressing cells were ciliated cells, not normal MUC5B+ secretory (club) cells or goblet cells. Notably, the nose contained the highest percentage of ACE2-expressing ciliated cells in the proximal airways (Figure 4G). The higher nasal ACE2 expression-level findings were confirmed by qPCR data comparing nasal to bronchial airway epithelia. qPCR data also revealed that ACE2 amounts further waned in the more distal bronchiolar and alveolar regions. Importantly, these ACE2 expression patterns were paralleled by high SARS-CoV-2 infectivity of nasal epithelium with a gradient in infectivity characterized by a marked reduction in the distal lung (bronchioles and alveoli) (Figures 6A and 6B).
Multiple aspects of the variability in SARS-CoV-2 infection of respiratory epithelia were notable in these studies. First, significant donor variations in virus infectivity and replication efficiency were observed. Notably, the variability was less in the nose than lower airways. The reason(s) for the differences in lower airway susceptibility are important but remain unclear (Cockrell et al., 2018). We identified variations in ACE2 receptor expression (Figures 4A–4D) but not numbers of ciliated cells as potential variables (Figure 6D). Second, variation in infectivity of a single cell type, i.e., the ciliated cell, was noted with only a fraction of ciliated cells having access to virus infected at 72 h (Figure 6A). Third, the dominant secretory cell, i.e., the MUC5B+ club cell, was not infected in vitro or in vivo, despite detectable ACE2 and TMPRSS2 expression (Figures 4G-4I). Collectively, these data suggest that measurements of ACE2 and TMPRSS2 expression do not fully describe cell infectivity and that a description of other variables that mediate susceptibility to infection, including the innate immune system(s), is needed (Menachery et al., 2014).
The ACE2 receptor gradient in the normal lung raised questions focused on the initial sites of respiratory tract virus infection, the mechanisms that seed infection into the deep lung, and the virus-host interaction networks that attenuate or augment intra-regional virus growth in the lung to produce severe disease, especially in vulnerable patients experiencing chronic lung or inflammatory diseases (Guan et al., 2020, Leung et al., 2020).
We speculate that nasal surfaces might be the dominant initial site for SARS-CoV-2 respiratory tract infection (Wölfel et al., 2020). First, SARS-CoV-2 RNA has been detected in aerosol particles in the range of aerodynamic sizes exhaled during normal tidal breathing (Liu et al., 2020, Papineni and Rosenthal, 1997). Aerosol deposition and fomite mechanical delivery deposition modeling suggest that aerosols containing virus inhaled by naive subjects achieve the highest density of deposition, i.e., highest MOI per unit surface area, in the nose (Booth et al., 2005, Farzal et al., 2019, Teunis et al., 2010). Second, the relatively high ACE2 expression in nasal specimens and the parallel high infectivity of the HNE cultures suggests the nasal cavity is a fertile site for early SARS-CoV-2 infection. Nasal infection likely is dominated by ciliated cells in the superficial epithelium, not nasal submucosal glands. Third, the nose is exposed to high but variable loads of environmental agents, producing a spectrum of innate defense responses. Hence, a portion of the variability of the clinical syndrome of COVID-19 might be affected by environmentally driven variance of nasal infectivity (Wu et al., 2020b).
Another aspect of the variability of the COVID-19 syndrome is the variable incidence and severity of lower lung disease. It is unlikely SARS-CoV-2 is transmitted to the lung by hematogenous spread, as demonstrated by the absence of infection of MVE cells and by previous reports that indicate airway cultures are difficult to infect from the basolateral surface (Sims et al., 2005, Wölfel et al., 2020). Theoretically, infection could be transmitted directly to lower lung surfaces by microaerosol inhalation with deposition on and infection of alveolar surfaces mediated in part by the high ACE2 binding affinity reported for SARS-CoV-2 (Shang et al., 2020, Wrapp et al., 2020). However, given the low amounts of ACE2 expression in alveolar cells in health, the correlated poor infectivity in vitro, and the absence of a homogeneous pattern radiographically, the importance of this route remains unclear (Santarpia et al., 2020).
In contrast, it is well-known that an oral-lung aspiration axis is a key contributor to many lower airways infectious diseases (Dickson et al., 2016, Esther et al., 2019, Gaeckle et al., 2020, Odani et al., 2019, Phillips et al., 2015). Nasal secretions are swept from the nasal surface rostrally by mucociliary clearance and accumulate in the oral cavity at a rate of ∼0.5 mL/h where they are admixed with oropharyngeal or tonsillar fluid (Eichner et al., 1983, Pandya and Tiwari, 2006). Especially at night, it is predicted that a bolus of relatively high titer virus is aspirated into the deep lung, either via microaspiration or as part of gastro-esophageal reflex-associated aspiration, sufficient to exceed the threshold PFU/unit surface area needed to initiate infection (Amberson, 1954, Gleeson et al., 1997, Huxley et al., 1978). Note, our data that tracheas exhibited significant viral infection in vivo suggest that small-volume microaspiration could also seed this site. Tracheal-produced virus could then also accumulate in the oropharynx via mucus clearance for subsequent aspiration into the deep lung (Quirouette et al., 2020). Oropharyngeal aspirates also contain enzymes and/or inflammatory mediators that might condition alveolar cells for infection. Aspiration of SARS-CoV-2 into the lung is consistent with the patchy, bibasilar infiltrates observed by chest CT in COVID-19 (Xu et al., 2020). Notably, robust microaspiration and gastro-esophageal aspiration are observed frequently in subjects who are at risk for more severe COVID-19 lower respiratory disease, e.g., older, diabetic, and obese subjects (Pan et al., 2020a, Phillips et al., 2015). Finally, our autopsy studies demonstrated patchy, segmental or subsegmental disease, consistent with aspiration of virus into the lung from the oropharynx.
These speculations describing the early pathogenesis of SARS-CoV-2 upper and lower respiratory tract disease are consistent with recent clinical observations. The data from Wölfel et al. (2020) in COVID-19-positive subjects support the concept of early infection in the upper respiratory tract (0–5 d) followed by subsequent aspiration and infection of the lower lung. These authors focused on the oropharynx as a potential site of the early virus propagation. As noted above, however, a nasal-oropharyngeal axis also exists, which has two implications. First, the nasal surfaces could seed the oropharynx for infection. Second, it is likely that oropharyngeal secretions reflect a mixture of local secretions admixed with a robust contribution of nasal mucus and virus.
Animal model data are also compatible with the scenario of aspiration-induced focal SARS-CoV-2 lung disease. The data of Rockx et al. (2020) noted focal lung disease after combined intranasal versus intratracheal dosing with SARS-CoV-2 in cynomolgus monkeys. Notably, other findings in this model phenocopied our observations of human disease, e.g., early nasal shedding of virus, infection of nasal ciliated cells, and infection of AT2 and likely AT1 cells. Perhaps more definitive data describing nasal cavity seeding of the lower lung by microaspiration emanate from the studies of Richard et al. (2020). These investigators demonstrated in ferret models that genetically marked virus delivered to the nasal cavity more efficiently transmitted infection to the lower lungs than a virus with a distinct genetic marker delivered directly into the lungs.
In addition to identifying possible microaspiration risk factors associated with COVID-19 disease severity in the elderly, diabetic, and obese, our studies provide insights into variables that control disease severity in subjects at risk because of pre-existing pulmonary disease (Leung et al., 2020, Sajuthi et al., 2020). For example, ACE2 expression was increased in the lungs of CF patients excised at transplantation. A major cytokine that produces the muco-inflammatory CF airways environment, IL-1β, was associated in vitro with increased ACE2 expression (Chen et al., 2019). The clinical outcome of increased ACE2 expression in CF is not yet known. The simple prediction is that increased ACE2 expression might be associated with more frequent or severe SARS-CoV-2 disease in CF populations. However, increased ACE2 expression is reported to be associated with improved lung function by negatively regulating ACE and the angiotensin II and the angiotensin II type 1a receptor (AT1a) in models of alveolar damage (pulmonary edema) and bacterial infection (Imai et al., 2005, Jia, 2016, Keeler et al., 2018, Kuba et al., 2005, Sodhi et al., 2019). Consequently, CF subjects might exhibit reduced severity of disease once acquired. Data describing outcomes of COVID-19 in the CF populations should emerge soon (Colombo et al., 2020).
Our autopsy studies also provide early insights into the variable nature of the severity and pathogenesis related to post-COVID-19 lung health or function (Atri et al., 2020, Kollias et al., 2020, Magro et al., 2020). Our study has identified another feature of COVID-19, i.e., the accumulation of apparently aberrantly secreted MUC5B in the alveolar region. Accumulation of MUC5B in the peripheral (alveolar) lung is characteristic of subjects who develop IPF, and polymorphisms in the MUC5B promoter associated with IPF have been reported (Evans et al., 2016). Future studies of the long-term natural history of SARS-CoV-2 survivors, in combination with studies delineating the cell types responsible for MUC5B secretion (AT2 versus airway cells) and genetics, e.g., MUC5B polymorphisms, might aid in understanding the long-term favorable versus fibrotic outcomes of COVID-19 disease (Chan et al., 2003, Rogers et al., 2018).
Our study also provides a SARS-CoV-2 infectious full-length cDNA clone for the field. Several strategies have been developed to construct stable coronavirus molecular clones, including the bacterial artificial chromosome (BAC) (Almazán et al., 2000, González et al., 2002) and vaccinia viral vector systems (Casais et al., 2001). In contrast, our in vitro ligation method solves the stability issue by splitting unstable regions and cloning the fragmented genome into separate vectors, obviating the presence of a full-length genome (Yount et al., 2000). Our in vitro ligation strategy has generated reverse genetic systems for at least 13 human and animal coronaviruses and produced hundreds of mutant recombinant viruses (Beall et al., 2016, Menachery et al., 2015, Scobey et al., 2013, Xie et al., 2020, Yount et al., 2003). In contrast to other reports (Thao et al., 2020), reporter recombinant SARS-CoV-2 viruses generated herein replicated to normal WT amounts in continuous cell lines, allowing for robust ex vivo studies in primary cultures.
Using this infectious clone, we generated a high-throughput luciferase reporter SARS-CoV-2 assay for evaluation of viral nAbs. In line with previous reports (Tian et al., 2020, Wrapp et al., 2020), our data show that several SARS-CoV RBD-binding nAbs fail to neutralize SARS-CoV-2, suggesting distant antigenicity within the RBD domains between the two viruses. Although more samples are needed, early convalescent sera demonstrated ∼1.5 log variation in neutralizing titers at ∼day 30 after infection, demonstrating a need to fully understand the kinetics, magnitude, and durability of the neutralizing antibody response after a primary SARS-CoV-2 infection. The detection of low-level SARS-CoV-2 cross-neutralizing antibodies in 2003 SARS-CoV serum samples is consistent with recent studies (Hoffmann et al., 2020, Walls et al., 2020), suggesting that existence of common neutralizing epitopes between the two CoVs. Interestingly, convalescent COVID-19 sera failed to cross-neutralize SARS-CoV in vitro, suggesting cross-neutralizing antibodies might be rare after SARS-CoV-2 infection. The location of these epitopes is unknown. The nLuc recombinant viruses described herein will be powerful reagents for defining the antigenic relationships between the Sarbocoviruses, the kinetics and durability of neutralizing antibodies after natural infection, and the breadth of therapeutic neutralizing antibodies and vaccine countermeasures (Wang et al., 2019).
In summary, our studies have quantitated differences in ACE2 receptor expression and SARS-CoV-2 infectivity in the nose (high) versus the peripheral lung (low). These studies should provide valuable reference data for future animal model development and expand the pool of tissues, e.g., nasal, for future study of disease pathogenesis and therapy. Although speculative, if the nasal cavity is the initial site mediating seeding of the lung via aspiration, these studies argue for the widespread use of masks to prevent aerosol, large droplet, and/or mechanical exposure to the nasal passages. Complementary therapeutic strategies that reduce viral titer in the nose early in the disease, e.g., nasal lavages, topical antivirals, or immune modulation, might be beneficial. Finally, our studies provide key reagents and strategies to identify type-specific and highly conserved neutralizing antibodies that can be assessed most easily in the nasal cavity as well as in the blood and lower airway secretions.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal human MUC5B | Santa Cruz | RRID: AB_2282256 |
| Mouse monoclonal human MUC5AC | Invitrogen | RRID: AB_10978001 |
| Rabbit polyclonal SARS coronavirus nucleocapsid | Invitrogen | RRID: AB_1087200 |
| Mouse monoclonal anti-acetylated tubulin | Sigma-Aldrich | RRID: AB_609894 |
| Rabbit polyclonal prosurfactant protein C | Sigma-Aldrich | RRID: AB_91588 |
| Goat polyclonal AGER | R&D Systems | RRID: AB_354628 |
| Rat monoclonal anti-tubulin | Millipore | RRID: AB_2210391 |
| Goat polyclonal anti-GFP | AbCam | RRID: AB_305643 |
| Rabbit polyclonal anti-GFP | AbCam | RRID: AB_305564 |
| Alexa Fluor phalloidin 647 | Invitrogen | RRID: AB_2620155 |
| Alexa Fluor phalloidin 555 | Invitrogen | Cat#A34055 |
| Hoechst 33342 | Invitrogen | Cat#H3570 |
| Goat anti-CCSP | Sigma-Aldrich | Cat#ABS1673 |
| Alexa Fluor 488-AffiniPure Donkey Anti-Goat IgG (H+L) (min X Ck,GP,Sy Hms,Hrs,Hu,Ms,Rb,Rat Sr Prot) antibody | Jackson ImmunoResearch | RRID: AB_2336933 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Invitrogen | RRID: AB_162543 |
| Donkey anti-Rat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Invitrogen | RRID: AB_2535795 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | RRID: AB_162542 |
| Alexa Fluor 488-AffiniPure Donkey Anti-Rabbit IgG (H+L) antibody | Jackson ImmunoResearch | RRID: AB_2313584 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Invitrogen | RRID: AB_2536180 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | RRID: AB_2535864 |
| S230 | UNC protein core facility | N/A |
| S230.15 | UNC protein core facility | N/A |
| S227.14 | UNC protein core facility | N/A |
| S227.9 | UNC protein core facility | N/A |
| MERS-27 | UNC protein core facility | N/A |
| m336 | UNC protein core facility | N/A |
| EDE1-C10 | UNC protein core facility | N/A |
| anti-SARS-CoV N protein | Invitrogen | Cat#PA1-41098 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 WA1 isolate | Natalie J. Thornburg, CDC | GenBank: MT020880 |
| icSARS-CoV-2-WT | This paper | GenBank: MT461669 |
| icSARS-CoV-2-GFP | This paper | GenBank: MT461670 |
| icSARS-CoV-2-nLuc-GFP | This paper | GenBank: MT461671 |
| Biological Samples | ||
| Human nasal, tonsil, and lung samples from CF and non-CF subjects | Marsico Lung Institute, UNC | See Table S1 for a list of donors |
| Human nasal and lung samples from healthy volunteers | NHLBI | See Table S1 for a list of donors |
| Human lung histology sections from COVID-19 subjects | University of New Mexico, New York Presbyterian Hospital | See STAR Methods |
| SARS patient serum samples (Toronto) | University Health Network, MaRS Center, Canada | IRB#:UHN REB 03-0250 |
| COVID-19 serum samples | UNC Hospital | IRB#:20-1141 |
| Mouse serum anti SARS-CoV-2 spike | This paper | N/A |
| Mouse serum anti SARS-CoV-2 nucleocapsid | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant human IL1β protein | R&D Systems | Cat#201-LB-005 |
| Recombinant human IL13 protein | R&D Systems | Cat#213-ILB-005 |
| Recombinant human IFNβ protein | R&D Systems | Cat#8499-IF-010 |
| Hydrocortisone | Sigma | Cat#H0888 |
| EGF | Invitrogen | Cat#PHG0313 |
| Insulin | Sigma | Cat#I5500 |
| Amphotericin B | Fisher Scientific | Cat#BP264550 |
| Gentamincin | GIBCO | Cat#15710-064 |
| Cholera toxin | Sigma | Cat#C8052 |
| Y-27632 | Enzo life Science | Cat#ALX-270-333-M025 |
| TRIzol Reagent | ThermoFisher | Cat#15596026 |
| Critical Commercial Assays | ||
| Direct-zol RNA Miniprep | ZYMO Research | Cat#R2051 |
| iScript™ Reverse Transcription Supermix for RT-qPCR | BIO-RAD | Cat#1708840 |
| SsoAdvanced Universal Probes Supermix | BIO-RAD | Cat#1725280 |
| RNAScope Multiplex Fluorescent Reagent Kit v2 | ACD | Cat#323100 |
| RNAScope 2.5 HD Duplex Reagent Kit | ACD | Cat#322430 |
| RNAScope 2.5 HD Reagent Kit-RED | ACD | Cat#322350 |
| RNAScope probe FOXJ1 (channel 2) | ACD | Cat#476351-C2 |
| RNAScope probe MUC5B (channel 2) | ACD | Cat#449888-C2 |
| RNAScope probe ACE2 (channel 1) | ACD | Cat#848151 |
| RNAScope probe ACE2 (channel 2) | ACD | Cat#848151-C2 |
| RNAScope probe TMPRSS2 (channel 1) | ACD | Cat#470341 |
| RNAScope probe SARS-CoV-2, S gene encoding the spike protein (channel 1) | ACD | Cat#848561 |
| RNAScope probe SARS-CoV-2, Antisense strand of the S gene (channel 1) | ACD | Cat#845701 |
| RNAScope probe SFTPC (channel 2) | ACD | Cat#452561-C2 |
| RNAScope probe HOPX (channel 1) | ACD | Cat#423001 |
| Vector® TrueVIEW® Autofluorescence Quenching Kit | Vector Laboratories | Cat#SP-8400 |
| Taqman probe TBP | Fisher Scientific | Cat#Hs99999910_m1 |
| Taqman probe GAPDH | Fisher Scientific | Cat#Hs02758991_g1 |
| Taqman probe ACE2 | Fisher Scientific | Cat#Hs01085333_m1 |
| Taqman probe TMPRSS2 | Fisher Scientific | Cat#Hs01122322_m1 |
| Nano-Glo Luciferase Assay | Promega | Cat#N1130 |
| QIAprep Spin Mini-prep Kit | QIAGEN | Cat#27106 |
| ExpiFectamine 293 transfection kit | Thermo | Cat#A14526 |
| NorthernMax-Gly Kit | Invitrogen | Cat#AM1946 |
| QIAquick Gel Extraction kit | QIAGEN | Cat#28706 |
| mMESSAGE mMACHINE T7 transcription kit | ThermoFisher | Cat#AM1344 |
| Chemiluminescent Nucleic Acid Detection Module | ThermoFisher | Cat#89880 |
| Oligotex mRNA Mini Kit | QIAGEN | Cat#70022 |
| Deposited Data | ||
| icSARS-CoV-2 WT genomic sequence | GenBank | MT461669 |
| icSARS-CoV-2-GFP genomic sequence | GenBank | MT461670 |
| icSARS-CoV-2-nLuc-GFP genomic sequence | GenBank | MT461671 |
| Experimental Models: Cell Lines | ||
| Simian kidney Vero | ATCC | Cat#CCL81 |
| Simian kidney Vero E6 | ATCC | Cat#CRL1586 |
| LLC-MK | ATCC | Cat#CCL-7 |
| UNCNN2TS | Marsico Lung Institute, UNC | N/A |
| Primary nasal cells | Marsico Lung Institute, UNC | N/A |
| Human bronchial epithelium | Marsico Lung Institute, UNC | N/A |
| Human alveolar type II pneumocytes | Marsico Lung Institute, UNC | N/A |
| Human primary lung microvascular endothelial cells | Marsico Lung Institute, UNC | N/A |
| Human primary lung fibroblasts | Marsico Lung Institute, UNC | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: BALB/c | Jackson Labs | Cat#000651 |
| Oligonucleotides | ||
| Leader forward primer: 5′- GTTTATACCTTCCCAGGT AACAAACC −3′ |
This paper | N/A |
| M gene reverse primer: 5′- AAGAAGCAATGAAGTA GCTGAGCC −3′ |
This paper | N/A |
| N gene primer: 5′-GTAGAAATACCATCTTGGACT GAGATC −3′ |
This paper | N/A |
| RT-PCR primer: 5′-GCTTCTGGTAATCTATTACTAG ATAAACG-3′ |
This paper | N/A |
| RT-PCR primer: 5′- AAGACATCAGCATACTCCTG ATTAGG −3′ |
This paper | N/A |
| biotin-labeled oligomer: 5′- BiodT/GGCTCTGTTGGGA ATGTTTTGTATGCG/BiodT-3′ |
This paper | N/A |
| Recombinant DNA | ||
| 7 plasmids of icSARS-CoV-2 WT | This paper | N/A |
| 1 plasmid encoding icSARS-CoV-2-nLuc-GFP reporter | This paper | N/A |
| 1 plasmid encoding icSARS-CoV-2-GFP reporter | This paper | N/A |
| Software and Algorithms | ||
| QuantStudio 6 Flex System | ThermoFisher Scientific | Cat#4485697 |
| QuantStudio Software v1.3 | ThermoFisher Scientific | https://thermofisher.com |
| GraphPad Prism 8 | GraphPad | https://graphpad.com |
| Olyvia V3.1.1 | Olympus | https://olympus-lifescience.com |
| Adobe Photoshop | Adobe | http://www.adobe.com/nl/products/photoshop.html |
| R version 3.5.1 | R Foundation | https://www.r-project.org/ |
| Other | ||
| T4 DNA Ligase | NEB | Cat#M0202S |
| BsmBI | NEB | Cat#R0580 |
| SacI | NEB | Cat#R0156S |
| PrimeSTAR GXL HiFi DNA polymerase | TaKaRa | Cat#RF220Q |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ralph S. Baric (rbaric@email.unc.edu).
Materials Availability
Material and reagents generated in this study will be made available upon installment of a material transfer agreement (MTA).
Data and Code Availability
Genomic sequences of recombinant viruses icSARS-CoV-2-WT, icSARS-CoV-2-GFP and icSARS-CoV-2-nLuc-GFP, which were generated in this study, have been deposited to GenBank (Accession # MT461669 to MT461671).
Experimental Model and Subject Details
Human Subjects
Excess surgical pathology tissues were obtained from the University of North Carolina (UNC) Tissue Procurement and Cell Culture Core under protocol #03-1396 approved by the UNC Biomedical IRB. Informed consent was obtained from tissue donors or authorized representatives. Cystic fibrosis lung tissue was obtained from donors undergoing transplantation, while human lungs from previously healthy individuals unsuitable for transplantation were obtained from Carolina Donor Services (Durham, NC), the National Disease Research Interchange (Philadelphia, PA), or the International Institute for Advancement of Medicine (Edison, NJ). Upper airway tissues were obtained from subjects undergoing elective surgeries. Excised tissue specimens were dissected and fixed in 10% neutral buffered formalin for 48 h followed by paraffin-embedding. The paraffin blocks were cut to produce 5 μm serial sections for RNA-ISH and immunohistochemistry. For qRT-PCR for ACE2 and TMPRSS2 expression in nasal and bronchial epithelial cells, nasal and bronchial epithelial cells were obtained from matched healthy volunteers by nasal scraping and bronchoscopic brush-biopsy under the National Heart, Lung, and Blood Institute IRB-approved protocol #07-H-0142. For single cell-RNA in situ hybridization (scRNA-ISH), human bronchial epithelial cells were obtained from the left main bronchus of healthy non-smoker volunteers by bronchoscopic brush-biopsy under the UNC Biomedical IRB-approved protocol #91-0679. Nasal surface epithelial cells were isolated from the resected nasal tissues as previously described (Fulcher and Randell, 2013, Okuda et al., 2019). After the isolation of nasal surface epithelial cells, the remaining nasal tissues were micro-dissected to isolate submucosal glands under the light microscope. Donor demographics was shown in Tables S1 and S2.
Tissue blocks or cut sections obtained from four COVID-19 autopsy lungs were obtained from Drs. Ross. E. Zumwalt (University of New Mexico) and Steven Salvatore and Alain Borczuk (New York Presbyterian Hospital). Donor demographics were described as below.
Donor 1. 40-year-old, male. Medical history: Diabetes mellitus. Clinical course: This donor had upper respiratory infection (URI) symptoms three days before he was found dead at home. No intubation was conducted. Postmortem testing of the lung was positive for SARS-CoV-2.
Donor 2. 64-year-old, male. Medical history: Diabetes mellitus, diabetic nephropathy, hepatitis C, heart failure, and coronary artery disease. Clinical course: This donor was transferred to ER because of fever and respiratory distress. Nasal swab was positive for SARS-CoV-2. He died five h after urgent care. No intubation was conducted.
Donor 3. 95-year-old, female. Medical history: Hypertension, hyperlipidemia, Alzheimer’s disease. Clinical course: This donor was transferred to ER because of respiratory distress. Nasal swab was positive for SARS-CoV-2. She died 48 h after urgent care. No intubation was conducted.
Donor 4. 69-year-old, male. Medical history: acute myeloid leukemia, type 2 diabetes mellitus. Clinical course: This donor was admitted to the hospital because of respiratory distress eight days after initial URI symptoms and diagnosis of SARS-CoV-2. He died five days post admission. No intubation was conducted.
Primary Cell Culture
Primary human nasal epithelial cells (HNE) were collected from healthy volunteers by curettage under UNC Biomedical IRB-approved protocols (#11-1363 and #98-1015) after informed consent as previously described (Kesic et al., 2011, Knowles et al., 2014). Briefly, superficial scrape biopsies were harvested from the inferior nasal turbinates under direct vision through a 9 mm reusable polypropylene nasal speculum (Model 22009) on an operating otoscope with speculum (Model 21700). Both nostrils were scraped 5 times without anesthesia using a sterile, plastic nasal curette (Arlington Scientific). Nasal cells were expanded using the conditionally reprogrammed cell (CRC) method (Gentzsch et al., 2017) or in Pneumacult EX Plus media (Stem Cell Technologies) (Speen et al., 2019) and then cultured on porous Transwell (Corning) supports in Pneumacult air liquid interface (ALI) media (Stem Cell Technologies). Human bronchial epithelial [large airway epithelial (LAE)] and bronchiolar [small airway epithelial (SAE)] cells, human alveolar type II pneumocytes (AT2), and human primary lung microvascular endothelial cells (MVE) and fibroblasts (FB) were isolated from freshly excised normal human lungs obtained from transplant donors with lungs unsuitable for transplant under IRB-approved protocol (#03-1396), as previously described (Fulcher and Randell, 2013, Okuda et al., 2019).
Cell Lines
Simian kidney cell lines Vero (ATCC # CCL81), Vero E6 (ATCC # CRL1586), and LLC-MK (ATCC# CCL-7) were purchased from ATCC and preserved in our laboratory. The Vero-furin cell line was reported previously (Mukherjee et al., 2016). LLC-MK cells expressing TMPRSS2 were generated in our laboratory. A novel immortalized nasal cell line (UNCNN2TS) was created by lentiviral overexpression of Bmi-1 and hTERT (Fulcher et al., 2009) in primary nasal cells, and subsequent lentiviral addition of SV40 T antigen (pBSSVD2005 was a gift from David Ron, Addgene plasmid # 21826). UNCNN2T cells are grown and infected in EpiX media (Propagenix).
Virus strains
Clinical SARS-CoV-2 isolate WA1 strain was provided by Dr. Natalie J. Thornburg at the U.S. Centers for Disease Control and Preventive (CDC). The virus was isolated from the first US COVID-19 patient identified in Washington state (GenBank Accession#: MT020880). Recombinant CoVs icSARS-CoV-Urbani, icSARS-CoV-GFP, icSARS-CoV-nLuc and icMERS-CoV-nLuc were generated in our laboratory as described previously (Scobey et al., 2013, Yount et al., 2003). Briefly, the strategy to synthesize full-length cDNA clones for SARS-CoV-Urbani and MERS-CoV was identical to the method reported herein, but with different restriction sites and junctions. The GFP and nLuc reporters were inserted into the accessory ORF7a of the icSARS-CoV-Urbani clone, whereas the nLuc reporter gene was introduced into the accessory ORF5a of the icMERS-CoV clone. Virus stocks were propagated on Vero E6 cells in minimal essential medium containing 10% fetal bovine serum (HyClone) and supplemented with penicillin/kanamycin (Gibico). Virus plaques were visualized by neutral red staining at two days post-infection. The UNC Institutional Biosecurity Committee and the National Institute of Allergy and Infectious Disease (NIAID) have approved the SARS-CoV-2 molecular clone project. All viral infections were performed under biosafety level 3 (BSL-3) conditions at negative pressure, and Tyvek suits connected with personal powered-air purifying respirators.
Human serum samples
SARS serum samples were obtained from SARS convalescent patients from University Health Network, MaRS Center, Toronto, Canada that had a confirmed SARS infection under IRB-approved protocol (#UHN REB 03-0250). COVID-19 serum samples were also provided as coded material and were deemed “not human subject research” by the UNC School of Medicine Biomedical IRB (#20-1141).
Method Details
Primary human cell culture and infection
Nasal cells were expanded using the conditionally reprogrammed cell (CRC) method (Gentzsch et al., 2017) or in Pneumacult EX Plus media (Stem Cell Technologies) (Speen et al., 2019) and then cultured on porous Transwell (Corning) supports in Pneumacult air liquid interface (ALI) media (Stem Cell Technologies). Human LAE and SAE cells were cultured as previously described (Fulcher and Randell, 2013, Okuda et al., 2019). Briefly, Isolated LAE and SAE cells were co-cultured with mitomycin-treated 3T3 J2 cells on collagen-coated tissue culture plastic dishes in DMEM media supplemented with 10 μM Y-27632 (Enzo Life Science). At 70%–90% confluence, LAE and SAE cells were passaged and sub-cultured for expansion. P2 LAE and SAE cells were transferred to human placental type IV collagen-coated, 0.4 μm pore size Millicell inserts (Millipore, PICM01250). The LAE and SAE cells were seeded at a density of 2.8 × 105 cells/cm2 and cultured in UNC ALI media. Upon confluence, cells were maintained at an ALI by removing apical media and providing UNC ALI media to the basal compartment only. Medium was replaced in the basal compartment twice a week, and the apical surfaces were washed with PBS once a week. After 28 days, LAE and SAE cells were utilized for SARS-CoV-2 recombinant viruses infection. Human type II pneumocytes (AT2) were prepared and cultured on porous supports as previously described (Bove et al., 2010). The AT2 cells are grown in DMEM with P/S and 10% FBS and switched to 4% FBS 24 h prior to infection. Cells were studied within three days and after five days, as they transdifferentiate into type I pneumocyte (AT1)-like cells. For serum-free and feeder-free AT2 cell cultures (mixed AT1/AT2 culture), human lung pieces (∼2 gm) were washed twice with PBS containing 1% Antibiotic-Antimycotic and cut into small pieces. Visible small airways and blood vessels were carefully removed to avoid clogging. Then samples were digested with 30 mL of enzyme mixture (collagenase type I: 1.68 mg/mL, dispase: 5U/mL, DNase: 10 U/mL) at 37°C for 45 min with rotation. The cells were filtered through a 100 μm strainer and rinsed with 15 mL PBS through the strainer. The supernatant was removed after centrifugation at 450x g for 10 min and the cell pellet was resuspended in red blood cell lysis buffer for five minutes, washed with DMEM/F12 containing 10% FBS and filtered through a 40 μm strainer. To purify human AT2 cells, approximately two million total lung cells were resuspended in SF medium and incubated with Human TruStain FcX (BioLegend) followed by incubation with HTII-280 antibody (Terrace Biotech). The cells were washed with PBS and then incubated with anti-mouse IgM microbeads. The cells were loaded into LS column (Miltenyi Biotec) and labeled cells collected magnetically. HTII-280+ human AT2 cells (1-3 × 103) were resuspended in culture medium. Serum-free feeder free medium and AT2 differentiation medium will be described elsewhere (S.V. and PRT et al., currently under revision in Cell Stem Cell). Culture plates were coated with Cultrex reduced growth factor basement membrane extract, Type R1 and cultured for five days followed by changing medium to AT2 differentiation medium for additional five days.
Human primary lung microvascular endothelial cells (MVE) and fibroblasts (FB) were grown as previously described (Scobey et al., 2013). For MVE cells, peripheral lung tissue minus the pleura was minced, digested with dispase/elastase, and cells were grown in EGM-2 media plus FBS (Lonza). Two or three rounds of CD31 bead purification (Dynabeads; Life Technologies) resulted in > 95% CD31-positive cells by flow cytometry that were used between passages 5 and 10. FBs were obtained by finely mincing distal human lung tissue and plating on scratched type 1/3 collagen-coated dishes in Dulbecco’s modified Eagle medium with high glucose (DMEMH) media plus 10% FBS, antibiotics, and antimycotics. Cells were released using trypsin/EDTA and subcultured in DMEMH, 10% FBS and P/S. The subcultured cells were elongated, spindly and negative for CD31 and pan-cytokeratin by flow cytometry and immunofluorescence, respectively.
icSARS-CoV-2-GFP virus infections were performed using well differentiated air-liquid interface (ALI) cultures of five donor specimens of human nasal epithelial (HNE) and large airway epithelial (LAE) cells using an MOI of three. Small airway epithelial (SAE) cell ALI cultures were created as previously described (Okuda et al., 2019). Paired LAE / SAE cells were inoculated with a SARS-CoV-2 clinical isolate, icSARS-CoV-2-WT, and icSARS-CoV-2-GFP, as well as wild-type icSARS-CoV-Urbani and icSARS-CoV-GFP on LAE, using an MOI of 0.5 for each virus. Transwell-cultured primary cells were inoculated with 200ul of virus via the apical surface and allowed to incubate at 37°C for two h. Following incubation, virus was removed, and cells were washed twice with 500ul PBS. Cells were returned to 37°C for the remainder of the experiment and observed for fluorescent signal, when appropriate, every 12-24 h. 100ul PBS was added to the apical surface of each culture and allowed to incubate for 10 min at 37°C in order to obtain an apical wash sample, at time points for analysis of viral replication by plaque assay. At the last time point, cells were lysed with 500ul TRIzol reagent (Invitrogen) to obtain total final RNA for analysis.
Primary human bronchial epithelial cell culture and cytokines exposure
Primary human LAE cells from normal donors (obtained from donors without previously known pulmonary diseases) were cultured on the human placenta collagen IV (Sigma Cat#C-7521) coated transwell (Corning Cat#3460) under air-liquid interface (ALI) condition. The apical surface was washed with PBS, and ALI medium (Fulcher et al., 2009) was replaced only in the basal compartment two-three times per week, and cells were cultured under ALI conditions for four weeks to allow full differentiation. Exposure with recombinant human cytokines was administrated started at the 5th week after ALI culture. All recombinant human cytokines (IL1β at 10 ng/mL, IL13 at 10ng/mL, and IFNβ at 10ng/m of final concentration in ALI media) were added to basolateral side of ALI media, and media were changed after three days supplied with freshly diluted cytokines. Cells were collected at day five – day seven for RNA isolation.
Whole-mount immunostaining and imaging
Well-differentiated mock or icSARS-CoV-2-GFP-infected LAE ALI cultures were fixed twice for 10 min in 4% formaldehyde in PBS and washed and stored in PBS. The GFP signal was enhanced by staining with anti-GFP antibody (Abcam ab6556; 0.5 ug/mL), a Venezuelan equine encephalitis virus (VEEV)-like replicon particle-immunized mouse antiserum against SARS-CoV-2 N protein (1:4000 dilution) and polyclonal rabbit anti-SARS-CoV N protein (Invitrogen PA1-41098, 0.5 ug/mL) using species-specific secondary antibodies as previously described (Ghosh et al., 2018). The cultures were also imaged for α-tubulin (Millipore MAB1864; 3ug/mL), MUC5AC (ThermoScientific 45M1; 4 ug/mL), MUC5B [polyclonal rabbit against a MUC5B peptide (MAN5BII), 1:1000] (Thornton et al., 2000), and CCSP (Sigma 07-623; 1:2000) as indicated. Filamentous actin was localized with phalloidin (Invitrogen A22287), and DNA with Hoechst 33342 (Invitrogen). An Olympus FV3000RS confocal microscope in Galvo scan mode was used to acquire 5-channel Z stacks by 2-phase sequential scan. Representative stacks were acquired with a 60X oil objective (xyz = 212um x 212um x ∼25um), and are shown as Z-projections or single-slice, XZ cross sections to distinguish individual cell features and to characterize the infected cell types. A 20X objective was used to acquire 2D, single-channel, apical snapshots of nine fields (636 um × 636 um; combined area = 3.64mm2), selected in evenly spaced grids across each sham infected donor culture, and ImageJ was used to measure the relative apical culture surface covered by multiciliated cells.
Immunohistochemistry
Immunohistochemical staining was performed on COVID-19 autopsy lung sections according to a protocol as previously described (Okuda et al., 2019). Briefly, paraffin-embedded sections were baked at 60c°C for 2–4chours, and deparaffinized with xylene (2 changes × 5 min) and graded ethanol (100% 2 × 5 min, 95% 1 × 5 min, 70% 1 × 5 min). After rehydration, antigen retrieval was performed by boiling the slides in 0.1cM sodium citrate pH 6.0 (3 cycles with microwave settings: 100% power for 6.5cmin, 60% for 6cmin, and 60% for 6cmin, refilling the Coplin jars with distilled water after each cycle). After cooling and rinsing with distilled water, quenching of endogenous peroxidase was performed with 0.5% hydrogen peroxide in methanol for 15cmin, slides washed in PBS, and blocked with 4% normal donkey serum, for an h at RT. Primary antibody (MUC5AC: 45M1, 1:1000, MUC5B: H300, 1:1000, SARS-CoV-2 nucleocapsid: 1:500, Anti-SARS mouse antiserum: 1:4000, Acetylated-α-tubulin: 1:1000, AGER: 1:400) were diluted in 4% normal donkey serum in PBST and incubated over night at 4c°C. Mouse and rabbit gamma globulin was used as an isotype control at the same concentration as the primary antibody. Sections were washed in PBST and secondary antibodies (biotinylated donkey anti-rabbit IgG, at 1:200 dilution in 4% normal donkey serum in PBST for chromogenic DAB staining for MUC5B, Alexa Fluor 488 donkey anti-rabbit IgG, at 1:1000 dilution and Alexa Fluor 594 donkey anti-mouse IgG, at 1:1000 dilution for fluorescent staining) were applied for 60cmin at RT. After washing in PBST, the Vector® TrueVIEW Autofluorescence Quenching Kit (Vector laboratories) was used to reduce background staining, and glass coverslips were placed over tissue sections with the ProLong Gold Antifade Reagent with DAPI (Invitrogen) for fluorescent staining. For chromogenic DAB staining, slides were incubated with avidin-peroxidase complex according to the manufacturer’s instructions (Vectastain kit, Vector laboratories), washed, incubated with the chromogenic substrate (Immpact Novared, Vector laboratories) and counterstained with Fast Red. Coverslipped slides were scanned and digitized using an Olympus VS120 whole slide scanner microscope with a 40X/60X 0.95 NA objective and Olympus confocal microscope with a 40X 0.6 NA or 60X 1.4 NA objective.
Cell dissociation for single cell-RNA in situ hybridization (scRNA-ISH)
Fresh bronchoscopically brush-biopsied human main bronchial epithelial cells, nasal surface epithelial and submucosal gland cells isolated from the resected nasal tissues were incubated with Accutase solution for 30 min at 37°C. The Accutase-treated cells were centrifuged (450 g, 2 min, 4°C) and then incubated with 10 mL HBSS (Ca+, Mg+) buffer containing DNase I (0.1 mg/mL) (Roche #10104159001) and collagenase IV (1 mg/mL) (GIBCO #17104-019) for 10 min and 30 min for bronchial/nasal surface epithelial cell and nasal submucosal gland cell isolation, respectively at 37°C with intermittent agitation. Nasal submucosal glands were micro-dissected from the nasal tissues under microscopy. The tissues were centrifuged (450 g, 2 min, 4°C) and then incubated with 10 mL HBSS (Ca+, Mg+) buffer containing DNase I (0.1 mg/mL) and collagenase IV (1 mg/mL) for 30 min at 37°C with intermittent agitation followed by additional incubation with Trypsin-EDTA (Final concentration: 0.125%, GIBCO #25200-056) for 20 min at 37°C. After incubation, enzymes were inactivated by adding 500 μL fetal bovine serum. Dissociated cells were filtered through a 40-μm cell strainer, centrifuged (450 g, 2 min, 4°C) and resuspended in PBS, adjusted to 105 cells/mL. Cell viability was examined by trypan blue dye exclusion. Single cell suspension was cytocentrifuged (55 g, 4 min, StatSpin CytoFuge2, Beckman Coulter) and fixed in 10% NBF for 30 min at room temperature. The cytocentrifuged cells were washed with PBS three times and then dehydrated with graded ethanol (50% 1 min, 70% 1 min, 100% 1 min). The slides were stored in 100% ethanol at −20°C until future use for scRNA-ISH.
RNA in situ hybridization
RNA-ISH was performed on cytocentrifuged single cells using the RNAscope Multiplex Fluorescent Assay v2, and on paraffin-embedded 5 μm tissue sections using the RNAscope 2.5 HD Reagent Kit and RNAscope 2.5 HD Duplex Reagent Kit according to the manufacturer’s instructions (Advanced Cell Diagnostics). Cytospin slides were rehydrated with graded ethanol (100% 1 min, 70% 1 min, 50% 1 min), permeabilized with PBS + 0.1% Tween 20 (PBST) at RT for 10 min, incubated with hydrogen peroxide (Advanced Cell Diagnostics) at RT for 10 min, followed by incubation with 1:15 diluted protease III at RT for 10 min. Tissue sections were deparaffinized with xylene (2 changes × 5 min) and 100% ethanol (2 changes × 1 min), and then incubated with hydrogen peroxide for 10 min, followed by target retrieval in boiling water for 15 min, and incubation with Protease Plus (Advanced Cell Diagnostics) for 15 min at 40°C. Slides were hybridized with custom probes at 40°C for 2 h, and signals were amplified according to the manufacturer’s instructions. The stained sections were scanned and digitized using an Olympus VS120 light or fluorescent microscope with a 40X 1.35 NA objective and Olympus confocal microscope with a 40X 0.6 NA or 60X 1.4 NA objective.
Calculation of frequency of ACE2 and TMPRSS2-positive cells in distinct anatomical airway regions as identified by scRNA-seq
Normalized log-transformed count+1 gene x cell matrix and meta-data were downloaded from https://www.genomique.eu/cellbrowser/HCA/, which represent 77,969 cells that passed quality control. Expression of ACE2 and TMPRSS2 were extracted from the matrix, and the number of cells with log normalized count > 0 were calculated.
RNA isolation and gene expression analysis by Taqman Assays
For qRT-PCR for ACE2 and TMPRSS2 expression in different airway regions, surface epithelial cells were isolated from freshly excised normal human lungs obtained from transplant donors by gentle scraping with a convex scalpel blade into F12 medium, excluding submucosal glands. Following centrifugation (450 g, 5 min, 4°C), the pelleted epithelial cells were resuspended in 1 mL of TRI Reagent (Sigma). Micro-dissected small airways and peripheral lung parenchyma were homogenized in 1 mL of TRI Reagent using a tissue homogenizer (Bertin Technologies). Debris was pelleted from the TRI Reagent by centrifugation, and the supernatant was used for RNA analysis.
The HBE cells growing on the transwell membrane were collected by excision of the whole membrane together with the cells using razor blade and lysed in TRI Reagent at 37°C shaker for 30 min. Total RNA was purified from the TRI Reagent lysates using the Direct-Zol RNA miniprep Kit (Zymo Research, cat#R2051), and examined by NanoDrop One Spectrophotometer (ThermoFisher) for its quality and quantity. 1 μg of total RNA was reverse transcribed to cDNA by iScript Reverse Transcription Supermix (BioRad, Cat#1708840) at 42°C for one h. Quantitative RT-PCR was performed using Taqman probes (Applied BioSystems) with SsoAdvanced Universal Probes Supermix (Bio-Rad, cat#1725280) on QuantStudio6 Real-time PCR machine (Applied Biosystem). The house-keeping gene used for normalization of gene expression for in vitro cultured HBE was TATA-binding protein (TBP) gene. See Key Resources Table for detailed information about primers/probes.
Assembly of SARS-CoV-2 WT and reporter cDNA constructs
Seven cDNA fragments covering the entire SARS-CoV-2 WA1 genome were amplified by RT-PCR using PrimeSTAR GXL HiFi DNA polymerase (TaKaRa). Junctions between each fragment contain non-palindromic sites BsaI (GGTCTCNˆNNNN) or BsmBI (CGTCTCNˆNNNN) with unique four-nucleotide cohesive ends. Fragment E and F contains two BsmBI sites at both termini, while other fragments harbor BsaI sites at the junction. Four-nucleotide cohesive ends of each fragment are indicated in Figure 1A. To assist the transcription of full-length viral RNA, we introduced a T7 promoter sequence into the upstream of fragment A, as well as a 25nt poly-A tail into the downstream of the fragment G. Each fragment was cloned into high-copy vector pUC57 and verified by Sanger sequencing. A silent mutation T15102A was introduced into a conserved region in nsp12 in plasmid D as a genetic marker. To enhance the efficiency of recovering SARS-CoV-2 virus in the cell culture, a sgRNA-N construct, encoding a 75nt leader sequence, N gene, 3′UTR, and a 25nt poly-A tail, was assembled under the control of a T7 promoter. Two reporter viruses, one containing GFP and the other harboring, a GFP-fused nLuc gene, were generated by replacing the ORF7 gene with the reporter genes.
Generation of full-length RNA transcript and recovery of recombinant viruses
Seven genomic cDNA fragments were digested with appropriate endonucleases, resolved on 0.8% agarose gels, excised and purified using a QIAquick Gel Extraction kit (QIAGEN). A full-length genomic cDNA was obtained by ligating seven fragments in an equal molar ratio with T4 DNA ligase (NEB). We then purified the ligated cDNA with chloroform and precipitated it in isopropanol. The full-length viral RNA or SARS-CoV-2 sgRNA-N were synthesized using the T7 mMESSAGE mMACHINE T7 transcription kit (Thermo Fisher) at 30°C for 4 h. The full-length SARS-CoV-2 transcript and sgRNA-N were mixed and electroporated into 8 × 106 of Vero E6 cells. The cells were cultured as usual in the medium for two to three days.
PCR of leader-containing sgRNAs
Viral replication in the electroporated cells was evaluated by amplification of leader sequence-containing sgRNAs. A forward primer targeting the leader sequence (5′- GTTTATACCTTCCCAGGTAACAAACC −3′) was paired with a reverse primer targeting M gene (5′- AAGAAGCAATGAAGTAGCTGAGCC −3′) or N gene (5′- GTAGAAATACCATCTTGGACTGAGATC −3′).
Identification of the genetic marker
To confirm that the introduced T15102A mutation exists in the recombinant viruses, viral RNA was extracted using TRI Reagent (Thermo Fisher). A 1579 bp fragment in nsp12 of each virus was amplified by RT-PCR using primer pair 5′- GCTTCTGGTAATCTATTACTAGATAAACG-3′ and 5′- AAGACATCAGCATACTCCTGATTAGG −3′. The fragment was subjected to Sanger sequencing or digested with SacI enzyme (NEB).
Northern Blot Analysis
Vero E6 cells were infected with SARS-CoV-2 isolate, icSARS-CoV-2-WT, icSARS-CoV-2-GFP or icSARS-CoV-2-GFP-nLuc at an MOI of 1. At 24 h post-infection, we extracted the total cellular RNA using TRIzol Reagent (Thermo Fisher). Poly A-containing messenger RNA was isolated from the total RNA using an Oligotex mRNA Mini Kit (QIAGEN). Messenger RNA (0.6-0.7 μg) was separated on an agarose gel and transferred to BrightStar-Plus membrane using a NorthernMax-Gly Kit (Invitrogen). Blots were hybridized with a biotin-labeled oligomer (5′- BiodT/GGCTCTGTTGGGAATGTTTTGTATGCG/BiodT-3′), then detected using a Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher) using the iBright Western Blot Imaging System (Thermo Fisher).
Generation of SAR-CoV-2 S protein-immunized mouse serum
The SAR-CoV-2 S and N genes was cloned into pVR21 3526 to generate virus replicon particles (VRPs), as previously described (Agnihothram et al., 2018). Briefly, SARS-CoV-2 S or N genes were inserted into pVR21, a vector encoding the genome of a VEEV strain 3526. The SARS-CoV-2-S-pVR21 construct, a plasmid containing the VEEV envelope glycoproteins, and a plasmid encoding the VEEV capsid protein were used to generate T7 RNA transcripts. The RNA transcripts were then electroporated into BHK cells. VRPs were harvested 48 h later and purified via high-speed ultra-centrifugation. Two groups of 10-week-old BALB/c mice (Jackson Labs) were then inoculated with the VRPs via footpad injection then boosted with the same dose once four weeks later. Serum samples were collected at 2 weeks post-boosting and were mixed together.
Monoclonal antibody large-scale production
SARS-specific S230, S230.15, S227.14, S227.9 IgG, MERS-specific MERS-27, m336 IgG, and a Dengue-specific EDE1-C10 IgG antibody variable heavy and light chain genes were obtained, codon-optimized for human mammalian cell expression, and cloned into heavy and light-chain variable-gene-expressing plasmids encoding a human IgG1 Fc region as described previously (Martinez et al., 2020). One hundred μg of each variable heavy and light chain plasmids were co-transfected using an ExpiFectamine 293 transfection kit in Expi293F (Thermo) cells at 2.5 million cells/mL in 1L flasks in suspension. Transfected cell supernatants were harvested two days later, and the soluble antibody was purified using Pierce protein A beads (Thermo) followed by fast protein liquid chromatography (FPLC). MAbs were buffer exchanged with sterile 1XPBS. Purified mAbs were quality controlled by western blotting and Coomassie blue staining to confirm mAb purity.
MERS-CoV, SARS-CoV, and SARS-CoV-2 neutralization assays
Recombinant viruses icMERS-CoV-nLuc, icSARS-CoV-nLuc, and icSARS-CoV-2-nLuc-GFP were tittered in Vero E6 cells to obtain a relative light units (RLU) signal of at least 20X the cell only control background. Vero E6 cells were plated at 20,000 cells per well the day prior in clear bottom black-walled 96-well plates (Corning 3904). Neutralizing antibody serum samples were tested at a starting dilution of 1:20 and mAb samples were tested at a starting dilution 50 μg/mL and were serially diluted 4-fold up to eight dilution spots. icMERS-CoV-nLuc, icSARS-CoV-nLuc, and icSARS-CoV-2-nLuc-GFP viruses were diluted and were mixed with serially diluted antibodies. Antibody-virus complexes were incubated at 37°C with 5% CO2 for 1 h. Following incubation, growth media was removed, and virus-antibody dilution complexes were added to the cells in duplicate. Virus-only controls and cell-only controls were included in each neutralization assay plate. Following infection, plates were incubated at 37°C with 5% CO2 for 48 h. After the 48 h incubation, cells were lysed, and luciferase activity was measured via Nano-Glo Luciferase Assay System (Promega) according to the manufacturer specifications. MERS-CoV, SARS-CoV, and SARS-CoV-2 neutralization titers were defined as the sample dilution at which a 50% reduction in RLU was observed relative to the average of the virus control wells.
Quantification and Statistical Analysis
For comparison of gene expression in response to cytokine exposure versus control (PBS) with one culture per code in each group, we performed Wilcoxon matched-pairs signed rank test by Graphpad Prism 8 built in function. For comparison of gene expression in response to cytokine exposure versus control (PBS) with more than one culture per code in each group, the linear mixed-effect models analysis were performed. The relative mRNA expression from Taqman assays were analyzed with linear mixed-effect models using the R package Ime4 (Bates et al., 2015), with treatment as fixed effect and code as random-effect factors. Statistical significance were evaluated with the R lmerTest package (Kuznetsova et al., 2017), using the Satterthwarte’s degrees of freedom method. Multiple post hoc comparisons of subgroups were performed using the R multcomp package (Hothorn et al., 2006). For cell count data, generalized linear mixed-effect models (glmer) with Poisson distribution was used. Wilcoxon rank sum test was used to determine the statistical significance between unpaired two groups in Figures 2D, 2F, 2H, and 6 Bvi using Graphpad Prism 8. One-way ANOVA followed by Tukey test was used to determine the statistical significance between groups in Figure 6Bv using Graphpad Prism 8. The “n” numbers for each experiment are provided in the text and figures. p < 0.05 was considered statistically significant.
Co-localization of ACE2 mRNA with marker-genes and quantification
RNA-ISH was performed on cytocentrifuged single cells using RNAscope Multiplex Fluorescent Assay v2, as described above, to assess colocalization of ACE2 mRNA and airway epithelial cell markers, including FOXJ1 (ciliated cells) and MUC5B (secretory cells). ACE2 probe (channel 1) was combined with each of airway epithelial cell marker (channel 2). The stained cytospin slides were scanned and digitized using an Olympus VS120 whole slide scanner microscope with a 40X 0.9 numerical aperture objective. Using Fiji software (Schindelin et al., 2012), quantification for colocalization was performed in the scanned images by an investigator blinded to slide identification. To calculate the occurrence of ACE2 + cells in preparations, ACE2 + cells and total cells were manually counted, and the frequency calculated. For quantitative co-localization analysis of ACE2 with airway epithelial marker-defined cells, the number of visible ACE2 signals (dots) was manually counted in each airway epithelial cell-marker-positive cell. The ACE2 + signals were quantitated in 200 FOXJ1 + or MUC5B + cells per subject. One or more dot signals defined an ACE2 + positive cell, while airway-epithelial-marker-positive cells were defined as cells expressing 10 or more dot signals for the epithelial marker.
Quantification of ACE2 and TMPRSS2 gene expression in tonsillar surface epithelium
Human tonsil tissue sections were analyzed for ACE2 and TMPRSS2 expression using RNA-ISH. Tonsillar surface epithelial regions with positive RNA ISH signals (4 to 8 regions per donor) were selected for quantification. Signal counts were normalized to the number of cells as determined by DAPI nuclear stain in each region. For quantification, the stratified epithelial layer was divided into two layers: 1) surface (flattened epithelial) layer and 2) basal (cuboidal epithelial) layer.
Acknowledgments
We would like to acknowledge funding sources from the National Allergy and Infectious Disease (NIAID), National Institution of Health (NIH) (U19-AI100625 , R01-AI089728, and U01-AI14964) to R.S.B. and the National Heart, Lung, and Blood Institute (NHLBI), NIH (UH3-HL123645, P01-HL110873, R01-HL136961, P30-DK065988-13, and P01-HL108808) to R.C.B. K.O. is funded by the Cystic Fibrosis Foundation (OKUDA10I0) and a research grant from Cystic Fibrosis Research Incorporation. D.R.M. is funded by NIH NIAID T32 AI007151 and a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award. T.K. is funded by a Senior Research Training Fellowship (RT-57362) of American Lung Association. A.J.K. is support by the National Center for Advancing Translational Sciences, NIH, through grant KL2TR002490. P.R.T. received a Pathways to Independence award from the NHLBI/NIH (R00HL127181 and R01HL146557) that partially supported this study. V.S. is supported by a postdoc fellowship from Regeneration Next Initiative at Duke University. We thank N.J. Thornburg at the CDC for providing us the SARS-CoV-2 clinical isolate WA1 strain. We are grateful for the technical support of Y. Escobar for HNE cultures and Lisa Morton for qPCR assays and to S. Weiss, Y. Park, J. Kuruc, and the UNC Blood Donor Center for COVID-19 serum sample preparation. The UNC Animal Histopathology & Laboratory Medicine Core is supported in part by an NCI Center Core Support Grant (5P30CA016086-41) to the UNC Lineberger Comprehensive Cancer Center. We thank E.C. Roe for assisting manuscript editing. Finally, we are grateful for the donors of primary cells and sera who made this study possible.
Author Contributions
Conceptualization R.C.B., R.S.B, and S.H.R.; Investigation: Y.J.H., K.O., C.E.E., D.R.M., T.A., K.D.3, T.K., R.L., B.L.Y., T.M.M., G.C., K.N.O., A.G., L.V.T., S.R.L., L.E.G., A.S., H.D., R.G.,S.N., L.S., L.F., W.K.O., and S.H.R.; Contribution to research materials: A.L.B., N.I.N., M.C., C.C., D.J.K., A.D.S., D.M.M., A.M., L.B., R.Z., F.J.M., S.P.S., A.B., P.R.T., V.S., A.K., I.J., and S.H.R., Writing – original draft preparation: Y.J.H.; Writing – review and editing: R.C.B., R.S.B., S.H.R., and W.K.O.; Visualization: Y.J.H., K.O., C.E.E., D.R.M., T.A., and T.K.; Funding acquisition: R.C.B. and R.S.B.
Declaration of Interests
The authors declare no competing financial interests.
Published: May 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.05.042.
Supplemental Information
References
- Agnihothram S., Menachery V.D., Yount B.L., Jr., Lindesmith L.C., Scobey T., Whitmore A., Schäfer A., Heise M.T., Baric R.S. Development of a broadly accessible Venezuelan equine encephalitis virus replicon particle vaccine platform. J. Virol. 2018 doi: 10.1128/JVI.00027-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar J.A., Tremblay B.J.-M., Mansfield M.J., Woody O., Lobb B., Banerjee A., Chandiramohan A., Tiessen N., Dvorkin-Gheva A., Revill S. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. bioRxiv. 2020 doi: 10.1101/2020.04.07.030742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán F., González J.M., Pénzes Z., Izeta A., Calvo E., Plana-Durán J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberson J.B. A clinical consideration of abscesses and cavities of the lung. Bull. Johns Hopkins Hosp. 1954;94:227–237. [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atri D., Siddiqi H.K., Lang J., Nauffal V., Morrow D.A., Bohula E.A. COVID-19 for the cardiologist: a current review of the virology, clinical epidemiology, cardiac and other clinical manifestations and potential therapeutic strategies. JACC Basic Transl. Sci. 2020 doi: 10.1016/j.jacbts.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B., Walker S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015;67:1–48. [Google Scholar]
- Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. MBio. 2016;7 doi: 10.1128/mBio.01451-15. e01451–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth T.F., Kournikakis B., Bastien N., Ho J., Kobasa D., Stadnyk L., Li Y., Spence M., Paton S., Henry B. Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J. Infect. Dis. 2005;191:1472–1477. doi: 10.1086/429634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher R.C. Muco-obstructive lung diseases. N. Engl. J. Med. 2019;380:1941–1953. doi: 10.1056/NEJMra1813799. [DOI] [PubMed] [Google Scholar]
- Bove P.F., Grubb B.R., Okada S.F., Ribeiro C.M., Rogers T.D., Randell S.H., O’Neal W.K., Boucher R.C. Human alveolar type II cells secrete and absorb liquid in response to local nucleotide signaling. J. Biol. Chem. 2010;285:34939–34949. doi: 10.1074/jbc.M110.162933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann D., Tsukahara T., Weinreb C., Logan D.W., Datta S.R. Non-neural expression of SARS-CoV-2 entry genes in the olfactory epithelium suggests mechanisms underlying anosmia in COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.03.25.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsana L., Sonzogni A., Nasr A., Rossi R., Pellegrinelli A., Zerbi P., Rech R., Colombo R., Antinori S., Corbellino M. Pulmonary post-mortem findings in a large series of COVID-19 cases from Northern Italy. medRxiv. 2020 doi: 10.1101/2020.04.19.20054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Thiel V., Siddell S.G., Cavanagh D., Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 2001;75:12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC COVID-19 Response Team Preliminary estimates of the prevalence of selected underlying health conditions among patients with coronavirus disease 2019 United States, February 12-March 28, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:382–386. doi: 10.15585/mmwr.mm6913e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.S., Zheng J.P., Mok Y.W., Li Y.M., Liu Y.N., Chu C.M., Ip M.S. SARS: prognosis, outcome and sequelae. Respirology. 2003;8(Suppl):S36–S40. doi: 10.1046/j.1440-1843.2003.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Sun L., Kato T., Okuda K., Martino M.B., Abzhanova A., Lin J.M., Gilmore R.C., Batson B.D., O’Neal Y.K. IL-1β dominates the promucin secretory cytokine profile in cystic fibrosis. J. Clin. Invest. 2019;129:4433–4450. doi: 10.1172/JCI125669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell A.S., Johnson J.C., Moore I.N., Liu D.X., Bock K.W., Douglas M.G., Graham R.L., Solomon J., Torzewski L., Bartos C. A spike-modified Middle East respiratory syndrome coronavirus (MERS-CoV) infectious clone elicits mild respiratory disease in infected rhesus macaques. Sci. Rep. 2018;8:10727. doi: 10.1038/s41598-018-28900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo C., Burgel P.R., Gartner S., van Koningsbruggen-Rietschel S., Naehrlich L., Sermet-Gaudelus I., Southern K.W. Impact of COVID-19 on people with cystic fibrosis. Lancet Respir. Med. 2020;8:e35–e36. doi: 10.1016/S2213-2600(20)30177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez M., Zaragosi L.-E., Truchi M., Garcia S.R., Arguel M.-J., Lebrigand K., Paquet A., Pee’r D., Marquette C.-H., Leroy S. A single-cell atlas of the human healthy airways. bioRxiv. 2019 doi: 10.1101/2019.12.21.884759. [DOI] [PubMed] [Google Scholar]
- Dickson R.P., Erb-Downward J.R., Martinez F.J., Huffnagle G.B. The microbiome and the respiratory tract. Annu. Rev. Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante M.A., Kurtenbach S., Sargi Z.B., Harbour J.W., Choi R., Kurtenbach S., Goss G.M., Matsunami H., Goldstein B.J. Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat. Neurosci. 2020;23:323–326. doi: 10.1038/s41593-020-0587-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner H., Behbehani A.A., Hochstrasser K. [Diagnostic value of nasal secretions, current state: normal values. 1] Laryngol Rhinol Otol (Stuttg) 1983;62:561–565. [PubMed] [Google Scholar]
- Esther C.R., Jr., Muhlebach M.S., Ehre C., Hill D.B., Wolfgang M.C., Kesimer M., Ramsey K.A., Markovetz M.R., Garbarine I.C., Forest M.G. Mucus accumulation in the lungs precedes structural changes and infection in children with cystic fibrosis. Sci. Transl. Med. 2019;11:eaav3488. doi: 10.1126/scitranslmed.aav3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C.M., Fingerlin T.E., Schwarz M.I., Lynch D., Kurche J., Warg L., Yang I.V., Schwartz D.A. Idiopathic pulmonary fibrosis: a genetic disease that involves mucociliary dysfunction of the peripheral airways. Physiol. Rev. 2016;96:1567–1591. doi: 10.1152/physrev.00004.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzal Z., Basu S., Burke A., Fasanmade O.O., Lopez E.M., Bennett W.D., Ebert C.S., Jr., Zanation A.M., Senior B.A., Kimbell J.S. Comparative study of simulated nebulized and spray particle deposition in chronic rhinosinusitis patients. Int. Forum Allergy Rhinol. 2019;9:746–758. doi: 10.1002/alr.22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher M.L., Gabriel S.E., Olsen J.C., Tatreau J.R., Gentzsch M., Livanos E., Saavedra M.T., Salmon P., Randell S.H. Novel human bronchial epithelial cell lines for cystic fibrosis research. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009;296:L82–L91. doi: 10.1152/ajplung.90314.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher M.L., Randell S.H. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol. Biol. 2013;945:109–121. doi: 10.1007/978-1-62703-125-7_8. [DOI] [PubMed] [Google Scholar]
- Gaeckle N.T., Pragman A.A., Pendleton K.M., Baldomero A.K., Criner G.J. The oral-lung axis: the impact of oral health on lung health. Respir. Care. 2020 doi: 10.4187/respcare.07332. respcare.07332. [DOI] [PubMed] [Google Scholar]
- Gentzsch M., Boyles S.E., Cheluvaraju C., Chaudhry I.G., Quinney N.L., Cho C., Dang H., Liu X., Schlegel R., Randell S.H. Pharmacological rescue of conditionally reprogrammed cystic fibrosis bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 2017;56:568–574. doi: 10.1165/rcmb.2016-0276MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Coakley R.C., Mascenik T., Rowell T.R., Davis E.S., Rogers K., Webster M.J., Dang H., Herring L.E., Sassano M.F. Chronic e-cigarette exposure alters the human bronchial epithelial proteome. Am. J. Respir. Crit. Care Med. 2018;198:67–76. doi: 10.1164/rccm.201710-2033OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson K., Eggli D.F., Maxwell S.L. Quantitative aspiration during sleep in normal subjects. Chest. 1997;111:1266–1272. doi: 10.1378/chest.111.5.1266. [DOI] [PubMed] [Google Scholar]
- González J.M., Pénzes Z., Almazán F., Calvo E., Enjuanes L. Stabilization of a full-length infectious cDNA clone of transmissible gastroenteritis coronavirus by insertion of an intron. J. Virol. 2002;76:4655–4661. doi: 10.1128/JVI.76.9.4655-4661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Liang W.H., Zhao Y., Liang H.R., Chen Z.S., Li Y.M., Liu X.Q., Chen R.C., Tang C.L., Wang T., China Medical Treatment Expert Group for COVID-19 Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur. Respir. J. 2020;55:2000547. doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T., Bretz F., Westfall P., Heiberger R.M. 2006. Simultaneous inference for general linear hypotheses. http://cran.r-project.org/web/packages/multcomp/index.html. [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley E.J., Viroslav J., Gray W.R., Pierce A.K. Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am. J. Med. 1978;64:564–568. doi: 10.1016/0002-9343(78)90574-0. [DOI] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre G. The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses. 2019;11:837. doi: 10.3390/v11090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H. Pulmonary angiotensin-converting enzyme 2 (ACE2) and inflammatory lung disease. Shock. 2016;46:239–248. doi: 10.1097/SHK.0000000000000633. [DOI] [PubMed] [Google Scholar]
- Keeler S.P., Agapov E.V., Hinojosa M.E., Letvin A.N., Wu K., Holtzman M.J. Influenza A virus infection causes chronic lung disease linked to sites of active viral RNA remnants. J. Immunol. 2018;201:2354–2368. doi: 10.4049/jimmunol.1800671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesic M.J., Simmons S.O., Bauer R., Jaspers I. Nrf2 expression modifies influenza A entry and replication in nasal epithelial cells. Free Radic. Biol. Med. 2011;51:444–453. doi: 10.1016/j.freeradbiomed.2011.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles M.R., Ostrowski L.E., Leigh M.W., Sears P.R., Davis S.D., Wolf W.E., Hazucha M.J., Carson J.L., Olivier K.N., Sagel S.D. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am. J. Respir. Crit. Care Med. 2014;189:707–717. doi: 10.1164/rccm.201311-2047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollias A., Kyriakoulis K.G., Dimakakos E., Poulakou G., Stergiou G.S., Syrigos K. Thromboembolic risk and anticoagulant therapy in COVID-19 patients: emerging evidence and call for action. Br. J. Haematol. 2020 doi: 10.1111/bjh.16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A., Brockhoff P.B., Christensen R.H.B. lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 2017;82:1–26. [Google Scholar]
- Leung J.M., Yang C.X., Tam A., Shaipanich T., Hackett T.L., Singhera G.K., Dorscheid D.R., Sin D.D. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur. Respir. J. 2020;55:2000688. doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020 doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 2020 doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D.R., Tu J.J., Kumar A., Mangold J.F., Mangan R.J., Goswami R., Giorgi E.E., Chen J., Mengual M., Douglas A.O. Maternal broadly neutralizing antibodies can select for neutralization-resistant, infant-transmitted/founder HIV variants. MBio. 2020 doi: 10.1128/mBio.00176-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA. 2005;102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Dinnon K.H., 3rd, Yount B.L., Jr., McAnarney E.T., Gralinski L.E., Hale A., Graham R.L., Scobey T., Anthony S.J., Wang L. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 2020 doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Eisfeld A.J., Schäfer A., Josset L., Sims A.C., Proll S., Fan S., Li C., Neumann G., Tilton S.C. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. MBio. 2014;5 doi: 10.1128/mBio.01174-14. e01174–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: The world should face the reality. Environ. Int. 2020;139:105730. doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossel E.C., Wang J., Jeffers S., Edeen K.E., Wang S., Cosgrove G.P., Funk C.J., Manzer R., Miura T.A., Pearson L.D. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology. 2008;372:127–135. doi: 10.1016/j.virol.2007.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Sirohi D., Dowd K., Chen C., Diamond M., Kuhn R., Pierson T. Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology. 2016;497:33–40. doi: 10.1016/j.virol.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odani K., Tachibana M., Tamashima R., Tsutsumi Y. Herpes simplex virus pneumonia: importance of aspiration etiology. Case Rep. Pathol. 2019;2019:7623576. doi: 10.1155/2019/7623576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K., Chen G., Subramani D.B., Wolf M., Gilmore R.C., Kato T., Radicioni G., Kesimer M., Chua M., Dang H. Localization of secretory mucins MUC5AC and MUC5B in normal/healthy human airways. Am. J. Respir. Crit. Care Med. 2019;199:715–727. doi: 10.1164/rccm.201804-0734OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K., Kobayashi Y., Dang H., Nakano S., Barbosa Cardenas S.M., O’Neal V.K., Kato T., Chen G., Gilmore R.C., Chua M. Regional Regulation of CFTR and Ionocyte Expression in Normal Human Airways. (abstract) Pediatr. Pulmonol. 2019;54:S173. [Google Scholar]
- Ota C., Ng-Blichfeldt J.P., Korfei M., Alsafadi H.N., Lehmann M., Skronska-Wasek W., M De Santis M., Guenther A., Wagner D.E., Königshoff M. Dynamic expression of HOPX in alveolar epithelial cells reflects injury and repair during the progression of pulmonary fibrosis. Sci. Rep. 2018;8:12983. doi: 10.1038/s41598-018-31214-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Chen D., Xia Y., Wu X., Li T., Ou X., Zhou L., Liu J. Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect. Dis. 2020;20:410–411. doi: 10.1016/S1473-3099(20)30114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya V.K., Tiwari R.S. Nasal mucociliary clearance in health and disease. Indian J. Otolaryngol. Head Neck Surg. 2006;58:332–334. doi: 10.1007/BF03049581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papineni R.S., Rosenthal F.S. The size distribution of droplets in the exhaled breath of healthy human subjects. J. Aerosol Med. 1997;10:105–116. doi: 10.1089/jam.1997.10.105. [DOI] [PubMed] [Google Scholar]
- Phillips L.K., Deane A.M., Jones K.L., Rayner C.K., Horowitz M. Gastric emptying and glycaemia in health and diabetes mellitus. Nat. Rev. Endocrinol. 2015;11:112–128. doi: 10.1038/nrendo.2014.202. [DOI] [PubMed] [Google Scholar]
- Quirouette C., Younis N.P., Reddy M.B., Beauchemin C.A.A. A mathematical model describing the localization and spread of influenza A virus infection within the human respiratory tract. PLoS Comput. Biol. 2020;16:e1007705. doi: 10.1371/journal.pcbi.1007705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., van den Brand J.M.A., Bestebroer T.M., Lexmond P., de Meulder D., Fouchier R.A.M., Lowen A.C., Herfst S. Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat. Commun. 2020;11:766. doi: 10.1038/s41467-020-14626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020:eabb7314. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A.J., Solus J.F., Hunninghake G.M., Baron R.M., Meyer N.J., Janz D.R., Schwartz D.A., May A.K., Lawson W.E., Blackwell T.S., Ware L.B. MUC5B promoter polymorphism and development of acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2018;198:1342–1345. doi: 10.1164/rccm.201801-0140LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajuthi S.P., DeFord P., Jackson N.D., Montgomery M.T., Everman J.L., Rios C.L., Pruesse E., Nolin J.D., Plender E.G., Wechsler M.E. Type 2 and interferon inflammation strongly regulate SARS-CoV-2 related gene expression in the airway epithelium. bioRxiv. 2020 doi: 10.1101/2020.04.09.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia J.L., Rivera D.N., Herrera V., Morwitzer M.J., Creager H., Santarpia G.W., Crown K.K., Brett-Major D., Schnaubelt E., Broadhurst M.J. Transmission potential of SARS-CoV-2 in viral shedding observed at the University of Nebraska Medical Center. medRxiv. 2020 doi: 10.1101/2020.03.23.20039446. [DOI] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims A.C., Baric R.S., Yount B., Burkett S.E., Collins P.L., Pickles R.J. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 2005;79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi C.P., Nguyen J., Yamaguchi Y., Werts A.D., Lu P., Ladd M.R., Fulton W.B., Kovler M.L., Wang S., Prindle T., Jr. A dynamic variation of pulmonary ACE2 is required to modulate neutrophilic inflammation in response to Pseudomonas aeruginosa lung infection in mice. J. Immunol. 2019;203:3000–3012. doi: 10.4049/jimmunol.1900579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speen A.M., Hoffman J.R., Kim H.H., Escobar Y.N., Nipp G.E., Rebuli M.E., Porter N.A., Jaspers I. Small molecule antipsychotic aripiprazole potentiates ozone-induced inflammation in airway epithelium. Chem. Res. Toxicol. 2019;32:1997–2005. doi: 10.1021/acs.chemrestox.9b00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunis P.F., Brienen N., Kretzschmar M.E. High infectivity and pathogenicity of influenza A virus via aerosol and droplet transmission. Epidemics. 2010;2:215–222. doi: 10.1016/j.epidem.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Thao T.T.N., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020 doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Thornton D.J., Gray T., Nettesheim P., Howard M., Koo J.S., Sheehan J.K. Characterization of mucins from cultured normal human tracheobronchial epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000;278:L1118–L1128. doi: 10.1152/ajplung.2000.278.6.L1118. [DOI] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Rosen O., Wang L., Turner H.L., Stevens L.J., Corbett K.S., Bowman C.A., Pallesen J., Shi W., Zhang Y. Structural definition of a neutralization-sensitive epitope on the MERS-CoV S1-NTD. Cell Rep. 2019;28:3395–3405.e6. doi: 10.1016/j.celrep.2019.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicht O., Li W., Willems L., Meuleman T.J., Wubbolts R.W., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014;88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.M., Norton A., Young F.P., Collins D.W. Airborne transmission of severe acute respiratory syndrome coronavirus-2 to healthcare workers: a narrative review. Anaesthesia. 2020 doi: 10.1111/anae.15093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu X., Nethery R.C., Sabath B.M., Braun D., Dominici F. Exposure to air pollution and COVID-19 mortality in the United States. medRxiv. 2020 doi: 10.1101/2020.04.05.20054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yu C., Qu J., Zhang L., Jiang S., Huang D., Chen B., Zhang Z., Guan W., Ling Z. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur. J. Nucl. Med. Mol. Imaging. 2020;47:1275–1280. doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Prabakaran P., Du L., Shi W., Feng Y., Wang Y., Wang L., Li W., Jiang S., Dimitrov D.S., Zhou T. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 2015;6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zhang S., Jiang L., Cui Y., Li D., Wang D., Wang N., Fu L., Shi X., Li Z. Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27. Sci. Rep. 2015;5:13133. doi: 10.1038/srep13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X., Hensley L.E., Prabakaran P., Rockx B., Sidorov I.A. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. USA. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequences of recombinant viruses icSARS-CoV-2-WT, icSARS-CoV-2-GFP and icSARS-CoV-2-nLuc-GFP, which were generated in this study, have been deposited to GenBank (Accession # MT461669 to MT461671).