Abstract
Background
There is increasing evidence for the benefit of poly ADP ribose polymerase (PARP) inhibitors in a subset of high-grade serous ovarian carcinoma (HGSC) patients, especially those with homologous recombination (HR)-deficient tumors. However, new treatment strategies, such as immune checkpoint inhibition, are required for patients with HR-proficient tumors.
Methods
A total of 80 cases of HGSC were analyzed in this study. Whole exome and RNA sequencing was performed for these tumors. Methylation arrays were also carried out to examine BRCA1 and RAD51C promoter methylation status. Mutations, neoantigen load, antigen presentation machinery, and local immune profile were investigated, and the relationships of these factors with clinical outcome were also analyzed.
Results
As expected, the numbers of predicted neoAgs were lower in HR-proficient (n=46) than HR-deficient tumors (n=34). However, 40% of the patients with HR-proficient tumors still had higher than median numbers of neoAgs and better survival than patients with lower numbers of neoAgs. Incorporation of human leukocyte antigen (HLA)-class I expression status into the survival analysis revealed that patients with both high neoAg numbers and high HLA-class I expression (neoAghiHLAhi) had the best progression-free survival (PFS) in HR-proficient HGSC (p=0.0087). Gene set enrichment analysis demonstrated that the genes for effector memory CD8 T cells, TH1 T cells, the interferon-γ response, and other immune-related genes, were enriched in these patients. Interestingly, this subset of patients also had better PFS (p=0.0015) and a more T-cell-inflamed tumor phenotype than patients with the same phenotype (neoAghiHLAhi) in HR-deficient HGSC.
Conclusions
Our results suggest that immune checkpoint inhibitors might be an alternative to explore in HR-proficient cases which currently do not benefit from PARP inhibition.
Keywords: immunology, tumours, tumor microenvironment
Background
Around 50% of high-grade serous ovarian carcinomas (HGSC) have deficiencies in homologous recombination (HR) pathways.1 2 Poly ADP ribose polymerase (PARP) inhibitors theoretically target tumors with HR-deficiencies. They are currently approved for use as a maintenance therapy in those patients with platinum-sensitive recurrent ovarian cancer who had recently responded to platinum-based chemotherapy regardless of HR status. They are also approved as a monotherapy for the treatment of patients with BRCA mutation-associated advanced ovarian cancer after treatment with multiple chemotherapies.3 Subgroup analyses from the phase III Nova (niraparib) and ARIEL3 (rucaparib) trials demonstrated the dramatic efficacy of PARP inhibitors in HGSC patients with HR-deficient tumors. In contrast, the efficacy was rather limited for HR-proficient tumors.3 4 Therefore, there is a need to improve outcomes of HGSC patients with HR-proficient tumors. New treatment modalities, such as immunotherapy, are urgently required.
Tumors exhibit multiple somatic mutations in the course of development. Mutational burden varies across different types of human cancers.5 Neoantigens derived from such tumor-specific mutations are good potential targets for effective antitumor immune responses because they are foreign to the immune system.6–8 Recent reports document that a clinical benefit of immune checkpoint inhibitors (ICI) was more likely to be achieved in melanoma and lung cancer patients with tumors harboring abundant neoantigens,9–12 although it is becoming increasingly clear that patients with high mutation burden do not always have clinical benefits by ICI possibly due to many mechanisms dampening antitumor immune responses in the tumor microenvironment. In contrast, the efficacy of ICI has been limited in cancers such as HGSC with a lower tumor mutation burden (TMB) and thus fewer potential neoantigens. A phase II trial of pembrolizumab for ovarian cancer yielded a response rate for HGSC of only 8.0%.13 Nonetheless, a small number of patients obviously do benefit from ICI and experience durable responses.14 Therefore, in those types of cancers, stricter criteria for patient selection would be desirable. Other than the TMB, antigen presentation machinery, interferon (IFN)-γ signatures and combinations of those factors might be employed for this purpose.
In the present study, we investigated the status of neoantigen load and immunologic characteristics of HGSCs, especially focusing on HR-proficient cancer using integrated molecular analysis to determine which tumors would be the best candidates for immunotherapy.
Methods
Sample description and preparation
Genomic DNA and total RNA were extracted from frozen tumor samples after cryostat sectioning, using DNA and AllPrep DNA/RNA Mini Kits (Qiagen, Hilden, Germany). Genomic DNA was isolated from matched peripheral blood samples using QIAamp DNA Mini Kits (Qiagen). Eighty HGSC samples were analyzed in this study.
Whole-exome sequencing, read mapping and detection of somatic mutations
Paired tumor and blood genomic DNA libraries were constructed according to the protocol provided with the KAPA Hyper Prep Kit (Kapa Biosystems). Whole-exome capture was performed with the SureSelect Human All Exon kit V.4 and V.5 (Agilent Technologies) following the manufacturer's protocols. We sequenced exome capture libraries on the HiSeq 2000 platform according to the manufacturer's instructions, and 2×100 bp paired-end reads were generated. Image analysis and base calling were performed using the Illumina pipeline with default settings.15 Exome reads were independently mapped to the human genome (GRCh37/hg19) using Burrows-Wheeler Aligner and Novoalign software. Reads with a minimal editing distance to the reference genome were taken to represent optimal alignments. Bam files were then locally realigned with short-read micro re-aligner (SRMA). Normal-tumor pair bam files were processed using the in-house genotyper karkinos (https://sourceforge.net/projects/karkinos/). OxoG artifacts were removed by the D-ToxoG program.16 RefSeq gene annotation was performed by ANNOVAR.17
RNA-seq
RNA sequencing was performed as previously described18 for 80 HGSC samples that had RNA of sufficient quality and quantity. An RNA-sequencing library was prepared using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) according to the manufacturer’s protocol. Briefly, 1 µg of total RNA was purified using oligo dT magnetic beads and poly A+RNA were fragmented at 94°C for 2 min. complementary DNA (cDNA) was synthesized using SuperScript II (Invitrogen) and adapter-ligated cDNA was amplified with 12 cycles of PCR. Each library was sequenced using HiSeq 2000, loading four libraries per lane of the flowcell, which produced an average of 59.2 million reads of 101-cycle reads for each sample. RNA-sequencing reads were aligned to a human transcriptome database (University of California Santa Cruz (UCSC) genes) and the reference genome (GRCh37/hg19) using the STAR algorithm (V.2.5.2a).19 Finally, fragments per kilobase of exon per million fragments mapped (FPKM) values were calculated for each UCSC gene while considering strand-specific information.
Methylation array
DNA bisulfite conversion was performed with EZ DNA Methylation Kits (Zymo Research)for tumor genomic DNA (500 ng). All samples were run with Infinium HumanMethylation450 BeadChip Kits (Illumina), which target >4,50,000 methylation sites, according to the manufacturer's protocol. To determine BRCA1 and RAD51C promoter methylation, eight and 15 probes for the promoter regions in the corresponding genes respectively were used as reported previously.20
Immune-related gene expression analyses
Gene set enrichment analysis (GSEA) was performed with GSEA V.3.0 software with default parameters. The immune cell-related gene sets provided by Angelova et al were used and the association with each immune cell gene set was represented by a normalized enrichment score.21 An immune cell type was considered significantly enriched when the familywise error rate (FWER, p value) was <0.05. Gene sets for ‘immune_responses’ in the gene ontology (GO) biological process (BP) retrieved from c5 in the Molecular Signature Database (MSigDB) V.7.0 and hallmark gene sets (MSigDB V.7.0) were used for the analysis.22
To quantify tumor-infiltrating immune cells (TICs), we analyzed RNA-seq data with CIBERSORT algorithm with default parameters.23 Twenty-two immune cell subtypes were analyzed.
HLA typing and MHC class I epitope binding prediction from whole exome and RNA sequencing data
Human leukocyte antigen (HLA)-typing was performed on the 80 patients from whole exome sequencing data of normal tissues or peripheral blood mononuclear cells (PBMCs) using HLA typing software (Omixon target HLA) (online supplementary table S1). Mutated peptides derived from missense mutations were used for major histocompatibility complex (MHC) class I binding prediction as previously described.24 25 The missense, insertion/deletion and non-stop mutations in genes with low expression that had FPKM values<1 were eliminated. Long peptides containing the predicted mutation or of wild-type were assessed using the Immune Epitope Database and Analysis Resource (http://www.iedb.org/) offline; 9-mer and 10-mer peptides were selected, each predicted to bind to a specific HLA allele for each patient. Mutated peptides (9-mer and 10-mer) which had an IC50 value below 500 nM were regarded as candidate neoepitopes from missense mutations (online supplementary table S2) and insertion/deletion (indel) and non-stop mutations (online supplementary table S3). We defined one neoantigen as a mutation producing any number of HLA-A, HLA-B, and HLA-C-restricted potential neoepitopes (online supplementary table S4).
jitc-2019-000375supp001.pdf (21.3KB, pdf)
jitc-2019-000375supp002.pdf (481.2KB, pdf)
jitc-2019-000375supp003.pdf (210.7KB, pdf)
jitc-2019-000375supp004.pdf (168.2KB, pdf)
Statistical analysis
Overall survival (OS) times were calculated as the number of days from surgery to death, or the last time the patient was known to be alive. Progression-free survival (PFS) times were calculated as the time elapsed between surgery and tumor progression or death. Survival was plotted according to the Kaplan-Meier method. The log rank test was used to examine the significance of differences in survival between groups. Comparison of results was by an unpaired, two-tailed Student t-test using GraphPad Prism 5 (GraphPad Software). A value of p<0.05 was considered significant.
Results
Patient characteristics
In total, 80 cases of HGSC were analyzed in this study. The clinicopathological characteristics of these patients including age, stage, residual tumor after surgery, and platinum-sensitivity are summarized in table 1. All patients received standard chemotherapy such as carboplatin plus paclitaxel; no patients received PARP inhibitors or immune checkpoint blockade such as programmed cell death-1/programmed cell death-ligand 1 (PD-1/PD-L1) blockade therapies.
Table 1.
Clinicopathological features of 80 high-grade serous ovarian carcinomas
| Patient characteristics | All patients (n). | HR-deficient (n). (%) | HR-proficient (n). (%) | P value | |
| 80 | 34 | 46 | |||
| Age | ≥55 | 52 | 17 (50) | 35 (76) | 0.0192 |
| <55 | 28 | 17 (50) | 11 (24) | ||
| Stage | I | 3 | 0 (0) | 3 (7) | 0.165 |
| II | 7 | 2 (6) | 5 (11) | ||
| III | 50 | 20 (59) | 30 (65) | ||
| IV | 20 | 12 (35) | 8 (17) | ||
| Residual tumor | Yes | 63 | 27 (79) | 36 (78) | 1 |
| No | 17 | 7 (21) | 10 (22) | ||
| Platinum-sensitivity* | Sensitive | 30 | 19 (60) | 11 (31) | 0.0039 |
| Partial sensitive | 16 | 8 (25) | 8 (22) | ||
| Resistant | 19 | 3 (9) | 16 (44) | ||
*The patients who refused or dropped out of chemotherapy (n=3) and who had no disease progression (n=12) were excluded.
†Significant values are underlined.
HR, homologous recombination.
Classification of HGSC patients based on HR status using NGS and methylation arrays
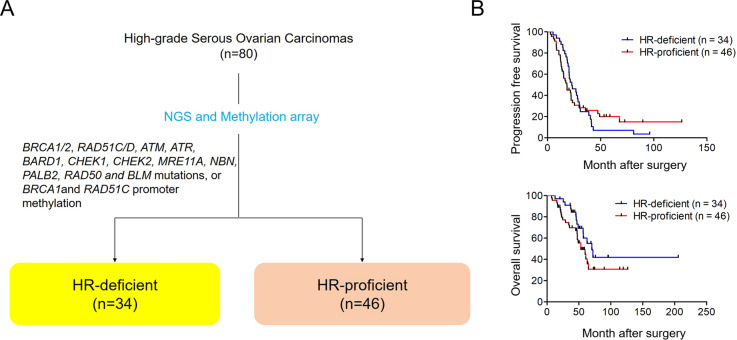
HGSC patients with HR-deficient tumors may derive more clinical benefit from treatment with PARP inhibitors, relative to patients with HR-proficient tumors. Therefore, we here focused on HR-proficient patients for whom this treatment modality is less effective. We first determined the HR-deficient or HR-proficient status of the 80 HGSC patients using exome sequencing and methylation arrays. We defined tumors with BRCA1/2, RAD51C/D, ATM, ATR, BARD1, CHEK1, CHEK2, MRE11A, NBN, PALB2, RAD50 and BLM mutations, or BRCA1 and RAD51C promoter methylation, as HR-deficient in this study. In this way, we classified 34 (43%) of patients as having HR-deficient tumors (figure 1A and table 1). Older patients (≥55 years) tended to have HR-proficient tumors but younger patients’ tumors were more likely to be HR-deficient. Although platinum-sensitivity is significantly higher in HR-deficient than HR-proficient tumors (table 1), there was no difference in PFS or OS between the patients in this cohort (figure 1B).
Figure 1.
(A) Classification based on HR status in 80 HGSCs using NGS and methylation arrays. (B) Kaplan-Meier progression-free and overall survival of 34 patients with HR-deficient tumors and 46 with HR-proficient tumors. HGSC, high-grade serous ovarian carcinoma; HR, homologous recombination; NGS, next-generation sequencing.
Predicted neoantigens and HLA expression in 80 HGSCs
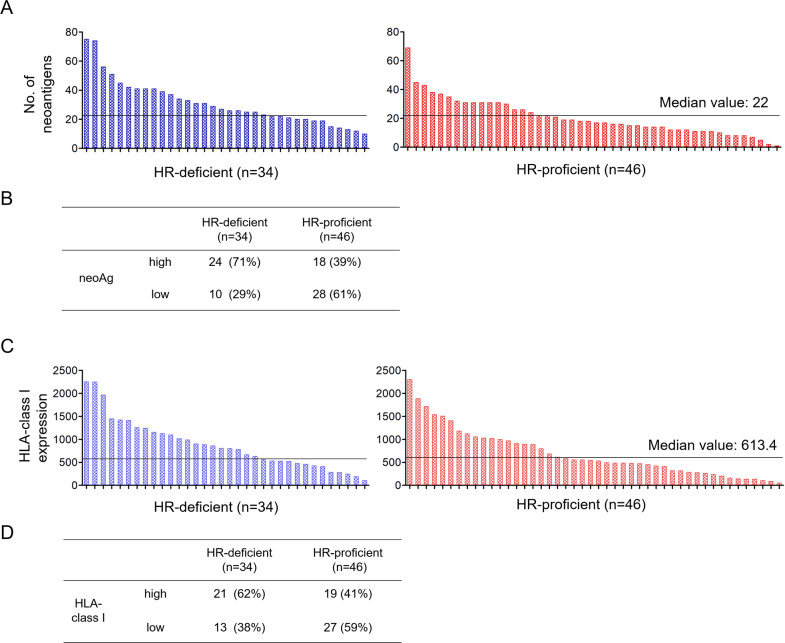
Neoantigens derived from somatic mutations are major targets for antitumor immune responses. Recent reports documented that HR-deficient tumors have higher neoantigen loads than HR-proficient HGSCs.26 Therefore, we surveyed our cohort of 80 HGSC and compared the number of predicted neoantigens derived from non-synonymous mutations including missense, indel and non-stop mutations in HR-deficient versus HR-proficient tumors. The number of non-synonymous mutations from whole-exome sequencing data ranged from nine to 286 (median value: 82) (online supplementary table S1), suggesting that most patients in this cohort were categorized into low/medium TMB group, as previously reported.5 We next identified candidate neoepitopes derived from non-synonymous mutations in genes with FPKM values≥1 in each tumor based on MHC class I binding prediction scores (IC50 <500 nM) according to NetMHCpan V.2.8, as previously reported24 25 (online supplementary tables S2 and S3). We defined antigenic mutations potentially generating one or more neoepitopes as predicted neoantigens (online supplementary table S4). The number of predicted neoantigens ranged from one to 75 (median number, 22) in these 80 cases (online supplementary table S1). As expected, this was lower in HR-proficient than in HR-deficient tumors (Student t-test, p=0.0018) (figure 2A). However, 39% of patients with HR-proficient tumors still had numbers of neoantigens higher than the median values of all 80 cases, falling into the neoAghi group within this HGSC cohort (figure 2B).
Figure 2.
Neoantigen load and HLA-class I expression in HR-deficient and HR-proficient HGSC. (A) The number of predicted neoantigens derived from missense, insertion/deletion (indel) and non-stop mutations was determined based on MHC class I binding prediction scores (IC50 <500 nM) and their gene expressions (FPKM ≥1) in HR-deficient and HR-proficient tumors (median, 22). (B) Patients were stratified according to their higher or lower than median numbers of predicted neoAgs, designated neoAghi (with ≥22) or neoAglo (with <22). (C) Mean FPKM values for HLA-A, HLA-B, and HLA-C (HLA FPKM) for HR-deficient and HR-proficient tumors (median, 613.4). (D) Patients were stratified according to their higher or lower than median values, designated HLAhi (≥613.4) or HLAlo (<613.4). FPKM, fragments per kilobase of exon per million fragments mapped; HGSC, high-grade serous ovarian carcinoma; HLA, human leukocyte; HR, homologous recombination; NS, not significant.
HLA expression is also important for antitumor immune responses because neoepitopes are recognized by antigen-specific T cells as peptide-HLA complexes on the cell surface. It is widely accepted that tumors can evade antitumor immune responses by downregulating MHC class I expression, as well as by antigen loss. A subset of HGSC cases with decreased expression of MHC class I has been reported to have worse clinical outcomes.27 Therefore, we evaluated the expression of HLA-A, HLA-B and HLA-C using RNA-seq data in HR-deficient and HR-proficient tumors. The mean FPKM values for HLA-A, HLA-B, and HLA-C ranged from 54.1 to 2300.4 (median, 613.4) in all 80 cases (figure 2C), but there was no significant difference between HR-deficient and HR-proficient patients (p=0.1122). Nonetheless, 41% of patients with HR-proficient tumors did have higher than median MHC class-I expression (HLAhi, above the median value of 613.4), whereas >60% of HR-deficient tumors were HLAhi (figure 2D).
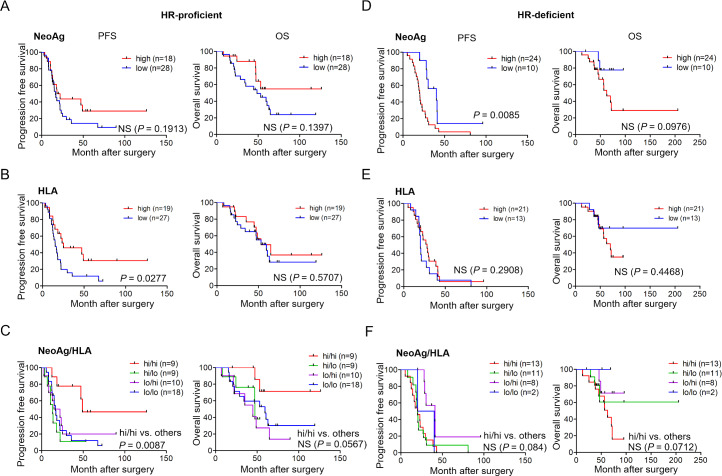
Neoantigen load and HLA-class I expression together identify a subgroup of patients with HR-proficient, but not HR-deficient, tumors that has superior survival
We next investigated neoantigen load and HLA-class I expression, and their relevance for prognosis in HR-proficient HGSC (n=46). There was no difference between the neoAghi group (n=18) and neoAglo group (n=28) in clinical outcomes (both PFS and OS) (log rank test, p=0.1913 and p=0.1397, respectively) (figure 3A). We also investigated HLA-class I expression and its relevance for clinical outcome. The HLAhi group (n=19) had a significantly more favorable PFS but not OS, relative to the HLAlo group (n=27) (log rank test, p=0.0277 and p=0.5707, respectively) (figure 3B).
Figure 3.
Prognosis according to the neoantigen load, HLA-class I expression or both in patients with HR-proficient or HR-deficient tumors. Kaplan-Meier PFS and OS of 46 HR-proficient HGSC patients stratified according to neoAg number (A), HLA-class I expression (B), and the combination of both neoAg load and HLA expression (C). Kaplan-Meier PFS and OS of 34 HR-deficient HGSC patients stratified according to neoAg number (D), HLA-class I expression (E), and both (F). Low, high designates values below or above the median. Statistical analysis was by log-rank testing. HGSC, high-grade serous ovarian carcinoma; HLA, human leukocyte antigen; HR, homologous recombination; NS, not significant; OS, overall survival; PFS, progression-free survival.
We further stratified these patients according to a combination of the number of neoantigens and the levels of HLA-class I expression. For this, patients were divided into four groups: (1) neoAghi/HLAhi tumors, (2) neoAghi/HLAlo, (3) neoAglo/HLAhi and (4) neoAglo/HLAlo. As shown in figure 3C, patients with neoAghi/HLAhi tumors had notably longer PFS but not OS than others (log rank test, p=0.0087 and 0.0567, respectively). The same analysis was applied to HR-deficient HGSC (n=34). However, patients in the neoAghi (n=24) (figure 3D), HLAhi (n=21) (figure 3E), or neoAghi/HLAhi groups (n=13) (figure 3F) did not have longer PFS or OS. Therefore, these results show that patients with neoAghi/HLAhi tumors have a better clinical course relative to the other neoAg/HLA groups, but only if their tumors are HR-proficient.
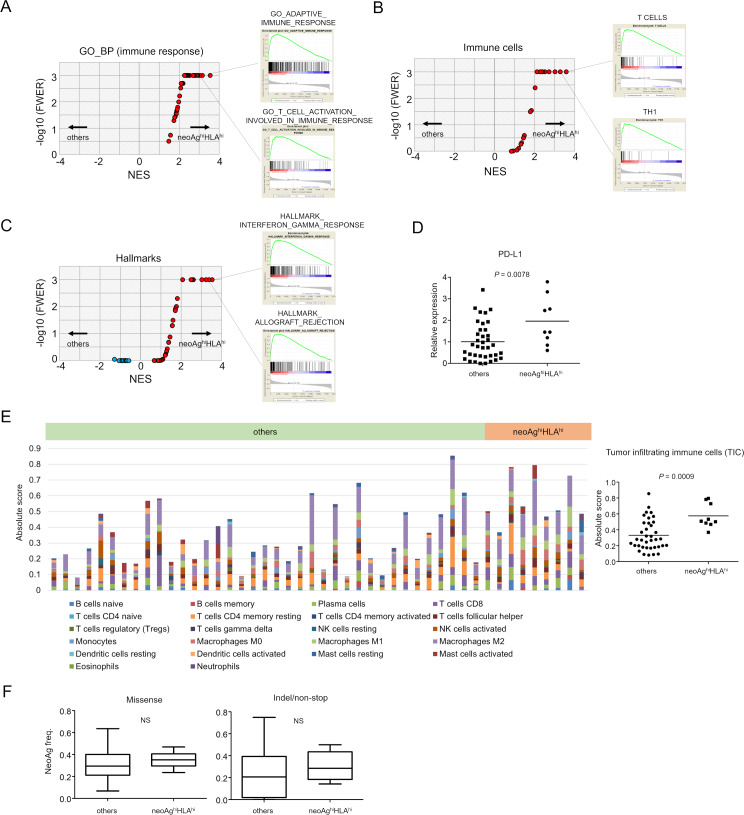
A subgroup of HR-proficient neoAghi/HLAhi tumors exhibits an immunologically ‘hot’ phenotype
To investigate whether the better prognosis of the HR-proficient neoAghi/HLAhi subgroup relative to all other HR-deficient subgroups is related to immune parameters in the tumor microenvironment, we next compared immunologic signatures between neoAghi/HLAhi (n=9) and the others (n=37) using RNA-seq. We performed GSEA. We used the 72 gene sets for ‘immune_responses’ in the GO BP retrieved from c5 in the MSigDB V.7.0. We found 42 out of 72 gene sets such as ‘GO_ADAPTIVE_IMMUNE_RESPONSE’ and ‘GO_T_CELL_ACTIVATION_INVOLVED_IN_IMMUNE_RESPONSE’ were significantly enriched in the neoAghi/HLAhi group (figure 4A) (online supplementary table S5). We also found that the gene sets for ‘T cells’ (FWER, p<0.001), ‘Th1’ (FWER, p<0.001), ‘TGD’ (FWER, p<0.001), ‘effector memory CD8’ (FWER, p<0.001), and certain others in 28 immune cell subpopulations (gene sets provided by Angelova et al) (figure 4B) (online supplementary table S6) and ‘HALLMARK_ALLOGRAFT_REJECTION’ (FWER, p<0.001), ‘HALLMARK INTERFERON GAMMA RESPONSE’ (FWER, p<0.001) in hallmark gene sets were significantly enriched in the neoAghi/HLAhi group (figure 4C) (online supplementary table S7). CD274 (PD-L1) expression, a known candidate biomarker for ICIs, was also higher in the neoAghi/HLAhi subgroup with statistical significance (Student t-test, p=0.0078) (figure 4D). We also tried to estimate TICs by CIBERSORT.23 Different types of immune cells with antitumor or protumor functions were observed as illustrated by absolute immune fraction scores (figure 4E left and online supplementary figure S1). Absolute scores of TICs were significantly higher in the neoAghiHLAhi tumors (Student t-test, p=0.0009) (figure 4E right).
Figure 4.
Gene expression analysis of neoAghiHLAhi versus others in HR-proficient tumors. RNA-seq analysis of 46 specimens showing gene expression between patients with neoAghiHLAhi tumors (n=9) and all others (n=37). Volcano plots demonstrating enrichment of gene sets in each group. The x-axis indicates the NES; the y-axis indicates −log10 (FWER, p value) calculated from the GSEA using ‘immune_responses’ in the GO_BP from c5 in the MSigDB V.7.0 (A), immune cell-related gene sets21 (B) and hallmark gene sets (C). Gene sets showing FWER, p<0.001 are plotted as p=0.001: y-axis is 3. Representative enrichment plots are also shown on the right side. (D) CD274 (PD-L1) expression by neoAghiHLAhi and other tumors. (E) Twenty-two immune cell subtypes were analyzed in 46 HR-proficient tumors by CIBERSOT (left) and the absolute scores of tumor-infiltrating immune cells in neoAghiHLAhi tumors (n=9) and all others (n=37) were shown (right). (F) Neoantigen frequencies, the ratios of neoantigens per missense mutations (left) or indel/non-stop mutations (right), are shown. BP, biological process; GO, gene ontology; FWER, familywise error rate; GSEA, gene set enrichment analysis; HLA, human leukocyte antigen; HR, homologous recombination; MSigDB, Molecular Signature Database; NES, normalized enrichment score; NS, not significant.
jitc-2019-000375supp005.pdf (21.2KB, pdf)
jitc-2019-000375supp006.pdf (19.2KB, pdf)
jitc-2019-000375supp007.pdf (21.5KB, pdf)
jitc-2019-000375supp008.pdf (1.5MB, pdf)
jitc-2019-000375supp009.pdf (1.5MB, pdf)
It has been shown that certain cancers known to be immunogenic exhibit preferential depletion of neoantigenic mutations within the totality of mutations in the tumor.25 28 29 To test whether the same may apply in an immunogenic subgroup of HGSC, we compared the ‘neoantigen frequency’ (the ratio of neoantigen per somatic mutation) between neoAghi/HLAhi (n=9) and the others (n=37). However, there were no significant differences in the ratios of neoantigens per missense or indel/non-stop mutations (figure 4F).
Comparison of immunologic signatures between HR-proficient and HR-deficient neoAghi/HLAhi tumors
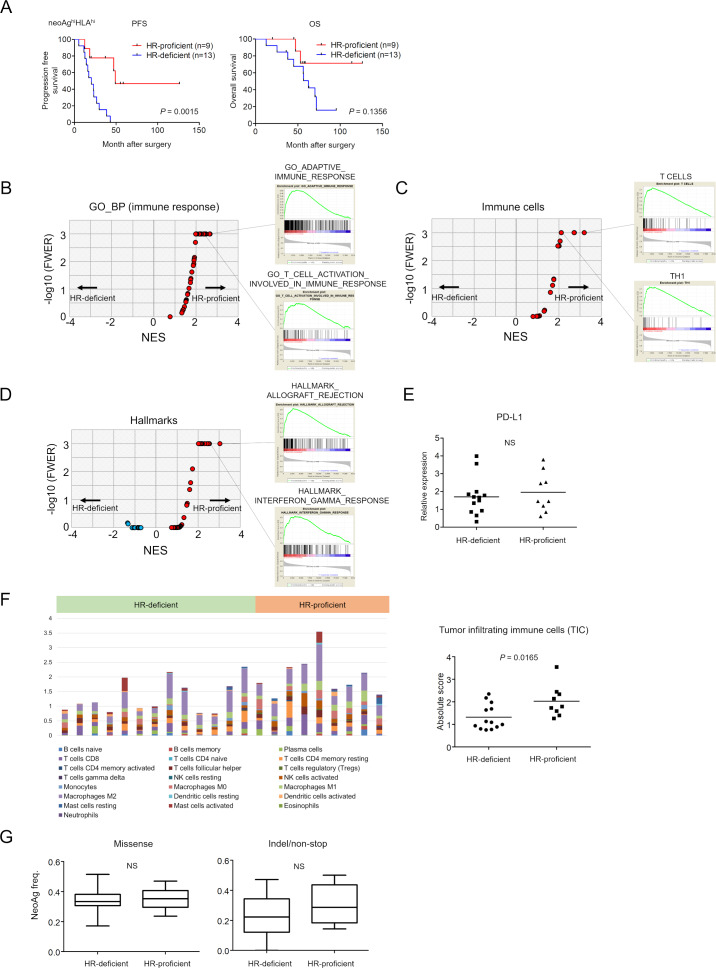
The subgroup of patients with neoAghi/HLAhi HR-proficient tumors had a better clinical outcome than patients with tumors of the same phenotype but with HR-deficiency, both for PFS and OS (log rank test, p=0.0015 and p=0.1356, respectively) (figures 3C, F and 5A). To investigate what causes this difference in clinical benefit between HR-proficient and HR-deficient patients despite their being in the same ‘high neoantigen load and high HLA expression’ subgroup, we next compared immune-related gene expression between them. We performed the same analysis as in figure 4, In GSEA using 72 gene sets for ‘immune_responses’ in GO BP from c5 in MSigDB V.7.0, we found 29 out of 72 gene sets such as ‘GO_ADAPTIVE_IMMUNE_RESPONSE’ and ‘GO_T_CELL_ACTIVATION_INVOLVED_IN_IMMUNE_RESPONSE’ were significantly enriched in the HR-proficient group (figure 5B and online supplementary table S8). We also found that the gene sets for ‘T cells’ (FWER, p<0.001), ‘Th1’ (FWER, p<0.0001), ‘TGD’ (FWER, p<0.001), ‘effector memory CD8’ (FWER, p<0.001), and others in 28 immune cell subpopulations were significantly enriched in patients with HR-proficient tumors (figure 5C) (online supplementary table S9). Furthermore, ‘HALLMARK_ALLOGRAFT_REJECTION’ (p<0.001), ‘HALLMARK_INTERFERON_ALPHA_RESPONSE’ (FWER, p<0.001) and ‘HALLMARK_INTERFERON_GAMMA_RESPONSE’ (FWER, p<0.001), in hallmark gene sets were significantly enriched in HR-proficient tumors (figure 5D) (online supplementary table S10). There was no statistically significant difference between HR-proficient and HR-deficient tumors for CD274 (PD-L1) expression (figure 5E). We estimated TICs by CIBERSORT again. Different types of immune cells with antitumor or protumor functions were observed (figure 5F left and online supplementary figure S2) and absolute scores of TICs were significantly higher in the HR-proficient tumors (Student t-test, p=0.0165) (figure 5F right).
Figure 5.
Survival and gene expression analysis of HGSC patients with HR-proficient or HR-deficient neoAghiHLAhi tumors. (A) Kaplan-Meier PFS and OS curves of 22 neoAghiHLAhi HGSC patients stratified according to HR-proficiency (n=9) versus HR-deficiency (n=13). RNA-seq analysis of 22 neoAghiHLAhi HGSC specimens showing gene expression between patients with HR-proficient (n=9) or HR-deficient (n=13) tumors. Volcano plots demonstrating enrichment of gene sets in each group. The x-axis indicates the NES; the y-axis indicates −log10 (FWER, p value) calculated from the GSEA using ‘immune_responses’ in the GO_BP from c5 in the MSigDB V.7.0 (B), immune cell-related gene sets21 (C) and hallmark gene sets (D). Gene sets showing FWER, p<0.001 are plotted as p=0.001: y-axis is 3. Representative enrichment plots are also shown on the right side. (E). CD274 (PD-L1) expression by HR-proficient and HR-deficient tumors. (F) Twenty-two immune cell subtypes were analyzed in 22 neoAghiHLAhi HGSC tumors by CIBERSOT (left) and the absolute scores of tumor-infiltrating immune cells in HR-proficient (n=9) and HR-deficient (n=13) tumors were shown (right). (G) Neoantigen frequencies, the ratios of neoantigens per missense mutations (left) or indel/non-stop mutations (right), are shown. BP, biological process; GO, gene ontology; GSEA, gene set enrichment analysis; FWER, familywise error rate; HGSC, high-grade serous ovarian carcinoma; HLA, human leukocyte antigen; HR, homologous recombination; MSigDB, Molecular Signature Database; NES, normalized enrichment score; NS, not significant; OS, overall survival; PFS, progression-free survival.
jitc-2019-000375supp010.pdf (22KB, pdf)
jitc-2019-000375supp011.pdf (19.4KB, pdf)
jitc-2019-000375supp012.pdf (21.5KB, pdf)
jitc-2019-000375supp013.pdf (1.4MB, pdf)
We again compared the ‘neoantigen frequency’ (the ratio of neoantigen per somatic mutation) between HR-proficient (n=9) and HR-deficient (n=13) tumors. However, there were no significant differences in the ratios of neoantigens per missense or indel/non-stop mutations (figure 5G).
Discussion
PARP inhibition has recently yielded encouraging results in the treatment of HR-deficient HGSCs, but is less effective in HR-proficient disease. New treatment modalities such as immunotherapy are urgently required for HR-proficient HGSC. Here, we have compared HR-proficient and HR-deficient HGSC regarding neoantigen load and HLA expression in order to determine whether these might represent biomarker candidates for predicting patient response to checkpoint blockade. We have identified a subgroup of HGSC patients whose HR-proficient tumors have a high neoantigen load and high HLA-class I expression (an ‘immunologically hot’ phenotype). This was associated with a better prognosis. These patients might therefore be candidates for treatment with ICI rather than PARP inhibitors.
Neoantigens derived from somatic mutations are well-recognized as good targets for T cells both over the natural course of tumor development and under several cancer therapies.30–35 However, unlike melanoma or non-small cell lung cancer, the TMB and thus neoantigen load in HGSC is not high.5 A recent report confirmed that, as expected, HR-deficient ovarian cancers have a higher TMB and more neoantigens than HR-proficient tumors.26 The former had greater T-cell infiltration and are therefore considered candidates for ICI. However, a biomarker analysis of the KEYNOTE-100 study, which evaluated the efficacy of pembrolizumab in ovarian cancer patients, found that neither BRCA nor HR status predicted the response, but that PD-L1 expression and a T-cell gene expression profile did.36 Our data support these previous findings26 of higher mutation and neoantigen burden in HR-deficient than HR-proficient tumors. Additionally, however, we show that a subset of the latter also manifested a strong immune response and had a better prognosis. Thus, patients with HR-proficient tumors which nonetheless possessed a neoAghiHLAhi immune signature also had a better prognosis. This may be because HR-proficient tumors with varying driver genes have different immunologic profiles.
Previous reports suggested that BRCA1 mutations or loss of BRCA1 function contributes to weaker Type I and II IFN responses, leading to reduced antigen presentation function and/or poor apoptosis of tumor cells.37–39 In fact, in the present study, we found that genes for Type I and II IFN responses and cell cycle checkpoints were lower in HR-deficient relative to HR-proficient patients in the neoAghiHLAhi group. Indeed, it was mostly the case that BRCA1 mutations or functional impairment of BRCA1 was observed in our HR-deficient patients, which may thus be related to poorer immune responses and poor apoptosis of tumor cells, resulting in an unfavorable prognosis in certain individuals even in the neoAghiHLAhi group. In addition, reduced antigen presentation function due to somatic mutations or loss of heterozygosity in HLA genes that might be induced by HR-deficiency should be investigated.
Patients with HR deficiency in our study have relatively abundant mutations compared with those with HR-proficiency, however, neoAghiHLAhi group in HR-deficient patients fell into immunologically cold phenotype possibly due to the mechanisms as mentioned above. Our data may explain as one example why all the patients with high TMB do not have clinical efficacy from standard and available immunotherapy agents now in clinical development. There could be different immunosuppressive mechanisms that would inhibit an immunologic response in different types of cancer.
There are several limitations to this study. First, we focused solely on neoAgs. We may therefore have missed other important types of antigen. Other antigens acting as antitumor immune targets, including cancer-testis antigens, viral-derived antigens, or other ovarian cancer-associated antigens may also play a role, but were beyond the scope of the present study. Second, this study was carried out mostly based on in silico analyses. Therefore, neoAg expression and its relevance for antigen-specific T-cell responses needs to be further investigated using tumor infiltrating lymphocytes (TILs) or PBMCs in addition to immune-related gene expression analysis.
In conclusion, we investigated the neoantigen landscape and immunologic features of HGSCs, especially focusing on HR-proficient tumors, and identified a subgroup of patients that may be potential candidates for immunotherapy.
Conclusions
ICI might be an alternative to explore in HR-proficient cases which currently do not benefit from PARP inhibition.
Footnotes
HM and KH contributed equally.
Contributors: HM and KH performed the described studies, analyzed data, and prepared the manuscript. SY and HA performed whole exome and RNA sequencing. KH, TK, YT, YK, RY, TN, and MM analyzed the data. KO, YI, KA, AY, AN, and KF provided clinical suggestions. KK advised on study design. All authors read and approved the final manuscript.
Funding: The work was supported by a Grant-in-Aid for P-CREATE by Japan Agency for Medical Research and Development (Kosei Hasegawa) under grant numbers 16cm0106502h0001; a Grant-in-Aid for Scientific Research of the Ministry of Education, Culture, Sports, Science and Technology of Japan (Hirokazu Matsushita, Kosei Hasegawa, Kazuhiro Kakimi) under grant numbers 16K07162, 16K11152, and 16H04708.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: HGSC tumors and peripheral blood from the same patients were collected by the University of Tokyo Hospital and Saitama Medical University International Medical Center following the approval of the institutional review boards and patients' written informed consent (ID: G3531 and 13-098, respectively).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available in a public, open access repository. The data sets generated and analyzed during the current study are available in the Japanese Genotype-Phenotype Archive under accession number JGAS00000000209.
References
- 1.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, et al. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov 2015;5:1137–54. 10.1158/2159-8290.CD-15-0714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey MK, Pothuri B. Homologous recombination deficiency (HRD) testing in ovarian cancer clinical practice: a review of the literature. Gynecol Oncol Res Pract 2017;4:4. 10.1186/s40661-017-0039-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med 2016;375:2154–64. 10.1056/NEJMoa1611310 [DOI] [PubMed] [Google Scholar]
- 4.Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:1949–61. 10.1016/S0140-6736(17)32440-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava PK. Neoepitopes of cancers: looking back, looking ahead. Cancer Immunol Res 2015;3:969–77. 10.1158/2326-6066.CIR-15-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher TN, Hacohen N. Neoantigens encoded in the cancer genome. Curr Opin Immunol 2016;41:98–103. 10.1016/j.coi.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 8.Ward JP, Gubin MM, Schreiber RD. The role of neoantigens in naturally occurring and therapeutically induced immune responses to cancer. Adv Immunol 2016;130:25–74. 10.1016/bs.ai.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbone DP, Reck M, Paz-Ares L, et al. First-Line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 2017;376:2415–26. 10.1056/NEJMoa1613493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matulonis UA, Shapira-Frommer R, Santin AD, et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Ann Oncol 2019;30:1080–7. 10.1093/annonc/mdz135 [DOI] [PubMed] [Google Scholar]
- 14.Hamanishi J, Mandai M, Ikeda T, et al. Safety and antitumor activity of anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. J Clin Oncol 2015;33:4015–22. 10.1200/JCO.2015.62.3397 [DOI] [PubMed] [Google Scholar]
- 15.Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014;46:583–7. 10.1038/ng.2984 [DOI] [PubMed] [Google Scholar]
- 16.Costello M, Pugh TJ, Fennell TJ, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 2013;41:e67. 10.1093/nar/gks1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomura M, Mukasa A, Nagae G, et al. Distinct molecular profile of diffuse cerebellar gliomas. Acta Neuropathol 2017;134:941–56. 10.1007/s00401-017-1771-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patch A-M, Christie EL, Etemadmoghadam D, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015;521:489–94. 10.1038/nature14410 [DOI] [PubMed] [Google Scholar]
- 21.Angelova M, Charoentong P, Hackl H, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol 2015;16:64. 10.1186/s13059-015-0620-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberzon A, Birger C, Thorvaldsdóttir H, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karasaki T, Nagayama K, Kuwano H, et al. Prediction and prioritization of neoantigens: integration of RNA sequencing data with whole-exome sequencing. Cancer Sci 2017;108:170–7. 10.1111/cas.13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsushita H, Hasegawa K, Oda K, et al. The frequency of neoantigens per somatic mutation rather than overall mutational load or number of predicted neoantigens per se is a prognostic factor in ovarian clear cell carcinoma. Oncoimmunology 2017;6:e1338996. 10.1080/2162402X.2017.1338996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strickland KC, Howitt BE, Shukla SA, et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016;7:13587–98. 10.18632/oncotarget.7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariya T, Hirohashi Y, Torigoe T, et al. Prognostic impact of human leukocyte antigen class I expression and association of platinum resistance with immunologic profiles in epithelial ovarian cancer. Cancer Immunol Res 2014;2:1220–9. 10.1158/2326-6066.CIR-14-0101 [DOI] [PubMed] [Google Scholar]
- 28.Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mlecnik B, Bindea G, Angell HK, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016;44:698–711. 10.1016/j.immuni.2016.02.025 [DOI] [PubMed] [Google Scholar]
- 30.Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012;482:400–4. 10.1038/nature10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nejo T, Matsushita H, Karasaki T, et al. Reduced neoantigen expression revealed by longitudinal Multiomics as a possible immune evasion mechanism in glioma. Cancer Immunol Res 2019;7:1148–61. 10.1158/2326-6066.CIR-18-0599 [DOI] [PubMed] [Google Scholar]
- 32.Robbins PF, Lu Y-C, El-Gamil M, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med 2013;19:747–52. 10.1038/nm.3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carreno BM, Magrini V, Becker-Hapak M, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015;348:803–8. 10.1126/science.aaa3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017;547:217–21. 10.1038/nature22991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017;547:222–6. 10.1038/nature23003 [DOI] [PubMed] [Google Scholar]
- 36.Ledermann JA, Shapira-Frommer R, Santin A, et al. LBA36Association of PD-L1 expression and gene expression profiling with clinical response to pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Ann Oncol 2018;29:viii728 10.1093/annonc/mdy424.043 [DOI] [PubMed] [Google Scholar]
- 37.Andrews HN, Mullan PB, McWilliams S, et al. BRCA1 regulates the interferon gamma-mediated apoptotic response. J Biol Chem 2002;277:26225–32. 10.1074/jbc.M201316200 [DOI] [PubMed] [Google Scholar]
- 38.Ouchi T, Lee SW, Ouchi M, et al. Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-gamma target genes. Proc Natl Acad Sci U S A 2000;97:5208–13. 10.1073/pnas.080469697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buckley NE, Hosey AM, Gorski JJ, et al. BRCA1 regulates IFN-gamma signaling through a mechanism involving the type I IFNs. Mol Cancer Res 2007;5:261–70. 10.1158/1541-7786.MCR-06-0250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2019-000375supp001.pdf (21.3KB, pdf)
jitc-2019-000375supp002.pdf (481.2KB, pdf)
jitc-2019-000375supp003.pdf (210.7KB, pdf)
jitc-2019-000375supp004.pdf (168.2KB, pdf)
jitc-2019-000375supp005.pdf (21.2KB, pdf)
jitc-2019-000375supp006.pdf (19.2KB, pdf)
jitc-2019-000375supp007.pdf (21.5KB, pdf)
jitc-2019-000375supp008.pdf (1.5MB, pdf)
jitc-2019-000375supp009.pdf (1.5MB, pdf)
jitc-2019-000375supp010.pdf (22KB, pdf)
jitc-2019-000375supp011.pdf (19.4KB, pdf)
jitc-2019-000375supp012.pdf (21.5KB, pdf)
jitc-2019-000375supp013.pdf (1.4MB, pdf)