Abstract
The half-life of an mRNA is an important parameter contributing to the steady-state level of the mRNA. Rapid changes in mRNA levels can result from decreasing the half-life of an mRNA. Establishing the detailed pathway of mRNA degradation for a particular class of mRNAs requires the ability to isolate mRNA degradation intermediates. High-throughput sequencing provides a method for detecting these intermediates. Here we describe a method for determining the intermediates in 3’ to 5’ degradation. Characterizing these intermediates requires not only determining the precise 3’ end of the molecule to a single nucleotide resolution, but also the ability to detect and characterize any untemplated nucleotides present on the intermediates. We achieve this by ligating a known sequence to all the 3’ termini in the cell, and then sequence the 3’ termini and the ligated linker to identify any alterations to the genomic reference sequence. We have applied this method to characterize the intermediates in histone mRNA metabolism, allowing us to deduce the pathway of 3’ to 5’ degradation. This method can potentially be applied to any RNA, and we discuss possible strategies for extending the method to include simultaneous determination of the 3’ and 5’ end of the same RNA molecule.
INTRODUCTION
The final step in metabolism of an RNA molecule is degradation of the RNA. Since the steady-state level of an RNA is a balance of the rate of synthesis and the rate of degradation, the half-life of an RNA is as important as its rate of synthesis in determining the level of any RNA molecule in the cell. Messenger RNAs have a wide range of half-lives in eukaryotes, ranging from a few minutes to many hours, or even days. Not only does the half-life of an mRNA contribute to its overall level in the cell, but if the cell needs to rapidly reduce the level of an mRNA, the only way to do that is to rapidly degrade the mRNA. Most mRNAs are protected from degradation by specialized elements at the ends of the RNA; a cap at the 5’ end of the mRNA, and a poly(A) tail at the 3’ end of the mRNA, each of which can be bound by specific proteins. Animal replication-dependent histone mRNAs are the only known cellular mRNAs that do not end in a polyA tail. In animal cells the replication-dependent histone mRNAs are capped at the 5’ end (as are all RNA polymerase II transcripts), but instead of ending in a poly(A) tail, they end in a conserved stemloop sequence which is bound by the stemloop binding protein (SLBP) [1].
There are multiple potential pathways for degrading an mRNA [2–4]. The three major pathways involve degradation starting at one of the two ends or by cleavage in the body of the mRNA. Degradation from the 3’ end involves deadenylation followed by 3’ to 5’ degradation by the exosome; degradation from the 5’ end starts with decapping (either before or after deadenylation) followed by 5’ to 3’ degradation by XRN1. Endonucleolytic cleavage can involve cleavage by a specific endonuclease, siRNA mediated cleavage by the Ago protein in the RISC complex [5], or cleavage by SMG6 as part of the NMD mechanism [6, 7], followed by subsequent exonucleolytic degradation of the fragments. These fragments could be deadenylated and/or decapped as well. An mRNA can also be degraded from both ends simultaneously (e.g. 5’ to 3’ by Xrn1 and 3’ to 5’ by the exosome). Histone mRNA molecules that likely arise by simultaneous degradation from the two ends of the mRNA have been identified by circular PCR [8].
mRNAs also can transition between actively translating forms and inactive forms of the mRNA, which are not translatable [9]. These include deadenylated mRNAs (which end in a short oligo(A) tail), which in animal cells is too small to bind PABP. Some mRNAs, like the ferritin mRNAs, may bind a specific protein in the 5’ UTR that blocks translation and this binding is reversible by addition of iron [10]. Deadenylation of an mRNA, resulting in a short oligo(A) tail which cannot bind PABP, inactivates translation of the mRNA and may promote binding of the Lsm1–7 complex as well as translational inhibitors such as Dhh1 (Rck/p54) helicase in animals) [9]. The inactive forms of mRNAs can be subsequently reactivated, by removal of inhibitory proteins or miRNAs, together with cytoplasmic polyadenylation. Untranslated mRNAs may accumulate in stress granules [11], at the synapse in neurons [12], in the p granules in early embryos, where they may be bound by specific sets of proteins. Binding of the RISC/miRNA complex to an mRNA blocks its translation, and stimulates deadenylation, which does not always lead to degradation of the mRNA. There are also examples of proteins that bind to the 3’ UTR inhibiting translation. Regulation of translation is the most important regulatory mechanism in cells that aren’t synthesizing RNAs, including oocytes, early embryos, spermatids, and nerve cells, where translation at the synapse is tightly regulated. Part of this regulation can involve modification of the mRNA, e.g. histone mRNAs are oligoadenylated in Xenopus oocytes and eggs [13, 14], which is one factor that blocks their translation.
MODIFICATION OF THE 3’ END OF RNAs
While most RNA molecules are a faithful copy of the DNA template, there are many RNAs where the 3’ end can be modified by addition of non-templated nucleotides. These include the polyadenylation of mRNA in eukaryotic cells, the addition of the CCA end to many tRNAs, either when the CCA is not templated in the DNA, or if it is lost from the tRNA, and oligoadenylation of the 3’ end of many defective RNAs by the TRAMP complex as a quality control mechanism in cells. Many RNAs can also be uridylated, including miRNAs and pre-miRNAs, as well as polyadenylated mRNAs, which are often uridylated after deadenylation to shorten the polyA tail [15, 16]. In bacteria, the 3’ ends of mRNAs are polyadenylated by poly A polymerase, and can also have random extended tails added by polynucleotide phosphorylase [17]. Both these 3’ extensions promote degradation of the mRNAs.
Histone mRNAs are the only known mRNAs in animal cells that are not polyadenylated, ending instead in a conserved stemloop (Fig 1). Histone mRNAs are also cell-cycle regulated being present only in S-phase when DNA is being replicated. Regulated degradation of histone mRNA is an important regulated step in histone mRNA metabolism. Histone mRNAs are rapidly degraded when DNA replication is inhibited during S-phase and also at the end of S-phase when DNA replication is completed [1]. We found several years ago that histone mRNAs are uridylated at the 3’ end when histone mRNAs are being degraded (Fig. 1)[8]. We subsequently showed that there is also uridylation to maintain the proper length of the 3’ end of histone mRNA during S-phase before degradation begins [18, 19]. Because of the natural coordination of degradation of multiple histone mRNAs, they provide an excellent system for identifying degradation intermediates.
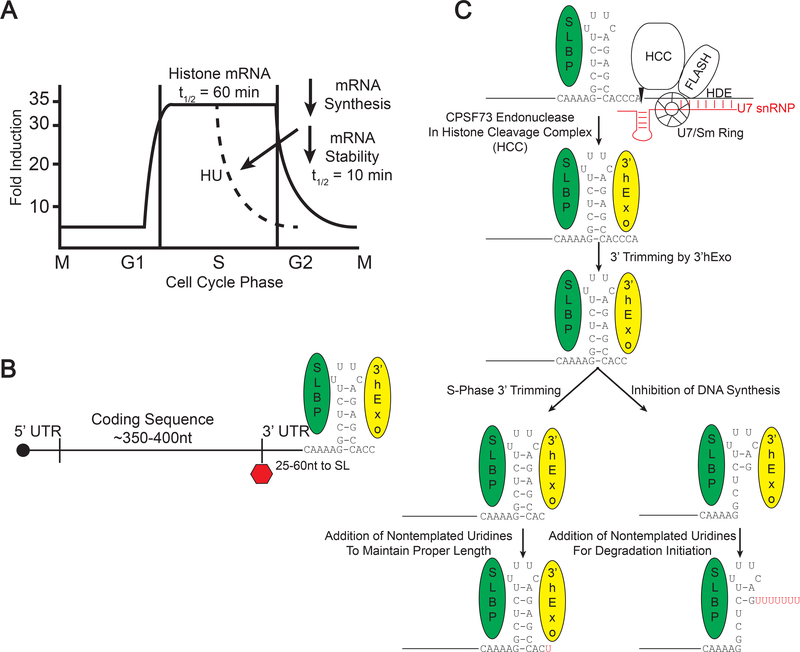
Fig. 1. Histone mRNA metabolism.
A. Cell cycle regulation of histone mRNA. Histone mRNAs accumulate at high levels only in S-phase. When DNA synthesis is inhibited in S-phase, histone mRNAs are rapidly degraded. Histone mRNAs are also rapidly degraded at the end of S-phase, and histone mRNA biosynthesis is down-regulated.
B. Structure of the histone mRNP. Histone mRNAs are capped and end in a conserved stemloop sequence at the 3’ end which binds to both SLBP and 3’hExo. C. Histone mRNA is formed by an endonucleolytic cleavage 5 nts after the stemloop, which requires SLBP, that binds the stemloop and U7 snRNP complexed with FLASH and the Histone Cleavage Complex (HCC) which contains the endonuclease CPSF73. After processing, 3’hExo binds and trims 2 nts from the 3’ end. In the cytoplasm during S-phase, there may be some trimming of the 3’ end, and the 3’ end is restored to 3 nts after the stemloop by uridylation. When DNA synthesis is inhibited, 3’hExo can degrade into the stemloop, where there are long U-tails added to the degradation intermediates.
STUDYING mRNA DEGRADATION
Measuring mRNA half-life requires that one determine the change in concentration of the mRNA as a function of time after stopping synthesis of that mRNA. Ideally one would do this by stopping biosynthesis of a specific mRNA and then watch its decay, without changing the metabolism of many other mRNAs. There are several reporter systems that allow one to do this. One system, pioneered by Shyu and Belasco, is to use the Fos promoter to drive expression of the mRNA [20]. When Fos expression is activated there is a burst of FOS mRNA synthesis and then Fos transcription is repressed as a result of Fos accumulation. The mRNA then decays in the absence of any new synthesis. A second approach is similar and uses the TET-on system [21]. Stable cell lines expressing the TET repressor are constructed (and some are commercially available), and expression of mRNA can be initiated by adding tetracycline/or a similar promoter (e.g. estrogen responsive promoters). A more recent (and more global) approach to mRNA half-life is to “pulse-label” cells with a modified nucleoside (e.g. thiouridine [22, 23] or bromouridine [24, 25] and then chase with uridine). The RNA containing the modified nucleotide is purified and subjected to RNA-Seq analysis. This allows determination of relative half-lives of all the RNA species, and is certainly the current best method to measure the half-lives of different RNA species, with minimal disruption to the metabolism of the cell. Determining the precise structure of mRNA degradation intermediates is more difficult. The above approaches allow you to detect some mRNA degradation intermediates, for example the formation and ultimate disappearance of deadenylated mRNAs, but detailed information on the precise sequences at the 3’ ends of the mRNAs cannot be determined without high-throughput sequencing, using strategies specifically designed to determine the ends of the RNA molecule.
HISTONE mRNAs
We have studied the metabolism of replication-dependent histone mRNAs. These mRNAs are tightly cell-cycle regulated and are present in high concentrations only in S-phase. Instead of a polyA tail they end in a conserved stemloop, consisting of a 26 nt sequence a six base stem and four base loop, and the 5 nts flanking the stemloop. Histone mRNAs also lack introns. A critical regulatory step in cell-cycle regulation of histone mRNA is rapid degradation of histone mRNA at the end of S-phase. Histone mRNAs are also rapidly degraded when DNA replication is inhibited in S-phase. Alteration of histone mRNA half-life is a mechanism the cell uses to rapidly adjust histone mRNA levels to provide appropriate amounts of histone mRNA for synthesizing the histone proteins necessary to package the newly replicated DNA.
There are multiple histone genes encoding each of the 5 histone proteins in mammals. These genes are not repeated but each gene has a unique sequence, particularly in the 5’ and 3’ UTR [26]. There are 10–15 distinct genes for each of the four nucleosomal core histones, and 5 histone genes encoding histone H1 all of which are coordinately degraded. Histone mRNAs make up about 5% of the total mRNAs in S-phase cells.
The conserved 26 nts sequence at the 3’ end of the mRNA is the cis element determining the regulation of the half-life of histone mRNA [27]. Translation is required for histone mRNA degradation, and the position of the stemloop relative to the stop codon is also critical for regulating degradation [28, 29]. The stemloop is normally present starting 20–60 nts after the stop codon. If the stemloop is moved to 200 nts after the stop codon or replaced by a polyA tail, then the mRNA is no longer rapidly degraded when DNA replication is inhibited or at the end of S-phase. The protein SLBP binds to the 3’ end of all histone mRNAs, and no other cellular mRNAs [30]. SLBP is required for histone pre-mRNA processing in the nucleus and remains with the mRNA as it is exported to the cytoplasm, and is essential for histone mRNA translation [31]. Thus, like polyadenylated mRNAs, the 3’ and 5’ ends of the RNAs interact with the translation initiation factors, creating a circular mRNP ribosome complex. This complex is the initial substrate on which mRNA degradation is initiated.
Early studies by Jeff Ross and coworkers suggested that histone mRNA degradation initiated at the 3’ end, and they identified a polysome-associated 3’ to 5’ exonuclease which would partially degrade into the stemloop [32–34]. This protein was almost certainly 3’hExo, which was subsequently purified as a protein that bound to the histone 3’ end together with SLBP [35]. RNAi experiments implicated several factors in histone mRNA degradation, including Upf1, the NMD factor; Lsm1, part of the Lsm1–7 ring, which binds the oligo(A) tail on deadenylated mRNA; and the exosome, a processive 3’ to 5’ exonuclease. There was a smaller effect of knockdown of enzymes involved in 5’ to 3’ decay, dcp2 or xrn1 [8]. These RNAi experiments certainly aren’t definitive, since knockdown of 3’hExo had no effect on histone mRNA degradation [8, 35], although subsequent knockout of 3’hExo showed it was essential for histone mRNA degradation [36].
In our earliest attempts to isolate some histone mRNA degradation intermediates we used circular-RT-PCR [8]. This method assumes the mRNAs have been decapped (or cleaved) and have a 5’ phosphate on the 5’ end. The mRNA intermediate can then be circularized with RNA ligase, and detected by qRT-PCR, and the PCR products cloned and sequenced, which allows identification of the 5’ and 3’ end of the mRNA. These sequences revealed the surprising finding that there were a number of degradation intermediates which ended in oligo(U) tails, most of which were in the stemloop (Fig 1). The 5’ ends of these intermediates were also partially degraded, indicating that the RNA molecules might be being degraded simultaneously 3’ to 5’ and 5’ to 3’. Priming cDNA with oligo(A) primers confirmed that the histone mRNA degradation intermediates containing oligo(U) tails were present only after inhibition of DNA replication or at the end of S-phase [8]. The yield of these intermediates was low and only a few molecules (~100) containing oligo(U) tails were obtained over several experiments, not enough to elucidate a pathway of degradation. The oligo(A) priming experiments also did not allow us to determine the length of the oligo(A) tail and restricted us to identifying tails that were at least 3 nts long.
We developed the method described below to determine the 3’ ends of histone mRNAs by high-throughput sequencing and to simultaneously identify any non-templated nts present at the 3’ end. This method makes use of the approaches developed by Tuschl and coworkers [37] and Hammond [38] to study miRNAs and pre-miRNAs. A modification of this method allows us to enrich for transcripts which have specific non-templated nts at the 3’ end, by using a cDNA primer which contains 3 additional nts (e.g. 3 A’s to identify oligo(U) tails).
Protocol for Preparation of RNA Libraries Using EnD-Seq-AppEnD Analysis of 3’ Termini
Overview
The goal of the EnD-Seq protocol is to determine the precise 3’ end of an RNA molecule, including any non-templated nucleotides that may be present on the 3’ end. A synthetic linker (chosen to not cross-react with other sequences in the organism being analyzed) is ligated to the 3’ end of all cellular RNAs. To prevent ligation of cellular RNA molecules to each other, the linker is chemically modified by addition of App to the 5’ end allowing ligation to occur in the absence of ATP. The linker also ends in an amino group rather than an OH, preventing ligation of the 3’ end of the linker. cDNA is synthesized from the modified RNA using the reverse complement of the linker as a primer. Subsequent amplification can be used to prepare sequencing libraries, and the 3’ ends are sequenced using standard sequencing protocols on an Illumina MiSeq. Following sequencing the 3’ ends can be mapped to the genome using the AppEnd informatics programs [18] which identifies both the template 3’ end of the RNA and any additional non-templated nts at the 3’ end (Fig 2A).
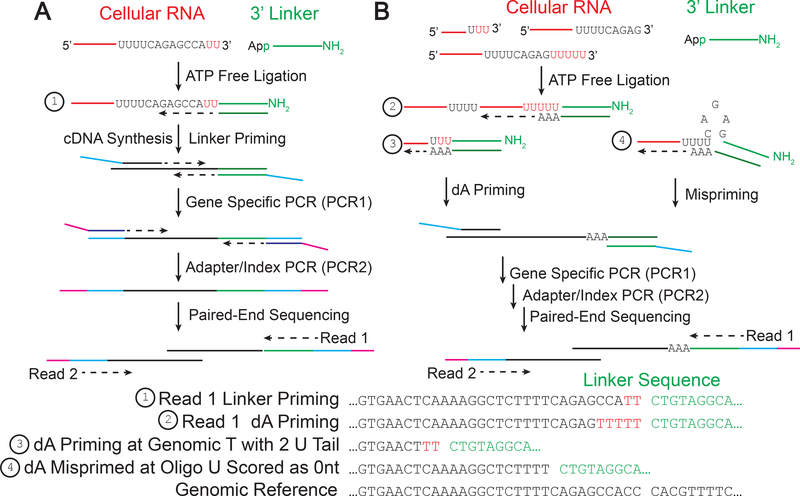
Fig. 2. Overview of the EnD-Seq/AppEnd workflow.
A. Total cellular RNA (or RNA from any subcellular fraction of interest) is ligated to a pre-activated synthetic linker sequence which is added to the 3’ OH on all RNAs. Note if the 3’ end is further modified (e.g. 2′Omethyl or phosphorylated) those ends will not be ligated. An oligonucleotide complementary to the linker is used to prime cDNA synthesis via reverse transcription. Specific sets of mRNAs can be amplified by PCR to specifically amplify the 3’ ends of the mRNAs of interest. The linker and gene specific primers are used to initially amplify the cDNA and a second round of PCR is used to add indices for each library and adapters for deep sequencing. Read 1 includes the linker sequence and 3’ terminus of the RNA and Read 2 includes the library indices and the gene sequence. The reads are aligned to a reference genome, the linker sequence is identified using the AppEnd algorithm [18]. Any nontemplated nucleotides on the 3’ termini (as small as one nt) will be identified as nts between the last template nucleotide and the start of the linker (example 1).
B. One can also select for specific non-templated nts (e.g. 3 uridines) at the 3’ end by adding 3 A’s to the 3’ end of the cDNA primer, followed by amplification of the cDNA for sequencing as in panel A. Note that this will allow one to determine the length of the oligo(U) tail (example 2). Note that if one primes at a template U or U’s, the AppEnd algorithm will identify only the nontemplated nts as part of the tail (example 3). We have observed mispriming at stretches of U’s, and these will be scored as no U tails (example 4). This method does an excellent job of identifying non-templated oligo(U) tails >2, and any shorter tails identified need to be interpreted cautiously.
Activation of the linker
A synthetic oligonucleotide containing a 5’ phosphate and 3’ amino group (to prevent ligation of the linker to other phosphorylated nucleotides) can be purchased from .a variety of vendors. An activated linker can be purchased from NEB (Cat. # S1315S) but if you are doing a large number of experiments and need flexibility in linker design it is cost-effective to synthesize your own. An alternative approach to the chemical approach described below is to use the 5’ DNA adenylation kit from NEB (M2611A) to synthesize an adenylated linker starting with a 5’ phosphorylated DNA, and an RNA ligase. The advantages of this kit are that it is easy to perform and adenylation efficiency is high, and it does not require further purification of the adenylated oligonucleotide. The kit can synthesize enough adenylated oligonucleotide for 5–10 libraries. Companies that provide synthetic oligonucleotides (e.g. Integrated DNA Technologies) will also sell activated oligonucleotides for a substantially higher cost. Note that the actual sequence of the linker is not important, as long as it will not prime cDNA synthesis of other mRNAs. The linker made by NEB (Cat. # S1315S) worked satisfactorily in a preliminary experiment.
The chemical activation of the linker requires the synthesis of ImpA (adenosine 5’-phosphorimidazolide) by exchange coupling an imidazole group onto the 5’ phosphate of 5’AMP. The ImpA molecule can then be coupled to the 5’phosphate (pN) on the linker, resulting in AppN as the 5’ end of the linker. This is the activated product of the first step of enzymatic ligation reactions. We use a 26 nt linker, which contains a sequence designed to amplify the cDNA for sequencing with Illumina primers, and also include the linker sequence on the 3’ end of all sequencing reads, an essential part of the procedure to accurately map the 3’ end as well as nontemplated nucleotides.
Generation of cDNA
The adenylated Linker is ligated to cellular RNA samples using a special T4 RNA ligase (Truncated Mutant K227Q) that ligates the activated AppNn oligonucleotide to the 3’ end of RNA in the absence of ATP. Once ligated, the linker sequence can be used as a primer site for reverse transcription. Using Super Script III and a primer specific to the 3’ Linker sequence, reverse transcribe the RNA to cDNA. Take a portion of the cDNA and use it for the first step of PCR amplification using the linker sequence as the 3’ primer and a gene specific 5’ primer, in our case a primer for the various histone genes. We have found that we can use multiple gene specific primers per PCR reaction to increase the number of RNAs analyzed in each experiment. Following the first round of PCR (PCR1), we continue with the second round of PCR (PCR2). The second round of PCR is to add on sequencing handles/adapters and barcodes for the MiSeq.
After amplification, we confirm library quality using a Bioanalyzer (Agilent 2100 Bioanalyzer) and quantify using a QuBit (Ver. 2.0 Fluorimeter). We sequence the libraries on a MiSEQ. Note that you have to add a custom primer to the MiSeq chip for this sequencing protocol. We have used a variety of MiSeq chips for sequencing, and prefer the chips that give 2 × 75 or 2 × 150 nt reads. The shorter MiSeq kit (V3–150) has consistently provided 30 million reads per kit, greatly increasing both the depth and multiplexing capacity of libraries. After sequencing, we analyze the libraries using the AppEnD workflow [18], which can be accessed at GitHub: https://github.com/jw156605/append.
DETAILED PROTOCOL
Synthesis of ImpA
This synthesis involves the use of flammable solvents and oxidizing agents and appropriate care should be taken during the synthesis. This is a straightforward synthesis for any chemistry laboratory with experience in organic chemical synthesis.
The following protocol has been adapted from the method of Hafner and coworkers [37]. Special care is taken to keep everything anhydrous. Failure to do so results in drastically lower yields of ImpA. If the product is stored at −20°C as described below, it is good for at least a year, and provides enough IMP to synthesis at least 1 micromole of adenylated oligonucleotide.
All steps were conducted in a fume hood.
Dry two 50mL glass flasks with a 140°C drying oven. Move flasks to fume hood and flush with argon gas. Seal flasks with septum and flush further with argon gas via a balloon and needle along with a second needle to allow air and excess argon out of the flasks. Allow the flasks to cool to room temperature.
Suspend 174 mg of 5’ AMP free acid (Sigma-Aldrich A2252) in 15 mL of anhydrous dimethylformamide (Sigma-Aldrich 227056) in one of the round-bottom flasks. The AMP will not dissolve entirely.
In the other flask, dissolve 262 mg of triphenylphosphine (Sigma-Aldrich T84409), 220 mg of 2,2′-dipyridyldisulfide (also found as Aldrithiol-2, Sigma-Aldrich 143049) and 170 mg of imidazole (Sigma-Aldrich I5513) in 15 mL of anhydrous dimethylformamide and 0.9 mL of triethylamine (Sigma-Aldrich T0886).
Add the AMP solution/suspension dropwise to the vigorously stirring triphenylphosphine containing solution at 4°C. Stir the reaction for another 3.5 hours at room temperature, keeping the flask sealed with a rubber septum. The 5’ AMP from the dimethylformamide solution/suspension will dissolve completely and the solution should turn to a clear yellow-green color.
Precipitate the ImpA by adding the reaction mixture dropwise into a 500 mL beaker containing a vigorously stirred solution of 1.1 g of sodium perchlorate (Sigma-Aldrich SX0692), 110 mL anhydrous acetone (Sigma-Aldrich 650501), and 55 mL anhydrous diethyl ether (Sigma-Aldrich 296082). Precipitate will appear immediately and the solution will become cloudier as the whole reaction mixture is added.
Stop stirring to allow the precipitate to settle to the bottom of the beaker. After 1–2 hours, decant as much as possible of the supernatant without dislodging the precipitate. Once most of the supernatant is decanted, use a pipette to remove as much of the clear supernatant as possible. If precipitate is agitated, let sit for another 30 minutes to settle before pipetting.
Once the volume has been reduced to about 20 mL, resuspend the precipitate in the residual supernatant and transfer to two dried 30 mL Corex glass centrifugation tubes. Rinse the beaker with two small volumes (5 mL each) of acetone and combine the wash solutions with the suspension already transferred to Corex tubes. Collect precipitate by centrifugation at 3000 rcf for 10 minutes
Pour off the supernatant and wash the pellet twice by resuspending it with 20 mL acetone in the Corex tubes, followed by 5 minute centrifugation at 3000 rcf. The pellet should be white and supernatant clear.
Resuspend the pellet in 10 mL anhydrous diethylether and immediately collect it again by centrifugation at 3000 rcf for 20 minutes. Pour off the diethylether supernatant. Seal the centrifugation tubes with a septum and insert two needles. Use a balloon of argon gas to carefully flush the tube with argon to remove excess air, and then remove the balloon. Let the pellet dry overnight in a dessicator filled with DrieRite.
Quickly transfer the flaky white powder to two small glass vials (one for each Corex tube) and use the same method as above to carefully exchange the air with a balloon of argon gas via a rubber septum. Seal with a screw-top lid and determine the mass of the product (minus initial weight of screw-top vial) to determine percent yield. Our yields were typically about 75%. Wrap the lid with parafilm and place in glass container of DrieRite. Seal the glass container with more parafilm and store at −20°C. We have successfully used it a year after synthesis when stored as described.
The purity of the product can be monitored by thin-layer chromatography as described by Hafner and coworkers [37].
Adenylation of Linker Sequence with ImpA
For adenylation we use a 300:1 molar ratio of ImpA:Linker and a 150:1 molar ratio of MgCl2:linker. Typically we first weigh the ImpA rapidly, to avoid it picking up water, by adding to a tared microfuge tube. The exact amounts do not need to be precise, since the volume of linker can be adjusted appropriately. We have observed identical results with 150:1 and 300:1 molar ratios of ImpA:Linker. The linkers were synthesized by IDT, 5’ phosphorylated and 3’Amino modified (/5Phos/ and /3Ammo/). We purchased 250 nmole of linker with standard desalting. A similar adenylated linker is available from NEB (Cat. #S1315). Note that the NEB linker is only 17 nt long, but is identical to the 5’ end of our linker sequence. All the suggested RT and PCR primers will work with the NEB linker..
Add about 13.6 mg solid ImpA (~30 μMol) to a microfuge tube. We have used 6–14 mg with similar results. The important parameter is to have ImpA in great molar excess to the linker)
Add 85 μL of 1.25mM Linker Oligonucleotide dissolved in nuclease free water (~100 nmol) to the ImpA in a 1.5mL microcentrifuge tube.
Add 16 μL of 1M MgCl2 in nuclease free water (16 μMol) to the microfuge tube.
Add 399 μL H2O to give a final volume of 500 μL. The ImpA dissolves readily in the water.
Incubate 50°C 3 hours in water bath in tube sealed with parafilm
Add 500 microliters of Formamide based RNA Dye without Xylene Cyanol (98% deionized Formamide, 0.025% SDS, 0.5mM EDTA pH 8.0, 0.025% Bromophenol Blue) and store at −20°C until ready to purify on the gel.
- Prepare a Medium (195 mm × 160 mm gel, 2 mm thick) sized Urea PAGE Gel 19% Acrylamide-1% bisacrylamide.
- 24g Urea
- 25mL 40% Acrylamide Solution (National Diagnostics AccuGel 19:1 40%)
- 5mL 10x TBE Buffer (500mM Tris, 500mM Boric Acid, 10mM EDTA pH 8)
- Add H2O to 50 mL total volume
- Add 500μL 10% APS (Ammonium Persulfate m/v)
- 50μL TEMED
- Dissolve urea in the acrylamide-buffer solution (a small amount of additional water may be required to fully dissolve urea) then continue to add water to a total volume of 50 ml, add APS and then immediately add TEMED to initiate polymerization, and cast gel (polymerizes in less than 1 minute).
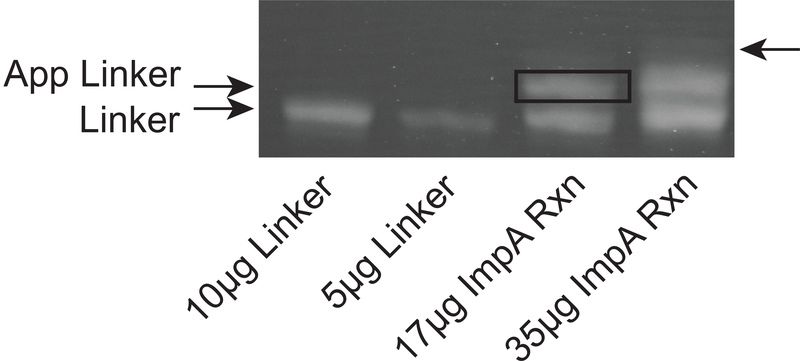
Run a control of Linker (not adenylated) to confirm the 1nt size shift along with 500uL of reaction (may take multiple gels to purify the whole reaction, depending on the yield and well size). We use (1.5. mm thick) combs with 1 cm teeth and load about 40–50 microliters (about 35–40 micrograms oligonucleotide) per well (see Fig. 3).
Run gel at 450 volts for 4–5 hours (or until bromophenol blue dye runs off) and then run 30 minutes longer after dye runs off. If dye runs off in less time or more time, reduce or extend the extra run time respectively.
Stain gel in ethidium bromide and 1× TBE solution (enough EtBr to produce a pale yellow color,20–100μL per Liter of 1×TBE) for 20 minutes rocking at room temperature. Destain 1 hour in 1×TBE rocking at room temperature.
Place gel on UV box to visualize bands using long wave UV light. Alternatively one can directly view the two linkers by UV-shadowing with a 254 nM light. Two bands of similar intensity should be apparent in the adenylation reaction, only one band in the negative control (Fig. 3). Cut a slice of gel containing the slower migrating (top) band in the adenylation reaction taking care not to cut the bottom band. If another, third, faint band appears above the top, do not take that band either.
Pass excised bands in gel through a 1mL syringe (no needle attached) to purée the gel into a fine slush. Add 4mL TE buffer (10mM Tris Base, 1mM EDTA, pH 8.0) and incubate overnight at 4°C rotating in a 15mL polypropylene conical tube. Seal with parafilm to prevent leaks.
Prepare GenElute tubes (Sigma-Aldrich GenElute Agarose Spin Columns 56500) by rinsing with 100μL TE buffer (use 6 tubes and the eluates will later be combined).
Let the gel/TE mixture sit upright for 5 minutes so the gel pieces settle to the bottom.
Load TE supernatant (minimize any small gel fragments that are taken up into pipette tip) onto GenElute tubes. Spin 18k rcf for 10 minutes at room temperature.
Collect all run through in one 15mL conical tube; this solution has the adenylated linker.
To concentrate the activated oligonucleotide, add 1 eq. volume of 2-Butanol to solution and sit at room temperature for 5 minutes. Two liquid layers should form. Remove the top (butanol) later and discard. Note the volume of the bottom (aqueous) layer should be slightly reduced.
Repeat the above step (adjusting for what 1 eq. vol is each time) until final volume of the bottom (aqueous) layer is 250–500μL (approximately 3–5 times) and transfer the bottom (aqueous) layer to a 1.5 mL microcentrifuge tube. This contains all the adenylated linker.
Extract the aqueous solution containing the adenylated linker with an equal volume Phenol:Chloroform (1:1) and shake vigorously.
Centrifuge the solution at 15K rcf for 15 minutes and carefully remove the top layer into a fresh microcentrifuge tube.
Add an equal volume of isopropanol and 5μL of Glycoblue reagent (15 mg/mL Invitrogen AM9515 glycogen with dye) to the top layer and precipitate (overnight at −20°C or 30 minutes to 1 hour at −80°C).
Recover the adenylated linker by centrifugation at 15K rcf for 30 minutes at 4°C. Remove the supernatant and add 200μL of cold 75% ethanol solution to resuspend pellet.
Centrifuge at 15K rcf for 15 minutes at 4°C, remove the supernatant and air dry for 2 minutes at room temperature.
Add 30μL nuclease free water and quantify the purified adenylated linker using a Nanodrop.
Fig. 3.
Purification of the activated linker. Following the reaction of the linker with ImpA to form AppLinker, the activated linker is purified away from the starting linker using a preparative 20% polyacrylamide gel. The first two lanes are the 26nt linker sequence (10 and 5 micrograms). In the next two lanes are 17 and 35 micrograms of the ImpA reaction. The bottom band is the unadenylated linker, and the upper band is the adenylated linker which is excised. We typically load about 35–40 micrograms, and obtain sufficient purity for library preparation. There is often a third, fainter band (arrow) above the two major bands which we do not elute.
RNA Preparation
Note that it is essential that the RNA is not contaminated with any DNA or DNase (and obviously RNase). RNA can be purified either from whole cells or from particular cell fractions (nuclei, cytoplasm, polyribosomes) after cell fractionation.
Whole cell RNA is isolated from whole cells by treating a 10 cm plate with 1mL Trizol reagent (Invitrogen/Fisher Sci. 15596026) after removal of media and washing with Trisbuffered saline (TBS 0.14M NaCl, 10mM Tris, pH 7.6). Don’t use PBS since residual phosphate can precipitate with ethanol after the RNA preparation. Suspension cells can be harvested by centrifugation, washed with TBS, and then suspended in TBS (5–10×106 Cells/250 μL), and the suspension added to 750 μL of Trizol.
Incubate cells incubated with Trizol for ~15 minutes at 4°C rotating.
Add 200μL chloroform per 1 ml of Trizol, vortex briefly, and centrifuge at 15K rcf for 15 minutes at 4°C.
Carefully remove the upper layer and add 1 volume of isopropanol. Precipitate 30 minutes at −80°C or overnight at −20 °C.
Spin RNA down 30 minutes 15K rcf at 4°C and remove sup.
Add 500 μL 75% ethanol to wash the pellet and spin down pellet 15 minutes at 15K rcf 4°C.
Remove supernatant and allow to air dry for 2 minutes. Remove any excess ethanol that collected at the bottom. Add nuclease free water to resuspend the RNA based on amount of cells harvested. We use 80μL water to resuspend a confluent well of cells in a 6-well plate. The typical yield is 20–40 micrograms per well of a 6 well plate.
Quantify RNA using a Nanodrop and run 1 μg of RNA on a 1% agarose gel to confirm quality and concentration. There should be clear 18S and 28S rRNA bands and no smearing indicative of degradation. If you are working with very small amounts of RNA then you can analyze the RNA on a Qubit (Ver. 2.0 Fluorometer) or on an Agilent Bioanalyzer. Note at this point the RNA may still be contaminated with small amounts of nucleotides, and the absorbance readings don’t necessarily reflect the total amount of RNA.
- Treat the RNA with DNase to remove all traces of DNA. If we have substantial amounts of RNA, we generally DNase treat 10–20 μg of RNA. We can start with about 1 μg of DNase treated RNA for a total cell RNA library prep; 5 μg if we are going to do a ribominus prep.
- 10μL 10x RQ1 DNase Buffer (Promega M6101)
- 10μL RQ1 DNase (Promega M6101)
- 10–20 μg RNA
- H2O up to final volume of 100μL
Incubate at 37°C for 30 minutes in a water bath
Add 400 μL of Phenol:Chloroform (1:1) to reaction and centrifuge at 15K rcf for 15 minutes at 4°C
Remove the top layer and then an equal volume of isopropanol. Store at −80°C 30 minutes or at −20 °C overnight to precipitate RNA.
Centrifuge 30 minutes at 15K rcf 4°C, remove sup
Wash pellet with cold 75% ethanol and centrifuge for 15 minutes at 15K rcf 4°C, remove supernatant and air dry 2 minutes
Resuspend in 20 μL nuclease free water and quantify using a Nanodrop. Check the quality of the DNAse treated RNA on a Qubit (Ver. 2.0 Fluorometer).
Ligation of Adenylated Linker to RNA
The goal of this step is to add to the linker to the 3’ end of every RNA molecule in the sample. Note that ligation requires a free 2′ and 3’ hydroxyl. RNAs ending in a 2′, 3’ or cyclic phosphate (e.g. 5’ tRNA halves) or in a 2′O-methyl nucleotide (some miRNAs) will not be ligated. If we are going to do a Ribominus preparation, we still ligate to total RNA prior to carrying out the depletion.
- Ligation Reaction: (final volume 10 μl)
- 1–1.5 μg RNA treated with DNase
- 500ng Adenylated Linker (about 50 picomoles)
- Add Nuclease Free Water up to total volume of 8 μL
- Incubate at 65°C 15 minutes to denature the RNA, then put on ice for 3 minutes
- 1 μL10x T4 Ligase Buffer (NEB M0351S)
- 1 μL T4 RNA Ligase 2, Truncated K227Q (NEB M0351S)
- Incubate at 16°C overnight (up to 24 hours)
- Reaction can be stored at −20°C at this point.
Purify ligated RNA by adding 90 μL water and 100 μL Phenol:Chloroform (1:1)
Shake vigorously and spin 12k rcf for 10min 4°C.
Remove the top layer. Add 100 μL Chloroform, shake vigorously, and spin 12k rcf for 10 min 4°C
Remove the top layer and add an equal volume of isopropanol and 1 μL Glycoblue {15 μg, Invitrogen AM9515)
Precipitate at −80°C for 30 minutes
Centrifuge at 15K rcf for 30min at 4°C, remove supernatant.
Wash with 200 μL 75% ethanol, centrifuge at 15K rcf for 15 min at 4°C, remove supernatantand air dry 2 min
Remove any excess ethanol that collected and add resuspend RNA in 10 μL water. Store at −20°C until ready for Reverse Transcription. We generally transcribe the entire ligation into cDNA within a week after preparation.
cDNA Synthesis
Add 5 μL of 10μM RT Primer specific to the Linker sequence to the 10 μL of ligated RNA from the previous step
Incubate at 65°C for 15 minutes to denature, then on ice for 3 minutes
Add 1 μL 10mM dNTPs (NEB N0447S)
Add 5 μL 5× RT Buffer (Invitrogen 18080093)
Add 1 μL 0.1M DTT (Invitrogen 18080093)
Add 2 μL water
Add 1 μL SuperScript III (Invitrogen 18080093)
Mix by pipetting and incubate at 50°C for 1 hour followed by 65°C for 20 minutes to inactivate SuperScript III
Store cDNA (25 μL total) at until ready for PCR amplification of library. We have stored cDNAs for 6 months to a year at −20°C for 6 months to a year.
PCR 1 (Library Amplification)
- PCR Reaction
- 5 μL of cDNA (20% of total Reverse Transcription reaction)
- 10 μL Q5 5x Buffer (NEB M0493S)
- 1 μL 10mM dNTPs
- 1 μL 10μM Linker Specific Reverse Primer
- 1 μL 10μM Gene Specific Forward Primer (or 0.5 μL of two gene specific primers) Note one can multiplex as many gene-specific primers as you want at this step. The gene specific primers will each have an Illumina primer sequence at the 5’ end.
- 0.5 μL Q5 Hot-Start DNA Polymerase (NEB M0493S)
- 31.5 μL of water to bring volume up to 50 μL
- Cycle Details
- 95°C 3 minutes Heated Lid
- 95°C 15 seconds
- Annealing Temp (56°C for Histone primers) 15 seconds
-
72°C 15 secondsRepeat steps 2–4 for 15 cycles total
- 72°C 2 minutes
- 16°C Hold
- PCR Cleanup
- Add 40 μL of Axygen PCR Mag Bead (Axygen/Fisher Sci. 14–223-151 ) slurry to PCR1
- Mix 10 times with pipetting
- Let sit at Room Temperature for 10 minutes
- Place on magnetic stand 5 minutes, remove supernatant
- Add 200 μL 100% ethanol to the beads and resuspend thoroughly
- Place on magnetic stand 5 minutes, remove supernatant
- Add 200 μL 100% ethanol to the beads and resuspend thoroughly
- Place on magnetic stand 5 minutes, remove supernatant
- Air dry 7–10 minutes, until Mag Bead pellet is no longer shiny, but has not cracked yet.
- Add 20 μL water and resuspend thoroughly
- Place on magnetic stand 5 minutes
- Carefully remove the supernatant and save for next step. Be careful not to take any of the Mag Bead pellet
- Use a Qubit (Ver. 2.0 Fluorometer) High Sensitivity dsDNA kit to quantify PCR1 of library for quality control
- Note that the readings at this step may be very low. There is no need to add more cycles to PCR1 unless the subsequent PCR2 step does not give any DNA. For example, a <0.1ng/μL reading does not mean PCR1 has failed if at least one library has a reading above 0.1ng/μL. We typically will make 5–10 libraries at once.
PCR 2 (Sequencing Adapter Addition)
You can do only a single PCR if you design the initial PCR1 primers with miSeq adapters included. In this case the oligonucleotides will be about 100 nts for each index you have synthesized. We elected to use a second PCR step to reduce the size of the primers. Please ask if interested about the longer primers
- PCR Reaction
- 15 μL PCR1 (about 2 ng maximum)
- 1 μL 10mM dNTPs
- 1 μL Distal Primer (10μM)
- 1 μL IR Indexing Primer (10μM) This is different for each library being made
- 0.5 μL Q5 Hot Start DNA Polymerase
- 10 μL Q5 5x Buffer
- 21.5 μL H2O to final volume of 50 μl
- Cycle Details
- 95°C 3 minutes Heated Lid
- 95°C 15 seconds
- 56°C 15 seconds
-
72°C 15 secondsRepeat steps 2–4 for 15 cycles total
- 72°C 2 minutes
- 16°C Hold
- PCR Cleanup
- Identical to protocol for PCR1 Cleanup
- Use a Qubit to quantify PCR2 of library
- Readings should be in the 10–50ng/μL range for ~500bp amplicon library preparations. If less than 5ng/μL, it may be necessary to concentrate the amplified DNA
Library Quality Control
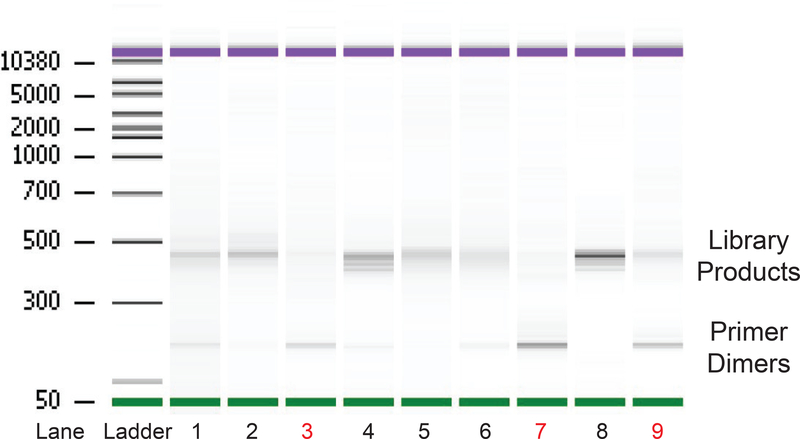
After recording concentrations of your library on the Qubit, load ~10ng onto a Bioanalyzer DNA Chip (Fig. 4). Ideally you should see only a clear band or clusters of bands about 450bp for the histone mRNAs. Note that the band is heterogenous, both because each histone mRNA has a slightly different length 3’ UTR and the amplified sample will contain all the degradation products as well as remaining full length RNA from multiple histone genes. A band around 120 indicates you have primer dimers, which likely occurred during the second PCR amplification. We will often simply repeat both the PCR1 and PCR2 with the remaining cDNA which usually results in a successful library. Small amounts of this band (5% of total) do not interfere with successful library sequencing. The bead cleanup details provided are a strict bead wash that gets rid of most primer dimers.
Fig. 4.
Quality Control of EnD-Seq Libraries. The final libraries are analyzed on a Agilent Bioanalyzer. About 10 ng of each sample (determined by the Cubit reading) is loaded in each lane. Shown is a standard DNA Agilent Chip. All the libraries we plan to multiplex are run on the same Chip. The expected products are about 450 nts for full-length histone mRNAs, with shorter lengths for the degradation intermediates. Primer dimers from PCR2 are about 120 nts and are a common contaminant. The left lane is the provided ladder for determining library size. Lanes 2, 4, 5, and 8 excellent libraries with minimal primer dimers. Lanes 1 and 6 have some primer dimer contamination, but gave good results on sequencing. Lanes 3 and 9 have excessive amounts of primer dimer. This is generally corrected by reamplifying the cDNA. Lane 7 is a failed library, which may require starting from a new reverse transcription of the ligated RNA.
MiSeq Run
We initially used the 300 v2 MiSeq kit which now typically yield about 20 million reads. With the introduction of the 150 v3 MiSeq kit, we now typically obtain 30 million reads per flow cell, and sufficient read length to map the products. Prepare the libraries by diluting each of your libraries (cleaned PCR2 reaction) to 2nM and follow the MiSeq preparation guidelines for the 2 nM library prep (not the 4 nM). Add 1 μL of each library to a small PCR tube to pool them all together (minimum of 5 μL final, but there is no maximum) and then being denaturing the libraries according to the MiSeq guidelines. The final pooled library concentration should be ~9–10 pM. 9 or 10pM library pools should be acceptable. If preparing libraries from a small number of target RNAs (as we do for the histone mRNAs), an additional library (with indices that do not conflict with your own) will be required in the pool to provide diversity in the sequencing reaction. Otherwise the miSEq Chip may fail. Add 1 μL of this library (diluted to 2 nM) to the pool. We typically use a library from another species or a RNA-Seq library to provide the diversity. The diversity library can be as low as 7% of the total MiSeq run. Add the PhiX DNA (provided by Illumina) to the libraries during the sample loading steps as indicated in the miSeq protocol so that the PhiX is 15–30% of the total input.
After injecting the pooled library samples into Port 1 (sample port), be sure to add the primer that recognizes the EnD-Seq libraries into Port 12 (Read 1 Primer Mix port). Use a 10 μL pipette with thin gel loading tips to add the primer. The final concentration of the primer should be 0.5 μM (3 μL of a 100 μM stock of the sequencing primer).
DATA ANALYSIS
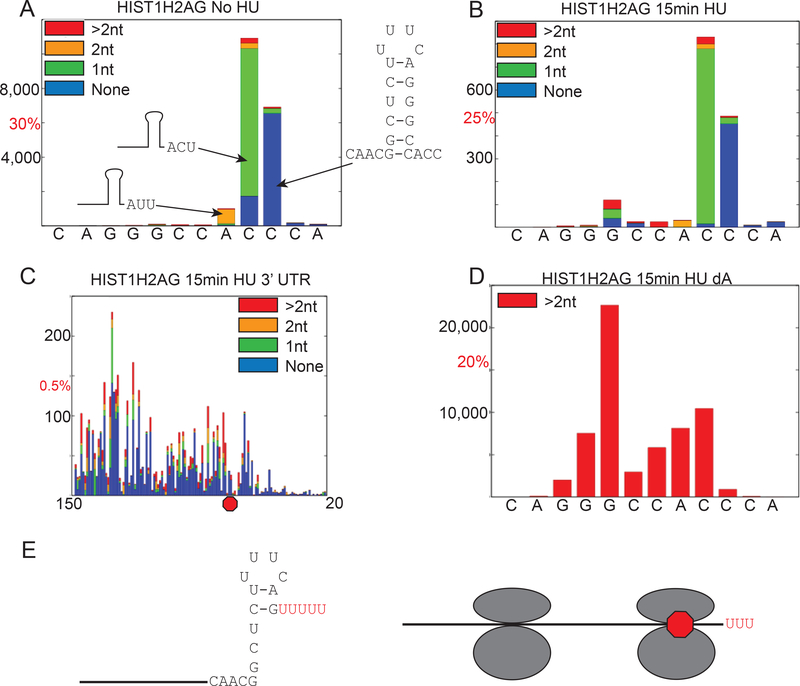
After running on the MiSeq, the libraries are analyzed using the AppEnD workflow [18]. To display the results, each nucleotide is assigned an index, with the 3’ terminus of processed RNA being assigned index zero, which is plotted on the X-axis. The Y axis indicates how many sequences end with that nucleotide as the final template nucleotide. The stacked bar graph demonstrates whether there is no non-templated tails, one nontemplated nucleotide, two nontemplated nucleotides, or 2+ nontemplated nucleotides. In Fig. 5A, we show the 3’ ends of histone mRNA in S-phase. Note that the template nts in the mRNA end at three different sites, but they all extend three nts past the stemloop, with the differences made up by non-templated uridines (Fig. 5A). Thus the last 3 nts are AUU, ACU or ACC with ACC being completely templated. Also shown are libraries prepared from synchronized cells treated with hydroxyurea (HU) focusing on two type of degradation intermediates: the intermediates in the stemloop (Fig. 5B), and the intermediates in the 3’ UTR through the stop codon (Fig. 5C). After 15 minutes of treatment with hydroxyurea, we can see a build-up of degradation intermediates in the stemloop and the 3’ UTR (Fig 5B, C).
Fig. 5.
Visualization of the Results. The AppEnd algorithm includes mapping to the genome giving the position of the last templated nucleotide, and determining the length and composition of any non-templated 3’ tails on each read [18]. These can be used to create bar graphs. We typically plot bar graphs showing the reads with 0 tails, 1 nt tails, 2 nt tails and greater than 2 nt tails as a function of position in the mRNA. A. S-Phase cells obtained after double thymidine block and aligned to the 3’ termini of histone HIST1H2AG mRNA. Index 0 corresponds to the last nt after processing. The actual number of reads is shown on the Y-axis, and the number corresponding to the indicated percentage of total reads is also shown. Plotting the percentage of total reads allows one to normalize between different libraries. Note the major 3’ ends are ACC, ACU and AUU, each extending 3 nts after the base of the stem. B. S-phase cells were treated with HU for 15 min to inhibit DNA replication, and the results plotted as in panel A. Note that the major 3’ ends are the same since only a small fraction of the mRNA has been degraded. There are additional intermediates in the stem loop which contain a high fraction of tails >2 nts. C. The data from the same library as in panel B is plotted for nts 20–150 (with the stemloop removed to show the intermediates). This shows there are very few intermediates present between the stop codon and the stem loop. Starting 15–20 nts after the stop codon there are large number of intermediates, many of which are uridylated, suggesting that the exosome has encountered the terminating ribosome, which stalls degradation. D. The same RNA sample as in panel B was primed with a linker ending in three A’s. Reads with >2 nt nontemplated tails are shown. Note that the most abundant RNAs with long tails are in the stem, and the short U tails at the 3’ end are no longer detected. E. Structure of kinetic intermediates in histone mRNA degradation, uridylated RNAs terminating in the stemloop (left) after initial degradation by 3’hEXO and intermediates 15 nts from the terminating stop codon bound by the terminating ribosome resulting from degradation pausing waiting for removal of the ribosome.
When we prime using the dA primer (3 A’s on the Reverse Transcription primer) to identify only longer uridine tails found during degradation, we see an enrichment of these species in the stemloop (Fig. 5D) and there are very few long tails on the RNAs ending after the stemloop. The intermediates in the stem are the predominant species. Note that we see a substantial number of artifactual reads when we use the dA primer, including internal priming at the 4 U’s in the loop (Fig. 2B), but these are removed during the analysis since they do not contain 3 non-templated nucleotides.
From this data we can deduce that the pathway of histone mRNA degradation 3’ to 5’ has two major “kinetic” intermediates. We see the same spectrum of intermediates when we analyze samples shortly after initiation of degradation (<10% of histone mRNA degraded) and when 60–70% of the histone mRNA has been degraded [39]. This suggests that the rate limiting step in degradation (not surprisingly) is initiation of degradation, which in this case is degradation into the stemloop by 3’hExo (Fig. 1D), and that individual molecules are then rapidly degraded. This results in accumulation of a steady-state distribution of intermediates which is maintained until the mRNA is degraded. If there are slow steps in degradation these will appear as an accumulation of reads at particular sites. We see two kinetic intermediates in histone mRNA degradation. 1. Accumulation of uridylated intermediates after degradation 4 to 5 nts into the stem and 2. Accumulation of intermediates 15 nts 3’ of the stop codon, which are also uridylated (Fig. 5E). Intermediate 1 likely accumulates because SLBP is still bound to the 3’ end, and must be removed prior to further degradation. Intermediate 2 likely results from the exosome reaching the stalled terminating ribosome. There are intermediates throughout the coding region likely due to other ribosomes bound to histone mRNA. Before degradation can proceed further the ribosome must be removed, likely by Dom 34 [39].
CONCLUSIONS
This approach can potentially give information about the 3’ end of every RNA in the cell. Since degradation intermediates are likely to be present in very low concentrations, selecting a subset of mRNAs to analyze is the only possible way to obtain sufficient number of degradation intermediates to deduce the 3’ to 5’ degradation pathway of an RNA. Note this method gives no information about the 5’ end of the degradation intermediates. The fact that we were able to isolate some molecules by circular RT-PCR that were partially degraded from both ends suggests that decapping and 5’ to 3’ degradation also plays a role in histone mRNA degradation, and it is not possible to determine the relative importance of the two pathways. We have shown that some of these degradation intermediates are capped [39], suggesting that 5’ to 3’ degradation and 3’ to 5’ degradation is not occurring simultaneously on all the histone mRNA molecules. These 3’ to 5’ degradation intermediates can be isolated from polyribosomes, consistent with degradation initiating and continuing on polyribosomal bound mRNP which has a stalled ribosome at the termination codon [39].
The degradation intermediates we can characterize by this approach are limited to molecules that can be amplified by using the primer in the coding region chosen for amplification. This is about 270 nts from the 3’ end of the mRNA (about 200 nts from both the start codon and 200 nts from the stop codon) in our experimental design. It is potentially possible to isolate intact histone mRNA degradation intermediates (e.g. by using a biotinylated antisense oligonucleotide identical to the primer for second strand cDNA synthesis) to isolate all molecules containing a central sequence of the histone mRNA in the coding region. If one first ligates the 3’ linker onto the total RNA sample and then isolates the RNA using a biotinylated nucleotide, then one can ligate a different primer onto the 5’ end, using protocols to identify 5’ phosphorylated ends [40, 41], or capped ends [42]. This would allow us to determine whether decapping occurs extensively on either intermediate 1 or on intermediates in the coding region.
This same approach can potentially be used to identify degradation intermediates on any mRNA. Elmar Wahle used a similar approach to detect degradation intermediates in Drosophila heat shock mRNA and observed non-templated A’s on some of the intermediates in the 3’ UTR which promote heat-shock mRNA degradation [43].
ACKNOWLEDGMENTS
This work was supported by NIH grant GM29832 to W.F.M. We thank Walter J. Wever for assistance with ImpA synthesis and Chelsea K. Raulerson for assistance with the data analysis. We also thank Drs. Mike Slevin and Patrick Lackey for advice in library preparation, and Dr. Josh Welch (U. Michigan) for advice on data analysis and development of the AppEnd analysis platform.
Footnotes
SEQUENCING DATA
The sequencing data in this paper are deposited in GEO: Accession numbers GSE68471 (Fig. 5A–D).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Marzluff WF, Wagner EJ, Duronio RJ, Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail, Nat Rev Genet 9(11) (2008) 843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Parker R, Song H, The enzymes and control of eukaryotic mRNA turnover, Nat Struct Mol Biol 11(2) (2004) 121–7. [DOI] [PubMed] [Google Scholar]
- [3].Schoenberg DR, Maquat LE, Regulation of cytoplasmic mRNA decay, Nat Rev Genet 13(4) (2012) 246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schoenberg DR, Mechanisms of endonuclease-mediated mRNA decay, Wiley Interdiscip Rev RNA 2(4) (2011) 582–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ, Argonaute2 is the catalytic engine of mammalian RNAi, Science 305(5689) (2004) 1437–41. [DOI] [PubMed] [Google Scholar]
- [6].Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH, SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells, Nat Struct Mol Biol 16(1) (2009) 49–55. [DOI] [PubMed] [Google Scholar]
- [7].Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E, SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan, RNA 14(12) (2008) 2609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mullen TE, Marzluff WF, Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5’ to 3’ and 3’ to 5’, Genes Dev 22(1) (2008) 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Coller J, Parker R, General translational repression by activators of mRNA decapping, Cell 122(6) (2005) 875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gray NK, Hentze MW, Iron regulatory protein prevents binding of the 43S translation preinitiation complex to ferritin and eALAS mRNAs, EMBO J 13(16) (1994) 3882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Buchan JR, Parker R, Eukaryotic stress granules: the ins and outs of translation, Mol Cell 36(6) (2009) 932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ, The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression, Cell 130(1) (2007) 179–91. [DOI] [PubMed] [Google Scholar]
- [13].Ballantine JE, Woodland HR, Polyadenylation of histone mRNA in Xenopus oocytes and embryos, FEBS Lett 180(2) (1985) 224–8. [DOI] [PubMed] [Google Scholar]
- [14].Sánchez R, Marzluff WF, The oligoA tail on histone mRNA plays an active role in translational silencing of histone mRNA during Xenopus oogenesis, Mol. Cell. Biol (24) (2004) 2513–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chang H, Lim J, Ha M, Kim VN, TAIL-seq: genome-wide determination of poly(A) tail length and 3’ end modifications, Mol Cell 53(6) (2014) 1044–52. [DOI] [PubMed] [Google Scholar]
- [16].Rissland OS, Norbury CJ, Decapping is preceded by 3’ uridylation in a novel pathway of bulk mRNA turnover, Nat Struct Mol Biol 16(6) (2009) 616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kushner SR, mRNA decay in prokaryotes and eukaryotes: different approaches to a similar problem, IUBMB Life 56(10) (2004) 585–94. [DOI] [PubMed] [Google Scholar]
- [18].Welch JD, Slevin MK, Tatomer DC, Duronio RJ, Prins JF, Marzluff WF, EnD-Seq and AppEnD: sequencing 3’ ends to identify nontemplated tails and degradation intermediates, RNA 21(7) (2015) 1375–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lackey PE, Welch JD, Marzluff WF, TUT7 catalyzes the uridylation of the 3’ end for rapid degradation of histone mRNA, RNA 22 (2016) 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shyu A-B, Greenberg ME, Belasco JG, The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways, Genes Dev 3 (1989) 60–72. [DOI] [PubMed] [Google Scholar]
- [21].Chen CY, Yamashita Y, Chang TC, Yamashita A, Zhu W, Zhong Z, Shyu AB, Versatile applications of transcriptional pulsing to study mRNA turnover in mammalian cells, RNA 13(10) (2007) 1775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rutkowski AJ, Dolken L, High-Resolution Gene Expression Profiling of RNA Synthesis, Processing, and Decay by Metabolic Labeling of Newly Transcribed RNA Using 4-Thiouridine, Methods Mol Biol 1507 (2017) 129–140. [DOI] [PubMed] [Google Scholar]
- [23].Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, Ameres SL, Thiol-linked alkylation of RNA to assess expression dynamics, Nat Methods 14(12) (2017) 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yamada T, Imamachi N, Onoguchi-Mizutani R, Imamura K, Suzuki Y, Akimitsu N, 5’ Bromouridine IP Chase (BRIC)-Seq to Determine RNA Half-Lives, Methods Mol Biol 1720 (2018) 1–13. [DOI] [PubMed] [Google Scholar]
- [25].Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Magnuson B, Wilson TE, Ljungman M, Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA, Methods 67(1) (2014) 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ, The human and mouse replication-dependent histone genes, Genomics 80(5) (2002) 487–98. [PubMed] [Google Scholar]
- [27].Pandey NB, Marzluff WF, The stemloop structure at the 3’ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability, Mol. Cell. Biol 7 (1987) 4557–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Graves RA, Pandey NB, Chodchoy N, Marzluff WF, Translation is required for regulation of histone mRNA degradation, Cell 48 (1987) 615–626. [DOI] [PubMed] [Google Scholar]
- [29].Kaygun H, Marzluff WF, Translation termination is involved in histone mRNA degradation when DNA replication is inhibited, Mol Cell Biol 25(16) (2005) 6879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brooks L 3rd, Lyons SM, Mahoney JM, Welch JD, Liu Z, Marzluff WF, Whitfield ML, A multiprotein occupancy map of the mRNP on the 3’ end of histone mRNAs, RNA 21(11) (2015) 1943–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sanchez R, Marzluff WF, The stemloop binding protein is required for efficient translation of histone mRNA in vivo and in vitro, Molecular and Cellular Biology 22(20) (2002) 7093–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ross J, Kobs G, H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3’ terminus and proceeds 3’ to 5’, J. Mol. Biol 188 (1986) 579–593. [DOI] [PubMed] [Google Scholar]
- [33].Ross J, Peltz SW, Kobs G, Brewer G, Histone mRNA degradation in vivo: the first detectable step occurs at or near the 3’ terminus, Mol. Cell. Biol 6 (1986) 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Caruccio N, Ross J, Purification of a human polyribosome-associated 3’ to 5’ exonuclease, J. Biol. Chem 269 (1994) 31814–31821. [PubMed] [Google Scholar]
- [35].Dominski Z, Yang X, Kaygun H, Marzluff WF, A 3’ exonuclease that specifically interacts with the 3’ end of histone mRNA, Molecular Cell 12 (2003) 295–305. [DOI] [PubMed] [Google Scholar]
- [36].Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V, Eri1 degrades the stemloop of oligouridylated histone mRNAs to induce replication-dependent decay, Nat Struct Mol Biol 20(1) (2013) 73–81. [DOI] [PubMed] [Google Scholar]
- [37].Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T, Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing, Methods 44(1) (2008) 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Newman MA, Mani V, Hammond SM, Deep sequencing of microRNA precursors reveals extensive 3’ end modification, RNA 17(10) (2011) 1795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Slevin MK, Meaux S, Welch JD, Bigler R, Miliani de Marval PL, Su W, Rhoads RE, Prins JF, Marzluff WF, Deep sequencing shows multiple oligouridylations are required for 3’ to 5’ degradation of histone mRNAs on polyribosomes, Mol Cell 53(6) (2014) 1020–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Willmann MR, Berkowitz ND, Gregory BD, Improved genome-wide mapping of uncapped and cleaved transcripts in eukaryotes--GMUCT 2.0, Methods 67(1) (2014) 64–73. [DOI] [PubMed] [Google Scholar]
- [41].Yu X, Willmann MR, Anderson SJ, Gregory BD, Genome-Wide Mapping of Uncapped and Cleaved Transcripts Reveals a Role for the Nuclear mRNA Cap-Binding Complex in Cotranslational RNA Decay in Arabidopsis, Plant Cell 28(10) (2016) 2385–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nechaev S, Fargo DC, Dos SG, Liu L, Gao Y, Adelman K, Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila, Science 327(5963) (2010) 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Harnisch C, Cuzic-Feltens S, Dohm JC, Gotze M, Himmelbauer H, Wahle E, Oligoadenylation of 3’ decay intermediates promotes cytoplasmic mRNA degradation in Drosophila cells, RNA 22(3) (2016) 428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]