Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019, causing a respiratory disease (coronavirus disease 2019, COVID-19) of varying severity in Wuhan, China, and subsequently leading to a pandemic. The transmissibility and pathogenesis of SARS-CoV-2 remain poorly understood. We evaluate its tissue and cellular tropism in human respiratory tract, conjunctiva, and innate immune responses in comparison with other coronavirus and influenza virus to provide insights into COVID-19 pathogenesis.
Methods
We isolated SARS-CoV-2 from a patient with confirmed COVID-19, and compared virus tropism and replication competence with SARS-CoV, Middle East respiratory syndrome-associated coronavirus (MERS-CoV), and 2009 pandemic influenza H1N1 (H1N1pdm) in ex-vivo cultures of human bronchus (n=5) and lung (n=4). We assessed extrapulmonary infection using ex-vivo cultures of human conjunctiva (n=3) and in-vitro cultures of human colorectal adenocarcinoma cell lines. Innate immune responses and angiotensin-converting enzyme 2 expression were investigated in human alveolar epithelial cells and macrophages. In-vitro studies included the highly pathogenic avian influenza H5N1 virus (H5N1) and mock-infected cells as controls.
Findings
SARS-CoV-2 infected ciliated, mucus-secreting, and club cells of bronchial epithelium, type 1 pneumocytes in the lung, and the conjunctival mucosa. In the bronchus, SARS-CoV-2 replication competence was similar to MERS-CoV, and higher than SARS-CoV, but lower than H1N1pdm. In the lung, SARS-CoV-2 replication was similar to SARS-CoV and H1N1pdm, but was lower than MERS-CoV. In conjunctiva, SARS-CoV-2 replication was greater than SARS-CoV. SARS-CoV-2 was a less potent inducer of proinflammatory cytokines than H5N1, H1N1pdm, or MERS-CoV.
Interpretation
The conjunctival epithelium and conducting airways appear to be potential portals of infection for SARS-CoV-2. Both SARS-CoV and SARS-CoV-2 replicated similarly in the alveolar epithelium; SARS-CoV-2 replicated more extensively in the bronchus than SARS-CoV. These findings provide important insights into the transmissibility and pathogenesis of SARS-CoV-2 infection and differences with other respiratory pathogens.
Funding
US National Institute of Allergy and Infectious Diseases, University Grants Committee of Hong Kong Special Administrative Region, China; Health and Medical Research Fund, Food and Health Bureau, Government of Hong Kong Special Administrative Region, China.
Introduction
Several coronaviruses infect the human respiratory tract, and usually cause mild disease; however, the beta coronaviruses severe acute respiratory syndrome-associated coronavirus (SARS-CoV) and Middle East respiratory syndrome-associated coronavirus (MERS-CoV) cause severe zoonotic respiratory disease. SARS emerged in 2002 in Guangdong province, China, and caused an epidemic leading to 8096 cases and 774 deaths globally in more than 25 countries across five continents, but was contained through public health interventions. MERS-CoV transmits from dromedary camels to humans, sometimes leading to clusters of human-to-human transmission, especially within health-care facilities. To date, within health-care facilities, with 2519 cases, with 866 deaths across 27 countries, have been confirmed as of January, 2020.1
In December, 2019, the novel coronavirus SARS-CoV-2 caused an outbreak of respiratory illness (coronavirus disease 2019; COVID-19) in Wuhan, China. Within 5 months, the disease burden and fatalities have surpassed both SARS and MERS, with more than 2 million confirmed cases and more than 150 000 deaths reported globally, as of April 19, 2020.2 WHO declared this outbreak a pandemic on March 11, 2020. Although the virus appears to be more transmissible than either SARS or MERS, disease severity is variable—from asymptomatic to fatal—and case fatality appears to be substantially lower than both SARS and MERS.3
Research in context.
Evidence before this study
We searched PubMed without language restriction for studies published from database inception until March 9, 2020, with the terms “SARS-CoV-2” or ”novel coronavirus” and “virus tropism” or “respiratory tract” or “ocular” or “conjunctiva” or “innate immunity” or “cytokine”, and found no relevant articles pertaining to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). To our knowledge, there have been no reports on infection, replication competence, tropism, and pathogenesis of the novel coronavirus SARS-CoV-2, in comparison with other respiratory pathogens including SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), 2009 pandemic influenza H1N1 virus (H1N1pdm), and highly pathogenic avian influenza H5N1 virus (H5N1), in human respiratory tract or extrapulmonary organs.
Added value of this study
We report that the conjunctival epithelium and the conducting airways appear to be potential portals of infection of SARS-CoV-2. Both SARS-CoV and SARS-CoV-2 replicated comparably in the alveolar epithelium, but SARS-CoV-2 replicated more extensively than SARS-CoV in bronchial epithelium, which might explain the increased transmissibility of the virus. SARS-CoV-2 was a less potent inducer of proinflammatory cytokines than H5N1, MERS-CoV, or H1N1pdm.
Implications of all the available evidence
SARS-CoV-2 replicates better than other human coronaviruses, such as SARS-CoV, but not as well as H1N1pdm in ex-vivo cultures of the human bronchus. The conjunctiva is an additional portal of infection. These findings are relevant to understanding transmission for infection prevention and control. Extrapulmonary routes of infection by SARS-CoV-2 should be further studied and validated in animal models.
Modes of transmission and pathogenesis have been key knowledge gaps. The virus is assumed to be primarily transmitted by large respiratory droplets, but there has been no direct evidence for this hypothesis. Identifying the organs and cell types that are permissive to implantation and virus replication will help to understand the portals by which the infection can be established. To our knowledge, to date there has been only one autopsy study reporting a patient dying after testing positive for SARS-CoV-2 using needle core samples and one histopathological study on two patients undergoing lobectomy.4, 5 Although these studies have provided information on the histopathology of COVID-19, they did not include characterisation of SARS-CoV-2 virus tropism by immunohistochemistry, and therefore do not shed light on virus tropism in the early stages of the infection process.4, 5 Thus, data from experimental infection of ex-vivo cultures of the respiratory tract are needed.
We have previously used ex-vivo cultures of the human lung, conducting airways, and ocular conjunctiva to investigate tropism of epidemic viruses such as the 2009 pandemic influenza H1N1 (H1N1pdm) virus and MERS-CoV.6, 7 Furthermore, when compared with patients with COVID-19 who were not in the intensive care unit (ICU), patients in the ICU had higher plasma concentrations of proinflammatory cytokines such as interferon gamma-induced protein 10 (IP-10), monocyte chemoattractant protein 1 (MCP-1), and tumour necrosis factor α (TNF-α).8 It is therefore important to investigate the role of innate host responses in pathogenesis.
We aimed to compare virus tropism and replication competence of SARS-CoV-2 virus with SARS-CoV, MERS-CoV, and H1N1pdm viruses in ex-vivo cultures of human bronchus and lung. The potential for SARS-CoV-2 to infect extrapulmonary tissues was also assessed using ex-vivo cultures of human conjunctiva and in-vitro cultures of human colorectal adenocarcinoma cell lines. The innate immune responses to SARS-CoV-2 were investigated in infected human alveolar epithelial cells and macrophages, and compared with those of highly pathogenic avian influenza (HPAI) H5N1 virus.
Methods
SARS-CoV-2 isolation
VeroE6 cells were used for virus isolation and were cultured in DMEM and 10% FCS. Cultured cell monolayers were maintained in their respective medium. The original clinical specimen was collected from the nasopharyngeal aspirate and throat swab of a patient (young adult male) with confirmed COVID-19 in Hong Kong in January, 2020, and was diluted (1:10) with DMEM supplemented with 2% FCS before adding to cells. After incubation at 37°C for 1·5 h, medium was topped-up to 1 mL with fresh culture medium. The cells were incubated at 37°C and observed daily for cytopathic effect (CPE). The culture supernatant was examined for presence of virus RNA by rtPCR.
All experiments were done in a biosafety level 3 facility at the School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong. Ethics approval of the use of human tissues was granted by the institutional review board of the University of Hong Kong and the hospital authority (Hong Kong West; institutional review board approval numbers UW 20–167 and UW19–802).
Viruses
In addition to the SARS-CoV-2 isolate (BetaCoV/Hong Kong/VM20001061/2020), we used: SARS-CoV (strain HK39849), isolated from a patient admitted to hospital with SARS infection in Hong Kong in 2003, MERS-CoV (prototype human MERS-CoV EMC strain, provided by Prof R Fouchier, Erasmus University Medical Center, Rotterdam, the Netherlands); the HPAI H5N1 virus (A/Hong Kong/483/1997), isolated from a fatal human infection in Hong Kong; and the 2009 H1N1pdm virus (A/Hong Kong/415742/2009), isolated from a patient in Hong Kong. SARS-CoV-2, SARS-CoV, and MERS-CoV viruses were passaged in VeroE6 cells and virus stock was aliquoted and titrated to determine 50% tissue culture infection dose (TCID50) in VeroE6 cells. Influenza viruses were passaged in Madin-Darby canine kidney (MDCK) cells and virus stock was aliquoted and titrated to determine TCID50 in MDCK cells.
Ex-vivo cultures and infection of human respiratory tract and conjunctiva
Fresh non-tumour conjunctiva (n=3), bronchus (n=5), and lung (n=4) tissues were obtained from patients aged 44–85 years undergoing elective surgery in Department of Ophthalmology and Surgery of Queen Mary Hospital (Pok Fu Lam, Hong Kong, China) from January to March, 2020, and were removed as part of clinical care but surplus for routine diagnostic requirements, as described previously.6, 9 Sampling of tissues was defined by convenience by the pathologists. The donor characteristics are listed in the appendix (p 7). Fragments of human tissues were infected with each virus at 5 × 105 TCID50 per mL for 1 h at 37°C for bronchus and lung tissues or 33°C for conjunctival tissues. Bronchus and lung tissues were infected with SARS-CoV-2, SARS-CoV, MCoV and H1N1pdm, whereas conjunctival tissues were infected with SARS-CoV-2 and SARS-CoV because of the small amount of tissue available. Mock-infected tissue—ie, tissue samples from the same specimens infected with medium without virus served as negative controls. The explants were washed three times with PBS and placed in culture medium (F-12K nutrient mixture with L-glutamine, and antibiotics) with or without a sterile surgical pathology sponge to establish an air–liquid interface condition in 24-well culture plates in a 37°C or 33°C incubator with 5% CO2. Infectious viral titres in culture supernatants were assessed at 1, 24, 48, 72 and 96 hours post-infection (hpi) by titration in VeroE6 or MDCK cells. Bronchus and lung tissues were fixed at 96 hpi and conjunctival tissues were fixed at 48 hpi in 10% formalin and processed for immunohistochemistry staining.
Viral titres in tissue culture wells without tissues were harvested at 1, 24, 48, and 72 hpi for titration by TCID50 assay to define the thermal inactivation of the virus in the absence of replication.
In-vitro culture and infection of alveolar epithelial cells, macrophage, and colorectal carcinoma cells
Primary human alveolar epithelial cells (AECs) and peripheral blood monocyte-derived macrophages were isolated from three donors and used for infection, as previously described,9 and human colorectal carcinoma cells (Caco-2; ATCC HTB-37) purchased from American Type Culture Collection were cultured in MEM with 10% FCS. AECs, macrophages, and Caco-2 cells were infected with SARS-CoV-2, SARS-CoV, MERS-CoV, H1N1pdm, and H5N1 viruses, either at a multiplicity of infection (MOI) of 0·1 for viral replication kinetics, or at an MOI of 2 for the analysis of cytokines (TNF-α and interleukin 6 [IL-6]), chemokines (IP-10, regulated on activation, normal T cell expressed and secreted [RANTES], and MCP-1), and angiotensin-converting enzyme 2 (ACE2) expression. Mock-infected cells served as negative controls. Viral titres in supernatant were determined by TCID50 assay. Cell lysates were collected at 24 hpi or 48 hpi, or both, for mRNA expression of SARS-CoV ORF1b,10 MERS-CoV UpE,7 influenza matrix gene, cytokines, chemokines, and ACE2 using rtPCR. Methods of culture, infection, and analysis are detailed in the appendix (pp 1–3).
Immunohistochemistry staining
The fixed paraffin-embedded ex-vivo cultures of human tissues were stained with cell type-specific markers (MUC5AC, ThermoFisher, Waltham, MA, USA; acetylated α-tubulin, Santa Cruz, Dallas, TX, USA; CC10, Protein-tech, Rosemont, IL, USA; p63a, Cell Signaling Technology, Danvers, MA, USA; pan cytokeratin AE1/AE3 and CD68, Dako, Agilent Technologies, Santa Clara, CA, USA), SARS-CoV-2 and SARS-CoV nucleoprotein (4D11),11 MERS-CoV nucleoprotein,7, 12 and influenza nucleoprotein (HB65, EVL anti-influenza nucleoprotein, subtype A).9, 13 Methods of immunohistochemistry staining are detailed in the appendix (p 3).
Statistical analysis
Experiments with the human ex-vivo cultures and in-vitro cultures of AECs and Caco-2 cells were done independently using tissue from least three different donors, each in duplicate or triplicate. Results shown in figures are mean (SD). Area-under-curve (AUC) was calculated by integrating infectious virus titres at 24–96 hpi in ex-vivo bronchus tissues, lung tissues, AECs, and macrophages; 24–48 hpi in conjunctiva tissues and 24–72 hpi in Caco-2 cells. The differences in log10-transformed viral titres and quantitative cytokine and chemokine mRNA between viruses and over time were compared using two-way ANOVA followed by a Bonferroni multiple-comparison test using GraphPad Prism, version 7.0. Differences were considered significant at a p value of less than 0·05.
Role of the funding source
The funders had no role in study design, data collection, analysis, or interpretation of the data, or in the writing of the report. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
Results
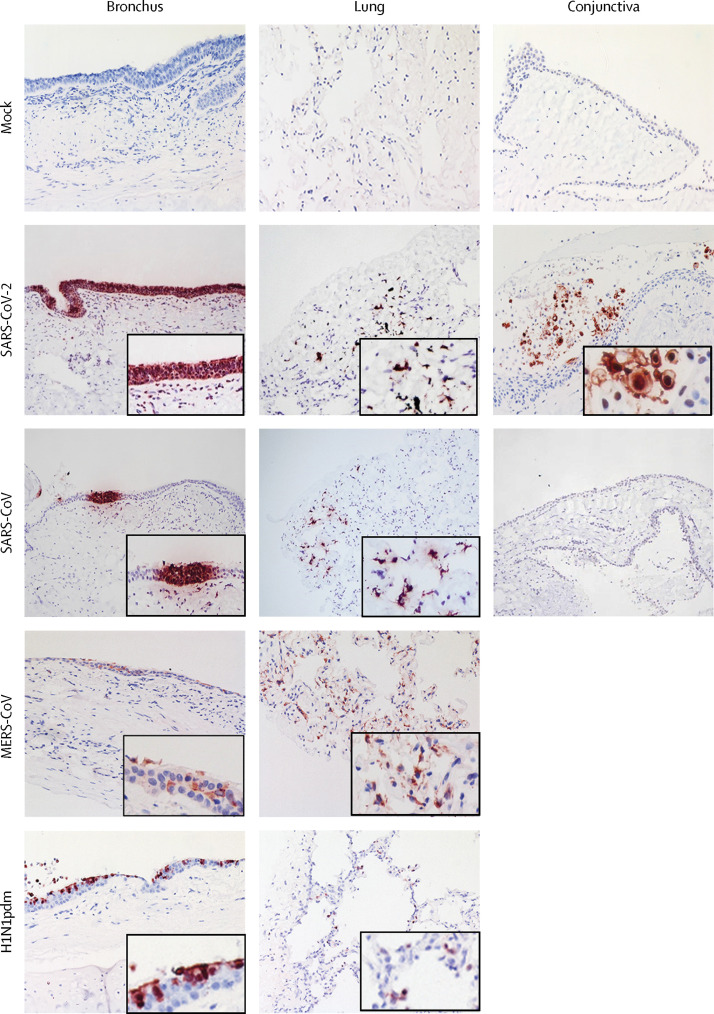
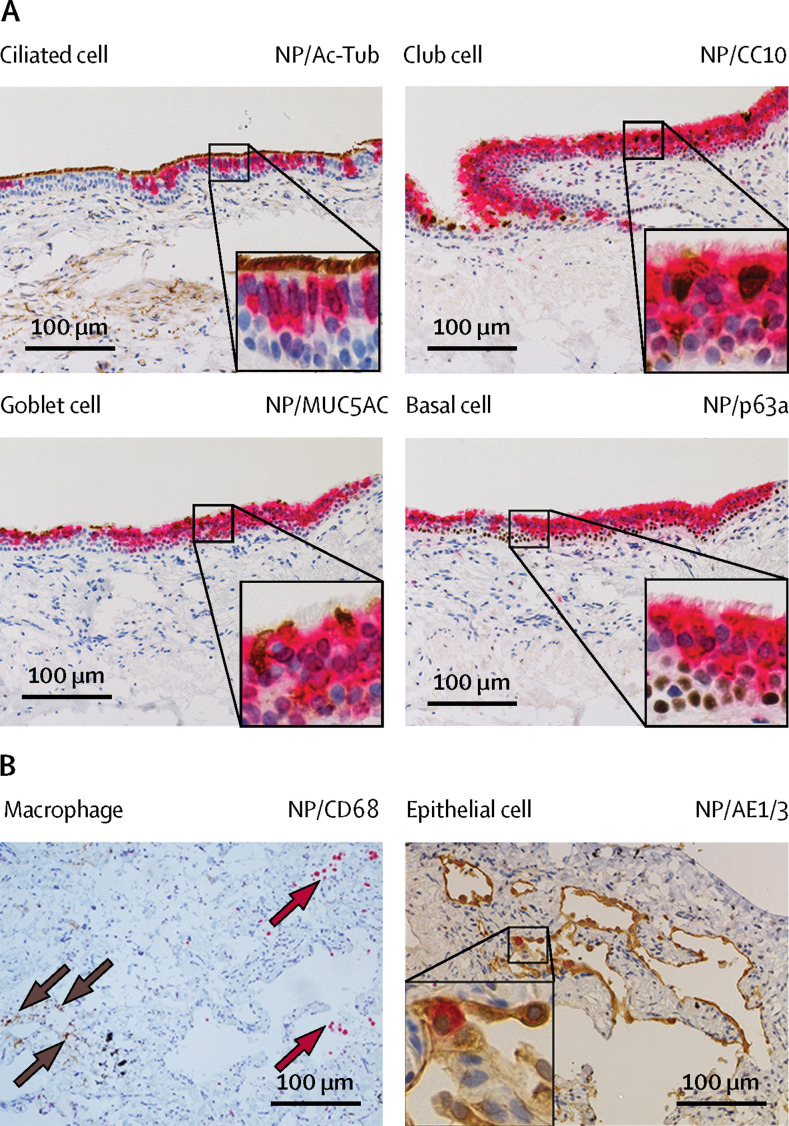
Immunohistochemistry staining showed that SARS-CoV-2 extensively infected bronchial epithelium (figure 1 ), with infection observed in ciliated cells, non-ciliated mucus secreting (goblet) cells, and club cells, but not basal cells (figure 2 ). Similar tissue tropism was observed with the bronchus infected with MERS-CoV, SARS-CoV, and H1N1pdm immunohistochemical staining, although the viral antigen staining was more extensive with SARS-CoV-2 (figure 1). In lung tissues, similar viral antigen staining was observed with SARS-CoV-2 as for SARS-CoV and H1N1pdm, but all were less extensive than that with MERS-CoV. In the lung parenchyma, there was positive antigen staining for SARS-CoV-2 in the spindled, morphologically epithelial type 1 pneumocytes (Figure 1, Figure 2). Double staining showed no colocalisation of viral antigen in macrophages (figure 2). There was no evidence of infection of vascular endothelium in the blood vessels of the lung, as was previously seen with MERS-CoV.12
Figure 1.
Tissue tropism of SARS-CoV-2 and SARS-CoV viruses in ex-vivo cultures of human respiratory tract and conjunctiva
Ex-vivo cultures of human bronchus and lung were infected with mock, SARS-CoV-2, SARS-CoV, MERS-CoV, and H1N1pdm viruses and the tissues were fixed with formalin at 96 hpi. Conjunctiva tissues were infected with mock, SARS-CoV-2, and SARS-CoV, and the tissues were fixed with formalin at 48 hpi. Paraffin-embedded sections were subjected to immunohistochemical staining with a monoclonal antibody against the SARS-CoV nucleoprotein, MCoV nucleoprotein, and influenza nucleoprotein. Positive cells are brown. Inset images are 200x magnification. SARS-CoV=severe acute respiratory syndrome-associated coronavirus. MERS-CoV=Middle East respiratory syndrome-associated coronavirus. H1N1pdm=2009 pandemic influenza H1N1.
Figure 2.
Cellular tropism of SARS-CoV-2 in ex-vivo cultures of human bronchus and lung
Ex-vivo explant cultures of (A) human bronchus and (B) lung were infected with SARS-CoV-2. At 96 hpi the tissues were fixed in formalin, embedded in paraffin, and immunohistochemically stained (brown) for indicated cell markers: (A) Ac-Tub-positive for ciliated cells, MUC5AC-positive for secretory goblet cells, CC10-positive for club cells, p63a-positive for basal cells; and (B) AE1/3 for epithelial cells (brown) and CD68 for macrophages (red), and a monoclonal antibody against the SARS-CoV-2 nucleoprotein (red). SARS-CoV-2-infected cells are identified with brown arrows, and CD68-positive cells are identified with red arrows. The images are representative of three individual donors. NP=nucleoprotein. Ac-Tub=acetyl-α-tubulin. MUC5AC=mucin 5AC. SCGB1A1=secretoglobin family 1A member 1. CC10=club cell protein 10. p63a=p63-alpha. AE1/3= cytokeratin AE1/AE3. CD68=cluster of differentiation 68.
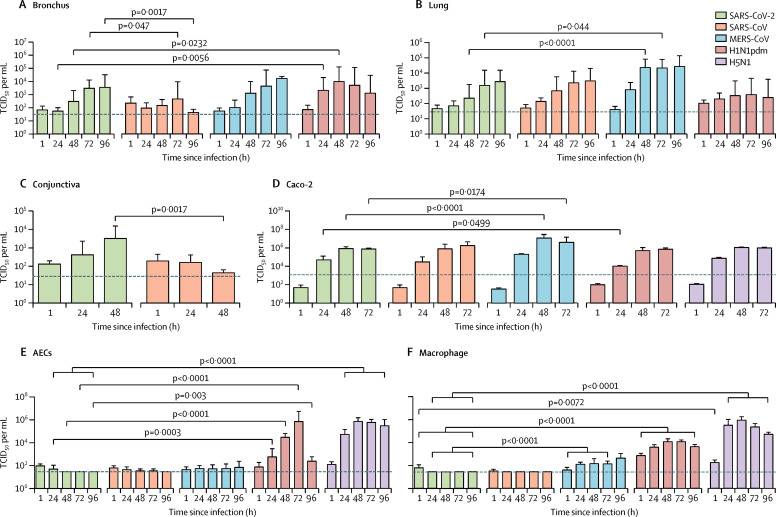
SARS-CoV-2 replicated in ex-vivo cultures of the human bronchus with a two log10 increase in TCID50 from 24 h to 96 h (figure 3 ). SARS-CoV-2 replicated similarly to MERS-CoV at all-timepoints, had lower titres than the pandemic H1N1pdm virus in bronchus at 24 hpi and 48 hpi, and replicated to significantly higher titres than SARS-CoV at 72 and 96 hpi (figure 3). In the absence of cells for virus replication, thermal inactivation of all viruses with input titres of 5 × 102 TCID50 per mL led to negligible residual infectious virus after 24 h incubation at 37°C (appendix p 6). AUC from 24 h to 96 h post infection was calculated from the data in figure 3, and is shown in the appendix (appendix p 5). The AUC of SARS-CoV-2 in bronchus explants was lower than that of H1N1pdm and a higher, but non-significant, AUC was observed when compared with SARS-CoV (appendix p 5). In lung explants, SARS-CoV-2 titres were similar to H1N1pdm and SARS-CoV, but had a lower replication competence than MERS-CoV at 48 hpi and 72 hpi in the lung (figure 3), supported further by the AUC of SARS-CoV-2 in lung explants compared with other viruses (appendix p 5).
Figure 3.
Viral replication kinetics of SARS-CoV-2 and SARS-CoV viruses in ex-vivo cultures of human respiratory tract, conjunctiva, Caco-2 cells, AECs and macrophages
(A and B) Human ex-vivo cultures of bronchus (n=5) and lung (n=4), were infected with 5 × 105 TCID50 per mL at 37°C. (C) Human ex-vivo cultures of conjunctiva (n=3) were infected with 5 × 105 TCID50 per mL at 33°C. (D) Caco-2 cells (n=3), (E) AECs (n=3) and (F) macrophages (n=3) were infected with the indicated viruses at a MOI of 0·1 cultured at 37°C for 72 h. Culture supernatants were harvested at the indicated times and virus titres were measured by TCID50 assay. Bar charts show the mean (SD). The horizontal dotted line denotes the limit of detection in the TCID50 assay. Statistical significances compared with SARS-CoV-2 are shown and statistical significances with exact p values compared among other viruses are listed in the appendix (p 8). TCID50=50% tissue culture infection dose. SARS-CoV=severe acute respiratory syndrome-associated coronavirus. MERS-CoV=Middle East respiratory syndrome-associated coronavirus. H1N1pdm=2009 pandemic influenza H1N1. H5N1=highly pathogenic avian influenza H5N1 virus.
SARS-CoV-2 can be detected in patients' tears,14 conjunctiva, and anal swabs;15 thus, it is important to study the potential for experimental infection of extrapulmonary tissues. Human conjunctival explant cultures were more extensively infected by SARS-CoV-2 than by SARS-CoV, as shown in immunohistochemical staining (figure 1) and higher infectious viral titres at 48 hpi (figure 3; appendix p 5). Thermal inactivation of the two coronaviruses showed that there are negligible infectious viruses after 24 h incubation at 33°C with a remaining viral titre of 5 × 102 TCID50 per mL (appendix p 6). We also infected human colorectal carcinoma (Caco-2) cells with MERS-CoV, SARS-CoV, SARS-CoV-2, H1N1pdm and H5N1. Infectious viral titres of all viruses increased by more than four log10 differences from 1 h to 72 h incubation in the Caco-2 cells. SARS-CoV-2 had a similar replication competence to SARS-CoV, H1N1pdm and H5N1 but was lower than MERS-CoV (figure 3; appendix p 5).
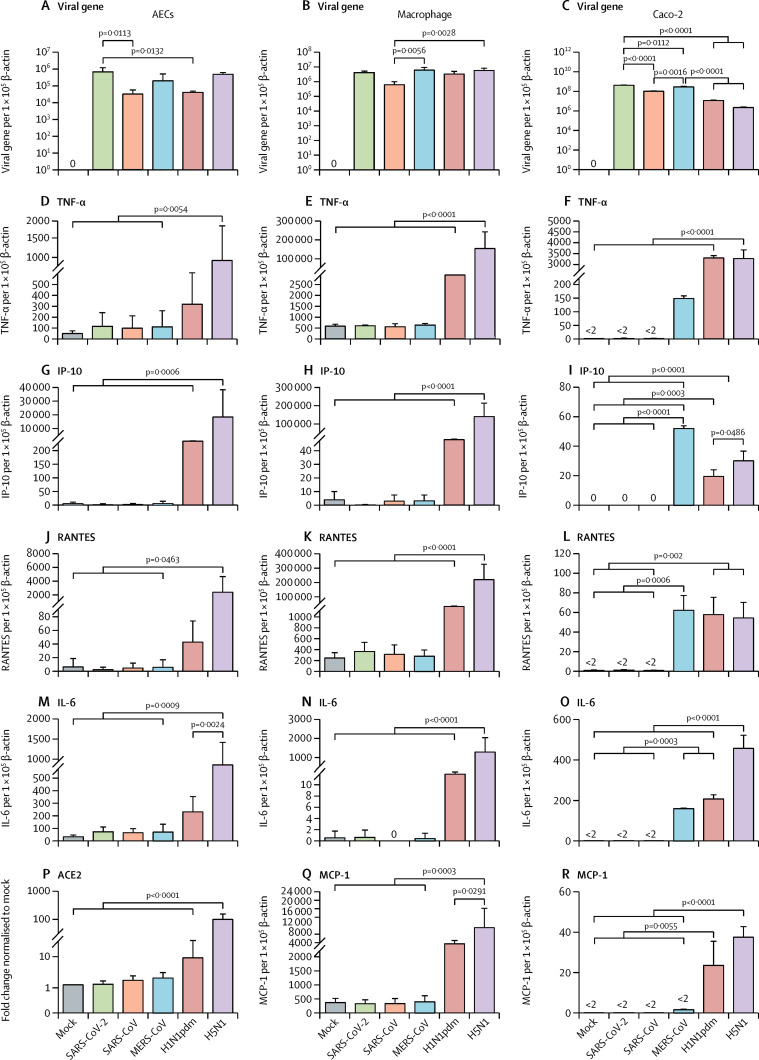
We assessed viral replication, proinflammatory cytokine responses, and chemokine responses in AECs, human peripheral blood-derived macrophages, and Caco-2 cells; we compared the response elicited by SARS-CoV-2 to that of SARS-CoV, MERS-CoV, H1N1pdm, and H5N1 (figure 4 ). High levels of viral gene mRNA expression were detected in AECs, macrophages at 24 hpi, and Caco-2 cells at 48 hpi (figure 4), with differences between the viruses. However, productive viral replication of SARS-CoV-2 was detected only in Caco-2 cells (figure 3; appendix p 5) with no robust virus replication observed in AECs or macrophages (appendix p 5). There was less induction of proinflammatory cytokines and chemokines (TNF-α, IP-10, RANTES, IL-6, and MCP-1) by SARS-CoV-2, SARS-CoV, and MERS-CoV in AECs and macrophages than by H5N1. In Caco-2 cells, both SARS-CoV-2 and SARS-CoV induced less intense cytokine responses than did H1N1pdm and H5N1, whereas MERS-CoV induced modest level of most cytokines and chemokines except for high levels for IP-10 in Caco-2 cells (figure 4).
Figure 4.
Viral gene, cytokine, chemokine, and ACE2 expression profile of SARS-CoV-2
Expression of the mRNA of viral genes (SARS-CoV-2 and SARS-CoV ORF1b gene; MERS-CoV UpE gene; influenza matrix gene), TNF-α, IP-10 and RANTES, IL-6, and ACE2 in alveolar epithelial cells (n=3), human macrophages (n=3) at 24 hpi, and Caco-2 cells (n=3) at 48 hpi. ACE2 is shown only for AECs and MCP-1 is shown for macrophages and Caco-2 cells. Graphs show mean mRNA copies expressed per 1 × 105 β-actin copies from three independent experiments. Data are mean (SD). Statistical significances with exact p values compared among other viruses are shown. SARS-CoV=severe acute respiratory syndrome-associated coronavirus. MERS-CoV=Middle East respiratory syndrome-associated coronavirus. TNF-α=tumour necrosis factor alpha. IP10=interferon gamma-induced protein 10. RANTES=regulated on activation, normal T cell expressed and secreted. IL-6=interleukin 6. ACE2= angiotensin-converting enzyme-2. AECs=alveolar epithelial cells. Caco-2=human colorectal carcinoma cells. MCP-1=monocyte chemoattractant protein 1. H1N1pdm=2009 pandemic influenza H1N1. H5N1=highly pathogenic avian influenza H5N1 virus.
We detected upregulation of ACE2 receptor mRNA in H1N1pdm and H5N1 influenza-infected AECs (figure 4); however, in human macrophages, ACE2 expression was low and this remained similar after infection with influenza and all tested coronaviruses (data not shown).
Discussion
We report the replication competence and cellular tropism of SARS-CoV-2 in the human respiratory tract and in extrapulmonary explant tissue and cells. The conjunctival epithelium and conducting airways appear to be potential portals of infection of SARS-CoV-2. Both SARS-CoV and SARS-CoV-2 replicated similarly in the alveolar epithelium, but SARS-CoV-2 replicated extensively in bronchial epithelium, which might explain the robust transmission of this pandemic coronavirus. SARS-CoV-2 was a less potent inducer of proinflammatory cytokines than H5N1, H1N1pdm, or MERS-CoV viruses.
SARS-CoV-2 replicated more efficiently than SARS-CoV in ex-vivo cultures of human bronchus, with peak viral titres at 96 hpi (Figure 1, Figure 3). Replication of SARS-CoV-2 was similar to MERS-CoV and less efficient than H1N1pdm. The lower AUC of SARS-CoV-2 might be partly attributable to differences at 24 hpi, at which time H1N1pdm already showed high virus titres although SARS-CoV-2 titres were low and only increased at 48 hpi. MERS-CoV can be transmitted among humans, mainly in health-care settings. The robust replication competence of SARS-CoV-2 in human bronchus might explain its efficient transmission efficiency among humans. In human lung, SARS-CoV-2 and H1N1pdm replicated to similar titres, but both were lower than that of MERS-CoV. The viral titres of SARS-CoV-2 at 24 hpi and 48 hpi were significantly lower than that of H1N1pdm. This observation is compatible with the longer incubation period of SARS-CoV-2 than of H1N1pdm. Virus shedding before symptom onset16, 17 probably contributes to the difficulty of controlling the spread of SARS-CoV-2 by public health interventions—much more so than with SARS-CoV.
Immunohistochemistry of ex-vivo cultures of the human bronchus showed SARS-CoV-2 infection of ciliated, non-ciliated mucus secreting (goblet), and club cells. Because there were no reliable antibodies for double staining which could distinguish between type 1 and 2 cells in the alveoli, we used the morphology of cells expressing virus antigen together with epithelial cell markers to conclude that type 1-like alveolar epithelium was being infected, similar to observations in patients infected with SARS-CoV11 and consistent with a study in macaques infected with SARS-CoV-2.18, 19 Type 1 pneumocytes, flattened in shape, are crucial in the process of gas exchange between the alveoli and the capillaries; their intercellular connections via occluding junctions prevent the leakage of tissue fluid into the alveolar air space. Damage to the type 1 pneumocytes might account for the acute lung injury of severe cases of COVID-19. Type 2 alveolar epithelial cells are important in producing surfactant proteins A and D and are crucial in reconstituting damaged type 1 alveolar epithelium, and loss of type 2 pneumocytes would lead to impairment of the repair and regeneration processes after alveolar epithelium damage. However, we still do not have evidence for SARS-CoV-2 infection of type 2 pneumocytes. Although the tropism of SARS-CoV-2 infection in the alveoli is similar to that seen with SARS-CoV,11 the extent of infection of the bronchus is significantly greater than with SARS-CoV and H1N1pdm.
Although individual genetic variability might result in heterogeneity of phenotypes in ex-vivo explant cultures, it is noted that five separate donors yielded comparable trends in SARS-CoV-2 replication kinetics in the human bronchus. However, more heterogeneity was observed in the ex-vivo explants of lungs (data not shown) and whether genetic or other host factors, such as the presence of ACE2, HAT, and transmembrane protease serine 2 (TMPRSS2) play a role in susceptibility to infection of ex-vivo human respiratory cultures should be addressed.
SARS-CoV-2 uses the same cell entry receptor (ACE2) and cellular protease (TMPRSS2) as SARS-CoV for cell entry.20, 21 The expression of ACE2 is relatively low in human bronchus compared with lung parenchyma.22, 23 However, unlike SARS-CoV, the viral titres of SARS-CoV-2 increased by a two log10 increase up to 96 h incubation in both the human bronchus and lung tissues. Besides TMPRSS2, human airway trypsin-like protease (HAT) activates SARS-CoV spike protein by cleavage at the S1/S2 site to promote viral-cell fusion.22 The presence of the insert sequence of SerProArgArg in the S1/S2 protease cleavage site in SARS-CoV-2, but not in SARS-CoV, causes the formation of an extended loop, which is more suitable for protease recognition than in SARS-CoV.24 Because there is a strong positive staining of TMPRSS2 and HAT in human bronchial tissues,22 and the evidence that trypsin treatment can overcome low-level expression of ACE2 receptor on target cells to activate spike protein of SARS-CoV,22 HAT might play a more important role than TMPRSS2 in upper respiratory replication of SARS-CoV-2. This finding is in line with the higher replication competence of SARS-CoV-2 than SARS-CoV in human ex-vivo bronchus explants, and suggests that protease inhibitors of TMPRSSs are potential therapeutic candidates for COVID-19.
We showed that ACE2 mRNA expression was significantly upregulated in alveolar epithelial cells after influenza A virus infection, with H5N1 having a more pronounced effect than H1N1pdm in vitro. If replicated in a larger sample, this upregulation could suggest that recent exposure to influenza virus might worsen the outcome of COVID-19 through upregulation of the ACE2 receptor in human respiratory epithelium. By contrast, ACE2 expression might also offer protective effects during acute lung injury as shown for SARS.25 Therefore, the role of ACE2 expression during influenza infection should be defined, and its implications on susceptibility to and severity of SARS-CoV-2 infection should be investigated.
Infection of extrapulmonary sites by SARS-CoV-2 has been supported by detection of the virus in tears and in anal swabs14, 15 and the detection and isolation of SARS-CoV-2 in tears, anal swabs, and stool specimens.15, 26 We showed infection and productive replication of SARS-CoV-2 in conjunctiva and in a colorectal carcinoma epithelial cell line, which might support these reports. Similar replication competence was observed in SARS-CoV-infected Caco-2 cells compared with SARS-CoV-2. The demonstration of infection of conjunctiva suggests that the eye might be an additional route of infection, an observation that is crucially relevant for infection prevention and control. Productive infection of the colorectal carcinoma epithelial cell line hints at virus infection and replication in the gastrointestinal tract, and might indicate the possibility of oral-faecal transmission as an additional route. These multiple routes of transmission by SARS-CoV-2 might explain the rapid global spread of COVID-19.
Higher plasma concentrations of proinflammatory cytokines and chemokines, including IP-10, MCP-1, and TNF-α, have been found in patients with COVID-19 in the ICU when compared with non-ICU patients.8 However, such studies cannot clarify whether these elevated cytokines and chemokines are a major driver of pathology or simply a reflection of the more severe lung damage that has taken place. Because ex-vivo cultures are heterogeneous in cell type and because susceptibility to virus and the infecting dose (MOI) cannot be precisely controlled, we compared the innate immune response to SARS-CoV-2 infection with that to H5N1, H1N1pdm, SARS-CoV, and MERS-CoV infections in Caco-2 cells (the only in-vitro system in which productive virus replication was observed). There was little induction of proinflammatory cytokines in Caco-2 cells by SARS-CoV-2 or SARS-CoV, even less so than MERS-CoV, whereas H1N1pdm and H5N1 viruses induced even higher levels of proinflammatory cytokine than MERS-CoV (with the exception of IP-10). The insufficiency of productive replication of SARS-CoV-2 in AECs is in line with lack of replication in human adenocarcinoma cells (A549).27 In summary, proinflammatory cytokine responses might contribute modestly to the pathogenesis and severity of human COVID-19, as was seen with MERS-CoV,28 and contrast with those observed in H5N1.29
The absence of robust virus replication in AECs in vitro, despite detection of infection in AECs in ex-vivo lung explant cultures, is reminiscent of observations during previous research into SARS-CoV.30 In-vitro culture of primary human lung epithelium is associated with a loss of permissiveness to both SARS-CoV and SARS-CoV-2, possibly via downregulation of the ACE2 receptor, which was ten times lower than that in Caco-2 cells (data not shown), or the absence of crucial proteases for the cleavage of the spike protein during isolation and in-vitro culture, or both. The experimental findings from the Caco-2 model could not be directly compared with clinical observation; however, the human gastrointestinal tract comprises a complex mixture of cell types with differing gene expression profiles between intestinal epithelium, mucosal epithelium, and the crypt–villus axis. Nevertheless, Caco-2 cells are readily available to most research laboratories as a simple and reproducible experimental tool allowing interlaboratory comparisons of findings.
A limitation of our study is the absence of data on explant cultures of nasopharyngeal tissues because of the unavailability of such tissues for ex-vivo infection experiments. The nasopharynx is crucial for virus for SARS-CoV-2 replication and one that is highly relevant for virus transmission. Restricted availability of conjunctival tissue allowed infection with only two of the coronaviruses. Additionally, cytokines were analysed only at 24 hpi (AECs and macrophages) and 48 hpi (Caco-2 cells). Replication of our study with increased sample size, more cytokines, and detection of secreted cytokine proteins would shed light on the pathogenesis of SARS-CoV-2.
Our findings support the notion that SARS-CoV-2 can transmit between humans via droplets being deposited in the airways or eyes and via fomite transmission when infectious virus is introduced to the eyes via contaminated hands. Replication of SARS-CoV-2 in colorectal cells suggests that the virus might also spread via the oral-faecal route.
Acknowledgments
Acknowledgments
Rachel Ching, Eric Lau, Rita Lai, Cassia Lin, Hin-Wo Yeung, Daniel Chu (School of Public Health, University of Hong Kong, Hong Kong, China), and Dora Kwong (Department of Clinical Oncology, University of Hong Kong, Hong Kong, China) provided the human upper respiratory tract tissues. Kevin Fung (Department of Pathology, The University of Hong Kong, Hong Kong, China) provided technical support, and Ron Fouchier (Erasmus University Medical Center, Rotterdam, Netherlands) provided the MERS-CoV. We acknowledge research funding from the US National Institute of Allergy and Infectious Diseases (NIAID) under Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract no. HHSN272201400006C and the Theme Based Research Scheme (Ref: T11–705/14N, T11-712/19-N), Hong Kong Special Administrative Region and a Commissioned Grant from the Health and Medical Research Fund (Ref: HKS-18-B03), Food and Health Bureau, Government of Hong Kong Special Administrative Region. The funding sources had no roles in the writing of the manuscript or the decision to submit it for publication. The authors have not been paid to write this article by a pharmaceutical company or other agency. The authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Declaration of interests
We declare no competing interests.
Contributors
KPYH isolated SARS-CoV-2, and designed and coordinated the study, analysed and interpreted the results, did experiments and wrote the Article. M-CC, K-CN, CHTB, JCWH, MMTN, and DITK did experiments, and analysed and interpreted results. RAPMP isolated SARS-CoV-2. KCS provided human conjunctiva tissue. S-WT provided and technically supported the human respiratory epithelial cells. LLMP developed quantitative PCR assays. MP designed the study, analysed and interpreted results and critically reviewed the manuscript. JMN designed the study, analysed and interpreted results from immunohistochemical staining and critically reviewed the manuscript. MCWC designed and was responsible for overall coordination of the study, analysed and interpreted the results, and wrote the Article.
Supplementary Material
References
- 1.WHO. MERS monthly summary, November 2019. 2019 (accessed Feb 24, 2020).
- 2.WHO COVID-19 dashboard. 2019. https://covid19.who.int/
- 3.Chen J. Pathogenicity and transmissibility of 2019-nCoV—a quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Z, Shi L, Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian S, Hu W, Niu L, Liu H, Xu H, Xiao SY. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J Thorac Oncol. 2020 doi: 10.1016/j.jtho.2020.02.010. published online Feb 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan MC, Chan RW, Yu WC. Tropism and innate host responses of the 2009 pandemic H1N1 influenza virus in ex vivo and in vitro cultures of human conjunctiva and respiratory tract. Am J Pathol. 2010;176:1828–1840. doi: 10.2353/ajpath.2010.091087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu DKW, Hui KPY, Perera RAPM. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc Natl Acad Sci USA. 2018;115:3144–3149. doi: 10.1073/pnas.1718769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Wang Y, Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hui KP, Chan LL, Kuok DI. Tropism and innate host responses of influenza A/H5N6 virus: an analysis of ex vivo and in vitro cultures of the human respiratory tract. Eur Respir J. 2017;49 doi: 10.1183/13993003.01710-2016. [DOI] [PubMed] [Google Scholar]
- 10.Chu DKW, Pan Y, Cheng SMS. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicholls JM, Butany J, Poon LL. Time course and cellular localization of SARS-CoV nucleoprotein and RNA in lungs from fatal cases of SARS. PLoS Med. 2006;3:e27. doi: 10.1371/journal.pmed.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mok CK, Lee HH, Lestra M. Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol. 2014;88:3568–3576. doi: 10.1128/JVI.02740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui KPY, Ching RHH, Chan SKH. Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir Med. 2018;6:846–854. doi: 10.1016/S2213-2600(18)30236-4. [DOI] [PubMed] [Google Scholar]
- 14.Xia J, Tong J, Liu M, Shen Y, Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J Med Virol. 2020;92:589–594. doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, Du RH, Li B. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou L, Ruan F, Huang M. SARS-CoV-2 Viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munster VJ, Feldmann F, Williamson BN. Respiratory disease and virus shedding in rhesus macaques inoculated with SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.03.21.001628. published online March 21. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockx B, Kuiken T, Herfst S. Comparative pathogenesis of COVID-19, MERS and SARS in a non-human primate model. Science. 2020 doi: 10.1126/science.abb7314. published online April 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou P, Yang XL, Wang XG. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M, Kleine-Weber H, Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertram S, Glowacka I, Müller MA. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J Virol. 2011;85:13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertram S, Heurich A, Lavender H. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong Meng HC, Zhang H, Kang Z. The insert sequence in SARS-CoV-2 enhances spike protein cleavage by TMPRSS. bioRxiv. 2020 doi: 10.1101/2020.02.08.926006. published online Feb 16. (preprint). [DOI] [Google Scholar]
- 25.Imai Y, Kuba K, Rao S. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Chen C, Zhu S. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) 2020. http://weekly.chinacdc.cn/en/article/id/ffa97a96-db2a-4715-9dfb-ef662660e89d [PMC free article] [PubMed]
- 27.Harcourt J, Tamin A, Lu X. Severe acute respiratory syndrome coronavirus 2 from patient with 2019 novel coronavirus disease, United States. Emerg Infect Dis. 2020 doi: 10.3201/eid2606.200516. published online March 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan RW, Chan MC, Agnihothram S. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013;87:6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung CY, Poon LL, Lau AS. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–1837. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 30.Mossel EC, Wang J, Jeffers S. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology. 2008;372:127–135. doi: 10.1016/j.virol.2007.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.