Abstract
Arabidopsis bHLH-type transcription factors—BRASSINOSTEROID INSENSITIVE 1-EMS-SUPPRESSOR 1 (BES1) and BRASSINAZOLE RESISTANT 1 (BZR1)—play key roles in brassinosteroid (BR) signaling. By contrast, the functions of the other four BES1/BZR1 homologs (BEH1–4) remain unknown. Here, we describe the detailed expression profiles of the BES1/BZR1 family genes. Their expressions were distinct regarding growth-stage dependence and organ specificity but exhibited some overlaps as well. Furthermore, their mRNA levels mostly remained unchanged responding to seven non-BR phytohormones. However, BEH1 and BEH2 were downregulated by brassinolide, suggesting a close association with the BR function. Additionally, BEH4 was ubiquitously expressed throughout the life of the plant but displayed some expression preference. For instance, BEH4 expression was limited to guard cells and the adjacent pavement cells in the leaf epidermis and was induced during growth progression in very young seedlings, suggesting that BEH4 is specifically regulated in certain contexts, although it is almost constitutively controlled.
Keywords: BES1/BZR1 homologs (BEH), brassinosteroids, brassinolide, β-glucuronidase (GUS), stomata
Introduction
Plant growth regulators (PGRs) are agrochemicals (natural or synthetic) that affect plant metabolic reactions at low concentrations, thereby modulating the growth, development, and stress responses of plants.1) To date, PGRs have been applied in modern agriculture to increase the yield and market value of crops. Brassinosteroids (BRs) play beneficial roles for plants, such as enhancing growth and improving stress tolerance, but have not yet been employed as PGRs in Japan, despite numerous field trials conducted in the 1980s and 1990s.2) To employ BRs effectively in agricultural practices, their molecular mechanism needs to be further elucidated.
The molecular mechanisms of BR signaling in Arabidopsis thaliana have been outlined.3,4) BRs are perceived by the plasma membrane receptor complex containing two leucine-rich repeat (LRR) receptor protein kinases: BRASSINOSTEROID INSENSITIVE 1 (BRI1) and BRASSINOSTEROID INSENSITIVE 1–associated receptor kinase 1 (BAK1). Intracellular signals generated upon ligand binding are transduced from the cell surface to the nucleus via multiple phosphorelay reactions. Downstream of BR signaling, two related basic helix–loop–helix (bHLH) transcription factors, BRI1-EMS-SUPPRESSOR 1 (BES1) and BRASSINAZOLE-RESISTANT 1 (BZR1), are activated upon their dephosphorylated status caused by the combined action of an inactivated GSK3/SHAGGY-like kinase, BRASSINOSTEROID INSENSITIVE 2 (BIN2), and an activated PROTEIN PHOSPHATASE 2A (PP2A). The activated BES1 and BZR1 transcriptionally regulate thousands of their target genes.5,6) In contrast, these two transcription factors are phosphorylated by the BIN2 kinase when endogenous BR levels are low or depleted, and their phosphorylated forms are further subjected to several reactions, including cytoplasmic sequestration by 14-3-3 and BRZ-SENSITIVE-SHORT HYPOCOTYL1 (BSS1) proteins,7,8) proteasome-mediated degradation,9,10) and/or reduced binding activity to the target promoter DNA.7,11) As mentioned above, BR signaling seems to be tightly and BR-dependently controlled by the elaborate mechanisms. In addition, BES1, BZR1, and BIN2 reportedly function as integration nodes between the BR pathway and other signaling pathways.12,13)
The Arabidopsis genome has four homologous genes of BES1 and BZR1—BEH1–4 (BES1/BZR1 homolog 1–4). They constitute a small gene family containing six members. In contrast to the huge number of studies elucidating BES1 and BZR1, only a few studies have been reported for the other homologs. For instance, Yin et al. (2005) demonstrated the BR-dependent dephosphorylation of BEH1–4 proteins, implying their association with BR signaling.14) Five BES1/BZR1 members other than BEH1were recently determined to bind with MAX2, a positive regulator in strigolactone signaling, suggesting a novel hormone crosstalk of BRs with strigolactone.15) However, it has not been validated whether BEH homologs are truly involved in BR signaling or how they function in the pathway if that is the case. Therefore, for a comprehensive understanding of BR signaling and its crosstalk with different pathways, it is necessary to further characterize the molecular and physiological properties of BEH1–4 proteins, in addition to those of BES1 and BZR1.
In the present study, we carefully examined the expression of BEH1–4 homologs together with BES1 and BZR1 as the first step toward the above-mentioned goal, to narrow down their functions from numerous conceivable possibilities. This report explains that their expressions were different by a large degree but that they somewhat overlapped at the organ and developmental-stage levels. Furthermore, we found that BEH1 and BEH2 responded to exogenous brassinolide (BL), the most bioactive BR. Additionally, we demonstrated histochemically that BEH4 was expressed almost ubiquitously throughout the life of the plant, with a certain degree of expression preference.
Materials and Methods
1. Chemicals
All chemicals were purchased from Fujifilm Wako Pure Chemical Corp., Japan, unless otherwise described. The following phytohormones were used for this study: brassinolide (BL; Brassino Co. Ltd., Toyama, Japan), 3-indoleacetic acid (IAA; Nacalai Tesque Inc., Kyoto, Japan), 6-benzyladenine (BA), gibberellin A3 (GA3; Kyowa Hakko Kogyo Co. Ltd., Tokyo, Japan), abscisic acid (ABA), 1-amino-1-cyclopropanecarboxylic acid (ACC) as an ethylene precursor, methyl jasmonate (JA), and salicylic acid (SA). The biosynthesis inhibitors brassinazole (Brz) and abamineSG (ASG) were used to reduce the endogenous BRs and ABA, respectively. These chemicals were dissolved in dimethyl sulfoxide (DMSO), except for ACC, which was dissolved in water. Drugs dissolved in DMSO were added to the media, not to exceed a final concentration of 0.1% of DMSO.
2. Plants and growth conditions
Plants used for this study are the Arabidopsis thaliana wild-type Columbia (Col-0) ecotype and the transgenic Col-0 lines harboring the BEH4::GUS gene. Sterilization and sowing on a solid medium of Arabidopsis seeds were performed according to Tanaka et al. (2003) with minor modifications: the sodium hypochlorite concentration used for sterilization was reduced from 5 to 2.5%; a half-strength (1/2) MS medium containing 0.32% gellan gum was used instead of an original MS medium with 0.5% gellan gum.16) The age of Arabidopsis seedlings was set as ‘day 0’ when cultivation commenced. When cultivated in an air-conditioned plant growth room, seeds were sown directly on vermiculite in a 6.5 or 7.5 cm diameter plastic pot and were grown at 24°C under light conditions of 16L:8D.
3. Plasmid construction and plant transformation
Construction of the BEH4::GUS gene was performed as described below. First, an approx. 1.3 kb DNA fragment containing a stretch of the promoter and 5′-untranslated region of the BEH4 gene was PCR-cloned from Arabidopsis genomic DNA with a set of primers (Table 1) using a KOD DNA polymerase (Toyobo Co. Ltd., Osaka, Japan). The PCR fragment was then sub-cloned in a SmaI site of pUC118 and was sequenced to confirm that its nucleotides perfectly matched those registered in the DDBJ database.17) The fragment was excised with XbaI and BamHI designed within the primers and was recloned into the 5′ region of a promoterless GUS gene in a binary vector, pBI101-Hm, using the corresponding restriction sites.18) Agrobacterium tumefaciens EHA105 harboring the resulting plasmid was subjected to the floral-dip method for Arabidopsis transformation (Col-0).19) Nine independent transgenic lines were obtained, all of which showed a similar staining pattern of GUS activity; line 5 was primarily used in this study.
Table 1. Sequence of PCR-primers and their annealing temperatures used for this study.
| Name | Locus | Sequence | A.T.a) | Experiment |
|---|---|---|---|---|
| ACT2 | At3g18780 | 5′-CCGCTCTTTCTTTCCAAGC-3′ | 55 | qRT-PCR |
| 5′-CCGGTACCATTGTCACACAC-3′ | ||||
| EF1aA4 | At5g60390 | 5′-GAGCCCAAGTTTTTGAAGA-3′ | 55 | |
| 5′-CTAACAGCGAAACGTCCCA-3′ | ||||
| BES1 | At1g19350 | 5′-ACCCGTTTTATGCGGTGTCT-3′ | 57 | |
| 5′-AGCCGGAGCATGGAACTG-3′ | ||||
| BZR1 | At1g75080 | 5′-AACAGCCATTCTCTGCCTCTATG-3′ | 60 | |
| 5′-TGAGGCGCAGGTTTCACA-3′ | ||||
| BEH1 | At3g50750 | 5′-CCTCCGTCGCCGACATT-3′ | 57 | |
| 5′-CGGCTTCACCGACACATCTA-3′ | ||||
| BEH2 | At4g36780 | 5′-CGATAACAACGAGGTTCTTAAAGCT-3′ | 57 | |
| 5′-TGGTGCCATCGTCTTCGA-3′ | ||||
| BEH3 | At4g18890 | 5′-TCCAAGCTTCCCTTCTTCCA-3′ | 60 | |
| 5′-GGGCTTCGAGCCAATGG-3′ | ||||
| BEH4 | At1g78700 | 5′-CGGAGAGCAATCGCAGCTA-3′ | 57 | |
| 5′-AATGCTTCGGAAGCTCGTAATT-3′ | ||||
| BES1 | At1g19350 | 5′-TGCGGCGAAGATTTATACTGG-3′ | 55 | semi-RT-PCR |
| 5′-TCCAATCCTTCCTTCCGACA-3′ | ||||
| BZR1 | At1g75080 | 5′-TTTCGAGGGGTTGGTTGTTGGTTTT-3′ | 60 | |
| 5′-ATGCCATTTGGGTTTGCCTAGTTGT-3′ | ||||
| BEH1 | At3g50750 | 5′-TGCCTCTCTTTTCTCCTCGTG-3′ | 56 | |
| 5′-TTAAACCACGATATTAACCTAGCCG-3′ | ||||
| BEH2 | At4g36780 | 5′-CTTCAGACTCACACACACACCC-3′ | 55 | |
| 5′-TCCCATTCATTCCGGACATTC-3′ | ||||
| BEH3 | At4g18890 | 5′-CGTGTTATTTTTCCAAATTCCGGTG-3′ | 54 | |
| 5′-AGCAAACGACTTTGATTCTTCTCTG-3′ | ||||
| BEH4 | At1g78700 | 5′-TGAATTAGCTTGAGTTTAGCTTCG-3′ | 54 | |
| 5′-AGCTGTGTTTTAACTTTGAGCA-3′ | ||||
| BEH4 pro | 5′-CCTCTAGACGACTAGGATATATAGCAACCCGC-3′ | Construction of BEH4::GUS | ||
| 5′-GATCTTAGCTGCGATTGCTC-3′ | ||||
| 5′-CCGGATCCACTACTCTCTGTTTCTTCTTCC-3′ | ||||
| GUS | 5′-ATGCGTCACCACGGTGATAT-3′ | |||
a) Annealing temperature (°C)
4. Chemical treatments
For qRT-PCR, Arabidopsis seedlings were treated with phytohormones (0.1 µM for BL; 1 µM for the other seven hormones) and a BR biosynthesis inhibitor (5 µM for Brz) according to Tanaka et al. (2005).20) For histochemical GUS staining, ABA and abamineSG were applied to BEH4::GUS transgenic seeds as described below. After cold treatment on 1/2 MS solid medium, the imbibed seeds were transferred to the same medium containing either 1 µM ABA or 50 µM abamineSG. They were cultured under continuous light at 24°C for the defined periods.
5. RNA extraction and RT-PCR
Total RNA was extracted from Arabidopsis plants according to the procedures described in Tanaka et al. (2005) and was subjected to RT-PCR analyses.20) qRT-PCR analysis was performed using a real-time PCR system (ABI 7300; Applied Biosystems, Foster City, CA, USA) whereby cDNA synthesis kits (ReverTra Ace; Toyobo Co. Ltd., Osaka, Japan) and Green Master Mix (Power SYBR; Thermo Fisher Scientific Inc.) were used for cDNA synthesis and quantitative PCR, respectively. Semi-RT-PCR was conducted as previously described by Tanaka et al. (2005).20) Primer sequences and annealing temperatures for the genes are presented in Table 1.
6. GUS staining
Histochemical β-glucuronidase (GUS) assays were performed according to Yoshimitsu et al. (2011).18) However, when older plants were stained, acetone was pretreated to partially disrupt cuticle barriers at the epidermis to enhance the permeability of plant bodies to 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-Gluc). Briefly, after the plant materials were immersed in 90% acetone for five minutes, they were washed three times with reverse-osmosis water to remove the organic solvent, followed by GUS staining. Photographs were taken using a digital camera (Power Shot S5 1S; Canon Inc.).
7. Statistical analysis
qRT-PCR was repeated biologically at least two times and was repeated technically three times. Statistical analyses were performed using a Student’s t-test or one-way ANOVA combined with either Tukey’s or Dunnett’s post-test, with significance assigned at p<0.05.
Results
1. Expressions of BES1/BZR1 family genes with developmental-stage dependence and organ specificity
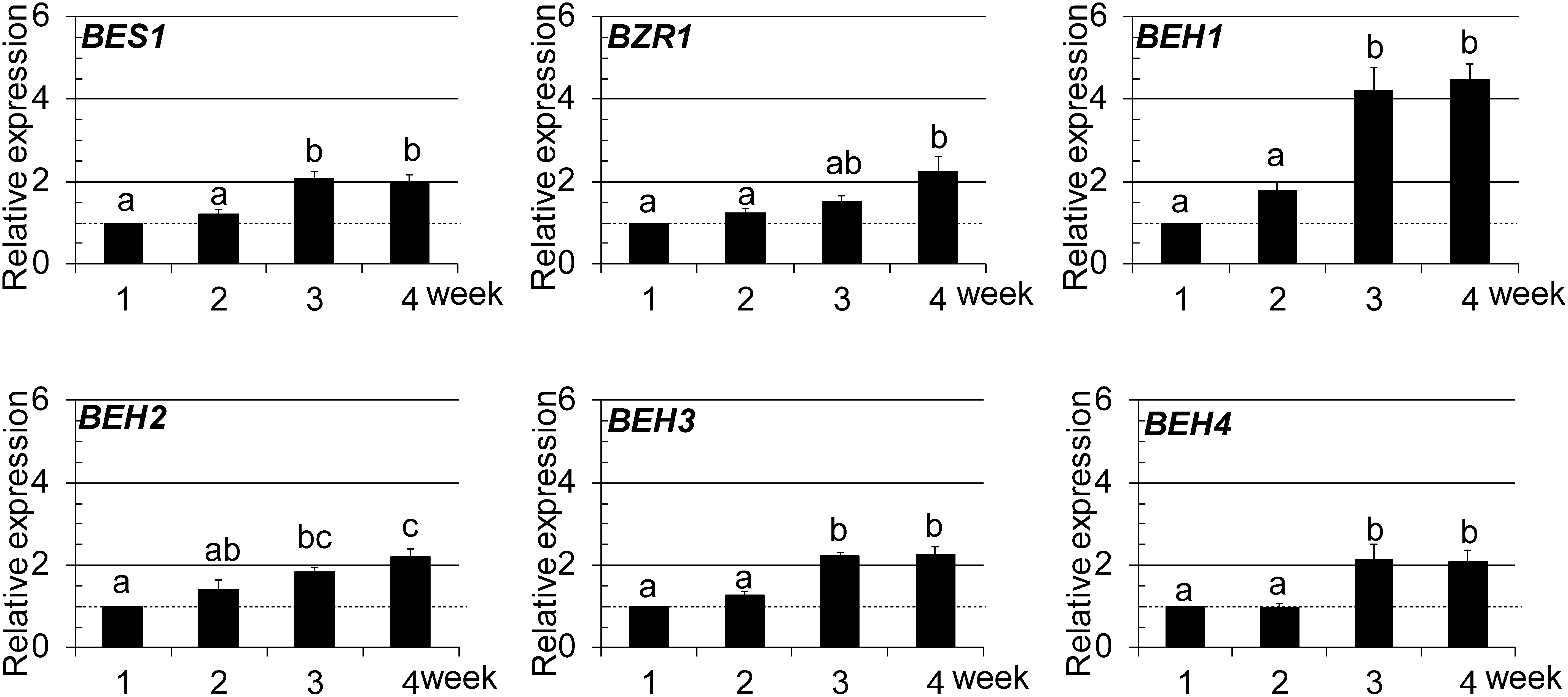
We first performed quantitative RT-PCR (qRT-PCR) to determine the expression of the six BES1/BZR1 family members at different growth stages of Arabidopsis plants from 1 to 4 weeks (wk) after sowing. As presented in Fig. 1, the expression levels of the six members were almost constant in the early stages (1–2 wk). Then they displayed gradually increased expression along with their growth. Among them, BEH1 expression in the late stages (3–4 wk) was nearly four times higher than that at 1 wk, whereas the other five were roughly two times higher. We also compared the expression levels between the BES1/BZR1 family members in 2-wk-old seedlings using semi-RT-PCR analysis, which revealed that five members other than BEH1 were nearly equally expressed and that the expression of BEH1 was a half to a quarter lower than that of the others (Supplemental Fig. S1). To summarize, the absolute mRNA levels of BES1/BZR1 family members were similar in the late growth stages, although their mRNA levels increased differently along with plant growth.
Fig. 1. Growth-dependent expression of BES1/BZR1 family members was examined using qRT-PCR. In the graphs, data are shown as relative values of the means with standard error (SE) (set 1 for 1-wk-old), following normalization by that of EF1aA4. Statistical analyses were conducted using Tukey’s test (p<0.05).
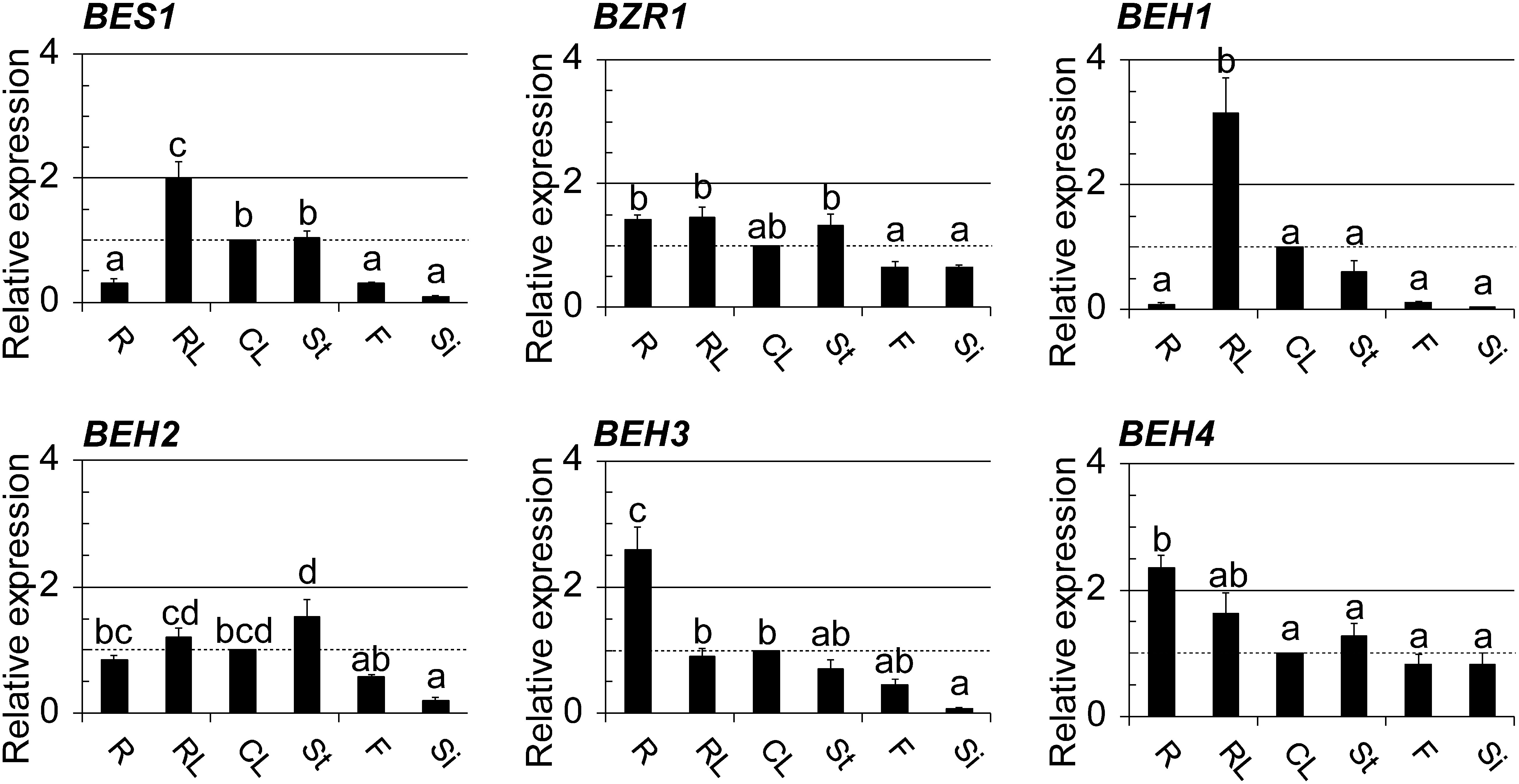
We then examined the mRNA levels of BES1/BZR1 members in different organs using qRT-PCR to determine their organ specificity. As presented in Fig. 2, their expression patterns differed, with some overlap. They were classified into two groups based on their expression patterns. The first group included BES1, BEH1, and BEH3: their expressions were different in different organs. In contrast, organ specificity was not as evident in the second group, which included BZR1, BEH2, and BEH4. In the first group, BEH1 and BEH3 were most strongly expressed in rosette leaves and roots, respectively, implying the higher requirements of BEH1 and BEH3 proteins in the corresponding organs as compared with the other organs. In addition, the expression profiles of BES1 and BEH1 were similar. They were the most strongly expressed in rosette leaves, moderately in cauline leaves and inflorescence stems, and weakly in roots, flowers, and siliques, thus implying their redundant or cooperative role at the organ level. In contrast, the three members in the second group were nearly equally expressed in all organs. BEH2 expression in siliques was exceptionally lower than that in the other organs tested, although significance was not found at the 5% level compared to that in flowers. BZR1 and BEH4 in this group were apparently equally expressed between the organs. It is interesting that the two genes were expressed preferentially in the sexual organs, such as flowers and siliques, compared to the other four genes.
Fig. 2. Organ-specific expression of BES1/BZR1 family members was examined using qRT-PCR; the used organs were roots (R), rosette leaves (RL), cauline leaves (CL), and stems (St) prepared from 4-wk-old plants as well as flowers (F) and siliques (Si) from 8-wk-old plants. Graphed data are shown as the relative values of the means with SE (set 1 for CL). The presentation style of the graphs is identical to those in Fig. 1.
2. Response of BES1/BZR1 family genes to different hormones at the mRNA level
Reportedly, BRs interact with other phytohormones such as auxin,18) gibberellin,21) ethylene,22) abscisic acid,23) and strigolactone.15) Therefore, we examined whether eight hormones, including BRs, affect the expressions of BES1/BZR1 family members in an attempt to find a novel hormone crosstalk between BRs and other hormones at the mRNA level. As depicted in Fig. 3, their mRNA levels did not change much upon administration of the seven hormones other than BRs. Exceptionally, ABA and GA3 slightly upregulated some of the members: BES1 (ABA), BEH1 (ABA and GA3), and BEH4 (ABA). However, the two hormones did not upregulate them beyond two times that of each control.
Fig. 3. qRT-PCR was used to evaluate the effects of phytohormones on the expression of BES1/BZR1 family members in 2-wk-old seedlings. In the graphs, data are shown as relative values of the means with SE (set 1 for the control treatment with 0.1% DMSO used as a solvent), following normalization by that of ACT2. Statistical analyses were conducted using Dunnett’s test (*p<0.05, **p<0.01).
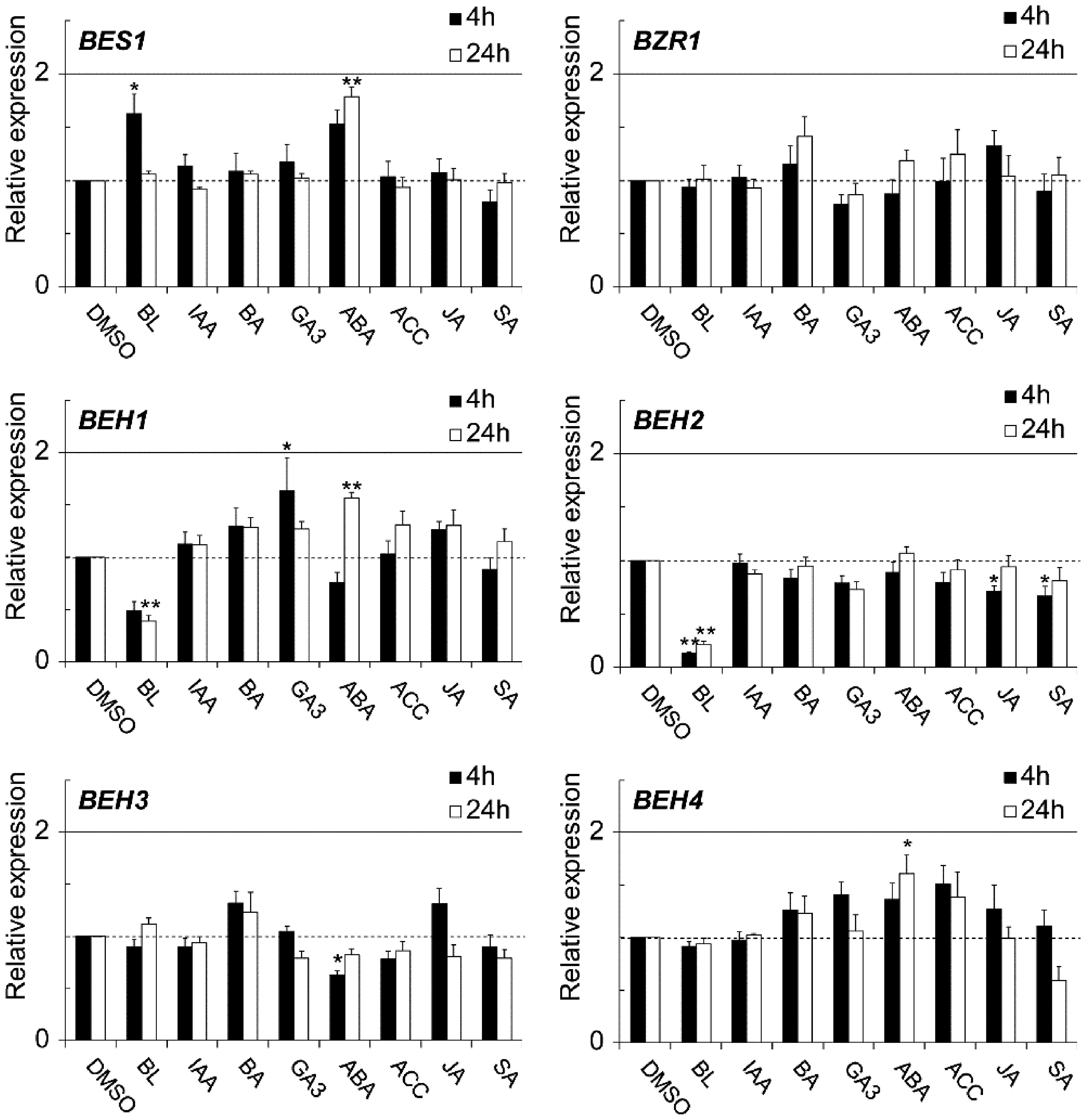
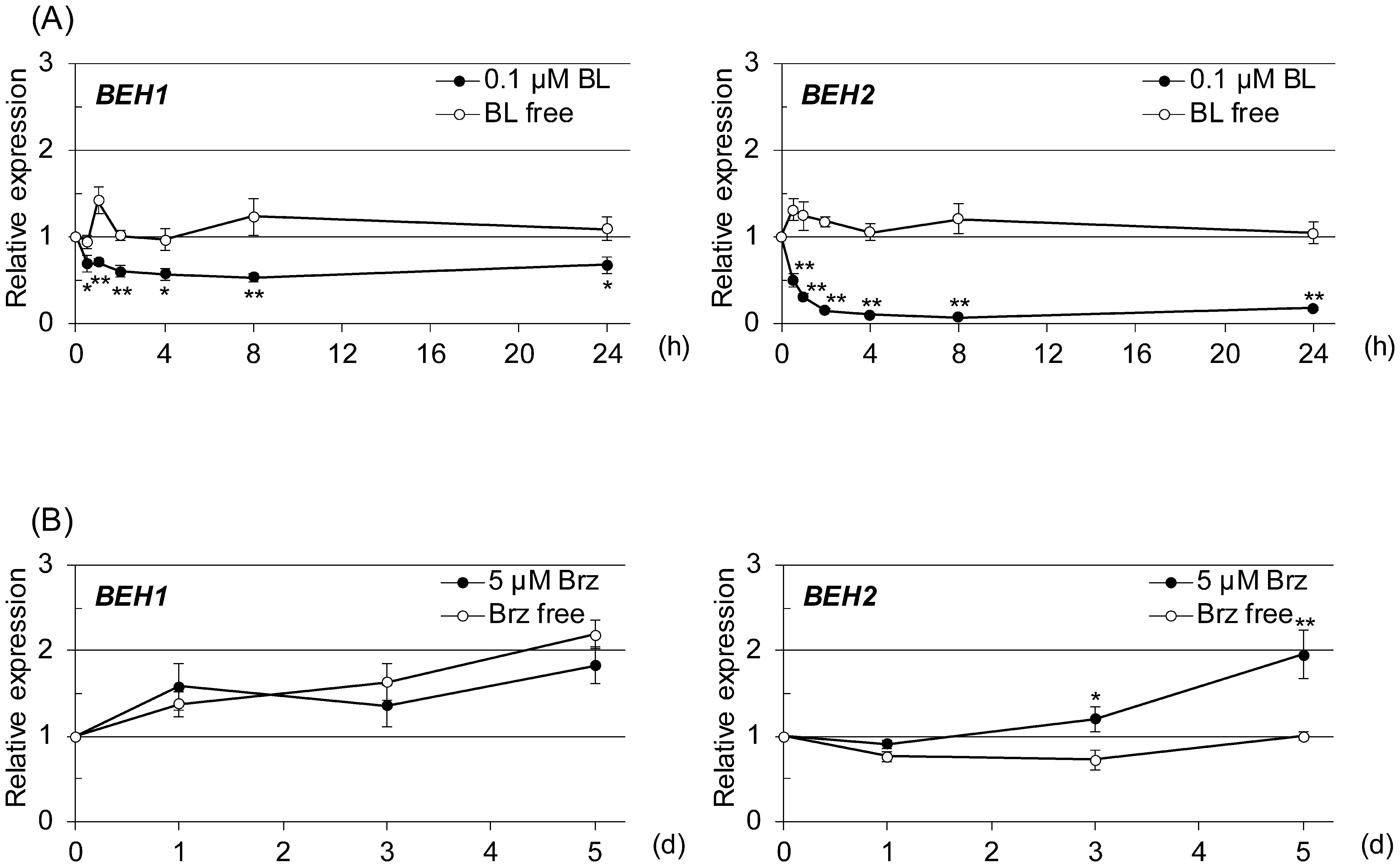
In contrast, BL, a representative compound of bioactive BRs, markedly downregulated BEH1 and BEH2 and slightly upregulated BES1 (less than twofold at 4 hr). The BEH2 mRNA level was reduced to nearly one-fifth at 4 hr and 24 hr by BL when compared with the control. Additionally, the BEH1 mRNA level was reduced nearly by half that of the control. Next, we conducted a time-course study to confirm and further investigate the kinetics of the BL-triggered reduction of BEH1 and BEH2 expressions. In this experiment, we pretreated 2-wk-old seedlings with Brassinazole (Brz), a specific inhibitor of BR biosynthesis, for 2 days, followed by BL treatment, because we reasoned that reducing endogenous BR levels would increase the sensitivity of our experiment for BL-mediated downregulation.20) As depicted in Fig. 4A, the expression levels of BEH1 and BEH2 were markedly reduced by BL throughout the experiment. The BEH2 mRNA level was reduced immediately to almost half that at the initial time after 0.5 hr of BL treatment, reaching one-tenth by 4 hr. This level continued until 24 hr. Similarly, BEH1 mRNA was decreased to half by 2 hr BL treatment and was maintained at the same level until 24 hr. We further examined whether Brz affects their expressions. The results revealed that BEH2 expression was increased time dependently by Brz, although BEH1 expression was not changed much (Fig. 4B), suggesting that at least BEH2 is regulated by the fluctuation of endogenous BR contents. Next, we searched known cis-elements recognized by BES1 and BZR1, a BR responsive element (BRRE: CGT GT/CG) and E-box (CANNTG) on the 5′-upstream regions (up to 2000 bp from the initiation codon) of BEH1 and BEH2 genes to further confirm the relation to BR signaling. As shown in Supplemental Table S1, BEH1 and BEH2 had many of these elements. Furthermore, BRRE and the G-box (CAC GTG), a type of E-box, were more enriched in BEH2 (seven for BRRE and five for the G-box) than in BEH1 (two each for BRRE and the G-box), while the total E-box numbers were similar to each other. These observations imply that the two genes were downregulated by BR through the actions of BES1 and BZR1 but differently.
Fig. 4. qRT-PCR was used to evaluate the time-dependent response of BEH1 and BEH2 to an increased or decreased BR level in 2-wk-old seedlings. (A) Brassinolide (BL, 0.1 µM) was given to plants for the indicated number of hours following 2 days’ pretreatment with 5 µM brassinazole (Brz), a BR biosynthesis inhibitor. (B) Brz (5 µM) was applied directly to the seedlings for the indicated number of days. In the graphs, data are shown as relative values of the means with SE (set 1 for time 0), following normalization by that of ACT2. Statistical analyses were performed with Student’s t-test (*p<0.05, **p<0.01).
3. BEH4 expression at tissue and cellular levels
Although the expression profiles of six BES1/BZR1 family members of Arabidopsis were largely disclosed at the organ and developmental-stage levels along with hormone responsiveness (Figs. 1–3), those at the tissue and cellular levels remain unknown. Therefore, we examined the BEH4 expression histochemically using a GUS reporter, because BEH4 expression was apparently rather ubiquitous and constitutive throughout the life of the plant, implying that BEH4 plays more general roles than the others.
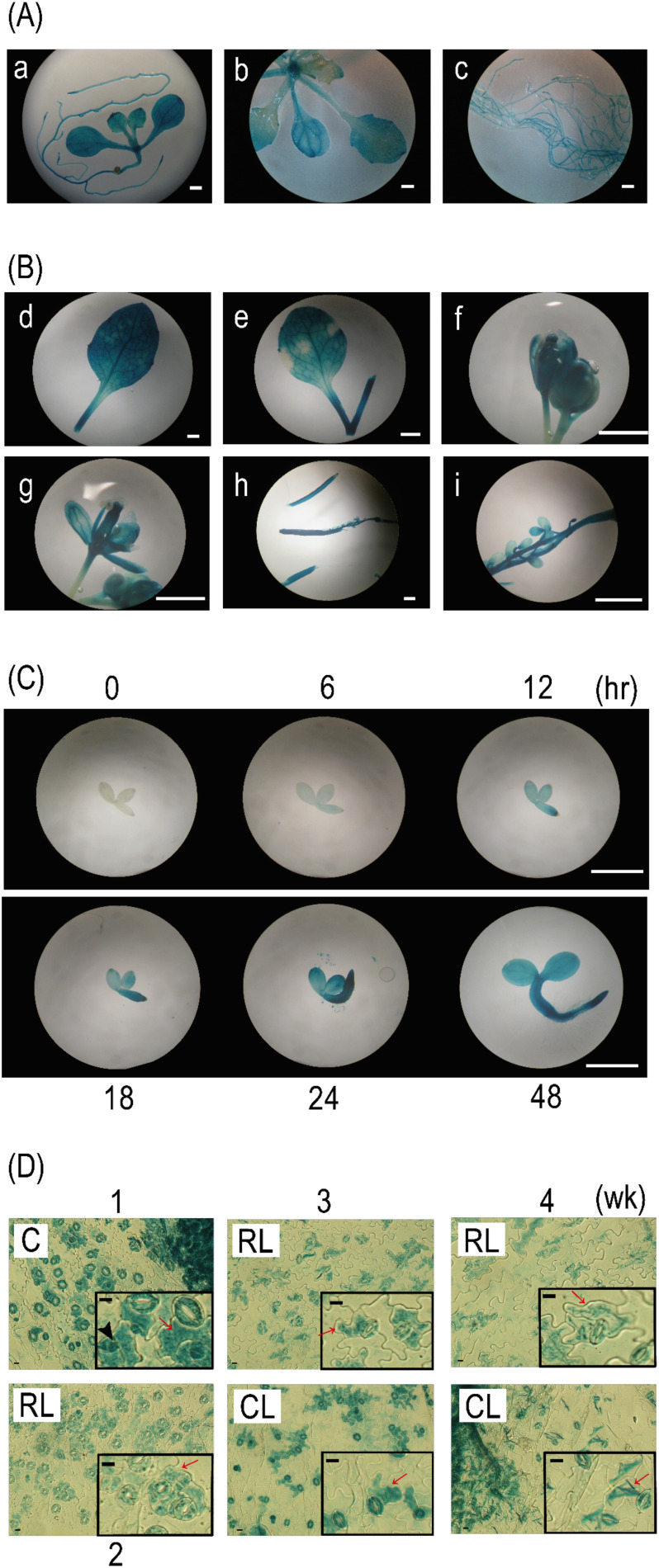
Consistent with the result of the qRT-PCR analysis (Figs. 1 and 2), all organs in BEH4::GUS plants were almost fully stained with GUS activity from the early growth stages (1 and 2 wk) to the late growth stages (3 wk and later) (Fig. 5A and B), demonstrating that BEH4 expression is ubiquitous. However, expression preferences were observed at the tissue level. For instance, root tips in early growth stages (Fig. 5A, a and c) and styles (Fig. 5B, g) and ovule stalks (Fig. 5B, i) in late growth stages were more strongly stained than other parts. In contrast, no GUS staining was detected in early seedlings immediately after commencing the culture (Fig. 5C). Faint GUS staining was detected in the whole body of the seedling 6 hr after cultivation. It strengthened over time (from 12 to 48 hr), particularly near the root apex. However, GUS staining was not detected in the root apex (12 hr) or root hairs (48 hr). Preferential expression of BEH4 was also observed in leaf epidermal tissues. As presented in Fig. 5D, most guard cells (GCs) were GUS-stained, irrespective of leaf type (cotyledonous, rosette, or cauline leaves) or developmental stage (1–4 wk). Furthermore, pavement cells (PCs) located adjacently to GCs were frequently stained, whereas other PCs located apart from GCs were not. Additionally, smaller cells that existed separately from GCs were often stained in cotyledons, which may be in the stomatal lineage. Together, the histochemical analyses suggest that BEH4 is ubiquitously expressed throughout the life of the plant, but also preferentially in some contexts.
Fig. 5. BEH4::GUS plants were subjected to histochemical GUS staining at different stages: seedlings in an early growth stage (A, 1 wk and 2 wk), several organ parts excised from adult plants in a late growth stage (B, 4 wk or later), and seedlings in a very early stage (C, 0–48 hr). Leaf epidermal tissues (D) that were peeled off from the cotyledon (C, 1 wk), rosette leaf (RL, 2–4 wk), and cauline leaf (CL, 3 wk and 4 wk) of BEH4::GUS plants were subjected to GUS staining. The lowercase letters in A and B indicate a 1-wk-old whole seedling (a); above-ground organs (b) and roots (c) of 2-wk-old seedlings; a 4-wk-old rosette leaf (d); a 4-wk-old cauline leaf (e); a 5-wk-old flower bud (f); an open flower (g); an immature silique (h); and a developing seed in a silique (i). Insets in Fig. 5D present magnified images of the epidermal tissues. White bars and black bars represent 1 mm and 10 µm, respectively. Red arrows and black arrowheads indicate pavement cells (PCs) located adjacently to guard cells (GCs) and smaller cells located away from GCs, respectively.
4. Influence of ABA on BEH4 expression in young seedlings
As described above, BEH4 expression was induced along with seedling growth in very early growth stages (Fig. 5C). BR and ABA reportedly function in an antagonistic manner during germination and seedling establishment.24) Therefore, we searched cis-elements for an ABA response and found two putative ABA responsive elements (ABRE) and six RAV1-A binding site motifs (RAV1-A) in the BEH4 promoter region (Fig. 6A; Table 2). Additionally, in our qRT-PCR using 2-wk-old seedlings, BEH4 was slightly upregulated (approx. 1.7 times) at 24 hr after ABA addition, compared to the mock control (Fig. 3). Then we examined whether ABA regulates BEH4 expression at very early growth stages using BEH4::GUS plants. As depicted in Fig. 6B, seedling growth was severely retarded when 1 µM ABA was applied. The morphology of seedlings treated with ABA for 5 d was similar to that of the mock-treated seedlings for 1 d; exogenous ABA likely suppressed the growth progression. Under this condition, ABA delayed GUS staining when compared with that of mock-treated seedlings, although the staining pattern remained almost unchanged (Fig. 6B). Consistent with this observation, the GUS staining patterns of the seedlings treated with ABA for 1–5 days were quite similar to those of the mock-treated seedlings for 12–24 hr (Figs. 5C and 6B). In contrast, 50 µM abamineSG neither affected the seedling growth nor the GUS staining (Fig. 6B). In total, our results imply that ABA affects BEH4 expression in terms of time, although how it regulates BEH4 remains to be determined.
Fig. 6. The response of BEH4 to increased or decreased ABA levels was examined histochemically. (A) The triangles on a schematic drawing of the BEH4 gene and its 5′-flanking sequence show the positions of ABRE-like and RAV-like elements in both strands of the DNA double helix. The element sequences are presented in Table 2. Box, exon; thin line, intron; thick line, 5′-flanking (promoter) sequence; UTR, either 5′- or 3′-untranslated regions of BEH4. (B) BEH4::GUS transgenic seedlings were subjected to histochemical GUS staining; following the treatment of either ABA (1 µM) or abamineSG (50 µM), a specific inhibitor of ABA synthesis DMSO (0.1%) was applied to plants as a mock treatment. Scale bars represent 1 mm.
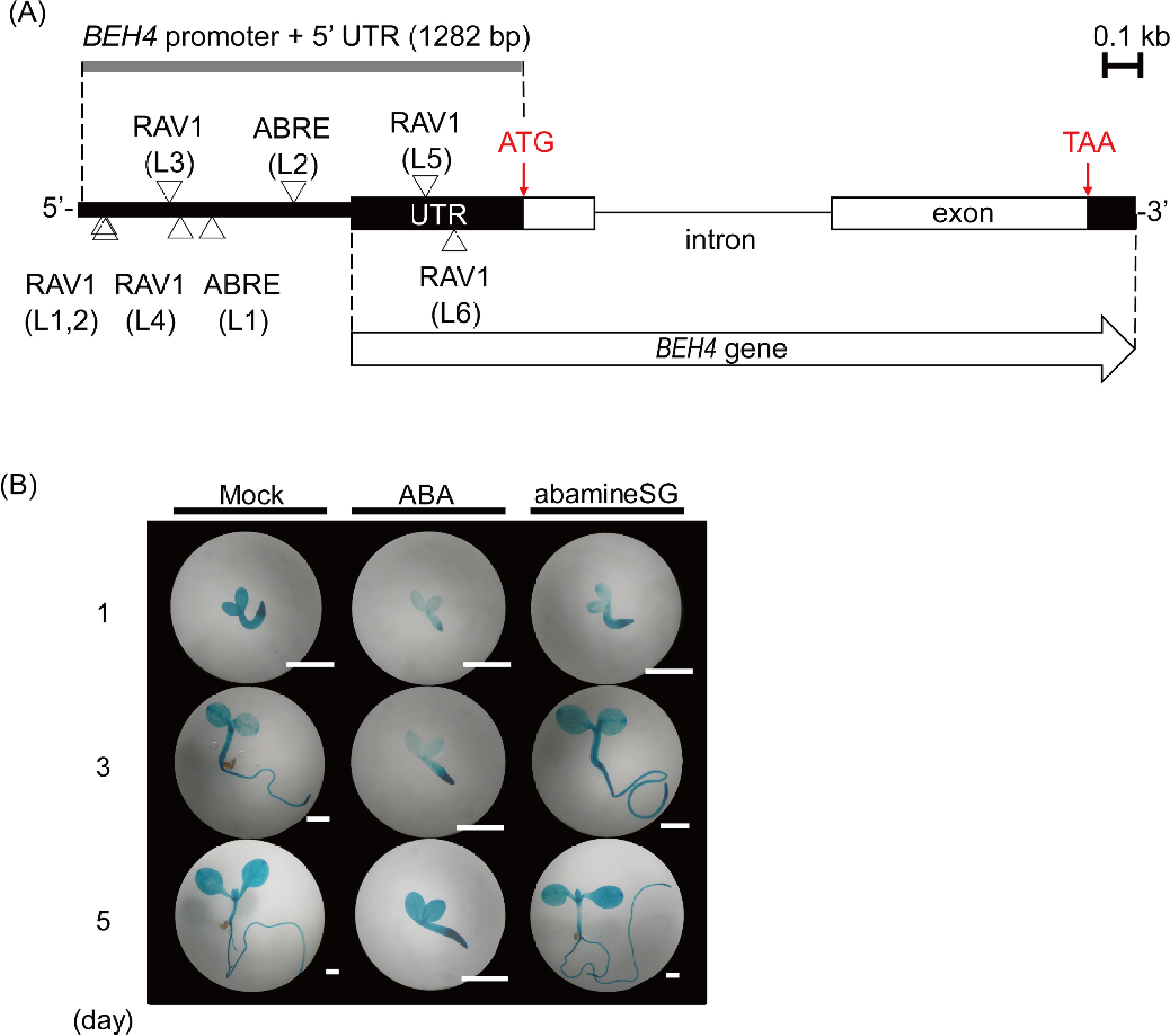
Table 2. ABRE-like and RAV-1-like motifs for ABA response found in BEH4 promoter.
| Consensus/like | cis-Element | Sequence |
|---|---|---|
| consensus | ABRE consensus | PyACGTG / TC |
| like | ABRE L1 | CACGTGGC |
| like | ABRE L2 | GCACGTGGTA |
| consensus | RAV1-A binding site motif | CAACA |
| RAV1-A L1 | CAACA | |
| RAV1-A L2 | CAACA | |
| RAV1-A L3 | CAACA | |
| RAV1-A L4 | CAACA | |
| RAV1-A L5 | CAACA | |
| RAV1-A L6 | CAACA |
Discussion
BES1 and BZR1 transcription factors play crucial roles in BR signaling and BR crosstalk with intrinsic cues and extrinsic signals.12,13) However, investigations of BES1/BZR1 homologous proteins BEH1–4 have lagged behind those of the two reference proteins except for the BR-induced dephosphorylation (BEH1–4)14) and the crosstalk (BEH2, BEH3, and BEH4) with strigolactone signaling through their interaction with MAX2.15) Although Chen et al. (2019) most recently demonstrated that BES1/BZR1 family proteins function redundantly and indispensably in BR signaling, suggesting the possible involvement of BEH homologs in this process,25) our knowledge of the functions of the four homologs is still limited. Therefore, to further elucidate their functions, we carefully examined BEH1–4 expression, as the expression analyses are thought to be effective to obtain a lot of valuable information for speculating on their functions. Herein, we discuss their potential roles based on their expression profiles.
All six BES1/BZR1 family genes were expressed during the 1–4 wk period (Fig. 1). Moreover, their mRNA levels in the late growth stages (3–4 wk) were higher than those in the early growth stages (1–2 wk). The vegetative-to-reproductive transition occurred at 2–3 wk under our light condition (16L:8D) (Fig. 5). Therefore, their expression tendency may be associated with growth-mode switching. Together, all members are regarded as having various physiological roles throughout the life of the plant. Their higher expressions in the later growth stages imply that they play more roles in the reproductive phase.
All BES1/BZR1 members were differentially expressed in individual organs, with some overlap (Fig. 2). Among them, BEH1 expression is noteworthy because it was rarely expressed in the sexual organs, such as flowers and siliques, even though overall BEH1 expression increased greatly along with growth progression (Fig. 1). This finding suggests that BEH1 may play physiological roles in organs other than the sexual organs that are directly committed to producing offspring seeds. In contrast, BZR1 and BEH4 were preferentially expressed in flowers and siliques when compared with the other members (Fig. 2). Moreover, BEH4 was found to express in both pistils and stamens of mature flowers (Fig. 5B). Jiang et al. (2013) reported that BRs transcriptionally control seed size and shape: BZR1 regulates several genes acting on the development of the integument, endosperm, and embryo.26) Most recently, the mutants of BES1/BZR1 family genes of a higher order than the quadruple mutant harboring a beh4 mutation were proven not to produce seeds, partly due to impaired anther development.25) Thus, the previous reports together with our results imply that BEH4 may also contribute to the birth of the next generation, similar to BZR1. Taken together, our qRT-PCR suggests that BES1/BZR1 family proteins play specific physiological roles in individual organs during different growth stages by controlling their expression, although they likely have similar molecular functions that can be assumed from their structural similarity27) and genetic redundancy.25,28)
BRs reportedly interact with other phytohormones in different processes related to hormone biosynthesis, inactivation and degradation, and signaling.3,13,24,29) However, our qRT-PCR analysis failed to identify a novel crosstalk at the mRNA level for BES1/BZR1 family members. Their expressions were affected only slightly by seven hormones, except for BRs (Fig. 3). This might be an expected consequence, because earlier studies have demonstrated that BES1 and BZR1 genes involved in BR signaling are modulated mostly at the protein level by phosphorylation,14,30) nucleus/cytoplasm shuttling,7,8,31,32) proteasome-mediated degradation,9,10) and DNA binding activity control.7,11) Therefore, interactions between BRs and other hormones via BES1/BZR1 family members may occur at the protein level. However, the BR-mediated downregulation of BEH1 and BEH2 is noteworthy. As described earlier, BR-dependent dephosphorylation supports the association of BEH1–4 with the BR pathway.14) Our finding that BL reduced the mRNA levels of the two homologs reinforces their close relation with BR signaling. It is also interesting that BEH1 mRNA was decreased by BL but not affected by Brz, whereas BEH2 mRNA was decreased and increased by BL and Brz, respectively (Figs. 3 and 4), which suggests that BRs control their expressions differently. In our earlier study,20) similar observations were made; the four BR-specific biosynthesis genes (DWF4, CPD, BR6ox1, and ROT3) were affected by both BL and Brz, while the sterol synthesis gene (DWF7) and BR inactivation–related gene (BAS1) were affected only by BL. The former four genes encode key enzymes in the central part of BR synthesis, so they must be more tightly controlled than others to quickly restore and optimize BR levels in a given context. BEH2 may be regulated by a mechanism similar to that for the four BR-specific biosynthesis genes, whereas BEH1 may be controlled similarly to DWF7 and BAS1, although BAS1 is upregulated by BL. Analogously, BEH2 might have a more important task in BR signaling than BEH1 has.
How are BEH1 and BEH2 expressions negatively regulated by BR? Previous studies describe BRRE elements (CGT GC/TG) as being generally enriched in BR-repressed genes, while E-boxes (CANNTG) are in BR-induced genes,5,6) which are recognized by both BES1 and BZR1 transcription factors. Sun et al. (2010) further claimed that the G-box (CAC GTG), a type of E-box, is also enriched in the BR-repressed genes.6) In the motif search analysis, we found that BEH1 and BEH2 had many of these BR-related cis-elements in their promoter regions (Supplemental Table S1). Furthermore, more BRRE and G-box elements existed in BEH2 than in BEH1, leading to the notion that the number of these elements may determine the difference in BR responses between the two homologs: BEH2 mRNA was more rapidly and strongly reduced by BL, compared with BEH1 (Fig. 4). Moreover, Nosaki et al. (2018) recently demonstrated that atypical bHLH transcription factors, BES1 and BZR1, can bind to both NN-BRRE (NNCGT G) and the G-box in vitro, while typical ones such as PIF4, BEE1, and BIM1 preferentially bind to the G-box.33) Their observation may support our assumption that BR-mediated downregulation of BEH1 and BEH2 is achieved through the binding of BES1/BZR1 transcription factors to these elements, but in a different manner. However, further analyses are necessary to obtain the precise mechanism underpinning the BR-mediated expression of BEH1 and BEH2.
BEH4 expression is also interesting because it was rather constantly expressed among the six members (Figs. 1–3), implying that BEH4 is associated with general roles such as housekeeping functions. Our histochemical analysis supports this notion by showing that BEH4::GUS transgenic plants were almost fully GUS stained throughout their entire lifecycle (Fig. 5A and B). However, detailed observations uncovered a preference in BEH4 expression at the tissue and cellular level. In this respect, BEH4 expression was restricted to guard cells (GCs) and their surrounding pavement cells (PCs) in the leaf epidermis in all stages (1–4 wk, Fig. 5D). In addition, small cells existing separately from GCs were stained at 1 wk (Fig. 5D), which might include stomata lineage cells such as meristemoid mother cells, meristemoid cells, stomatal lineage ground cells, and guard mother cells.34) BEH4 expression in leaf epidermal tissues is curious because BRs reportedly suppress stomatal development by inactivating the BIN2 kinase, which acts as a positive regulator of stomatal development35) as well as a negative regulator of BR signaling.4) Additionally, Lau et al. (2014) reported that SPCHLESS (SPCH), a downstream transcription factor in the stomata differentiation pathway, binds to the BEH4 promoter to enhance its expression in Arabidopsis mutants with a meristemoid-enriched phenotype, implying the involvement of BEH4 in this process.36) BEH4 expression in mature GCs is noteworthy because BRs antagonistically regulate ABA-mediated stomatal closure via competitive use of BAK1 proteins.37) Although we conjectured the involvement of BEH4 in stomatal development and movement based on its peculiar expression profile in epidermal tissues, whether BEH4 plays roles in these processes remains undetermined. If not, BEH4 may still be a useful marker to address the mechanism underlying both processes. Furthermore, BEH4 expression was altered drastically during seed germination and subsequent seedling establishment (Fig. 5C). In this context, ABA primarily suppresses seed germination, whereas GAs and BRs promote germination and seedling establishment.38) Which phytohormone controls BEH4 expression in very early growth stages? Among three, ABA is a promising candidate for regulating BEH4 during this period, because GA3 and BRs did not affect BEH4 expression, whereas ABA slightly upregulated BEH4 (Fig. 3). However, the attempt to address this question presented us with a result that was difficult to interpret: BEH4 expression was delayed upon ABA administration but was not changed in a spatial manner (Fig. 6B). Consequently, answering this question must be left to future research.
Collectively, we describe here several unique and interesting features in expression for six BES1/BZR1 family members. Their expression profiles provide a basic platform to further elucidate the molecular and physiological roles of the BES1/BZR1 family, which then could be used to exploit a novel application of BRs as PGRs and to employ these genes as targets for crop breeding in the future.
Acknowledgments
We thank Dr. Tadao Asami of Tokyo University and Dr. Shigeo Yoshida of the Kihara Institute for Biological Research for kindly providing Brz and abamineSG. We are also grateful to Dr. Hisashi Iwai of Kagoshima University and Dr. Akihiro Suzuki of Saga University for helpful discussions and suggestions.
Electronic supplementary materials
The online version of this article contains supplementary materials (Supplemental Fig. S1 and Table S1), which are available at http://www.jstage.jst.go.jp/browse/jpestics/.
Conflicts of Interest
Declarations of interest: none
Supplementary Data
References
- 1).A. Basra: “Plant Growth Regulators in Agriculture and Horticulture: Their Role and Commercial Uses,” CRC Press, 2000.
- 2).A. Sakurai, T. Yokota and S. D. Clouse (eds.): “Brassonosteroids Steroidal Plant Hormones,” Springer, Tokyo, 1999.
- 3).Y. Chung and S. Choe: Crit. Rev. Plant Sci. 32, 396–410 (2013). [Google Scholar]
- 4).J. Y. Zhu, J. Sae-Seaw and Z. Y. Wang: Development 140, 1615–1620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).X. Yu, L. Li, J. Zola, M. Aluru, H. Ye, A. Foudree, H. Guo, S. Anderson, S. Aluru, P. Liu, S. Rodermel and Y. Yin: Plant J. 65, 634–646 (2011). [DOI] [PubMed] [Google Scholar]
- 6).Y. Sun, X. Y. Fan, D. M. Cao, K. He, W. Tang, J. Y. Zhu, J. X. He, M. Y. Bai, S. Zhu, E. Oh, S. Patil, T. W. Kim, H. Ji, W. H. Wong, S. Y. Rhee and Z. Y. Wang: Dev. Cell 19, 765–777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).S. S. Gampala, T. W. Kim, J. X. He, W. Tang, Z. Deng, M. Bai, S. Guan, S. Lalonde, Y. Sun, J. M. Gendron, H. Chen, N. Shibagaki, R. J. Ferl, D. Ehrhardt, K. Chong, A. L. Burlingame and Z. Y. Wang: Dev. Cell 13, 177–189 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).S. Shimada, T. Komatsu, A. Yamagami, M. Nakazawa, M. Matsui, H. Kawaide, M. Natsume, H. Osada, T. Asami and T. Nakano: Plant Cell 27, 375–390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).M. Yang, C. Li, Z. Cai, Y. Hu, T. Nolan, F. Yu, Y. Yin, Q. Xie, G. Tang and X. Wang: Dev. Cell 41, 47–58.e44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).J. X. He, J. M. Gendron, Y. Yang, J. Li and Z. Y. Wang: Proc. Natl. Acad. Sci. U.S.A. 99, 10185–10190 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).G. Vert and J. Chory: Nature 441, 96–100 (2006). [DOI] [PubMed] [Google Scholar]
- 12).Q. F. Li, J. Lu, J. W. Yu, C. Q. Zhang, J. X. He and Q. Q. Liu: Biochim. Biophys. Acta. Gene Regul. Mech. 1861, 561–571 (2018). [DOI] [PubMed] [Google Scholar]
- 13).T. Nolan, J. Chen and Y. Yin: Biochem. J. 474, 2641–2661 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Y. Yin, D. Vafeados, Y. Tao, S. Yoshida, T. Asami and J. Chory: Cell 120, 249–259 (2005). [DOI] [PubMed] [Google Scholar]
- 15).Y. Wang, S. Sun, W. Zhu, K. Jia, H. Yang and X. Wang: Dev. Cell 27, 681–688 (2013). [DOI] [PubMed] [Google Scholar]
- 16).K. Tanaka, Y. Nakamura, T. Asami, S. Yoshida, T. Matsuo and S. Okamoto: J. Plant Growth Regul. 22, 259–271 (2003). [DOI] [PubMed] [Google Scholar]
- 17).http://www.ddbj.nig.ac.jp/
- 18).Y. Yoshimitsu, K. Tanaka, W. Fukuda, T. Asami, S. Yoshida, K. Hayashi, Y. Kamiya, Y. Jikumaru, T. Shigeta, Y. Nakamura, T. Matsuo and S. Okamoto: PLoS One 6, e23851 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).M. Narusaka, T. Shiraishi, M. Iwabuchi and Y. Narusaka: Plant Biotechnol. 27, 349–351 (2010). [Google Scholar]
- 20).K. Tanaka, T. Asami, S. Yoshida, Y. Nakamura, T. Matsuo and S. Okamoto: Plant Physiol. 138, 1117–1125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).M. Y. Bai, J. X. Shang, E. Oh, M. Fan, Y. Bai, R. Zentella, T. Sun and Z. Y. Wang: Nat. Cell Biol. 14, 810–817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).B. Lv, H. Tian, F. Zhang, J. Liu, S. Lu, M. Bai, C. Li and Z. Ding: PLoS Genet. 14, e1007144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).J. Gui, S. Zheng, C. Liu, J. Shen, J. Li and L. Li: Dev. Cell 38, 201–213 (2016). [DOI] [PubMed] [Google Scholar]
- 24).S. Saini, I. Sharma and P. K. Pati: Front. Plant Sci. 6, 950 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).L. G. Chen, Z. Gao, Z. Zhao, X. Liu, Y. Li, Y. Zhang, X. Liu, Y. Sun and W. Tang: Mol. Plant 12, 1408–1415 (2019). [DOI] [PubMed] [Google Scholar]
- 26).W. B. Jiang, H. Y. Huang, Y. W. Hu, S. W. Zhu, Z. Y. Wang and W. H. Lin: Plant Physiol. 162, 1965–1977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).J. Zhao, P. Peng, R. J. Schmitz, A. D. Decker, F. E. Tax and J. Li: Plant Physiol. 130, 1221–1229 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).M. Saito, Y. Kondo and H. Fukuda: Plant Cell Physiol. 59, 590–600 (2018). [DOI] [PubMed] [Google Scholar]
- 29).S. P. Choudhary, J. Q. Yu, K. Yamaguchi-Shinozaki, K. Shinozaki and L. S. P. Tran: Trends Plant Sci. 17, 594–605 (2012). [DOI] [PubMed] [Google Scholar]
- 30).S. Xue, J. Zou, Y. Liu, M. Wang, C. Zhang and J. Le: Int. J. Mol. Sci. 20, 2339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Y. Yin, Z. Y. Wang, S. Mora-Garcia, J. Li, S. Yoshida, T. Asami and J. Chory: Cell 109, 181–191 (2002). [DOI] [PubMed] [Google Scholar]
- 32).Z. Y. Wang, T. Nakano, J. Gendron, J. He, M. Chen, D. Vafeados, Y. Yang, S. Fujioka, S. Yoshida, T. Asami and J. Chory: Dev. Cell 2, 505–513 (2002). [DOI] [PubMed] [Google Scholar]
- 33).S. Nosaki, T. Miyakawa, Y. Xu, A. Nakamura, K. Hirabayashi, T. Asami, T. Nakano and M. Tanokura: Nat. Plants 4, 771–776 (2018). [DOI] [PubMed] [Google Scholar]
- 34).L. R. Lee and D. C. Bergmann: J. Cell Sci. 132, jcs228551 (2019).31028153 [Google Scholar]
- 35).T. W. Kim, M. Michniewicz, D. C. Bergmann and Z. Y. Wang: Nature 482, 419–422 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).O. S. Lau, K. A. Davies, J. Chang, J. Adrian, M. H. Rowe, C. E. Ballenger and D. C. Bergmann: Science 345, 1605–1609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Y. Shang, C. Dai, M. M. Lee, J. M. Kwak and K. H. Nam: Mol. Plant 9, 447–460 (2016). [DOI] [PubMed] [Google Scholar]
- 38).K. Shu, X. Liu, Q. Xie and Z. He: Mol. Plant 9, 34–45 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.