Abstract
UDP-glucuronosyltransferases 1 A (UGT1A) enzymes are capable of detoxifying a broad range of endo- and xenobiotic compounds, which contributes to antioxidative effects, modulation of inflammation and cytoprotection. In the presence of low-function genetic UGT1A variants fibrosis development is increased in various diseases. This study aimed to examine the role of common UGT1A polymorphisms in NASH. Therefore, htgUGT1A-WT mice and htgUGT1A-SNP mice (carrying a common human haplotype present in 10% of the white population) were fed a high-fat Paigen diet for 24 weeks. Serum aminotransferase activities, hepatic triglycerides, fibrosis development and UGT1A expression were assessed. Microscopic examination revealed higher hepatic fat deposition and a significant induction of UGT1A gene expression in htgUGT1A-WT mice. In agreement with these observations, lower serum aminotransferase activities and lower expression levels of fibrosis-related genes were measured in htgUGT1A-SNP mice. This was accompanied by reduced PPARα protein levels in htgUGT1A-WT but not in SNP mice. Our data demonstrate a protective effect of a UGT1A SNP haplotype, leading to milder hepatic steatosis and NASH. Higher PPARα protein levels in animals with impaired UGT1A activity are the likely result of reduced glucuronidation of ligands involved in PPARα-mediated fatty acid oxidation and may lead to the observed protection in htgUGT1A-SNP mice.
Subject terms: Metabolic diseases, Risk factors, Non-alcoholic fatty liver disease
Introduction
The growing number of patients with non-alcoholic fatty liver disease (NAFLD) renders this condition one of the most important and common liver disorder worldwide and thus to a significant medical challenge. It is estimated that around 30% of the general American population is affected by NAFLD and more than 85% of these individuals are morbidly obese1,2. The prevalence for NAFLD is closely related to obesity and its associated comorbidities and thus leads to an increased mortality rate3,4. In view of the high numbers of obese individuals, it is likely that the global incidence of patients with NAFLD will continue to increase. Apart from dietary habits further influences such as gender, age, ethnicity and genetic disorders have been identified as additional risk factors for NAFLD5, which has the potential to progress to more severe non-alcoholic steatohepatitis (NASH), cirrhosis and hepatocellular carcinoma (HCC)6,7. The molecular events include lobular inflammation, oxidative stress, hepatocellular apoptosis and fibrosis leading to NASH in approximately one quarter of NAFLD individuals8,9.
The development of a variety of liver diseases is almost invariably associated with a deregulation of nuclear receptor activation and changes in hepatic enzyme expression patterns. Previous studies have shown that the presence of NAFLD modulates the transcriptional activity of phase I and II enzymes10,11. Altered expression levels of hepatic UDP-glucuronosyltransferase 1a (Ugt1a) genes have been detected in (ob/ob) mice12. Moreover, Hardwick and colleagues identified variations of UGT1A expression levels in human liver samples during various stages of NAFLD, but their role in NAFLD progression remain unclear13. UGT1A enzymes are localized in the inner membrane of the endoplasmatic reticulum and contribute to cytoprotection by catalysing the detoxification of a broad array of endo- and exogenous compounds. These include therapeutic drugs, environmental xenobiotics, reactive metabolites, bilirubin, bile acids, dietary fatty acids and other eicosanoids14–16. UGT1A proteins catalyse the covalent conjugation with glucuronic acid rendering lipophilic target substrates water soluble and inactive thereby facilitating biliary or renal elimination17. The presence of single nucleotide polymorphisms (SNPs) in the promoter and coding regions modifies the function of UGT1A genes18. Among more than 100 identified SNPs, which lead to varying degrees of UGT1A function and expression, the Gilbert syndrome-associated UGT1A1*28 variant probably represents the best studied UGT1A polymorphism19. Individuals homozygous for UGT1A1*28 exhibit a ~70% lower UGT1A1 promoter activity20. Genetic UGT1A variants, commonly present in individuals with Gilbert syndrome, have been associated with several liver diseases including HCC and a more severe fibrosis development in patients with hepatitis B and C21,22. Based on these findings we designed experiments expecting that enhanced UGT1A expression confers a protective effect during hepatic steatosis, NASH development and, as a consequence, in the progression to liver fibrosis. Therefore, the aim of the study was to elucidate the role of UGT1A polymorphisms for NASH progression and determine the histopathological consequences for the liver. To this end, humanized transgenic (htg) UGT1A wild type (WT) and htgUGT1A-SNP mice, containing a human haplotype of 10 common occurring UGT1A SNPs, were used. Since this SNP haplotype is present in approx. 10% of the white population, our study further allows a risk assessment of NASH progression for a large proportion of the human population. Moreover, special interest was given to the nuclear receptor biology of farnesoid X receptor (FXR) and its downstream target peroxisome proliferator-activated receptor alpha (PPARα), which was shown to be downregulated in patients with fatty livers23. Both nuclear receptors have been identified as promising therapeutic targets for the treatment of NAFLD due to their ability to control a broad range of hepatic functions involved in lipid and glucose metabolism, inflammation and fibrogenesis24,25. Therefore, potential molecular mechanisms leading to the deregulation of FXR and PPARα activation possibly arising as a consequence of altered UGT1A activity in htgUGT1A-SNP mice are discussed and compared to the results of human population studies observed during NAFLD and NASH.
Results
Aggravated liver injury and inflammation in htgUGT1A-WT mice
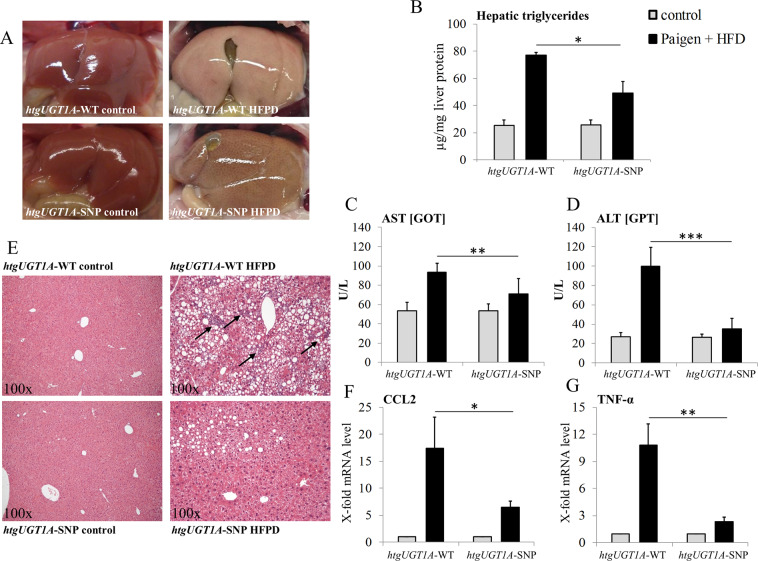
To elucidate the role of UGT1A enzymes in diet-induced liver injury, htgUGT1A-WT and SNP mice were fed with a high-fat Paigen diet (HFPD) for 24 weeks and analysed for differences regarding hepatic lipid deposition, triglyceride levels and serum aminotransferase activities. Contrary to our initial hypothesis, the macroscopic examination of the livers revealed higher fat deposition in htgUGT1A-WT mice (Fig. 1A). Measurement of hepatic triglycerides (Fig. 1B) confirmed the visual impression and revealed significantly higher triglyceride levels in htgUGT1A-WT animals (WT: 77.1 µg/mg; SNP: 49.0 µg/mg). In comparison to control diet fed animals, both mouse lines showed marked elevations of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) activities after HFPD exposition (Fig. 1C,D). Interestingly, significantly lower AST (24%) and ALT (65%) levels were detected in in mice carrying the UGT1A SNP variant.
Figure 1.
Differential effects of 24 weeks high-fat Paigen diet (HFPD) in htgUGT1A-WT and SNP mice. (A) Macroscopic liver depictions after abdominal opening. Livers of htgUGT1A-WT mice fed with HFPD showed a higher degree of steatosis as those of their SNP counterparts. (B) Hepatic triglyceride content expressed as µg/mg liver protein. In the presence of SNPs significantly lower levels of triglycerides were detected. (C) Representative sections of hematoxylin and eosin (H&E) stained liver tissue (magnification 100×). Hepatocytes of htgUGT1A-WT mice showed more fat accumulation and a higher proportion of infiltrated inflammation cells (black arrows). (D,E) Liver injury was assessed by serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) activities. Mice carrying the UGT1A SNP haplotype had significantly lower AST and ALT levels. (F,G) Gene expression levels of the pro-inflammatory markers C-C chemokine ligand 2 (CCL2) and tumour necrosis factor alpha (TNF-α). Induction of the transcriptional activation was significantly lower in htgUGT1A-SNP mice indicating a higher degree of inflammation. Each column represents the mean ± standard deviation. *p < 0.05; **p < 0.01; ***p < 0.001.
Hepatic steatosis was further analysed by haematoxylin-eosin (H&E) histological staining (Fig. 1E). In agreement with the differential hepatic lipid incorporation, HFPD-treated htgUGT1A-WT mice not only showed a higher proportion of steatotic hepatocytes, but also cellular ballooning that was almost exclusively observed in mice carrying the human wild type UGT1A gene locus. Moreover, an advanced degree of liver inflammation, indicated by the massive infiltration of inflammatory cells, was observed in htgUGT1A-WT mice suggesting the development of NASH (Fig. 1E black arrows). This observation was supported by significant differences in transcriptional activation of the proinflammatory marker genes C-C chemokine ligand 2 (CCL2) and tumour necrosis factor alpha (TNF-α) (Fig. 1F,G). In htgUGT1A-WT mice, a 17.4-fold CCL2 upregulation and a 10.8-fold TNF-α mRNA induction was detected, whereas in the presence of SNPs transcriptional activation of CCL2 (6.4-fold) and TNF-α (2.4-fold) was significantly less evident.
Attenuated hepatic fibrosis in htgUGT1A-SNP mice
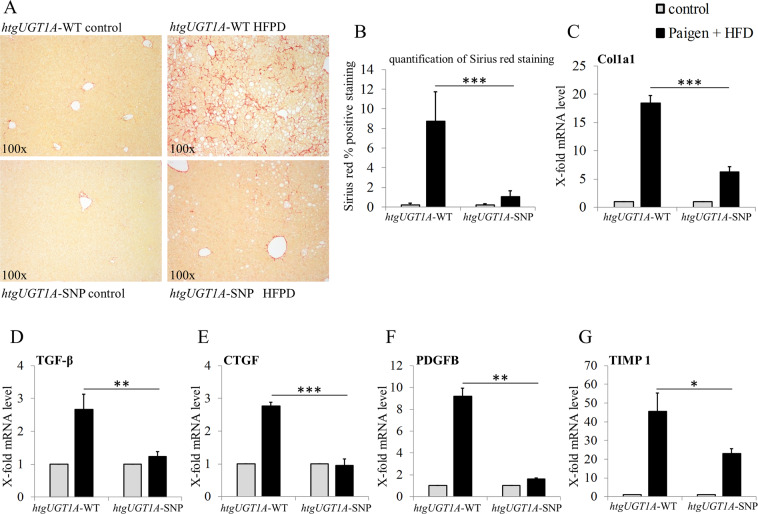
With the intention of determining differences in fibrosis development and hepatic collagen deposition, computational quantification of Sirius red staining and gene expression analysis of profibrotic biomarkers was evaluated. Histological staining showed a higher content of fibrillar collagens, and thus fibrosis development, in htgUGT1A-WT mice (Fig. 2A). Computational quantification of Sirius red staining further supported the histological finding and detected 8.4-fold higher percentage portion of red coloured fibrillar collagens in htgUGT1A-WT mice (Fig. 2B). Moreover, a 3.0-fold higher transcriptional activation of collagen type 1 alpha 1 (Col1a1) in htgUGT1A-WT mice supported the results at a molecular level and confirmed the differences in fibrosis development between both mouse lines (Fig. 2C).
Figure 2.
Assessment of liver fibrosis in htgUGT1A-WT and SNP mice after high-fat Paigen diet (HFPD) treatment. (A) Representative hepatic sections of histological Sirius red staining and (B) computational quantification of Sirius red stained areas. Hepatic collagen deposition was significantly lower in htgUGT1A-SNP mice. (C–G) Gene expression levels of the profibrotic biomarkers collagen type 1 alpha 1 (Col1a1), transforming growth factor beta (TGF-β), connective tissue growth factor (CTGF), platelet-derived growth factor subunit B (PDGFB) and tissue inhibitor metalloprotease 1 (TIMP 1) as indicators for severity of liver fibrosis. Transcriptional activation of all depicted fibrosis marker genes was significantly lower in htgUGT1A-SNP mice. In case of TGF-β and CTGF, minor transcriptional inductions were detected in HFPD-treated htgUGT1A-SNP mice. Graphs are expressed as means ± standard deviation. *p < 0.05; **p < 0.01; ***p < 0.001.
Manifestation of liver fibrosis is usually associated with an increased expression of cytokines, chemokines and various key genes influencing fibrogenesis. In htgUGT1A-WT mice, HFPD administration caused significantly higher transcriptional activation of the profibrotic markers transforming growth factor beta (TGF-β), connective tissue growth factor (CTGF), platelet-derived growth factor subunit B (PDGFB) and tissue inhibitor metalloprotease 1 (TIMP1), whereas mRNA induction was reduced (TIMP1 and PDGFB) or absent (TGF-β and CTGF) in the presence of SNPs (Fig. 2D–G). In combination, these data suggest a protective effect of a common UGT1A SNP haplotype during diet-induced steatohepatitis, resulting in attenuated hepatic fibrosis and inflammation.
Increased UGT1A expression in htgUGT1A-WT mice
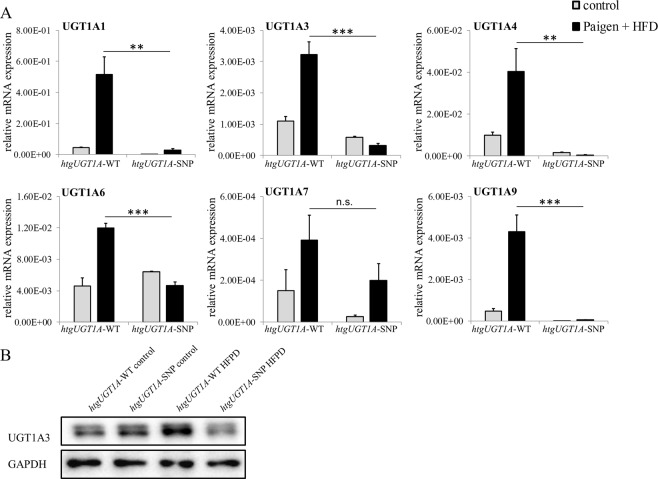
To further evaluate the potential mechanisms responsible for the observed protective effects in htgUGT1A-SNP mice, hepatic UGT1A expression was determined in both animal models (Fig. 3A). In the livers of htgUGT1A-WT mice, HFPD led to a significant upregulation of mRNA expression of all investigated UGT1A genes. In contrast and expectedly, significantly lower transcriptional activation was measured in htgUGT1A-SNP mice. Attention was given to the expression of UGT1A3 representing the only UGT1A isoform capable of glucuronidating bile acids, which in turn are key regulators of nuclear receptors involved in glucose and lipid metabolism26,27. In line with the detected mRNA expression results, hepatic UGT1A3 protein quantity was markedly increased in HFPD treated htgUGT1A-WT mice (Fig. 3B).
Figure 3.
Hepatic UGT1A regulation in htgUGT1A-WT and SNP mice after high-fat Paigen diet (HFPD) treatment. (A) Hepatic mRNA expression levels of UGT1A isoforms relative to mouse β-actin. In htgUGT1A-WT mice, HFPD treatment led to a significant transcriptional upregulation of all depicted UGT1A isoforms, whereas in the presence of SNPs UGT1A induction was reduced (UGT1A1 and UGT1A9) or absent and remained below those observed in WT carriers. (B) Western blot analysis of hepatic UGT1A3 protein quantity. Higher protein amount was detected in htgUGT1A-WT mice. Graphs are expressed as means ± standard deviation. n.s. not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
Reduced expression of FXR and PPARα in HFPD fed htgUGT1A-WT mice
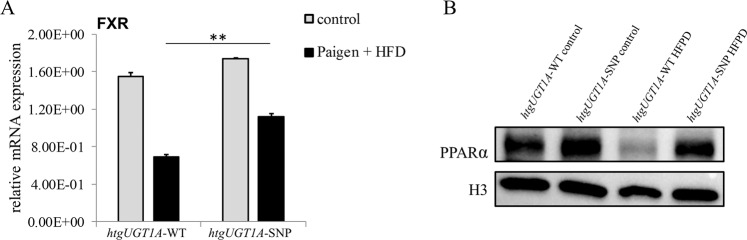
FXR is known to play a crucial role in mediating effects of bile acids during NAFLD. After diet-induced liver injury, a significant decrease of the FXR mRNA expression was detected in both mouse lines (Fig. 4A). Interestingly, the degree of inhibition reached 58% in htgUGT1A-WT mice, compared to only 36% measured in htgUGT1A-SNP mice. PPARα is a downstream target of FXR and a well-known mediator of hepatic fatty acid oxidation28. The nuclear translocation of PPARα protein was downregulated in HFPD fed htgUGT1A-WT mice whereas PPARα activation remained unchanged in htgUGT1A-SNP mice (Fig. 4B). These results demonstrate a differential effect of UGT1A SNPs for the expression of nuclear receptors involved in cellular protection.
Figure 4.
Hepatic mRNA expression and nuclear protein quantity of nuclear receptors in htgUGT1A-WT and SNP mice after high-fat Paigen diet (HFPD) treatment. (A) Enhanced inhibition of farnesoid X receptor (FXR) mRNA expression level was detected in htgUGT1A-WT mice (58%) compared to htgUGT1A-SNP mice (36%). (B) Western blot analysis of nuclear peroxisome proliferator-activated receptor alpha (PPARα) protein levels. Significantly reduced PPARα protein amount was measured in HFPD-treated htgUGT1A-WT, in contrast to unchanged levels in the presence of SNPs. Graphs are expressed as means ± standard deviation. *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
Effects of altered UGT1A expression on the pathogenesis of NAFLD and the associated consequences for the pathology of the liver have not been experimentally analysed. Contrary to our expectations and our original hypothesis, the results demonstrate that increased UGT1A expression does not protect against NASH progression in a humanized UGT1A animal model of NAFLD. The data suggest a protective effect of a common low-function UGT1A SNP haplotype in NAFLD/NASH. HtgUGT1A-SNP mice exhibited milder hepatic steatosis, significantly lower levels of hepatic triglycerides and a less pronounced elevation of AST and ALT levels compared to htgUGT1A-WT mice. Furthermore, a reduced deposition of fibrillar collagens and decreased expression levels of profibrotic and proinflammatory marker genes underscore an attenuated process of liver fibrosis in htgUGT1A-SNP mice. This was accompanied by the significant upregulation of UGT1A expression levels in htgUGT1A-WT mice, compared to a reduced or absent induction in mice carrying the low-function SNP variant.
To date, contradictory results have been published in various cohort-studies investigating an association between genetic UGT1A variants and human NAFLD. In line with our study, a decreased risk of paediatric NAFLD has been reported in 234 obese Taiwanese children associated with a low-activity UGT1A1 gene variant29. Similarly, a case-control study with 641 adult patients suspected to have NAFLD reported an inverse association between unconjugated hyperbilirubinemia and the histopathological severity of liver damage in NASH30. In contrast, genome-wide association studies and other genetic studies of human NAFLD have failed to find an association between UGT1A1 polymorphisms and NAFLD31. A likely explanation for the inconsistent data reported in these studies may involve the presence of UGT1A polymorphisms found in other isoforms than UGT1A1. Since many SNPs exist in linkage-disequilibrium with each other, the inter-individual UGT1A sequence variation is highly variable22. Previously, we were able to show that 76% of homozygous UGT1A1*28 carriers, a genotype that is associated with elevated bilirubin levels, were simultaneously homozygous for 9 other UGT1A polymorphisms32. Therefore, a combination of multiple polymorphisms in distinct UGT1A isoforms is suggested to be responsible for the observed hepatoprotective effects in this study and maybe also for the differential results in human population studies only considering polymorphisms in the UGT1A1 gene.
A molecular mechanism potentially triggering the UGT1A SNP-associated protective effects could include the impaired glucuronidation of molecules that function as ligands for nuclear receptors (i.e. FXR or PPARα) involved in lipid homeostasis. A specific example is the UGT1A-mediated glucuronidation of dietary fatty acids, such as arachidonic acids and its metabolites 20-hydroxyeicosatetraenoic acid (20-HETE) and leukotriene B4 (LTB4), which have been previously identified as potent PPARα activators33 and as UGT1A3 or UGT1A9 substrates14,34,35. In this context, Little et al. suggested that UGT1A enzymes may function as modulators for the availability of fatty acids to act as ligands for nuclear receptors. Therefore, the decreased UGT1A expression in many different isoforms, as present in htgUGT1A-SNP mice, may lead to higher intracellular levels of ligands involved in a broad array of lipid homeostasis signalling pathways. The impaired capacity of htgUGT1A-SNP mice to eliminate potential FXR or PPARα ligands may improve the PPARα-mediated mitochondrial fatty acid oxidation, hepatic fatty acid uptake, peroxisomal beta-oxidation or other crucial processes of lipid metabolism and leads to the lower degree of hepatic steatosis. Moreover, polymorphisms in the specific UGT1A3 isoform, which is the major enzyme in the UGT1A gene locus capable to catalyse the glucuronidation of bile acids, are likely to also contribute to the beneficial effects and mediated independently of the UGT1A1*28 promoter polymorphism. Bile acid levels were shown to be elevated in patients with NASH36 and have been identified as FXR inducers37. Due to higher UGT1A3 expression in htgUGT1A-WT animals potentially leading to increased glucuronidation of FXR-inducing bile acids, unaffected UGT1A3 activity is suggested to reduce FXR activation leading to an impaired FXR-induced PPARα-mediated fatty acid oxidation. Reduced FXR expression as well as lower levels of nuclear PPARα protein in htgUGT1A-WT mice underline this hypothesis and lead to the conclusion that the differential regulation of both nuclear receptors is likely to be directly linked to the differential expression of UGT1A genes. Although glucuronidation of bile acids has previously been demonstrated in mice38, it is important to note that bile acid glucuronidation only constitutes a relevant detoxification pathway in humans. In mice, however, it is predominantly converted into muricholic acid and only minute amounts are present39,40. Even though the relative occurrence of this conjugation reaction is minor in mice, the proposed physiological role is likely to occur in humans. In addition, the FXR downstream targets small heterodimer partner and PPARα have previously been shown to initiate the downregulation of the lipogenic master regulator sterol regulatory element-binding protein 1c (SREBP-1c)28,41. Therefore, increased FXR expression in htgUGT1A-SNP mice might be associated with reduced SREBP-1c activation and consequently with a downregulation of de novo lipogenesis.
As a consequence, the combination of two or more polymorphisms in different UGT1A isoforms may be a more accurate indicator for the occurrence of UGT1A SNP-associated liver protection during NAFLD or NASH in human population studies. Along with PPARα and FXR, other nuclear receptors such as constitutive androstane receptor, pregnane X receptor, liver X receptor and hepatocyte nuclear factor 4 have also been implicated in the pathogenesis of NAFLD24 while also being involved in the transcriptional regulation of UGT1A genes42. Therefore, polymorphisms in UGT1A genes may ultimately lead to various metabolic changes in patients with NAFLD and large-scale OMICS analysis might be necessary to get a full picture of the transcriptomic and proteomic alterations associated with this common UGT1A-SNP genotype. However, we cannot fully exclude that the observed effects are influenced by the murine Ugt1a enzymes which are also expressed in htgUGT1A-WT and SNP mice. This leads to an increased glucuronidation capacity in both mouse lines, which is lower in htgUGT1A-SNP mice. Therefore, further studies with Ugt1a knockout mice simultaneously carrying the human UGT1A-WT or UGT1A-SNP transgene would be needed to exclude the effects of non-physiologically high levels of glucuronidation.
In conclusion, our data indicate a protective effect of a Gilbert syndrome-associated UGT1A haplotype leading to milder hepatic steatosis during the development of NASH. Higher expression of UGT1A enzymes was observed in htgUGT1A-WT mice, while htgUGT1A-SNP mice showed lower serum aminotransferase levels and reduced hepatic collagen deposition. Due to the decreased PPARα protein and lower FXR expression levels in htgUGT1A-WT mice, we hypothesize that increased UGT1A expression may lead to the facilitated elimination of potential ligands involved in lipid homeostasis.
Methods
Animal model and experimental design
For animal experiments, previously described 8–10 week-old htgUGT1A-WT and SNP mice were used32,43. As both mouse lines contain the human UGT1A transgene in addition to the murine Ugt1a gene locus, the glucuronidation capacity of both animal models is likely above the physiological levels in non-transgenic C57Bl/6 mice, which is, due to the presence of 10 common UGT1A polymorphisms, significantly lower in htgUGT1A-SNP mice. Male mice of each genotype were divided into two groups consisting of six to eight animals. Both groups were either fed a regular chow or a HFPD for 24 weeks. The HFPD was purchased from the Altromin GmbH & Co. KG Company (Seelenkamp, Germany) and contained a raw fat content of 42% in which fat contributes 70% of total energy requirements. This diet is further characterized by the addition of 1.25% cholesterol and 0.5% sodium cholate. All mice had ad libitum access to water and chow and were kept at 22 °C with a 12 hour day/night cycle in the Central Animal Facility of the University Hospital Bonn. 10 days before HFPD administration htgUGT1A mice received a 4% raw fat containing diet to adapt animals to the change of dietary components. All experiments were performed and in accordance to the “German Animal-Protection Law” and the relevant guidelines of the Local Institutional Animal Care unit of our university (Haus für experimentelle Therapie, Bonn, Germany) and approved by the relevant North Rhine-Westphalian state-agency for Nature, Environment and Consumer Protection (LANUV, Germany) under the file reference LANUV 84-02.04.2016.A483.
Tissue collection and biochemical analysis
After HFPD treatment, animals were sacrificed for organ and blood collection. For biochemical analysis of serum ALT and AST activities, the collected blood was centrifuged at 4.800 rpm for 10 min to remove blood cells. The supernatant was stored at −20 °C until analyzation by means of a Fuji DRI-CHEM NX500i serum analyser.
Organ samples were immediately snap-frozen in liquid nitrogen and stored at −80 °C until use. The right lateral lobe was separated before, fixed in 4% paraformaldehyde for three days, subsequently embedded in paraffin and then used for pathohistological examinations.
Histological staining and computational analysis
Paraffin-embedded liver sections were trimmed into 2 µm thick slices, and stained in 0.1% Sirius red solution (DirectRed 80 in saturated picric acid) for detection of fibrous collagen tissue. The Sirius red positive stained area was quantified using ImageJ software (U.S. National Institutes of Health; http://rsb.info.nih.gov/ij/) and shown as percentage positive staining of the total section area. Images were analysed from four randomly selected images (magnification 100×) of each animal and were averaged. H&E staining was applied to visualize lipid-droplets within hepatocytes, cellular ballooning and infiltration of inflammatory cells according to a standard protocol procedure with minor modifications44.
Triglyceride measurement
Hepatic triglyceride levels were photometrically determined using the Triglyceride Colorimetric Assay Kit (Cayman Chemicals). 300 mg snap-frozen liver tissue was hydrolysed according to manufacturer’s instructions and analysed in duplicates using a MultiSkan GO microplate reader at 540 nm.
Gene expression analysis
RNA isolation from snap-frozen liver samples, cDNA synthesis and gene expression analysis by qPCR were performed as described previously43. The assays listed below were purchased from Thermo Scientific and used for quantification of inflammatory and fibrosis-related gene expression levels. These include C-C chemokine ligand 2 (CCL2; Mm00441442_m1), tumour necrosis factor alpha (TNF-α; Mm00443260_g1), collagen type 1 alpha 1 (Col1a1; Mm00801666_g1), transforming growth factor beta (TGF-β; Mm01178820_m1), platelet-derived growth factor subunit B (PDGFB; Mm00440677_m1) connective tissue growth factor (CTGF; Mm01192933_g1) and tissue inhibitor metalloprotease 1 (TIMP1; Mm01341361_m1). Expression levels were normalized relative to mouse beta-actin and expressed as fold mRNA levels compared to untreated htgUGT1A-WT or SNP mice.
Western blot analysis
60 mg of frozen liver tissue was homogenized in RIPA extraction buffer containing protease inhibitor cocktail and subsequently incubated for 1 h on a shaking plate at 4 °C. After centrifugation (13.000 rpm, 10 min, 4 °C), the supernatant was used for Western blot analysis. Nuclear extraction tissue specimens were prepared using Nuclear Extraction Kit (Abcam) according to manufacturer’s instructions. For SDS-PAGE separation, 30 µg of protein was boiled at 95 °C for 5 min in Laemmli sample buffer, separated on a 10% acrylamide gel and blotted onto a nitrocellulose membrane via electrotransfer using the Trans-Blot®-Turbo transfer system. Incubation with primary antibodies (anti UGT1A3, Abnova H00054659-M02, anti GAPDH, Santa Cruz sc-32233, anti PPARα, Santa Cruz sc-398394 and anti H3 Abcam (ab12079)) was carried out in 5% dry milk. Appropriate secondary antibodies (Santa Cruz, sc-516102 and sc-2054) were used and protein was visualized by chemoluminescence with the use of ChemiDOC MP imaging system (Supplementary Figure S1,S2).
Statistical analysis
Data are expressed as mean ± SD determined by one-way analysis of variance followed by Students t-test to define significance. A pool of six to eight mice in each HFPD group was analysed (four mice per control group); p values below 0.05 were considered as statistically significant.
Supplementary information
Acknowledgements
The study was supported by grants from the Deutsche Forschungsgemeinschaft project STR 493/8-1 (C.P.S.).
Author contributions
S.L. performed experiments, analyses and interpretation of data and drafted the manuscript. S.K. contributed to analyses, drafted the manuscript and supervised the study. S.P. performed experiments and contributed to data analyses. C.P.S. performed administrative support, contributed to data interpretation and analysis, obtained funding, supervised and designed the study and revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-65481-4.
References
- 1.McCullough AJ. Epidemiology of the metabolic syndrome in the USA. J. Dig. Dis. 2011;12:333–40. doi: 10.1111/j.1751-2980.2010.00469.x. [DOI] [PubMed] [Google Scholar]
- 2.Younossi ZM, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 3.Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment. Pharmacol. Ther. 2011;34:274–85. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 4.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. Jama. 2012;307:491–7. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- 5.Perumpail BJ, et al. Clinical epidemiology and disease burden of nonalcoholic fatty liver disease. World J. Gastroenterol. 2017;23:8263–8276. doi: 10.3748/wjg.v23.i47.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu. Rev. Pathol. 2010;5:145–71. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 7.Sanyal AJ. NASH: A global health problem. Hepatol. Res. 2011;41:670–4. doi: 10.1111/j.1872-034x.2011.00824.x. [DOI] [PubMed] [Google Scholar]
- 8.Marra F, Gastaldelli A, Svegliati Baroni G, Tell G, Tiribelli C. Molecular basis and mechanisms of progression of non-alcoholic steatohepatitis. Trends Mol. Med. 2008;14:72–81. doi: 10.1016/j.molmed.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Kleiner DE, Makhlouf HR. Histology of Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis in Adults and Children. Clin. Liver Dis. 2016;20:293–312. doi: 10.1016/j.cld.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher CD, et al. Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug. Metab. Dispos. 2009;37:2087–94. doi: 10.1124/dmd.109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwick RN, Fisher CD, Canet MJ, Lake AD, Cherrington NJ. Diversity in antioxidant response enzymes in progressive stages of human nonalcoholic fatty liver disease. Drug. Metab. Dispos. 2010;38:2293–301. doi: 10.1124/dmd.110.035006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Kulkarni SR, Li L, Slitt AL. UDP-glucuronosyltransferase expression in mouse liver is increased in obesity- and fasting-induced steatosis. Drug. Metab. Dispos. 2012;40:259–66. doi: 10.1124/dmd.111.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardwick RN, et al. Altered UDP-glucuronosyltransferase and sulfotransferase expression and function during progressive stages of human nonalcoholic fatty liver disease. Drug. Metab. Dispos. 2013;41:554–61. doi: 10.1124/dmd.112.048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little JM, Williams L, Xu J, Radominska-Pandya A. Glucuronidation of the dietary fatty acids, phytanic acid and docosahexaenoic acid, by human UDP-glucuronosyltransferases. Drug. Metab. Dispos. 2002;30:531–3. doi: 10.1124/dmd.30.5.531. [DOI] [PubMed] [Google Scholar]
- 15.Strassburg CP. Pharmacogenetics of Gilbert’s syndrome. Pharmacogenomics. 2008;9:703–15. doi: 10.2217/14622416.9.6.703. [DOI] [PubMed] [Google Scholar]
- 16.Bock KW. Functions and transcriptional regulation of adult human hepatic UDP-glucuronosyl-transferases (UGTs): mechanisms responsible for interindividual variation of UGT levels. Biochem. Pharmacol. 2010;80:771–7. doi: 10.1016/j.bcp.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 17.Strassburg Christian P., Kalthoff Sandra. Metabolism of Drugs and Other Xenobiotics. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2012. UDP-Glucuronosyltransferases; pp. 67–116. [Google Scholar]
- 18.Strassburg CP, Nguyen N, Manns MP, Tukey RH. Polymorphic expression of the UDP-glucuronosyltransferase UGT1A gene locus in human gastric epithelium. Mol. Pharmacol. 1998;54:647–54. [PubMed] [Google Scholar]
- 19.Strassburg CP, Kalthoff S, Ehmer U. Variability and function of family 1 uridine-5′-diphosphate glucuronosyltransferases (UGT1A) Crit. Rev. Clin. Lab. Sci. 2008;45:485–530. doi: 10.1080/10408360802374624. [DOI] [PubMed] [Google Scholar]
- 20.Bosma PJ, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s syndrome. N. Engl. J. Med. 1995;333:1171–5. doi: 10.1056/nejm199511023331802. [DOI] [PubMed] [Google Scholar]
- 21.Tang KS, Lee CM, Teng HC, Huang MJ, Huang CS. UDP-glucuronosyltransferase 1A7 polymorphisms are associated with liver cirrhosis. Biochem. Biophys. Res. Commun. 2008;366:643–8. doi: 10.1016/j.bbrc.2007.11.125. [DOI] [PubMed] [Google Scholar]
- 22.Strassburg CP. Gilbert-Meulengracht’s syndrome and pharmacogenetics: is jaundice just the tip of the iceberg? Drug. Metab. Rev. 2010;42:168–81. doi: 10.3109/03602530903209429. [DOI] [PubMed] [Google Scholar]
- 23.Francque S, et al. PPARalpha gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J. Hepatol. 2015;63:164–73. doi: 10.1016/j.jhep.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Trauner M, Halilbasic E. Nuclear receptors as new perspective for the management of liver diseases. Gastroenterology. 2011;140(1120–1125):e1–12. doi: 10.1053/j.gastro.2011.02.044. [DOI] [PubMed] [Google Scholar]
- 25.Han Chang. Update on FXR Biology: Promising Therapeutic Target? International Journal of Molecular Sciences. 2018;19(7):2069. doi: 10.3390/ijms19072069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erichsen TJ, et al. Regulation of the human bile acid UDP-glucuronosyltransferase 1A3 by the farnesoid X receptor and bile acids. J. Hepatol. 2010;52:570–8. doi: 10.1016/j.jhep.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Halilbasic E, Claudel T, Trauner M. Bile acid transporters and regulatory nuclear receptors in the liver and beyond. J. Hepatol. 2013;58:155–68. doi: 10.1016/j.jhep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pineda Torra I, et al. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol. Endocrinol. 2003;17:259–72. doi: 10.1210/me.2002-0120. [DOI] [PubMed] [Google Scholar]
- 29.Lin YC, Chang PF, Hu FC, Chang MH, Ni YH. Variants in the UGT1A1 gene and the risk of pediatric nonalcoholic fatty liver disease. Pediatrics. 2009;124:e1221–7. doi: 10.1542/peds.2008-3087. [DOI] [PubMed] [Google Scholar]
- 30.Hjelkrem M, Morales A, Williams CD, Harrison SA. Unconjugated hyperbilirubinemia is inversely associated with non-alcoholic steatohepatitis (NASH) Aliment. Pharmacol. Ther. 2012;35:1416–23. doi: 10.1111/j.1365-2036.2012.05114.x. [DOI] [PubMed] [Google Scholar]
- 31.DiStefano JK, et al. Genome-wide analysis of hepatic lipid content in extreme obesity. Acta Diabetol. 2015;52:373–82. doi: 10.1007/s00592-014-0654-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehmer U, et al. Gilbert syndrome redefined: a complex genetic haplotype influences the regulation of glucuronidation. Hepatology. 2012;55:1912–21. doi: 10.1002/hep.25561. [DOI] [PubMed] [Google Scholar]
- 33.Bock KW. Roles of human UDP-glucuronosyltransferases in clearance and homeostasis of endogenous substrates, and functional implications. Biochem. Pharmacol. 2015;96:77–82. doi: 10.1016/j.bcp.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 34.Bushee JL, Liang G, Dunne CE, Harriman SP, Argikar UA. Identification of saturated and unsaturated fatty acids released during microsomal incubations. Xenobiotica. 2014;44:687–95. doi: 10.3109/00498254.2014.884253. [DOI] [PubMed] [Google Scholar]
- 35.Jarrar YB, et al. Determination of major UDP-glucuronosyltransferase enzymes and their genotypes responsible for 20-HETE glucuronidation. J. Lipid Res. 2014;55:2334–42. doi: 10.1194/jlr.m051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aranha MM, et al. Bile acid levels are increased in the liver of patients with steatohepatitis. Eur. J. Gastroenterol. Hepatol. 2008;20:519–25. doi: 10.1097/meg.0b013e3282f4710a. [DOI] [PubMed] [Google Scholar]
- 37.Lew JL, et al. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J. Biol. Chem. 2004;279:8856–61. doi: 10.1074/jbc.m306422200. [DOI] [PubMed] [Google Scholar]
- 38.Zhou X, et al. PPARalpha-UGT axis activation represses intestinal FXR-FGF15 feedback signalling and exacerbates experimental colitis. Nat. Commun. 2014;5:4573. doi: 10.1038/ncomms5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alnouti Y, Csanaky IL, Klaassen CD. Quantitative-profiling of bile acids and their conjugates in mouse liver, bile, plasma, and urine using LC-MS/MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008;873:209–17. doi: 10.1016/j.jchromb.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi S, et al. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 2016;57:2130–2137. doi: 10.1194/jlr.m071183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuchs CD, Claudel T, Scharnagl H, Stojakovic T, Trauner M. FXR controls CHOP expression in steatohepatitis. FEBS Lett. 2017;591:3360–3368. doi: 10.1002/1873-3468.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu DG, Meech R, McKinnon RA, Mackenzie PI. Transcriptional regulation of human UDP-glucuronosyltransferase genes. Drug. Metab. Rev. 2014;46:421–58. doi: 10.3109/03602532.2014.973037. [DOI] [PubMed] [Google Scholar]
- 43.Kalthoff S, Landerer S, Reich J, Strassburg CP. Protective effects of coffee against oxidative stress induced by the tobacco carcinogen benzo[alpha]pyrene. Free. Radic. Biol. Med. 2017;108:66–76. doi: 10.1016/j.freeradbiomed.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Itagaki H, Shimizu K, Morikawa S, Ogawa K, Ezaki T. Morphological and functional characterization of non-alcoholic fatty liver disease induced by a methionine-choline-deficient diet in C57BL/6 mice. Int. J. Clin. Exp. Pathol. 2013;6:2683–96. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.