Abstract
The Transatlantic Slave Trade transported more than 9 million Africans to the Americas between the early 16th and the mid-19th centuries. We performed a genome-wide analysis using 6,267 individuals from 25 populations to infer how different African groups contributed to North-, South-American, and Caribbean populations, in the context of geographic and geopolitical factors, and compared genetic data with demographic history records of the Transatlantic Slave Trade. We observed that West-Central Africa and Western Africa-associated ancestry clusters are more prevalent in northern latitudes of the Americas, whereas the South/East Africa-associated ancestry cluster is more prevalent in southern latitudes of the Americas. This pattern results from geographic and geopolitical factors leading to population differentiation. However, there is a substantial decrease in the between-population differentiation of the African gene pool within the Americas, when compared with the regions of origin from Africa, underscoring the importance of historical factors favoring admixture between individuals with different African origins in the New World. This between-population homogenization in the Americas is consistent with the excess of West-Central Africa ancestry (the most prevalent in the Americas) in the United States and Southeast-Brazil, with respect to historical-demography expectations. We also inferred that in most of the Americas, intercontinental admixture intensification occurred between 1750 and 1850, which correlates strongly with the peak of arrivals from Africa. This study contributes with a population genetics perspective to the ongoing social, cultural, and political debate regarding ancestry, admixture, and the mestizaje process in the Americas.
Keywords: African diaspora, Transatlantic Slave Trade, admixture dynamics, mestizaje
Introduction
The Transatlantic Slave Trade was an international enterprise involving Brazilian, British, Danish, Dutch, French, German, Portuguese, Spanish, and Swedish traders. They brought over 9 million Africans to the Americas between the early 16th and the mid-19th centuries. African regions of origin included far away locations as Senegambia is from Tanzania. Destiny ports in the Americas were also distant as Boston is from Buenos Aires (Thomas 1999; Eltis 2008; Gomes 2019). The Transatlantic Slave Trade shaped the genetic structure of American continent populations (Alves-Silva et al. 2000; Carvalho-Silva et al. 2001; Salzano and Bortolini 2001; Tishkoff et al. 2009; Bryc et al. 2010; Moreno-Estrada et al. 2013; Campbell et al. 2014; Kehdy et al. 2015; Baharian et al. 2016; Mathias et al. 2016; Rotimi et al. 2016; Ongaro et al., 2019). Although most genetic studies have estimated the overall African ancestry in the Americas, a finer genomic and geographic analysis is needed to infer how different African groups contributed to North-, Central-, South-American, and Caribbean populations and to estimate these contributions. The geopolitical factors that permeated the African Diaspora have been seldom discussed at a continental scale, despite its potential influence on the genetic structure of populations.
Formal integration of genetic and demographic data has historical and solid root of more than 50 years in human population genetics (Cavalli-Sforza et al. 2013), but this kind of analysis has become rare in the era of human population genomics. In particular, a formal comparison of information from demographic history records of the Transatlantic Slave Trade with inferences based on genomic diversity of current populations from Africa and the Americas has not been performed. Here, we perform a joint systematic analysis of genetic data and historical records of the Transatlantic Slave Trade to address the following questions: 1) Is there a correspondence between the geographic origin of specific African populations of the Diaspora and specific destinations in the Americas?; 2) Was intercontinental admixture dynamics in the Americas associated with the dynamics of arrivals of African slaves?; 3) Considering the geographic extension and the massive demographic magnitude of the African Diaspora, as well as the level of between-populations genetic differentiation in the African regions of origin of slaves, did the Transatlantic Slave Trade lead to a higher, similar or lower level of between-population differentiation of the African gene pool in the Americas?
Results and Discussion
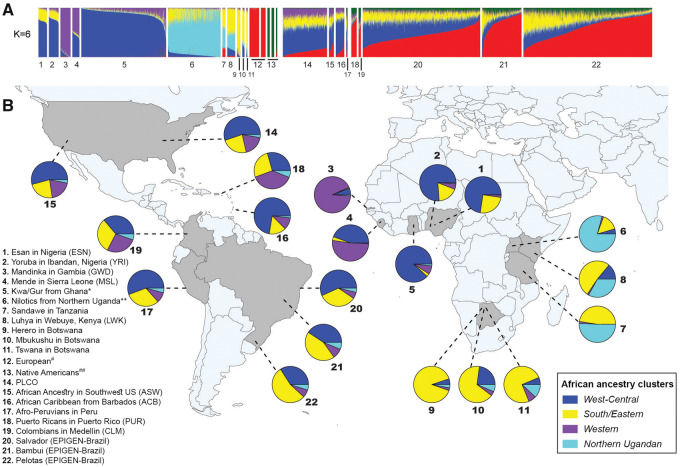
We combined genome-wide data from 25 populations: 9 admixed from the Americas, 11 Africans, 2 Europeans, and 3 Native Americans and created a data set of 6,267 unrelated individuals with >10% of African ancestry (fig. 1A and B, supplementary fig. S2, table S1, and sections S1 and S2, Supplementary Material online). Using ADMIXTURE (Alexander et al. 2009), we identified two continental (European and Native American) and four African-specific ancestry clusters, named based on their association with geographic regions (supplementary table S1, Supplementary Material online, represented by different colors in fig. 1): 1) West-Central African (blue), 2) Western African (purple), and 3) South/East African (yellow), which are prevalent in the Americas, as well as 4) Northern Ugandan (cyan), which accounts for a very low proportion of African ancestry in the Americas. Hereafter, whereas in African individuals, the proportions of ADMIXTURE ancestry clusters are relative to their whole genome ancestry (fig. 1A, supplementary table S1, Supplementary Material online), in American continent individuals, these proportions are relative to the sum of the four African ancestry clusters (fig. 1B). We also estimated haplotype-based population admixture proportions from different African regions (Lawson et al. 2012; Hellenthal et al. 2014) relative to the total contribution of African populations (fig. 2A and B, supplementary fig. S3, tables S3 and S4, Supplementary Material online).
Fig. 1.
Ancestry analysis of African and admixed populations of the Americas inferred using ADMIXTURE (K = 6). (A) Vertical bar plot showing the total African, European, and Native American proportions of the ancestry clusters (supplementary fig. S1 and section S2.6.1, Supplementary Material online). (B) Percentages of subcontinental African ancestry clusters. For admixed populations of the American continent these percentages are relative to the total African ancestry (i.e., the sum of the four African-associated clusters: West-Central, Western, Southern/Eastern, Northern Ugandan). The arrows on the map represent the regions from where the samples were collected. *The Kwa/Gur data set includes approximately 35 ethno-linguistic groups, predominantly from the Kwa and Gur Niger-Congo linguistic group (Gouveia et al. 2019). **The Nilotics data set includes predominantly three ethno-linguistic groups in Northern Uganda (Langi, Acholi, and Lugbara) from the Nilotic linguistic group (Gouveia et al. 2019) ; #the Europeans are: Iberian Population in Spain (IBS) and Utah residents with Northern and Western European ancestry (CEU), in this order in the ADMIXTURE bar plot; ##The Native Americans are: Shimaa, Ashaninka, and Aymara, respectively from Borda V et al. (2019); the PLCO (Prostate, Lung, Colorectal, and Ovarian Cancer Screening) data comprised African-Americans from East United States.
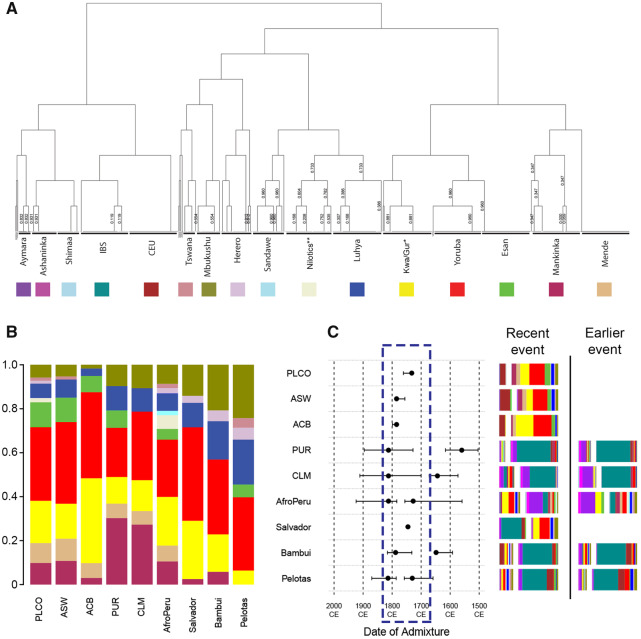
Fig. 2.
Haplotype-based clustering of parental individuals and admixture inferences for admixed American continent populations. (A) fineSTRUCTURE tree of parental individuals. *The Kwa/Gur data set includes approximately 35 ethno-linguistic groups, predominantly from the Kwa and Gur linguistic group (Gouveia et al. 2019). **The Nilotics data set includes predominantly three ethno-linguistic groups from Northern Uganda (Langi, Acholi, and Lugbara) of the Nilotic linguistic group (Gouveia et al. 2019). (B) Subcontinental contributions relative to the total African ancestry in admixed populations inferred by the MIXTURE MODEL (supplementary section S2.3, Supplementary Material online). (C) GLOBETROTTER inference of admixture events for each admixed population. Inferred date(s) and 95% confidence intervals are represented by dots and horizontal lines in the graph. Dashed rectangle in the admixture dates plots highlights the most dynamic period for admixture. Beside the dating graph, we represented the inferred admixing sources (bars) for recent and earlier events. Bar size represents the genetic contribution of the source. Each color corresponds to the proportion of each parental population contribution. CEU, Utah Residents (CEPH) with Northern and Western Ancestry-United States, IBS, Iberian population in Spain; CLM, Colombians from Medellin; PUR, Puerto Ricans from Puerto Rico; ACB, African Caribbeans in Barbados; ASW, African Americans in Southwest United States; PLCO, African Americans from East United States.
Ancestry Correspondence between African and Admixed American Continent Populations, and the Influence of Geography and Geopolitics
The West-Central Africa-associated ancestry cluster is the most prevalent African cluster in the Americas, including African-Caribbean from Barbados (72% of the total African ancestry), Northeastern Brazilians (57%), Afro-Peruvians (56%), and US African-Americans (54–55%) (blue in fig. 1B, supplementary table S1 and section S2.1, Supplementary Material online). Moreover, haplotype-based analysis (Lawson et al. 2012; Hellenthal et al. 2014) reveals a higher contribution in the Americas from Yoruba-like and Esan-like populations (from Nigeria, mean: 38%) than from Kwa/Gur-like populations (from Ghana, mean: 18%) (fig. 2A and B, supplementary tables S3 and S4, Supplementary Material online).
The Western Africa-associated ancestry cluster has its highest proportions in Puerto Ricans (38% of the total African ancestry), Colombians (27%), and US African-Americans (19–20%, purple in fig. 1B, supplementary table S1, Supplementary Material online), whereas Brazilians have the lowest proportion (<9%), limited to a Mandinka-like (Gambia) contribution and with no Mende-like (Sierra Leone) contribution (fig. 2A and B, supplementary tables S3 and S4, Supplementary Material online).
The South/East Africa-associated ancestry cluster, in contrast, shows its highest proportion in South and Southeast Brazil (44% and 54% of total African ancestry, respectively) (yellow in fig. 1B, supplementary table S1, Supplementary Material online). Haplotype-based methods (Lawson et al. 2012; Hellenthal et al. 2014) identified two different sources of gene flow associated with the South/Eastern Africa ancestry cluster: one from Mbukushu-like populations (Botswana, Western Bantu speakers from Southern Africa, 20–24% to South/Southeast Brazil) and one from Luhya-like populations (Kenya, Eastern Bantu speakers from Eastern Africa, 17–20% to South/Southeast Brazil, fig. 2A and B, supplementary tables S3 and S4, Supplementary Material online). Western- and Eastern-Bantu speakers historically correspond to the two streams of the Bantu migrations in the last 4,000–2,500 years (Tishkoff et al. 2009; Busby et al. 2016; Patin et al. 2017).
Pioneering mitochondrial DNA studies of the first decade of this century showed how African Bantu-associated haplotypes were more frequent in South America than in Central- and North-America (Salas et al. 2004; Hünemeier et al. 2007; Gonçalves et al. 2008). However, our approach, based on a genome-wide data set and a larger number of studied individuals, allows for finer geographic inferences and estimates of genome-wide admixture proportions from the different African regions, adding new layers of knowledge to our understanding of the African Diaspora.
This emerging portrait of the African ancestry in the Americas suggests an influence of geography and geopolitics. Geographical factors include: 1) the latitudinal proximity between Western Africa and Caribe-Central/North America, as well as between South/East Africa and Southern Brazil, 2) the winds and ocean currents, that shaped two navigation systems: the North-Atlantic, with voyages mostly to North America, and the South-Atlantic, with voyages predominantly to Brazil (Domingues da Silva 2008). Indeed, West-Central Africa- and Western Africa-associated ancestry clusters are more commonly observed in northern latitudes, whereas the South/East Africa-associated ancestry cluster is more evident in southern latitudes.
Differently, the Portugal possessions in the Americas (Brazil) and its influence in South and East African coasts (current Angola and Mozambique) (Klein 1987) exemplify the geopolitical factors that affected, in particular, the distribution of the South/East Africa-associated ancestry cluster. Although the Portuguese Crown had earlier privileged relations with the kingdoms of Benin in nowadays Nigeria, it later extended its influence to Bantu-speaking areas such as Congo/Angola and Mozambique (Coelho et al. 2009). Indeed, Portuguese–Brazilian slave trade routes departed from Luanda and Cabinda (Angola) and from Zanzibar (Tanzania) and Inhambane (Mozambique) during 18th and 19th centuries (Eltis 2008). The abolition of slavery by the British in 1807, who controlled the North Atlantic route, also led Portuguese traders to prefer routes in the South Atlantic (Versiani 2008). Therefore, geography (intercontinental distances and climatic factors affecting transatlantic navigation) and geopolitics (European colonial influences and possessions) influenced the geographic and linguistic diversity of African emigrants as well as favored the regional differentiation of African ancestry in the Americas.
The Dynamics of African Admixture in the Americas with Europeans and Native Americans Accompanied the Dynamics of Arrivals of African Slaves
Remarkably, linkage-disequilibrium-based inference (Hellenthal et al. 2014) shows that all the studied admixed populations of the Americas exhibit the signature of an intensification of intercontinental admixture in the interval from 1750 to 1850 (fig. 2C, supplementary table S5 and section S3, Supplementary Material online), revealing a continental trend. This trend is consistent with results by Baharian et al. (2016), focused in the United States and by Fortes-Lima et al. (2017) focused on French Guiana and Suriname isolated populations and on Colombia and Rio de Janeiro. Importantly, this time interval matches or is immediately subsequent to regional peaks of number of slaves arriving from Africa to United States, Barbados, Puerto Rico, and Brazil (supplementary fig. S4, Supplementary Material online). Thus, we reveal that in most of the Americas, the arrival of the largest contingent of Africans between 1700 and 1850 (supplementary fig. S4, Supplementary Material online) was almost synchronic with intensive intercontinental admixture, a process that was also characterized by positive ancestry-based assortative mating (Kehdy et al. 2015).
The African Gene Pool Is More Homogenous Between-Populations in the Americas Than in Africa
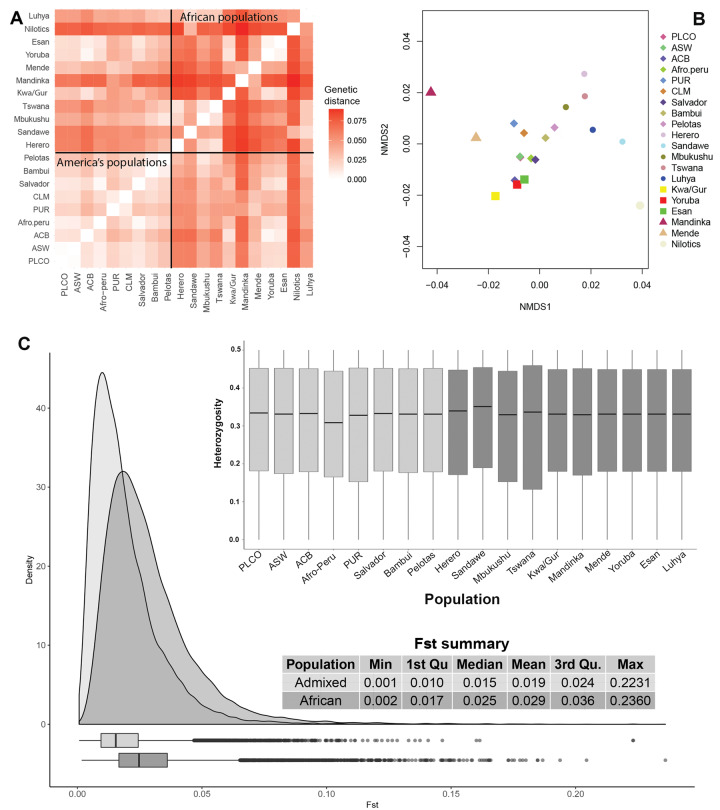
Figure 1B suggests that African ancestry clusters are more homogeneously distributed between admixed American continent populations than between the African populations that contributed to the Transatlantic Slave Trade. Considering only the African gene pool, the largest differentiation, measured by the African-Specific Genetic Distance (ASGD, see Materials and Methods section, fig. 3, supplementary section S4, and fig. S5, Supplementary Material online), is observed between African populations (mean: 0.057, mean excluding populations with marginal contribution to the Americas [Nilotics and Sandawe: 0.53]), followed by differentiation between African versus America’s populations (mean: 0.043) and between populations of the Americas (mean: 0.018, 32% of the ASGD between African populations) (Wilcoxon test, P < 10−6 for the three pairwise comparisons, fig. 3A). Corroborating these results, our approach based on local ancestry (see Materials and Methods and supplementary section S4.2, Supplementary Material online) showed that: 1) the between-populations differentiation of the African gene pool in the American continent populations (single-nucleotide polymorphisms, SNPs mean FST = 0.02) is two-thirds of the value observed between the African populations that contributed to the African Diaspora (i.e., excluding the Nilotics, SNPs mean FST = 0.03, P < 10−16 for comparison between the distributions, fig. 3C); and 2) the within-population genetic diversity (i.e., mean heterozygosity across SNPs) is not lower in the African segments of American continent population than in African populations (Kruskal–Wallis P = 0.53, fig. 3C). Thus, on average, chromosomal fragments of African origin from different populations are more similar in the Americas than in Africa, despite the very similar within-population African genetic diversity in the Americas and Africa (fig. 3C).
Fig. 3.
Pairwise genetic distances of the African gene pool between populations of the American continent and Africa. (A) Heatmap Matrix and (B) multidimensional scaling of the African gene pool genetic distances. We used solid squares, triangles, and circles to represent populations associated with WCA, West-Central Africa; SEA, South/East Africa; WA, Western Africa ancestry clusters. CLM, Colombians from Medellin; PUR, Puerto Ricans from Puerto Rico; ACB, African Caribbeans in Barbados; ASW, Americans of African ancestry in South western United States; PLCO, African-Americans from Eastern United States. (C) SNPs FST distributions between: 1) African populations that contributed to the African Diaspora (dark gray) and 2) American continent populations (gray), considering only chromosome fragments of African origin; and the within-population African genetic heterozygosity in the Americas and Africa. The CLM population was not included in this analysis because it did not have enough SNPs inferred as being of African origin.
To better understand this pattern of between-populations homogenization of the African gene pool in the Americas we compared: 1) proportions of West-Central Africa-, Western Africa-, and South-East Africa-associated ADMIXTURE ancestry clusters (fig. 1) with 2) expected proportion of these ancestry clusters, estimated considering both the proportions of arrivals from different African locations (fig. 4, supplementary tables S6 and S7, section S5, Supplementary Material online) and the ADMIXTURE ancestry clusters composition of those African locations of the origin of the Diaspora. We performed these comparisons for the geographic regions represented in our data set for which there are also historical demography records of origin and destination of Africans (Eltis 2008).
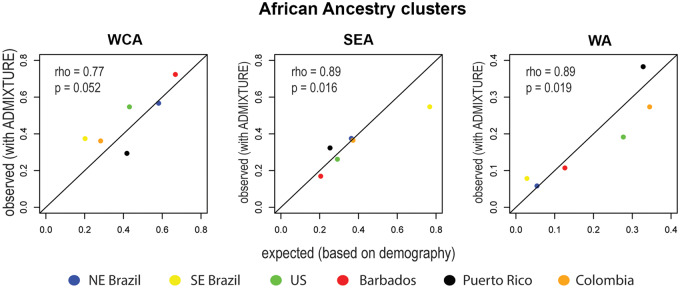
Fig. 4.
Observed and expected proportions of genomic African ancestry clusters in the Americas. We compared 1) the observed proportions of genomic African ancestry clusters (inferred using ADMIXTURE [Alexander et al. 2009]) in the vertical axis, with 2) expected proportions of genomic African ancestry clusters, estimated based on demographic historical records from the African Voyages Database1, in the horizontal axis (see supplementary table S3, Supplementary Material online). rho, Spearman’s coefficients of correlation; p, p value significance. The significance was evaluated using randomization tests of 10,000 replications. WCA, West-Central Africa; SEA, South/East Africa; WA, Western Africa.
Although we recognize that an accurate estimation of the expected proportions of ancestry based on the number of disembarks from different regions from Africa is a complex task, here we formally attempt to integrate demographic and genetics data from the African populations of origin of the Diaspora to obtain such estimation. The assumptions of our approach are shared by several population genetics methods: 1) that the current ancestry compositions of African populations are good proxies of real sources of the African Diaspora located in the same geographic areas, and 2), that the migration from Africa to the Americas occurred in a unique migration event.
Overall, for New World admixed populations, the proportions of South-Eastern African and Western African ancestry clusters are highly correlated with the expected ancestry based on the numbers of arrivals to Americas ports and departures from African ports (Spearman rho = 0.89, P = 0.02). However, for the West-Central African ancestry cluster the correlation does not reach significance (fig. 4). For the entire American continent, we observe an excess of the observed individual proportions of West-Central Africa ancestry cluster (47.7% observed vs. 40% expected, being this a conservative estimation of the difference, supplementary section S5, Supplementary Material online, P = < 2.2e−16), mainly determined by Southeastern Brazil and the US populations (supplementary table S8, Supplementary Material online). The poorest concordance between observed and expected ancestries is observed in Southeastern Brazil, that presents more of the West-Central African ancestry cluster (37%) than expected (20%) (P = < 2.2e−16) and complementarily, less of the South/East African ancestry cluster than expected based on arrivals (55% observed vs. 76% expected, P < 2.2 × 10−16). The US population also shows an excess of the West-Central African ancestry cluster (54.7% observed vs. 43.1% expected, P < 3.33−16), compensated by a deficit of the Western African ancestry cluster (19.1% observed vs. 27.7% expected, P < 3.33−16). Therefore, the between-population homogenization of the African gene pool in the Americas is partly explained by the excess of the West-Central Africa ancestry cluster in Southeast Brazil and in the United States (fig. 4). The higher between-population homogeneity of the African gene pool in the Americas by reducing population stratification, contributes to a more statistical power of genetic association studies involving individuals with African ancestry from different populations of the Americas.
In general, the limitations of our study derive from: 1) the smaller sample sizes of the non-Brazilian samples with respect to Brazilians, except for the United States, from where we included a fair number of Afro-Americans (n = 524) from the PLCO cohort; 2) the lack of a Central America sample; 3) the use of SNP-array data that contain an ascertainment bias. Even considering these limitations, our results are based on observed general patterns and not on results based on a specific population and our within- and between-populations estimates of genetic diversity are consistently calculated from genomic fragments of African origin, and therefore, the possible effect of the ascertainment bias is the same in African and American continent populations.
In conclusion, genetic data trace the African genetic roots of admixed individuals of the Americas to a broad geographic extension (from Western Africa to East Africa), associated with a high linguistic diversity (Niger Kordofanian non-Bantu and Western- and Eastern-Bantu language speakers). Considering the level of between-populations genetic differentiation in the African regions of origin of slaves, historical facts that homogenized the between-populations component of genetic diversity in the Americas have predominated over facts that tend to maintain or increase it. This latter group of facts includes geographic (i.e., intercontinental distances and maritime winds/currents) and geopolitical factors (i.e., specific European colonial influences and possessions and the abolition of the slavery by British in 1807), that shaped an association of Western African ancestry with northern latitudes and South/East African ancestry with southern latitudes. Contrastingly, the following combination of facts, that occurred in Africa and the Americas, associated with the African Diaspora, have contributed to gene flow between individuals with different African ancestries and therefore, to the between-populations homogenization of the African gene pool in the Americas: 1) the heterogeneous contribution via the Transatlantic Slave Trade of the different African regions to the Americas, 2) despite their specific European origins, traders/vessels transported slaves, frequently illegally, to different American continent ports (Klein 1987, 2010; Eltis 2008); 3) forced amalgamation, which is the preference of slave owners for slaves from different geographic and linguistic origins, so that they could not understand each other and thus, reducing the risk of riots (Olcott 1838); and 4) the role of islands in the Americas such as Jamaica and Barbados, which centralized parts of arrivals of African slaves and redistributed them to different parts of the Americas (Thomas 1999) and also ports/islands in Africa with similar roles. Other factors that contributed to the between-population homogenization of the African gene pool may be related to more general demographic trends of admixed populations of the Americas, and are not necessarily and specifically related to the African Diaspora. Importantly, by combining genetic and demographic data, we show that the between-population homogenization of the African gene pool in the Americas is partly explained by the excess of the West-Central Africa ancestry cluster (the most prevalent in the Americas) in the United States and Southeast Brazil with respect to demographic expectations, which suggests a spread of this ancestry in the American continent. Interestingly, in most of the Americas, the arrival of the largest contingent of Africans between 1700 and 1850 was almost synchronic with the intensification of intercontinental admixture, which implies that this time interval was critical to shape the structure of the African gene pool in the New World. This study, by dissecting and estimating the African ancestry proportions in different populations of the Americas, and inferring the dynamics of biological admixture, contributes with a population genetics perspective to the ongoing social, cultural and political debate regarding ancestry, admixture, and mestizaje and the different perceptions of race in the Americas (Clinton 2001; Wade et al. 2014).
Materials and Methods
Database and Population Structure Analyses
We analyzed a final data set of 6,267 unrelated individuals from Africa and the Americas (with more than 10% of African ancestry) for 533,242 SNPs (supplementary table S1, Supplementary Material online). We inferred population structure and admixture using ADMIXTURE (Alexander et al. 2009) and Principal Component Analysis (Price et al. 2006) for unlinked SNPs. Presented results are based on ADMIXTURE runs with K = 6 because it corresponds to the lower cross-validation error. We inferred haplotypes using the SHAPEIT2 software (Delaneau et al. 2012). Haplotype-based analyses were performed using ChromoPainter and fineSTRUCTURE (Lawson et al. 2012). The admixture contributions from the different African regions were inferred using GLOBETROTTER (Hellenthal et al. 2014). Demographic information of embarked and disembarked African slaves was obtained from the African Voyages database (https://www.slavevoyages.org/; last accessed February 20, 2020).
African-Ancestry Genetic Distance
The genetic differentiation between populations considering only the African gene pool was estimated using two strategies. First, we conceived the African-ancestry genetic distance (AAGD, supplementary section S4.1, Supplementary Material online), based on: 1) the mean proportions of the subcontinental African ancestry clusters from each population based on ADMIXTURE results (K = 6) (supplementary table S1, Supplementary Material online). In the case of the population of the Americas, these proportions were with respect to the total African ancestry. 2) The FST between the African ancestry clusters estimated by the ADMIXTURE software (Alexander et al. 2009) (supplementary table S2, Supplementary Material online). AAGD between two populations is given by the sum of the Euclidean genetic distances between each pair of subcontinental ancestries weighted by the FST (in sensu ADMIXTURE [Alexander et al. 2009]) between the ancestry clusters. Specifically, considering two populations (A and B) with C ancestry clusters (, and ), the African-ancestry genetic distance is calculated as:
where Ac and Bc are the ancestry proportions of the ADMIXTURE cluster c in the populations A and B, with respect to the total African ancestry.
Our second strategy (supplementary section S4.1, Supplementary Material online) to measure the genetic differentiation between populations considering only the African gene pool (from chromosome fragments of African origins), consisted in comparing the distribution of continental SNPs-FST, estimated as (Wright 1943, 1949) (supplementary section S4.1, Supplementary Material online):
where is the minor-allele frequency of the SNP i in the population j, p is a vector with allele frequencies of an SNP for all the considered populations, var(p) denotes the between-population variance of pi and p¯ denotes the mean of pi across populations. Allele frequencies in the African populations were those that, on the basis of our results, contributed to the African Diaspora (i.e., conservatively excluding Nilotics). For the American continent populations, allele frequencies of the African gene pool were estimated from a minimum of 20 chromosome fragments of African origin, as inferred using RFMix (Maples et al. 2013). Analogously, within-population diversity for the African gene pool was estimated by the i-SNP-heterozygosity for the j-population, as:
Estimating the Expected Proportions of African Ancestry Clusters Based on Demography
We used data available in the African Voyages database (Eltis 2008) to estimate the expected ancestry in a specific destiny proxy of the Americas, by considering the proportion of individuals from each embarkation major region (representing the ancestry origin proxies), that arrived in specific ports of disembarkation in the Americas (supplementary table S7, Supplementary Material online). To avoid the unrealistic assumption that the individuals from the African embarkation major regions have a homogenous ancestry, we calculated the weighted expected ancestry. This was estimated by taking into account the proportion of WCA, WA, SEA genomic ancestry clusters estimated in selected current African populations from the embarkation major regions that contributed to the African genomic pool in the Americas (supplementary table S7, Supplementary Material online and figs. 1A and 2B): 1) Kwa/Gur and Yoruba populations for the WCA proportion; 2) Mandinka (GWD) and Mende (MSL) for the WA proportion; and 3) Mbukushu and Luhya (LWK) for the SEA proportion. Exception were the Brazilian populations, in which we used only GWD population to obtain the WA proportion, since the Mende (MSL) did show contribution to the Brazilian populations (fig. 2A and B, supplementary tables S3 and S4, Supplementary Material online).
Thus, we calculated the weighted expected proportion of the i-ancestry cluster () for each proxy-destiny population as:
where n is the number of African population proxies of origin (p); ep is the expected ancestry of the population proxies of origin p based on the proportion of individuals arrived from each of the n African populations, with respect to the total of individuals disembarked in the population; op,i is the observed proportion of the ADMIXTURE ancestry cluster i of population proxies of origin p. To assess the correlation between observed and expected ancestries, we applied the Spearman correlation test, implemented in the R and the significance was evaluated using 10,000 randomization tests (supplementary section S5, Supplementary Material online).
Flowcharts of the performed analyses are available in the EPIGEN Scientific Workflow (Magalhães et al. 2018) website (http://ldgh.com.br/scientificworkflow; last accessed February 20, 2020). Masterscripts are available under request from the authors for academic purposes. Details of Materials and Methods section are in the Supplementary information.
Data Availability
EPIGEN-Brazil data are deposited at the European Nucleotide Archive (PRJEB9080 [ERP010139]), accession number EGAS00001001245, under EPIGEN Committee Controlled Access mode. The Nilotics and Kwa/Gur data sets are deposited in dbGaP at phs001705.v1.p1 and phs000838.v1.p1, respectively. The Botswana and Tanzania data sets from Sarah Tishkoff Lab are available at dbGaP accession number phs001396.v1.p1 and SRA BioProject PRJNA392485.
Supplementary Material
Acknowledgments
We thank Sergio D. Pena, Marcia Beltrame, Rosangela Loschi, Eduardo F. Paiva, Fabricio Santos, Renan Souza, Claudio Struchiner, Ricardo Santos, and Garrett Hellenthal for advice, discussions and criticisms. This work was supported by the Brazilian Ministry of Health (Department of Science and Technology from the Secretaria de Ciência, Tecnología e Insumos Estratégicos) through Financiadora de Estudos e Projetos (FINEP) to the EPIGEN-Brazil Initiative. The EPIGEN-Brazil investigators were also supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior of the Brazilian Ministry of Education (CAPES Agency). E.T.S., M.H.G., V.B., T.P.L., and M.F.L.C. were supported by Brazilian National Research Council (CNPq), Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG), and Pró-Reitoria de Pesquisa da Universidade Federal de Minas Gerais. M.H.G. performed part of this study as CAPES-PDSE fellow, V.B. was a CAPES-PEC-PG fellow. M.L.S. was a TWAS-CNPq PhD fellow. MHG is supported by the Intramural Research Program of the National Institutes of Health in the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is supported by the National Human Genome Research Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, the Center for Information Technology, and the Office of the Director at the National Institutes of Health (1ZIAHG200362). Tishkoff Laboratory is funded by the National Institutes of Health (1R01DK104339-0 and 1R01GM113657-01). EMBLEM is funded by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute (NCI) (HHSN261201100063C and HHSN261201100007I) and, in part, by the Intramural Research Program, National Institute of Allergy, and Infectious Diseases (SJR), National Institutes of Health, Department of Health and Human Services. Bioinformatics support was provided by the Sagarana HPC cluster, CPAD-ICB-UFMG, Brazil.
Author Contributions
The project was conceived by M.H.G. and E.T.S. M.H.G. assembled data sets. M.H.G., V.B., T.P.L., R.G.M., M.M.A., G.S.A., N.M.A., F.S.G.K., M.M., W.C.S.M., L.A.M., M.R.R., F.R.-S., H.P.S.A., M.L.S., M.O.S., G.S.S., C.Z. analyzed genetic data. R.L. and R.Z. performed laboratory experiments. E.T.S. supervised bioinformatic and statistical analyses. M.D., R.H.G., H.G., A.C.P., M.F.L.C., M.L.B., B.L.H., S.M.M., S.J.C., S.A.T., and M.Y. contributed with data. M.H.G., M.C.B., V.B., A.W.B., M.Y., S.A.T. contributed to data interpretation. M.H.G., V.B., and E.T.S. wrote the manuscript. All authors read the manuscripts and contributed with suggestions.
References
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Silva J, da Silva Santos M, Guimarães PE, Ferreira AC, Bandelt HJ, Pena SD, Prado VF.. 2000. The ancestry of Brazilian mtDNA lineages. Am J Hum Genet. 67(2):444–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharian S, Barakatt M, Gignoux CR, Shringarpure S, Errington J, Blot WJ, Bustamante CD, Kenny EE, Williams SM, Aldrich MC, et al. 2016. The Great Migration and African-American genomic diversity. PLoS Genet. 12(5):e1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borda V, Alvim I, Aquino MM, Silva C, Soares-Souza GB, Leal TP, Scliar MO, Zamudio R, Zolini C, Padilla C, et al. 2020. The genetic structure and adaptation of Andean highlanders and Amazonian dwellers is influenced by the interplay between geography and culture. bioRxiv [Internet]:2020.01.30.916270. Available from: https://www.biorxiv.org/content/10.1101/2020.01.30.916270v2, last accessed February 20, 2020. [Google Scholar]
- Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo J-M, Wambebe C, Tishkoff SA, et al. 2010. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci U S A. 107(2):786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby GB, Band G, Si Le Q, Jallow M, Bougama E, Mangano VD, Amenga-Etego LN, Enimil A, Apinjoh T, Ndila CM, et al. 2016. Admixture into and within sub-Saharan Africa. Elife 5:pii:e15266. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Hirbo JB, Townsend JP, Tishkoff SA.. 2014. The peopling of the African continent and the diaspora into the new world. Curr Opin Genet Dev. 29:120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Silva DR, Santos FR, Rocha J, Pena SD.. 2001. The phylogeography of Brazilian Y-chromosome lineages. Am J Hum Genet. 68(1):281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Moroni A, Zei G.. 2013. Consanguinity, inbreeding, and genetic drift in Italy (MPB-39). Oxford, UK: Princeton University Press. Available from: 10.1515/9781400847273 [DOI]
- Clinton WJ. 2001. Erasing America’s color lines. The New York Times [Internet]. [cited 2019 Feb 19]; Section4, Page17. Available from: https://www.nytimes.com/2001/01/14/opinion/erasing-america-s-color-lines.html. Accessed February 20, 2020. [Google Scholar]
- Coelho M, Sequeira F, Luiselli D, Beleza S, Rocha J.. 2009. On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola. BMC Evol Biol. 9:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury J-F.. 2012. A linear complexity phasing method for thousands of genomes. Nat Methods 9(2):179–181. [DOI] [PubMed] [Google Scholar]
- Domingues da Silva DB. 2008. The Atlantic slave trade to Maranhão, 1680–1846: volume, routes and organisation. Slavery Abol. 29(4):477–501. [Google Scholar]
- Eltis D. 2008. A brief overview of the trans-Atlantic slave trade. Voyages: the trans-Atlantic slave trade database: http://www.slavevoyages.org [Internet]. [cited 2019 Feb 19]; 1:1-11. Available from: http://www.redemaosdadas.org/wp-content/uploads/2014/02/HIST211-1.3.3-TransAtlanticSlaveTrade.pdf. Accessed February 20, 2020.
- Fortes-Lima C, Gessain A, Ruiz-Linares A, Bortolini M-C, Migot-Nabias F, Bellis G, Moreno-Mayar JV, Restrepo BN, Rojas W, Avendaño-Tamayo E, et al. 2017. Genome-wide ancestry and demographic history of African-descendant Maroon communities from French Guiana and Suriname. Am J Hum Genet. 101(5):725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes L. 2019. Escravidão—Vol. 1: do primeiro leilão de cativos em Portugal até a morte de Zumbi dos Palmares. Rio de Janeiro: Globo Livros. [Google Scholar]
- Gonçalves VF, Carvalho CMB, Bortolini MC, Bydlowski SP, Pena S.. 2008. The phylogeography of African Brazilians. Hum Hered. 65(1):23–32. [DOI] [PubMed] [Google Scholar]
- Gouveia MH, Bergen AW, Borda V, Nunes K, Leal TP, Ogwang MD, Yeboah ED, Mensah JE, Kinyera T, Otim I, et al. 2019. Genetic signatures of gene flow and malaria-driven natural selection in sub-Saharan populations of the “endemic Burkitt Lymphoma belt”. PLoS Genet. 15(3):e1008027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, Falush D, Myers S.. 2014. A genetic atlas of human admixture history. Science 343(6172):747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hünemeier T, Carvalho C, Marrero AR, Salzano FM, Pena SDJ, Bortolini MC.. 2007. Niger-Congo speaking populations and the formation of the Brazilian gene pool: mtDNA and Y-chromosome data. Am J Phys Anthropol. 133(2):854–867. [DOI] [PubMed] [Google Scholar]
- Kehdy FSG, Gouveia MH, Machado M, Magalhães WCS, Horimoto AR, Horta BL, Moreira RG, Leal TP, Scliar MO, Soares-Souza GB, et al. 2015. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci U S A. 112(28):8696–8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HS. 1987. A demografia do tráfico atlântico de escravos para o Brasil. Estud Econ. 17:129–149. [Google Scholar]
- Klein HS. 2010. The Atlantic slave trade. Cambridge: Cambridge University Press. [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, Falush D.. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8(1):e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhães WCS, Araujo NM, Leal TP, Araujo GS, Viriato PJS, Kehdy FS, Costa GN, Barreto ML, Horta BL, Lima-Costa MF, et al. 2018. EPIGEN-Brazil initiative resources: a Latin American imputation panel and the scientific workflow. Genome Res. 28(7):1090–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples B, Gravel S, Kenny E, Bustamante C.. 2013. RFMix: A Discriminative Modeling Approach for Rapid and Robust Local-Ancestry Inference. Am J Hum Genet. 93(2):278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias RA, Taub MA, Gignoux CR, Fu W, Musharoff S, O'Connor TD, Vergara C, Torgerson DG, Pino-Yanes M, Shringarpure SS, et al. 2016. A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat Commun. 7:12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, Gignoux CR, Ortiz-Tello PA, Martínez RJ, Hedges DJ, Morris RW, et al. 2013. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 9(11):e1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olcott C. 1838. Two lectures on the subjects of slavery and abolition. Massillon: Massillon, Ohio. [Google Scholar]
- Ongaro L, Scliar M O, Flores R, Raveane A, Marnetto D, Sarno S, Gnecchi-Ruscone GA, Alarcón-Riquelme ME, Patin E, Wangkumhang Pet al. . 2019. The Genomic Impact of European Colonization of the Americas. Curr Biol. 29(23):3974–3986.e4. [DOI] [PubMed] [Google Scholar]
- Patin E, Lopez M, Grollemund R, Verdu P, Harmant C, Quach H, Laval G, Perry GH, Barreiro LB, Froment A, et al. 2017. Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science 356(6337):543–546. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D.. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38(8):904–909. [DOI] [PubMed] [Google Scholar]
- Rotimi CN, Tekola-Ayele F, Baker JL, Shriner D.. 2016. The African diaspora: history, adaptation and health. Curr Opin Genet Dev. 41:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Richards M, Lareu M-V, Scozzari R, Coppa A, Torroni A, Macaulay V, Carracedo A.. 2004. The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet. 74(3):454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzano FM, Bortolini MC.. 2001. The evolution and genetics of Latin American populations by Francisco M. Salzano. Cambridge: Cambridge University Press. [Google Scholar]
- Thomas H. 1999. The slave trade: the story of the Atlantic slave trade: 1440–1870. New York: Simon and Schuster Paperbacks. [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo J-M, Doumbo O, et al. 2009. The genetic structure and history of Africans and African Americans. Science 324(5930):1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versiani FR. 2008. D. João VI e a (não) abolição do tráfico de escravos para o Brasil In: Committee of IX BRASA, editors. Brasa IX Proceedings. New Orleans: Brazilian Studies Association; p. 27–29. [Google Scholar]
- Wade P, López-Beltrán C, Restrepo E, Ventura-Santos R.. 2014. Mestizo genomics. North Carolina: Duke University Press. [Google Scholar]
- Wright S. 1943. Isolation by distance. Genetics 28:114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1949. The genetical structure of populations. Ann Eugen. 15(1):323–354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
EPIGEN-Brazil data are deposited at the European Nucleotide Archive (PRJEB9080 [ERP010139]), accession number EGAS00001001245, under EPIGEN Committee Controlled Access mode. The Nilotics and Kwa/Gur data sets are deposited in dbGaP at phs001705.v1.p1 and phs000838.v1.p1, respectively. The Botswana and Tanzania data sets from Sarah Tishkoff Lab are available at dbGaP accession number phs001396.v1.p1 and SRA BioProject PRJNA392485.