Abstract
Antarctic cryptoendolithic communities are self-supporting borderline ecosystems spreading across the extreme conditions of the Antarctic desert and represent the predominant life-form in the ice-free areas of McMurdo Dry Valleys, accounted as the closest terrestrial Martian analogue. Components of these communities are highly adapted extremophiles and extreme-tolerant microorganisms, among the most resistant known to date. Recently, studies investigated biodiversity and community composition in these ecosystems but the metabolic activity of the metacommunity has never been investigated. Using an untargeted metabolomics, we explored stress-response of communities spreading in two sites of the same location, subjected to increasing environmental pressure due to opposite sun exposure, accounted as main factor influencing the diversity and composition of these ecosystems. Overall, 331 altered metabolites (206 and 125 unique for north and south, respectively), distinguished the two differently exposed communities. We also selected 10 metabolites and performed two-stage Receiver Operating Characteristic (ROC) analysis to test them as potential biomarkers. We further focused on melanin and allantoin as protective substances; their concentration was highly different in the community in the shadow or in the sun. These results clearly indicate that opposite insolation selected organisms in the communities with different adaptation strategies in terms of key metabolites produced.
Introduction
The Antarctic cryptoendolithic communities are microbial ecosystems that dominate the biology of most ice-free areas in Continental Antarctica. They were described for the first time in the McMurdo Dry Valleys, Southern Victoria Land [1], the largest ice-free area of the continent. The McMurdo Dry Valleys are a nearly pristine environment largely undisturbed and uncontaminated by humans and show remarkable peculiarities, representing an important analogue for the conditions of ancient Earth and Mars and a model environment for astrobiological studies [2–7]. These ice-free areas, dominated mostly by oligotrophic mineral soil and rocky outcrops [8, 9], due to the harshest conditions such as low temperatures (always below the freezing point) [10], strong and rapid thermal fluctuations, dryness, oligotrophy and high UV radiation, are dominated by cryptic microbial life-forms dwelling inside rocks [11]. For their unique geological and biological peculiarities, the McMurdo Dry Valleys, as a whole, are now designated as an ASMA (Antarctic Specially Managed Area), to assist planning and coordination of activities, improving cooperation between parties and minimise environmental impacts [12] and include five different ASPA (Antarctic Specially Protected Areas) to protect outstanding environments that require specific permits for entry. There, the prohibitive conditions are incompatible with active life on rock surfaces, and endolithic adaptation confers to microbes the chance to exploit a more protected niche inside rock porosity characterized by a milder and more stable microclimate [13–15]. Many different typologies of endolithic microbial communities have been described but the most complex and widespread in these areas are surely the cryptoendolithic lichen-dominated communities [1]. These are complex and self-supporting assemblages of phototrophic and heterotrophic microorganisms, including Bacteria, Chlorophyta and both free-living and lichen-forming fungi [1, 16] and, living at the edge of their physiological adaptability, represent the only chance for survival before extinction [11, 17].
The effect of global warming is dramatically intense at the Poles [18] and disturbance due to climate change may have irreversible effects on the weak equilibrium of these highly adapted microbial communities [19–21], but sensitive to any external perturbation [22]. Thus, improving our knowledge on functional capacity and multiple stress-responses of these ultimate ecosystems will give us important clues for monitoring and predicting any future variation due to climate change [19, 23]. Recent studies investigate the biodiversity of the McMurdo Dry Valleys [16, 24–27] and provided new insights on how the combination of environmental parameters of altitude and sea distance influence the distribution and the settlement of cryptoendolithic colonization [19–21, 28–29]. The key effect of the sun exposure in shaping the composition and abundance of functional groups of fungi in the Antarctic endolithic communities has been recently proved [19–21] as the northern exposed surfaces present considerably more favourable conditions than southern facing rocks [30–32] that, being in the shadow, may be too cold to allow biological activity.
Indeed, the capability to maintain biological activity depends on sufficient insolation on the rock surfaces allowing an efficient photosynthetic process; moreover, temperature became warmer for metabolic activity due the increasing of aw as consequence of melting of snow [33].
Antarctic cryptoendolithic microorganisms have to face several stresses simultaneously and they need to concurrently develop survival strategies to address external conditions. A conspicuous number of studies have been performed on the adaptation strategies on black fungi isolated from these communities, resulting highly resistant in terms of extremes of temperatures, acidity, osmotic stress and salinity, dehydration and irradiation [34–39]; besides, the stress-response mechanisms of whole metacommunity has not yet been investigated.
In this study, for the first time to our knowledge, metabolomics was successfully applied to Antarctic cryptoendolithic lichen-dominated communities to define adaptation strategies adopted by these communities to survive in these harshest conditions. We found different microbes’ response to sun exposure by modulating specific pathways; in particular, among the candidates to be considered biomarkers, the precursor metabolites of melanin and allantoin pathways were the most affected by sun exposure and we may consider these pathways to be directly involved to response to the environmental pressure.
Materials and methods
Samples collection
Sandstones were collected in triplicate at Finger Mt. (1720 m a.s.l., McMurdo Dry Valleys, Southern Victoria Land, Continental Antarctica) both from north (77°45’0.93"S 160°44'45.2"E) and south (77°45’10"S 160°44'44.39.7"E) exposed surfaces by Laura Selbmann during the XXXI Italian Antarctic Expedition (Dec. 2015-Jan. 2016) (Fig 1). Samples have been collected in the frame of Italian National Antarctic Research Program (PNRA) projects. Sampling permits have been obtained for activity in Special Managed Areas (ASMA) and Special Protected Areas (ASPA) in compliance with the "Protocol on Environmental Protection to the Antarctic Treaty" Annex V, art.7.
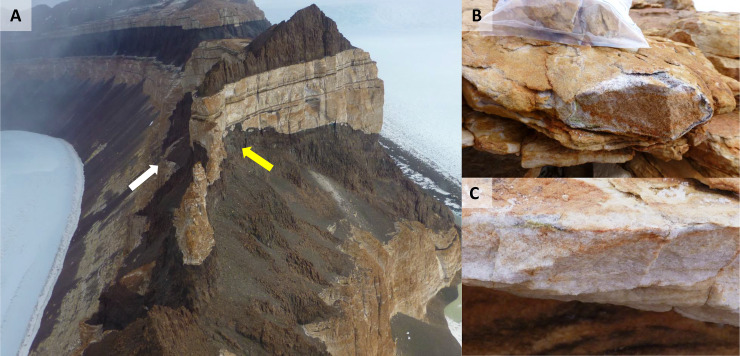
Fig 1.
A) Finger Mt. landscape: the yellow arrow indicates the north exposed surface, while the white arrow indicates the south exposed surface; B) north exposed sandstone rock; C) south exposed sandstone rock.
The presence of endolithic colonization was assessed by direct observation in situ using magnification lenses. Rocks were excised using a geological hammer, placed in sterile bags and shipped at -20°C at the at University of Tuscia (Italy) where have been preserved at -20°C in the Mycological Section of the Italian Antarctic National Museum (MNA), until processing.
Metabolites extraction
Classical metabolomics extraction protocol was adapted to rock sample. The frozen samples were powdered in liquid nitrogen. Ultrasonic crushing was performed at a low temperature, two times for 30 min. One gram of each rock was added to 3000 μl of a chloroform/methanol/water (1:3:1 ratio) solvent mixture stored at -20°C. Briefly, samples were vortexed for 5 min and left on ice for 2 h for total protein precipitation. Solutions were then centrifuged for 15 min at 15,000×g. and were dried to obtain visible pellets. Finally, the dried samples were re-suspended in 0.1 mL of RNA/DNAse free water, 5% formic acid and transferred to glass autosampler vials for LC/MS analysis. Extraction was performed in triplicate.
Ultra-high-performance liquid chromatography
Twenty uL of each extracted supernatant sample were injected into an ultra-high-performance liquid chromatography (UHPLC) system (Ultimate 3000, Thermo): samples were loaded onto a Reprosil C18 column (2.0mm× 150 mm, 2.5 μm-DrMaisch, Germany) for metabolite separation. Chromatographic separations were made at flow rate of 0.2 ml/min. A 0–100% linear gradient of solvent A (ddH2O, 0.1% formic acid) to B (acetonitrile, 0.1% formic acid) was employed over 20 min, returning to 100% A in 2 min and holding solvent A for a 6-min post time hold. The UHPLC system was coupled online with a Q Exactive mass spectrometer (Thermo Fisher, Rockford, IL) scanning in full MS mode (2 μ scans) at resolution of 70,000 in the 67 to 1,000 m/z range, with 3.8 kV spray voltage, 40 sheath gas, and 25 auxiliary gas. The system was operated in positive ion mode. Calibration was performed before each analysis against positive or negative ion mode calibration mixes (Thermo Fisher) to ensure error of the intact mass within the sub ppm range. Metabolite assignments were performed using MAVEN v5.2 [40]. Each replicate was analysed separately and a p-value < 0.01 was used to infer significance for all abundance comparisons between sets of triplicates.
Data elaboration and statistical analysis
Replicate files were processed through MAVEN v5.2, enabling rapid and reliable metabolites quantitation from multiple reaction-monitoring data or high-resolution full-scan mass spectrometry data. Mass Spectrometry chromatograms were created for peak alignment, matching and comparison of parent and fragment ions with tentative metabolite identification within a 2-p.p.m. mass-deviation range between the observed and expected results against an imported KEGG database [41]. To visualize the number of significant changes m/z values between metabolites datasets, a Volcano plot were created using the MetaboAnalyst 4.0 software (http://metpa.metabolomics.ca/). Raw data were normalized by sum and auto-scaling in order to increase the importance of low-abundance ions without significant amplification of noise. This type of plot displays the fold change differences and the statistical significance for each variable. The log of the fold change is plotted on the x-axis so that changes in both directions (up and down) appear equidistant from the centre. The y-axis displays the negative log of the p-value from a two-sample t-test. False discovery rate (FDR) [42] were used for controlling multiple testing.
Metabolites subject to major changes were displayed and Receiver Operator Characteristic (ROC) curves for each of them was calculated using the same software to evaluate potential metabolites to be considered as biomarkers. A ROC diagram plots the true positive rate (sensitivity) of a test on the y-axis against the false positive rate (100-specificity) on the x-axis, yielding the ROC area under the curve (AUC). Within this model, the ROC curves used 5-fold cross validation. AUC can be interpreted as the probability that a test or a classifier will rank a randomly chosen positive instance higher than a randomly chosen negative one. If all positive samples are ranked before negative ones (i.e. a perfect classifier), the AUC is 1.0. An AUC of 0.5 is equivalent to randomly classifying subjects as either positive or negative (i.e. the classifier is of no practical utility).
Metabolic pathways were displayed with Graphpad Prism v5.01 (Graphpad Software Inc) and statistical analyses were performed with the same software (p < 0.05). Data are presented as mean ± SD. Differences were considered statistically significant at *p < 0.05 and further stratified to **p < 0.01, respectively.
Results
Shaping in metabolites composition
An untargeted metabolomics analysis was performed on six differently exposed rocks and more than 10,000 peaks per sample were referred to the KEGG database; among them, 2,807 metabolites were analysed more precisely and identified.
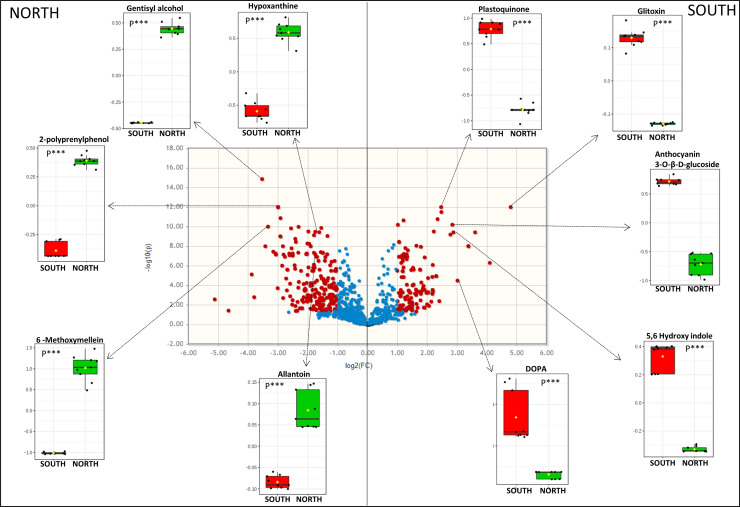
The significant discriminating metabolites were identified using the Volcano plot analysis (Fig 2); the univariate analysis identified significant accumulation of specific metabolites; most of them were expressed in rocks collected in south exposure, while few others were overexpressed in north exposed rocks.
Fig 2. Volcano plot analysis of metabolic changes Finger Mt. north and Finger Mt. south.
Each point on the volcano plot was based on both p-value and fold-change values, and in this study these two values were set at 0.05 and 2.0, respectively. The points which satisfy the condition p < 0.05 and fold change > 2.0 appear in red and are marker candidates, whereas the others appear in blue that are not significant. The potential biomarkers between experimental groups were annotated with their matched metabolite names. Bar plots show the original values (mean +/- SD). Differences were considered statistically significant at *p < 0.05 further stratified to **p < 0.01 and ***p < 0.001 respectively.
Based on the specific criteria, 206 metabolites were significantly upregulated in Antarctic cryptoendolithic communities of rocks exposed to the north whereas 125 were significantly upregulated in the south exposed ones (Fig 2). S1 Table summarized all metabolites identified considering the following parameters: p-value, FC (fold change), log2 (FC fold change), and -log10 (p-value).
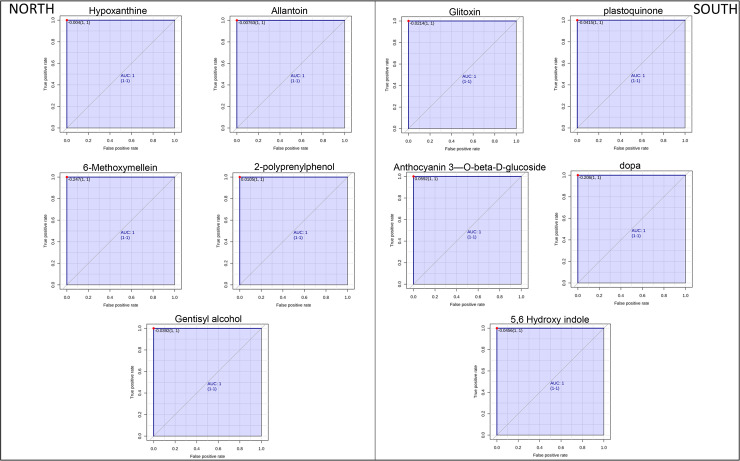
Candidate markers were selected, using a Receiver Operating Characteristic (ROC) curve analysis, by examining the volcano plot and considering a fold change threshold of 2 and p-value < 0.05. Five significantly upregulated metabolites in the north-exposed communities (Allantoin, Hypoxanthine, 6-Methoxymellein, 2-Polyprenylphenol and Gentisyl alcohol) and five metabolites increased significantly in those facing south (Fig 2) (Gliotoxin, Plastoquinone, Anthocyanin 3-O-β-D-glucoside, 5–6 Dihydroxyindole and L-DOPA) showed an AUC = 1 that represented a perfect ROC curve according to the accepted classification of biomarkers [43] (Fig 3).
Fig 3. Area under the receiver-operating characteristic curves (AUROC) of the significant metabolite (with AUROC value > 0.9) identified 5 metabolites (Allantoin, Hypoxanthine, 6-Methoxymellein, 2-Polyprenylphenol and Lomefloxacin hydrochloride) as significantly elevated in Finger Mt. north samples and 5 metabolites (Gliotoxin, Plastoquinone, Anthocyanin 3-O-β-D-glucoside, 5–6 Dihydroxyindole and L-DOPA) significantly elevated in Finger Mt. south samples.
They are candidate biomarkers with excellent value (AUC = 1; CI = 1−1).
Based on ROC analysis, changes in Allantoin (northern rock surface) and L-DOPA (southern rock surface) expression were further analysed to clarify their roles in the Antarctic cryptoendolithic communities.
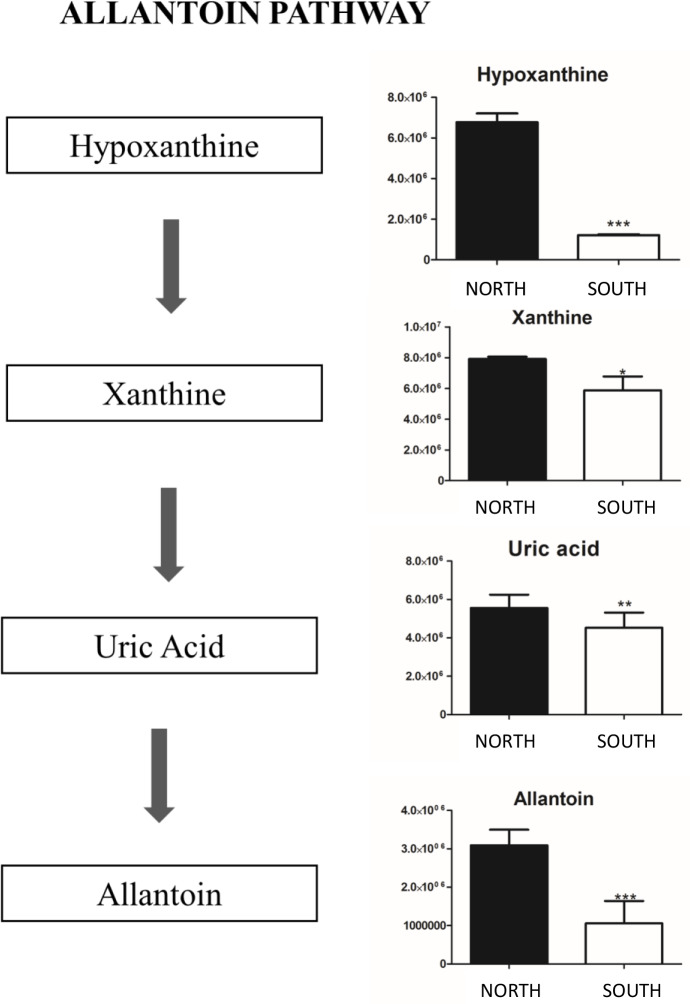
Allantoin pathway
Fig 4 showed the significant changes observed in the intermediates allantoin biosynthesis.
Fig 4. Allantoin pathway.
Total amount of allantoin pathway intermediates appears to be increased in Finger Mt. north. Metabolites are expressed as the mean ± SD concentration over Finger Mt. north *p < 0.05, **p < 0.01 against Finger Mt. south.
Biosynthesis starts from purine metabolism degradation (Hypoxanthine and Xanthine) and the intermediate Uric Acid. All allantoin pathway intermediates were stable and more abundant in the north exposure; while, an intermediates fluctuation was observed in the south samples. The amount of uric acid and allantoin decrease significantly in south samples (p < 0.05).
Melanin pathway
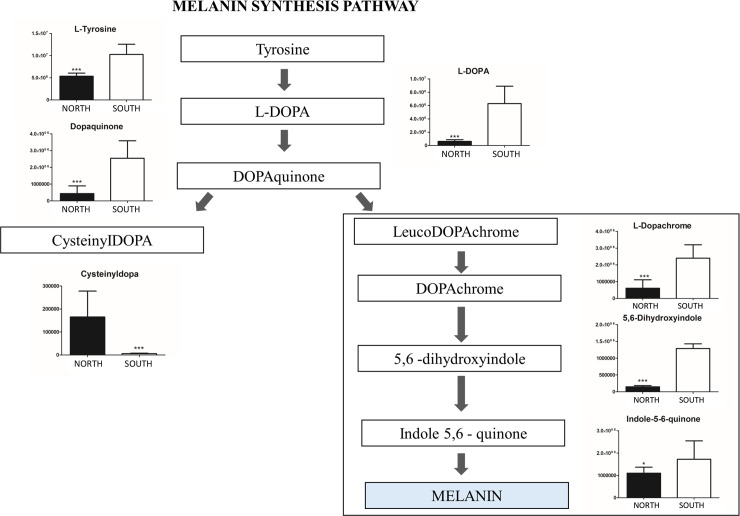
In Fig 5 the melanin biosynthetic pathway is reported. Melanin is produced starting from oxidation of amino acid L-Tyrosine. With our mass spectrometry techniques, it is possible to identify the tyrosine metabolism intermediates. The rate-limiting initial step in the biosynthesis of melanin is the hydroxylation of tyrosine to L-3,4-dihydroxyphenylalanine (DOPA) and its immediate subsequent oxidation to DOPAquinone (DQ). Dopaquinone went through instantaneous intramolecular nonenzymatic cyclization forming leucochrome, which is rapidly oxidized by dopaquinone to dopachrome at the level of DOPAquinone point of switch. The pathway lead to the synthesis of melanin instead of cysteinyl DOPA. Since all the intermediates (leucoDOPAchrome, DOPAchrome, 5–6 dihydroxy indole, Indole 5,6 quinone) reached the maximum level in south exposed samples (p < 0.05), we have considered the melanin biosynthesis significantly enhanced in southern exposed rocks.
Fig 5. Melanin synthesis pathway.
The conversion of tyrosine to melanin involves several steps and we found overall up-regulation of Finger Mt. south intermediates whereas Cysteinyl DOPA decreases, confirming that samples from south increase the synthesis of melanin. Metabolites are expressed as the mean ± SD concentration over Finger Mt. north *p < 0.05, **p < 0.01 against Finger Mt. south.
Discussion
Understanding the mechanisms driving functional and ecological processes in the extremes remains a major challenge [44–46], particularly since environmental stressors are often associated with diminished ecosystem capacity and functionality [47–50]. This is particularly true in hot and cold desert ecosystems such as the Antarctic Dry Valleys (Continental Antarctica) are accounted as the coldest and most hyper-arid desert on Earth and the closest Mars analogue on Earth [5, 6].
Metabolites are the result of both biological and environmental factors, and provide great potential to bridge knowledge of genotype and phenotype. In contrast to targeted metabolomics, non-targeted approaches offer the potential to determine biomarkers. The idea that untargeted mass spectrometry (MS)-based metabolomics analysis will result in a large list of ‘identified’ small molecules that can be mapped to networks and pathways is often assumed [51].
In this study, we considered rocks collected at opposite exposure, accounted among the principal driving factor for settlement and shaping stress-response in cryptoendolithic life-style; indeed, recently, sun exposure was found to be critical in shifting community composition, taxon abundance, and distribution of functional groups of fungi in Antarctic endolithic communities [21, 28], shaping these ecosystems more than other abiotic parameters previously considered (e.g. altitude). Besides, the differences in the ability to respond to stress between northern and southern exposed communities, if any, have never been investigated.
In our study, the concentration and composition of metabolites presented a clear pattern of correlation across different exposed surfaces of the same mountain, where the changes in the microbial community metabolic profiles following sunlight deprivation were evident. Overall, the remarkable variability observed across exposures indicated that this environmental parameter can be considered as driving factor shaping the stress-response strategies in the Antarctic cryptoendolithic communities. Indeed, through the Volcano Plots, we highlighted the differential abundance of metabolites influenced by this parameter. In total, we identified 206 metabolites that were significantly expressed in north exposed rocks and 125 in southern samples, as shown in Fig 2 (p < 0.01, S1 Table), where conditions are more severe. The ability to thrive at temperature and conditions, such as those associated to southern-exposed rocks, requires a vast array of adaptations to maintain the metabolic rates to guarantee active life; thus, microbial communities may have evolved diverse and tougher mechanisms to successfully exploit southern surface where conditions are harsher compared with north exposed faces [52, 53], by up or down-regulating the expression of a significant number of genes encoding cold-shock proteins (CSPs) process namely cold-shock response. Genes that are strongly up-regulated in response to cold exposure include a number of cold-induced RNA helicases, molecular chaperones, heat shock proteins and genes associated with sugar transport and metabolism [54, 55] and genes encoding antioxidative enzymes such as catalases and superoxide dismutase [56].
Recent studies reported an up-regulations of a highly diverse set of metabolic features to cope cold-associated stresses; for instance, in an Antarctic alga Chlamydomonas sp., RNA helicase genes were up-regulated after low temperature exposure [57, 58], while, photosynthetic metabolism in Chaetoceros neogracile, isolated from Antarctic ocean, was found highest at 4°C than at room temperature [59]; earlier, a protease was found to be produced in higher amount at low temperature in the Antarctic yeast Candida humicola [60]. However, given the many different adaptive mechanisms that are used by different psychrophiles, many other strategies for survival at cold temperatures are still to be discovered [61].
Additionally, the volcano plot analysis, coupled with Receiver Operating Characteristics (ROC) curves analysis, was employed to explore the chemical markers that contributed most to the difference in profiles across sampled area, determining which compounds were of interest. Among the detected metabolites, we produced a hit of metabolites specimens and identified at least 10 metabolites that were specifically correlated to a specific condition. The ROC curves (Fig 3), determined that allantoin, hypoxanthine, 6-methoxymellein, 2-polyprenylphenol, and Gentisyl alcool were the most sensitive and specific biomarkers for north exposed rocks (AUC = 1), while l-dopa, 5-6-dihydroxindole, gliotoxin, plastoquinone, and anthocyanin were unique for south exposed microbial communities (AUC = 1). All these metabolites are intermediates of crucial pathways responsible for stress-adaptation: among the north-related biomarkers, 6-methoxymellein has been reported to be involved in UV-radiation response in plant systems [62]; 2-polyprenylphenol (member of the class of phenols and precursor of ubiquinone) plays an important role in plant resistance and in plant growth and development, through the biosynthesis compounds that act as antioxidants [63]. Their synthesis is generally triggered in response to biotic/abiotic stresses and especially under salt stress conditions; they take part in defence responses during infection, excessive sun exposure, injuries and heavy metal stress [64].
Instead, among biomarkers responsible for stress-response in south-exposed communities, gliotoxin is involved in protecting Aspergillus fumigatus against oxidative stress against the reactive oxygen species (ROS) [65, 66]; indeed, gliotoxin is an epipolythiodioxopiperazine (ETP), a toxic secondary metabolites made only by fungi with molecular mass 326 Da and contains a disulphide bridge which can undergo repeating cleavage and reformation, thereby resulting in a potent intracellular redox activity [67]. Plastoquinone participates in the metabolism of various important chemical compounds, acting as antioxidants, involved in plant response to salinity stress [68, 69]. Anthocyanins are well-known water‐soluble pigments found in all plant tissues throughout the plant kingdom, involved in UV-B radiation, water stress and particularly cold temperatures [70], as reported in Arabidopsis [71, 72]. Indeed, McKown et al. [73] suggested a sort of commonality between anthocyanin biosynthesis and freezing tolerance as Arabidopsis mutants deficient in freezing tolerance were unable to accumulate anthocyanins, resulting in an increased tolerance of cool temperatures.
We further focused on allantoin and hypoxanthine over expressed in north, and DOPA and 5–6 hydroyndole over expressed in south since they can play a role in different stress responses in the endolithic communities of the two-opposite exposed surfaces. The allantoin plays an important protective role to excessive sun exposure; this pathway resulted over- expressed in the north exposed communities, where climatic conditions are more buffered and allow a successful settlement of endolithic colonization. Besides, the degree of insulation is much higher than in the south and photosynthetic organisms such as cyanobacteria and algae need to be protected by UV irradiation. Allantoin is also produced in plants, providing protective function by blocking UV-B before it reaches cells, the consequences of UV-B exposure such as repairing damaged DNA [74]. Metabolomics studies revealed allantoin as a major purine metabolite in Arabidopsis thaliana, Oryza sativa and other species under various stress conditions such as drought [75–77], high salinity [78, 79], low temperature [80] and nutrient constraints [81–83], environmental characteristics that are exacerbated in Antarctic ice-free areas.
Conversely, the south exposed surface is in more prohibitive climatic conditions and in the shadow over the entire year, with limited possibility for metabolic activity and the need of a super-protective shield is of utmost importance for the survival of the community as a whole. In this environment, melanin pathway resulted more expressed that in the northern surface. Melanins are ancient biological pigments found in all kingdoms of life and, in particular in the fungal kingdom, are present across all phyla with remarkable physicochemical properties, allowing them to perform diverse functions in biological systems [84–87]. Mainly, two pathways of melanin synthesis are found in fungi: most of them synthesize melanin via the DHN (1,8-dihydroxynaphthalene) pathway, whereas few species are able to produce melanin via l-DOPA (3,4-dihydroxyphenylalanine) in a pathway that resembles mammalian melanin biosynthesis [88]. Recently, it was found evidence for the production of a water-soluble pigment similar to pyomelanin by Penicillium chrysogenum [89]. Despite the wide number of melanotic species and biosynthetic pathways for melanin production, the exact melanins produced by the Antarctic cryptoendolithic fungi are still unknown; besides, it has been suggested that all fungal melanins, regardless of their precursor, may share similar physicochemical properties [90]. Some fungal species, constitutively melanized are referred to as black, dematiaceous, micro colonial or meristematic fungi [34] with a worldwide distribution, typically colonizing harsh environmental niches not suitable to most life forms such as saltpans, hydrocarbon-contaminated sites, exposed bare rocks and monuments, icy habitats, deserts, solar panels and building roofs [35, 91–96]. This group of fungi is also invariably present in Antarctic cryptoendolithic communities [20, 97–100] where they form a black barrier just above the photobiont’s stratification, supplying protection to algae by dissipating excessive sunlight [101].
These results are confirmed in a recent study where the authors showed that black fungi were invariably predominant in communities sampled in southern exposed rocks [21]. Actually, the highly melanized cell wall in black fungi plays a crucial protective role also to other stresses in addition to sunlight and UV radiation such as low temperature, high osmotic pressure, oxidative stresses, low water activity and nutrient availability [102–107] that can be more intense in south exposed surfaces.
Conclusions
This first contribution allowed us starting to discern the functionality of these unique microbial communities, demonstrating how the metabolic response shifts across variations due to sun exposure, and giving insights on the capabilities of these ultimate ecosystems to sustain their growth and survival in such harsh environments. We validated application of an untargeted metabolomics method that provides optimal identification of a wide range of metabolites to detect metabolic changes in the main pathways, determining which products are being released into the environment. We conclude that further investigations on changes to the metabolic profiles may have potential for use as early indicators, to forecast the impact of Global Change that is challenging for the structure and functioning of cryptoendolithic ecosystems in Antarctic deserts.
Supporting information
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
L.S. and C.C. wish to thank the Italian National Program for Antarctic Researches (PNRA) for funding sampling campaigns and researches in Italy in the frame of the PNRA projects. The Italian Antarctic National Museum (MNA) is kindly acknowledged for financial support to the Mycological Section on the MNA for preserving Antarctic rock samples, herein analysed, stored in the Culture Collection of Fungi from Extreme Environments (CCFEE), University of Tuscia, Italy. The authors wish to thank Italian Space Agency (ASI n. 2018-6-U0) for co-funding the BIOSIGN-Microfossils project.
References
- 1.Friedmann EI. Endolithic microorganisms in the Antarctic cold desert. Science. 1982;215:1045–1053. 10.1126/science.215.4536.1045 [DOI] [PubMed] [Google Scholar]
- 2.Horowitz NH, Cameron RE, and Hubbard JS. Microbiology of the dry valleys of Antarctica. Science. 1972;176(4032):242–245. 10.1126/science.176.4032.242 [DOI] [PubMed] [Google Scholar]
- 3.Wynn-Williams DD, and Edwards HGM. Proximal analysis of regolith habitats and protective biomolecules in situ by laser Raman spectroscopy: overview of terrestrial Antarctic habitats and Mars analogs. Icarus. 2000;144(2):486–503. 10.1006/icar.1999.6307 [DOI] [Google Scholar]
- 4.Onofri S, Selbmann L, Zucconi L, and Pagano S. Antarctic microfungi as models for exobiology. Planet Space Sci. 2004;52:229–237. 10.1016/j.pss.2003.08.019 [DOI] [Google Scholar]
- 5.Doran PT, Lyons WB, and McKnight DM. (Eds.). (2010) Life in Antarctic deserts and other cold dry environments: astrobiological analogs (Vol. 5). Cambridge University Press; 10.1017/S0954102011000502 [DOI] [Google Scholar]
- 6.Tamppari LK, Anderson RM, Archer PD, Douglas S, Kounaves SP, McKay CP, et al. Effects of extreme cold and aridity on soils and habitability: McMurdo Dry Valleys as an analogue for the Mars Phoenix landing site. Antarct Sci. 2012;24(3):211–228. 10.1017/S0954102011000800 [DOI] [Google Scholar]
- 7.Heldmann JL, Pollard W, McKay CP, Marinova MM, Davila A, Williams KE, et al. The high elevation Dry Valleys in Antarctica as analog sites for subsurface ice on Mars. Planet Space Sci. 2013;85:53–58. 10.1016/j.pss.2013.05.019 [DOI] [Google Scholar]
- 8.Thomas DSG. Arid zones: their nature and extent In: Thomas DSG (ed) Arid zone geomorphology, 2nd edn Wiley, Chichester, pp. 3–12. 10.1002/9780470710777.ch1 [DOI] [Google Scholar]
- 9.Ugolini FC, and Bockheim JG. Antarctic soils and soil formation in a changing environment: a review. Geoderma. 2008;144(1–2):1–8. 10.1016/j.geoderma.2007.10.005 [DOI] [Google Scholar]
- 10.Doran PT, McKay CP, Clow GD, Dana GL, Fountain AG, Nylen T, et al. Valley floor climate observations from the McMurdo Dry Valleys, Antarctica, 1986–2000. JGR-Atmospheres. 2002;107:(D24). 10.1029/2001JD002045 [DOI] [Google Scholar]
- 11.Nienow JA, and Friedmann EI. Terrestrial lithophytic (rock) communities In: Friedmann E.I. (ed) Antarctic Microbiology. Wiley-Liss; New York: pp. 343–412. [Google Scholar]
- 12.SCAR (2004). SCAR Bulletin 155. Polar Rec (Gr Brit) 40:371–382. 10.1017/S0032247404003948 [DOI] [Google Scholar]
- 13.Friedmann EI, and Koriem AM. Life on Mars: how it disappeared (if it was ever there). Adv Space Res. 1989;9(6):167–172. 10.1016/0273-1177(89)90224-x [DOI] [PubMed] [Google Scholar]
- 14.Kappen L. Plant activity under snow and ice, with particular reference to lichens. Arctic. 1993;46(4)297–302. [Google Scholar]
- 15.Wierzchos J, and Ascaso C. Life, decay and fossilisation of endolithic microorganisms from the Ross Desert, Antarctica. Polar Biol. 2001;24(11):863–868. 10.1007/s003000100296 [DOI] [Google Scholar]
- 16.de la Torre JR, Goebel BM, Friedmann E, and Pace NR. Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valleys. Antarctica. Applied Environ Microbiol. 2003;69:3858–3867. 10.1128/aem.69.7.3858-3867.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedmann EI, and Ocampo R. Endolithic blue-green algae in dry valleys-primary producers in Antarctic desert ecosystem. Science. 1976;193:1247–1249. 10.1126/science.193.4259.1247 [DOI] [PubMed] [Google Scholar]
- 18.Bekryaev RV, Polyakov IV, and Alexeev VA. Role of polar amplification in long-term surface air temperature variations and modern Arctic warming. J Clim. 2010;23(14):3888–3906. 10.1175/2010JCLI3297.1 [DOI] [Google Scholar]
- 19.Selbmann L, Onofri S, Coleine C, Buzzini P, Canini F, and Zucconi L. Effect of environmental parameters on biodiversity of the fungal component in the lithic Antarctic communities. Extremophiles. 2017;21(6):1069–1080. 10.1007/s00792-017-0967-6 [DOI] [PubMed] [Google Scholar]
- 20.Coleine C, Stajich JE, Zucconi L, Onofri S, Pombubpa N, Egidi E, et al. Antarctic cryptoendolithic fungal communities are highly adapted and dominated by Lecanoromycetes and Dothideomycetes. Frontiers Microbiol. 2018;9:1392 10.3389/fmicb.2018.01392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleine C, Zucconi L, Onofri S, Pombubpa N, Stajich JE, and Selbmann L. Sun exposure shapes functional grouping of fungi cryptoendolithic Antarctic communities. Life. 2018;8(2):19 10.3390/life8020019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selbmann L, Isola D, Fenice F, Zucconi L, Sterflinger K, and Onofri S. Potential extinction of Antarctic endemic fungal species as a consequence of Global Warming. Sci Total Environ. 2012;438:127–134. 10.1016/j.scitotenv.2012.08.027 [DOI] [PubMed] [Google Scholar]
- 23.Hogg ID, and Wall DH. Global change and Antarctic terrestrial biodiversity. Polar Biol. 2011;34:1625–1627. 10.1007/s00300-011-1108-9 [DOI] [Google Scholar]
- 24.Cary SC, McDonald IR, Barrett JE, and Cowan DA. On the rocks: the microbiology of Antarctic Dry Valley soils. Nature Rev Microbiol. 2010;8:129–138. 10.1038/nrmicro2281 [DOI] [PubMed] [Google Scholar]
- 25.Wei ST, Lacap-Bugler DC, Lau MC, Caruso T, Rao S, de Los Rios A, et al. Taxonomic and functional diversity of soil and hypolithic microbial communities in Miers Valley, McMurdo Dry Valleys, Antarctica. Front Microbiol. 2016;7:1642 10.3389/fmicb.2016.01642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Archer SD, de los Ríos A, Lee KC, Niederberger TS, Cary SC, Coyne KJ, et al. Endolithic microbial diversity in sandstone and granite from the McMurdo Dry Valleys, Antarctica. Polar Biol. 2017;40(5):997–1006. 10.1007/s00300-016-2024-9 [DOI] [Google Scholar]
- 27.Lee J, Cho J, Cho YJ, Cho A, Woo J, Lee J, et al. The latitudinal gradient in rock-inhabiting bacterial community compositions in Victoria Land, Antarctica. Sci Total Environ. 2019;657:731–738. 10.1016/j.scitotenv.2018.12.073 [DOI] [PubMed] [Google Scholar]
- 28.Coleine C, Stajich JE, Pombubpa N, Zucconi L, Onofri S, Canini F, et al. Altitude and fungal diversity influence the structure of Antarctic cryptoendolithic Bacteria communities. Environ Microbiol Rep. 2019;11(5):718–726. 10.1111/1758-2229.12788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coleine C, Pombubpa N, Zucconi L, Onofri S, Stajich JE, and Selbmann L. Endolithic fungal species markers for harshest conditions in the McMurdo Dry valleys, Antarctica. Life. 2020;10(2),13 10.3390/life10020013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deegenaars ML, and Watson K. Heat shock response in psychrophilic and psychrotrophic yeast from Antarctica. Extremophiles. 1998;2(1):41–50. 10.1007/s007920050041 [DOI] [PubMed] [Google Scholar]
- 31.Friedmann EI. Microorganisms in Antarctic desert rocks from dry valleys and Dufek Massif. Antarctic J US. 1977;12(4):26–29. [Google Scholar]
- 32.Friedmann EI, and Weed R. Microbial trace-fossil formation, biogenous, and abiotic weathering in the Antarctic cold desert. Science 1987;236(4802):703–705. 10.1126/science.11536571 [DOI] [PubMed] [Google Scholar]
- 33.McKay CP, and Friedmann EI. The cryptoendolithic microbial environment in the Antarctic cold desert: temperature variations in nature. Polar Biol. 1985;4(1):19–25. 10.1007/BF00286813 [DOI] [PubMed] [Google Scholar]
- 34.Sterflinger K. Black yeasts and meristematic fungi: ecology, diversity and identification In Biodiversity and Ecophysiology of Yeasts (pp. 501–514). Springer, Berlin, Heidelberg: 10.1007/3-540-30985-3_20 [DOI] [Google Scholar]
- 35.Sterflinger K, Tesei D, and Zakharova K. Fungi in hot and cold deserts with particular reference to microcolonial fungi. Fungal Ecol. 2012;5(4):453–462. 10.1016/j.funeco.2011.12.007 [DOI] [Google Scholar]
- 36.Gorbushina AA, Whitehead K, Dornieden T, Niesse A, Schulte A, and Hedges JI. Black fungal colonies as units of survival: hyphal mycosporines synthesized by rock-dwelling microcolonial fungi. Can J Botany. 2003;81(2):131–138. 10.1139/b03-011 [DOI] [Google Scholar]
- 37.Dadachova E, Bryan RA, Huan X, Moadel T, Schweitzer AD, Aisen P, et al. Ionizing radiation changes the electronic properties of melanin and enhances the growth of melanized fungi. PloS One. 2007;2(5):e457 10.1371/journal.pone.0000457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onofri S, Zucconi L, Selbmann L, de Hoog S, de los Rios A, Ruisi S, et al. Fungal associations at the cold edge of life In: Seckbach J. (ed) Algae and cyanobacteria in extreme environments. Springer; Netherlands: 2007; pp. 735–757. 10.1007/978-1-4020-6112-7_40 [DOI] [Google Scholar]
- 39.Onofri S, Barreca D, Selbmann L, Isola D, Rabbow E, Horneck G, et al. Resistance of Antarctic black fungi and cryptoendolithic communities to simulated space and Martian conditions. Stud Mycol. 2008;61:99–109. 10.3114/sim.2008.61.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clasquin MF, Melamud E, and Rabinowitz JD. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Current Protocols in Bioinformatics. 2012;14:11–14. 10.1002/0471250953.bi1411s37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, and Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, and Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. J Royal Stat Soc. 1995;57(1):289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 43.Andersen MB, Kristensen M, Manach C, Pujos-Guillot E, Poulsen SK, Larsen TM, et al. Discovery and validation of urinary exposure for different plant foods by untargeted metabolomics. Anal Bioanal Chem. 2014;406:1829–44. 10.1007/s00216-013-7498-5 [DOI] [PubMed] [Google Scholar]
- 44.Casanueva A, Tuffin M, Cary C, and Cowan DA. Molecular adaptations to psychrophily: the impact of ‘omic’ technologies. Trends Microbiol. 2010;18:374–381. 10.1016/j.tim.2010.05.002 [DOI] [PubMed] [Google Scholar]
- 45.Andrei AŞ, Banciu H.L, and Oren A. Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol Lett. 2012;330:1–9. 10.1111/j.1574-6968.2012.02526.x [DOI] [PubMed] [Google Scholar]
- 46.Makhalanyane TP, Valverde A, Birkeland NK, Cary SC, Tuffin IM, and Cowan DA. Evidence for successional development in Antarctic hypolithic bacterial communities. ISME J. 2013; 7:2080–2090. 10.1038/ismej.2013.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petchey OL, McPhearson PT, Casey TM, and Morin PJ. Environmental warming alters food-web structure and ecosystem function. Nature. 1999;402(6757):69 10.1038/47023 [DOI] [Google Scholar]
- 48.Schimel J, Balser TC, and Wallenstein M. Microbial stress‐response physiology and its implications for ecosystem function. Ecology. 2007;88(6):1386–1394. 10.1890/06-0219 [DOI] [PubMed] [Google Scholar]
- 49.Harrison JP, Gheeraert N, Tsigelnitskiy D, and Cockell CS. The limits for life under multiple extremes. Trends Microbiol. 2013; 21:204–212. 10.1016/j.tim.2013.01.006 [DOI] [PubMed] [Google Scholar]
- 50.Ferrenberg S, Reed SC, and Belnap J. Climate change and physical disturbance cause similar community shifts in biological soil crusts. Proc Natl Acad Sci USA. 2015;112:12116–12121. 10.1073/pnas.1509150112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, and McLean JA. Untargeted metabolomics strategies—challenges and emerging directions. J Am Soc Mass Spectr. 2016;27(12):1897–1905. 10.1007/s13361-016-1469-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’amico S, Collins T, Marx JC, Feller G, and Gerday C. Psychrophilic microorganisms: challenges for life. EMBO Rep. 2006;7(4):385–389. 10.1038/sj.embor.7400662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Margesin R, and Miteva V. Diversity and ecology of psychrophilic microorganisms. Res Microbiol. 2011; 162(3):346–361. 10.1016/j.resmic.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 54.Gao H, Yang ZK, Wu L, Thompson DK, and Zhou J. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol. 2006;188:4560–4569. 10.1128/JB.01908-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Z, Yu H, Li L, Hu S, and Dong X. The genome and transcriptome of a newly described psychrophilic archaeon, Methanolobus psychrophilus R15, reveal its cold adaptive characteristics. Environ Microbiol Rep. 2012;4:633–641. 10.1111/j.1758-2229.2012.00389.x [DOI] [PubMed] [Google Scholar]
- 56.Chattopadhyay MK. The link between bacterial radiation resistance and cold adaptation. J Biosci. 2002;27:(02). 10.1007/bf02703760 [DOI] [PubMed] [Google Scholar]
- 57.An M, Mou S, Zhang X, Ye N, Zheng Z, Cao S, et al. Temperature regulates fatty acid desaturases at a transcriptional level and modulates the fatty acid profile in the Antarctic microalga Chlamydomonas sp. ICE-L. Bioresour Technol. 2013;134:151–157. 10.1016/j.biortech.2013.01.142 [DOI] [PubMed] [Google Scholar]
- 58.Liu C, and Huang X. Transcriptome-wide analysis of DEAD-box RNA helicase gene family in an Antarctic psychrophilic alga Chlamydomonas sp. ICE-L. Extremophiles. 2015;19:921–931. 10.1007/s00792-015-0768-8 [DOI] [PubMed] [Google Scholar]
- 59.Hwang Y, Jung G, and Jin E. Transcriptome analysis of acclimatory responses to thermal stress in Antarctic algae. Biochem Biophys Res Commun. 2008;367:635–641. 10.1016/j.bbrc.2007.12.176 [DOI] [PubMed] [Google Scholar]
- 60.Ray MK, Devi KU, Kumar GS, and Shivaji S. Extracellular protease from the Antarctic yeast Candida humicola. Appl Environ Microbiol. 1992;58:1918–1923. PMC195704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Maayer P, Anderson D, Cary C, and Cowan DA. Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 2014;15(5):508–517. 10.1002/embr.201338170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mercier J, Arul J, and Julien C. Effect of food preparation on the isocoumarin, 6-methoxymellein, content of UV-treated carrots. Food Res Int. 1994;27(4):401–404. 10.1016/0963-9969(94)90196-1 [DOI] [Google Scholar]
- 63.Liu M, and Lu S. Plastoquinone and ubiquinone in plants: Biosynthesis, physiological function and metabolic engineering. Front Plant Sci. 2016; 7:1898 10.3389/fpls.2016.01898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D’Souza MR, and Devaraj VR. Biochemical responses of Hyacinth bean (Lablab purpureus) to salinity stress. Acta Physiol Plantarum. 2010;32(2):341–353. 10.1007/s11738-009-0412-2 [DOI] [Google Scholar]
- 65.Scharf DH, Remme N, Heinekamp T, Hortschansky P, Brakhage AA, and Hertweck C. Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J Am Chem Soc. 2010;132(29):10136–10141. 10.1021/ja103262m [DOI] [PubMed] [Google Scholar]
- 66.Schrettl M, Carberry S, Kavanagh K, Haas H, Jones GW, O'Brien J, et al. Self-protection against gliotoxin—a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 2010;6(6):e1000952 10.1371/journal.ppat.1000952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi HS, Shim JS, Kim JA, Kang SW, and Kwon HJ. Discovery of gliotoxin as a new small molecule targeting thioredoxin redox system. Biochem Biophys Res Commun. 2007;359(3):523–528. 10.1016/j.bbrc.2007.05.139 [DOI] [PubMed] [Google Scholar]
- 68.Takahashi S, and Murata N. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 2008;13(4):178–182. 10.1016/j.tplants.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 69.Suzuki N, Koussevitzky SHAI, Mittler RON, and Miller GAD. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012;35(2):259–270. 10.1111/j.1365-3040.2011.02336.x [DOI] [PubMed] [Google Scholar]
- 70.Chalker‐Scott L. Environmental significance of anthocyanins in plant stress responses. Photochem Photobiol. 1999;70(1):1–9. 10.1111/j.1751-1097.1999.tb01944.x [DOI] [Google Scholar]
- 71.Graham TL. Flavonoid and flavonol glycoside metabolism in Arabidopsis. Phys Chem Earth. 1998;36:135–144. 10.1016/S0981-9428(98)80098-3 [DOI] [Google Scholar]
- 72.Leyva A, Jarillo JA, Salinas J, and Martinez-Zapater JM. Low temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol. 1995;108:39–46. 10.1104/pp.108.1.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKown R, Kuroki G, and Warren G. Cold responses of Arabidopsis mutants impaired in freezing tolerance. J Exp Bot. 1996;47:1919–1925. 10.1093/jxb/47.12.1919 [DOI] [Google Scholar]
- 74.Smirnoff N. Plant resistance to environmental stress. Curr Opin Biotechnol. 1998;9(2):214–219. 10.1016/s0958-1669(98)80118-3 [DOI] [PubMed] [Google Scholar]
- 75.Oliver MJ, Guo L, Alexander DC, Ryals JA, Wone BWM, and Cushman JC. A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. The Plant Cell. 2011;23:1231–1248. 10.1105/tpc.110.082800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silvente S, Sobolev AP, and Lara M. Metabolite adjustments in drought tolerant and sensitive soybean genotypes in response to water stress. PLoS One. 2011;7:e38554 10.1371/journal.pone.0038554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yobi A, Wone BW, Xu W, Alexander DC, Guo L, Ryals JA, et al. Metabolomic profiling in Selaginella lepidophylla at various hydration states provides new insights into the mechanistic basis of desiccation tolerance. Mol Plant. 2013;6:369–385. 10.1093/mp/sss155 [DOI] [PubMed] [Google Scholar]
- 78.Kanani H, Dutta B, and Klapa MI. Individual vs. combinatorial effect of elevated CO 2 conditions and salinity stress on Arabidopsis thaliana liquid cultures: comparing the early molecular response using time-series transcriptomic and metabolomic analyses. BMC Syst Biol. 2010;4(1):177 10.1186/1752-0509-4-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang WS, Zhao XQ, Li M, Huang LY, Xu JL, Zhang F, et al. 2016. Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. Journal of Experimental Botany. 2016;67:405–419. 10.1093/jxb/erv476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, et al. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004;136(4):4159–4168. 10.1104/pp.104.052142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, et al. Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 2005;138(1):304–318. 10.1104/pp.104.053793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rose MT, Rose TJ, Pariasca-Tanaka J, Yoshihashi T, Neuweger H, Goesmann A, et al. Root metabolic response of rice (Oryza sativa L.) genotypes with contrasting tolerance to zinc deficiency and bicarbonate excess. Planta. 2012;236:959–973. 10.1007/s00425-012-1648-4 [DOI] [PubMed] [Google Scholar]
- 83.Coneva V, Simopoulos C, Casaretto JA, El-kereamy A, Guevara DR, Cohn J, et al. Metabolic and co-expression network-based analyses associated with nitrate response in rice. BMC Genomics. 2014;15(1):1056 10.1186/1471-2164-15-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cordero RJ. Melanin for space travel radioprotection. Environ Microbiol. 2017. a;19(7):2529–2532. 10.1111/1462-2920.13753 [DOI] [PubMed] [Google Scholar]
- 85.Cordero RJ, and Casadevall A. Functions of fungal melanin beyond virulence. Fungal Biol Rev. 2017. b;31(2):99–112. 10.1016/j.fbr.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cordero RJ, Vij R, and Casadevall A. Microbial melanins for radioprotection and bioremediation. Microb Biotechnol. 2017. c;10(5):1186 10.1111/1751-7915.12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cordero RJ, Robert V, Cardinali G, Arinze ES, Thon SM, and Casadevall A. Impact of yeast pigmentation on heat capture and latitudinal distribution. Curr Biol. 2018;28(16):2657–2664. 10.1016/j.cub.2018.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eisenman HC, and Casadevall A. Synthesis and assembly of fungal melanin. Appl Microbiol Biotechnol. 2012;93(3):931–940. 10.1007/s00253-011-3777-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vasanthakumar A, DeAraujo A, Mazurek J, Schilling M, and Mitchell R. Pyomelanin production in Penicillium chrysogenum is stimulated by L-tyrosine. Microbiology. 2015;161(6):1211–1218. 10.1099/mic.0.000030 [DOI] [PubMed] [Google Scholar]
- 90.Fogarty RV, and Tobin JM. Fungal melanins and their interactions with metals. Enzyme Microb Technol. 1996;19(4):311–317. 10.1016/0141-0229(96)00002-6 [DOI] [PubMed] [Google Scholar]
- 91.Adams BJ, Bardgett RD, Ayres E, Wall DH, Aislabie J, Bamforth S, et al. Diversity and distribution of Victoria Land biota. Soil Biol Biochem. 2006;38:3003–3018. 10.1016/j.soilbio.2006.04.030 [DOI] [Google Scholar]
- 92.Abdel-Hafez SII. Studies on soil mycoflora of desert soils in Saudi Arabia. Mycopathologia. 1994;80:3e8. [Google Scholar]
- 93.Gunde-Cimerman N, Zalar P, de Hoog S, and Plemenitaš A. Hypersaline waters in salterns–natural ecological niches for halophilic black yeasts. FEMS Microbiol Ecol. 2000;32(3):235–240. 10.1111/j.1574-6941.2000.tb00716.x [DOI] [PubMed] [Google Scholar]
- 94.Gunde-Cimerman N, Sonjak S, Zalar P, Frisvad JC, Diderichsen B, and Plemenitaš A. Extremophilic fungi in arctic ice: a relationship between adaptation to low temperature and water activity. Phys Chem Earth. 2003;28(28):1273–1278. 10.1016/j.pce.2003.08.056 [DOI] [Google Scholar]
- 95.Selbmann L, Zucconi L, Isola D, and Onofri S. Rock black fungi: excellence in the extremes. From the Antarctic to space. Curr Genet. 2015;61:335–345. 10.1007/s00294-014-0457-7 [DOI] [PubMed] [Google Scholar]
- 96.Ruibal C, Selbmann L, Avci S, Martin-Sanchez P, and Gorbushina A. Roof-Inhabiting Cousins of Rock-Inhabiting Fungi: Novel Melanized Microcolonial Fungal Species from Photocatalytically Reactive Subaerial Surfaces. Life. 2018;8(3):30 10.3390/life8030030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Selbmann L, De Hoog GS, Mazzaglia A, Friedmann EI, and Onofri S. Fungi at the edge of life: cryptoendolithic black fungi from Antarctic desert. Stud Mycol. 2005;51(1):1–32. 10.3114/sim.2008.61.10 [DOI] [Google Scholar]
- 98.Selbmann L, De Hoog G.S, Zucconi L, Isola D, Ruisi S, van den Ende AG, et al. Drought meets acid: three new genera in a dothidealean clade of extremotolerant fungi. Stud Mycol. 2008;61:1–20. 10.3114/sim.2008.61.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Selbmann L, Onofri S, Zucconi L, Isola D, Rottigni M, Ghiglione C, et al. Distributional records of Antarctic fungi based on strains preserved in the Culture Collection of Fungi from Extreme Environments (CCFEE) Mycological Section associated with the Italian National Antarctic Museum (MNA). MycoKeys. 2015;10:57 10.3897/mycokeys.10.5343 [DOI] [Google Scholar]
- 100.Egidi E, de Hoog GS, Isola D, Onofri S, Quaedvlieg W, de Vries M, et al. Phylogeny and taxonomy of meristematic rock-inhabiting black fungi in the Dothideomycetes based on multi-locus phylogenies. Fungal Div 2014;65:127–165. 10.1007/s13225-013-0277-y [DOI] [Google Scholar]
- 101.Selbmann L, Egidi E, Isola D, Onofri S, Zucconi L, de Hoog GS, et al. Biodiversity, evolution and adaptation of fungi in extreme environments. Plant Biosystems-An International Journal Dealing with all Aspects of Plant Biology. 2013;147(1):237–246. 10.1080/11263504.2012.753134 [DOI] [Google Scholar]
- 102.Dadachova E, and Casadevall A. Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin. Curr Opin Chem Biol. 2008;11(6):525–531. 10.1016/j.mib.2008.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Onofri S, de Vera JP, Zucconi L, Selbmann L, Scalzi G, Venkateswaran KJ, et al. Survival of Antarctic cryptoendolithic fungi in simulated Martian conditions on board the International Space Station. Astrobiology. 2015;15(12):1052–1059. 10.1089/ast.2015.1324 [DOI] [PubMed] [Google Scholar]
- 104.Onofri S, Selbmann L, Pacelli C, de Vera J, Horneck G, Hallsworth J, et al. Integrity of the DNA and cellular ultrastructure of cryptoendolithic fungi in space or Mars conditions: a 1.5-year study at the International Space Station. Life. 2018;8(2):23 10.3390/life8020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Onofri S, Selbmann L, Pacelli C, Zucconi L, Rabbow E, and de Vera JP. Survival, DNA, and Ultrastructural Integrity of a Cryptoendolithic Antarctic Fungus in Mars and Lunar Rock Analogs Exposed Outside the International Space Station. Astrobiology. 2019;19(2):170–182. 10.1089/ast.2017.1728 [DOI] [PubMed] [Google Scholar]
- 106.Selbmann L, Isola D, Zucconi L, and Onofri S. Resistance to UV-B induced DNA damage in extreme-tolerant cryptoendolithic Antarctic fungi: detection by PCR assays. Fungal Biol. 2011;115:937–944. 10.1016/j.funbio.2011.02.016 [DOI] [PubMed] [Google Scholar]
- 107.Selbmann L, Pacelli C, Zucconi L, Dadachova E, Moeller R, de Vera JP, et al. Resistance of an Antarctic cryptoendolithic black fungus to radiation gives new insights of astrobiological relevance. Fungal Biol. 2018;122(6):546–554. 10.1016/j.funbio.2017.10.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.