Abstract
Inflammation is a major risk factor of morbidity and mortality in older adults. Although its precise etiology is unknown, low-grade inflammation in older adults is commonly associated with increased intestinal epithelial permeability (leaky gut) and abnormal (dysbiotic) gut microbiota. The increasing older population and lack of treatments to reduce aging-related microbiota dysbiosis, leaky gut, and inflammation culminates in a rise in aging-related comorbidities, constituting a significant public health concern. Here, we demonstrate that a human-origin probiotic cocktail containing 5 Lactobacillus and 5 Enterococcus strains isolated from healthy infant gut prevented high-fat diet–induced (HFD-induced) microbiota dysbiosis, leaky gut, inflammation, metabolic dysfunctions, and physical function decline in older mice. Probiotic-modulated gut microbiota primarily reduced leaky gut by increasing tight junctions, which in turn reduced inflammation. Mechanistically, probiotics modulated microbiota in a way to increase bile salt hydrolase activity, which in turn increased taurine abundance in the gut that stimulated tight junctions and suppressed gut leakiness. Furthermore, in Caenorhabditis elegans, taurine increased life span, reduced adiposity and leaky gut, and enhanced physical function. The results suggest that such probiotic therapies could prevent or treat aging-related leaky gut and inflammation in the elderly.
Keywords: Gastroenterology, Microbiology
Keywords: Glucose metabolism, Innate immunity, Macrophages
A human-origin probiotic cocktail improved metabolic dysfunction, gut microbiota, and taurine production, which increased tight junctions and reduced leaky gut and inflammation in older mice.
Introduction
The number of older adults is increasing worldwide, and aging increases the risk of comorbidities, such as obesity, diabetes, cancer, and cardiovascular diseases (1, 2). Although many preventative or therapeutic targets to ameliorate such individual aging-related comorbidities have been known, common pathways that involve all of these comorbidities and their amenable interventions in older adults remain elusive. Increased low-grade inflammation is a major risk factor for morbidity and mortality in older adults (3, 4) and a hallmark of aging-related comorbidities. Although the precise etiology of increased inflammation in older adults is not known, emerging evidence suggests that abnormally higher intestinal epithelial permeability (“leaky gut”) is associated with increased inflammation in older adults (3, 5, 6). Higher gut permeability leads to influx of antigens, endotoxins, pathogens, and other proinflammatory substances from gut into the blood and lymphatic circulation, leading to systemic inflammation. However, increased inflammation also disrupts intestinal epithelial barriers and induces leaky gut (7, 8). Therefore, the link between leaky gut and inflammation is bidirectional and remains correlative. Leaky gut and inflammation are commonly associated with poor health of older adults, specifically in individuals with obesity, diabetes, or reduced physical function (5). Abnormal (dysbiotic) gut microbiota could account for leaky gut and inflammation in older adults (3, 9), since dysbiotic microbiota can produce metabolites that interact with the host’s intestinal epithelial cells to reduce epithelial barrier proteins like zonulin-1 (Zo1), occludin (Ocln), and claudins, resulting in disrupted cell-to-cell connections, which compromises intestinal permeability (10, 11). Therefore, maintaining a healthy gut microbiota can benefit the health of older adults by reducing leaky gut and inflammation.
Probiotics are live microorganisms, which — when administered in adequate amounts — confer a health benefit on the host (12). Probiotics are one of the most accepted strategies to modulate the gut microbiota and normalize dysbiosis (13–17); therefore, we posit that probiotic therapy can maintain healthy gut microbiota and reduce leaky gut and inflammation. Several preclinical studies have shown that some specific probiotic strains may improve intestinal permeability homeostasis (18, 19) and inflammation (20, 21). Multiple studies also suggest that probiotics are beneficial for several human diseases, including obesity and diabetes (13–17, 21–23); however, the origin of such probiotics remains largely unknown. Probiotics isolated from a human body source are recommended to be used in clinical practice. However, well-defined human-origin probiotics exhibiting beneficial effects on aging-related leaky gut and inflammation are not well known; hence, this warrants development of human-origin probiotics. In addition, it remains largely unknown how probiotics reduce leaky gut and inflammation, specifically in older adults, and the majority of evidence accumulated to date remains correlative. For example, while some reports show that probiotics act primarily on immune cells to suppress inflammation and improve intestinal barriers, others show that probiotics primarily enhance intestinal barriers, which in turn helps suppress inflammation (24, 25). Determining whether probiotics-microbiota interactions primarily improve intestinal permeability or immune cells will not only pave the way to understand the mechanisms, but it will also provide opportunities to further improve their therapeutic potential. Moreover, probiotics primarily modulate gut microbiota, which produce an array of metabolites; however, which specific metabolite mediates the effect of a specific probiotic is not well defined. Finding new microbial metabolites can open avenues to develop them as small molecule therapy to ameliorate leaky gut and inflammation in older adults.
Similar to humans, older mice show higher leaky gut, inflammation, microbiome dysbiosis, glucose intolerance, insulin resistance, and physical functions compared with young counterparts (26–32). By using this model, we studied a multistrain probiotic cocktail isolated from infant gut (33) and defined its functional role in aging-related metabolic dysfunctions, leaky gut, and inflammation in the approximately 80-week-old C57BL/6J mice (equivalent to > 65 years human age). We have systematically pinpointed physiological, cellular, and molecular mechanisms by which such probiotic therapy modulates the microbiota/metabolite/leaky gut/inflammation axis.
Results
Probiotics feeding prevents HFD-induced metabolic dysfunction and physical function decline in older mice.
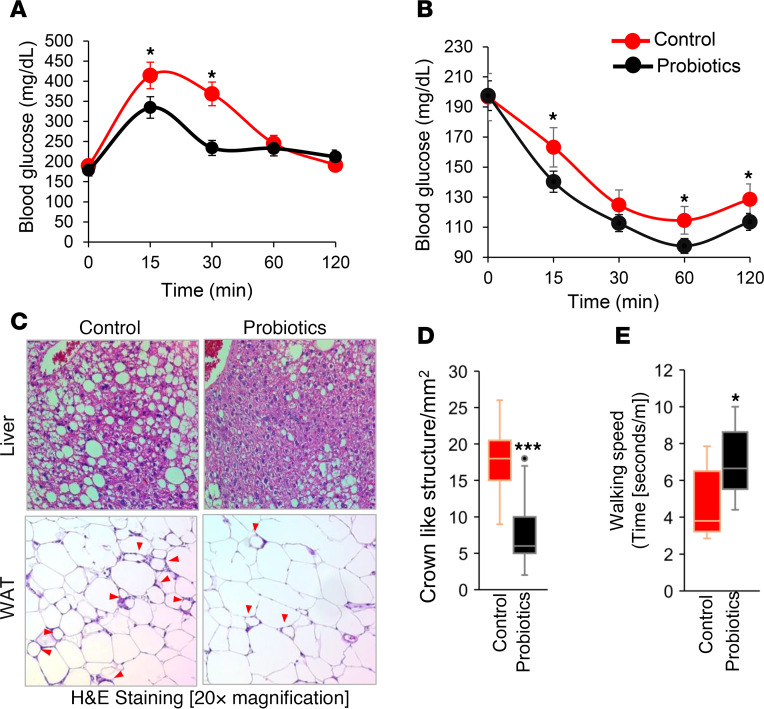
Older adults are more susceptible to obesity- and type 2 diabetes–related metabolic dysfunctions like glucose intolerance, insulin resistance, hepatic steatosis, and inflammation in adipose tissues upon high calorie/fat intake (34–37). Here, we investigated whether our probiotic cocktail can protect such high-fat diet–induced (HFD-induced) dysfunctions in older mice. Interestingly, 10 weeks of probiotic cocktail feeding significantly increased blood glucose clearance in oral glucose tolerance test (OGTT) (Figure 1A) and improved insulin sensitivity upon exogenous insulin administration during insulin tolerance test (ITT) (Figure 1B) in HFD-fed older mice compared with their age- and sex-matched controls. These data suggest that probiotic feeding prevented HFD-induced glucose metabolism dysfunctions in older mice. However, no significant differences were observed on body weight and weight gain, food and water intake, or energy expenditure in probiotic-fed group compared with their controls (Supplemental Figure 1, A–G; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.132055DS1). This indicates that the probiotics cocktail was well tolerated and enhanced metabolic functions of older mice. Interestingly, fat accumulation in liver (hepatic steatosis) was significantly attenuated, and crown-like structures in white adipose tissue (WAT), an inflammation marker, were significantly lower in probiotic-fed older mice than in controls (Figure 1, C and D), indicating that probiotic feeding reduced fat accumulation and inflammation in metabolic organs like liver and WAT.
Figure 1. Probiotics feeding prevents HFD-induced metabolic dysfunctions in older mice.
(A and B) Ten weeks of probiotics improved glucose tolerance and enhanced insulin sensitivity in older obese mice, measured by oral glucose tolerance test (A) and insulin tolerance test (B) (n = 6 in control and n = 8 in probiotics groups; *P < 0.05, 2-way ANOVA). (C) Representative images of H&E staining of liver (upper panels) showing reduced fat accumulation and white adipose tissue (WAT; lower panels) showing reduced adipocyte size, along with reduced inflammation (indicated by crown-like structures; red arrows) in probiotics fed mice (n = 8) compared with their controls (n = 6). (D) Crown-like structures are graphed. (E) Probiotic-fed older obese mice (n = 8) exhibited higher physical function presented as walking speed compared with their age- and sex-matched HFD-fed controls (n = 6). Values are mean of n = 6–8 mice in each group, and data are shown as mean ± SEM. *P < 0.05, and ***P < 0.001 by 2-way ANOVA with Bonferroni’s correction (A and B) and Student’s t test (D and E).
Obese older adults face higher decline in physical function, such as reduced walking speed that is commonly associated with poor health outcomes and increased mortality in older adults (38–40). Interestingly, probiotic-fed obese older mice had higher walking speed than controls (Figure 1E), suggesting that probiotic therapy attenuated physical function decline in older obese mice. Altogether, these results indicate that probiotic therapy prevented HFD-induced metabolic derangements like glucose intolerance, insulin resistance, hepatic steatosis, and inflammation in WAT and improved physical function of older mice.
Probiotic therapy beneficially modulates gut microbiota in obese older mice.
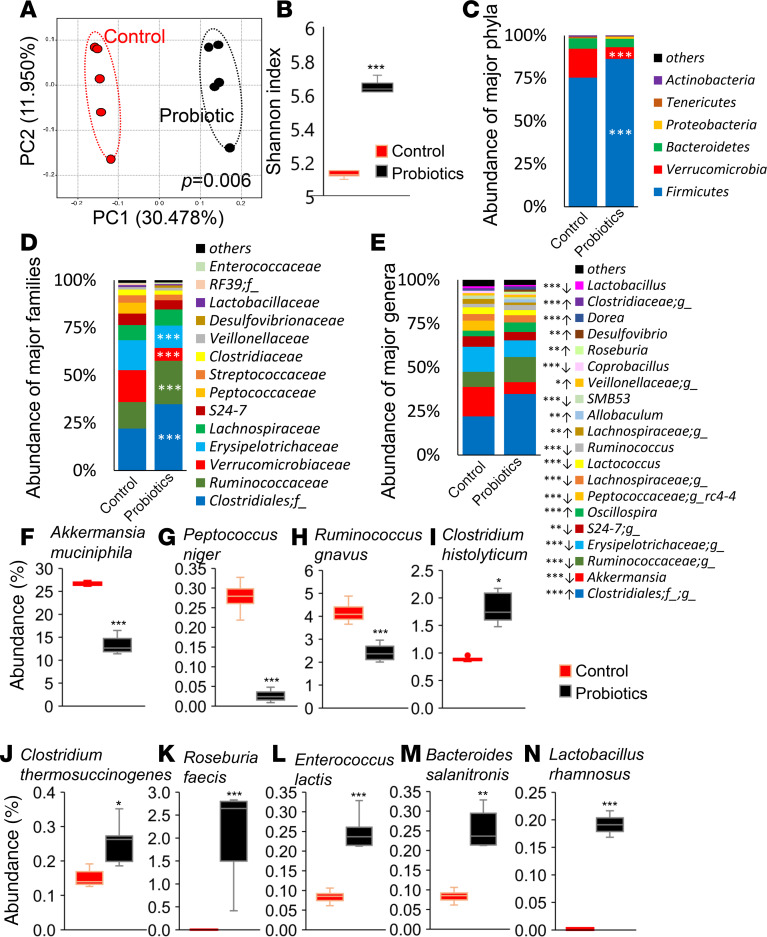
The primary action of probiotics is posited through modulating the gut microbiota, and we found that feeding this human-origin probiotic cocktail significantly changed microbial composition in the gut of older mice (Figure 2). Reduced microbial diversity measured by α-diversity (microbial diversity within the sample) and β-diversity (microbial diversity among the samples) are known indicators of dysbiosis (41), and interestingly, feeding this probiotic cocktail enhanced microbial diversity, as indicated by increased Shannon index (α-diversity) and by a significantly distinct clustering of β-diversity observed in principal coordinate analysis (PCoA) (Figure 2, A and B). Other α-diversity indices like phylogenetic diversity (PD) whole tree, Chao1, and number of operational taxonomic units (OTUs) remain significantly unchanged in probiotic-fed mice and their control mice, but trends were toward higher α-diversity in probiotic-fed mice compared with controls (Supplemental Figure 2, A–C). The abundance of phylum Firmicutes, family Rumminoccocaceae, and an unidentified family of order Clostridiales was significantly increased, while abundance of phylum Verrucomicrobia and families Verrucomicrobiaceae and Erysipelotrichaceae was decreased in probiotic-fed mice compared with controls (Figure 2, C and D). Specifically, probiotic feeding promoted the abundance of Clostridiales;f_;g_, Oscilliospira, Allbacullum, Rosiburia, Desulfovibrio, and Dorea, while it suppressed the population of Akkermansia, S24-7, Peptococcacea;g_rc4-4, Lachnospiraceae;g_, Lactococcus, Ruminococcus, SMB53, Coprobacillus, and Lactobacillus (Figure 2E). Similar bacterial phyla, families, and genera appeared during our Linear discriminatory analysis effect size (LEfSe) analysis (Supplemental Figure 2, D and E). Interestingly, among top 10 significantly changed bacterial species due to probiotics feeding, the abundance of Akkermansia muciniphila, Peptococcus niger, and Ruminicoccus gnavus significantly decreased, while that of Clostridium histolyticum (C. histolyticum), C. thermosuccinogenes, Roseburia faecis, Enterococcus lactis, Bacteroides salanitronis, and Lactobacillus rhamnosus was significantly increased in probiotic-fed obese older mice compared with their controls (Figure 2, F–N). These results demonstrate that older HFD-fed mice that received the probiotic cocktail developed a significantly distinct gut microbiota signature — enriched with beneficial commensals — that was associated with improvement in the metabolic health of older obese mice.
Figure 2. Probiotic therapy beneficially modulates gut microbiome in older obese mice.
(A–E) Gut microbiome signature in terms of β-diversity (A), α-diversity (Shannon index) (B), and abundance of major phyla (C), families (D), and genera (E) were significantly changed in probiotic-treated HFD-fed older mice (n = 5) compared with their controls (n = 5). (F–N) Specifically, probiotic therapy decreased Akkermansia muciniphila (F), Peptococcus niger (G), and Ruminicoccus gnavus (H) and increased C. histolyticum (I), C. thermosuccinogenes (J), Roseburia faecis (K), Enterococcus lactis (L), Bacteroides salanitronis (M), and Lactobacillus rhamnosus (N). Values are mean of n = 5 in each group, and data are shown as mean ± SEM. *P < 0.05; **P < 0.01, and ***P < 0.001 by PERMANOVA (A), unpaired 2-tailed Student’s t test (F–N), and 1-way ANOVA (B–E).
Probiotics feeding reduces inflammation and leaky gut markers in HFD-fed older mice.
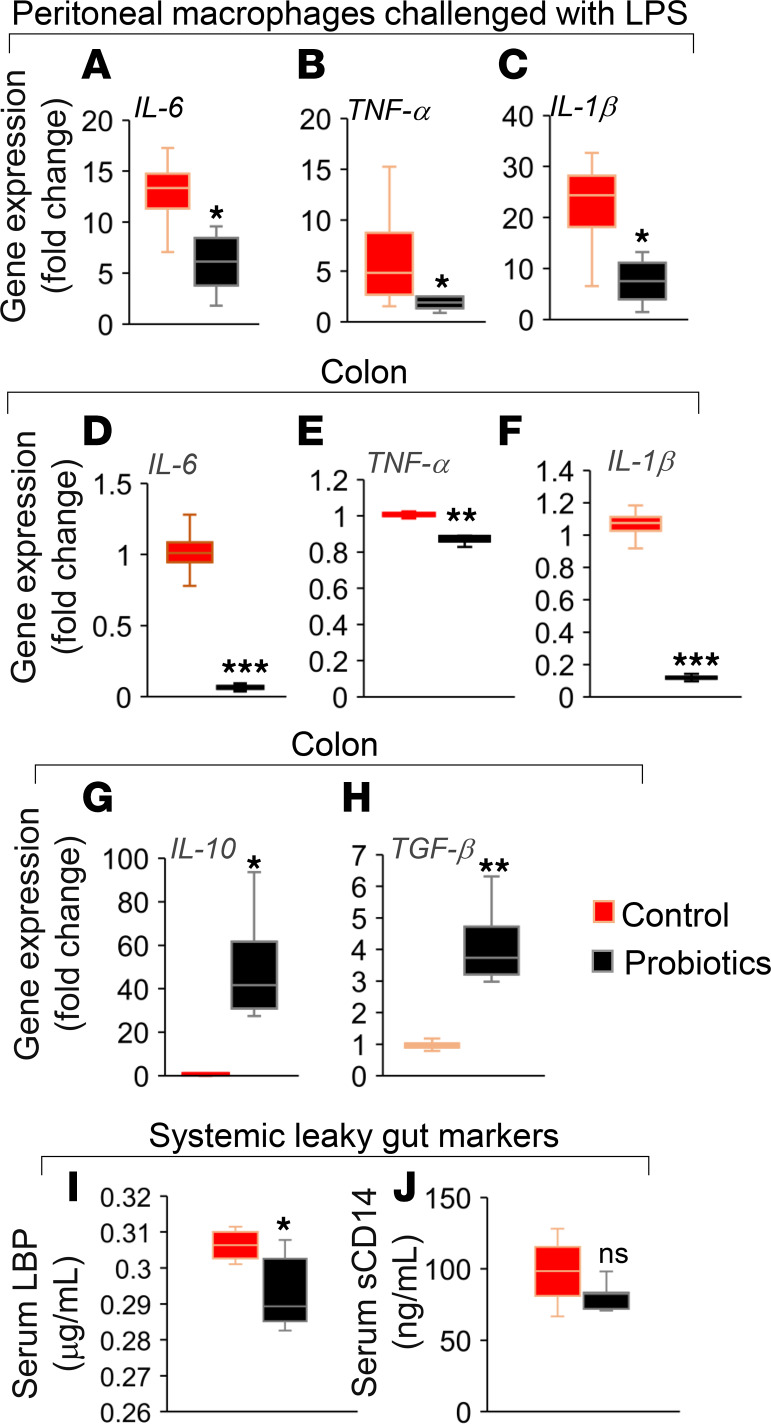
Low-grade inflammation is a major risk factor of metabolic dysfunctions, poor health, and high mortality in older adults (4, 42). Herein, we found that the probiotics feeding significantly decreased the expression of proinflammatory markers like IL-6, TNF-α, and IL-1β in LPS-treated primary macrophages isolated from the peritoneal cavities of older mice compared with their controls (Figure 3, A–C), indicating that the probiotic feeding suppressed the inflammatory responses in the mucosal lymphatic macrophages present in peritoneal cavity. Similarly, the expression of IL-6, TNF-α, and IL-1β was decreased, while antiinflammatory markers like IL-10 and TGF-β expression was increased in the colon tissues of probiotic-fed mice (Figure 3, D–H), suggesting that probiotic feeding also suppresses the inflammation in the intestinal milieu. These results demonstrate that probiotic therapy reduced inflammation in intestinal tissues and distant immune cells like peritoneal lymphatic macrophages.
Figure 3. Probiotics treatment reduces inflammation in peritoneal macrophages and intestine of older obese mice.
(A–C) LPS-induced inflammatory response in terms of mRNA expression of IL-6 (A), TNF-α (B), and IL-1β (C) was reduced in primary macrophages isolated from peritoneal cavity of older HFD-fed mice treated with probiotics (n = 8) compared with their controls (n = 6). (D–H) In addition, the expression of proinflammatory markers such as IL-6 (D), TNF-α (E), and IL-1β (F) were decreased, while antiinflammatory genes like IL-10 (G) and TGF-β (H) mRNA expressions were increased in the colon of probiotic-fed older obese mice (n = 8) compared with their controls (n = 6). (I and J) In addition, systemic leaky gut markers such as LPS binding protein (LBP) (I) and soluble CD14 (J) were reduced in the serum of probiotic-fed older obese mice (n = 7) compared with their controls (n = 6). Values are mean of n = 6–8 in each group, and data are shown as mean ± SEM. *P < 0.05; **P < 0.01, ***P < 0.001 by Student t-test (A–J).
Increased inflammation in older adults is often associated with increased leaky gut indicated by system markers such as LPS binding protein (LBP) and soluble CD14 (sCD14) in circulation/blood that are linked with poor health outcomes (3, 5). Interestingly, we found that the feeding of our human-origin probiotic cocktail significantly reduced LBP levels in the serum of older mice compared with their controls (Figure 3I). The sCD14 levels were also decreased in the serum of probiotic-fed older mice but did not achieve statistical significance (Figure 3J). These results indicated that the probiotics feeding reduces leaky gut markers that are linked with reduced inflammation in intestine and peritoneal macrophages.
Probiotics feeding enhances expression of tight junction proteins in obese older mice.
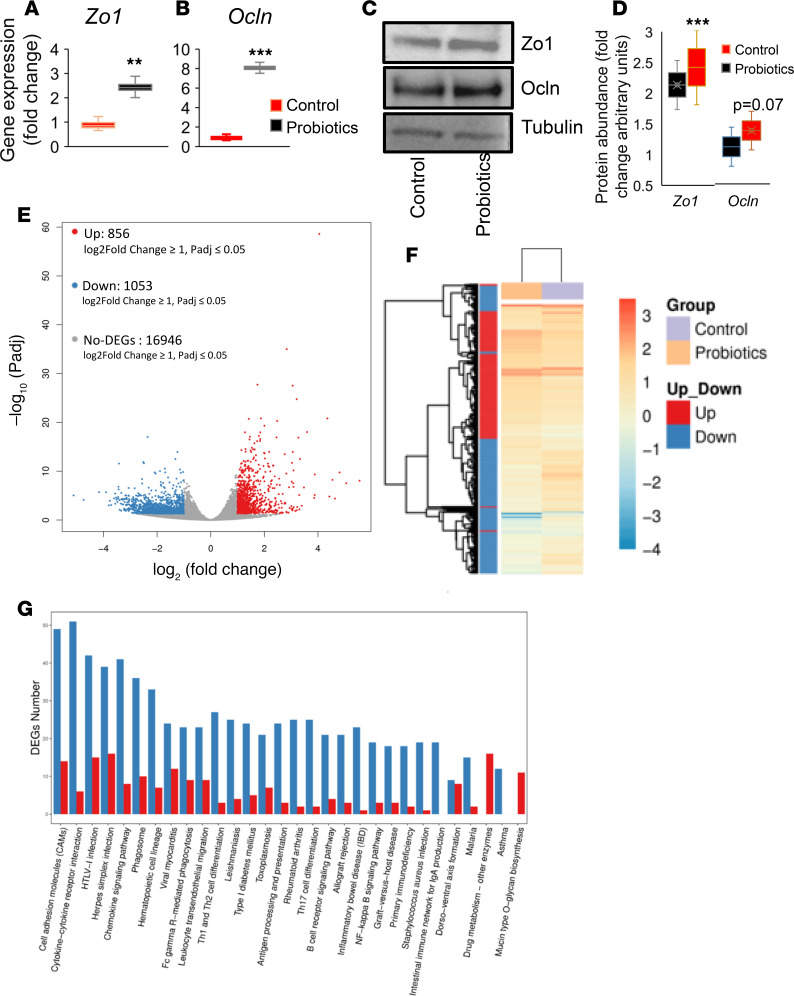
Abnormalities in the gut epithelium, specifically the downregulation of tight junction proteins, are associated with compromised intestinal epithelial permeability (leaky gut) and increased inflammation in obesity and aging (3, 41). We demonstrated that the probiotic therapy significantly increased the mRNA expression of tight junction proteins such as Zo1 and Ocln in the intestinal tissues of older obese mice (Figure 4, A and B). The protein expression of Zo1 increased, while marginal increase with no statistical significance in Ocln protein was seen (Figure 4, C and D). This suggests that probiotic therapy improved the intestinal epithelial integrity that is linked to reduced inflammation in local intestinal tissues, as well as systemically, as corroborated by the decrease in peritoneal macrophages. Interestingly, global and unbiased gene expression analysis by RNA sequencing (RNAseq) revealed that the probiotics feeding significantly changed the expression of a large number of genes and pathways. Specifically, probiotics feeding significantly upregulated around 856 genes — while downregulating around 1053 genes in the gut of older mice compared with their controls (Figure 4E) — that were very distinctly clustered among these groups (Figure 4F). Furthermore, pathway analyses revealed that the cell adhesion molecules (CAMs) and cytokine pathways were the top 2 largely affected pathways in the intestinal tissues of probiotic-fed obese older mice in comparison with their controls (Figure 4G). These results demonstrate that the probiotic therapy enhanced cell adhesion pathways and particularly increased tight junction proteins in the intestine of older obese mice that were associated with decreased inflammation.
Figure 4. Probiotics treatment increases expression of tight junctions in the intestine of older obese mice.
(A and B) The mRNA expression of tight junction proteins like Zonulin-1 (Zo1) (A) and Occludin (Ocln) (B) were significantly increased in colon of probiotics fed older mice (n = 8) compared with their controls (n = 6). (C and D) Western blot analysis shows that Zo1 protein expression was significantly increased, while Ocln showed marginal increase in the colon tissues of probiotic-treated older mice (n = 7) compared with their controls (n = 6). (E and F) Global gene expression using RNAseq analysis revealed that probiotic feeding significantly increased around 856 genes while it decreased 1053 genes that were distinctly clustered in the probiotic-treated (n = 7) group versus controls (n = 6). (G) Pathway analysis of deferentially expressed genes (DEGs) shows that cell adhesion and cytokine (immune) pathways were more affected by probiotics treatment compared with their controls. Values are mean of n = 6–7 each group, and data are shown as mean ± SEM. **P < 0.01 and ***P < 0.001. Student t test (A, B, D) and random forest analysis (E) were used, as well as hierarchical clustering between samples using hclust, with diagrams drawn with ggplot2 (F) and differential expression of genes (DEGs) (G) were completed using R programs.
Probiotics primarily act on intestinal tight junctions instead of immune cells.
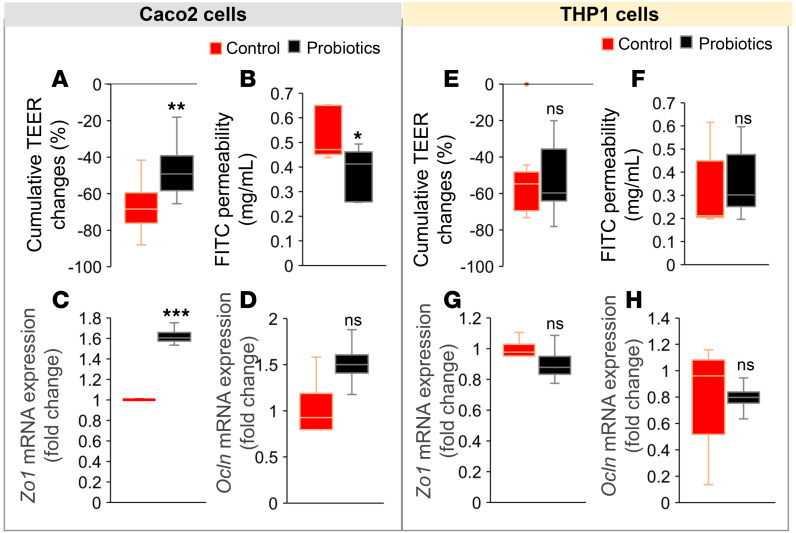
We demonstrate that the probiotic therapy improved metabolic functions and gut microbiota that are associated with reduced inflammation and increased expression of tight junction proteins; however, it is not known whether (a) probiotic-modulated gut microbiota primarily act on intestinal epithelium by improving tight junctions to reduce inflammation or vice versa or (b) these changes in tight junctions and inflammation are parallel events. We used a coculture system of human intestinal epithelial cells (Caco-2; in upper chamber) and human macrophages (THP-1–differentiated macrophages; in lower chamber) to test the effect of probiotics cecal conditioned media (CCM), which was prepared from probiotic-fed mouse cecal contents compared with control CCM. When Caco-2 cells were exposed to CCM of probiotic-fed mice, we observed significantly reduced intestinal epithelial permeability measures like transepithelial electrical resistance (TEER) and FITC-dextran diffusion in the Caco-2 monolayer compared with the CCM of control mice (Figure 5, A and B). However, no significant changes in TEER and FITC diffusion of the Caco-2 monolayer were observed when cocultured macrophages were exposed to both CCM (Figure 5, E and F). Similarly, the expression of tight junction proteins (Zo1 and Ocln) mRNA was significantly increased in Caco-2 monolayer treated with CCM of probiotic-fed mice compared with their controls (Figure 5, C and D), while no significant changes were observed in these measures in Caco-2 cells cocultured with CCM-treated macrophages (Figure 5, G and H). These results demonstrate that the probiotic-modulated gut microbiota primarily act on intestinal epithelial cells to increase the expression of tight junction proteins, resulting in decreased epithelial permeability, which in turn reduced inflammation in local tissues and peripheral immune cells.
Figure 5. Probiotics primarily act on intestinal tight junctions versus immune cells.
(A–C) Treatment of Caco-2 cell monolayers (cultured with THP1 cells) with cecal conditioned media (CCM) from probiotic-fed mice shows significantly less changes in transepithelial electrical resistance (TEER) (A) with reduced FITC-dextran (4 kDa) (B) and increased Zo1 (C) and Ocln (D) mRNA and protein expressions compared with control CCM–treated cells. (E–H) No significant changes were observed in these measures when cocultured THP-1 cells were treated with CCM prepared from probiotics and control mouse cecal contents. Values are mean of 2–3 repeated experiments done in triplicate, and CCM was prepared from cecal contents of n = 6 controls and n = 7 probiotic-treated mice. Data are shown as mean ± SEM. **P < 0.01 ***P < 0.001 by ANOVA with Bonferroni’s corrections and Student’s t test.
Probiotics feeding significantly modulates metabolites and increases taurine abundance in the gut of HFD-fed older mice.
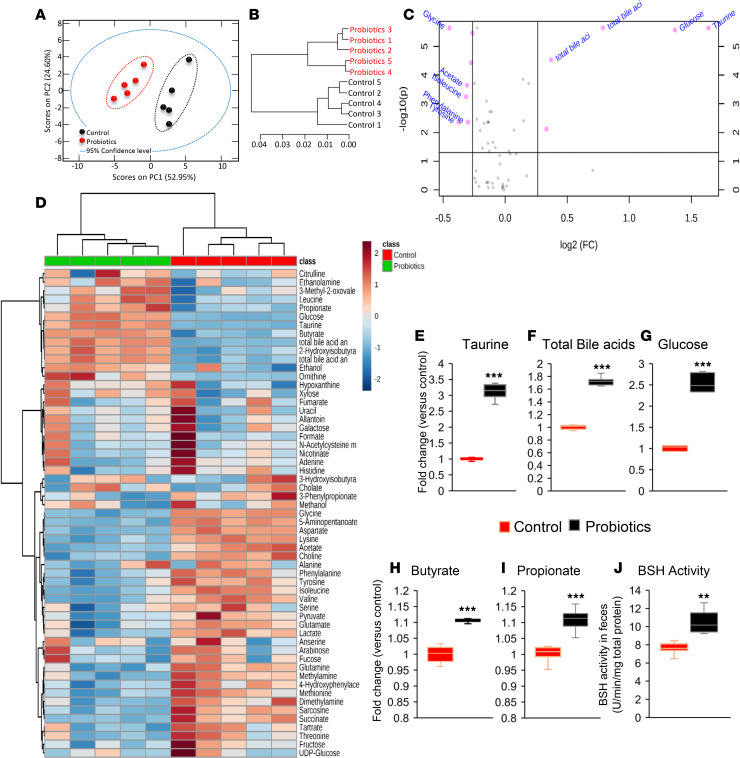
To determine the missing link by which probiotic-modulated gut microbiota improves intestinal tight junctions and inflammation in obese older mice, we tested the hypothesis that gut microbiota–derived metabolites may be involved in these improvements. Therefore, we performed untargeted-unbiased global metabolomics in the feces of these mice (43). Interestingly, the metabolites from probiotic-fed mice were clustered distinctly from their controls (Figure 6, A and B). In addition, the volcano plot suggests that major metabolites like taurine, glucose, and total bile acids (Figure 6, E–G) (44), as well as glycine, acetate, isoleucine, phenylalanine, and tyrosine, were significantly abundant in the gut of probiotic-fed compared with control older obese mice (Figure 6C). Furthermore, the heatmap in Figure 6D also shows that various metabolites in terms of fold change are significantly distinct in probiotic-fed versus control mice. Altogether, these results indicate that probiotics feeding significantly changed metabolites in the gut of older mice.
Figure 6. Probiotic therapy modulates gut metabolome and increases taurine production by enhancing bile salt hydrolase (BSH) activity in the gut of older HFD-fed mice.
(A–C) Untargeted-unbiased metabolomics analyses show that the production of distinct metabolites shown by principal component analysis (PCA) (A), group clustering (B), and fold change abundance enrichment (C) were significantly changed in probiotic-fed old mice (n = 5) compared with their controls (n = 5). (D) Similarly, metabolite heatmap shows differential clusters that are significantly changed in probiotic-fed older mice feces compared with controls. (E–I) Abundance of taurine (E) and total bile acids (F), glucose (G), butyrate (H) and propionate (I) was significantly increased in the feces of probiotic-fed older mice (n = 5) compared with their controls (n = 5). (H) Probiotics feeding significantly enhanced bile salt hydrolase (BSH) activity in the gut of older mice. Values are mean of n = 5 control and n = 5 probiotics group, and data are shown as mean ± SEM. **P < 0.01 and ***P < 0.001. PLS-tool box in MatLab (A and B) and Welch t-test (E–J) were used.
A pathway analysis plotted using pathway enrichment analysis and pathway impact values further indicated that the major Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/) pathways significantly affected after probiotics feeding were taurine and hypotaurine metabolism (Supplemental Figure 3). The abundance of taurine, total bile acids, glucose, butyrate, and propionate were significantly increased in the gut of probiotic-fed older mice compared with their controls (Figure 6, E–I). The highest increase (fold change, 3.9) in taurine levels in the feces of probiotic-fed older mice (Figure 6E) suggested that taurine is linked with improvements in intestinal tight junctions, inflammation, and metabolic, physical, and cognitive functions in older mice fed with HFD. However, the mechanism of how taurine levels increased in the feces of probiotic-fed mice is not known. Taurine is a modified amino acid produced in the liver and released by conjugation to bile acids (bile salts) such as taurocholate in the gut through bile juice (45, 46). These taurine-carrying bile salts are deconjugated by a bacterial enzyme, bile salt hydrolase (BSH) (47). Therefore, we tested a hypothesis that probiotics feeding may have increased BSH activity in the gut microbiota, resulting in increased deconjugation of taurine from bile acids like taurocholate. Interestingly, the BSH activity was significantly increased in the feces of probiotic-fed older mice compared with their controls (Figure 6J), suggesting that probiotics changed the gut microbiota in a way that increased BSH activity, which resulted in higher deconjugation of tauro-bile salts to release more taurine in the gut. Most of the strains in our probiotics cocktail show BSH activity (Supplemental Figure 4, A and B), suggesting that feeding of the probiotic cocktail increased BSH activity in the gut of older mice, which may lead to increased deconjugation of free taurine from bile salts.
Taurine restores intestinal integrity and improves health indicators in C. elegans.
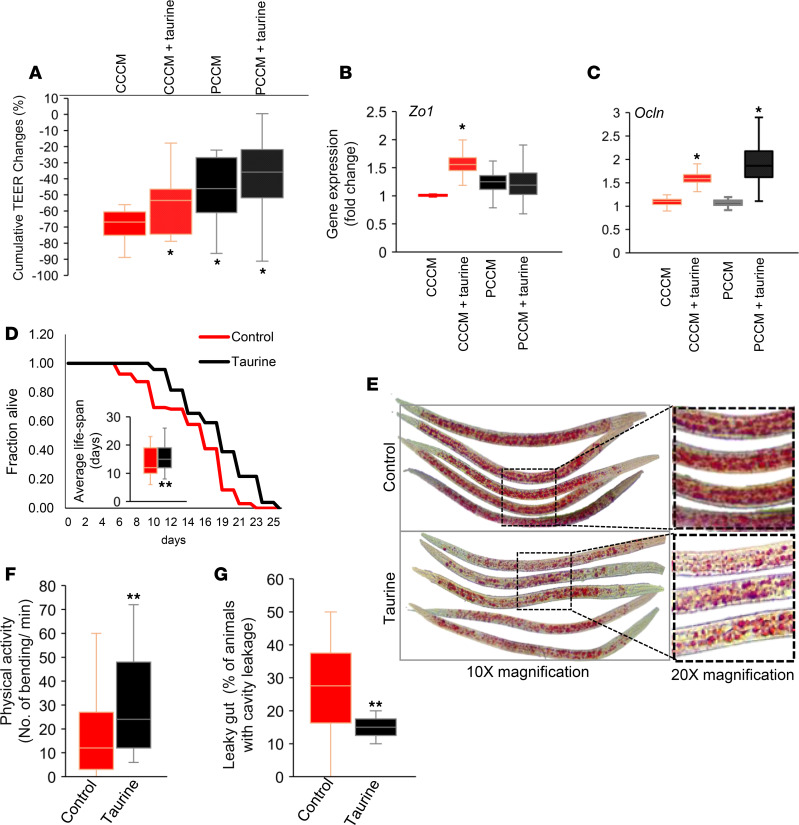
To further determine whether increased taurine levels in the gut contributes to increasing tight junctions, thus reducing intestinal epithelial permeability, we supplemented taurine (200 μM) in the CCM and incubated it with Caco-2 cell’s monolayer. Interestingly, taurine supplementation significantly reduced decline in TEER and increased the expression of Zo1 and Ocln mRNA (Figure 7, A–C, and Supplemental Figure 5), indicating that taurine increases intestinal epithelial barrier integrity to reduce gut permeability by increasing the expression of tight junction proteins. Furthermore, to determine the impact of taurine feeding on life-span in Caenorhabditis elegans (C. elegans), we showed that taurine feeding extended the life span of C. elegans by > 2 days, which is equivalent of > 10–12 years of human age (Figure 7D), suggesting that taurine exhibits antiaging effects. In addition, taurine feeding also reduced fat accumulation and intestinal permeability (measured by using Smurf assay; ref. 48) (Supplemental Figure 6) in older C. elegans (Figure 7, E–G). This further reveals that increased taurine in the gut is beneficial to ameliorate aging-related dysfunctions like adiposity and leaky gut, which in turn improves healthy life-span.
Figure 7. Taurine significantly decreases epithelial permeability, increases tight junction proteins, reduces fat accumulation, and reduces gut leakiness in C. elegans.
(A–C) Taurine supplementation in cecal conditioned media (CCM) in control significantly reduced changes in TEER of Caco-2 cells monolayer (A) and significantly increased expression of tight junction protein Zo1 (B) and Ocln (C) expression. Values are mean of 2–3 repeated experiments done in triplicate. (D) Taurine treatment increased the life span of C. elegans. Kaplan-Meier curves were generated using survival data in days for each worm (n = 150–195 worms in each group), and statistical significance was calculated using the log-rank test. (E) Composite image of multiple worms. (F and G) Taurine treatment also increased physical activity (F) and intestinal permeability (leaky gut) using Smurf assay (G) in n = 120-150 worms in each group. Values are mean of 2–3 trials of cell culture experiments done in triplicate (A–C) and n = 120-195 worms (D–G). Data are shown as mean ± SEM. *P < 0.05, **P < 0.01 by 2-way ANOVA with Bonferroni’s corrections (A), Student’s t test (B, C, F, G), and log-rank (D).
Discussion
In this study, we demonstrate that a probiotic cocktail, which we isolated from the infant gut (33), has beneficial effects to prevent HFD-induced gut dysbiosis, leaky gut, and inflammation, along with metabolic abnormalities and decline in physical function of older mice. This study also untangles that the probiotic-modulated gut microbiota primarily act on intestinal epithelia to reduce leaky gut, followed with inflammation, instead of action on immune cells. This study also comprehensively demonstrates how a well-tolerated human origin probiotic therapy modulates gut microbiota to enhance the abundance of beneficial metabolites like taurine, which reduces leaky gut and improves healthy life span. As a large portion of the world’s population gets older, the demand for interventions to address aging-related comorbidities like obesity, diabetes, hepatic steatosis, and movement decline increases. Because these comorbidities are commonly associated with dysbiotic gut microbiome, leaky gut, and inflammation (3), interventions that modulate the gut microbiota and resolve dysbiosis are good candidate approaches.
Older adults have an even higher susceptibility for such comorbidities when exposed to higher caloric intake than younger counterparts (49, 50). However, the underlying biology of this susceptibility in the context of a HFD and preventive strategies are poorly understood. Here, we demonstrated that a human-origin probiotic therapy prevented HFD-induced metabolic dysfunctions — namely glucose intolerance, insulin resistance, hepatic steatosis, and fat tissue inflammation — in older mice. Reduced physical function and increased inflammation are major risk factors of increased morbidity and mortality of obese older adults (38–40), and probiotic therapy reduced both of these risk factors. Our findings indicate that such probiotic therapies could be beneficial for preventing aging-related metabolic dysfunctions, physical decline, and inflammation, and they may promote healthy aging.
Emerging evidence suggests that the gut of older adults harbors dysbiotic microbiota, which is associated with poor health outcomes (3, 51–53). We observed that probiotic therapy of human-origin multistrain probiotic cocktail beneficially modulated gut microbiota in older obese mice. Although several other probiotics and commensal strains are also known to modulate gut microbiota and have shown beneficial effects on host health (3, 54, 55), their mechanisms of actions are not clear. Some probiotic strains are known to modulate immune cells to suppress inflammation, whereas others have been shown to enhance the intestinal barrier to reduce inflammation by reducing nonspecific leakiness of inflammatory substances from gut to immune sites (18, 19). Nevertheless, these results remain correlative, as leaky gut induces inflammation and increased inflammation also promotes derangements in intestinal barrier functions to increase leakiness (20, 21). It is largely unknown whether probiotic-modulated gut microbiota primarily act on immune cells to suppress inflammation or act on intestinal barriers. Our results demonstrating that the probiotic modulated gut microbiota primarily act on intestinal epithelial permeability — resulting in reduced inflammation, instead of acting on immune cells to reduce inflammation — untangled this cause versus effect on intestinal epithelial versus inflammation mechanism of action. However, such effects might be probiotics formulation and strain dependent; therefore, further comprehensive studies are warranted. Altogether, these results demonstrate that such probiotic therapies can improve aging-related microbiota dysbiosis, leaky gut, and inflammation, and a decoupled mechanism of action can strengthen the ways to devise better microbiota manipulators in reducing leaky gut, and thereby inflammation, in older adults.
Although earlier studies used certain probiotics by either exposing bacterial cells to intestinal epithelial and immune cells or their cell-free supernatants to determine their impact on intestinal epithelial permeability (56–58), how probiotic-mediated changes in gut microbiota and its metabolites impact host system were not declared. Here, we used the CCM as a tool to define this. The use of CCM is innovative because CCM mimics in vivo conditions, where microbial cells and larger ingredients of microbiota are filtered out, while microbiota ingredients like metabolites are preserved and can interact with host cells. Using this system, our studies established the cause versus effect relationship of leaky gut and inflammation, which is affected by probiotic-microbiota action, and we demonstrated how host-microbe interactions contribute to ameliorate leaky gut and inflammation in older and obese gut.
To further understand how probiotic-induced changes in gut microbiota improved intestinal integrity, we tested the hypothesis that probiotics induced the production of microbial metabolites to impact intestinal permeability. Metabolites produced by gut microbiota are the major functional moieties that interact with host cells and can stimulate epithelial cells to preserve intestinal barrier homeostasis (11). Herein, we observed that taurine abundance was significantly increased in the gut of probiotic-fed older obese mice, using a global metabolomics approach (59). Taurine is one of the most abundant modified amino acids present in the human body (60). In liver, taurine conjugates with bile acids to produce bile salts like taurocholate (61). Taurine bile salts are released in bile into the upper gastrointestinal tract. Although 95% of bile salts are reabsorbed into the small intestine (61), a microbial enzyme called BSH can deconjugate bile salts by breaking covalent bonds between taurine and bile acids, resulting in increased taurine and bile acids in the lower gut or feces. We demonstrated that probiotic therapy significantly enhanced the BSH activity in the feces and increased taurine and bile acids in feces of probiotic-fed older obese mice. Our findings support the notion that probiotic therapy increases BSH activity, which in turn increases bile salt deconjugation to release taurine. Since the probiotic strains included in this cocktail have demonstrated BSH activity, this further suggests that probiotic bacterial cells supplied the BSH activity in the gut of probiotic-fed older mice and resulted in increased taurine through bile deconjugation. However, it is also possible that probiotics feeding modulated the intrinsic microbiota in a way to either increase BSH-expressing bacterial numbers, increase BSH activity, or both. Further studies are needed to pinpoint the proportional source of BSH activity.
Taurine is known to have beneficial effects to modulate several cellular and physiological functions, such as hepatoprotective, antidiabetic, antiobese, and many more health-promoting properties (60, 62, 63). We demonstrate that taurine exhibits antiaging effect to extend life span of C. elegans, which is in agreement with other reports that taurine deficiency accelerate death in mice and has beneficial effects against aging-related disorders (60, 64). However, the mechanisms remain largely unknown. Our results demonstrate that taurine decreased intestinal permeability and increased the expression of tight junction proteins in intestinal epithelia (Caco-2 cell monolayers), suggesting that increased taurine abundance in the gut increases intestinal epithelial integrity by enhancing tight junctions to reduce leaky gut and inflammation. In addition, taurine feeding also reduced the fat accumulation and improved physical function in older C. elegans, and this suggests that taurine-mediated reduction in the inflammation may help reduce fat accumulation as well as enhance skeletal muscle functions to maintain higher physical function. Taurine increases functions and mass of skeletal muscles (65, 66), and skeletal muscle–specific depletion of taurine enhances senescence in the muscle cells and increased mortality in mice (64), further supporting our findings that increased taurine can be beneficial for improving metabolic and physical functions in older adults. Other independent studies also show that taurine supplementation in drinking water reduces dextran sulfate sodium–induced colitis (67), as well as improves tight junctions in the lung epithelia (68). This suggests that taurine may be responsible for action of this probiotic therapy in improving aging-related metabolic dysfunctions, leaky gut, and inflammation in older mice; however, the precise mechanism of action by which taurine reduces these dysfunctions and the signaling pathways involved in such effects of taurine remain largely unknown. Although here we studied the taurine because it was largely increased metabolite in probiotic-treated older mice, we also observed significant increases in the abundance of total bile acids propionate and butyrate, and these molecules are also known to benefit intestinal barrier functions (69–72). These molecules, along with taurine, may have played beneficial roles in improving whole phenotype of older mice, and further studies to determine their individual and/or synergistic effects in combinations on leaky gut and inflammation are needed. However, the current study provides an advancement in our understanding of how a multistrain probiotic cocktail can be effective to induce changes in the microbiota and its metabolites in the gut of elderly and how it can be proposed as an alternative biotherapy for aging-related leaky gut and inflammation. In addition, discovery of new microbial metabolites like taurine can provide ways to ameliorate aging-related leaky gut and inflammation, either as a single small molecule or combinations.
In conclusion, this study provides evidence that a potentially novel human-origin probiotic cocktail prevents HFD-induced gut microbiota dysbiosis, intestinal permeability, and inflammation and that it improved metabolic and physical functions in older mice. It also dismantles that the probiotic-modulated microbiota primarily enhances intestinal epithelial integrity — thereby suppressing the inflammation and not the other way around, meaning suppressing inflammation to reduce leaky gut — and it explains cause versus effect relationship of leaky gut versus inflammation. In addition, the study further elucidates the mechanisms by which probiotic therapy enhances BSH activity of the gut microbes in older obese mice, resulting in enhanced bile salt deconjugation and increased levels of taurine in the gut. Higher taurine results in decreased intestinal permeability by enhancing the expression of tight junctions. Furthermore, taurine feeding also demonstrates antiaging effects in C. elegans by ameliorating aging-related decline in physical function, adiposity, and leaky gut. Altogether, these findings suggest that such human-origin probiotic strains and cocktails could ameliorate age-related leaky gut and inflammation, hence opening up novel avenues to explore and exploit the clinical utilities of such therapeutic regimens.
Methods
Probiotics cultures and formulation.
We used 5 Lactobacillus and 5 Enterococcus strains, which were isolated from human infant gut and characterized for their genetic identity by DNA sequencing; their safety profile, such as absentia of pathogenic islands in their genome and antibiotics resistance; and their probiotic attributes, such as acid bile tolerance, epithelial cell adhesion, and their antibacterial activities, as we described earlier (33). To prepare the probiotic cocktail used in this study, freshly cultured bacterial cells were counted, and equal numbers were pooled to prepare stocks of 20% glycerol media that were stored in –80°C. Stock cultures were freshly thawed each day and dissolved in drinking water to feed mice from the experimental group, while an equal amount of glycerol was supplemented in drinking water of control mice, as described below.
Animals.
Older C57BL/6J (>78 weeks) male mice were purchased from the Jackson Laboratory and acclimatized in a temperature-controlled room (22°C) on a 12-hour light/dark cycle for 2 weeks before any treatment. At the age of 80 weeks, mice were randomized in 2 groups; (a) control and (b) probiotics (n = 5–8 in each group), as described elsewhere (73). The second group (probiotics) was fed a cocktail of 10 live probiotic strains suspended in the drinking water (1 × 109 CFU/mL), while an equal amount of glycerol was suspended in the water of the first group (control). Both groups were fed a HFD (60% energy by fat; catalog D12492; Research Diets). Food and water were given ad libitum, and water was freshly changed every day. Food and water intake was measured daily. Body weight of mice was measured weekly. After 10 weeks of treatments, mice were euthanized and tissues were collected for further analyses.
GTT and ITT.
For OGTT, 12-hour overnight–fasted mice were orally given glucose solution (2.5 g/kg body weight) and for ITT, 4-hour fasted mice were given the insulin (Humulin, Eli Lilly) solution i.p. at the rate of 0.75U/kg body weight. The blood glucose was measured before (0 minutes) and after 15, 30, 60, and 120 minutes of glucose/insulin administration, using Truetrack glucometer (Nipro Diagnostics), as also described in our earlier studies (15, 41, 74).
Histology.
Liver and WAT (gonadal) were fixed in 10% formalin, embedded in paraffin blocks, and cut in 0.5 μm–thick sections that were stained with H&E and imaged with AmScope microscope (Irvine) on 20× magnification using 9–megapixel digital camera. Crown-like structures in WAT were counted using ImageJ software (NIH).
Indirect calorimetry.
To measure whole body energy expenditure in live animals, older mice were individually housed in metabolic chambers of TSE PhenoMaster caging systems (Chesterfield) with free access to food and water. The energy expenditure and resting metabolic rates were calculated from the volume of oxygen consumption (VO2) and carbon dioxide production (VCO2), that also allow to determine the ratio between fat and glucose utilization for energy expenditure in terms of resting exchange ratio (RER). All the parameters were measured continuously and simultaneously for 48 hours, as also described in our earlier studies (74, 75).
Measurements of physical function in mice.
Physical function measures such as exercise endurance and walking speed were performed in blinded groups, as described before (73).
Gut microbiota analysis.
The gut microbiome was examined, as per our previously described methods (33, 73, 76–79). In brief, bacterial genomic DNA was isolated from 200 mg of freshly frozen collected feces using Qiagen DNA Stool Mini Kit (Qiagen). The V4 hypervariable region of the of 16S rRNA gene was PCR amplified using the universal primers 515 (barcoded) and 806R and was sequenced using MiSeq Illumina platform (Illumina Miseq Reagent Kit v3). The obtained sequences were demultiplexed, quality filtered, clustered, and taxonomically assigned against Greengenes data base with RDP-classifier using Quantitative Insights into Microbial Ecology (QIIME, version 1.9.1) software package (80, 81) according to our previously described workflow and parameters (41, 73, 76, 78). The α-diversity was computed using QIIME, and the β-diversity was analyzed using PCoA of the unweighted and weighted Unifrac distance (using EMPeror, version 0.9.3-dev). Bacterial taxonomy assignment was calculated within QIIME using default settings to compare the bacterial diversity and abundance between the different groups. LEfSe was applied on phylum, family, and genus level data on Galaxy platform (82), in order to identify discriminative features (unique bacterial taxa) that drive differences in experimental versus control groups.
Measurements of systemic leaky gut markers.
To determine the impact of probiotics feeding on systemic leaky gut, markers such as LBP (catalog HK205-01, Hycult Biotech) and sCD14 (catalog MC140, R&D Systems) were measured in serum using ELISA kits and following manufacturer instructions.
Peritoneal macrophage culture and stimulation.
Peritoneal macrophages were harvested from mice by flushing the peritoneal cavity with cold PBS. The peritoneal cells were cultured in RPMI-1640 media (MilliporeSigma) containing 100 U/mL penicillin and 100 μg/mL streptomycin (Thermo Fisher Scientific). After 2 hours of incubation, floating cells were removed by washing with PBS and adherent macrophages were treated with or without 100 ng/mL LPS for 6 hours. Total RNA was extracted, and gene expression was measured using real-time PCR, as described below.
Real-time PCR.
Total RNA was extracted from tissues and cells using RNeasy kit (Qiagen) and was reverse transcribed to cDNA using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Expression of Zo1, Ocln, IL-6, TNF-α, IL-1β, IL-10, and TGF-β (primer details in Supplemental Table 1) mRNAs were quantified using real-time PCR reaction in Applied Biosystems. The expression of 18S was used as an internal control. All the quantitative PCR (qPCR) analyses were run in triplicate, and results were expressed as fold changes calculated using the ΔΔCt method, as we published earlier (15, 41, 74, 76).
Western blotting.
Total proteins were extracted from intestinal tissues and cell lines using a cell lysis buffer (10 mM Tris, pH 7.6; 1 mM EDTA; and 1% Triton X-100) supplemented with protease inhibitors (Roche). Equal amounts of total protein for each sample were separated on SDS-PAGE gel (10%–20%) and transferred to PVDF membranes. Membranes were incubated with primary antibodies (Supplemental Table 2) and secondary antibodies, followed with chemiluminescent detection and imaging using PXi machine and GeneSys software (SynGene Frederick). Tubulin was used as loading control in each gel.
RNAseq analysis.
Global and unbiased gene expression analysis was done in intestinal (ileum) tissues and was performed using Beijing Genomic Institute protocols and pipelines of RNAseq. The RNAseq library was prepared for sequencing using standard Illumina protocols. After quality control, clean reads were analyzed for the quantification of read numbers mapped to each gene using HTSeq v0.6.1. Differentially expressed genes (DEGs) were defined using q < 5%. Gene ontology and KEGG enrichment analyses were implemented for the DEGs by the GOseq R package and KOBAS software.
Metabolomics analysis.
Fecal samples were extracted using a method described by Gratton et al. (83) with slight changes. The extracted water samples were mixed with phosphate buffer (pH 7.4) containing 10% D2O and 0.1 mM trimethylsilyl propionate (TSP). NMR experiments were carried out on a Bruker Ascend 400 MHz high-resolution NMR (Bruker Biospin) using a 1-D first increment of a NOESY (noesygppr1d)with water suppression and a 4-second recycle delay. All NMR spectra were phased and referenced to TSP in TopSpin 4.06 (Bruker BioSpin). The NMR spectra were analyzed in Amix 3.9 (Bruker Biospin), and a manual pattern was created using the metabolites peaks range determined by Chenomx 8.4 (Chenomx Inc.) to extract the metabolites peak intensities. Total intensity normalization was applied before further data analysis.
Preparation of CCM.
The snap-frozen cecum from control and probiotic-fed mice that were collected at the time of euthanasia and stored in –80oC were crushed in liquid nitrogen using a mortar and pestle. The obtained fine powder was suspended in the cold DMEM media (25 mg cecal content in 10 mL media) and was kept shaking at 4°C for 1 hour on a shaker in cold room at 200 rpm. The resulted suspension was centrifuged for 10 minutes at 3000 g and filtered 2 times through a 0.45-μm filter and 2 times through 0.22-μm filter in sterile condition under a fume hood. The 1:10 dilution of obtained cecal conditional media (CCM) was used to treat cells.
Coculture transwell system and measuring TEER.
Caco-2 cells (HTB-37, ATCC) were seeded on tissue culture polycarbonate membrane filters (pore size 3.0 μm) in Corning 12-well Transwell plates (catalog CLS3402) at a seeding density of 2 × 104 cells/cm2 (70%–80% confluence). The culture medium on both the donor and the acceptor compartment was changed every second day. The cells were left to differentiate for 21 days after seeding, with monitoring of TEER values using an EVOM2 Epithelial Voltometer (WPI). The THP-1 cells (TIB-202, ATCC) were cultured in RPMI-based CCM substituted with 10% FBS. For coculture experiments, 2 million THP-1 cells were seeded in each well of a fresh 12-well plate and differentiated with PMA (100 nM) following a 24-hour rest period. The macrophage-differentiated THP-1 cells were cocultured in Transwell plates (12-well Corning transwell cell culture inserts) with Caco-2 monolayer cells. The TEER of these were measured, and the treatments were applied in the upper or lower compartments, which contain Caco-2 and THP-1 cells, respectively. The treatments were applied on Caco-2 cells directly on the upper compartment, while — to treat the THP-1 cells — CCM treatments were given for 2 hours, followed with washing and substitution with fresh media and coculturing with Caco-2 cells.
FITC-dextran permeability assay in Caco-2 cell monolayers.
After an 8-hour treatment with CCMs of 21-day-old monolayers of Caco-2 cells, FITC-dextran 4 (FITC-4; 4 kDa) solution (1 mg/mL) was added on the apical (upper) side of the monolayers, and the levels of FITC-4 in basolateral (lower) side were determined using a fluorescence 96-well plate reader. The excitation and emission wavelengths were 400 nm and 535 nm, respectively.
BSH activity in the feces.
Frozen, fecal samples (250 mg) were crushed in liquid nitrogen, suspended in 5 mL PBS, and settled on ice for 10 minutes. Bacterial cells were pelleted from the supernatant by centrifuging at 7000 g for 15 minutes at 4°C. The washed bacterial cells were sonicated for 60 seconds, followed by centrifugation at 9700 g for 15 minutes. A total of 4 mL of 0.1M sodium phosphate buffer, pH 6.0, containing 10 mM DTT, 500 μL of a 200 mM taurodeoxycholic acid (sodium salt) (Cayman, 15935), and 500 μL of cell-free extract were mixed and incubated at 37°C for 30 minutes. Then, a sample (100 μL) was taken and 200 μL of 15% (w/v) trichloroacetic acid was added to terminate the reaction. The sample was centrifuged (9700 g for 15 minutes), and 200 μL of the supernatant was added to 1.8 mL of ninhydrin reagent (5 mg ninhydrin, 1.2 mL glycerol, and 0.7 mL 0.5M pH 5.5 sodium citrate buffer). The mixture was vortexed and boiled for 14 minutes. After subsequent cooling, the absorbance at 570 nm was determined using taurine as a standard. One unit of BSH activity (U/mL) was defined as the amount of enzyme that liberated 1 μmol of taurine from the substrate per minute. Protein concentration was determined by the BCA kit method (Thermo Fisher Scientific), with BSA as a standard. All experiments were repeated twice.
BSH activity in probiotics cultures.
We have performed BSH activity in individual probiotic strains using 2 methods. The soft plate method (84) entailed the following: MRS soft plate was prepared using MRS broth (55 g/L), bile salts (0.3%, w/v), CaCl2 (0.375 g/L), and agar (0.75%, w/v) (Thermo Fisher Scientific). The overnight cultures of 10 probiotic strains (5 μL for each) were dropped on the soft agar and cultivated anaerobically at 37°C for 72 hours. The positive BSH activity was indicated with visible halos around the colony (Supplemental Figure 4A). The second method was the solid plate method (85); BSH activity was also screened on MRS solid plate with MRS broth (55 g/L), sodium salt of taurodeoxycholic acid (0.5% w/v; MiliporeSigma), CaCl2 (0.375 g/L), and agar (0.75%, w/v). After overnight cultivation, 10 strains were scarpered onto the plates and further cultivated for 72 hours anaerobically. The precipitation surrounding the colonies indicated the BSH activity (Supplemental Figure 4B).
C. elegans life span assay.
WT N2 strain of C. elegans was cultured by standard procedures on nematode growth media (NGM) complete agar media and E. coli OP50. The life span screening assay was carried out using liquid medium in 96-well plates according to protocol described by Solis and Petrascheck (86). In brief, 10–17 age-synchronized C. elegans (L1 larva) were cultured in S-complete media containing Ampicillin, Carbenicillin, and Amphotericin B (for inhibiting microbial contamination) and 100 μg/mL fluorodeoxyuridine (FUDR; for blocking fertilization) in 96-well plates. C. elegans were fed (3 × 107 cfu/mL) E. coli OP50 with or without taurine (200 μM). The number of live worms was counted daily on the basis of body movement, and the fraction of animals alive was calculated.
Oil Red O staining in C. elegans.
Treated worms were collected, settled with gravity, and washed with PBS before fixing with 1.5 mL cold (–20°C) methanol for 5 minutes. After adding 1 mL PBS with 0.1% Tween-20, worms were centrifuged (1200 g, 2 minutes) and then washed (2×) with PBS containing 0.1% Tween-20. Worms were stained for 30 minutes in Oil Red O solution prepared according the protocol (87). The worms were washed once with PBS after removal of the dye; then, they were mounted directly on a glass slide with a 2% agarose pad and imaged using an AmScope microscope at 4× and 10×.
Measurement of leaky gut in C. elegans using Smurf assay.
This assay was performed as protocol described by Gelino et al. (48). In brief, animals were raised as described above for life span assays with 200 μM taurine in comparison with nontreated worms as control. Animals were suspended for 3 hours in liquid cultures of standard OP50 bacteria (grown overnight) mixed with blue food dye (Spectrum FD&C Blue #1 PD110, 5.0% w/v in water) (Spectrum Chemical). Animals were then placed onto an unseeded NG P6 plate. After the NG plates were dry enough to pick worms off, we transfered worms onto the glass slides and fixed with 2% agarose gel to image the presence or absence of blue food dye in the body cavity using a Nikon Eclipse TE300 microscope under 4× and 10× magnification. For each time point, 3 or more independent experiments were carried out, each with 10–15 animals per treatment.
Physical activity assay in C. elegans.
Animals were picked into M9 buffer on a microscope slide. The number of leftward and rightward body bends in 10 seconds was counted. One leftward and 1 rightward bend were considered as 1 stroke, and counting was repeated 5 times in 50–100 animals in each group. The numbers were multiplied by 6 to give the movement rate per minute, as described elsewhere (88).
Statistics.
Statistical analyses were performed using 2-tailed t test and 2-way ANOVA. For repeated measures data (GTT, ITT, and TEER changes), significance was determined by 2-way ANOVA for repeated measures with Bonferroni’s correction. α-Diversity indices and bacterial abundance between the 2 groups were compared using unpaired 2-tailed Student’s t test and 1-way ANOVA. Differences in β-diversity were tested by permutational multivariate analysis of variance (PERMANOVA), a permutation-based multivariate ANOVA to a matrix of pairwise distance to partition the intergroup and intragroup distance. Hierarchical clustering and heatmaps based on average linkage on Euclidean distance depicting the patterns of abundance and log values were constructed within R using the heatmap.2 and ggplots packages. LEfSe was used to identify unique bacterial taxa that drive differences in probiotic versus control group mice (82). RNAseq data analyses used built-in statistical pipelines in Beijing Genomics Institute’s (BGI’s) statistical analyses considering 2-group, multiple measures (gene) analyses to determine statistical significance (P < 0.05). Principal component analysis (PCA) was applied using all features of the NMR spectra with PLS-tool box (Eigenvector Research Inc.) in Matlab (MathWorks). Welch t test was applied for statistical significance analysis for metabolites in Amix 3.9 (Bruker Biospin) and a FDR was applied to control the family-wise error. The heatmaps, volcano plot, pathway analysis, and dendrogram were carried out in MetaboAnalyst 3.0. For C. elegans life span data. Kaplan-Meier curves were generated using survival duration in days for each worm using the log-rank test. Unless otherwise stated, all the values presented herein are mean ± SEM. P < 0.05 was considered statistically significant unless specified.
Study approval.
All the procedures performed in animals were approved by the IACUC of the Animal Resource Program of the Wake Forest School of Medicine.
Author contributions
SA and SW performed experiments in the study; RN performed gut microbiome analyses; SJ performed energy expenditure studies; BW performed metabolomics studies; XZ and ZW performed peritoneal macrophage experiments; AR performed Western blots; SPM helped in analyses of RNAseq data; KK helped in physical function studies; and HY conceived the idea, organized and supervised the whole study, and compiled the manuscript. All the authors contributed in manuscript draft revisions and agree with the contents and interpretations.
Supplementary Material
Acknowledgments
Authors are thankful for the support provided by the NIH grants R01HL142930 (KK) and R01HL132035 (XZ), a grant from the Pepper Older Americans for Independence Center (P30AG21332), and grants from the Department of Defense funding W81XWH-18-1-0118, W81XWH-19-1-0236, and R01AG018915 (HY), as well funds and services provided from the Center for Diabetes, Obesity and Metabolism, Wake Forest Baptist Medical Center, and the National Center for Advancing Translational Sciences (NCATS). The NIH-funded Wake Forest Clinical and Translational Science Institute (WF CTSI) was funded through grant award number UL1TR001420.
Version 1. 04/17/2020
In-Press Preview
Version 2. 05/07/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(9):e132055.https://doi.org/10.1172/jci.insight.132055.
Contributor Information
Shokouh Ahmadi, Email: ahmadi.shokouh@gmail.com.
Shaohua Wang, Email: shaowang@wakehealth.edu.
Ravinder Nagpal, Email: rnagpal@wakehealth.edu.
Bo Wang, Email: bwang1@ncat.edu.
Shalini Jain, Email: sjain@wakehealth.edu.
Atefeh Razazan, Email: arazazan@wakehealth.edu.
Sidharth P. Mishra, Email: spmishra@wakehealth.edu.
Xuewei Zhu, Email: xwzhu@wfubmc.edu.
Zhan Wang, Email: zhawang@wakehealth.edu.
Kylie Kavanagh, Email: kkavanag@wakehealth.edu.
Hariom Yadav, Email: hyadav@wakehealth.edu.
References
- 1.Harper S. Economic and social implications of aging societies. Science. 2014;346(6209):587–591. doi: 10.1126/science.1254405. [DOI] [PubMed] [Google Scholar]
- 2.Sato Y, Yanagita M. The unprecedented era of aging. Inflamm Regen. 2019;39:15. doi: 10.1186/s41232-019-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagpal R, et al. Gut microbiome and aging: Physiological and mechanistic insights. Nutr Healthy Aging. 2018;4(4):267–285. doi: 10.3233/NHA-170030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung HY, et al. Molecular inflammation: underpinnings of aging and age-related diseases. Ageing Res Rev. 2009;8(1):18–30. doi: 10.1016/j.arr.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kavanagh K, Hsu FC, Davis AT, Kritchevsky SB, Rejeski WJ, Kim S. Biomarkers of leaky gut are related to inflammation and reduced physical function in older adults with cardiometabolic disease and mobility limitations. Geroscience. 2019;41(6):923–933. doi: 10.1007/s11357-019-00112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buford TW. (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome. 2017;5(1):80. doi: 10.1186/s40168-017-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luissint AC, Parkos CA, Nusrat A. Inflammation and the Intestinal Barrier: Leukocyte-Epithelial Cell Interactions, Cell Junction Remodeling, and Mucosal Repair. Gastroenterology. 2016;151(4):616–632. doi: 10.1053/j.gastro.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui W, et al. Tumor necrosis factor alpha increases epithelial barrier permeability by disrupting tight junctions in Caco-2 cells. Braz J Med Biol Res. 2010;43(4):330–337. doi: 10.1590/S0100-879X2010007500020. [DOI] [PubMed] [Google Scholar]
- 9.Spychala MS, et al. Age-related changes in the gut microbiota influence systemic inflammation and stroke outcome. Ann Neurol. 2018;84(1):23–36. doi: 10.1002/ana.25250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh R, et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun. 2019;10(1):89. doi: 10.1038/s41467-018-07859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yadav H, Jain S, Bissi L, Marotta F. Gut microbiome derived metabolites to regulate energy homeostasis: how microbiome talks to host. Metabolomics. 2016;6(2):e150 [Google Scholar]
- 12.Hill C, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 13.Kumar M, et al. Cancer-preventing attributes of probiotics: an update. Int J Food Sci Nutr. 2010;61(5):473–496. doi: 10.3109/09637480903455971. [DOI] [PubMed] [Google Scholar]
- 14.Nagpal R, Kumar A, Kumar M, Behare PV, Jain S, Yadav H. Probiotics, their health benefits and applications for developing healthier foods: a review. FEMS Microbiol Lett. 2012;334(1):1–15. doi: 10.1111/j.1574-6968.2012.02593.x. [DOI] [PubMed] [Google Scholar]
- 15.Yadav H, Lee JH, Lloyd J, Walter P, Rane SG. Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J Biol Chem. 2013;288(35):25088–25097. doi: 10.1074/jbc.M113.452516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra V, Shah C, Mokashe N, Chavan R, Yadav H, Prajapati J. Probiotics as potential antioxidants: a systematic review. J Agric Food Chem. 2015;63(14):3615–3626. doi: 10.1021/jf506326t. [DOI] [PubMed] [Google Scholar]
- 17.Mishra SP, et al. Probiotics and Prebiotics for the Amelioration of Type 1 Diabetes: Present and Future Perspectives. Microorganisms. 2019;7(3):E67. doi: 10.3390/microorganisms7030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ait-Belgnaoui A, et al. Prevention of gut leakiness by a probiotic treatment leads to attenuated HPA response to an acute psychological stress in rats. Psychoneuroendocrinology. 2012;37(11):1885–1895. doi: 10.1016/j.psyneuen.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 19.Krumbeck JA, et al. Probiotic Bifidobacterium strains and galactooligosaccharides improve intestinal barrier function in obese adults but show no synergism when used together as synbiotics. Microbiome. 2018;6(1):121. doi: 10.1186/s40168-018-0494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chander AM, Yadav H, Jain S, Bhadada SK, Dhawan DK. Cross-Talk Between Gluten, Intestinal Microbiota and Intestinal Mucosa in Celiac Disease: Recent Advances and Basis of Autoimmunity. Front Microbiol. 2018;9:2597. doi: 10.3389/fmicb.2018.02597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16(10):605–616. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- 22.George Kerry R, Patra JK, Gouda S, Park Y, Shin HS, Das G. Benefaction of probiotics for human health: A review. J Food Drug Anal. 2018;26(3):927–939. doi: 10.1016/j.jfda.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petschow B, et al. Probiotics, prebiotics, and the host microbiome: the science of translation. Ann N Y Acad Sci. 2013;1306:1–17. doi: 10.1111/nyas.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gitter AH, Bendfeldt K, Schmitz H, Schulzke JD, Bentzel CJ, Fromm M. Epithelial barrier defects in HT-29/B6 colonic cell monolayers induced by tumor necrosis factor-alpha. Ann N Y Acad Sci. 2000;915:193–203. doi: 10.1111/j.1749-6632.2000.tb05242.x. [DOI] [PubMed] [Google Scholar]
- 25.Guttman JA, Samji FN, Li Y, Vogl AW, Finlay BB. Evidence that tight junctions are disrupted due to intimate bacterial contact and not inflammation during attaching and effacing pathogen infection in vivo. Infect Immun. 2006;74(11):6075–6084. doi: 10.1128/IAI.00721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 27.Cani PD, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 28.Rohr MW, Narasimhulu CA, Rudeski-Rohr TA, Parthasarathy S. Negative Effects of a High-Fat Diet on Intestinal Permeability: A Review. Adv Nutr. 2020;11(1):77–91. doi: 10.1093/advances/nmz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad R, Rah B, Bastola D, Dhawan P, Singh AB. Obesity-induces Organ and Tissue Specific Tight Junction Restructuring and Barrier Deregulation by Claudin Switching. Sci Rep. 2017;7(1):5125. doi: 10.1038/s41598-017-04989-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thevaranjan N, et al. Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host Microbe. 2017;21(4):455–466.e4. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott KA, et al. Revisiting Metchnikoff: Age-related alterations in microbiota-gut-brain axis in the mouse. Brain Behav Immun. 2017;65:20–32. doi: 10.1016/j.bbi.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 32.van der Lugt B, et al. Integrative analysis of gut microbiota composition, host colonic gene expression and intraluminal metabolites in aging C57BL/6J mice. Aging (Albany NY) 2018;10(5):930–950. doi: 10.18632/aging.101439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagpal R, et al. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci Rep. 2018;8(1):12649. doi: 10.1038/s41598-018-30114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cetin DC, Nasr G. Obesity in the elderly: more complicated than you think. Cleve Clin J Med. 2014;81(1):51–61. doi: 10.3949/ccjm.81a.12165. [DOI] [PubMed] [Google Scholar]
- 35.Fakhouri TH, Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity among older adults in the United States, 2007-2010. NCHS Data Brief. 2012;(106):1–8. [PubMed] [Google Scholar]
- 36.Samper-Ternent R, Al Snih S. Obesity in Older Adults: Epidemiology and Implications for Disability and Disease. Rev Clin Gerontol. 2012;22(1):10–34. doi: 10.1017/S0959259811000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kritchevsky SB. Taking Obesity in Older Adults Seriously. J Gerontol A Biol Sci Med Sci. 2017;73(1):57–58. doi: 10.1093/gerona/glx228. [DOI] [PubMed] [Google Scholar]
- 38.Stenholm S, et al. Association of Physical Activity History With Physical Function and Mortality in Old Age. J Gerontol A Biol Sci Med Sci. 2016;71(4):496–501. doi: 10.1093/gerona/glv111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warraich HJ, et al. Physical Function, Frailty, Cognition, Depression, and Quality of Life in Hospitalized Adults ≥60 Years With Acute Decompensated Heart Failure With Preserved Versus Reduced Ejection Fraction. Circ Heart Fail. 2018;11(11):e005254. doi: 10.1161/CIRCHEARTFAILURE.118.005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landi F, et al. Impact of physical function impairment and multimorbidity on mortality among community-living older persons with sarcopaenia: results from the ilSIRENTE prospective cohort study. BMJ Open. 2016;6(7):e008281. doi: 10.1136/bmjopen-2015-008281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagpal R, Newman TM, Wang S, Jain S, Lovato JF, Yadav H. Obesity-Linked Gut Microbiome Dysbiosis Associated with Derangements in Gut Permeability and Intestinal Cellular Homeostasis Independent of Diet. J Diabetes Res. 2018;2018:3462092. doi: 10.1155/2018/3462092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fülöp T, Larbi A, Witkowski JM. Human Inflammaging. Gerontology. 2019;65(5):495–504. doi: 10.1159/000497375. [DOI] [PubMed] [Google Scholar]
- 43.Lin H, An Y, Hao F, Wang Y, Tang H. Correlations of Fecal Metabonomic and Microbiomic Changes Induced by High-fat Diet in the Pre-Obesity State. Sci Rep. 2016;6:21618. doi: 10.1038/srep21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ijare OB, et al. Simultaneous quantification of glycine- and taurine-conjugated bile acids, total bile acids, and choline-containing phospholipids in human bile using 1H NMR spectroscopy. J Pharm Biomed Anal. 2010;53(3):667–673. doi: 10.1016/j.jpba.2010.05.028. [DOI] [PubMed] [Google Scholar]
- 45.Enright EF, Joyce SA, Gahan CG, Griffin BT. Impact of Gut Microbiota-Mediated Bile Acid Metabolism on the Solubilization Capacity of Bile Salt Micelles and Drug Solubility. Mol Pharm. 2017;14(4):1251–1263. doi: 10.1021/acs.molpharmaceut.6b01155. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Augustin O, Sanchez de Medina F. Intestinal bile acid physiology and pathophysiology. World J Gastroenterol. 2008;14(37):5630–5640. doi: 10.3748/wjg.14.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foley MH, O’Flaherty S, Barrangou R, Theriot CM. Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019;15(3):e1007581. doi: 10.1371/journal.ppat.1007581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gelino S, et al. Intestinal Autophagy Improves Healthspan and Longevity in C. elegans during Dietary Restriction. PLoS Genet. 2016;12(7):e1006135. doi: 10.1371/journal.pgen.1006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kain V, et al. Obesogenic diet in aging mice disrupts gut microbe composition and alters neutrophil:lymphocyte ratio, leading to inflamed milieu in acute heart failure. FASEB J. 2019;33(5):6456–6469. doi: 10.1096/fj.201802477R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunes-Souza V, César-Gomes CJ, Da Fonseca LJ, Guedes Gda S, Smaniotto S, Rabelo LA. Aging Increases Susceptibility to High Fat Diet-Induced Metabolic Syndrome in C57BL/6 Mice: Improvement in Glycemic and Lipid Profile after Antioxidant Therapy. Oxid Med Cell Longev. 2016;2016:1987960. doi: 10.1155/2016/1987960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi J, Hur TY, Hong Y. Influence of Altered Gut Microbiota Composition on Aging and Aging-Related Diseases. J Lifestyle Med. 2018;8(1):1–7. doi: 10.15280/jlm.2018.8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu Y, et al. Gut dysbiosis is associated with the reduced exercise capacity of elderly patients with hypertension. Hypertens Res. 2018;41(12):1036–1044. doi: 10.1038/s41440-018-0110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buford TW, et al. Composition and richness of the serum microbiome differ by age and link to systemic inflammation. Geroscience. 2018;40(3):257–268. doi: 10.1007/s11357-018-0026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duncan SH, Flint HJ. Probiotics and prebiotics and health in ageing populations. Maturitas. 2013;75(1):44–50. doi: 10.1016/j.maturitas.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Gill HS, Rutherfurd KJ, Cross ML, Gopal PK. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am J Clin Nutr. 2001;74(6):833–839. doi: 10.1093/ajcn/74.6.833. [DOI] [PubMed] [Google Scholar]
- 56.De Marco S, et al. Probiotic Cell-Free Supernatants Exhibited Anti-Inflammatory and Antioxidant Activity on Human Gut Epithelial Cells and Macrophages Stimulated with LPS. Evid Based Complement Alternat Med. 2018;2018:1756308. doi: 10.1155/2018/1756308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao RK, Samak G. Protection and Restitution of Gut Barrier by Probiotics: Nutritional and Clinical Implications. Curr Nutr Food Sci. 2013;9(2):99–107. doi: 10.2174/1573401311309020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52(7):988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoyles L, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med. 2018;24(7):1070–1080. doi: 10.1038/s41591-018-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ripps H, Shen W. Review: taurine: a “very essential” amino acid. Mol Vis. 2012;18:2673–2686. [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang JYL. Bile acid metabolism and signaling in liver disease and therapy. Liver Res. 2017;1(1):3–9. doi: 10.1016/j.livres.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blacher E, Levy M, Tatirovsky E, Elinav E. Microbiome-Modulated Metabolites at the Interface of Host Immunity. J Immunol. 2017;198(2):572–580. doi: 10.4049/jimmunol.1601247. [DOI] [PubMed] [Google Scholar]
- 63.Levy M, et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 2015;163(6):1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ito T, Yoshikawa N, Inui T, Miyazaki N, Schaffer SW, Azuma J. Tissue depletion of taurine accelerates skeletal muscle senescence and leads to early death in mice. PLoS One. 2014;9(9):e107409. doi: 10.1371/journal.pone.0107409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spriet LL, Whitfield J. Taurine and skeletal muscle function. Curr Opin Clin Nutr Metab Care. 2015;18(1):96–101. doi: 10.1097/MCO.0000000000000135. [DOI] [PubMed] [Google Scholar]
- 66.Goodman CA, et al. Taurine supplementation increases skeletal muscle force production and protects muscle function during and after high-frequency in vitro stimulation. J Appl Physiol. 2009;107(1):144–154. doi: 10.1152/japplphysiol.00040.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Z, et al. Attenuation by dietary taurine of dextran sulfate sodium-induced colitis in mice and of THP-1-induced damage to intestinal Caco-2 cell monolayers. Amino Acids. 2008;35(1):217–224. doi: 10.1007/s00726-007-0562-8. [DOI] [PubMed] [Google Scholar]
- 68.Gordon RE, Heller RF, Del Valle JR, Heller RF. Membrane perturbations and mediation of gap junction formation in response to taurine treatment in normal and injured alveolar epithelia. Exp Lung Res. 1989;15(6):895–908. doi: 10.3109/01902148909069634. [DOI] [PubMed] [Google Scholar]
- 69.Hegyi P, Maléth J, Walters JR, Hofmann AF, Keely SJ. Guts and Gall: Bile Acids in Regulation of Intestinal Epithelial Function in Health and Disease. Physiol Rev. 2018;98(4):1983–2023. doi: 10.1152/physrev.00054.2017. [DOI] [PubMed] [Google Scholar]
- 70.Sarathy J, et al. The Yin and Yang of bile acid action on tight junctions in a model colonic epithelium. Physiol Rep. 2017;5(10):e13294. doi: 10.14814/phy2.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelly CJ, et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe. 2015;17(5):662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.VanHook AM. Butyrate benefits the intestinal barrier. Sci Sig. 2015;8(378):ec135. doi: 10.1126/scisignal.aac6198. [DOI] [Google Scholar]
- 73.Wang S, et al. Lipoteichoic acid from the cell wall of a heat killed Lactobacillus paracasei D3-5 ameliorates aging-related leaky gut, inflammation and improves physical and cognitive functions: from C. elegans to mice. Geroscience. 2020;42(1):333–352. doi: 10.1007/s11357-019-00137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yadav H, et al. Protection from obesity and diabetes by blockade of TGF-β/Smad3 signaling. Cell Metab. 2011;14(1):67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhuang L, et al. Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nat Commun. 2018;9(1):1796. doi: 10.1038/s41467-018-04127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahmadi S, et al. Prebiotics from acorn and sago prevent high-fat-diet-induced insulin resistance via microbiome-gut-brain axis modulation. J Nutr Biochem. 2019;67:1–13. doi: 10.1016/j.jnutbio.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagpal R, et al. Gut Microbiome Composition in Non-human Primates Consuming a Western or Mediterranean Diet. Front Nutr. 2018;5:28. doi: 10.3389/fnut.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagpal R, Neth BJ, Wang S, Craft S, Yadav H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment. EBioMedicine. 2019;47:529–542. doi: 10.1016/j.ebiom.2019.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagpal R, et al. Comparative Microbiome Signatures and Short-Chain Fatty Acids in Mouse, Rat, Non-human Primate, and Human Feces. Front Microbiol. 2018;9:2897. doi: 10.3389/fmicb.2018.02897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gratton J, et al. Optimized Sample Handling Strategy for Metabolic Profiling of Human Feces. Anal Chem. 2016;88(9):4661–4668. doi: 10.1021/acs.analchem.5b04159. [DOI] [PubMed] [Google Scholar]
- 84.DiMarzio M, Rusconi B, Yennawar NH, Eppinger M, Patterson AD, Dudley EG. Identification of a mouse Lactobacillus johnsonii strain with deconjugase activity against the FXR antagonist T-β-MCA. PLoS One. 2017;12(9):e0183564. doi: 10.1371/journal.pone.0183564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dashkevicz MP, Feighner SD. Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl Environ Microbiol. 1989;55(1):11–16. doi: 10.1128/AEM.55.1.11-16.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Solis GM, Petrascheck M. Measuring Caenorhabditis elegans life span in 96 well microtiter plates. J Vis Exp. 2011;(49):2496. doi: 10.3791/2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tserevelakis GJ, Megalou EV, Filippidis G, Petanidou B, Fotakis C, Tavernarakis N. Label-free imaging of lipid depositions in C. elegans using third-harmonic generation microscopy. PLoS One. 2014;9(1):e84431. doi: 10.1371/journal.pone.0084431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gaffney CJ, Bass JJ, Barratt TF, Szewczyk NJ. Methods to assess subcellular compartments of muscle in C. elegans. J Vis Exp. 2014;(93):e52043. doi: 10.3791/52043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.