COVID-19 is the disease caused by the Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) that originated from Wuhan, China in late 2019. It was classified as a global pandemic in March, 2020 by the World Health Organization (WHO). Finding effective, affordable treatments to this pandemic is of utmost importance.
Biotechnology platforms for the rapid high-throughput identification and prioritization of effective therapeutic candidates for multiple indications have the potential to significantly strengthen our response to pathogenic outbreaks and save countless lives [1–4]. In response to the current COVID-19 outbreak, numerous technologies, including those based on high throughput protein-protein complex pulldowns and network biology, have been applied to quickly screen and identify drug repurposing candidates that may be rapidly deployed to treat infected individuals without the need for full regulatory approval [1, 2, 5, 6].
We developed the Computational Analysis of Novel Drug Repurposing Opportunities (CANDO) platform for shotgun multitarget drug discovery, repurposing, and design [1, 2, 7], funded in part by a 2010 NIH Director’s Pioneer Award and previously described in Drug Discovery Today in 2014 [1], for precisely this type of pandemic scenario. The platform screens and ranks every existing human use drug for every disease/indication through large scale modelling and analysis of interactions between comprehensive libraries of drugs/compounds and protein structures. The interaction may be determined by any screening or docking method but the built-in ones using a fast bioinformatic docking protocol and the hierarchical fragment-based docking with dynamics protocol CANDOCK [8] are prioritized. The drug-proteome signature comparison and ranking approach used by the CANDO platform yields benchmarking accuracies of 20–40% for ~1500 indications relative to random control accuracies of 2–15%. Across twelve prospective in vitro validation studies, 58/163 (35%) top ranking predictions made using the CANDO platform had comparable or better activity relative to existing drugs across ten indications, and represent potential novel repurposed therapies for indications such as dengue, dental caries, diabetes, herpes, lupus, malaria, and tuberculosis [1, 2].
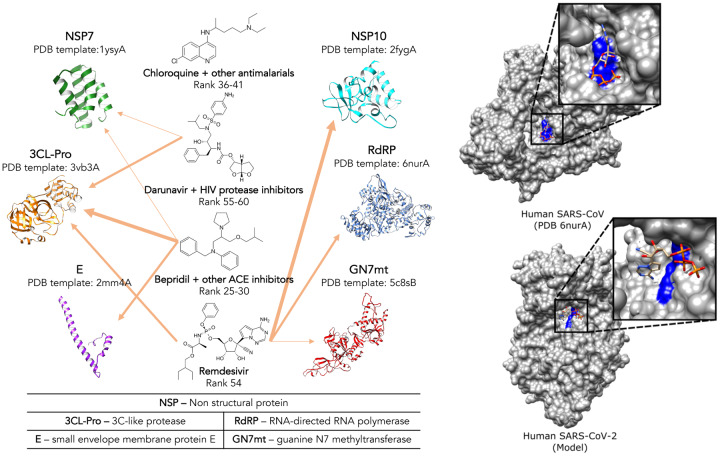
We used the CANDO platform to generate putative drug repurposing candidates against SARS-CoV-2 (Figure 1). The platform ranks a number of clinical trial candidates listed in Table 1 of Harrison [9] in the top 1% of predictions and provides relevant target and off-target interaction information for them.
Figure 1 – A selection of putative drug candidates of preclinical and clinical interest against SARS-CoV-2 and COVID-19 generated by the CANDO shotgun repurposing platform (left).
The orange arrows in decreasing thickness indicate the interaction score (1st, 5th, or 10th percentile) between the drug and predicted protein target. In the case of prodrug remdesivir, conversion to its active form diminishes its predicted interaction with the protease and greatly strengthens it with the RdRP: The top predicted poses of the active form of remdesivir docked to the solved and template-based model structures of the RdRP (right) from both SARS-CoV and SARS-CoV-2 using CANDOCK [8] indicate binding directly into the catalytic site (colored blue). The site consists of two adjacent aspartic acid residues, indicating that remdesivir disrupts RdRP function when it binds and is potentially effective against at least two different coronaviruses. Other interesting predictions from our March 16, 2020 round (http://protinfo.compbio.buffalo.edu/cando/results/covid19/) include ACE inhibitors at rank 25–30, remdesivir at rank 54, and darunavir and other HIV protease inhibitors at rank 55–60. A separate pipeline within CANDO based on drug-drug similarity to known SARS-CoV actives identified chloroquine and other antimalarials at rank 36–41, which may be effective via a host-based mechanism since no viral proteins are predicted to be strongly targeted. All of the highlighted candidates have been shown or are believed to have activity against SARS-CoV-2 and/or are undergoing clinical trials to demonstrate efficacy [9]. Additionally, the drugs at rank 1 and 14 (omacetaxine mepesuccinate and mycophenolate mofetil, not shown) were previously identified in experimental assays to be potent inhibitors of coronaviruses [10, 11]. Therefore, the other higher ranked drugs in our lists are also worth evaluating, with the potential payoff of choice, greater efficacy, and reduced cost for compassionate off-label use and/or in clinical trials. Shotgun repurposing platforms such as CANDO not only generates short lists of therapeutic candidates rapidly but may also provide mechanistic atomic level detail of relevant interactions between targets and repurposable drugs identified by us or by any other means (including serendipity and analysis of medical records).
We are currently in the process of undertaking in vitro validation of top ranked candidates as well as using EHR data to corroborate or negate predictions made by the platform. This pandemic highlights the importance of developing such robust shotgun repurposing platforms that not only make drug discovery more efficient by systematically evaluating multiple uses of a human ingestible drug but may also be rapidly deployed every time a new disease arises.
Three coronavirus outbreaks in two decades, including the current pandemic, indicates a necessity of preparation for the next one that may be more deadly and costly. The CANDO drug repurposing platform was originally funded and implemented for predicting drug leads for epidemics and pandemics. Sustained funding for shotgun drug repurposing biotechnology that have been benchmarked extensively to identify potential drugs for all diseases, such as CANDO, will prepare us for this eventuality while also providing us with an array of therapeutic solutions to help improve human health and quality of life.
Acknowledgments
This work was supported in part by a National Institute of Health Director’s Pioneer Award (DP1OD006779), a National Institute of Health Clinical and Translational Sciences Award (NCATS) (UL1TR001412), an NCATS ASPIRE design challenge award, a National Library of Medicine T15 Award (T15LM012495), a National Cancer Institute/Veterans Affairs Big Data-Scientist Training Enhancement Program Fellowship in Big Data Sciences, startup funds from the Department of Biomedical Informatics at the University at Buffalo, a start-up package from the Department of Chemistry at Purdue University, Ralph W. and Grace M. Showalter Research Trust award, the Integrative Data Science Initiative award, the Jim and Diann Robbers Cancer Research Grant for New Investigators award, and NIH NCATS ASPIRE Design Challenge awards to Gaurav Chopra. Additional support, in part by, a NCATS Clinical and Translational Sciences Award from the Indiana Clinical and Translational Sciences Institute (UL1TR002529), and the Purdue University Center for Cancer Research NIH grant P30 CA023168 are also acknowledged. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Footnotes
Teaser: Sustained research investment into shotgun, or “every disease”, drug discovery and repurposing platforms, already proving to be useful against the current COVID-19 pandemic, will better prepare us for inevitable future outbreaks.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Minie M., et al. , CANDO and the infinite drug discovery frontier. Drug discovery today, 2014. 19(9): p. 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chopra G., et al. , Combating ebola with repurposed therapeutics using the CANDO platform. Molecules, 2016. 21(12): p. 1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng F., et al. , Systems biology-based investigation of cellular antiviral drug targets identified by gene-trap insertional mutagenesis. PLoS computational biology, 2016. 12(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saberian N., et al. , A new computational drug repurposing method using established disease–drug pair knowledge. Bioinformatics, 2019. 35(19): p. 3672–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y., et al. , Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell discovery, 2020. 6(1): p. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon D.E., et al. , A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. BioRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangione W., et al. , cando.py: Open source software for predictive bioanalytics of large scale drug-protein-disease data. bioRxiv, 2020: p. 845545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fine J., et al. , CANDOCK: Chemical atomic network based hierarchical flexible docking algorithm using generalized statistical potentials. Journal of Chemical Information and Modeling, 2020. 60(3): p. 1509–1527. [DOI] [PubMed] [Google Scholar]
- 9.Harrison C., Coronavirus puts drug repurposing on the fast track. Nat Biotechnol, 2020. [DOI] [PubMed] [Google Scholar]
- 10.Shen L., et al. , High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. Journal of virology, 2019. 93(12): p. e00023–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyall J., et al. , Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrobial agents and chemotherapy, 2014. 58(8): p. 4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]