Abstract
Background
Serum biomarkers may inform and improve care in traumatic brain injury (TBI). We aimed to correlate serum biomarkers with clinical severity, care path and imaging abnormalities in TBI, and explore their incremental value over clinical characteristics in predicting computed tomographic (CT) abnormalities.
Methods
We analyzed six serum biomarkers (S100B, NSE, GFAP, UCH-L1, NFL and t-tau) obtained <24 h post-injury from 2867 patients with any severity of TBI in the Collaborative European NeuroTrauma Effectiveness Research (CENTER-TBI) Core Study, a prospective, multicenter, cohort study. Univariable and multivariable logistic regression analyses were performed. Discrimination was assessed by the area under the receiver operating characteristic curve (AUC) with 95% confidence intervals.
Findings
All biomarkers scaled with clinical severity and care path (ER only, ward admission, or ICU), and with presence of CT abnormalities. GFAP achieved the highest discrimination for predicting CT abnormalities (AUC 0•89 [95%CI: 0•87–0•90]), with a 99% likelihood of better discriminating CT-positive patients than clinical characteristics used in contemporary decision rules. In patients with mild TBI, GFAP also showed incremental diagnostic value: discrimination increased from 0•84 [95%CI: 0•83–0•86] to 0•89 [95%CI: 0•87–0•90] when GFAP was included. Results were consistent across strata, and injury severity. Combinations of biomarkers did not improve discrimination compared to GFAP alone.
Interpretation
Currently available biomarkers reflect injury severity, and serum GFAP, measured within 24 h after injury, outperforms clinical characteristics in predicting CT abnormalities. Our results support the further development of serum GFAP assays towards implementation in clinical practice, for which robust clinical assay platforms are required.
Funding
CENTER-TBI study was supported by the European Union 7th Framework program (EC grant 602150).
Keywords: Traumatic brain injury, Biomarkers, GFAP, Serum, Diagnostic, Computerized tomography, Clinical decision rule, Injury severity
Research in context.
Evidence before this study
Blood-based biomarkers hold potential for informing and substantially improving the clinical management of patients with traumatic brain injury (TBI). To date, however, only one biomarker (S100B) has been integrated into some national guidelines for triaging the need to perform computerized tomography scanning (CT scan) of the brain in patients with mild TBI. The superiority or incremental benefit of biomarkers beyond canonical clinical variables used in CT prediction rules has not convincingly been demonstrated. Moreover, uncertainty exists as to how single or multi-biomarker tests perform. We conducted a living systematic review and meta-analysis to quantify the ability of blood biomarkers with advanced analytical and clinical validity (GFAP, UCH-L1, NSE, S100B, t-tau and NFL) to predict the presence of traumatic abnormalities on head CT scanning in the acute clinical setting. We screened MEDLINE, Embase, EBM Reviews and Cochrane Library for relevant articles on October 25, 2016, and, subsequently updated the results by monitoring the literature every 3 months. Synthesis of these data indicated that only S100B had high sensitivity and negative predictive value (NPV) and could be used to rule out the need for acute CT scanning. The evidence to support use of other emerging markers was limited and insufficient to warrant clinical application. Recently, the ALERT-TBI trial (1959 participants), published in 2018, showed that GFAP and UCH-L1, in combination, discriminated between patients with and without CT abnormalities. Results from the TRACK-TBI study in the US, highlighted the potential of biomarkers as a screening tool for MRI abnormalities in patients with normal CT findings. However, the clinical utility of these biomarkers still remains uncertain. Further work is needed to determine the most effective and efficient biomarker or multimarker strategy for integration into clinical care.
Added value of this study
We use data from a well-characterized multicenter cohort of 2867 patients with TBI to provide the first evidence of medical utility of blood biomarkers beyond standard of care-based clinical characteristics. Our results support potential for their adoption into clinical use to inform and improve decision making in current practice. In particular, we corroborate and extend recent results regarding the diagnostic performance of GFAP, showing that GFAP captures the greatest discriminatory information, performing as well as a combination of all markers and adding value to clinical characteristics. We believe this is the largest study to simultaneously assess and compare the diagnostic performance of a panel of 6 biomarkers reflecting distinct types of injury and pathophysiological mechanisms, across a population of patients with a full range of TBI severities and wide range of injury patterns.
The assessment of the utility of biomarker measurements in individual care pathways, as defined in CENTER-TBI, allows us to explore their use in a range of contexts of care. This ensures the robustness and generalizability of our estimates derived from real-world patient population and clinical scenarios, while confirming the generation of actionable information for clinicians. Comparison of our results with those from other studies shows similar trends, but also highlights between-platform inconsistencies in assay calibration and reported biomarker values.
Implications of all the available evidence
This study provides the most exhaustive and comparable assessment to date of the six best-validated TBI biomarkers, demonstrating their potential utility in refining diagnosis, triage, injury characterization, and clinical care in TBI, beyond currently established clinical variables. We highlight the potential role of GFAP as part of a comprehensive triage strategy and consider it to be best positioned for implementation into medical practice and incorporation in clinical decision algorithms. Robust clinical use assay platforms are a prerequisite for such clinical implementation.
Alt-text: Unlabelled box
1. Introduction
The delivery of precision medicine for traumatic brain injury (TBI) requires objective tools to identify disease phenotypes and to guide clinical decisions [1]. Clinical assessment and computerized tomography (CT) of the head form the diagnostic cornerstone in clinical practice, but a need remains for more detailed disease classification utilizing a multidimensional approach. Moreover, indiscriminate use of CT, resulting in high costs, and increased recognition of risks of radiation exposure have called for more selective use of CT scanning in patients with milder forms of TBI [1,2]. Various clinical decision rules (CDR) have been developed for this purpose 3, 4, 5, 6, 7, but their adoption in clinical practice is variable.
A major focus of recent research has been on the potential of biomarkers to improve diagnosis and patient characterization, and enable tailored management [8]. Several publications have provided extensive evidence of analytical and early clinical validity of various biomarkers, and documented efforts to achieve regulatory clearance [9]. However, the development of clinical algorithms and guidelines which integrate biomarker measurements to inform decision-making has been inconsistent, partial and inconclusive [10]. S100 calcium-binding protein B (S100B), a biomarker of astroglial breakdown, has been implemented in the Scandinavian TBI Guidelines [11], but is seldom used outside the Nordic countries and with sub-optimal performance in real-world conditions [12]. The pivotal ALERT-TBI study showed high sensitivity for the combination of astroglial (glial fibrillary acidic protein [GFAP]) and neuronal (ubiquitin C-terminal hydrolase L1 [UCH-L1]) biomarker blood levels measured within 12 h after injury in triaging the need for CT scanning [13], but did not address the added value compared to clinical characteristics used in CDRs, or explore its value relative to S100B [14]. More in general, few studies have examined the incremental value of biomarkers beyond clinical characteristics.
As a consequence, uncertainty exists how biomarkers, either singly or in combination, can best improve existing decision-making and processes of care, creating a barrier to widespread implementation and adoption of these tests in medical practice. Nevertheless, blood-based biomarkers provide objective information, offer additional risk stratification and hold potential to inform personalized interventions.
The CENTER-TBI Core study (Collaborative European NeuroTrauma Effectiveness Research: www.center-tbi.eu) was designed to advance multimodal characterization and classification in TBI [15,16]. Within this unique framework, in which patients were stratified by care path, we aimed to determine the relation – and their relative performance – of a panel of biomarkers, assessed within 24 h of injury, with clinical severity, care pathways and presence of CT abnormalities across the entire injury spectrum of TBI. We further aimed to explore the incremental value of biomarkers compared to established clinical characteristics in predicting the presence of CT abnormalities.
2. Materials and methods
2.1. Study design and participants
The CENTER-TBI Core study is a prospective observational clinical and biomarker study of patients with TBI, conducted in 65 clinical sites from 17 European countries and Israel between December 19, 2014, and December 17, 2017. The study was registered with ClinicalTrials.gov (NCT02210221). Details of protocol and clinical data have been previously published [15,16]. In brief: patients with all severities of TBI presenting to a study center within 24 h of injury and scheduled for CT scanning were enrolled, stratified by care path (emergency department [ER], admission [Adm] and intensive care unit [ICU]). The only exclusion criterion was severe pre-existing neurological disorder.
The study was conducted in accordance with all relevant laws of the EU if directly applicable or of direct effect and all relevant laws of the country where the Recruiting sites were located, including but not limited to, the relevant privacy and data protection laws and regulations (the “Privacy Law”), the relevant laws and regulations on the use of human materials, and all relevant guidance relating to clinical studies from time to time in force including, but not limited to the ICH Harmonized Tripartite Guideline for Good Clinical Practice (CPMP/ICH/135/95) (“ICH GCP”) and the World Medical Association Declaration of Helsinki entitled “Ethical Principles for Medical Research Involving Human Subjects”. Informed Consent by the patients and/or the legal representative/next of kin was obtained for all patients and documented in the e-CRF. The use of the biological samples was in accordance with the terms of the informed consent. The list of sites, Ethical Committees, approval numbers and approval dates can be found on https://www.center-tbi.eu/project/ethical-approval.
The study is reported in accordance with the STROBE recommendations (see Supplementary material).
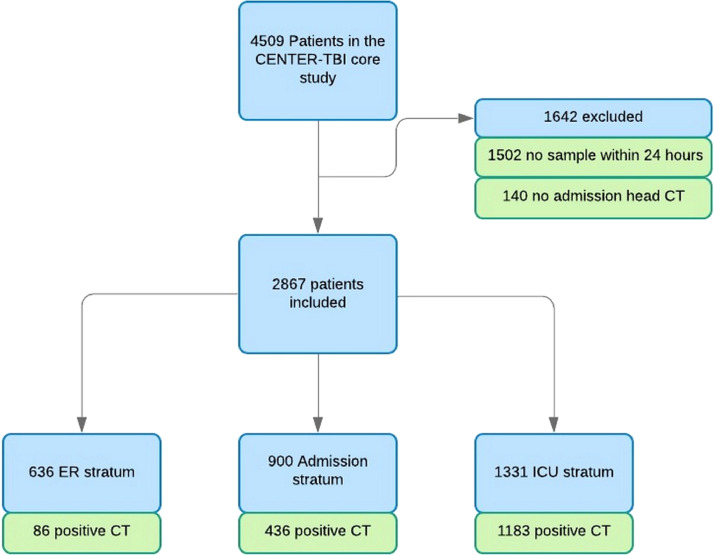
In this analysis, which was pre-specified in the Description of Work for CENTER-TBI, we focused on a cohort of patients in whom (1) blood sampling within 24 h of injury, and (2) an early CT scan were available (Fig. 1).
Fig. 1.
Flow chart for biomarker cohort in the CENTER TBI core study.
2.2. Procedures
Blood samples for determination of biomarkers were collected using gel-separator tubes for serum, and centrifuged within 60 minutes. Serum was processed, aliquoted (8 × 0·5 ml), and stored at −80 °C locally until shipment on dry ice to the central CENTER-TBI biobank (Pécs, Hungary).
We assayed six biomarkers: S100B, neuron-specific enolase (NSE), GFAP, UCH-L1, neurofilament protein-light (NFL), and total tau (t-tau). S100B and NSE were measured with a clinical-use automated system, using an electrochemiluminescence immunoassay kit (ECLIA) (Elecsys S100 and Elecsys NSE assays) run on the e 602 module of Cobas 8000 modular analyzer (Roche Diagnostics, Mannheim, Germany) at the University of Pécs (Pécs, Hungary). Serum GFAP, UCH-L1, NFL and t-tau were analyzed with an ultrasensitive immunoassay using digital array technology (Single Molecule Arrays, SiMoA)-based Human Neurology 4-Plex B assay (N4PB) run on the SR-X benchtop assay platform (Quanterix Corp., Lexington, MA) at the University of Florida (Gainesville, Florida). Unique aliquots were used for analyses on the two platforms to avoid repeated freeze-thaw cycles, and analyzed in one round of experiments using the same batch of reagents by qualified laboratory technicians blinded to clinical information. Replicate assays were performed on a subset of samples with a balanced distribution across strata. The percent of replicates performed were 4·3% for the SiMoA platform and 5·7% for the Roche platform – these numbers were selected to fit within the assaying logistics and assay work flow for the full assay runs on the respective platforms.
Clinical data, including variables used in clinical decision rules (CDRs) for triaging CT scanning (Supplementary Table 1), were collected using a web-based electronic case report form (eCRF), with variables coded in accordance with the Common Data Elements (CDE) scheme (https://commondataelements.ninds.nih.gov/).
All patients underwent head CT examinations according to local protocols. Imaging studies were transmitted to a central repository (Icometrix, Leuven) for structured reporting according to the NINDS TBI-CDEs. Central reviewers were blinded to clinical information, except gender, age, and care path. The presence of any traumatic intracranial abnormality on CT was considered a positive scan. Skull fractures in isolation were not considered as intracranial abnormality.
MR scans performed according to study protocol were obtained in a subgroup of 502 patients. We report on 152 of these patients who had a negative CT scan on presentation.
2.3. Statistical analysis
Baseline characteristics were summarized using standard descriptive statistics. Continuous variables are presented as median (interquartile range) and categorical variables as absolute frequencies and percentages. Bland–Altman plots were made for replicate assays. The coefficient of variation was calculated on the log transformed values and expressed as percentage.
Relations of biomarkers to clinical severity (GCS) and care path were displayed in tabular and graphical formats. Correlations between biomarkers were visualized by scatterplots and quantified using Spearman's rank correlation coefficients. Distributions of biomarker levels for patients with intracranial abnormalities were compared to those without abnormalities with Mann–Whitney U tests. We explored adjusting for multiple testing (Bonferroni, false discovery rate), but only reported these results when they changed the interpretation of the results.
Multiple imputation of missing characteristics was performed using the mice package [17], assuming a missing at random mechanism. Clinical characteristics with >50% completion, CT positivity and biomarkers were included in the imputation model. Predictive mean matching was used for continuous data, logistic regression for binary data, and polytomous regression for categorical data. Fifty imputed datasets were created, with results summarized according to Rubin's rules [18]. The diagnostic performance of biomarkers, separately and in combination to identify patients with positive CT findings was assessed with logistic regression. We allowed for non-linear effects of log transformed biomarkers using restricted cubic splines with 3 degrees of freedom. From these models, we derived estimates of the area under the ROC curves (AUC, or c-statistic). A bootstrap resampling procedure with 200 repetitions was applied to calculate 95% confidence intervals (CIs). Univariable and multivariable analysis, adjusting for clinical characteristics, was performed.
The clinical characteristics derived from CDRs were included in the multivariable analysis as they are presented in Supplementary Table 2. Continuous variables were included without categorization to fully capture the diagnostic information they contain. In case variables were non-informative (GCS in the GCS 15 sensitivity analysis, or depressed skull fracture in the ER stratum), they were excluded from the model.
The performance of biomarkers compared to clinical characteristics in the univariable analysis was explored by a bootstrapping procedure that included 1000 repetitions. The percentage of repetitions where a univariable model with the biomarker outperformed (higher c-statistic) a multivariable model with all clinical characteristics used in current CDRs was calculated.
Predictions based on clinical characteristics were compared to predictions with biomarkers added to the clinical variables and results visualized using reclassification plots [19].
Sensitivity analyses were carried out on patients with GCS 13–14, in those with GCS 15, with major extracranial injuries (defined as AIS>=3), and according to stratum (ER, admission, and ICU). In addition, AUCs were generated for samples collected at different times post-injury (in 6-h intervals) to explore possible influence of sampling time. Statistical analysis was performed using R (http://www.r-project.org, version 3.5.1) in RStudio (http://www.rstudio.com, version 1.1.456).
3. Results
3.1. Patient cohort and sampling
Data on 2867/4509 (64%) patients analyzed in the CENTER-TBI Core study were available for analysis of biomarkers in serum samples obtained within 24 h of injury (Fig. 1). The time between injury and sampling was shortest in the ER stratum (5·1 h; IQR [3·4–9·73]), in contrast to the admission (15·7 h; IQR [9·8–20·2]) and ICU (14·3 h; IQR [7·5–19·7]) strata (Supplementary Fig. 1). The median needle to freezer times was 1·08 h (IQR [0·92–1·33]), with no substantial differences across strata. Agreement between replicates of biomarker assessments was good for the clinical platform assays of S100B and NSE (CV: 7% for both on a log transformed scale), but poorer for the research-use only (RUO) assays of GFAP, UCH-L1, NFL and t-tau (CV: 22-30%) (Supplementary Fig. 2).
Clinical characteristics of the study cohort, differentiated by stratum, are summarized in Table 1 and the frequency of specific characteristics, contained in CDRs for predicting CT abnormalities in patients with mild TBI, presented in Supplementary Table 2. Characteristics of patients excluded (n=1642; see Fig. 1) were largely similar to those analyzed (n=2867), although the median GCS was lower (14 vs 15) and the percentage of non-reacting pupils higher (9·0% vs 5·3%) (Supplementary Table 3).
Table 1.
Characteristics of biomarker cohort (n=2867) in the CENTER-TBI core study.
| N complete | Overall (n=2867) | ER (n=636) | Admission (n=900) | ICU (n=1331) | |
|---|---|---|---|---|---|
| Demographic characteristics | |||||
| Age (median [IQR]) | 2867 | 49 [30, 66] | 48 [30, 65] | 53 [33, 68] | 48 [30, 64] |
| >65 years (%) | 735 (25·6) | 153 (24·1) | 266 (29·6) | 316 (23·7) | |
| Male sex (%) | 2867 | 1948 (67·9) | 354 (55·7) | 619 (68·8) | 975 (73·3) |
| Cause of injury | |||||
| Cause of injury | 2711 | ||||
| Road traffic incident (%) | 1098 (38·3) | 204 (32·1) | 295 (32·8) | 599 (45·0) | |
| Incidental fall (%) | 1264 (44·1) | 317 (49·8) | 436 (48·4) | 511 (38·4) | |
| Clinical presentation | |||||
| GCS baseline (median [IQR]) | 2775 | 15 [10, 15] | 15 [15, 15] | 15 [14, 15] | 10 [4, 14] |
| Severe (3–8) (%) | 601 (21·0) | 1 (0·2) | 7 (0·8) | 593 (44·6) | |
| Moderate (9–12) (%) | 222 (7·7) | 2 (0·3) | 28 (3·1) | 192 (14·4) | |
| Mild (13–14) (%) | 457 (15·9) | 38 (6·0) | 202 (22·4) | 217 (16·3) | |
| Mild (15) (%) | 1494 (52·1) | 589 (92·6) | 643 (71·4) | 262 (19·7) | |
| Pupillary reactivity | 2732 | ||||
| One pupil unreactive (%) | 97 (3·4) | 2 (0·3) | 14 (1·6) | 81 (6·1) | |
| Two pupils unreactive (%) | 144 (5·0) | 7 (1·1) | 4 (0·4) | 133 (10·0) | |
| Hypoxia (prehospital/ER) (%) | 2709 | 184 (6·4) | 1 (0·2) | 15 (1·7) | 168 (12·6) |
| Hypotension (prehospital/ER) (%) | 2735 | 177 (6·2) | 3 (0·5) | 12 (1·3) | 162 (12·2) |
| Any major extracranial injury (AIS >=3) (%) | 2867 | 1032 (36·0) | 22 (3·5) | 262 (29·1) | 748 (56·2) |
| CT characteristics | |||||
| Any intracranial abnormality at central reading (%) | 2867 | 1705 (59·5) | 86 (13·5) | 436 (48·4) | 1183 (88·9) |
| Biomarker (median [IQR]) | |||||
| S100B, µg/L | 2861 | 0·15 [0·08, 0·33] | 0·09 [0·05, 0·15] | 0·09 [0·06, 0·16] | 0·28 [0·16, 0·58] |
| NSE, ng/ml | 2858 | 17·08 [12·49, 25·85] | 14·02 [11·14, 18·09] | 14·22 [11·18, 19·39] | 23·14 [16·24, 34·10] |
| GFAP, ng/ml | 2850 | 3·14 [0·53, 15·07] | 0·30 [0·11, 0·94] | 1·51 [0·39, 5·28] | 12·92 [4·22, 34·92] |
| UCH-L1, pg/ml | 2846 | 94·74 [35·54, 307·12] | 35·48 [17·63, 63·34] | 51·44 [24·92, 109·66] | 274·35 [119·23, 622·66] |
| Tau, pg/ml | 2851 | 2·79 [1·23, 7·67] | 1·16 [0·71, 1·84] | 1·81 [1·05, 3·45] | 7·08 [3·21, 16·61] |
| NFL, pg/ml | 2849 | 18·55 [8·40, 49·74] | 7·90 [5·09, 13·29] | 12·88 [7·15, 24·55] | 42·88 [19·49, 104·59] |
3.2. Biomarker values by stratum and clinical severity
The median values of the six biomarkers displayed a clear association with injury severity (classified according to the GCS) and care path (Table 1, and Supplementary Table 4). Within the group of mild TBI (GCS 13–15), median values were higher in patients with a GCS of 13–14 compared to 15.
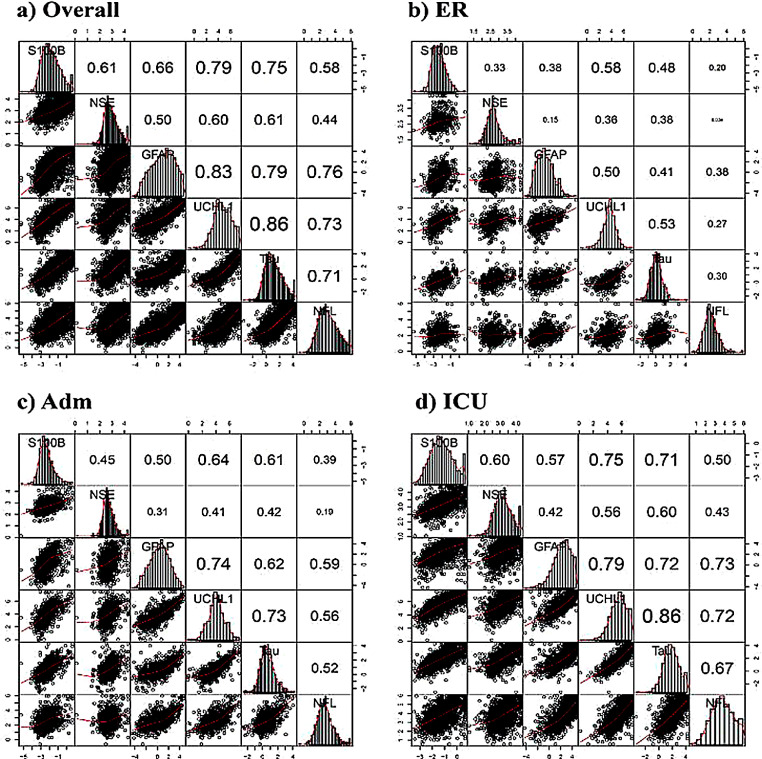
All biomarkers showed correlations across all strata, except for NSE in the ER stratum with NFL and GFAP (Fig. 2). Correlations were strongest in the ICU stratum, likely reflecting greater differences in case-mix. The strongest correlation was found between UCH-L1 (neuronal marker) and t-tau (axonal marker) varying from 0·53 (ER) to 0·86 (ICU). The correlation between GFAP and S100B, both glial markers, was relatively weak, varying between strata from 0·38 (ER) to 0·57 (ICU).
Fig. 2.
Correlation plots displaying associations between biomarkers in each stratum. The diagonal part with the name of the biomarker contains the distribution plot specific for the log-transformed biomarker. Scatter plots of correlations between biomarkers are presented below the diagonal, and Spearman correlation coefficients above the diagonal. The font size is indicative of the strength of correlation.
3.3. Biomarkers and traumatic intracranial CT abnormalities
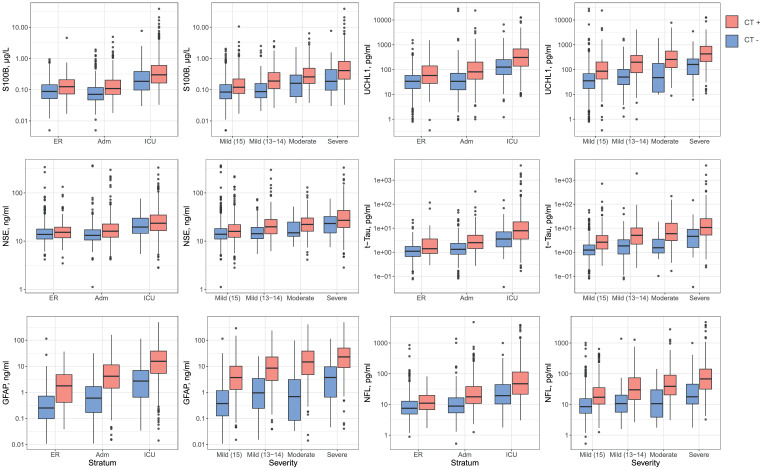
Biomarker levels were higher in patients with traumatic abnormalities on CT scanning compared to those without (Fig. 3 and Supplementary Table 5). Differences in biomarker levels between CT+ and CT- patients were greater when analyzed by clinical severity (GCS) than by care path. Differences were most pronounced for GFAP.
Fig. 3.
Biomarker values by stratum and by clinical severity, differentiated for the absence (blue) or presence (pink) of traumatic intracranial CT abnormalities.P-values of the Mann–Whitney U tests are presented in the Supplemental material, Table 11.
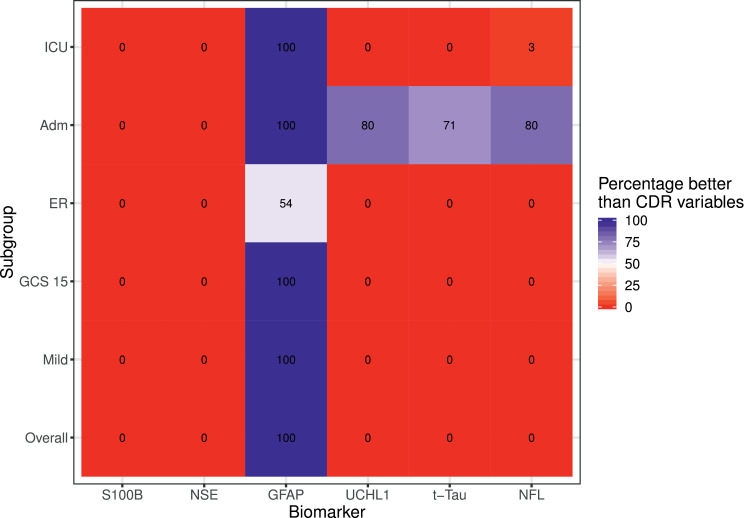
Univariable analysis confirmed that GFAP had the highest discrimination for predicting the presence of CT abnormalities (AUC 0·89 [95%CI: 0.87–0·90]), and performed as well, and even better in the admission stratum, as clinical characteristics (Supplementary Fig. 3 and Supplementary Table 6). Most other biomarkers showed substantial discrimination, but performed poorer than clinical characteristics. This finding was confirmed in the bootstrap analysis: the chance that GFAP outperformed clinical characteristics was >99% in all subgroups, except for the ER stratum (Fig. 4). In the admission stratum, UCH-L1, NFL and t-tau outperformed clinical characteristics in less than 95% of the bootstrap samples. Combining all biomarkers showed slightly higher discrimination compared to GFAP alone. We found no clear benefit of any other combination of biomarkers, including the combination of GFAP and UCH-L1 (Supplementary Fig. 4 and Supplementary Table 7).
Fig. 4.
Heat map demonstrating the discriminative ability of single biomarkers in comparison to a regression model that includes clinical characteristics contained in CT decision rules. The heat map summarizes the percentage of bootstrap replicates in which the model with the biomarker outperforms (higher c-statistic) the model with CT decision rule variables. The lower number of positive replicates for GFAP in the ER stratum may be due to lower number of events in this stratum (86/636 CT positive).
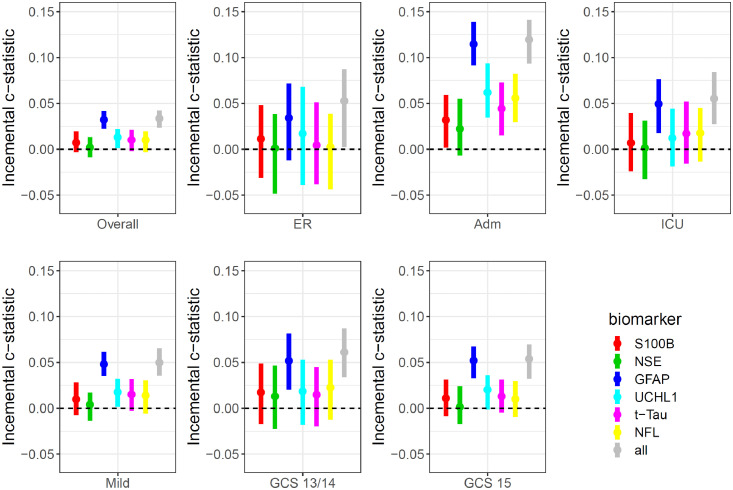
Multivariable analysis adjusted for clinical characteristics incorporated in CT rules confirmed that GFAP provided incremental discriminative ability over clinical characteristics (increase from 0·89 [95%CI: 0·88–0·90] to 0·92 [95%CI: 0·91–0·93] when GFAP was included; Fig. 5 and Supplementary Table 8). The incremental value was most pronounced in the admission stratum (increase in AUC from 0·72 [95%CI: 0·69–0·75] to 0·84 [95%CI: 0·81–0·86]), and was consistent in patients with a GCS of 15 (increase in AUC from 0·83 [95%CI: 0·80–0·85] to 0·88 [95%CI: 0·86–0·89]), and in those with a GCS of 13–14 (increase in AUC from 0·84 [95%CI: 0·80–0·88] to 0·90 [95%CI: 0·86–0·92]). Combinations of biomarkers showed no clear increase in discrimination compared to GFAP alone on multivariable analysis (Supplementary Fig. 5 and Supplementary Table 9).
Fig. 5.
Incremental discriminative ability of biomarkers to predict CT positivity. Plots show the difference in Area under the ROC curve (AUC) with 95% confidence intervals (bars) of logistic regression models combining age, time interval (injury to needle time) as well as clinical parameters included in current CT rules with and without biomarkers. Panels are presented for the overall sample (n=2867), according to stratum (ER, Admission, ICU), and for the mild (GCS 13–15; GCS 13–14; GCS 15) groups. Six biomarkers are considered separately and in combination (“all”). The dotted line indicates the predictive value of the clinical parameters and serves as a reference. The absolute values are presented in the Supplementary material, Table S7.
While profound differences in the performance of biomarkers were not observed across different time intervals within 24 h of injury, GFAP levels displayed the greatest incremental predictive power when measured 12–18 h post-injury. (Supplementary Fig. 6).
The incremental value of biomarkers was similar across patients with major extracranial injury and those without major extracranial injury (Supplementary Fig. 7).
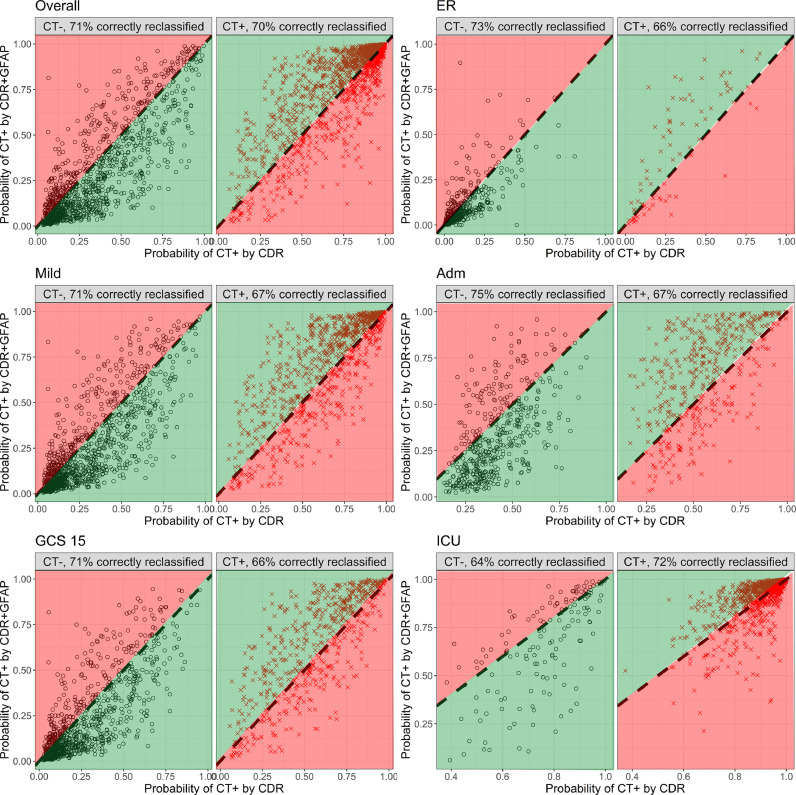
Reclassification plots confirmed the incremental value of GFAP across all strata compared to clinical characteristics (Fig. 6). Adding GFAP correctly reclassified 71% of patients with a negative CT scan, and 70% of the patients with a positive CT scan. The plots show that when added to the decision rule, GFAP levels often provide higher risk estimates to patients who had lower predicted risks by clinical characteristics. The same pattern, and similar extent of correct reclassification was seen when all biomarkers were combined (Supplementary Fig. 8).
Fig. 6.
Reclassification plots illustrating superior performance of GFAP compared to clinical characteristics in predicting traumatic intracranial CT abnormalities. GFAP is added to a logistic regression model that includes clinical parameters from current CT rules. The model with GFAP (y-axis), is compared to the model without GFAP (x-axis). Panels are presented for the overall sample (n=2867), for the strata (ER, Admission, ICU), and for the mild (GCS 13–15), and GCS 15 subgroups. Black dots represent patients with a negative CT scan, red crosses represent patients with a positive CT scan. The percentage of correct reclassification, indicated with green (higher probability of CT positivity if CT is positive, and lower probability of CT positivity if CT is negative), is displayed in the top of each plot.
We further explored the discriminative value of biomarkers for predicting abnormalities on MR imaging performed within 3 weeks of injury in a subgroup of 152 patients who had a negative CT scan on presentation and underwent subsequent MR imaging. Traumatic MR abnormalities were found in 44/152 cases. The estimates were uncertain, but GFAP showed the highest discriminative ability (c-statistic: 0·76, 95% CI: 0·67–0·85, Supplementary Fig. 9 and Supplementary Table 10).
4. Discussion
We studied the relation of serum biomarkers to clinical severity, care path and the presence of traumatic intracranial abnormalities on CT scanning. We found that all biomarkers studied scaled with injury severity, classified according to the GCS and with care path intensity. Within each of these stratifications, biomarkers were higher in patients with abnormalities on head CT imaging compared to those without. Serum GFAP levels in the first 24 h post-injury were highly predictive for CT positivity, outperforming other markers and adding value to clinical variables considered in contemporary CT decision rules. Combining results of all biomarkers did not clearly improve discrimination compared to GFAP alone.
To the best of our knowledge, this is the first large scale study across all injury severities, evaluating systematically a panel of 6 biomarkers, and quantifying their performance relative to clinical characteristics in predicting CT abnormalities in patients with mild TBI. Recently, Thelin et al. reported on the same panel of biomarkers in a cohort of 172 patients with TBI [20]. The focus of this study was, however, on prognosis. Other previous studies have shown relations of biomarkers to clinical severity and to the presence of CT abnormalities 21, 22, 23, 24. Most, however, mainly focused on one or two biomarkers and have seldom addressed performance of biomarkers relative to clinical characteristics. Moreover, a living systematic review identified serious problems in the design, analysis and reporting of many of the studies [8]. Until recently, the strongest evidence for a role in triaging CT scanning existed for S100B. This marker is incorporated in the Scandinavian guidelines for the management of moderate to minimal TBI [11]. Previously, a study including 397 patients with general trauma, of whom 209 had mild TBI, reported that GFAP performed better than S100B, in particular in patients with extracranial injuries [21]. Our results expand these earlier data, convincingly showing that GFAP outperforms S100B both in the overall group of mild TBI and in the subset of patients with a GCS of 15. However, we believe that a decision to replace S100B by GFAP in such guidelines may be premature. Assays for S100B are commercially available with high reproducibility. Currently, no GFAP assays are available as a commercialized clinical assay platform. Indeed, the platform we used for analysis of GFAP is a research-use-only (RUO) platform, and our replication assays showed substantial variation (28% on a log transformed scale). Factors that may have contributed to poor reproducibility include: not fully automated analyses, and that replicate samples had undergone a second freeze-thaw cycle. There is no evidence that refrigerated storage of samples for up to 72 h has a significant effect on GFAP values [25], but a decrease of GFAP levels has been reported in CSF after two freeze-thaw cycles [26].
The clear effects of GFAP and other biomarkers in the presence of assay heterogeneity speaks to the robustness of our findings. The role of biomarkers goes beyond triage for CT scanning in patients with mild TBI. Our results in this specific subgroup confirm the potential of GFAP to predict presence of MRI abnormalities in patients with normal CT findings after TBI [2].
4.1. Assay reproducibility and thresholds
Reliability and reproducibility of biomarker assays are fundamental for clinical implementation. Absolute GFAP levels in our study were much higher than those reported in the ALERT-TBI study, which used a different platform [13]. Whilst different reference values between platforms may be acceptable, insight into comparability of values obtained with different platforms is desirable. Variation between platforms also precludes the concept of determining a universal cut-off value. Moreover, cut-off values are generally derived from reference values, obtained from healthy controls, whilst “action thresholds” are needed in diseased patients, which may be very different from reference values [27]. Further, it should be recognized that any biomarker represents a continuous variable and that use of a threshold value leads to loss of information. We suggest that biomarkers may be combined with clinical characteristics for risk estimation, and then continuous values may be retained. We have hence refrained from suggesting threshold values, a decision which was reinforced by the variability in replicate assays.
4.2. GFAP versus a multi-marker approach
Contrary to our expectations, a multi-marker approach applying combinations of biomarkers did not increase the diagnostic value for CT positivity, compared to GFAP alone. GFAP showed similar discrimination as all biomarkers combined when analyzed versus clinical severity and care path. Nevertheless, these observations do not preclude potential usefulness of combinational approaches in terms of outcome prediction and/or tracking the disease process over time. Conceptually, different biomarkers should differentially reflect specific aspects of the disease process of TBI [21,28]. However, in this study we did not find any benefit of combining acute GFAP levels with UCH-L1 levels, a combination of (presumed) glial and neuronal markers used in the recent ALERT-TBI trial, which provided the basis for a recent FDA marketing authorization [9,13]. Our results provide no support for implementation of this combined assay into clinical practice. We do note, however, that a key difference between our study and ALERT-TBI is that the time window for blood sampling was 24 h in our study and 12 h in ALERT-TBI. This may be relevant as the half-life for UCH-L1 is short [29]. However, sensitivity analysis differentiated for time of sampling (Supplementary Fig. 6) did not show superior performance of UCH-L1 in the first six hours after trauma. The finding that a single marker approach (GFAP) may be sufficient in the acute phase to inform diagnosis and care path is of particular relevance for low and middle income countries (LMICs) and other austere environments, where even basic imaging is inaccessible, or too expensive and unevenly distributed, with limited opportunities for patient transfer.
4.3. Strengths and limitations
Strengths of our study include the large number of patients, analyses across all severities of TBI, the use of a comprehensive panel of biomarkers that addresses current clinical interest and the focus on the incremental value of biomarkers in predicting CT positivity compared to clinical characteristics used in CDRs. These strengths support the generalizability of our findings, obtained in the “real-world” situation of an observational study.
Several limitations of our study should be acknowledged:
First, our study should be considered as an exploratory diagnostic accuracy study [27], and was not designed to seek regulatory approval.
Second, we were able to analyze samples from 2867/4509 (64%) patients available in the CENTER-TBI database. Although baseline characteristics of the study cohort were very similar to those reported for the Core study (n=4509) [15], we here included slightly less severe patients.
Third, we utilized a research-use only (RUO) platform for assays of four biomarkers (GFAP, UCH-L1, NFL and t-tau), and coefficients of variation in replicate samples were relatively high.
Fourth, the inclusion criteria for CENTER-TBI included the intent to perform a CT scan. As a consequence, the patient population may have been biased towards inclusion of more patients with CT abnormalities. However, overall 40% of patients were CT negative (86% in ER, 52% in Admission and 11% in the ICU stratum).
Fifth, the reported interpretation of results is only valid for the biomarkers studied and cannot be extrapolated to other biomarkers. CENTER-TBI has prepared for facilitating legacy research on other markers or on clinical-use platform(s) by reserving a number of pristine aliquots for future studies.
Sixth, the permitted time window of 24 h may have affected the diagnostic accuracy of biomarkers with short circulating half-lives. Understanding the kinetics of such biomarkers may [30,31] inform optimization of time windows for improving diagnostic performance.
Seventh, we did not explore possible gender effects.
Eighth, we did not take the clustered structure of the data into account, because it was statistically not feasible to adjust both for clinical characteristics as well for between center variations.
Finally, as explained above, we deliberately refrained from attempting to identify action thresholds (cutoff values) in a post-hoc analysis.
In conclusion, each of the six investigated biomarkers scaled with the severity of TBI and with care path. GFAP serum levels obtained within 24 h post-injury predict CT positivity across the full range of injury severities. In patients with mild TBI and in patients with a GCS of 15, GFAP adds value to clinical characteristics and outperforms other markers, including S100B. No clear additional value for predicting CT positivity was found when combining GFAP with other biomarkers. Our evidence supports development of novel CT decision rules, combining serum GFAP with clinical characteristics, for triaging patients with mild TBI for CT scanning. To this purpose, validated clinical-use assays are required.
Declaration of Competing Interest
All authors declare support to the CENTER-TBI project by funding bodies and other organizations as listed in the acknowledgement section.
DKM reports grants from National Institute for Health Research (NIHR; UK), during the conduct of the study; grants, personal fees and non-financial support from GlaxoSmithKline, personal fees from Neurotrauma Sciences, personal fees from Lantmaanen AB, personal fees from Pressura, personal fees from Pfizer, outside the submitted work. AIRM declares consulting fees from PresSura Neuro, Integra Life Sciences and NeuroTrauma Sciences. EC, KA and AB report grants Higher Education Institutional Excellence Programme – Grant No. 20765-3/2018/FEKUTSTRAT, FIKP II/S, EFOP-3.6.2.-16-2017-00008, GINOP-2.3.2-15-2016-00048, and GINOP-2.3.3-15-2016-00032 and the Hungarian Brain Research Program 2.0 Grant No. 2017-1.2.1-NKP-2017-00002. KKWW is co-founder and shareholder of Gryphon Bio. and was co-founder and shareholder of Banyan Biomarkers. VFJN is supported by an Academy of Medical Sciences/The Health Foundation Clinician Scientist Fellowship. SR reports funding from the Wellcome Trust for a Clinician Ph.D. Fellowship.
BYG, FL, SM, EWS, JV, THvdV, HX, and ZY declare no competing interests.
Acknowledgments
Acknowledgments
CENTER-TBI study supported by the European Union 7th Framework program (EC grant 602150). Additional funding was obtained from the Hannelore Kohl Stiftung (Germany), from OneMind (USA), from Integra LifeSciences Corporation (USA) and from Neurotrauma Sciences (USA).
Role of funding statement
The funders had no role in the study design, collection, analysis and interpretation of data, nor in the writing of the report or in publication decisions. All authors had full access to the study data and the senior authors had final responsibility for the decision to submit for publication.
Contributors’ statement
All persons who meet authorship criteria are listed as authors, and all authors certify that they have participated in the concept, design, analysis, writing, or revision of the manuscript. All authors participated in the reported analyses and interpretation of results relevant to their domain of interest. EC, KA, AB, SM, AIRM and DKM prepared the draft manuscript and coordinated its finalization. EC, BYG and EWS performed the data extraction, statistical analyses and drafting of tables and figures. HX and ZY conducted biomarker assay (UCH-L1-GFAP-t-tau-NFL) analysis, HX, ZY and KKWW supervised data extraction and curation. SM and KA provided quality control of analytical methods. ThvdV and JV, FL, VFJN, SR performed the structured reporting and analyses of neuro-images. All authors approved the final manuscript.
Data sharing statement
Will individual participant data be available?
Yes, including data dictionary.
What other documents will be available?
Study protocol, analytic code and analysis scripts.
When will data be available?
Immediately following publication, conditional to approved study proposal; no end date.
With whom?
Researchers who provide a methodologically sound study proposal that is approved by the management committee.
For what type of analyses?
To achieve the aims in the approved proposal.
By what mechanism will data be made available?
Proposals may be submitted online https://www.center-tbi.eu/data. A Data Access Agreement is required, and all access must comply with regulatory restrictions imposed on the original study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102785.
Appendix. Supplementary materials
References
- 1.Maas A.I.R., Menon D.K., Adelson P.D. Traumatic brain injury: integrated approaches to improve prevention, clinical care, and research. Lancet Neurol. 2017;16(12):987–1048. doi: 10.1016/S1474-4422(17)30371-X. [DOI] [PubMed] [Google Scholar]
- 2.Yue J.K., Yuh E.L., Korley F.K. Association between plasma GFAP concentrations and MRI abnormalities in patients with CT-negative traumatic brain injury in the TRACK-TBI cohort: a prospective multicentre study. Lancet Neurol. 2019;18(10):953–961. doi: 10.1016/S1474-4422(19)30282-0. [DOI] [PubMed] [Google Scholar]
- 3.Stiell I.G., Wells G.A., Vandemheen K. The Canadian CT head rule for patients with minor head injury. Lancet. 2001;357(9266):1391–1396. doi: 10.1016/s0140-6736(00)04561-x. [DOI] [PubMed] [Google Scholar]
- 4.Foks K.A., van den Brand C.L., Lingsma H.F. External validation of computed tomography decision rules for minor head injury: prospective, multicentre cohort study in the Netherlands. BMJ. 2018;362:k3527. doi: 10.1136/bmj.k3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National_Clinical_Guideline_C._National_Clinical_Guidance_Centre. CG 176 . Vol. 2014. National Institute for Health and Care Excellence; 2014. (Head injury: triage, assessment, investigation and early management of head injury in children, young people and adults). [PubMed] [Google Scholar]
- 6.Smits M., Dippel D.W., Steyerberg E.W. Predicting intracranial traumatic findings on computed tomography in patients with minor head injury: the CHIP prediction rule. Ann Intern Med. 2007;146(6):397–405. doi: 10.7326/0003-4819-146-6-200703200-00004. [DOI] [PubMed] [Google Scholar]
- 7.Haydel M.J., Preston C.A., Mills T.J., Luber S., Blaudeau E., DeBlieux P.M.C. Indications for computed tomography in patients with minor head injury. N Engl J Med. 2000;343(2):100–105. doi: 10.1056/NEJM200007133430204. [DOI] [PubMed] [Google Scholar]
- 8.Mondello S., Sorinola A., Czeiter E. Blood-based protein biomarkers for the management of traumatic brain injuries in adults presenting to emergency departments with mild brain injury: a living systematic review and meta-analysis. J Neurotrauma. 2020 doi: 10.1089/neu.2017.5182. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.US_Food_and_Drug_Administration. Evaluation of automatic class III designation for Banyan Brain Trauma Indicator. https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN170045.pdf. 2019.
- 10.Mondello S., Hayes R.L. Biomarkers. Handb Clin Neurol. 2015;127:245–265. doi: 10.1016/B978-0-444-52892-6.00016-7. [DOI] [PubMed] [Google Scholar]
- 11.Unden J., Ingebrigtsen T., Romner B., Scandinavian Neurotrauma C. Scandinavian guidelines for initial management of minimal, mild and moderate head injuries in adults: an evidence and consensus-based update. BMC Med. 2013;11:50. doi: 10.1186/1741-7015-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minkkinen M., Iverson G.L., Kotilainen A.K. Prospective validation of the scandinavian guidelines for initial management of minimal, mild, and moderate head injuries in adults. J Neurotrauma. 2019;36(20):2904–2912. doi: 10.1089/neu.2018.6351. [DOI] [PubMed] [Google Scholar]
- 13.Bazarian J.J., Biberthaler P., Welch R.D. Serum GFAP and UCH-L1 for prediction of absence of intracranial injuries on head CT (ALERT-TBI): a multicentre observational study. Lancet Neurol. 2018;17(9):782–789. doi: 10.1016/S1474-4422(18)30231-X. [DOI] [PubMed] [Google Scholar]
- 14.Maas A.I.R., Lingsma H.F. ALERT-TBI study on biomarkers for TBI: has science suffered? Lancet Neurol. 2018;17(9):737–738. doi: 10.1016/S1474-4422(18)30275-8. [DOI] [PubMed] [Google Scholar]
- 15.Maas A.I., Menon D.K., Steyerberg E.W. Collaborative European NeuroTrauma Effectiveness Research in Traumatic Brain Injury (CENTER-TBI): a prospective longitudinal observational study. Neurosurgery. 2015;76(1):67–80. doi: 10.1227/NEU.0000000000000575. [DOI] [PubMed] [Google Scholar]
- 16.Steyerberg E.W., Wiegers E., Sewalt C. Case-mix, care pathways, and outcomes in patients with traumatic brain injury in CENTER-TBI: a European prospective, multicentre, longitudinal, cohort study. Lancet Neurol. 2019;18(10):923–934. doi: 10.1016/S1474-4422(19)30232-7. [DOI] [PubMed] [Google Scholar]
- 17.van Buuren S. CRC Press, Taylor & Francis Group; 2018. Flexible imputation of missing data. [Google Scholar]
- 18.Rubin D.B. Wiley; 2004. Multiple imputation for nonresponse in surveys. [Google Scholar]
- 19.Steyerberg E.W., Vedder M.M., Leening M.J. Graphical assessment of incremental value of novel markers in prediction models: From statistical to decision analytical perspectives. Biom J. 2015;57(4):556–570. doi: 10.1002/bimj.201300260. [DOI] [PubMed] [Google Scholar]
- 20.Thelin E., Al Nimer F., Frostell A. A serum protein biomarker panel improves outcome prediction in human traumatic brain injury. J Neurotrauma. 2019;36(20):2850–2862. doi: 10.1089/neu.2019.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papa L., Silvestri S., Brophy G.M. GFAP out-performs S100β in detecting traumatic intracranial lesions on computed tomography in trauma patients with mild traumatic brain injury and those with extracranial lesions. J Neurotrauma. 2014;31(22):1815–1822. doi: 10.1089/neu.2013.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czeiter E., Mondello S., Kovacs N. Brain injury biomarkers may improve the predictive power of the IMPACT outcome calculator. J Neurotrauma. 2012;29(9):1770–1778. doi: 10.1089/neu.2011.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mondello S., Linnet A., Buki A. Clinical utility of serum levels of ubiquitin C-terminal hydrolase as a biomarker for severe traumatic brain injury. Neurosurgery. 2012;70(3):666–675. doi: 10.1227/NEU.0b013e318236a809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon P.J., Panczykowski D.M., Yue J.K. Measurement of the glial fibrillary acidic protein and its breakdown products GFAP-BDP biomarker for the detection of traumatic brain injury compared to computed tomography and magnetic resonance imaging. J Neurotrauma. 2015;32(8):527–533. doi: 10.1089/neu.2014.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rezaii P.G., Grant G.A., Zeineh M.M. Stability of Blood biomarkers of traumatic brain injury. J Neurotrauma. 2019;36(16):2407–2416. doi: 10.1089/neu.2018.6053. [DOI] [PubMed] [Google Scholar]
- 26.Abdelhak A., Hottenrott T., Morenas-Rodríguez E. Glial activation markers in CSF and serum from patients with primary progressive multiple sclerosis: potential of serum GFAP as disease severity marker? Front Neurol. 2019;10:280. doi: 10.3389/fneur.2019.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bossuyt P.M., Olsen M., Hyde C., Cohen J.F. An analysis reveals differences between pragmatic and explanatory diagnostic accuracy studies. J Clin Epidemiol. 2020;117:29–35. doi: 10.1016/j.jclinepi.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Wang K.K., Yang Z., Zhu T. An update on diagnostic and prognostic biomarkers for traumatic brain injury. Exp Rev Mol Diagn. 2018;18(2):165–180. doi: 10.1080/14737159.2018.1428089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papa L., Brophy G.M., Welch R.D. Time course and diagnostic accuracy of glial and neuronal blood biomarkers GFAP and UCH-L1 in a large cohort of trauma patients with and without mild traumatic brain injury. JAMA Neurol. 2016;73(5):551–560. doi: 10.1001/jamaneurol.2016.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brophy G.M., Mondello S., Papa L. Biokinetic analysis of ubiquitin C-terminal hydrolase-L1 (UCH-L1) in severe traumatic brain injury patient biofluids. J Neurotrauma. 2011;28(6):861–870. doi: 10.1089/neu.2010.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch R.D., Ellis M., Lewis L.M. Modeling the kinetics of serum glial fibrillary acidic protein, ubiquitin carboxyl-terminal Hydrolase-L1, and S100B concentrations in patients with traumatic brain injury. J Neurotrauma. 2017;34(11):1957–1971. doi: 10.1089/neu.2016.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.