Abstract
Advanced colorectal cancer (CRC) is often accompanied by development of liver metastases (LMs) and skeletal muscle wasting (i.e., cachexia). Despite plaguing the majority of CRC patients, cachexia remains unresolved. By using mice injected with Colon-26 mouse tumors, either subcutaneously (s.c.; C26) or intrasplenically to mimic hepatic dissemination of cancer cells (mC26), here we aimed to further characterize functional, molecular, and metabolic effects on skeletal muscle and examine whether LMs exacerbate CRC-induced cachexia. C26-derived LMs were associated with progressive loss of body weight, as well as with significant reductions in skeletal muscle size and strength, in line with reduced phosphorylation of markers of protein anabolism and enhanced protein catabolism. mC26 hosts showed prevalence of fibers with glycolytic metabolism and enhanced lipid accumulation, consistent with abnormalities of mitochondrial homeostasis and energy metabolism. In a comparison with mice bearing s.c. C26, cachexia appeared exacerbated in the mC26 hosts, as also supported by differentially expressed pathways within skeletal muscle. Overall, our model recapitulates the cachectic phenotype of metastatic CRC and reveals that formation of LMs resulting from CRC exacerbate cancer-induced skeletal muscle wasting by promoting differential gene expression signatures.
Keywords: Cell Biology, Muscle Biology
Keywords: Colorectal cancer, Cytokines, Muscle
A model that recapitulates advanced colorectal cancer phenotypes demonstrates that liver metastases exacerbate cachexia, along with abnormal metabolism and altered muscle gene expression signatures.
Introduction
By the end of 2020, about 148,000 new cases of colorectal cancer (CRC) are expected to be diagnosed in the United States, and over 53,000 people will ultimately die from the disease, thus representing the second leading cause of death among all cancers (1). Notably, in 70% of CRC cases, the most serious impediment is the development of liver metastases (LMs), frequently accompanied by the onset of skeletal muscle wasting (i.e., cachexia) (2), a condition that cannot be rescued by conventional nutritional support (3–5). Along with loss of muscle mass, cachectic cancer patients experience loss of muscle strength, as well as cardiac and respiratory failure, altogether contributing to functional impairment and inability to withstand anticancer treatments (6–10). Despite its known debilitating influence on patient outcomes and survival, cachexia remains an understudied field, mainly because of minimal progress on the identification of new pathogenic mechanisms and treatments.
A useful tool to interrogate mechanisms of disease and test therapeutic interventions to combat diseases is the use of small animal models. Unfortunately, with respect to cancer cachexia, only a handful of mouse models are currently in use, and their poor molecular characterization represents a major limitation (11–13). Indeed, data on genetic and metabolic profiling are generally missing, with the exception of the Colon-26 (C26) allograft mouse model (14), which remains the most widely used model to study CRC cachexia (15). However, clinical relevance and translational capacity of the current circulating models used for the study of cachexia has been recently questioned. In fact, though cachexia can be detected already during the early stages of tumor progression, it is extremely prominent and highly correlated with advanced metastatic cancers (15), thus corroborating the need for preclinical models of metastatic cancer–induced cachexia. This need was also recently and elegantly discussed by Tomasin et al., highlighting the fact that the field is still lacking quality, well-characterized, and clinically translational models of metastatic cancer cachexia (15). In hopes of meeting this need and advancing the field of cachexia, we and others have employed potentially novel models of CRC-associated LMs, thus taking advantage of instrasplenic injections, which allow for rapid migration of tumor cells to the liver (16–20).
In the present study, we sought to thoroughly characterize the skeletal muscle in a model of colorectal LMs using the well-characterized C26 murine cell line. We also assessed the systemic metabolic effects of tumor hosts bearing LMs and examined whether the development of LMs could exacerbate the cachectic phenotype compared with the traditional C26 allograft model. Overall, we aimed at further establishing the importance of developing and using preclinical models of metastatic cancer to better understand cachexia induced by advanced CRC. Here, we demonstrate that metastatic CRC induces dramatic loss of skeletal muscle mass and perturbs energy metabolism. Furthermore, we provided evidence that colorectal LMs exacerbate loss of skeletal muscle mass and strength compared with s.c. C26 tumors, thereby highlighting the importance of using preclinical metastatic CRC models to better understand cachexia induced by advanced CRC.
Results
Formation of colorectal LMs leads to loss of body weight, muscle mass, and muscle strength.
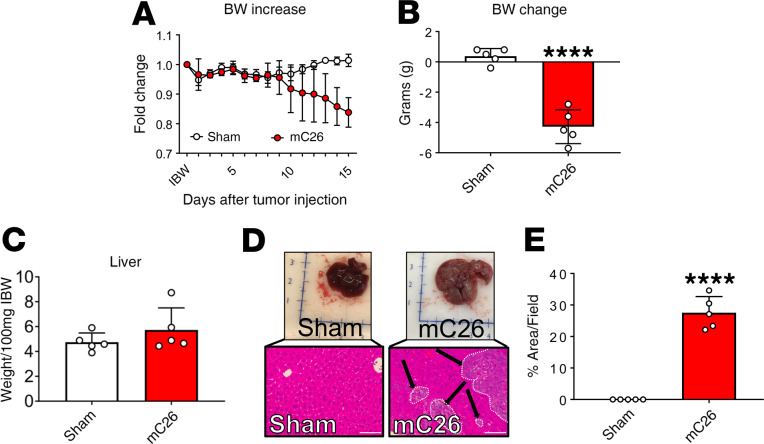
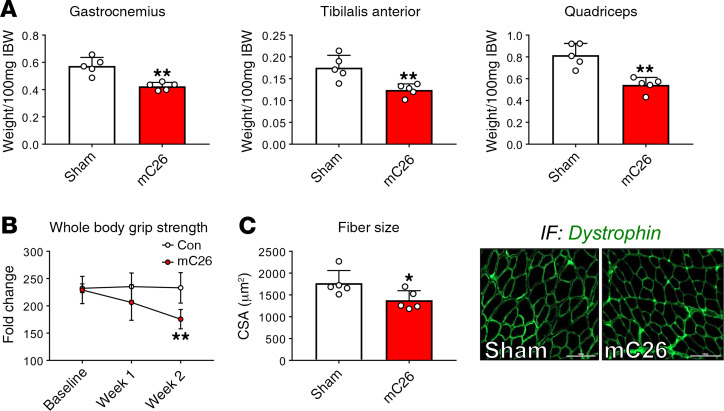
To assess the impact of colorectal LMs on skeletal muscle mass in vivo, CD2F1 male mice were intrasplenically injected with 2.5 × 105 C26 tumor cells (mC26). This procedure resulted in formation of LMs following liver dissemination via the portal circulation, without generating tumors in extrahepatic organs, such as spleen or lungs. Tumor-injected and sham-operated animals were monitored daily for body weight over the course of the experiment. Following tumor cell injection, mC26 hosts experienced progressive weight loss, which resulted in a 16% reduction compared with sham-operated animals (P < 0.0001; Figure 1, A and B). mC26 hosts saw a nonsignificant increase in liver size (+21%) compared with sham-operated animals, which can likely be attributed to the localization of C26 tumors within the liver (Figure 1, C–E). The loss of body weight was accompanied by wasting in several skeletal muscles, including the gastrocnemius (–26%, P < 0.01), tibialis anterior (–29%, P < 0.01), and quadriceps (–33%, P < 0.01) (Figure 2A). The loss of skeletal muscle mass in the mC26 hosts was paralleled by a 25% decline in whole body grip strength (P < 0.01; Figure 2B), as well as muscle atrophy, as indicated by reduced tibialis anterior cross-sectional area (CSA; –22%, P < 0.05) (Figure 2C).
Figure 1. mC26 tumor hosts experience a significant body weight (BW) reduction.
(A) BW curves in CD2F1 male mice (12 weeks old) intrasplenically injected with C26 tumor cells (250,000 cells/mouse in sterile PBS, mC26) or an equal volume of vehicle (Sham) (n = 5). (B) Net BW change (initial to final), expressed in grams. (C) Liver weights (normalized to initial body weight; IBW). (D) Representative whole liver and H&E staining of liver from Sham and mC26 mice. Black arrows indicate tumors, and images were taken at 20× magnification. Scale bars: 100 μm. (E) Quantification of relative tumor area within livers from Sham and mC26 mice. Data are expressed as mean ± SD. Two-tailed t tests were used to determine differences between Sham and mC26. Significance of the differences: ****P < 0.0001 versus Sham.
Figure 2. mC26 induces muscle atrophy and weakness.
(A) Muscle weights normalized to initial body weight (IBW) in CD2F1 male mice (12 weeks old) intrasplenically injected with C26 tumor cells (250,000 cells/mouse in sterile PBS, mC26) or an equal volume of vehicle (Sham) (n = 5). (B) Weekly whole body grip strength assessment (expressed in grams). (C) Cross-sectional area (CSA) of entire tibialis anterior muscles and representative CSA image of tibialis anterior muscle sections stained with anti-dystrophin antibody. Scale bars: 100 μm. Data are expressed as mean ± SD. Two-tailed t tests were used to determine differences between Sham and mC26. *P < 0.05, **P < 0.01 versus Sham.
mC26 hosts experience atrophic signaling within skeletal muscle.
To determine if the phenotypic reductions in skeletal muscle mass and weakness were mimicked by disruptions in markers of the anabolic/catabolic balance, we assessed multiple proteins previously implicated in progression of cancer cachexia (14, 21–23). We observed a significant increase in the phospho-STAT3/STAT3 ratio (+136%, P < 0.0001), which we have reported in other models of cancer-induced cachexia (Figure 3) (14, 23). On the other hand, we witnessed no significant changes in either ERK or p38 phosphorylation. Despite the unchanged phospho-AKT/AKT ratio, similar to ref. 23, we did observe reductions in mTOR phosphorylation (–23%, P < 0.05), also due to an increase in total mTOR content (+100%) (Figure 3). The reduction in the phospho-mTOR/mTOR ratio was further supported by reductions in its 2 downstream effectors, phospho-4EBP1 (–58%, P < 0.05) and phospho-p70S6K (–45%, P < 0.05) (Figure 3). Aside from suppressed markers of anabolic signaling, skeletal muscle from mC26 tumor hosts also experienced heightened markers of protein catabolism, including total protein ubiquitination (+142%, P < 0.01) and upregulated gene expression of the E3 ubiquitin ligases Atrogin-1 (+671%, P < 0.001), MuRF-1 (+2384, P < 0.05), and Fbxo31 (+593%, P < 0.001) (Figure 4, A and B).
Figure 3. mC26 disrupts skeletal muscle anabolism.
Representative Western blotting and quantification (expressed as fold change versus Sham) for phospho-STAT3, STAT3, phospho-ERK1/2, ERK1/2, phospho-p38, p38, phospho-AKT, AKT, phospho-mTOR, mTOR, phospho-4EBP1, and 4EBP1 (blot 1) and for phospho-p70S6K and p70S6K (blot 2) in the muscle of CD2F1 male mice (12 weeks old) intrasplenically injected with C26 tumor cells (250,000 cells/mouse in sterile PBS, mC26) or an equal volume of vehicle (Sham) (n = 5). Tubulin was used as loading control in both blots. Quantification of phospho/total protein ratios are reported as mean ± SD. Two-tailed t tests were used to determine differences between Sham and mC26. *P < 0.05, **P < 0.01, ****P < 0.0001 versus Sham.
Figure 4. Increased protein catabolism in mC26 mice.
(A) Representative Western blotting and quantification (expressed as fold change versus Sham) for total ubiquitin and tubulin in CD2F1 male mice (12 weeks old) intrasplenically injected with C26 tumor cells (250,000 cells/mouse in sterile PBS, mC26) or an equal volume of vehicle (Sham) (n = 5). Tubulin was used as loading control. (B) Gene expression levels for Atrogin-1, MuRF-1, and Fbxo31 ubiquitin ligases measured by quantitative real-time PCR. Gene expression was normalized to TBP levels. Data are expressed as mean ± SD. Two-tailed t tests were used to determine differences between Sham and mC26. *P < 0.05, **P < 0.01, ***P < 0.001 versus Sham.
mC26 disrupts skeletal muscle mitochondrial homeostasis.
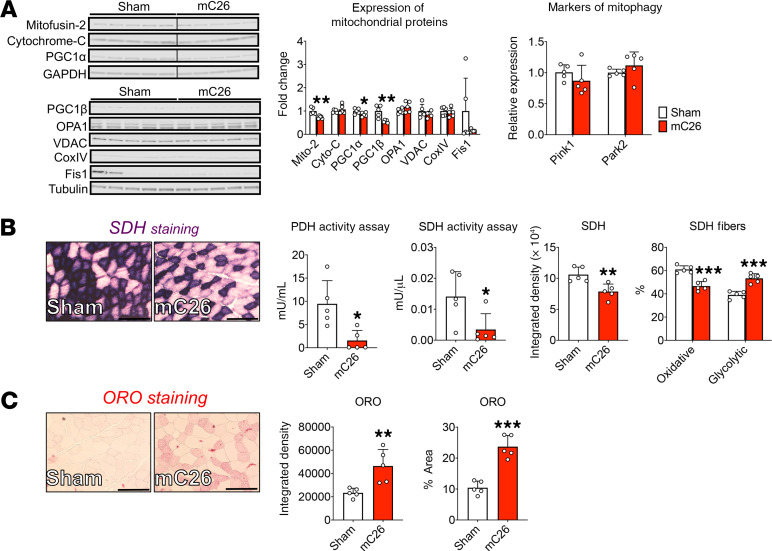
We have recently demonstrated that cachexia, as induced by either cancer or chemotherapy, is accompanied by reductions in various mitochondrial proteins required for fusion and biogenesis (23, 24). Therefore, we sought to determine whether disruptions of mitochondrial homeostasis were detected in the mC26 model of cachexia. Here, we demonstrate reductions of Mitofusin-2 (–27%, P < 0.01), PGC1α (–15%, P < 0.05), and PGC1β (–44%, P < 0.01) levels in the skeletal muscle of mC26 mice compared with Sham animals (Figure 5A). Meanwhile, we did not witness alterations in protein levels of Cytochrome-C, OPA1, VDAC, Cox IV, or Fis1 or in gene expression levels of Pink1 and Parkin2 (Figure 5A). In line with the reduction seen in mitochondrial proteins, we assessed and determined reduced enzymatic activity of both pyruvate dehydrogenase (PDH; –87%, P < 0.05) and succinate dehydrogenase (SDH; –81%, P < 0.05) (Figure 5B) in mC26 skeletal muscle. This was further complimented by SDH staining of the tibialis anterior muscle, which revealed a shift from oxidative to glycolytic metabolism (+14% glycolytic fibers) in mC26 hosts (Figure 5B). To further examine possible disruption of skeletal muscle energy metabolism, we performed Oil Red O (ORO) staining and found robust increases in intramuscular fat accumulation (integrated density, +99%, P < 0.01; percent area, +127%, P < 0.001) of mC26 skeletal muscle (Figure 5C).
Figure 5. Skeletal muscle of mC26 mice displays disrupted mitochondrial function.
(A) Representative Western blotting and quantification (expressed as fold change versus Sham) for Mitofusin-2, Cytochrome-C, and PGC1α (blot 1; GAPDH used as loading control), as well as for PGC1β, OPA1, VDAC, CoxIV, and Fis1 (blot 2; tubulin used as loading control) in the muscle of CD2F1 male mice (12 weeks old) intrasplenically injected with C26 tumor cells (250,000 cells/mouse in sterile PBS, mC26) or an equal volume of vehicle (Sham) (n = 5). Gene expression levels for Pink1 and Park2 measured by quantitative PCR and normalized to TBP levels. (B) Enzymatic activity for pyruvate dehydrogenase (PDH) and succinate dehydrogenase (SDH) and SDH staining and quantification on tibialis anterior muscles. (C) Oil Red O (ORO) staining and quantification of tibialis anterior muscles. Images were captured at a magnification of 20×. Scale bars: 100 μm. Data are expressed as means ± SD. Two-tailed t tests were used to determine differences between Sham and mC26. *P < 0.05, **P < 0.01, ***P < 0.001 versus Sham.
NMR metabolomics analysis revealed energy perturbations in mC26 hosts.
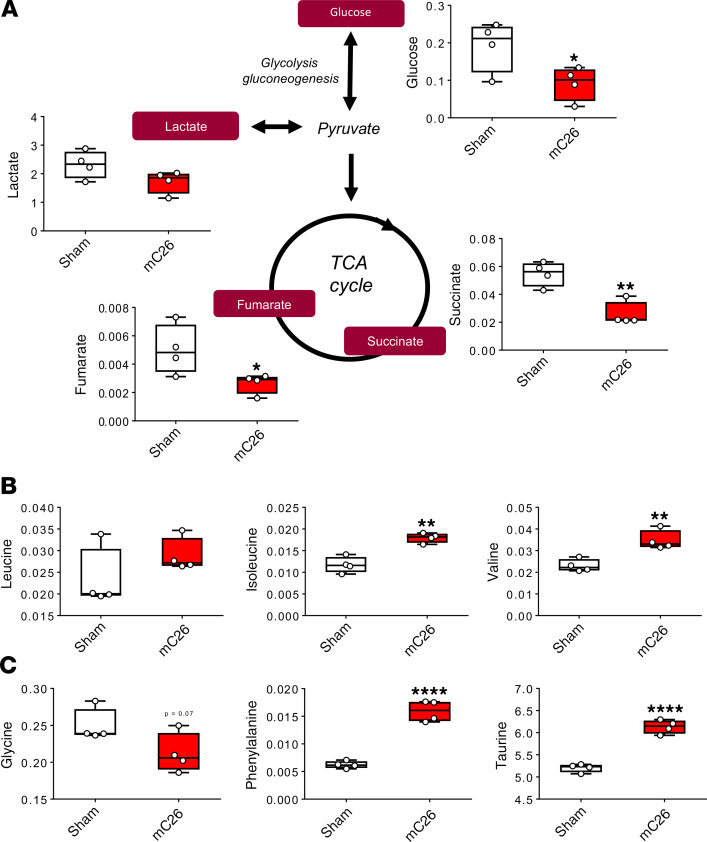
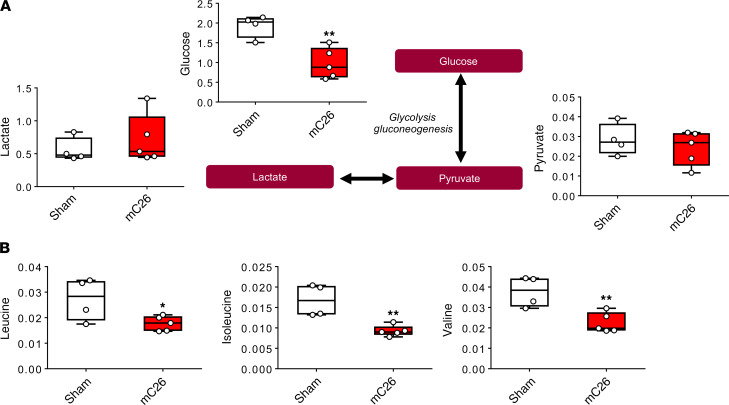
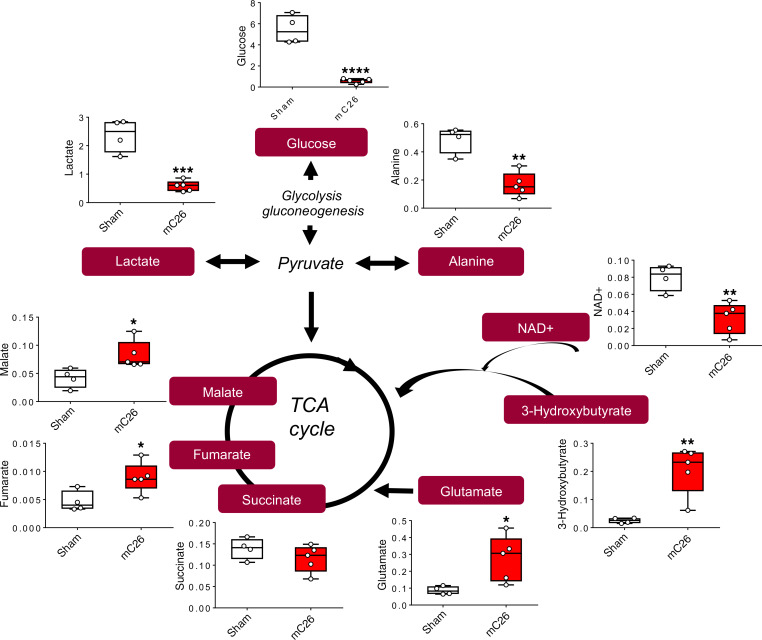
In line with our recent findings showing metabolite changes in skeletal muscle, plasma, and liver of mice bearing C26 allografts (25), we aimed to assess if similar perturbations occurred with the formation of C26 LMs. Nuclear magnetic resonance–based (NMR-based) metabolomics analysis of skeletal muscle revealed a significant reduction in glucose (–52%, P < 0.05) in the mC26 hosts (Figure 6A). Interestingly, despite the observed alterations in PDH enzyme activity (Figure 5B), skeletal muscle lactate was not significantly changed (Figure 6A). However tricarboxylic acid (TCA) cycle components succinate and fumarate were reduced by 53% (P < 0.01) and 47% (P < 0.05), respectively, indicating impaired TCA flux of mC26 skeletal muscle (Figure 6A). Interestingly, mC26 hosts displayed increases in isoleucine (+53%, P < 0.01), valine (+51%, P < 0.01), phenylalanine (+156%, P < 0.0001), and the amino acid derivative taurine (+18%, P < 0.0001) (Figure 6, B and C). In line with reductions in skeletal muscle glucose, mC26 hosts also displayed reductions in plasma glucose (–49%, P < 0.01), while plasma lactate and pyruvate were unchanged (Figure 7A). In contrast to skeletal muscle, serum branched chain amino acids (BCAAs) leucine (–35%, P < 0.05), isoleucine (–45%, P < 0.01), and valine (–40%, P < 0.01) were reduced in mC26 hosts (Figure 7B). Metabolomics analysis of the liver revealed a dramatic reduction in glucose (–89%, P < 0.0001) and glycogen (–95%, P < 0.0001) in mC26 hosts, which is in line with an increased systemic demand for glucose (Figure 8 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.136687DS1). mC26 hosts also displayed significant reductions in liver lactate (–75%, P < 0.001) and alanine (–66%, P < 0.01), likely reflecting an attempt to ramp up gluconeogenesis in order to increase glucose levels. An increase in the ketone body, 3-hydroxybutyrate (+714%, P < 0.01) along with a decrease in NAD+ (–60%, P < 0.01) are consistent with both fatty acid oxidation and an upregulation of gluconeogenesis. We also observed a marked increase in the anapleurotic substrate glutamate (+218%, P < 0.05) — and the TCA cycle intermediates fumarate (+92%, P < 0.05) and malate (+99%, P < 0.05) — in mC26 hosts, suggesting increased flux of liver TCA cycle (Figure 8).
Figure 6. Skeletal muscle of mC26 hosts display compromised metabolome.
(A) Representative diagram of glycolysis and tricarboxylic acid (TCA) cycle showing metabolite concentrations (mM) in skeletal muscle of CD2F1 male mice (12 weeks old) intrasplenically injected with C26 tumor cells (250,000 cells/mouse in sterile PBS, mC26) or an equal volume of vehicle (Sham) (n = 4). (B and C) Branch chain amino acid (leucine, isoleucine, and valine) concentrations expressed in mM (B) and glycine, phenylalanine, and taurine concentrations (in mM) (C) of sham and mC26 hosts. Data are expressed as means ± SD. Two-tailed t tests were used to determine differences between Sham and mC26. Significance of the differences: *P < 0.05, **P < 0.01, ****P < 0.0001 versus Sham.
Figure 7. mC26 mice exhibit systemic shifts in metabolism.
(A) Representative diagram of glycolysis showing metabolite concentrations (mM) in serum of CD2F1 male mice (12 weeks old) intrasplenically injected with C26 tumor cells (250,000 cells/mouse in sterile PBS, mC26) or an equal volume of vehicle (Sham) (n = 4–5). (B) Serum branch chain amino acid (leucine, isoleucine, and valine) concentrations (mM) of Sham and mC26 hosts. Data are expressed as means ± SD. Two-tailed t tests were used to determine differences between Sham and mC26. *P < 0.05, **P < 0.01 versus Sham.
Figure 8. mC26 hosts have altered liver metabolome.
Representative diagram of glycolysis and tricarboxylic acid (TCA) cycle showing metabolite concentrations (mM) in livers of CD2F1 male mice (12 weeks old) intrasplenically injected with C26 tumor cells (250,000 cells/mouse in sterile PBS, mC26) or an equal volume of vehicle (Sham) (n = 4-5). Data are expressed as means ± SD. Two-tailed t tests were used to determine differences between Sham and mC26. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus Sham.
mC26 hosts have marked cancellous bone loss.
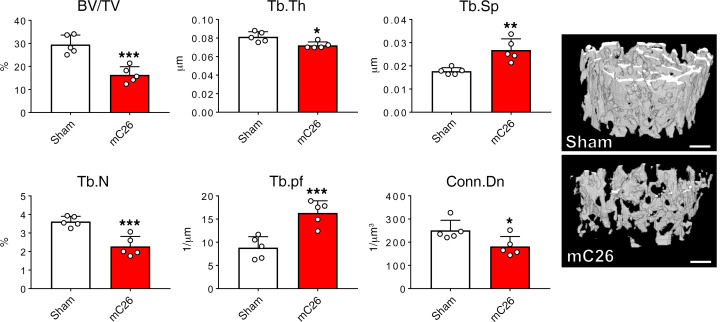
We and others have recently demonstrated that cancer- and chemotherapy-induced skeletal muscle wasting can also be accompanied by disruptions in bone homeostasis, as we previously showed in the ES-2 model of ovarian cancer and in the HT-29 and Apcmin/+ models of CRC (23, 26–28). Interestingly, the traditional C26 allograft model does not present with significant bone disruption (28). Therefore, we sought to examine whether the mC26 model drove bone loss in addition to skeletal muscle wasting. To examine cancellous bone morphometry, μCT was performed on femurs from Sham and mC26 tumor–bearing mice. mC26 hosts revealed marked reductions in femur cancellous bone. This was evidenced by robust loss of trabecular bone volume fraction (BV/TV; –45%, P < 0.001), trabecular thickness (Tb.th; –11%, P < 0.05), trabecular number (Tb.N; –37%, P < 0.01), and connectivity density (Conn.Dn; –28%, P < 0.05), while both trabecular separation (Tb.Sp; +51%, P < 0.01) and trabecular pattern factor (Tb.pf; +85%, P < 0.01) were increased (Figure 9).
Figure 9. Formation of colorectal liver tumors affects trabecular bone.
Representative 3-dimensional rendering of μCT scanned images and quantification of bone volume fraction (BV/TV), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), trabecular number (Tb.N), trabecular pattern factor (Tb.pf), and trabecular connectivity densitiy (Conn.Dn) of femur bones from CD2F1 male mice (12 weeks old) intrasplenically injected with C26 tumor cells (250,000 cells/mouse in sterile PBS, mC26) or an equal volume of vehicle (Sham) (n = 5). Scale bar: 1 mm. Data are expressed as means ± SD. Two-tailed t tests were used to determine differences between Sham and mC26. *P < 0.05, **P < 0.01, ***P < 0.001 versus Sham.
Formation of colorectal LMs appears to exacerbate skeletal muscle wasting.
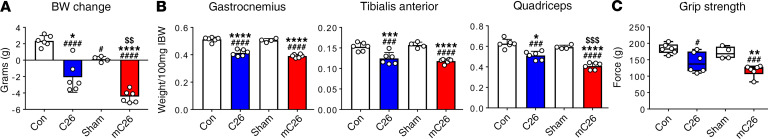
Since we observed exacerbated bone loss in the mC26 hosts not previously identified in allograft C26 tumor hosts, we sought to examine whether formation of colorectal LMs was also responsible for worsened skeletal muscle wasting. In order to interrogate this point, in a separate experiment, we transplanted CD2F1 male mice with either s.c. C26 allografts or intrasplenic injections to induce LMs (mC26). At time of sacrifice, mC26 animals saw greater reductions in body weight loss compared with the C26 hosts (–2.35 grams, –113%, P < 0.01) (Figure 10A). Moreover, mC26 mice consistently demonstrated greater reductions than C26 mice in skeletal muscle mass respective to their control littermates, as indicated by reductions in the weight of gastrocnemius (C26, –19%, P < 0.001 versus control; mC26, –23%, P < 0.0001 versus Sham), tibialis anterior (C26, –19%, P < 0.01 versus control; mC26, –25%, P < 0.0001 versus Sham), and especially quadriceps (C26, –18%, P < 0.001 versus control; mC26, –31%, P < 0.0001 versus Sham; mC26, –20%, P < 0.001 versus C26) (Figure 10). Interestingly, the hearts of mC26 hosts were also reduced to a greater extent compared with controls (–10%, P < 0.01) than C26 hosts (–7%, P < 0.05) (Supplemental Figure 2). This was accompanied by greater reductions in cardiac ANP and Myh7b gene expression in mC26 hosts compared with control (ANP, –61%, P < 0.001; Myh7b, –59%, P < 0.001) and Sham (ANP, –58%, P < 0.001; Myh7b, –62%, P < 0.001) animals than in C26 hosts (ANP, –49% versus control, P < 0.01, and –46% versus Sham, P < 0.05; Myh7b, –33% versus control, P < 0.05, and–38% versus Sham, P < 0.05) (Supplemental Figure 2). Moreover, whole body grip strength also revealed a greater reduction in mC26 (–33%, P < 0.01) than C26 (–22%, P < 0.05) mice when compared with their respective experimental controls (Figure 10).
Figure 10. Colorectal liver metastases exacerbate muscle wasting and weakness.
(A) Body weight (BW) change (vs. initial body weight) at time of sacrifice, expressed in grams; (B) muscle weights normalized to initial body weight (IBW); and (C) whole body grip strength assessment (expressed in grams) in CD2F1 male mice (8 weeks old) either injected with C26 tumor cells s.c. (1,000,000 cells/mouse in sterile PBS, C26) or intrasplenically (250,000 cells/mouse in sterile PBS, mC26). Control (Con for C26) and Sham (controls for mC26) were injected with an equal volume of vehicle (n = 4–6). Data are expressed as mean ± SD. One-way ANOVA was performed to determine differences between Con, C26, Sham, and mC26. #P < 0.05, ###P < 0.001, ####P < 0.0001 versus Con; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus Sham; $$P < 0.01, $$$P < 0.001 versus C26.
mC26 and C26 hosts have differentially expressed signaling networks.
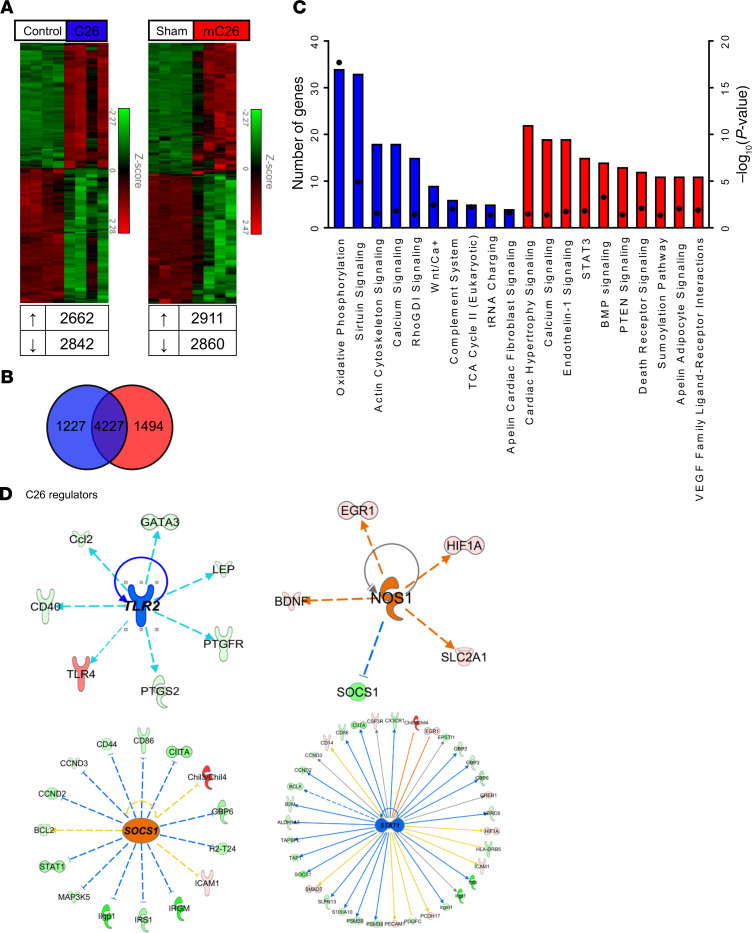
Since we observed an exacerbated cachectic phenotype in mC26 hosts, we investigated the divergence in gene expression signatures and its associated pathways within skeletal muscle of metastatic and nonmetastatic CRC. Next-Generation RNA sequencing (RNA-seq) analysis revealed a large population of commonly shared genes (>60%), between mC26 and C26, when compared with their respective controls. However, 1227 and 1494 differentially expressed genes were found to be unique to C26 and mC26, respectively (Figure 11, A and B). The differentially expressed genes were then used to run pathway and upstream regulator analysis, which revealed both common and differentially altered signaling pathways within C26 and mC26 hosts, as well as distinct upstream regulators (Figure 11, C–E). Interestingly, one of the only altered pathways similar between C26 and mC26 was calcium signaling, revealing that formation of LMs distinctly alters signaling within skeletal muscle. Identified upstream regulators were also distinct between the 2 groups, with TLR2 and SOCS1 identified in C26 skeletal muscle and NOS1 and STAT1 identified in mC26 skeletal muscle.
Figure 11. RNA sequencing analysis of skeletal muscle reveals differentially expressed genes in C26 and mC26 mice.
Next-Generation RNA sequencing was performed on whole RNA extracted from skeletal muscle of CD2F1 male mice (8 weeks old) either injected with C26 tumor cells s.c. (1,000,000 cells/mouse in sterile PBS, C26) or intrasplenically (250,000 cells/mouse in sterile PBS, mC26). Control (Con for C26) and Sham (controls for mC26) were injected with an equal volume of vehicle (n = 5–6). (A) Heatmap representing the full list of differentially expressed genes in both C26 and mC26 skeletal muscle. Cut-offs for genes were fold-change of 1.5 and a FDR of 0.05. (B) Differential expression analysis identified 4277 common genes between C26 and mC26, along with several unique genes. (C) Top 10 representative pathways: Analysis was performed using the unique genes from C26 (1227 genes) and mC26 (1494 genes). Square dots indicate –Log10 P value. Blue indicates the pathways identified in C26, and red indicates the pathways in mC26. (D and E) Upstream regulators identified in our differential expression analysis for C26 and mC26. Blue indicates inhibition, and orange indicates activation of the regulator.
Discussion
According to recent statistics, CRC represents the third most prevalent cancer in the United States and worldwide, and with an estimated 1 in 25 lifetime probability of developing CRC, it remains a major health concern (1). Cachexia, a devastating comorbidity in several types of cancer, including CRC (2), directly contributes to over 20% of cancer-related deaths and is often worsened by anticancer drugs (8–10, 24, 29, 30). We and others have discussed the importance of maintaining lean body mass to improve treatment tolerance and survival outcomes in cancer patients (31). Despite spanning several decades, research efforts have yielded minimal progress toward a cure for cachexia, likely also due to the limited availability of preclinical animal models. Indeed, it was recently discussed that minimal change in the use of clinically relevant and translational animal models for the study of cachexia has occurred over the past 10 years (15). This is particularly true when translating current animal models to metastatic cancer cachexia, especially with respect to CRC, in which the C26-bearing mouse remains the most widely used, published, and characterized model (15). Given that cachexia is typically observed in the more advanced CRC patients, usually burdened by LMs, it is clear that better and thoroughly characterized models of metastatic CRC cachexia are needed not only to better understand the disease at a mechanistic level, but also in hopes of counteracting the loss of lean mass in order to improve survival (2). In this paper, we sought to narrow this pressing gap in the literature by characterizing a metastatic model of CRC cachexia.
In order to mimic the cachectic phenotype of metastatic CRC cachexia, we employed an intrasplenic injection approach to disseminate LMs in vivo by using murine C26 colorectal tumor cells. This particular approach to induce LMs is an accepted state-of-the-art model commonly used in cancer biology and has emerged as a growing area of interest to study cachexia. Though this particular approach has been used previously to demonstrate muscle wasting associated with LMs (16, 18), minimal genetic, metabolic, or molecular alterations of skeletal muscle associated with metastatic CRC have been investigated. In the present study, we demonstrate that C26 LMs induce progressive loss of body weight (Figure 1), which was accompanied by severe atrophy of several skeletal muscles and a progressive decline in strength (Figure 2). Moreover, we demonstrate that the skeletal muscle wasting was accompanied by disruptions in skeletal muscle anabolism, catabolism, mitochondrial homeostasis, and energy metabolism.
We and others have implicated STAT3 in cancer-induced muscle wasting in several tumor models, including the C26 allograft and Apcmin/+ CRC models, as well as the Lewis lung carcinoma (LLC), B16 melanoma, and ES-2 ovarian cancer models (14, 21, 23, 32–38). In line with these findings, here we show a significant increase in STAT3 phosphorylation in mC26 hosts (Figure 3), showing that STAT3 may be a critical prognosticator of cancer-induced muscle atrophy in the presence of C26 metastatic tumors. In line with elevated STAT3 signaling, we also demonstrated heightened protein catabolism, as indicated by increased protein ubiquitination, as well as by elevated expression of the E3 ubiquitin ligases Atrogin-1, MuRF-1, and Fbxo31 (Figure 4), which have been previously reported in association with cachectic muscle, including cachexia associated with LM (18, 23, 39–41) (18, 23, 39–41). Interestingly, we did not observe alterations in the activation of several other proteins previously described to play roles in cancer cachexia, including ERK (Figure 3), previously shown to increase in the skeletal muscle of mice bearing C26 allografts (22). On the other hand, p38 and AKT phosphorylation were found unchanged (22), despite reduced phosphorylation of mTOR and its 2 downstream effectors, 4EBP1 and p70S6K (Figure 3). This is in line with the work of White et al. that showed reductions in mTOR, 4EBP1, and p70S6K in the Apcmin/+ model of CRC (42). Moreover, the previously mentioned work by Murphy et al. showed no change in phosphorylation of AKT with reduced phosphorylation of p70S6K in animals bearing LM (18).
Quite recently, the role of mitochondria in the sustainment of skeletal muscle mass in cancer cachexia has received much attention. Along this line, we and others have demonstrated that loss of mitochondrial proteins in skeletal muscle of mice bearing cancer or receiving chemotherapy is associated with both loss of muscle mass and strength (23, 24, 43, 44). Moreover, Brown et al. recently demonstrated that mitochondrial dysfunction may actually precede muscle wasting in the LLC model of cachexia, while Xi et al. was able to show that overexpression of Mitofusin-2 could partially sustain skeletal muscle mass in CRC (43, 44). In the present study, we showed that mC26 hosts have reduced levels of mitochondrial proteins, including Mitofusin-2, PGC1α, and PGC1β (Figure 5), in line with our previously published findings in mice bearing C26 allografts, whereas Cytochrome-C and OPA1 were unchanged (29). Consistent with such impaired mitochondrial homeostasis, we also observed marked reductions in SDH activity. This is also in line with previous findings from ours and other groups showing that cachectic animals bearing cancers or receiving chemotherapy had reduced muscle SDH activity (18, 23, 24).
Given the perturbed skeletal muscle mitochondria, we wanted to assess whether mC26 hosts also experienced impaired energy metabolism, both within the skeletal muscle and systemically. We have recently shown disruptions in skeletal muscle and systemic energy metabolism in animals bearing CRC and receiving chemotherapy (25, 45). Our present findings indicate an increased systemic demand in glucose metabolism, as reflected by reduced plasma and skeletal muscle glucose in mC26 hosts (Figures 6 and 7). This is also in line with our previous observations showing reduced plasma glucose in the C26 allograft model (25). Also similar to our previous study, here we show a reduction in circulating BCAAs, consistent with increased muscle catabolism and subsequent oxidation. Increases in BCAA oxidation have been observed in whole body and skeletal muscle in conditions such as sepsis and trauma, as well as after endotoxin or tumor necrosis factor treatment (46–48). Interestingly, in mC26 skeletal muscle, we show elevated isoleucine and valine, consistent with aggressive catabolism of skeletal muscle proteins. Interestingly, TCA intermediates succinate and fumarate are suppressed within mC26 skeletal muscle, implying reduced TCA flux. This is in line with reduced SDH and PDH enzyme activity (Figure 5B), also suggesting impaired oxidative metabolism. The impaired oxidative environment of skeletal muscle is consistent with a shift to glycolysis as the dominant energy-producing pathway (Figure 6A).
As the metabolome of both skeletal muscle and plasma of mC26 hosts revealed drastic impairments, we wanted to assess whether the liver was showing similar changes, especially given its robust metabolic flexibility in times of energy stress. Indeed, here we show that the high systemic demand for glucose demonstrated by plasma and skeletal muscle was also reflected by drastic reductions in liver glucose (Figure 8) and glycogen (Supplemental Figure 1). These alterations mimic prior data showing reduced liver glucose and glycogen in C26 hosts (25). The liver of mC26 hosts presented significant reductions in the levels of alanine and lactate, which is consistent with their use in gluconeogenesis (Figure 8) (25). A dramatic increase in the ketone body 3-hydroxybutyrate (+714%) is consistent with an increase in gluconeogenesis. Under these conditions, the oxaloacetate typically used to condense with Acetyl-CoA to form citrate and feed the TCA cycle is reduced to malate and continues on the gluconeogenic pathway. Therefore, the consequent backup of acetyl-CoA from fatty acid β-oxidation leads to the formation of 3-hydroxybutyrate. A decrease of NAD+ was observed, which is consistent with its consumption in β-oxidation. The TCA cycle intermediates malate and fumarate were markedly increased in the mC26 hosts, along with the anapleurotic substrate glutamate, suggesting an increase in TCA cycle activity. Significantly increased TCA flux was not clearly evident in the liver of C26 bearers, indicating that tumor infiltration of the liver may induce greater energy perturbations (25).
In line with the need for improved models of metastatic cancer cachexia to further our understanding of the disease, there is also a necessity for better characterization of multiple organs that are negatively affected by different types of tumors and chemotherapeutics (49). Indeed, examining this organ crosstalk within cachexia may provide important clues to teasing out the mechanisms that drive this morose disease. Our group and others have demonstrated that cancer-induced cachexia does not solely affect skeletal muscle, but that heart, fat, and bone are also impaired (23, 26, 28, 50). In particular, recent observations have implicated that abnormal muscle-bone crosstalk may play a significant role in cancer cachexia. For example, observations generated in several mouse models of CRC, including C26, HT-29, and Apcmin/+, revealed differential bone loss, despite consistent loss of skeletal muscle mass (28). Interestingly, cancellous bone in the femurs of C26 tumor hosts was generally maintained, whereas in the present study, the use of C26 tumor cells to induce LMs was sufficient to drive both skeletal muscle and bone loss (Figure 9), further indicating an exacerbation in bone phenotype with LMs. Our data also suggest that, since LMs often occur in advanced CRC patients, examination of bone mineral density should be warranted, especially considering that heightened bone loss can further contribute to muscle loss and weakness (50).
Given the finding that bone loss occurred in mC26 hosts, compared with a normal bone phenotype in mice bearing s.c. C26 tumors, we sought to understand whether skeletal muscle loss was also exacerbated in metastatic tumor–bearing hosts. In a follow-up experiment involving a direct comparison among mice bearing C26 allograft and mice carrying mC26 tumors, we demonstrated greater losses in body weight, skeletal muscle mass, and cardiac size, as well as whole body grip strength in the presence of LMs, with the quadriceps muscle 20% smaller in mC26 compared C26 tumor hosts (Figure 10 and Supplemental Figure 2). This exacerbated wasting occurred in the animals with CRC LMs, despite the fact that the mC26 hosts initially received fewer tumor cells compared with the C26 bearers (2.5 × 105 versus 1.0 × 106, respectively). This is important to note because it implicates the site of injection as a critical prognosticator for the development of cachexia, as also previously demonstrated in work by Chiba et al. (51). Nonetheless, that study did not take into exam the impact of LMs. To understand the pathways that may potentially lead to this exacerbation, we performed RNA-seq on quadriceps muscles. We found that over 60% of genes altered in mC26 and C26 hosts were similar, including known prognosticators of muscle wasting such as STAT3, MurRF-1, Atrogin-1, FBXO31, and PDK4 (14, 21, 39–41, 52). However, the fold-change elevation of these genes tended to be generally greater in mC26 hosts (STAT3, 5.6 versus 4.6; MuRF-1, 25 versus 20; Atrogin-1, 14 versus 13.9; FBX031, 6 versus 6.2; PDK4, 8.7 versus 5.7), which may in part explain the worsened muscle wasting. Follow-up pathway analysis performed using the differentially expressed genes identified several differentially regulated pathways, as well as differential upstream regulators within mC26 and C26 tumor hosts (Figure 11, D and E). Several of the modulated pathways, including calcium, sirtuin, STAT3, PTEN, and oxidative phosphorylation, have been implicated in muscle wasting diseases (14, 21, 23, 25, 53–55). The identified C26 regulator TLR2 has previously shown to mediate myotube atrophy, although in the present context, TLR2 was found to be downregulated (56). We also identified STAT1 as an upstream regulator within mC26 skeletal muscle. Interestingly, STAT1 has recently been implicated as a regulator of autophagy, a degradation process known to be upregulated in cachectic cancer patients and animal models of cachexia (57–59). Future studies will interrogate these pathways to better understand the mechanisms by which formation of LMs differentially alter skeletal muscle signaling.
Overall, our study clearly demonstrates that formation of LMs induces, and even aggravates, muscle atrophy induced by CRC. Though we examined and identified differential signaling networks within skeletal muscle of tumor hosts bearing LMs, investigation into how CRC metastases alter the liver endocrine function and how this may ultimately influence skeletal muscle wasting was not explored in the current study. Additionally, tumor burden in the mC26 model was crudely assessed using only histological analysis, thus possibly representing a limitation of our approach and preventing us from performing a direct comparison with tumor size in the C26 hosts. Given the search for more translationally relevant models, another limitation of the present study is the shortened nature in the development of cachexia. Though the goal of the study to examine cachexia in a context of CRC LM was achieved, future studies may consider using lower doses of C26 tumor cells or perhaps other CRC cells that may allow the progression of cachexia to extend beyond 2 weeks. Moreover, in the present study, we did not take into consideration whether the administration of chemotherapeutics further aggravate muscle wasting, especially considering that our lab and others have demonstrated that several anticancer compounds induce cachexia independently of their effects on tumor growth (24, 26, 27, 29, 30, 60). Lastly, another limitation of the current study was the focus on male animals, leaving out possible sex differences in response to LMs. As sexual dimorphism has been identified in other models of CRC, future studies should examine how LMs influence skeletal muscle in males versus females (33).
In conclusion, we have demonstrated that formation of C26 CRC LMs induces robust skeletal muscle atrophy. Skeletal muscle atrophy in mC26 hosts was accompanied by elevated protein catabolism, disrupted mitochondrial homeostasis, and perturbed skeletal muscle metabolism. Formation of C26 LMs also leads to systemic alterations in energy metabolism and impaired bone homeostasis. Moreover, formation of C26 LMs aggravates cachexia and induces differential gene expression within skeletal muscle compared CRC cachexia induced by C26 subcutaneous tumors. Overall, our study provides support for the use of in vivo metastatic models for the study of cancer cachexia.
Methods
Cell lines.
Before conducting animal work, C26 cells, provided by Donna McCarthy (Ohio State University, Columbus, Ohio, USA), were cultured in DMEM (Thermo Fisher Scientific) medium supplemented with 10% FBS (Thermo Fisher Scientific), 1% penicillin/streptomycin (Thermo Fisher Scientific), and 1% sodium pyruvate (Thermo Fisher Scientific) and maintained in a 5% CO2, 37°C humidified incubator. C26 cells were cultured, passaged, and trypsinized when subconfluent to be prepared for animal injection in sterile saline.
Animals.
For the colorectal LM model, we used methods previously employed by our lab and others (16–20, 61). Briefly, 12-week-old CD2F1 (ENVIGO) male mice were placed under anesthesia, and a side subcostal incision was made to carefully expose the spleen. Animals were then intrasplenically injected with 100 μL of saline containing 2.5 × 105 C26 tumor cells (mC26) or saline alone (Sham) over the period of 1 minute, followed by 2 minutes of hemostasis (n = 5/group). The intrasplenic injection approach allows for tumor cells to quickly enter the portal circulation, thus infiltrating the liver, without forming tumors in the spleen or other common metastatic sites of CRC, such as the lung. In a separate experiment 8-week-old CD2F1 male mice were either intrasplenically (mC26; 2.5 × 105) or s.c. (C26; 1.0 × 106) injected with C26 cells (n = 4–6/group) (62). Non–tumor bearing (control) and sham-operated (Sham) mice were used as controls. Mice were weighed daily and then euthanized under light isoflurane anesthesia. At the time of sacrifice, skeletal muscle tissues were harvested, weighed, snap-frozen in liquid nitrogen, and stored at −80°C for further studies. The tibialis anterior muscles were frozen in liquid nitrogen–cooled isopentane (Thermo Fisher Scientific) for histology, as previously described (23). All mouse carcasses, including a portion of the liver, were fixed for 2 days in 10% neutral buffered formalin (Thermo Fisher Scientific) and then transferred into 70% ethanol.
Whole-body grip strength assessment.
Whole-body grip strength was assessed using a commercially available automatic grip strength meter (Columbus Instruments) as previously shown (63). The absolute force (expressed in grams) was recorded over 5 measurements, with the top 3 measurements used for analysis. To further avoid habituation bias, animals were only tested once a week during the experimental period.
H&E staining.
To examine the formation of LM, fixed liver tissue was paraffin embedded and sectioned (10 μm) in preparation for H&E staining (23). Stained liver sections were then observed under an Axio Observer.Z1 motorized microscope (Zeiss), and 5× images were recorded for tumor infiltration assessment. Using ImageJ 1.43 software (NIH), images were assessed for the tumor area relative to liver area (expressed as a percentage).
Muscle CSA.
To assess skeletal muscle atrophy, 10 μm–thick cryosections of tibialis anterior muscles, taken at the midbelly, were processed for immunostaining as described previously (30). Briefly, sections were blocked for 1 hour at room temperature and incubated overnight at 4˚C with a dystrophin primary antibody (Developmental Studies Hybridoma Bank, Iowa City, Iowa, USA; MANDRA1[7A10]), followed by a 1-hour secondary antibody (AlexaFluor 594, A-11032, Thermo Fisher Scientific) incubation at room temperature. Entire dystrophin-stained sections were analyzed for CSA using Lionheart LX automated microscope (BioTek Instruments).
Western blotting.
Skeletal muscle protein extracts were obtained by homogenizing 50 mg of quadriceps muscle tissue in RIPA buffer (150 mM NaCl [Thermo Fisher Scientific], 1.0% NP-40 [Sigma-Aldrich], 0.5% sodium deoxycholate [Thermo Fisher Scientific], 0.1% SDS [Thermo Fisher Scientific], and 50 mM Tris [Thermo Fisher Scientific], pH 8.0) supplemented with inhibitor cocktails for proteases (Roche) and phosphatases (Thermo Fisher Scientific). Cellular debris was removed by centrifugation (15 minutes; 14,000 g; 4˚C), the supernatant was collected, and protein concentration was determined using the BCA protein assay method (Thermo Fisher Scientific). Protein extracts (30 μg) were then electrophoresed in 4%–15% gradient SDS Criterion TGX precast gels (Bio-Rad), followed by gel transfer to nitrocellulose membranes (Bio-Rad). Membranes were blocked with SEA BLOCK blocking reagent (Thermo Fisher Scientific) at room temperature for 1 hour, followed by an overnight incubation with diluted antibody in SEA BLOCK buffer (Thermo Fisher Scientific) containing 0.2% Tween-20 [Thermo Fisher Scientific] at 4°C with gentle shaking. After washing with PBS containing 0.2% Tween-20 (PBST), the membrane was incubated at room temperature for 1 hour with either anti–rabbit IgG (H+L) DyLight 800 (catalog 5151S) or anti–mouse IgG (H+L) DyLight 680 (5470S) secondary antibodies (Cell Signaling Technologies). Blots were then visualized and quantified using the Odyssey Infrared Imaging System (LI-COR Biosciences). Antibodies used were phospho-STAT3 (Tyr705, catalog 9145), STAT3 (catalog 12640), phospho-ERK1/2 (Thr202/Tyr204, catalog 4370), ERK1/2 (catalog 4695), phospho-p38 (Thr180/Tyr182, catalog 4511), p38 (catalog 9212), phospho-AKT (Ser473, catalog 4060), AKT (catalog 9272), phospho-mTOR (Ser2448, catalog 5536), mTOR (catalog 2983), phospho-4EBP1 (Thr37/46, catalog 2855), 4EBP1 (catalog 9644), phospho-p70S6K (Thr389, catalog 9234), p70S6K (catalog 9209), ubiquitin (catalog 3933), Mitofusin-2 (catalog 9482), Cytochrome C (catalog 11940), OPA-1 (catalog 80471), COX IV (catalog 4844), VDAC (catalog 4866), and GAPDH (catalog 97166) from Cell Signaling Technologies; PGC-1α (catalog AB3242) from MilliporeSigma; PGC-1β (catalog ab176328) from Abcam; FIS1 (catalog 10956-1-AP) from Proteintech; and α-tubulin (catalog 12G10) from Developmental Studies Hybridoma Bank. In general, phosphorylated protein levels were normalized to the expression of the respective total proteins, and tubulin or GAPDH were used as loading controls.
Quantitative PCR (qPCR).
RNA from quadriceps muscle was isolated using the mi RNeasy Mini Kit (Qiagen), following the protocol provided by the manufacturer. RNA was quantified using a Synergy H1 spectrophotometer (BioTek). Total RNA was reverse transcribed to cDNA using the Verso cDNA kit (Thermo Fisher Scientific). Transcript levels were measured by Real-Time PCR (Light Cycler 96, Roche), taking advantage of the TaqMan gene expression assay system (Invitrogen). Expression levels for Atrogin1 (Mm00499523_m1), MuRF1 (Mm01185221_m1), Fbxo31 (Mm00505343_m1), Pink1 (Mm00550827) Park2 (Mm00450187), ANP (Mm01255747), BNP (Mm01255770), and Myh7b (Mm1249941) were detected. Gene expression was normalized to TBP (Mm01277042_m1) levels using the standard 2−ΔΔCT methods.
PDH and SDH enzymatic activity.
The enzymatic activities of PDH and SDH were measured using Colorimetric Assay Kits (MAK183 and MAK197, respectively) from MilliporeSigma based on the manufacturer’s instructions. Briefly, 10 mg of quadriceps muscle was homogenized in 100 μL of ice-cold assay buffer, followed by centrifugation (5 minutes; 10,000 × g; 4°C). A total of 10 μL of sample supernatant was added to 96-well plates. PDH and SDH reaction mixes were added to appropriate wells, resulting in a colorimetric (450 nm for PDH and 600 nm for SDH) product proportional to the enzymatic activity. The absorbance was recorded by incubating the plate (37°C for PDH and 25°C for SDH) and taking measurements (450 nm and 600 nm) every 5 minutes for 30 minutes.
SDH staining.
Tibialis anterior muscles were cut into 10-μm cross-sections on a cryostat and incubated for 30 minutes at 37°C with 0.5 mg/mL nitroblue tetrazoliumand (MilliporeSigma), 50 mM Na-succinate (MilliporeSigma), and 0.08 mM phenazine methosulfate (Thermo Fisher Scientific) in PBS. Sections were then rinsed 3 times in deionized water, mounted with PBS-glycerol (MilliporeSigma), and photographed using an Axio Observer.Z1 motorized microscope (Carl Zeiss). Entire SDH-stained sections were quantified for integrated density, as well as total, oxidative, and glycolytic fiber number using ImageJ software (NIH).
ORO staining.
For ORO staining, tibialis anterior muscles were sectioned (10 μm) and immediately fixed in ice cold formaldehyde (3.7%; Thermo Fisher Scientific) for 1 hour. Sections were serially washed in Milli-Q water (MilliporeSigma) and stained in ORO working solution (prepared as previously described; ref. 64) for 45 minutes at room temperature in the dark. Following ORO staining, sections were again serially washed in Milli-Q water and then rinsed in running tap water for 10 minutes. Sections were mounted in 50% glycerol (in PBS) and photographed using an Axio Observer.Z1 motorized microscope (Zeiss). Entire ORO-stained sections were analyzed for signaling intensity and area of positive staining using ImageJ software.
Metabolomics analysis by NMR.
Plasma samples for NMR analysis were prepared by diluting 100 μL of plasma with 500 μL of a deuterated phosphate buffer solution (pH 7.4) containing 2,2,-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS; Thermo Fisher Scientific) with a final concentration of 0.5 mM to be used as a chemical shift and quantitation reference. The solution was filtered through a 10 kDa molecular weight cut-off filter (MilliporeSigma) to remove large proteins. Samples were then placed in 5-mm NMR tubes for analysis. Muscle and liver tissues for NMR analysis were prepared according to the methanol/chloroform water procedure as previously performed (25). Tissue samples of ~100 mg were used for all samples, but actual weights were recorded to normalize the data. NMR data were acquired on a Bruker Avance III 700 MHz NMR spectrometer with a TXI triple resonance probe operating at 25°C. Spectra were collected with a 1D NOESY pulse sequence covering 12 ppm. The spectra were digitized with 32,768 points during a 3.9-second acquisition time. The mixing time was set to 100 ms, and the relaxation delay between scans was set to 2.0 seconds. All data were then processed using Advanced Chemistry Development Spectrus Processor (version 2016.1). The spectra were zero filled to 65,536 points and apodized using a 0.3-Hz decaying exponential function, and they were fast Fourier transformed. Automated phase correction and first-order baseline correction were applied to all samples. Metabolite concentrations were quantified using the Chenomx NMR Suite (version 8.2). The DSS-d6 was used as a chemical shift and quantification reference for all spectra and was set to a chemical shift of 0.00 and a concentration of 500 μM. Quantitative fitting of each spectrum was carried out in batch mode, followed by manual adjustments to correct for errors arising from spectral overlap. For tissue samples, the final concentrations were normalized based on the weight of the tissue used to prepare each sample. The quantification of glycogen in liver tissue was carried out using a colorimetric Glycogen Assay Kit II (Abcam, ab16955), per manufacturer’s instructions.
μCT analysis of femur bone morphometry.
μCT scanning was performed to measure morphological indices of metaphyseal regions of femurs. After euthanasia, the left femurs were wrapped in saline-soaked gauze and frozen at –20°C until imaging. Bone samples were rotated around their long axes, and images were acquired using a Bruker Skyscan 1176 with the following parameters: pixel size, 9 μm3; peak tube potential, 50 kV; x-ray intensity, 500 μA; and 0.3° rotation step. Calibration of the grayscale levels was performed using a hydroxyapatite phantom. Based on this calibration and the corresponding standard curve generated, the equivalent minimum calcium hydroxyapatite level is 0.42 g/cm3. Raw images were reconstructed using the SkyScan reconstruction software (NRecon, Bruker) to 3-dimensional cross-sectional image data sets using a 3-dimensional cone beam algorithm. Structural indices were calculated on reconstructed images using the Skyscan CT Analyzer software (CTAn, Bruker). Trabecular bone was analyzed between 1.0 and 2.0 mm under the femoral distal growth plate using a threshold of 80–255. Trabecular parameters included BV/TV, Tb.N, Tb.Th, Tb.Sp, and Tb.pf.
RNA-seq.
Total RNA was first evaluated for its quantity, as well as quality, using Agilent Bioanalyzer 2100. For RNA quality, a RNA integrity number (RIN) of 7 or higher is desired. A total of 50 ng of total RNA was used. cDNA library preparation included mRNA purification/enrichment, RNA fragmentation, cDNA synthesis, ligation of index adaptors, and amplification, following the KAPA mRNA Hyper Prep Kit Technical Data Sheet, KR1352 – v4.17 (Roche). Each resulting indexed library was quantified, its quality was accessed by Qubit and Agilent Bioanalyzer, and multiple libraries were pooled in equal molarity. A total of 5 μL of 2 nM pooled libraries per lane were denatured, neutralized, and applied to the cBot for flow cell deposition and cluster amplification, before loading to HiSeq 4000 for 75 bp end sequencing (Illumina). Approximately 30M reads per library were generated. A Phred quality score (Q score) was used to measure the quality of sequencing. More than 90% of the sequencing reads reached Q30 (99.9% base call accuracy). The sequencing data were first assessed using FastQC (Babraham Bioinformatics) for quality control. Then, all sequenced libraries were mapped to the mouse genome (mm10) using STAR RNA-seq aligner (65) with the following parameter: “--outSAMmapqUnique 60”. The reads distribution across the genome was assessed using bamutils (from ngsutils) (66). Uniquely mapped sequencing reads were assigned to mm10 refGene genes using featureCounts (from subread) (67) with the following parameters: “-s 2 –p –Q 10”. Quality control of sequencing and mapping results was summarized using MultiQC (68). Genes with read count per million (CPM) < 0.5 in more than 4 of the samples were removed. The data were normalized using the TMM (trimmed mean of M values) method. Differential expression analysis was performed using edgeR (69). FDR was computed from P values using the Benjamini-Hochberg procedure. The heatmap was generated using Partek Flow genomic analysis software. Ingenuity pathway analysis (IPA; QIAGEN Inc.) software was used to identify the canonical pathways and upstream regulators (70). Pathways with P < 0.05 were considered significant. We only considered the upstream molecules that were differentially expressed in our data set. We considered the pathways that had any z-score. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE142455 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142455) (71).
Statistics.
Two-tailed t tests were used to determine differences between Sham and mC26 groups for Figures 1–9. One-way ANOVA was performed to determine differences between control, C26, Sham, and mC26. Post hoc comparisons were accomplished via a Tukey’s test, with statistical significance set a priori at P ≤ 0.05. All statistics were performed using GraphPad Prism 7.04, and all data are presented as means ± SD.
Study approval.
All studies were in compliance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and with the 1964 Declaration of Helsinki and its later amendments. Furthermore, the studies were conducted in line and with approval of the IACUC at Indiana University School of Medicine (IU IACUC protocol no. 11275).
Author contributions
JRH, LJN, and AB conceived and designed the experiments; JRH, LJN, and FP performed the in vivo experiments, grip strength analysis, and molecular characterization of cachexia; TMO analyzed the metabolomics data; JRH, AN, TAZ, and AB analyzed the RNA-seq data; and JRH, TMO, and AB wrote and edited the paper.
Supplementary Material
Acknowledgments
This study was supported by the Department of Surgery and the Department of Otolaryngology – Head & Neck Surgery at Indiana University, by grants from the National Cancer Institute (R01CA122596, R01CA194593), the Veterans Administration (1I01-BX004177-01), the Lustgarten Foundation, and the IU Simon Cancer Center (NIH P30CA082709) to TAZ, as well as by grants from the Showalter Research Trust, the V Foundation for Cancer Research (V2017-021), the American Cancer Society (Research Scholar Grant 132013-RSG-18-010-01-CCG) to AB. JRH was supported by a T32 Institutional Training Grant from NIH (AR065971). The RNA Sequencing was carried out in the Center for Medical Genomics at Indiana University School of Medicine, which is partially supported by the Indiana Genomic Initiative at Indiana University (INGEN); INGEN is supported in part by the Lilly Endowment, Inc. The no. 12G10 anti-tubulin monoclonal antibody (developed by Frankel J. and Nelsen E.M. at University of Iowa) and the MANDRA1(7A10) anti-dystrophin monoclonal antibody (developed by Morris G.E. at NE Wales Institute) were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, Iowa, USA.
Version 1. 04/16/2020
In-Press Preview
Version 2. 05/07/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(9):e136687.https://doi.org/10.1172/jci.insight.136687.
Contributor Information
Joshua R. Huot, Email: jrhuot@iu.edu.
Leah J. Novinger, Email: lnovinge@iu.edu.
Fabrizio Pin, Email: fpin@iu.edu.
Ashok Narasimhan, Email: ashnaras@iu.edu.
Teresa A. Zimmers, Email: zimmerst@iu.edu.
Andrea Bonetto, Email: abonetto@iu.edu.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Ruers T, Bleichrodt RP. Treatment of liver metastases, an update on the possibilities and results. Eur J Cancer. 2002;38(7):1023–1033. doi: 10.1016/S0959-8049(02)00059-X. [DOI] [PubMed] [Google Scholar]
- 3.Fearon K, et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 2011;12(5):489–495. doi: 10.1016/S1470-2045(10)70218-7. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 5.Thoresen L, et al. Nutritional status, cachexia and survival in patients with advanced colorectal carcinoma. Different assessment criteria for nutritional status provide unequal results. Clin Nutr. 2013;32(1):65–72. doi: 10.1016/j.clnu.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Bruggeman AR, Kamal AH, LeBlanc TW, Ma JD, Baracos VE, Roeland EJ. Cancer Cachexia: Beyond Weight Loss. J Oncol Pract. 2016;12(11):1163–1171. doi: 10.1200/JOP.2016.016832. [DOI] [PubMed] [Google Scholar]
- 7.Fearon KC, Glass DJ, Guttridge DC. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metab. 2012;16(2):153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Barton BE. IL-6-like cytokines and cancer cachexia: consequences of chronic inflammation. Immunol Res. 2001;23(1):41–58. doi: 10.1385/IR:23:1:41. [DOI] [PubMed] [Google Scholar]
- 9.Loberg RD, Bradley DA, Tomlins SA, Chinnaiyan AM, Pienta KJ. The lethal phenotype of cancer: the molecular basis of death due to malignancy. CA Cancer J Clin. 2007;57(4):225–241. doi: 10.3322/canjclin.57.4.225. [DOI] [PubMed] [Google Scholar]
- 10.Tisdale MJ. Mechanisms of cancer cachexia. Physiol Rev. 2009;89(2):381–410. doi: 10.1152/physrev.00016.2008. [DOI] [PubMed] [Google Scholar]
- 11.Aulino P, et al. Molecular, cellular and physiological characterization of the cancer cachexia-inducing C26 colon carcinoma in mouse. BMC Cancer. 2010;10:363. doi: 10.1186/1471-2407-10-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benny Klimek ME, Aydogdu T, Link MJ, Pons M, Koniaris LG, Zimmers TA. Acute inhibition of myostatin-family proteins preserves skeletal muscle in mouse models of cancer cachexia. Biochem Biophys Res Commun. 2010;391(3):1548–1554. doi: 10.1016/j.bbrc.2009.12.123. [DOI] [PubMed] [Google Scholar]
- 13.Mori M, et al. Cancer cachexia syndrome developed in nude mice bearing melanoma cells producing leukemia-inhibitory factor. Cancer Res. 1991;51(24):6656–6659. [PubMed] [Google Scholar]
- 14.Bonetto A, et al. STAT3 activation in skeletal muscle links muscle wasting and the acute phase response in cancer cachexia. PLoS One. 2011;6(7):e22538. doi: 10.1371/journal.pone.0022538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomasin R, Martin ACBM, Cominetti MR. Metastasis and cachexia: alongside in clinics, but not so in animal models. J Cachexia Sarcopenia Muscle. 2019;10(6):1183–1194. doi: 10.1002/jcsm.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huot JR, Novinger LJ, Pin F, Bonetto A. HCT116 colorectal liver metastases exacerbate muscle wasting in a mouse model for the study of colorectal cancer cachexia. Dis Model Mech. 2020;13(1):dmm043166. doi: 10.1242/dmm.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuruppu D, Christophi C, Bertram JF, O’Brien PE. Characterization of an animal model of hepatic metastasis. J Gastroenterol Hepatol. 1996;11(1):26–32. doi: 10.1111/j.1440-1746.1996.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 18.Murphy KT, Struk A, Malcontenti-Wilson C, Christophi C, Lynch GS. Physiological characterization of a mouse model of cachexia in colorectal liver metastases. Am J Physiol Regul Integr Comp Physiol. 2013;304(10):R854–R864. doi: 10.1152/ajpregu.00057.2013. [DOI] [PubMed] [Google Scholar]
- 19.Xu H, Zhang Y, Peña MM, Pirisi L, Creek KE. Six1 promotes colorectal cancer growth and metastasis by stimulating angiogenesis and recruiting tumor-associated macrophages. Carcinogenesis. 2017;38(3):281–292. doi: 10.1093/carcin/bgw121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu HK, et al. Suppression of colorectal cancer liver metastasis and extension of survival by expression of apolipoprotein(a) kringles. Cancer Res. 2004;64(19):7092–7098. doi: 10.1158/0008-5472.CAN-04-0364. [DOI] [PubMed] [Google Scholar]
- 21.Bonetto A, et al. JAK/STAT3 pathway inhibition blocks skeletal muscle wasting downstream of IL-6 and in experimental cancer cachexia. Am J Physiol Endocrinol Metab. 2012;303(3):E410–E421. doi: 10.1152/ajpendo.00039.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penna F, Costamagna D, Fanzani A, Bonelli G, Baccino FM, Costelli P. Muscle wasting and impaired myogenesis in tumor bearing mice are prevented by ERK inhibition. PLoS One. 2010;5(10):e13604. doi: 10.1371/journal.pone.0013604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pin F, et al. Growth of ovarian cancer xenografts causes loss of muscle and bone mass: a new model for the study of cancer cachexia. J Cachexia Sarcopenia Muscle. 2018;9(4):685–700. doi: 10.1002/jcsm.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barreto R, Waning DL, Gao H, Liu Y, Zimmers TA, Bonetto A. Chemotherapy-related cachexia is associated with mitochondrial depletion and the activation of ERK1/2 and p38 MAPKs. Oncotarget. 2016;7(28):43442–43460. doi: 10.18632/oncotarget.9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pin F, Barreto R, Couch ME, Bonetto A, O’Connell TM. Cachexia induced by cancer and chemotherapy yield distinct perturbations to energy metabolism. J Cachexia Sarcopenia Muscle. 2019;10(1):140–154. doi: 10.1002/jcsm.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barreto R, et al. ACVR2B/Fc counteracts chemotherapy-induced loss of muscle and bone mass. Sci Rep. 2017;7(1):14470. doi: 10.1038/s41598-017-15040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hain BA, Xu H, Wilcox JR, Mutua D, Waning DL. Chemotherapy-induced loss of bone and muscle mass in a mouse model of breast cancer bone metastases and cachexia. JCSM Rapid Commun. 2019;2(1):e00075. [PMC free article] [PubMed] [Google Scholar]
- 28.Bonetto A, et al. Differential Bone Loss in Mouse Models of Colon Cancer Cachexia. Front Physiol. 2016;7:679. doi: 10.3389/fphys.2016.00679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barreto R, Mandili G, Witzmann FA, Novelli F, Zimmers TA, Bonetto A. Cancer and Chemotherapy Contribute to Muscle Loss by Activating Common Signaling Pathways. Front Physiol. 2016;7:472. doi: 10.3389/fphys.2016.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huot JR, et al. Chronic Treatment with Multi-Kinase Inhibitors Causes Differential Toxicities on Skeletal and Cardiac Muscles. Cancers (Basel) 2019;11(4):E571. doi: 10.3390/cancers11040571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pin F, Couch ME, Bonetto A. Preservation of muscle mass as a strategy to reduce the toxic effects of cancer chemotherapy on body composition. Curr Opin Support Palliat Care. 2018;12(4):420–426. doi: 10.1097/SPC.0000000000000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baltgalvis KA, Berger FG, Pena MM, Davis JM, Muga SJ, Carson JA. Interleukin-6 and cachexia in ApcMin/+ mice. Am J Physiol Regul Integr Comp Physiol. 2008;294(2):R393–R401. doi: 10.1152/ajpregu.00716.2007. [DOI] [PubMed] [Google Scholar]
- 33.Hetzler KL, et al. Sex differences in the relationship of IL-6 signaling to cancer cachexia progression. Biochim Biophys Acta. 2015;1852(5):816–825. doi: 10.1016/j.bbadis.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narsale AA, et al. Liver inflammation and metabolic signaling in ApcMin/+ mice: the role of cachexia progression. PLoS One. 2015;10(3):e0119888. doi: 10.1371/journal.pone.0119888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pretto F, et al. Sunitinib prevents cachexia and prolongs survival of mice bearing renal cancer by restraining STAT3 and MuRF-1 activation in muscle. Oncotarget. 2015;6(5):3043–3054. doi: 10.18632/oncotarget.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puppa MJ, Gao S, Narsale AA, Carson JA. Skeletal muscle glycoprotein 130’s role in Lewis lung carcinoma-induced cachexia. FASEB J. 2014;28(2):998–1009. doi: 10.1096/fj.13-240580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silva KA, et al. Inhibition of Stat3 activation suppresses caspase-3 and the ubiquitin-proteasome system, leading to preservation of muscle mass in cancer cachexia. J Biol Chem. 2015;290(17):11177–11187. doi: 10.1074/jbc.M115.641514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmers TA, Fishel ML, Bonetto A. STAT3 in the systemic inflammation of cancer cachexia. Semin Cell Dev Biol. 2016;54:28–41. doi: 10.1016/j.semcdb.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwak KS, et al. Regulation of protein catabolism by muscle-specific and cytokine-inducible ubiquitin ligase E3alpha-II during cancer cachexia. Cancer Res. 2004;64(22):8193–8198. doi: 10.1158/0008-5472.CAN-04-2102. [DOI] [PubMed] [Google Scholar]
- 40.Milan G, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun. 2015;6:6670. doi: 10.1038/ncomms7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White JP, et al. The regulation of skeletal muscle protein turnover during the progression of cancer cachexia in the Apc(Min/+) mouse. PLoS One. 2011;6(9):e24650. doi: 10.1371/journal.pone.0024650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown JL, et al. Mitochondrial degeneration precedes the development of muscle atrophy in progression of cancer cachexia in tumour-bearing mice. J Cachexia Sarcopenia Muscle. 2017;8(6):926–938. doi: 10.1002/jcsm.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xi QL, et al. Mitofusin-2 prevents skeletal muscle wasting in cancer cachexia. Oncol Lett. 2016;12(5):4013–4020. doi: 10.3892/ol.2016.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Connell TM, Pin F, Couch ME, Bonetto A. Treatment with Soluble Activin Receptor Type IIB Alters Metabolic Response in Chemotherapy-Induced Cachexia. Cancers (Basel) 2019;11(9):E1222. doi: 10.3390/cancers11091222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck SA, Tisdale MJ. Nitrogen excretion in cancer cachexia and its modification by a high fat diet in mice. Cancer Res. 1989;49(14):3800–3804. [PubMed] [Google Scholar]
- 47.Cala MP, et al. Multiplatform plasma fingerprinting in cancer cachexia: a pilot observational and translational study. J Cachexia Sarcopenia Muscle. 2018;9(2):348–357. doi: 10.1002/jcsm.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sthoeger ZM, et al. The tolerogenic peptide hCDR1 downregulates pathogenic cytokines and apoptosis and upregulates immunosuppressive molecules and regulatory T cells in peripheral blood mononuclear cells of lupus patients. Hum Immunol. 2009;70(3):139–145. doi: 10.1016/j.humimm.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 49.Porporato PE. Understanding cachexia as a cancer metabolism syndrome. Oncogenesis. 2016;5:e200. doi: 10.1038/oncsis.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waning DL, et al. Excess TGF-β mediates muscle weakness associated with bone metastases in mice. Nat Med. 2015;21(11):1262–1271. doi: 10.1038/nm.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiba F, et al. The importance of tissue environment surrounding the tumor on the development of cancer cachexia. Int J Oncol. 2014;44(1):177–186. doi: 10.3892/ijo.2013.2180. [DOI] [PubMed] [Google Scholar]
- 52.Pin F, et al. PDK4 drives metabolic alterations and muscle atrophy in cancer cachexia. FASEB J. 2019;33(6):7778–7790. doi: 10.1096/fj.201802799R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen J, Nie X, Huang SY, Qin YQ, Pan LL, Wang XT. Neuromuscular electrical stimulation improves muscle atrophy induced by chronic hypoxia-hypercapnia through the MicroRNA-486/PTEN/FoxO1 pathway. Biochem Biophys Res Commun. 2019;509(4):1021–1027. doi: 10.1016/j.bbrc.2018.12.147. [DOI] [PubMed] [Google Scholar]
- 54.Toledo M, Busquets S, Ametller E, López-Soriano FJ, Argilés JM. Sirtuin 1 in skeletal muscle of cachectic tumour-bearing rats: a role in impaired regeneration? J Cachexia Sarcopenia Muscle. 2011;2(1):57–62. doi: 10.1007/s13539-011-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Regan JN, et al. Osteolytic Breast Cancer Causes Skeletal Muscle Weakness in an Immunocompetent Syngeneic Mouse Model. Front Endocrinol (Lausanne) 2017;8:358. doi: 10.3389/fendo.2017.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hahn A, et al. Serum amyloid A1 mediates myotube atrophy via Toll-like receptors. J Cachexia Sarcopenia Muscle. 2020;11(1):103–119. doi: 10.1002/jcsm.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aversa Z, et al. Autophagy is induced in the skeletal muscle of cachectic cancer patients. Sci Rep. 2016;6:30340. doi: 10.1038/srep30340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldberg AA, et al. Regulation of ULK1 Expression and Autophagy by STAT1. J Biol Chem. 2017;292(5):1899–1909. doi: 10.1074/jbc.M116.771584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Penna F, et al. Autophagy Exacerbates Muscle Wasting in Cancer Cachexia and Impairs Mitochondrial Function. J Mol Biol. 2019;431(15):2674–2686. doi: 10.1016/j.jmb.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 60.Hain BA, et al. Zoledronic Acid Improves Muscle Function in Healthy Mice Treated with Chemotherapy. J Bone Miner Res. 2020;35(2):368–381. doi: 10.1002/jbmr.3890. [DOI] [PubMed] [Google Scholar]
- 61.Brand MI, Casillas S, Dietz DW, Milsom JW, Vladisavljevic A. Development of a reliable colorectal cancer liver metastasis model. J Surg Res. 1996;63(2):425–432. doi: 10.1006/jsre.1996.0287. [DOI] [PubMed] [Google Scholar]
- 62.Bonetto A, Rupert JE, Barreto R, Zimmers TA. The Colon-26 Carcinoma Tumor-bearing Mouse as a Model for the Study of Cancer Cachexia. J Vis Exp. 2016;(117):54893. doi: 10.3791/54893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonetto A, Andersson DC, Waning DL. Assessment of muscle mass and strength in mice. Bonekey Rep. 2015;4:732. doi: 10.1038/bonekey.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peck B, Huot J, Renzi T, Arthur S, Turner MJ, Marino JS. Mice lacking PKC-θ in skeletal muscle have reduced intramyocellular lipid accumulation and increased insulin responsiveness in skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 2018;314(3):R468–R477. doi: 10.1152/ajpregu.00521.2016. [DOI] [PubMed] [Google Scholar]
- 65.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Breese MR, Liu Y. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics. 2013;29(4):494–496. doi: 10.1093/bioinformatics/bts731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 68.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krämer A, Green G, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.