Abstract
Liver cancer is the sixth most common cancer worldwide, and the third most common cause of cancer-related death. Hepatocellular carcinoma (HCC), which accounts for more than 90% of primary liver cancers, is an important public health problem. In addition to cirrhosis caused by hepatitis B viral (HBV) or hepatitis C viral (HCV) infection, non-alcoholic fatty liver disease (NAFLD) is becoming a major risk factor for liver cancer because of the prevalence of obesity. Non-alcoholic steatohepatitis (NASH) will likely become the leading indication for liver transplantation in the future. It is well recognized that gut microbiota is a key environmental factor in the pathogenesis of liver disease and cancer. The interplay between gut microbiota and liver disease has been investigated in animal and clinical studies. In this article, we summarize the roles of gut microbiota in the development of liver disease as well as gut microbiota-targeted therapies.
Keywords: Microorganism, Hepatocellular carcinoma (HCC), Non-alcoholic fatty liver disease (NAFLD), Non-alocholic steatohepatitis (NASH), Cirrhosis, Probiotics, Prebiotics, Synbiotics
1. Introduction
There are about 100 trillion (1014) microorganisms and approximately 2000 different bacterial species in the human digestive tract.1 The gut microbiota colonizes immediately after birth and plays an essential role in keeping the host healthy by assisting digestion, producing vitamins, generating bile acids, and modulating local and systemic immunity.2–5 Many factors, including diet, age, medication, illness, stress, and lifestyle, influence the gut microbiota community structure, which has an impact on disease development.6 It is important to note that genetic factors only contribute to 5–15% of most cancers. About 80% of cancers are caused by the environment or lifestyle.7 Emerging evidence reveals that the gut microbiota is a major environmental and etiological factor for liver disease development.8–12 In this review, we summarize publications on the topics of gut microbiota in liver disease development, as well as treatment, focusing on non-alcoholic fatty liver disease (NAFLD), non-alcoholic steatohepatitis (NASH), cirrhosis, and hepatocellular carcinoma (HCC).
We performed a literature search in PubMed for papers published within the past 10 years, using the keywords: microorganism, microbiota, bacteria, liver, liver disease, HCC, hepatocellular carcinogenesis, NAFLD, NASH, cirrhosis, probiotics, prebiotics, synbiotics, and their combinations.
2. Role of gut microbiota in liver diseases
2.1. Gut microbiota and NAFLD as well as NASH
NAFLD is a global public health problem because of the prevalence of obesity.13 NAFLD is a spectrum of chronic liver diseases, including simple steatosis, NASH, advanced fibrosis, cirrhosis, and HCC.6 Dysbiosis refers to unfavorable alteration of the microbiota. It is commonly characterized with a decreased ratio of autochthonous to nonautochthonous taxa. Increasing evidence indicates that gut dysbiosis has an important role in the development of NASH via regulation of inflammation, insulin resistance, bile acids, and choline metabolism.4,14–16
Data generated from human studies have established the relationship between gut microbiota and NAFLD (Table 1).14,17–24 In, 2013, Mouzaki et al.17 reported that NASH patients have significantly lower levels of Bacteroidetes compared to healthy individuals. Shen et al.18 showed similar findings. However, Wang et al.19 found that non-obese patients with NAFLD have significantly higher levels of Bacteroidetes and lower abundance of Firmicutes in addition to reduced diversity. In these non-obese patients with NAFLD, the depletion of Firmicutes included Lachnospiraceae, Ruminococcaceae, and Lactobacillaceae, which generated short-chain fatty acids (SCFAs).19 Additionally, a different Bacteroidetes abundance pattern in adolescents was revealed in a study conducted by Stanislawski et al.20 The abundance of Bacteroides showed a U-shaped pattern based on hepatic fat; both low and high abundances were associated with elevated hepatic fat, while a moderate level was associated with reduced hepatic fat.20 In addition, Bacteroides is associated with a high-fat diet (HFD).25 However, certain species of Bacteroides have protective roles in obesity.26
Table 1.
Gut microbiota alteration in NAFLD and NASH patients.
| Authors | Population | N | Comparison | Implicated microbiota |
Methodology | ||
|---|---|---|---|---|---|---|---|
| Phylum | Family | Genus | |||||
| Boursier et al.14 | F0/F1 fibrosis without NASH F0/F1 fibrosis with NASH |
20 10 |
NASH vs no NASH | Bacteroidetes Bacteroidetes |
Bacteroidaceae↑ Prevotellaceae ↓ |
Bacteroides↑ Prevotella↓ |
16S rRNA gene sequencing (Stool sample) |
| F ≥ 2 fibrosis without NASH F ≥ 2 fibrosis with NASH |
2 25 |
F ≥ 2 fibrosis vs F0/1 fibrosis | Bacteroidetes Bacteroidetes Firmicutes Firmicutes |
Bacteroidaceae↑ Prevotellaceae↓ Ruminococcaceae Erysipelotrichaceae↓ |
Bacteroides↑
Prevotella ↓ Ruminococcus↑ N/A |
||
| Mouzaki et al.17 | Steatosis patients NASH patients Healthy controls |
11 22 17 |
NASH vs Healthy NASH vs Steatosis |
Bacteroidetes ↓ Bacteroidetes ↓ Firmicutes |
N/A N/A Lachnospiraceae |
N/A N/A Clostridium coccoides↑ |
Quantitative real-time PCR (Stool sample) |
| Shen et al.18 | NAFLD patients Healthy controls |
25 22 |
NAFLD vs Healthy | Proteobacteria ↑ Fusobacteria ↑ Firmicutes Firmicutes Firmicutes Bacteroidetes↓ |
Enterobacteriaceae↑ N/A Lachnospiraceae ↑ Erysipelotrichaceae ↑ Streptococcaceae↑ Prevotellaceae ↓ |
Escherichia_Shigella
↑ N/A Lachnospiraceae_Incenae_Sedis ↑, Blautia↑ Clostridium_XVIII ↑ Streptococcus↑ Prevotella ↓ |
16S rDNAamplicon sequencing (Stool sample) |
| NAFLD patients with NASH NAFLD patients with fibrosis(F≥2) |
6 4 |
NASH vs no NASH F ≥ 2 fibrosis vs F0/F1 fibrosis |
Firmicutes Protenbacteria |
Lachnospiraceae ↑ Enterobacteriaceae ↑ |
Blautia
↑ Escherichia_Shigella ↑ |
||
| Wang et al.19 | NAFLD patients Healthy controls |
43 83 |
NAFLD vs Healthy | Bacteroidetes ↑ Firmicutes ↓ Proteobacteria (Gramnegative bacteria↑) |
N/A Lachnospiraceae ↑ Enterobacteriales |
N/A N/A Escherichia_Shigella ↑ |
454 pyrosequencing of thel6S rRNA V3 region (Stool sample) |
| Stanislawski et al.20 | Adolescents exposure to gestational diabetes mellitus during singleton pregnancies | 107 | HFF vs non HFF | Proteobacteria Bacteroidetes Proteobacteria Bacteroidetes Firmicutes |
Desulfovibrionaceae Prevotellaceae RF32 Bacteroidaceae Ruminococcaceae |
Bilophila↑ Paraprevotella ↑ Suturella ↑ RF32↑ Bacteroides (U-shaped pattern; ↑ or ↓) oscillospira ↓ |
16S rRNA gene sequencing (Stool sample) |
| Del Chierico et al.21 | NAFLD patients | 61 | NAFLD vs Healthy | Proteobacteria | Bradyrhizobiaceae | Bradyrhizobium ↑ | 454 pyrosequencing of the16S rRNA V1-V3 region (Stool sample) |
| Healthy controls | 54 | Firmicutes Firmicutes Actinobacteria Firmicutes Firmicutes Firmicutes Bacteroidetes |
unassigned unassigned Propionibacteriaceae Lachnospiraceae Ruminococcaceae Ruminococcaceae Rikenellaceae |
Anaerococcust
↑ Peptoniphilus↑ Propionibacterium acnes↑ Dorea ↑ Ruminococcus↑ oscillospira↑ Rikenellaceae ↓ |
|||
| Zhu et al.22 | NASH patients | 22 | NASH or Obese vs Healthy | Actinobacteria ↓ | Bifidobacteriaceae ↓ | Bifidobacterium ↓ | 16S rRNA pyrosequencing (Stool sample) |
| Obese patients | 25 | Bacteroidetes↑ | Bactemidaceae (−) | Bacteroides (−) | |||
| Healthy controls | 16 | Bacteroidetes↑ Bacteroidetes↑ Bacteroidetes ↑ Bacteroidetes↑ Firmicutes ↓ Firmicutes ↓ Firmicutes ↓ Firmicutes ↓ Firmicutes ↓ Firmicutes ↓ Firmicutes ↓ Firmicutes ↓ Firmicutes↓ Firmicutes↓ Firmicutes ↓ Firmicutes ↓ Firmicutes ↓ Firmicutes↓ Firmicutes ↓ Proteobacteria ↓ Proteobacteria ↓ Proteobacteria ↓ |
Porphyromonadaceae (−) Porphyromonadaceae Prevotellaceae ↑ Rikenellaceae ↑ Clostridiales family XI (−) Peptoniphilaceae Peptoniphilaceae Lachnospiraceae ↓ Clostridiaceae Lachnospiraceae Eubacteriaceae Lachnospiraceae Ruminococcaceae Ruminococcaceae ↓ Clostridiaceae Ruminococcaceae Veillonellaceae (-} Veillonellaceae (−) Veillonellaceae (−) Alcaligenaceae↓ Campylobacteraceae (−) Enterobacteriaceae↑ |
Parabacteroides (−) Porphyromonas (−) Prevotella ↑ Alistipes↑ Anaerococcus (−) Finegoldia (−) Peptoniphilus ↑ Blautia ↓ Closm’dlum (−) Coprococcus↓ Eubacteriutnm↓ Roseburia↓ Ruminococcus (−) Faecalibacterium (−1) Oscillospira ↓ Ruminococcus ↓ Acidaminococcus (−) Dialister (−) Megamonas (−) N/A Campylobacter (−) Escherichia ↑ |
|||
| Koniffkoff et al.23 | Mild/moderate NAFLD (Stage 0–2 fibrosis) | 72 | Stage 0–2 fibrosis vs Stage 3 or 4 fibrosis | Proteobacteria ↑ | N/A | N/A | Whole genome shotgun sequencing of DNA (Stool sample) |
| Advanced fibrosis (Stage 3 or 4 fibrosis) | 14 | Firmicutes ↓ | Eubacteriaceae | Eubacteriutn rectale ↓ | |||
| Firmicutes ↓ | Ruminococcaceae |
Ruminococcus obeum CAG:39↓, Ruminococcus obeum ↓ |
|||||
| Roma et al.24 | NAFLD | 30 | NAFLD vs Healthy | Proteobacteria | Kiloniellaceae ↑ | N/A | 16S rRNA gene pyrosequencing (Stool sampie) |
| Healthy controls | 30 | Proteobacteria Firmicutes Firmicutes |
Pasteurellaceae↑
Lactobacillaceae ↑ Lachnospiraceae ↑ |
N/A Lactobacillus↑ Robinsoniella ↑,Roseburia↑, Dorea ↑ |
|||
| Firmicutes Firmicutes Bacteroidetes |
Ruminococcaceae ↓ Veillonellaceae ↑ Porphyromonadaceae ↓ |
Oscillibacter↓ N/A N/A |
|||||
Comparison of condition A vs condition B:
signifies an increase in condition A relative to condition B.
signifies a decrease in condition A relative to condition B.
(−) signifies no changes in condition A relative to condition B.
Abbreviations: NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; HFF, hepatic fat fraction; N/A, not applicable.
Stanislawski et al.20 also found that Bilophila, Paraprevotella, Suturella, and RF32 have a positive relationship with hepatic fat, while Oscillospira and Varibaculum correlate negatively. The positive association between hepatic fat and Bilophila is accompanied by reduced Oscillospira. These results suggest that Bilophila might contribute to fatty liver, while Oscillospira might counteract its effects.21 The abundance of Bilophila wadworthia increases in response to a western diet or HFD.27,28 Bilophila wadworthia is also associated with T helper 1 (Thl)-mediated intestinal inflammation. Oscillospira is reduced in pediatric NAFLD and NASH.21,22 Reduced Oscillospira accompanied by increased 2-butanone has been identified as a gut microbiota signature of NAFLD onset. Increases in Ruminococcus and Dorea have been identified as gut microbiota signatures of NAFLD and NASH progression.21 Oscillospira is generally linked to leanness and health.23 Bilophila, Oscillospira, and Bacteroides are associated with diets high in animal products.29–31 In addition, increased levels of Lactobacillus and selected members of the Firmicutes (Lachnospiraceae; genera, Dorea, Robinsoniella, and Roseburia) have been observed in NAFLD patients.24 NAFLD patients and healthy subjects have a distinct intestinal microbiota community structure.
Further evidence shows that gut dysbiosis and altered metabolic function are linked with the severity of NAFLD. A study by Boursier et al.14 demonstrated that Bacteroides and Ruminococcus are associated with NASH and the severity of fibrosis. Patients with NASH and fibrosis severity F ≥ 2 have higher abundance of Bacteroides and lower abundance of Prevotella compared to those without NASH. Patients with F ≥ 2 fibrosis have higher abundance of Bacteroides and Ruminococcus and lower abundance of Prevotella compared with those with F0/F1 fibrosis.14 Patients with mild/moderate NAFLD have a higher abundance of Firmicutes, while patients with advanced fibrosis NAFLD have a higher abundance of Proteobacteria. Patients with advanced fibrosis have lower abundance of Ruminococcus obeum CAG: 39, Ruminococcus obeum, and Eubacterium rectale compared to those with mild/moderate NAFLD.32
Small intestinal bacterial overgrowth (SIBO) is defined as bacterial culture >105 CFU/ml in upper jejunal aspirate.33,34 SIBO has a direct relationship with the severity of liver disease. Many patients with chronic liver disease have dysbiosis with SIBO.3,35 SIBO in patients with NAFLD/NASH has an estimated prevalence of 39–85%.36–41 As a consequence of reduced intestinal motility and decreased bile acid production, SIBO has a role in NAFLD progression.42 Miele et al.37 have reported that SIBO is implicated in increased intestinal permeability and development of fatty liver. SIBO increases lipopolysaccharide (LPS) secretion and inflammation. Hepatic expression of Toll-like receptor 4 (TLR4), together with release of interleukin-8 (IL-8) induced by SIBO, promotes inflammation.4 SIBO increases endogenous ethanol and intestinal permeability, favoring LPS production and increased inflammation via TLR4 signaling.43,44 SIBO is considered as an independent risk factor for the severity of NAFLD and is essential for NAFLD to progress into NASH, followed by development of cirrhosis.15,19,38,45,46
Enteric dysbiosis or intestinal inflammation induced by HFD and dextran sulfate sodium significantly promotes liver fibrosis in mice with NASH.47 The inflammasome-mediated dysbiosis, including increased Prevotellaceae and Porphyromonadaceae families as well as the TM7 taxa, promote NAFLD progression in mouse models.6 Apart from providing bacterial byproducts and increasing intestinal permeability, the gut microbiota might also inhibit small intestinal secretion of fasting-induced adipocyte factor, resulting in increased hepatic triglyceride deposition.48 Antibiotic treatment or surgical removal of the bypassed section of the intestine can reverse SIBO and steatohepatitis.36,42 SIBO might be an important target for using antibiotics in treating NAFLD as well as NASH.49,50
Patients with liver cirrhosis and liver or colon cancer have reduced bile acid receptor farnesoid X receptor (FXR).51–53 Wan’s group has shown that the sex of an animal can affect the gut microbiota, which is implicated in the dissimilar development of steatosis in both western-diet-fed mice and FXR knockout (KO) mouse models according to sex.54 Decreased S24–7, in parallel with increased Bacteroidaceae, Rikeneilaceae, Lactobacillaceae, and Verrucomicrobiaceae, has been observed in wild-type female mice compared to their male counterparts. However, these sex differences are abolished in FXR KO mice, indicating that sex difference in steatosis is FXR dependent.54 Western-diet-fed male FXR KO mice develop advanced NASH with massive hepatic lymphocyte infiltration, and have decreased Firmicutes and increased Proteo-bacteria.55 Broad-spectrum as well as a Gram-negative coverage antibiotics are useful in treating NASH in male FXR KO mice, but are relatively ineffective when FXR K0 male mice are on a western diet.55 In the Proteobacteria, the relative abundance of Heli-cobacteraceae and Desulfovibrionaceae substantially increases because of FXR inactivation. Consistently, antibiotic-reduced hepatic inflammation is accompanied by their reduction. In contrast, Lactococcus, Lactobacillus, and Coprococcus have a protective effect in hepatic inflammation.55 The basic mechanisms of dysbiosis affecting liver disease are summarized in Fig. 1.
Fig. 1. The mechanisms by which gut microbiota affects liver health and diseases.
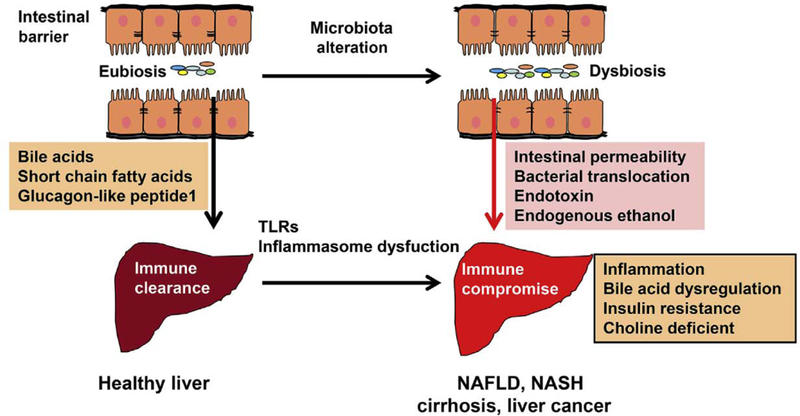
Under healthy condition, intestinal barrier and integrity prevent the entry of bacterial products, such as endotoxin, from the gut into the portal circulation. Liver immune cells rapidly clear the microbial products and bacteria passing though the gut barrier, thereby establishing immune tolerance without inflammation, Gut microbiota contributes to improving insulin sensitivity, reducing inflammation, and hepatic lipid accumulation via modulating the productions of bile acids, short-chain fatty acids, glucagon-like peptide 1, etc. Factors such as antibiotics, injury, infection, and high-fat diet can cause dysbiosis. Dysbiosis increases endogenous ethanol, endotoxin, and intestinal permeability, thereby leading the translocations of bacteria and bacterial metabolites from the intestine to the liver. Bacteria and their metabolites can activate the innate immune system via toll-like receptors and cause inflammation and subsequent liver damage. Moreover, dysbiosis-associated bile acid dysregulation increases insulin resistance, hepatic lipid accumulation, and inflammatory signaling. Furthermore, dysbiosis converts choling to trimethylamine, Which leads to choline deficiency. All these metabolites and factors contribute to liver diseases. Abbreviations: NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; TLRs, Toll-like receptors.
2.2. Gut microbiota and liver cirrhosis as well as cirrhosis-associated complications
2.2.1. Gut microbiota and liver cirrhosis
Liver cirrhosis is the end stage of chronic liver diseases and is characterized by fibrosis, abnormal hepatic architecture, and portal hypertension. Liver cirrhosis may lead to progressive hepatic failure and cancer. It has been shown that dysbiosis can affect clinical outcomes, including 90-day-hospitalization, organ failure, and death.56–58
Cirrhosis-associated gut dysbiosis is accompanied by reduced Bacteroidetes, increased Proteobacteria at the phylum level, and reduced Lachnospiraceae as well as increased Enterobacteriaceae and Veillonelaceae at the family level5. Potentially pathogenic overgrowth of Enterobacteriaceae is linked to the severity of cirrhosis and its complications, such as hepatic encephalopathy.5 Chen et al.59 demonstrated an increase in Proteobacteria and Fusobacteria, along with a decrease in Bacteroidetes and change in Firmicutes at the phylum level in fecal samples from cirrhotic patients. In addition, cirrhotic patients have increased fecal Entero-bacteriaceae, Veillonelaceae, and Streptococcaceae, and reduced Lachnospiraceae.59 Moreover, several commensal genera, such as, Coprococcus, Pseudobutyrivibrio, and Roseburia in the Lachnospiraceae family, are beneficial to the host via production of SCFAs.59
In 2014, Bajaj et al.56 compared fecal microbiota analysis in cirrhotic patients and healthy controls. They reported that the reduction of autochthonous taxa, including Lachnospiraceae, Ruminococcaceae, and Clostridiales XIV, and increase of non-autochthonous taxa including Staphylococcaceae, Enter-ococcaceae, and Enterobacteriaceae, are linked to liver failure and plasma LPS levels in cirrhosis patients. In addition, Enterobacteriaceae and endotoxemia are enriched in patients with alcoholic compared with non-alcoholic cirrhosis.56 Enterobacteriaceae are also frequently found in spontaneous bacterial peritonitis; an infection in decompensated cirrhosis.60 Enterobacteriaceae are more abundant in patients with decompensated cirrhosis compared to patients with compensated cirrhosis and healthy controls.51 The mucosal microbiota in the duodenum also differs markedly between cirrhotic patients and healthy controls.34 Based on the predicted metagenomes analyzed, pathways related to nutrient absorption are enriched in the duodenal microbiota of patients with cirrhosis, while bacterial proliferation and colonization, including bacterial motility proteins and secretory systems, are over-represented in control subjects.34
Bile acid pool size and composition are major regulators of microbiome structure.61,62 Increased primary bile acid, cholic acid (CA) can cause dysbiosis with a dramatic shift toward the Firmicutes, particularly Clostridium cluster XlVa and can increase production of deoxycholic acid (DCA).61,62 Cirrhosis-associated dysbiosis increases inflammation via metabolism, LPS, and translocation. Inflammation can suppress synthesis of bile acids in the liver.61,62 Secondary bile acids, which are generated by the Clostridiales cluster, are reduced in cirrhotic patients.63,64 Bile acids have an important role in the pathogenesis of cirrhosis,54,55,65 Reduced bile acid secretion facilitates oral microbiota migration to the distal gut and boosts SIBO. In contrast, activation of FXR stimulates bile acid excretion and induces production of antimicrobial peptides.66,67 The interaction between bile acids and microbiota plays an important role in cirrhosis.61,62 The data related to alteration of gut microbiota in cirrhotic patients are summarized in Table 2.34,56,57,59,64
Table 2.
Gut microbiota alteration in cirrhotic patients.
| Authors | Population | N | Comparison | Implicated microbiota |
Methodology | ||
|---|---|---|---|---|---|---|---|
| Phylum | Family | Genus | |||||
| Chen et al.34 | Cirrhotic patients with HBV | 24 | Cirrhosis vs Healthy | Actinobacteria Firmicutes |
Coriobacteriaceae Veillonellaceae |
Atopobium↑ Dialister↑, Veillonella↑ and Megasphera↑ |
16S rRNA gene pyrosequencing (Mucosa of the distal duodenum sample) |
| Cirrhotic patients with PBC | 6 | Proteobacteria | Pasteurellaceae | Hemophilus↓, Neisseria↓ and SR 1 genera incertae sedis↓ | |||
| Healthy controls | 28 | ||||||
| Bajaj et al.56 | Patients with liver cirrhosis | 219 | Cirrhosis vs Healthy | Firmicutes | Lachnospiraceae↓, Ruminococcaceae↓ and Clostridiales XIV ↓, |
N/A | Multi-tagged pyrosequencing (Stool sample) |
| Healthy controls | 25 | Firmicutes | Staphylococcaceae ↑, Enterococcaceae ↑ |
N/A | |||
| Proteobacteria Firmicutes Bacteroidetes |
Enterobacteriaceae↑ Veillonellaceae ↓, Porphyromonadaceae ↓ |
N/A N/A |
|||||
| Bajaj et al.57 | Patients with liver cirrhosis | 278 out of 335 | Hospitalized vs non Hospitalized | Bacteroidetes | Bacteroidaceae ↓ | N/A | 16S rRNA pyrosequencing (Stool sample) |
| Non hospitalized patients with liver cirrhosis within 90 days | 162 | Firmicutes | Clostridiales XIV↓, Lachnospiraceae↓, Ruminococcacae↓ |
N/A | |||
| Firmicutes Proteobacteria |
Enterococcaceae↓ Enterobacteriaceae ↓ |
N/A N/A |
|||||
| Hospitalized patients with liver cirrhosis | 94 | Bacteroidetes ↓ | BacterDidetes_Bacteroidaceae ↓, | N/A | |||
| Bacteroidetes↓ | Bacteroidetes_Porphyromonadaceae ↓ | N/A | |||||
| Firmicutes Firmicutes Firmicutes Firmicutes Firmicutes Proteobacteria Proteobacteria |
Firmicutes_Lactobacillaceae ↑ Firmicutes_Enterococcaceae ↑ Firmicutes_Clostridiales XIV↓ Firmicutes_Lachnospiraceae ↓ Firmicutes_Ruminococcaceae ↓ Proteobacteria-Enterobacteriaceae↑ Proteobacteria_Pasteurellaceae ↑ |
N/A N/A N/A N/A N/A N/A N/A N/A |
|||||
| Non DM | 191 | DM vs non DM | Bacteroidetes | Bacteroidetes_Bacteroidaceae ↓ | N/A | ||
| DM | 87 | Proteobacteria Firmicutes Firmicutes Firmicutes Actinobacteria |
Firmicutes_Eubacteriaceae ↓ Firmicutes_Ruminococcaceae ↑ Firmicutes_VeilIonellaceae↓ Firmicutes_Streptococcaceae↓ Actinobacteria_Streptomycetae ↓ |
N/A N/A N/A N/A N/A |
Mucosal sample | ||
| Firmicutes Bacteroidetes Fusobacteria |
Firmicutes_Clostridiacaeae ↓ Bacteroidetes_Prevotellaceae ↑ Fusobacteria_Fusobacteriaceae ↑ |
N/A N/A N/A |
|||||
| Chen et al.59 | Patients with liver cirrhosis Healthy controls |
36 24 |
Cirrhosis vs Healthy | Bacteroidetes↓ Proteobacteria ↓ Fusobacteria↓ Firmicutes Firraicutes Firmicutes |
Enterobacteriaceae ↑ N/A N/A Veillonellaceae↑ Streptococcaceae↑ Lachnospiraceae↓ |
N/A N/A N/A N/A N/A Coprococcus ↓, Pseudobutyrivibrio ↓ Roseburia ↓ |
The 16S rRNA V3 region pyrosequencing; Real-time PCR (Stool sample) |
| Kakiyama et al.64 | Early cirrhotics | 23 | Cirrhosis vs Healthy | Firmicutes | Lachonospiraceae↓, Ruminococcaceae ↓ |
N/A | Culture-independent multitagged-pyrosequencing (Stool sample) |
| Firmicutes | Lachnospiraceae | Blautia ↓ | |||||
| Advanced cirrhotics | 24 | Bacteroidetes | Rikenellaceae↓ | N/A | |||
| Healthy controls | 14 | Proteobacteria | Enterobacteriaceae ↑ | N/A | |||
Comparison of condition A vs condition B:
signifies an increase in condition A relative to condition B.
signifies a decrease in condition A relative to condition B.
Abbreviations: HBV, hepatitis B virus; PBC, primary biliary cirrhosis; DM, diabetes mellitus; N/A, not applicable.
2.2.2. Gut microbiota and complications associated with liver cirrhosis
Bacterial translocation (BT) plays a crucial role in the development of complications associated with hepatic cirrhosis.68 By inoculating an equal amount of Escherichia coli (E. coli) into small and large intestines, it was found that BT predominantly occurs in the small intestine.69 Consistently, the small intestine is a preferred site for BT in cirrhotic patients.70 In addition, BT is closely associated with SIBO as well as intestinal barrier injury in cirrhotic rats.71
Spontaneous bacterial peritonitis is a common complication of liver cirrhosis because bacterial infections occur in cirrhotic patients with ascites.72–74 Most of the bacteria in patients with spontaneous bacterial peritonitis are E. coli, Klebsiella pneumoniae, coagulase-negative Staphylococcus, and Enterococcus.74 E. coli is the predominant pathogen of spontaneous bacterial peritonitis. 72–74
Hepatic encephalopathy is a common complication of liver cirrhosis and a result of liver failure.75,76 Hepatic encephalopathy affects brain astrocytes, microglia, and neurons.75,76 A decrease in autochthonous bacteria and increase in Gram-negative bacteria are observed in cirrhotic patients with hepatic encephalopathy. It has been shown that elevated serum ammonia levels are linked to astrocytic impairment77 Moreover, ammonia-associated brain magnetic resonance imaging changes are associated with autochthonous taxa and Enterobacteriaceae, while white matter inflammatory changes are associated with oral taxa such as Porphyromonadaceae.77 Only mucosal and not fecal microbiota is altered significantly in patients with hepatic encephalopathy. The Firmicutes phylum, including Veillonella, Megasphaera, Bifidobacterium,.and Enterococcus, is highly enriched in hepatic encephalopathy, whereas Roseburia is more abundant in the non-hepatic encephalopathy group.73
2.2.3. Gut microbiota and liver transplantation
Liver transplantation is one option used to treat cirrhosis or cirrhosis-associated complications,78,79. Liver transplantation affects the recipient’s microbiota (Table 3).79–81 Gut microbiota diversity is increased after liver transplantation, but does not reach the levels in healthy controls.80 Alteration of Proteobacteria and Firmicutes links with improved cognitive level of patients with liver transplantation.80 In 2016, the Wan laboratory established the relationship between intestinal microbiota and expression of hepatic genes in regenerating the liver, using partial hepatectomy mouse models.82 Removal of two-thirds of mouse liver led to rapid changes in gut microbiota, with increased Bacteroidetes S24–7 and Rikenellaceae as well as decreased Firmicutes Clostridiales, Lach-nospiraceae, and Ruminococcaceae.82 The abundance of Rumino coccacea, Lachnospiraceae, and S24–7 was closely linked with liver metabolism and immune functions.82 Hepatic secondary bile acids are positively correlated with Firmicutes and negatively with Bacteroidetes, while tauro-conjugated bile acids show positive correlations with Bacteroidetes and negative correlations with Firmicutes.82 Priming mice with all-trans retinoic acid lowers the ratio of Firmicutes to Bacteroidetes and increases hydrophilic bile acids, which is linked with facilitated metabolism and enhanced cell proliferation in regenerating mouse livers.83
Table 3.
Gut microbiota alteration and liver transplantation.
| Authors | Population | N | Comparison | Implicated microbiota |
Methodology | ||
|---|---|---|---|---|---|---|---|
| Phylum | Family | Genus | |||||
| Sun et al.79 | Post-LT patients Healthy controls |
9 15 |
Post-LT vs Pre-LT | Proteobacteria Proteobacteria Actinobacteria Proteobacteria Proteobacteria Verrucomicrobia |
Pasteurellaceae Enterobacteriaceae Micromonosporaceae Desulfobacteraceae Eubacteriaceae Akkermansiaceae |
Actinobacillus
↓ Escherichia↓ and Shigella↓ Micromonosporaceae ↑ Desulfobacterales ↑ the Sarcina genus of Eubacteriaceae ↑ Akkermansia ↑ |
MiSeq-PE250 sequencing of the V4 region of 16S rRNA (Stool sample) |
| Bajaj et al.80 | Outpatient patients with cirrhosis on the LT list Healthy controls | 45 45 |
Improved cognition post-LT vs Pre-LT | Proteobacteria ↓ and Firmicutes ↑ | N/A | N/A | Multitagged sequencing; 16s rRNA (V1-V2) sequencing (Stool sample) |
| Not improved cognition after LT vs Healthy | Proteobacteria ↑ and Firmicutes ↓ | N/A | N/A | ||||
| post-LT vs Pre-LT | Firmicutes (−) | Ruminococcaceae ↑ and Lachnospiraceae ↑ |
N/A | ||||
| Bacteroidetes (−) | N/A | N/A | |||||
| Proteobacteria (−) | Enterobacteriaceae | Escherichia ↓, Salmonella↓ and Shigella ↓ | |||||
| Pre-LT patients vs Healthy Post-LT patients vs Healthy |
Proteobacteria (−) Firmicutes (−) Actinobacteria Bacteroidetes |
Enterobacteriaceae Ruminococcaceae ↑ and Lachnospiraceae ↑ Bifidobacteriaceae ↓ Bacteroidaceae↑ |
Escherichia, ↑
Shigella, ↑
Salmonella
↑
N/A N/A N/A |
||||
| Bajaj et al.81 | Patients with cirrhosis | 40 | Post-LT vs Pre-LT | Proteobacteria Proteobacteria Proteobacteria Actinobacteria Firmicutes Firmicutes Firmicutes Firmicutes Firmicutes Firmicutes Bacteroidetes |
Enterobacteriaceae Sutterellaceae Desulfovibrionales Bifidobacteriaceae Clostridiales Incertae Sedis XI Ruminococcaceae Clostridiales Incertae Sedis XIII Lachnospiraceae Streptococcaceae Clostridiaceae Rikenellaceae |
Shigella↓, Escherichia
↓, and Salmone↓ Suterella ↑ Bilophila↑ Bifidobacterium ↓ Desulfatibacter ↑, and Sporanaerobacter↑ ClostridiumlV↑, Osdiiibocte↑, Anaerovorax ↑ Anaerostipes↑, Clostridium XIVb ↑, Blautia↑ Roseburia↑, and Dorea↑ Streptococcus↑ Butyricicoccus↑, CtostridiumXIVa ↑ Alistipes↑ |
Multitagged sequencing (Stool sample) |
Comparison of condition A vs condition B:
signifies an increase in condition A relative to condition B.
signifies a decrease in condition A relative to condition B.
(−) signifies no changes in condition A relative to condition B.
Abbreviations: LT, liver transplantation; N/A, not applicable.
Bajaj et al.81 have reported the effect of liver transplantation on microbial composition and functionality in patients. Successful liver transplantation increases the microbial diversity accompanied by an increase in autochthonous and a decrease in potentially pathogenic taxa.81 The favorable changes in the gut microbiota also have the benefit of increasing fecal bile acids and urinary phenyl-acetylglutamine, accompanied with a reduction in serum ammonia and endotoxemia.81
2.2.4. Fungal dysbiosis and complications associated with liver cirrhosis
In addition to bacteria, microbiota includes archaea, protists, fungi, viruses, and bacteriophages.84 A recent study showed fungal dysbiosis in cirrhotic patients. Bajaj et al.85 have demonstrated a link between fungal and bacterial diversity in patients with liver cirrhosis, and Bacteroidetes/Ascomycota ratio can affect 90-day-hospitalization (Table 4). Moreover, Candida overgrowth and reduced intestinal fungal diversity are observed in patients with alcoholic cirrhosis (Table 4).86
Table 4.
Fungal dysbiosis in complications associated with liver cirrhosis.
| Authors | Population | N | Comparison | Implicated microbiota |
Methodology | ||
|---|---|---|---|---|---|---|---|
| Phylum | Family | Genus | |||||
| Bajaj et al.85 | Outpatients cirrhotics | 77 | Inpatients vs Outpatients | Basidiomycota↓ Ascomycota |
Bacteroidetes/Ascomycota ratio ↑ Saccharomycetaceae |
N/A Candida↑ |
Metagenomics (Stool sample) |
| Inpatients cirrhotics | 66 | After antibiotics vs before antibiotics | Ascomycota | Saccharomycetaceae | Candida↑ | ||
| Controls | 26 | Inpatients vs Outpatients | Ascomycota | Saccharomycetaceae | Candida↑ | ||
| Proteobacteria | Enterobacteriaceae↑ | N/A | |||||
| Firmicutes | Enterococcaceae↑ | N/A | |||||
| Outpatients vs Controls | Basidiomycota↓ | N/A | N/A | ||||
| Ascomycota | Saccharomycetaceae | Candida↑ | |||||
| Inpatients vs Controls | Proteobacteria | Enterobacteriaceae ↑ | N/A | ||||
| Firmicutes | Enterococcaceae↑ | N/A | |||||
| Outpatients on antibiotics | Ascomycota ↑ | Saccharomycetaceae | Candida↑ | ||||
| Proteobacteria | Pasteurellaceae↑ | N/A | |||||
| Yang et al.86 | Healthy individuals | 8 | Patients vs Healthy individuals | Ascomycota | Saccharomycetaceae | Candida↑ | lllumina MiSeq platform |
| Alcohol-dependent | 10 | sequencing of the V4 region of 16S rRNA (Stool sample) | |||||
| patients (nonprogressive liver disease) | |||||||
| Patients with alcoholic liver cirrhosis | 4 | ||||||
Comparison of condition A vs condition B:
signifies an increase in condition A relative to condition B.
signifies a decrease in condition A relative to condition B.
Abbreviation: N/A, not applicable.
2.2.5. Oral microbiota and liver cirrhosis
Oral microbiota contributes to the progression of liver diseases. Elevated oral Streptococcus and Veillonella are found in cirrhotic patients.87 Increased Enterobacteriaceae and Enterococcaceae, as well as reduced autochthonous bacteria, are found in patients with previous episodes of hepatic encephalopathy.87 Oral microbiota has a significant impact on duodenal microbiota. At the genus level, the most distinctive taxa found in cirrhotic patients and controls include Veillonella, Prevotella, Neisseria, and Haemophilus, which are commonly found in the oral cavity.88
Bajaj et al.89 performed a direct comparison of the salivary microbiome between healthy controls and patients with cirrhosis. Relative abundance of potentially pathogenic taxa (Prevotella and Fusobacteriaceae) increased whereas autochthonous taxa (Lach-nospiraceae and Ruminococcaceae) decreased in oral microbiota of cirrhotic patients with previous hepatic encephalopathy.83 Mi-crobes of oral origin can be present in the duodenum. Duodenal Prevotella and Fusobacterium are also increased significantly in, cirrhotic patients.34 Proton pump inhibitors increase the microbiota of oral origin in patients with cirrhosis.90 The removed pH barrier in the gastrointestinal tract allows the microbiota of oral origin to migrate along the gastrointestinal tract and even into feces.90 Certain oral bacteria can produce high levels of hydrogen sulfide (H2S) and methyl mercaptan (CH3SH).91 Higher proportions of Neisseria, Porphyromonas, and SRI are linked to H2S production that can damage deoxyribonucleic acid (DNA). Prevotella, Veillonella, Atopobium, Megasphaera, and Selenomonas are associated with production of CH3SH, which contributes to development of hepatic encephalopathy.91,92 Literature related to oral microbiota and liver disease is summarized in Table 5.34,87,89,90
Table 5.
Oral microbiota alteration in patients with cirrhosis.
| Authors | Population | N | Comparison | Impiicated microbiota |
Methodology | ||
|---|---|---|---|---|---|---|---|
| Phylum | Family | Genus | |||||
| Chen et al.34 | Cirrhotic patients | 30 | Patients vs Controls | Actinobacteria | Coriobacteriaceae | Atopobium↑ | 16S rRNA gene pyrosequencing |
| Firmicutes | Veillonellaceae | Dialister↑, Veillonella↑, and Megasphera↑ | (Mucosal from the distal duodenum sample) | ||||
| Healthy controls | 28 | Proteobacteria | Pasteurellaceae | Hemophilus ↓ | |||
| Proteobacteria | Neisseriaceae | Neisseria ↓ | |||||
| undefined | undefined | SR 1 genera incertae sedis↓ | |||||
| Qin et al.87 | Patients with cirrhosis | 98 | Patients vs Controls | Firmicutes | Streptococcaceae | Streptococcus↑ | Quantitative metagenomics (Stool sample) |
| Firmicutes | Veillonellaceae | Veillonella ↑ | |||||
| Healthy controls | 83 | Firmicutes | Enterococcaceae ↑ | N/A | |||
| Proteobacteria | Enterobacteriaceae↑ | N/A | |||||
| Bajaj et al.89 | Patients with cirrhosis without HE | 59 | Patients vs Controls | Bacteroidetes | Prevotellaceae | Prevotella ↑ | Quantitative metagenomics (Stool or saliva sample) |
| Fusobacteria | Fusobacteriaceae ↑ | N/A | |||||
| Patients with cirrhosis with previous HE | 43 | Firmicutes | Lachnospiraceae ↓ and | N/A | |||
| Ruminococcaceae ↓ | |||||||
| age-matched controls | 32 | Proteobacteria | Enterobacteriaceae↑ | N/A | |||
| Firmicutes | Enterococcaceae ↑ | N/A | |||||
| Bajaj et al.90 | Cirrhotic outpatients on PPI | 59 | PPI users vs Patients without PPI and Controls | Firmicutes | Streptococcaceae↑ | N/A | Multi-tagged sequencing (Stool sample) |
| Firmicutes | Lachnospiraceae↓ | N/A | |||||
| Cirrhotic outpatients not on PP1 | 78 | ||||||
| Healthy controls | 45 | ||||||
| Cirrhotic outpatients not on PPI | 15 | After vs Before PPI initiation | Bacteroidetes Firmicutes |
Porphyromonadaceae ↑ Streptococcaceae↑ |
N/A N/A |
||
| Patients with decompensated cirrhosis on chronic PPI |
15 | PPI withdrawal vs Pre-PPI therapy | Bacteroidetes | Porphyromonadaceae ↓ | N/A | ||
| Firmicutes | Streptococcaceae ↓, and Veillonellaceae↓ | N/A | |||||
Comparison of condition A vs condition B:
signifies an increase in condition A relative to condition B.
signifies a decrease in condition A relative to condition B.
Abbreviations: HE, hepatic encephalopathy; PP1, proton pump inhibitors; N/A, not applicable.
2.3. Gut microbiota and HCC
Gut microbes are implicated in liver carcinogenesis.5,93,94 Helicobacter species are important pathogens that may be directly involved in the occurrence of liver cancer, and are found in human HCC specimens.95 A human study has shown that Helicobacter is present in the liver of patients with primary liver carcinoma but not in controls without primary liver carcinoma.96 However, Helicobacter hepaticus (H. hepaticus) is not present in HCC patients with chronic hepatitis B or C.97
H. hepaticus infection promotes HCC in chemical and viral transgenic liver cancer models.98 However, HCV transgene or H. hepaticus exposure alone is not sufficient to initiate liver cancer.98 Moreover, increased risk of HCC is not dependent on translocation of H. hepaticus to the liver.98 Gut H. hepaticus colonization induces nuclear factor κ-light-chain-enhancer of activated B cell signaling, which activates innate and Th1-type adaptive immunity.98 Thus, H. hepaticus in the intestinal niche without translocation to the liver can change the immune signaling and play a synergistic role with chemical and viral carcinogenic factors.98
Gut dysbiosis is found in patients with liver cirrhosis and HCC as well as animal models using streptozotocin-HFD, diethylnitrosamine (DEN), or carbon tetrachloride (CCI4). Blooming of E. coli is found in cirrhotic patients who have HCC compared to those without HCC.99 In the C57BL/6J mouse model of NASH and HCC induced by streptozotocin-HFD, a significant increase of Atopobium spp., Bacteroides spp., Bacteroides vulgatus, Bacteroides acidifaciens, Bacteroides uniformis, Clostridium cocleatum, Clostridium xylanolyticum, and Desulfovibrio spp. is associated with disease progression.100 A significant reduction of Lactobacillus, Bifidobacterium, and Enterococcus species, along with increased E. coli and Atopobiumcluster, has been found in rat models of HCC induced by DEN.101 Moreover, Clostridium spp. are reduced in CCl4-induced liver carcinogenesis models.102 When DEN is used in combination with CCl4, gut sterilization or TLR4 deletion reduces tumor number and volume but does not affect tumor incidence, while continuous low-dose LPS administration increases tumor number and size.103
Changes in the microbiota of the tongue coating have been noted in patients with HCC.104 Moreover, enrichment of tongue Oribacterium and Fusobacterium could be microbial biomarkers of HCC.104 Microbial genes in the categories related to nickel/iron transport, amino acid transport, energy-producing systems, and metabolism differ in abundance between HCC patients and healthy controls.104
The steatohepatitis-inducing HFD (STHD-01) is a NASH-inducing HFD, which promotes HCC without chemical carcinogens.105 A recent study revealed that gut bacteria associated with secondary bile acid production promote STHD-01-induced HCC development that can be prevented by antibiotics.105,106 In addition, Prevotella and Oscilibacter, producers of anti-inflammatory metabolites, can inhibit carcinogenesis. This anti-cancer effect may result from increased regulatory T (Treg) cells and reduced, migration of Thl7 cells to the liver.94 The gut microbiota plays a key role in HCC development and can potentially be used to treat HCC. Literature related to microbiota alteration in human HCC and animal model of HCC is summarized in Tables 6 and 7,95–97,99–102,104 respectively.
Table 6.
Gut microbiota alteration in human HCC.
| Authors | Population | N | Comparison | Implicated microbiota | Methodology | ||
|---|---|---|---|---|---|---|---|
| Phylum | Family | Genus | |||||
| Nilsson et al. 95 | Liver specimens of patients with cholangiocarcinoma HCC human specimens Controls (liver tissue from patients with resected métastasés from colorectal cancers) |
14 16 20 |
HCC or cholangiocarcinoma specimens vs Controls | Proteobacteria | Helicobacteraceae | Helicobacter spp.↑ | PCR and DNA sequencing (HCC human specimens) |
| Huang et al.96 | HCC human specimens Controls without HCC |
20 16 |
HCC specimens vs Controls | Proteobacteria | Helicobacteraceae | Helicobacter pylori↑ | PCR, DNA sequencing, and immunostaining (Liver specimens) |
| Krüttgen et al.97 | Patients with viral-induced HCC Control patients |
14 11 |
Patients with viral-induced HCC vs Control patients | Proteobacteria | Helicobacteraceae | H. hepaticus (no exist) | PCR (Stool sample) |
| Grat et al.99 | Patients with HCC Non HCC patients |
15 15 |
HCC vs non HCC | Proteobacteria | Enterobacteriaceae | Escherichia coli ↑ | Culturing on enriching and selective agar media (Stool sample) |
| Lu et al.104 | Early liver carcinoma patients with cirrhosis Healthy controls |
35 25 |
HCC vs Healthy | Firmicutes Fusobacteria |
Lachnospiraceae Fusobacteriaceae |
Oribacterium changes Fusobacterium changes |
16S rRNA gene sequencing (Tongue coat sample) |
Comparison of condition A vs condition B:
signifies an increase in condition A relative to condition B.
Abbreviation: HCC, hepatocellular carcinoma.
Table 7.
Gut microbiota alteration in HCC animal models.
| Authors | Model | Agent | Comparison | Implicated microbiota |
Methodology | ||
|---|---|---|---|---|---|---|---|
| Phylum | Family | Genus | |||||
| Xie et al.100 | NASH-HCC C57BL/6J mouse model | STZ-HFD | NASH-HCC vs Controls | Actinobacteria Bacteroidetes |
Coriobacteriaceae Bacteroidaceae |
Atopobium spp.↑
Bacteroides spp. ↑ Bacteroides vulgcitus,↑ Bacteroides acidifaciens↑ and Bacteroides uniformis ↑ |
16S rDNA gene pyrosequencing (Stool sample) |
| Firmicutes | Clostridiaceae | Clostridium cocleatum, ↑ Clostridium and xylanolyticum ↑ | |||||
| Proteobacteria | Desulfovibrionaceae | Desulfovibrio spp.↑ | |||||
| Zhang et al.101 | Male Sprague-Dawley HCC rats | DEN | HCC rats vs Controls | Firmicutes Firmicutes Actinobacteria |
Lactobacillaceae Enterococcaceae Bifidobacteriaceae |
Lactobacillus
↓ Enterococcus ↓ Bifidobacterium ↓ |
16S rRNA based quantitative real-time PCR (Stool sample) |
| Gómez-Hurtado et al.102 | Female Balb/c fibrosis mice | CCI4 | Fibrosis mice vs Controls | Firmicutes Firmicutes Firmicutes |
Clostridiaceae Clostridiaceae Clostridiaceae |
Clostiidia spp.↓ Clostiidium coccoides ↓ Clostridium leptum ↓ |
Quantitative real-time PCR (Stool sample) |
Comparison of condition A vs condition B:
signifies an increase in condition A relative to condition B.
signifies a decrease in condition A relative to condition B.
Abbreviations: STZ-HFD, streptozotocin-high fat diet; DEN, diethylnitrosamine; HCC, hepatocellular carcinoma; CCL4 carbon tetrachloride.
3. Gut microbiota-targeted therapy
Dysbiosis contributes to the development of liver diseases. Thus, restructuring the gut microbiota community to establish eubiosis can be effective in preventing or treating liver diseases.
3.1. Probiotics
Probiotics are live microorganisms that provide health benefits for the host when consumed in adequate amounts.107 In addition to the beneficial effects on gastrointestinal diseases, probiotics also exert a beneficial effect in liver diseases.108–113
Li et al.83 reported that using Prohep for feeding reduces the liver tumor size in xenograft mouse models. Prohep consists of Lactobacillus rhamnosus (L. rhamnosus) GG, E. coli Nissle 1917, and heat-inactivated VSL#3. Prohep feeding increases the abundance of Prevotella and Oscillibacter and generates anti-inflammatory metabolites, which lead to reduced Th17 polarization and increased differentiation of Treg/Trl cells in the gut. In addition, L. rhamnosus GG protects mice from high-fructose-induced NAFLD and reduces cholesterol in HFD-fed mice.114,115 Lactobacillus casei shirota protects against NAFLD in multiple mouse NAFLD models via improved insulin sensitivity, reduced plasma LPS-binding protein, and inhibition of LPS/TLR4 signaling in the liver.116–118 Other probiotics such as Lactobacillus plantarum MA2, Lactobacillus plantarum NCU116, Lactobacillus johnsonii BS15, Lactobacillus reuteri GMNL-263, and Lactobacillus gasseri BNR17 also have protective roles in improving dyslipidemiaand NAFLD.119–122 Moreover, Bifidobacterium prevents fat accumulation and increases insulin sensitivity in HFD-fed rats.123 Probiotics of Bifidobacterium are superior to Lactobacillus acidophilus in decreasing hepatic fat accumulation.124
Probiotics of Clostridium butyricum MIYAIRI 588, a butyrate-producing bacterium, reduce hepatic lipid droplets and improve insulin sensitivity in rats with HFD-induced NAFLD.125 This strain also decreases hepatic lipids and LPS in rats with NAFLD induced by choline-deficient/L-amino acid-defined diet.126 Kumar et al.127 have demonstrated that probiotic-fermented milk and chlorophyllin significantly reduce the incidence of aflatoxin B1-associated HCC.
Although the health effects of probiotics are mainly obtained from animal studies, some consistent results have been generated in clinical studies. Administration of L. rhamnosus GG and a mixture of Lactobacillus bulgaricus and Streptococcus thermophiles has, beneficial effects on obese children with NAFLD.128,129 VSL#3 im, proves liver function and increases glucagon-like peptide 1 levels in obese children with NASH.130 Moreover, L. rhamnosus GG alters gut microbiota in patients with cirrhosis.131 Compared with placebo, L. rhamnosus GG increases the beneficial autochthonous Clostridiales Incertae Sedis XIV and Lachnospiraceae and reduces the abundance of Enterobacteriaceae and Porphyromonadaceae in patients with stable cirrhosis and minimal hepatic encephalopathy.131 Combination of Bifidobacterium longum and fructooligosaccharides (FOSs, a mixture of fermentable dietary fibers) improved minimal and overt hepatic encephalopathy in clinical studies.132,133 In addition, VSL#3 prevented hepatic encephalopathy in a randomized controlled clinical study.134 Compared with baseline, 3 months treatment with VSL#3 increased psychometric hepatic encephalopathy scores and reduced the levels of arterial ammonia, SIBO, and orocecal transit time.134 Over 6 months, VSL#3 treatment reduced the recurrence of hepatic encephalopathy in, cirrhotic patients compared with the placebo-treated controls.135 VSL#3 also decreased the hepatic venous pressure gradient, cardiac index, and heart rate, and increases systemic vascular resistance and mean arterial pressure in patients with cirrhosis and ascites.136 This indicates that VSL#3 improves the hepatic and systemic hemodynamics in patients with cirrhosis.136 A probiotics combination of eight strains of Lactobacillus, Bifidobacterium, and Streptococcus, is also effective in preventing secondary hepatic encephalopathy in patients with cirrhosis.137 However, in a study conducted by Solga et al.138 4 months supplementation with VSL#3 increased hepatic lipid content in four patients who already had steatosis. Another randomized double-blind study conducted by And reasen et al 139 revealed that 4 weeks intake of L acidophilus NCFM improved insulin sensitivity but did not affect systemic inflammatory response. Additionally, 6 weeks supplementation with L acidophilus did not change serum lipids in volunteers who had elevated cholesterol.140 More well-designed trials are needed to further study the effects of probiotics in preventing liver diseases.
3.2. Prebiotics
Prebiotics are food ingredients that selectively stimulate the growth or activity of beneficial microorganisms, such as bacteria and fungi.141,142 They can alter the composition and/or activity of gut microbiota. Prebiotics are useful in preventing NAFLD in laboratory animals and clinical studies.109,143–145
Prebiotics of FOSs prevent NAFLD via restoring the gut microbiota composition and intestinal epithelial barrier function, leading to reduced serum LPS, hepatic inflammation, and hepatic cholesterol content in NAFLD mice.146–148,157 FOS supplementation significantly reduces serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in NASH patients.143 Lactulose increases the growth of lactic acid bacteria and Bifidobacterium.149 It also decreases serum LPS and hepatic inflammation in rats with NASH.150 Chitin-glucan, a prebiotic from a fungal source, reduces hepatic triglyceride, body weight gain, and glucose intolerance via restoring clostridial cluster XlVa in HFD-induced obese mice.151 Treatment with isomaltooligosaccharides plus lycopene increases adipose tissue fat mobilization, reduces body weight gain, and improves insulin sensitivity in HFD-induced NAFLD mice.152 Prebiotics have great potential for prevention of liver disease through improving metabolism and the intestinal barrier, as well as reducing endotoxemia.
3.3. Synbiotics
Synbiotics refer to the combination of probiotics and prebiotics in a form of synergism.153 Synbiotics that consist of Lactobacillus paracasei B21060 plus arabinogalactan and FOSs reduce hepatic inflammation in diet-induced NAFLD.154 Supplementation with seven probiotics consisting of L. casei, L. rhamnosus, S. thermophilus. Bifidobacterium breve, L. acidophilus, B. longum, and L bulgaricus plus FOSs improves fasting blood glucose, serum triglycerides, and inflammatory cytokines in both lean and obese NAFLD patients.155,156 Compared to lifestyle intervention alone, synbiotics of B. longum plus FOSs have added benefits for NASH patients. This intervention reduces serum tumor necrosis factor α (TNFα), C-reactive protein, endotoxin, and AST.157
Milk oligosaccharides (MOs) selectively increase the growth of Bifidobacterium infantis (B. infantis). Synbiotics of B. infantis and MOs prevent occurrence of cancer-prone NASH in western-diet-fed FXR KO male mice. B. infantis and MOs increase G protein-coupled bile acid receptor 1 (also known as Takeda G protein-coupled receptor, TGR5)-regulated signaling, thereby generating beneficial effects. B. infantis and/or MO treatment also improves ileal SCFA, signaling in western-diet-fed FXR KO mice. Furthermore, MOs alone and B. infantis plus MOs inhibit the growth of genus Bilophila and reduce the abundance of bacterial genes including dissimilatory sulfite reductase (dsrA) and methyl coenzyme M reductase A (mcrA), which are increased in mice with NASH.159
3.4. Other approaches
3.4.1. Bacterial metabolite butyrate
Butyrate is generated by bacterial fermentation of non-digestible polysaccharides.15,160 Sodium butyrate treatment reduces inflammation and fat accumulation in diet-induced NAFLD, potentially via enriching beneficial bacteria Christensenellaceae, Blautia, and Lactobacillus.161 Additionally, butyrate supplementation reverses NASH via reducing hepatic β-muricholic acid (β-MCA) as well as DCA, which are implicated in the development of NASH in western-diet-fed FXR-KO mice.65,69 It has been shown that Lactobacillus and Bifidobacterium reduce adiposity and inflammation in NAFLD rats via butyrate production and butyrate receptor G-protein-coupled receptor 109A-regulated signaling.160 Butyrate and its synthetic derivative, N-(l-carbamoyl-2-phenyl-ethyl) butyramide, reduce the intracellular lipid accumulation and oxidative stress in diet-induced insulin-resistant obese mice.162 Furthermore, sodium butyrate has a protective role in NAFLD pathogenesis via increased duodenal melatonin synthesis, as well as decreased hepatic inducible nitric oxide synthase in fructose-induced NAFLD mice.163
3.4.2. Fecal microbiota transplantation
A randomized clinical trial in patients with cirrhosis and recurrent hepatic encephalopathy was conducted to compare the safety of fecal microbiota transplantation with no such intervention.164 Fecal microbiota transplantation reduced hospitalization and improved cognition and dysbiosis in patients with cirrhosis with recurrent hepatic encephalopathy, when compared with standard of care (SOC).164 Fecal microbiota transplantation has protective effects in rats with CCl4-induced hepatic encephalopathy.165 Fecal microbiota transplantation reduces intestinal permeability and improves the TLR response of the liver, leading to improved cognitive function and reduced liver function indexes.165
3.4.3. Diet
Diet is a contributing factor to liver diseases. Fructose-enriched diet alters liver metabolism and gut barrier function, increases endotoxemia, decreases Bifidobacterium and Lactobacillus, and eventually leads to NAFLD.166 Long-term fructose consumption increases lipogenic enzymes via activation of sterol regulatory element binding protein-lc (SRFBPlc) and carbohydrate responsive element binding protein (ChREBP).167 It promotes lipogenesis, hypertriglyceridemia, hepatic insulin resistance, and hepatic steatosis.167 A diet rich in fermented milk, vegetables, cereals, coffee, and tea contributes to a higher microbial diversity in patients with cirrhosis.168 Microbial diversity is an independent factor that reduces the risk of 90-day hospitalization.168
4. Conclusions and perspectives
Gut microbiota plays a pivotal role in the pathogenesis of metabolic liver diseases. Re-establishing eubiosis using probiotics, prebiotics, and synbiotics, as well as natural products, is a promising avenue to prevent and treat liver diseases and, potentially, liver cancer. Although bile acid and SCFA-regulated pathways can explain how diet through gut microbiota affects health and disease processes, other molecular links remain to be uncovered. With the advancement of sequencing technology as well as cultural techniques, specific bacterial species and microbial functions can be uncovered to establish a causal relationship. There is no doubt that metabolomics and epigenetic genomics are powerful tools to elucidate the underlying mechanism for disease processes, leading to innovative treatment strategies. The generated information should have an impact on personalized nutrition as well as precision medicine.
Acknowledgments
This work was supported by the USA National Institutes of Health (NIH), USA grants U01CA179582 and R01 CA222490. We also thank Michelle Nguyen (Department of Medical Pathology and Laboratory Medicine, University of California, Davis, Sacramento, CA, USA) and Mindy Huynh (Department of Dermatology, University of California, Davis, Sacramento, CA, USA) for reviewing the manuscript.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Minemura M, Shimizu Y. Gut microbiota and liver diseases. World J Gastroenterol. 2015;21:1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pennisi E Cancer therapies use a little help from microbial friends. Science. 2013;342:921. [DOI] [PubMed] [Google Scholar]
- 3.Wan MLY, El-Nezami H Targeting gut microbiota in hepatocellular carcinoma: probiotics as a novel therapy. Hepatobiliary Surg Nutr. 2018;7:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma J, Zhou Q, Li H. Gut microbiota and nonalcoholic fatty liver disease: insights on mechanisms and therapy. Nutrients. 2017:9 10.3390/nu9101124 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanduzzi Zamparelli M, Rocco A, Compare D. Nardone G The gut microbiota: a new potential driving force in liver cirrhosis and hepatocellular carcinoma. United European Gastroenterol J. 2017;5:944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henao-Mejia J, Elinav E. Jin C et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safe S, Papineni S, Chintharlapalli S. Cancer chemotherapy with indole-3-carbinol, bis(3’-indolyl)methane and synthetic analogs. Cancer Lett 2008;269:326–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wieland A Frank DN Harnke B Bambha K Systematic review: microbial dysbiosis and nonalcoholic fatty liver disease. Aliment Pharmacol Ther, 2015;42:1051–1063. [DOI] [PubMed] [Google Scholar]
- 9.Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut. 2016;65: 2035–2044. [DOI] [PubMed] [Google Scholar]
- 10.Fukui H Gut microbiome-based therapeutics in liver cirrhosis: basic consideration for the next step J Clin Transl Hepatol. 2017;5:249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acharya C, Bajaj JS. Gut microbiota and complications of liver disease. Gastroenterol Clin JV Am. 2017:46:155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henao-Mejia J Elinav E Thaiss CA, Flavell RA The intestinal microbiota in chronic liver disease. Adv Immunol. 2013;117:73–97 [DOI] [PubMed] [Google Scholar]
- 13.Doulberis M, Kotronis G. Gialamprinou D Kountouras J, Katsinelos P Non-alcoholic fatty liver disease: an update with special focus on the role of gut microbiota. Metabolism. 2017;71:182 197. [DOI] [PubMed] [Google Scholar]
- 14.Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quigley EM, Monsour HP. The gut microbiota and nonalcoholic fatty liver disease. Semin Liver Dis. 2015;35:262–269. [DOI] [PubMed] [Google Scholar]
- 16.Miura K Ohnishi H Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World J Gastroenterol. 2014:20:7381–7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mouzaki M, Comelli EM, Arendt BM, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. [DOI] [PubMed] [Google Scholar]
- 18.Shen F, Zheng RD, Sun XQ, Ding WJ, Wang XY, Fan JG. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int. 2017;16:375–381. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Jiang X, Cao M. et at Altered fecal microbiota correlates with liver biochemistry in nonobese patients with non-alcoholic fatty liver disease. Sri Rep. 2016;6:32002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanislawski MA. Lozupone CA, Wagner BD, et al. Gut microbiota in adoles cents and the association with fatty liver: the EPOCH study. Pediatr Res. 2018;84:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Del Chierico F, Nobili V, Vemocchi P, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017:65:451–464. [DOI] [PubMed] [Google Scholar]
- 22.Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. [DOI] [PubMed] [Google Scholar]
- 23.Konikoff T, Gophna U. Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016;24:523–524. [DOI] [PubMed] [Google Scholar]
- 24.Raman M, Ahmed I, Gillevet PM, et al. Fecal microbome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11:868–875 (el-e3). [DOI] [PubMed] [Google Scholar]
- 25.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walters WA. Xu Z Knight R Meta-analyses of human gut microbes associated with obesity and 1BD. FEBS Lett. 2014;588:4223–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devkota S, Wang Y. Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in 1110−/− mice. Nature. 2012;487: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Keefe SJ. Li jV, Lahti L, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu GD. Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David IA Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loomba R, Seguritan V, Li W, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metabol. 2017;25:1054–1062 (e5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckburg PB. Bik EM Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y Ji F Guo J, Shi D, Fang D Li L Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci Rep. 2016:6:34055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Compare D Coccoli P Rocco A, et al. Gut-liver axis: the impact of gut microbiota on non alcoholic fatty liver disease. Nutr Metabol Cardiovasc Dis. 2012;22:471–476. [DOI] [PubMed] [Google Scholar]
- 36.Wigg AJ, Roberts-Thomson IC Dymock RB, McCarthy PJ Grose RH, Cummins AG The role of small intestinal bacterial overgrowth, intestinal permeability, endotoxaemia, and tumour necrosis factor alpha in the pathogenesis of non-alcoholic steatohepatitis. Gut. 2001;48:206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miele L, Valenza V, La Torre G, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49:1877–1887. [DOI] [PubMed] [Google Scholar]
- 38.Sabaté JM. Jouët P, Harnois F, et al. High prevalence of small intestinal bacterial overgrowth in patients with morbid obesity: a contributor to severe hepatic steatosis. Obes Surg. 2008;18:371–377. [DOI] [PubMed] [Google Scholar]
- 39.Rafiei R, Bemanian M, Rafiei F, et al. Liver disease symptoms in non-alcoholic fatty liver disease and small intestinal bacterial overgrowth. Rom J Intern Med. 2018;56:85–89. [DOI] [PubMed] [Google Scholar]
- 40.Bures J , Cyrany J, Kohoutova D, et al. Small intestinal bacterial overgrowth syndrome. World J Gastroenterol. 2010;16:2978–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu H, Williams B, Schnabl B. Gut microbiota, fatty liver disease, and hepatocellular carcinoma. Liver Res. 2018;2:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henao-Mejia J, Elinav E, Thaiss CA, Licona-Limon P, Flavell RA. Role of the intestinal microbiome in liver disease. J Autoimmun. 2013;46:66–73. [DOI] [PubMed] [Google Scholar]
- 43.Bibbò S, laniro G, Dore MP, Simonelli C, Newton EE, Cammarota G. Gut microbiota as a driver of inflammation in nonalcoholic fatty liver disease. Mediat Injlamm. 2018;2018:9321643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saltzman ET, Palacios T, Thomsen M, Vitetta L Intestinal microbiome shifts, dysbiosis, inflammation, and non-alcoholic fatty liver disease. Front Microbiol. 2018;9:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sajjad A Mottershead M, Syn WK, Jones R, Smith S Nwokolo CU Ciprofloxacin suppresses bacterial overgrowth, increases fasting insulin but does not correct low acylated ghrelin concentration in non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2005;22:291–299. [DOI] [PubMed] [Google Scholar]
- 46.Arslan N Obesity, fatty liver disease and intestinal microbiota. World J Gastroenterol. 2014;20:16452–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gäbele E, Dostert K, Hofmann C, et al. DSS induced colitis increases portal LPS levels and enhances hepatic inflammation and fibrogenesis in experimental NASH. J Hepatol. 2011. ;55:1391–1399. [DOI] [PubMed] [Google Scholar]
- 48.Bäckhed F Manchester JK, Semenkovich CF Gordon JI Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007:104:979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferolla SM, Armiliato GN, Couto CA, Ferrari TC. The role of intestinal bacteria overgrowth in obesity-related nonalcoholic fatty liver disease. Nutrients. 2014;6:5583–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fialho A Thota P, McCullough AJ, Shen B Small intestinal bacterial overgrowth is associated with non-alcoholic fatty liver disease. J Gastrointestin Liver Dis. 2016;25:159–165. [DOI] [PubMed] [Google Scholar]
- 51.Lax S, Schauer G, Prein K, et al. Expression of the nuclear bile acid receptor/ farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis, int J Cancer. 2012;130:2232–2239. [DOI] [PubMed] [Google Scholar]
- 52.Liu N, Meng Z, Lou G, et al. Hepatocarcinogenesis in FXR−/− mice mimics human HCC progression that operates through HNFlα regulation of FXR expression. Mol Endocrinol. 2012;26:775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su H Ma C, Liu J, et al. Downregulation of nuclear receptor FXR is associated with multiple malignant clinicopathological characteristics in human hepatocellular carcinoma. Am J Physiol Gastrointest Liver Physiol. 2012;303: G1245–G1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheng L, Jena PK, Liu HX, et al. Gender differences in bile acids and microbiota in relationship with gender dissimilarity in steatosis induced by diet and FXR inactivation. Sci Rep. 2017;7:1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jena PK, Sheng L, Liu HX, et al. Western diet-induced dysbiosis in farnesoid X receptor knockout mice causes persistent hepatic inflammation after anti-biotic treatment. Am J Pathol. 2017;187:1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bajaj JS, Heuman DM, Hylemon PB, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bajaj JS. Betrapally NS, Hylemon PB, et al. Gut microbiota alterations can predict hospitalizations in cirrhosis independent of diabetes mellitus. Sci Rep. 2015:5:18559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Guo J, Qian G, et al. Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality J Gastroenterol Hepatol. 2015;30:1429–1437 [DOI] [PubMed] [Google Scholar]
- 59.Chen Y, Yang F, Lu H, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–572. [DOI] [PubMed] [Google Scholar]
- 60.Tandon P, Garcia-Tsao G. Bacterial infections, sepsis, and multiorgan failure in cirrhosis. Semin Liver Dis. 2008;28:26–42. [DOI] [PubMed] [Google Scholar]
- 61.Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microb. 2013;4:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Gut microbiota, cirrhosis, and alcohol regulate bile acid metabolism in the gut. Dig Dis. 2015;33:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vlahcevic ZR, Buhac I, Bell CC Jr, Swell L Abnormal metabolism of secondary bile acids in patients with cirrhosis. Gut. 1970;11:420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kakiyama G, Pandak WM, Gillevet PM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheng L Jena PK, Hu Y, et al. Hepatic inflammation caused by dysregulated bile acid synthesis is reversible by butyrate supplementation. J Pathol. 2017;243:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. [DOI] [PubMed] [Google Scholar]
- 67.Inagaki T Moschetta A, Lee YK, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA. 2006;103:3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wiest R, Garcia-Tsao G. Bacterial translocation (BT) in cirrhosis. Hepatology. 2005;41:422–433. [DOI] [PubMed] [Google Scholar]
- 69.Powell DW. Barrier function of epithelia. Am J Physiol. 1981. ;241 :G275–G288. [DOI] [PubMed] [Google Scholar]
- 70.Wiest R, Lawson M, Geuking M. Pathological bacterial translocation in liver cirrhosis. J Hepatol. 2014;60:197–209 [DOI] [PubMed] [Google Scholar]
- 71.Pardo A, Bartoli R, Lorenzo-Zuniga V, et al. Effect of cisapride on intestinal bacterial overgrowth and bacterial translocation in cirrhosis. Hepatology. 2000;31:858–863. [DOI] [PubMed] [Google Scholar]
- 72.Lutz P, Nischalke HD, Strassburg CP, Spengler U. Spontaneous bacterial peritonitis: the clinical challenge of a leaky gut and a cirrhotic liver. World J Hepatol. 2015;7:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bibi S, Ahmed W, Arif A, Khan F, Alam SE. Clinical, laboratory and bacterial profile of spontaneous bacterial peritonitis in chronic liver disease patients. J Coll Physicians Surg Pak. 2015;25:95–99. [PubMed] [Google Scholar]
- 74.Shi L, Wu D, Wei L, et al. Nosocomial and community-acquired spontaneous bacterial peritonitis in patients with liver cirrhosis in China: comparative microbiology and therapeutic implications. Sci Rep. 2017;7:46025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vilstrup H, Amodio P, Bajaj J, et al. Hepatic encephalopathy in chronic liver disease: 2014 practice guideline by the American Association for the study of liver diseases and the European Association for the study of the liver. Hepatology. 2014;60:715–735. [DOI] [PubMed] [Google Scholar]
- 76.Kang DJ, Betrapally NS, Ghosh SA, et al. Gut microbiota drive the development of neuroinflammatory response in cirrhosis in mice. Hepatology. 2016;64: 1232–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ahluwalia V, Betrapally NS, Hylemon PB, et al. Impaired gut-liver-brain axis in patients with cirrhosis. Sci Rep. 2016:6:26800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dunn W, O’Neil M, Zhao J, et al. Donor PNPLA3 rs738409 genotype affects fibrosis progression in liver transplantation for hepatitis C. Hepatology. 2014;59:453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun LY, Yang YS, Qu W, et al. Gut microbiota of liver transplantation recipients. Sci Rep. 2017;7:3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bajaj JS, Fagan A, Sikaroodi M, et al. Liver transplant modulates gut microbial dysbiosis and cognitive function in cirrhosis. Liver Transplant. 2017;23: 907–914. [DOI] [PubMed] [Google Scholar]
- 81.Bajaj JS, Kakiyama G, Cox 1J et al. Alterations in gut microbial function following liver transplant. Liver Transplant. 2018;24:752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu HX, Rocha CS, Dandekar S, Wan YJ. Functional analysis of the relationship between intestinal microbiota and the expression of hepatic genes and pathways during the course of liver regeneration. J Hepatol. 2016;64: 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu HX, Hu Y. Wan YJ Microbiota and bile acid profiles in retinoic acid-primed mice that exhibit accelerated liver regeneration. Oncotarget. 2Q16;7: 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bajaj JS. Liu EJ Kheradman R, et al. Fungal dysbiosis in cirrhosis. Gut. 2018;67: 1146–1154. [DOI] [PubMed] [Google Scholar]
- 86.Yang AM, Inamine T, Hochrath K, et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017;127:2829–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. [DOI] [PubMed] [Google Scholar]
- 88.Bik EM, Long CD, Armitage GC, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISMEJ. 2010;4:962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bajaj JS, Betrapally NS, Hylemon PB, et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62:1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bajaj JS, Acharya C, Fagan A. et al. Proton pump inhibitor initiation and withdrawal affects gut microbiota and readmission risk in cirrhosis. Am J Gastroenterol. 2018:113:1177–1186. [DOI] [PubMed] [Google Scholar]
- 91.Takeshita T, Suzuki N, Nakano Y, et al. Discrimination of the oral microbiota associated with high hydrogen sulfide and methyl mercaptan production. Sci Rep. 2012:2:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Al Mardini H, Bartlett K, Record CO. Blood and brain concentrations of mercaptans in hepatic and methanethiol induced coma. Gut. 1984;25:284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fatima N, Akhtar T, Sheikh N. Prebiotics: a novel approach to treat hepatocellular carcinoma. Chin J Gastroenterol Hepatol. 2017:2017:6238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li J, Sung CY, Lee N, et al. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Natl Acad Sri USA. 2016;113: E1306–E1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nilsson HO, Mulchandani R, Tranberg KG, Stenram li, Wadström T Helicobacter species identified in liver from patients with cholangiocarcinoma and hepatocellular carcinoma. Gastroenterology. 2001;120:323–324. [DOI] [PubMed] [Google Scholar]
- 96.Huang Y, Fan XG, Wang ZM, Zhou JH, Tian XF, Li N. Identification of Helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J Clin Pathol. 2004;57:1273–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krüttgen A, Horz HP. Weber-Heynemann J et al. Study on the association of Helicobacter species with viral hepatitis-induced hepatocellular carcinoma. Gut Microb. 2012;3:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fox JG, Feng Y, Theve EJ, et al. Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut. 2010;59: 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grat M, Wronka KM, Krasnodebski M, et al. Profile of gut microbiota associated with the presence of hepatocellular cancer in patients with liver cirrhosis. Transplant Proc. 2016;48:1687–1691. [DOI] [PubMed] [Google Scholar]
- 100.Xie G, Wang X, Liu P, et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget. 2016;7:19355–19366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang HL, Yu LX, Yang W, et al. Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarrinogenesis in rats J Hepatol. 2012;57:803–812. [DOI] [PubMed] [Google Scholar]
- 102.Gómez-Hurtado I, Santacruz A, Peiró G, et al. Gut microbiota dysbiosis is associated with inflammation and bacterial translocation in mice with CC14-induced fibrosis. PLoS One. 2011;6, e23037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dapito DH, Mencin A, Gwak GY, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu H, Ren Z, Li A, et al. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci Rep. 2016:6:33142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yamada S Takashina Y, Watanabe M, et al. Bile acid metabolism regulated by the gut microbiota promotes non-alcoholic steatohepatitis-associated hepatocellular carcinoma in mice. Oncotarget. 2018;9:9925–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yoshimoto S, Loo TM, Atarashi K. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. [DOI] [PubMed] [Google Scholar]
- 107.Hill C, Guarner F, Reid G, et al. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014:11:506–514. [DOI] [PubMed] [Google Scholar]
- 108.Brandi G, De Lorenzo S. Candela M, et al. Microbiota, NASH, HCC and the potential role of probiotics. Carcinogenesis. 2017;38:231–240. [DOI] [PubMed] [Google Scholar]
- 109.Tarantino G, Finelli C. Systematic review on intervention with prebiotics/probiotics in patients with obesity-related nonalcoholic fatty liver disease. Future Microbiol. 2015;10:889–902. [DOI] [PubMed] [Google Scholar]
- 110.Qamar AA. Probiotics in nonalcoholic fatty liver disease, nonalcoholic steatohepatitis, and cirrhosis. J Clin Gastroenterol. 2015;49:S28–S32. [DOI] [PubMed] [Google Scholar]
- 111.Xue L, He J, Gao N, et al. Probiotics may delay the progression of nonalcoholic fatty liver disease by restoring the gut microbiota structure and improving intestinal endotoxemia. Sci Rep. 2017:7:45176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cesaro C, Tiso A, Del Prete A, et al. Gut microbiota and probiotics in chronic liver diseases. Dig Liver Dis. 2011. ;43:431–438. [DOI] [PubMed] [Google Scholar]
- 113.Chen M, Wang MC, Ni R, et al. Role of probiotics in treatment of nonalcoholic fatty liver disease. Zhonghua Gan Zang Bing Za Zhi. 2017;25:77–80. [DOI] [PubMed] [Google Scholar]
- 114.Ritze Y, Bárdos G, Claus A, et al. Lactobacillus rhamnosus GG protects against non-alcoholic fatty liver disease in mice. PLoS One. 2014;9, e80169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim B, Park KY, Ji Y, Park S, Holzapfel W, Hyun CK. Protective effects of Lactobacillus rhamnosus GG against dyslipidemia in high-fat diet-induced obese mice. Biochem Biophys Res Commun. 2016;473:530–536. [DOI] [PubMed] [Google Scholar]
- 116.Okubo H, Sakoda H, Kushiyama A, et al. Lactobacillus casei strain Shirota protects against nonalcoholic steatohepatitis development in a rodent model. Am J Physiol Gastrointest Liver Physiol. 2013;305:G911–G918. [DOI] [PubMed] [Google Scholar]
- 117.Naito E, Yoshida Y, Makino K, et al. Beneficial effect of oral administration of Lactobacillus casei strain Shirota on insulin resistance in diet-induced obesity mice. J Appl Microbiol. 2011; 110:650–657. [DOI] [PubMed] [Google Scholar]
- 118.Wagnerberger S, Spruss A, Kanuri G, et al. Lactobacillus casei Shirota protects from fructose-induced liver steatosis: a mouse model. J Nutr Biochem. 2013;24:531–538. [DOI] [PubMed] [Google Scholar]
- 119.Wang Y, Xu N, Xi A, Ahmed Z, Zhang B, Bai X. Effects of Lactobacillus plantarum MA2 isolated from Tibet kefir on lipid metabolism and intestinal microflora of rats fed on high-cholesterol diet. Appl Microbiol Biotechnol. 2009:84:341–347. [DOI] [PubMed] [Google Scholar]
- 120.Li C, Nie SP, Zhu KX, et al. Lactobacillus plantarum NCU116 improves liver function, oxidative stress and lipid metabolism in rats with high fat diet induced non-alcoholic fatty liver disease. Food Funct. 2014;5:3216–3223. [DOI] [PubMed] [Google Scholar]
- 121.Xin J Zeng D Wang H, et al. Preventing non-alcoholic fatty liver disease through Lactobacillus johnsonii BS15 by attenuating inflammation and mitochondrial injury and improving gut environment in obese mice. Appl Microbiol Biotechnol. 2014;98:6817–6829. [DOI] [PubMed] [Google Scholar]
- 122.Kang JH, Yun SI, Park MH, Park JH, Jeong SY, Park HO. Anti-obesity effect of Lactobacillus gasseri BNR17 in high-sucrose diet-induced obese mice. PLoS One. 2013;8, e54617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen J, Wang R, Li XF, Wang RL. Bifidobacterium adolescentis supplementation ameliorates visceral fat accumulation and insulin sensitivity in an experimental model of the metabolic syndrome. Br J Nutr. 2012:107: 1429–1434. [DOI] [PubMed] [Google Scholar]
- 124.Xu RY, Wan YP, Fang QY, Lu W, Cai W. Supplementation with probiotics modifies gut flora and attenuates liver fat accumulation in rat nonalcoholic fatty liver disease model. J Clin Biochem Nutr. 2012;50:72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Seo M, Inoue I Tanaka M, et al. Clostridium butyricum MIYAIR1 588 improves high-fat diet-induced non-alcoholic fatty liver disease in rats. Dig Dis Sci. 2013;58:3534–3544. [DOI] [PubMed] [Google Scholar]
- 126.Endo H, Niioka M, Kobayashi N, Tanaka M, Watanabe T. Butyrate-producing probiotics reduce nonalcoholic fatty liver disease progression in rats: new insight into the probiotics for the gut-liver axis. PLoS One. 2013:8, e63388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kumar M, Verma V, Nagpal R, et al. Effect of probiotic fermented milk and chlorophyllin on gene expressions and genotoxicity during AFB1-induced hepatocellular carcinoma. Gene. 2011;490:54–59. [DOI] [PubMed] [Google Scholar]
- 128.Vajro P, Mandato C Licenziati MR. et al. Effects of Lactobacillus rhamnosus strain GG in pediatric obesity-related liver disease. J Pediatr Gastroenterol Nutr. 2011;52:740–743. [DOI] [PubMed] [Google Scholar]
- 129.Aller R, De Luis DA, Izaola O, et al. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: a double blind randomized clinical trial. Eur Rev Med Pharmacol Sci. 2011; 15:1090–1095. [PubMed] [Google Scholar]
- 130.Alisi A, Bedogni G, Baviera G, et al. Randomised clinical trial: the beneficial effects of VSL#3 in obese children with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2014;39:1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bajaj JS, Heuman DM, Hylemon PB, et al. Randomised clinical trial: Lactobacillus GG modulates gut microbiome, metabolome and endotoxemia in patients with cirrhosis. Aliment Pharmacol Ther. 2014;39:1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Malaguarnera M, Greco F, Barone G, Gargante MP, Toscano MA. Bifidobacterium longum with fructooligosaccharide (FOS) treatment in minimal hepatic encephalopathy: a randomized, double-blind, placebo-controlled study. Dig Dis Sci. 2007:52:3259–3265. [DOI] [PubMed] [Google Scholar]
- 133.Malaguarnera M, Gargante MP, Malaguarnera G, et al. Bifidobacterium combined with fructo-oligosaccharide versus lactulose in the treatment of patients with hepatic encephalopathy. Eur J Gastroenterol Hepatol. 2010;22: 199–206. [DOI] [PubMed] [Google Scholar]
- 134.Lunia MK, Sharma BC, Sharma P. Sachdeva S Srivastava S Probiotics prevent hepatic encephalopathy in patients with cirrhosis: a randomized controlled trial. Clin Gastroenterol Hepatol. 2014;12:1003–1008 (el). [DOI] [PubMed] [Google Scholar]
- 135.Dhiman RK, Rana B, Agrawal S, et al. Probiotic VSL#3 reduces liver disease severity and hospitalization in patients with cirrhosis: a randomized, controlled trial. Gastroenterology. 2014;147:1327–1337 (e3). [DOI] [PubMed] [Google Scholar]
- 136.Rincón D Vaquero J Hernando A, et al. Oral probiotic VSL#3 attenuates the circulatory disturbances of patients with cirrhosis and ascites. Liver Int. 2014;34:1504–1512. [DOI] [PubMed] [Google Scholar]
- 137.Agrawal A, Sharma BC. Sharma P Sarin SK, Secondary prophylaxis of hepatic encephalopathy in cirrhosis: an open-label, randomized controlled trial of lactulose, probiotics, and no therapy. Am J Gastroenterol. 2012; 107: 1043–1050. [DOI] [PubMed] [Google Scholar]
- 138.Solga SF, Buckley G, Clark JM, Horska A, Diehl AM. The effect of a probiotic on hepatic steatosis. J Clin Gastroenterol. 2008;42:1117–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Andreasen AS, Larsen N, Pedersen-Skovsgaard T, et al. Effects of Lactobacillus acidophilus NCFM on insulin sensitivity and the systemic inflammatory response in human subjects. Br J Nutr. 2010;104:1831–1838. [DOI] [PubMed] [Google Scholar]
- 140.Lewis SJ, Burmeister S. A double-blind placebo-controlled study of the effects of Lactobacillus acidophilus on plasma lipids. Eur J Clin Nutr. 2005;59: 776–780. [DOI] [PubMed] [Google Scholar]
- 141.Gibson GR, Hutkins R, Sanders ME, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491–502. [DOI] [PubMed] [Google Scholar]
- 142.Hutkins RW, Krumbeck JA, Bindels LB, et al. Prebiotics: why definitions matter. Curr Opin Biotechnol. 2016;37:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Daubioul CA, Horsmans Y, Lambert P, Danse E, Delzenne NM. Effects of oligofructose on glucose and lipid metabolism in patients with nonalcoholic steatohepatitis: results of a pilot study. Eur J Clin Nutr. 2005;59:723–726. [DOI] [PubMed] [Google Scholar]
- 144.Parnell JA, Raman M, Rioux KP. Reimer RA The potential role of prebiotic fibre for treatment and management of non-alcoholic fatty liver disease and associated obesity and insulin resistance. Liver Inc. 2012;32:701–711. [DOI] [PubMed] [Google Scholar]
- 145.Sawas T, Al Halabi S, Hemaez R, Carey WD, Cho WK. Patients receiving prebiotics and probiotics before liver transplantation develop fewer infections than controls: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2015;13:1567–1574 (e3). [DOI] [PubMed] [Google Scholar]
- 146.Cani PD, Possemiers S, Van de Wiele T. et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Matsumoto K, Ichimura M, Tsuneyama K, et al. Fructo-oligosaccharides and intestinal barrier Function in a methionine-choline-deficient mouse model of nonalcoholic steatohepatitis. PLoS One. 2017; 12, eOl 75406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pachikian BD, Essaghir A, Demoulin JB, et al. Prebiotic approach alleviates hepatic steatosis: implication of fatty acid oxidative and cholesterol synthesis pathways. Mol Nutr Food Res. 2013;57:347–359. [DOI] [PubMed] [Google Scholar]
- 149.Salminen S, Salminen E. Lactulose, lactic acid bacteria, intestinal microecology and mucosal protection. Scand J Gastroenterol Suppl. 1997;222:45–48. [DOI] [PubMed] [Google Scholar]
- 150.Fan JG, Xu ZJ, Wang GL Effect of lactulose on establishment of a rat nonalcoholic steatohepatitis model. World j Gastroenterol. 2005;11:5053–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Neyrinck AM, Possemiers S, Verstraete W. De Backer F, Cani PD Delzenne NM Dietary modulation of clostridial cluster XlVa gut bacteria (Roseburia spp.) by chitin-glucan fiber improves host metabolic alterations induced by high-fat diet in mice. J Nutr Biochem. 2012;23:51–59. [DOI] [PubMed] [Google Scholar]
- 152.Singh DP, Khare P, Zhu J, et al. A novel cobiotic-based preventive approach against high-fat diet-induced adiposity, nonalcoholic fatty liver and gut derangement in mice. Int J Obes. 2016;40:487–496. [DOI] [PubMed] [Google Scholar]
- 153.Pandey KR, Naik SR, Vakil BV. Probiotics, prebiotics and synbiotics-a review. J Food Sci Technol. 2015;52:7577–7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Raso GM. Simeoli R, lacono A, et al. Effects of a Lactobacillus paracasei B21060 based synbiotic on steatosis, insulin signaling and toll-like receptor expression in rats fed a high-fat diet. J Nutr Biochem. 2014;25:81–90. [DOI] [PubMed] [Google Scholar]
- 155.Mofidi F, Poustchi H, Yari Z, et al. Synbiotic supplementation in lean patients with non-alcoholic fatty liver disease: a pilot, randomised, double-blind, placebo-controlled, clinical trial. Br J Nutr. 2017;117:662–668. [DOI] [PubMed] [Google Scholar]
- 156.Asgharian A, Askari G, Esmailzade A, Feizi A, Mohammadi V. The effect of symbiotic supplementation on liver enzymes, C-reactive protein and ultrasound findings in patients with non-alcoholic fatty liver disease: a clinical trial. Int J Prev Med. 2016;7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Malaguamera M, Vacante M, Antic T, et al. Bifidobacterium longum with fructooligosaccharides in patients with non-alcoholic steatohepatitis. Dig Dis Sci. 2012;57:545–553. [DOI] [PubMed] [Google Scholar]
- 158.Jena PK, Sheng L, Nagar N, et al. Synbiotics Bifidobacterium infantis and milk oligosaccharides are effective in reversing cancer-prone non-alcoholic steatohepatitis using Western diet-fed FXR knockout mouse models. J Nutr Biochem. 2018;57:246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jena PK, Sheng L, Nagar N, et al. The effect of synbiotics Bifidobacterium Infantis and milk oligosaccharides on shaping gut microbiota community structure and NASH treatment Data. Brief. 2018;19:1025–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liang Y, Lin C, Zhang Y, Deng Y, Liu C, Yang Q. Probiotic mixture of Lactobacillus and Bifidobacterium alleviates systemic adiposity and inflammation in non-alcoholic fatty liver disease rats through Gprl09a and the commensal metabolite butyrate. Inflammopharmacology. 2018;26:1051–1055. [DOI] [PubMed] [Google Scholar]
- 161.Zhou D, Pan Q, Xin FZ, et al. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J Gastroenterol. 2017;23:60–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mollica MP, Mattace Raso G, Cavaliere G, et al. Butyrate regulates liver mitochondrial function, efficiency, and dynamics in insulin-resistant obese mice. Diabetes. 2017;66:1405–1418. [DOI] [PubMed] [Google Scholar]
- 163.Jin CJ, Engstler AJ, Sellmann C. et al. Sodium butyrate protects mice from the development of the early signs of non-alcoholic fatty liver disease: role of melatonin and lipid peroxidation. Br J Nutr. 2016;23:1–12. [DOI] [PubMed] [Google Scholar]
- 164.Bajaj JS, Kassam Z, Fagan A, et al. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: a randomized clinical trial. Hepatology. 2017;66:1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang WW, Zhang Y, Huang XB, You N, Zheng L, Li J. Fecal microbiota transplantation prevents hepatic encephalopathy in rats with carbon tetrachloride-induced acute hepatic dysfunction. World J Gastroenterol. 2017;23:6983–6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jegatheesan P, Beutheu S, Ventura G, et al. Effect of specific amino acids on hepatic lipid metabolism in fructose-induced non-alcoholic fatty liver disease. Clin Nutr. 2016;35:175–182. [DOI] [PubMed] [Google Scholar]
- 167.Herman MA, Samuel VT. The sweet path to metabolic demise: fructose and lipid synthesis. Trends Endocrinol Metab. 2016;27:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bajaj JS, Idilman R, Mabudian L, et al. Diet affects gut microbiota and modulates hospitalization risk differentially in an international cirrhosis cohort. Hepatology. 2018;68:234–247. [DOI] [PubMed] [Google Scholar]


