Abstract
The 2019 novel coronavirus (COVID-19) is a newly emerged strain that has never been found in humans before. At present, the laboratory-based reverse transcription-polymerase chain reaction (RT-PCR) is the main method to confirm COVID-19 infection. The intensification of the COVID-19 epidemic overwhelms limited clinical resources in particular, but not only, in developing countries, resulting in many patients not being tested for the infection and in large queues of potentially infected individuals waiting to be tested while providing a breeding ground for the disease. We describe here a rapid, highly sensitive, point-of-care, molecular test amenable for use at home, in the clinic, and at points of entry by minimally trained individuals and with minimal instrumentation. Our test is based on loop mediated isothermal amplification (COVID-19 LAMP) and for higher sensitivity on nested nucleic acid, two stage isothermal amplification (COVID-19 Penn-RAMP). Both tests can be carried out in closed tubes with either fluorescence or colorimetric (e.g., leuco crystal violet LCV) detection. COVID-19 LAMP performs on par with COVID-19 RT-PCR. COVID-19 RAMP has 10 fold better sensitivity than COVID-19 LAMP and COVID-19 RT-PCR when testing purified targets and 100 times better sensitivity than COVID-19 LAMP and COVID-19 RT-PCR when testing rapidly prepared sample mimics. Due to fortunate scarcity of COVID-19 infections in the USA, we were not able to test our assays and methods with patient samples. We hope that such tests will be carried out by colleagues in impacted countries. Our Closed-Tube Penn-RAMP has the potential to significantly reduce false negatives while being amenable to use with minimal instrumentation and training.
Graphical Abstract
Introduction
Coronaviruses are a large family of RNA viruses including HCoV-229E, OC43, NL63, and HKU1 that usually cause mild respiratory illnesses (1, 2) with the exceptions of the fatal Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) (3, 4) and Middle East Respiratory Syndrome Coronavirus (MERS) (5) of the last two decades. The 2019 novel coronavirus was discovered due to Wuhan Viral Pneumonia cases in 2019, and was named “COVID-19” by the World Health Organization (6). The COVID-19 is a newly emerged coronavirus that has never been found in humans before. Since December 2019, Wuhan City, Hubei Province has maintained surveillance of “influenza and related diseases,” and identified multiple cases of viral pneumonia with high mortality rate. The World Health Organization has classified the COVID-19 outbreak as “Public Health Emergency of International Concern” (7).
Reverse transcription-PCR (RT-PCR) kits have been rapidly developed for the qualitative detection of the COVID-19 in nasopharyngeal swabs, alveolar lavage fluid, sputum, and blood samples (8). However, RT-PCR tests require well-equipped laboratories and skilled personnel. The growing number of suspected cases exceeds the capacity of many hospitals, leaving many patients untested challenging to the control of the disease. A rapid, point -of –care molecular diagnostics for the COVID-19 is urgently needed.
We report here on simple closed-tube molecular tests for COVID-19 that can be carried out at home and in the clinic by minimally trained personnel without a need for sophisticated equipment. Using bioinformatics, we have identified highly conserved sequences in the COVID-19 and we have designed primers targeting the open reading frame 1ab (ORF1ab) gene of the COVID-19 RNA. Previously, we have shown that RT-LAMP successfully detects pathogen nucleic acids with either bioluminescent indicator and smartphone (9) or colorimetrically and with minimal instrumentation (10). Since many COVID-19 infected individuals are reported negative in the commonly used RT-PCR test, we address the need for a higher sensitivity test with our two-stage isothermal method, dubbed Penn-RAMP (11). We have originally developed Penn-RAMP to enable high level of multiplexing, but surreptitiously discovered that, in many cases, our two-stage method is 10 times more sensitive than LAMP and PCR when processing purified nucleic acids and 100 times more sensitive than LAMP and RT-PCR with minimally processed samples.
Since at the time of writing this paper, only a very small number of COVID-19 cases have been identified in the USA, we have not been able to test our assay with actual patient samples. Given the simplicity and promise of our test, we hope that colleagues in endemic regions will test our assays with actual samples.
Materials and Methods
LAMP Primer Design
Complete genome sequences of various COVID-19 (Table S1) were aligned and analyzed to identify conserved sequences using Clustal X (http://www.clustal.org/clustal2/) and then compared with sequences of other coronaviruses (Table S1). We selected to target the conserved sequence of ORF1ab because of its high homology among COVID-19s sequences and high divergence from all the other coronaviruses examined. Our LAMP primer set (Table 1) was designed with the PrimerExplorer V5 software (Eiken Chemical Co. Ltd) and primers’ specificity was verified with a BLAST search of the GenBank nucleotide database. The LAMP sequences were synthesized by Integrated DNA Technologies (IDT, Coralville, IA).
Table 1.
Sequences and concentrations of COVID-19 LAMP primers.
| Primer name | Sequence (5’ to 3’) | Concentration (μM) |
|---|---|---|
| F3 | TGCTTCAGTCAGCTGATG | 0.2 |
| B3 | TTAAATTGTCATCTTCGTCCTT | 0.2 |
| FIP | TCAGTACTAGTGCCTGTGCC- CACAATCGTTTTTAAACGGGT |
1.6 |
| BIP | TCGTATACAGGGCTTTTGACATCTA- TCTTGGAAGCGACAACAA |
1.6 |
| Loop F | CTGCACTTACACCGCAA | 0.8 |
| Loop B | GTAGCTGGTTTTGCTAAATTCC | 0.8 |
The COVID-19 mimic target
Synthesized DNA (Table S2, 619bp) containing the targeted sequence was obtained from IDT to mimic the COVID-19 target.
LAMP Assay
In addition to primers, the LAMP reaction mix contained 1 X Isothermal MasterMix (ISO-001, OptiGene, USA), 1 X EvaGreen® dye (Biotium, Inc.; excluded when colorimetric detection was used), and 1 μL of synthesized template. During lab-based testing, LAMP amplification was carried out with 10 μL reaction mix and monitored with a BioRad Thermal Cycler (BioRad, Model CFD3240) at 63°C for 50 minutes. Non-template controls (NTC) were included in each run to ensure absence of contamination.
Specificity of the newly developed LAMP primer set
RNAs of coronaviruses from different genera [Alphacoronavirus (PEDV and TGEV); Gammacoronavirus (IBV); and Deltacoronavirus (PDCoV)] that are available in our lab were used as negative controls to test the specificity of our newly developed LAMP assay.
qPCR Amplification
The gold standard RT-PCR currently used for routine tests of COVID-19 in Chinese laboratories was developed by the Chinese Center for Disease Control and Prevention (Chinese CDC) (12). The PCR primers and TaqMan probe for our qPCR test were synthesized by IDT (Table S3). Each qPCR reaction mix contained 1 X PrimeTime® Gene Expression Master Mix (IDT), 400 nM of each PCR primer, 250 nM of TaqMan probe, and 1 μL of synthesized COVID-19 target (Table S2). qPCR was carried out with a Thermal Cycler (BioRad, Model CFD3240) with a temperature profile of 95°C for 3 min, followed by 50 cycles of amplification (95°C for 15 s and 60°C for 1 min).
Closed-tube Penn-RAMP
Penn-RAMP consists of two isothermal amplification processes: Recombinase Polymerase Amplification (RPA) (38°C) and LAMP (63°C). We carry out the RPA process in the cap of a tube and the LAMP in the tube. The RPA reaction mix includes 480 nM of each LAMP F3 and B3 primers, 1X rehydrated twistAmp® Basic buffer (twistAmp® Basic kit, TwistDx Limited, Cambridge, UK), 14 mM MgAc. The sample with various target concentrations is inserted in the cap together with the RPA mix. The LAMP reaction mix is the same as described above, but without any target. The ratio of the volumes of RPA and LAMP reaction mixtures was kept at 1 : 9 to prevent inhibition of the LAMP reaction. Typically, we used RPA volume of 5 μL and LAMP volume of 45 μL. When a centrifuge is not available, it may be desirable to increase the LAMP volume to 70 μL (see below). After loading the tube cap with RPA mix and the tube itself with the LAMP mix, the tube was sealed and remained so throughout the duration of the process, protecting the work area from exposure to amplicons. The closed tube was incubated in our thermal cycler with the lid and block temperature at 38°C. After 15–20 min incubation, the tube was either centrifuged or flipped back and forth a few times to blend the RPA and LAMP reaction volumes. The tube was then incubated with the thermal cycler lid and block temperature at 63°C for 40 min with real time signal monitoring.
Rapid Sample Preparation
Since we have no access to COVID-19 samples, we tested sample collection with inactivated HIV virus particles. We collected nasal mucosa from healthy volunteers with nasal swabs; spiked the swabs with various concentrations of HIV particles and other pathogens; submerged the swabs in 3 mL purified water; and heated the water to 95°C and 65°C for various time. We determined the efficacy of our sample processing method by monitoring threshold amplification time as a function of sample prep conditions.
Colorimetric Detection of COVID-19
LCV solution containing 0.5 mM crystal violet (CV), 30 mM sodium sulfite, and 5 mM β-cyclodextrin was prepared and stored at −20 °C until use. For field use, LCV can be lyophilized for long shelf life at room-temperature (13). The first-stage RPA mix was carried out as described above. The second-stage LAMP reaction was performed with a Loopamp DNA amplification kit (Eiken Chemical Co., Ltd., Japan) that, in contrast to the Optigene kit, does not include fluorescent dye. To ease hand-mixing, we used 70 μL LAMP volume. The LAMP reaction mix included: 1.6 μM FIP and BIP, 0.8 μM LF and LB; 1 μL Bst DNA polymerase (provided in Loopamp DNA amplification kit, http://loopamp.eiken.co.jp/e/products/dna/); and 5.5 μL LCV solution. The reaction buffer was incubated at 63°C for up to 40 min. The LCV color change was observed at the end of the incubation process by naked eye and, if desired, it can be recorded with a smartphone.
RESULTS and DISCUSSION
Analytical Performance of COVID-19 LAMP Assay
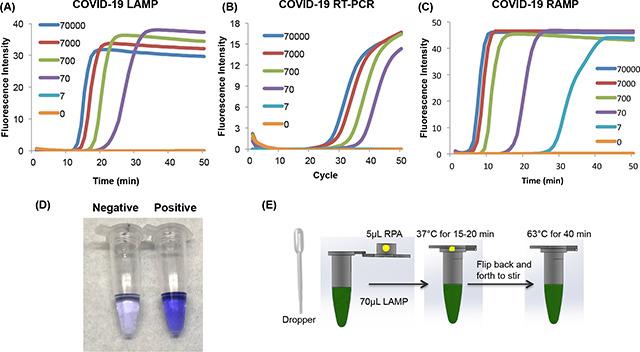
Our COVID-19 LAMP successfully detected fewer than 100 targets per reaction volume, comparable to COVID-19 PCR (Fig. 1) albeit with somewhat shorter time. The threshold times of our LAMP assay (Fig. 1A) correlated linearly (Fig. 1D) with template concentration, indicating that our assay is quasi-quantitative and can be used for disease progression monitoring. In contrast to PCR, our LAMP assay is incubated at a fixed temperature (63°C) greatly simplifying instrumentation and reducing power consumption. Indeed, the LAMP assay can be incubated with an exothermic chemical reaction in the absence of electrical power (9, 10).
Figure 1: Comparison of LAMP, RT-PCR, and Closed-Tube Penn-RAMP for COVID-19 Detection.
(A) LAMP, (B) PCR, (C) closed-tube Penn-RAMP detection of 70000, 7000, 700, 70, 7, and 0 (no template control) copies per reaction. The limits of detection of LAMP, PCR, and closed-tube Penn-RAMP are, respectively 70, 70, and 7 copies per reaction. The threshold time of (D) LAMP, threshold cycle of (E) PCR, and threshold time of (F) one-tube Penn-RAMP as functions of the log of nCoV-2019 target concentration (n = 3).
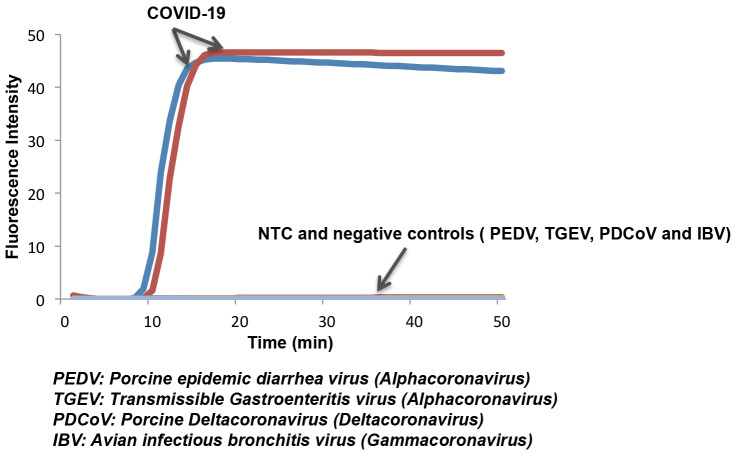
In addition to examining COVID-19 LAMP specificity in silico, we tested samples of other coronaviruses available in our lab such as alphacoronaviruses (PEDV and RGEV), Gammacoronavirus (IBV), and deltacoronavirus (PDCoV). Only samples containing COVID-19 exhibited a positive signal (Fig. 2). Our test does not exhibit any false positives.
Figure 2: LAMP primers for the COVID-19 are specific.
Only samples with COVID-19 nucleic acid show positive signal, while negative coronaviruses controls (PEDV, TGE, PDCoV, and IBV) and non-template control (NTC) do not show any signal during incubation.
Analytical Performance of COVID-19 Closed-Tube Penn-RAMP Assay
Our COVID-19 Closed-Tube Penn-RAMP Assay (Fig. 1) shows that as low as 7 copies of COVID-19 target can be readily detected by one-tube Penn-RAMP, while less than 70 copies of COVID-19 target cannot be detected by LAMP and RT-PCR, showing that the sensitivity of our one-tube Penn-RAMP is 10 times higher than LAMP and RT-PCR.
Currently, for the patients with low viral load, due to insufficient detection sensitivity of RT-PCR, many false negative results and so-called “relapse after negative”cases were reported (14). It is very likely our one-tube Penn-RAMP method can alleviate this situation with its higher sensitivity.
Home Test for COVID-19
Both COVID-19 LAMP and COVID-19 Closed-Tube Penn-RAMP can be carried out with minimal sample preparation at home and in settings with rudimentary equipment.
Rapid Sample Preparation
Minimally invasive sample collection is desired for home and field testing. We selected to use nasal swabs consistent with the common sampling method in endemic regions. In the absence of clinical COVID-19 samples, we contrived samples by spiking inactivated HIV viral particles into the nasal swab after nasal mucosa collection. The objective of the experiment is to determine whether we can minimize polymerase inhibition with minimal sample preparation such as eluting the swab into water and heating. We measured the amplification threshold time as a function of sample incubation time and incubation temperature and determined that incubation for over 15 min at temperature above 65°C is enough to lyse the virus and suppress inhibitors (Figure S1) with a relatively small increase in threshold time and reduced reproducibility (Figure S2). The rapid sample preparation may not be adequate for quantification; but is good enough to confirm infection. Rapid sample preparation with COVID-19 LAMP and COVID-19 PCR provided 100% sensitivity at 700 viral particles per reaction and 75% sensitivity at 70 viral copies per reaction (Table 2). In contrast, COVID-19 Closed-Tube Penn-RAMP provided 100% sensitivity at 7 virions per reaction. COVID-19 Closed-Tube Penn-RAMP appears to have much greater tolerance to inhibitors than either PCR or LAMP, performing with minimally prepared samples nearly as well as with purified samples.
Table 2:
Sensitivity of LAMP, qPCR, and Closed-Tube Penn-RAMP for the detection of rapidly prepared COVID-19 nasal swab samples (mimic)*.
| Spiked samples | COVID-19-LAMP assay | COVID-19-qPCR assay | COVID-19-RAMP assay |
|---|---|---|---|
| 70000 copies/reaction | 4/4 | 4/4 | 4/4 |
| 7000 copies/reaction | 4/4 | 4/4 | 4/4 |
| 70 copies/reaction | 3/4 | 3/4 | 4/4 |
| 7 copies/reaction | 0/4 | 0/4 | 4/4 |
| 0 copies/reaction | 0/4 | 0/4 | 0/4 |
The table documents the number of positive results normalized with the number of tests.
Colorimetric Detection of COVID-19
Visual detection does not require any instrumentation and therefore is desired for diagnostic systems for home use. The LCV dye is nearly colorless in the absence of dsDNA and deep violet in presence of dsDNA, enabling detection of amplicons. We included LCV in the LAMP reaction mix in our Closed-Tube Penn-RAMP assay (Fig. 3A). The process was carried out with 70 μL of LAMP mix and manual flipping of the tube (without any centrifugation), demonstrating successful instrument-free detection of as few as 100 targets per reaction.
Figure 3: Home Test for COVID-19.
(A) Visual detection of COVID-19 with our one-tube Penn-RAMP with LCV dye. Negative: 0 copies of COVID-19 synthesized DNA; Positive: 100 copies of synthesized DNA. (B) Sequence of operations of the home test. The reactions can be incubated either in a block heater or in a domestic oven with temperature control.
CONCLUSIONS
We report on a closed tube, single stage COVID-19 LAMP and COVID-19 Closed-Tube Penn-RAMP assays for use at home, in the clinic and at ports of entry. Our assays require minimal sample processing. The patient or health care provider collects a nasal sample with swab, elutes the swab in water and heats the water to above 65°C. The sample is then inserted into a tube for single stage amplification or two stage amplification (with greater sensitivity). The reaction tube is incubated at 63°C in the LAMP process and at 38°C and 63°C in the two stage Penn-RAMP process. The incubation can be carried out with simple instrumentation as no thermal cycling is needed. The amplification process can be monitored in real time with fluorescent dye, enabling quantification. In prior work, we have demonstrated that fluorescent dye can be excited with a smartphone flash and read with a smartphone camera (15). To simplify our system even further, we have demonstrated that we can detect test results colorimetrically with LCV dye that changes color from colorless in the absence of dsDNA to deep violet color in the presence of abundant dsDNA. This color change is visible to the naked eye and does not require any instrumentation. All reagents can be readily stored in the tube in dry form, providing long, refrigeration-free shelf life.
Although much simpler, our COVID-19 LAMP test performs on par with the conventionally used COVID-19 PCR, providing similar sensitivity. Our closed-tube, two stage Penn RAMP outperforms both PCR and LAMP providing 10-fold better sensitivity than LAMP alone when purified samples are used and 100-fold better sensitivity when minimally prepared samples are used. This added sensitivity is important since it has been reported that a significant number of COVID-19 patients test negative with the COVID-19 PCR test.
By necessity, our experiments were restricted to contrived samples as fortunately the number of COVID-19 infections in the USA is very small. We hope that scientists and medical personnel in endemic regions will be able to validate our test and put it into a good use.
Supplementary Material
Acknowledgment
The Fulbright Visiting Scholar Program supported Dr. Mohamed El-Tholoth. H.H.B. is supported, in part, by NIH grant R21 AI134594-01A1 to the University of Pennsylvania. J.S. is supported, in part, by NIH grant K01 1K01TW011190–01A1 to the University of Pennsylvania.
Footnotes
CONFLICT OF INTEREST
University of Pennsylvania has applied for a patent on Penn-RAMP with J.S. and H.H.B. listed as co-inventors.
REFERENCES
- 1.Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229e, hku1, nl63, and oc43 detected over 3 years using a novel multiplex real-time pcr method. J Clin Microbiol 2010;48:2940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng ZQ, Chen DH, Tan WP, Qiu SY, Xu D, Liang HX, et al. Epidemiology and clinical characteristics of human coronaviruses oc43, 229e, nl63, and hku1: A study of hospitalized children with acute respiratory tract infection in guangzhou, china. Eur J Clin Microbiol Infect Dis 2018;37:363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World health organization. (2003). Consensus document on the epidemiology of severe acute respiratory syndrome (sars). World health organization; Https://apps.Who.Int/iris/handle/10665/70863. [Google Scholar]
- 4.Smith RD. Responding to global infectious disease outbreaks: Lessons from sars on the role of risk perception, communication and management. Soc Sci Med 2006;63:3113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zumla A, Hui DS, Perlman S. Middle east respiratory syndrome. Lancet 2015;386:995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World health organization. (2020). Surveillance case definitions for human infection with novel coronavirus (ncov): Interim guidance v1, January 2020. World health organization; Https://apps.Who.Int/iris/handle/10665/330376. License: Cc by-nc-sa 3.0 igo. [Google Scholar]
- 7.WHO SotsmotIHRECrtoonc-nJ, 2020), https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov).
- 8.Who, novel coronavirus (2019-ncov) technical guidance: Laboratory testing for 2019-ncov in humans. Https://www.Who.Int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance.
- 9.Song JZ, Pandian V, Mauk MG, Bau HH, Cherry S, Tisi LC, Liu CC. Smartphone-based mobile detection platform for molecular diagnostics and spatiotemporal disease mapping. Anal Chem 2018;90:4823–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song JZ, Mauk MG, Hackett BA, Cherry S, Bau HH, Liu CC. Instrument-free point-of-care molecular detection of zika virus. Anal Chem 2016;88:7289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song JZ, Liu CC, Mauk MG, Rankin SC, Lok JB, Greenberg RM, Bau HH. Two-stage isothermal enzymatic amplification for concurrent multiplex molecular detection. Clin Chem 2017;63:714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinese CDC, specific primers and probes for detection 2019 novel coronavirus. Http://ivdc.Chinacdc.Cn/kyjz/202001/t20200121_211337.Html.
- 13.Miyamoto S, Sano S, Takahashi K, Jikihara T. Method for colorimetric detection of double-stranded nucleic acid using leuco triphenylmethane dyes. Anal Biochem 2015;473:28–33. [DOI] [PubMed] [Google Scholar]
- 14.Xie Xingzhi, Zhong Zheng, Zhao Wei, Zheng Chao, Wang Fei, Liu Jun. Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing https://pubs.Rsna.Org/doi/10.1148/radiol.2020200343.2020. [DOI] [PMC free article] [PubMed]
- 15.Liao SC, Peng J, Mauk MG, Awasthi S, Song JZ, Friedman H, et al. Smart cup: A minimally-instrumented, smartphone-based point-of-care molecular diagnostic device. Sensor Actuat B-Chem 2016; 229 : 232–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.