Abstract
Lipid homeostasis allows cells to adjust membrane biophysical properties in response to changes in environmental conditions. In the yeast Saccharomyces cerevisiae, a downward shift in temperature from an optimal reduces membrane fluidity, which triggers a lipid remodeling of the plasma membrane. How changes in membrane fluidity are perceived, and how the abundance and composition of different lipid classes is properly balanced, remain largely unknown. Here, we show that the levels of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], the most abundant plasma membrane phosphoinositide, drop rapidly in response to a downward shift in temperature. This change triggers a signaling cascade transmitted to cytosolic diphosphoinositol phosphate derivatives, among them 5-PP-IP4 and 1-IP7, that exert regulatory functions on genes involved in the inositol and phospholipids (PLs) metabolism, and inhibit the activity of the protein kinase Pho85. Consistent with this, cold exposure triggers a specific program of neutral lipids and PLs changes. Furthermore, we identified Pho85 as playing a key role in controlling the synthesis of long-chain bases (LCBs) via the Ypk1-Orm2 regulatory circuit. We conclude that Pho85 orchestrates a coordinated response of lipid metabolic pathways that ensure yeast thermal adaptation.
Keywords: Saccharomyces cerevisiae, low temperature, phosphoinositide, 1-IP7, sphingolipid, phospholipid, sphingoid bases, triacylglyceride, TORC2-Pkh1-Ypk1 signaling module, Orm2
1. Introduction
Lipids are essential components of eukaryotic membranes and the main determinants of their functionality. The wide range of lipid classes and species is the basis of the membrane architecture, facilitates the formation of microdomains and determines their specific association with proteins in signaling circuits [1,2]. Alterations in lipid composition compromise the plasma membrane properties and are responsible of dysfunctions in specific organelles that affect almost all cellular processes. Changes in temperature and other environmental conditions [e.g., hydrostatic pressure and salinity] affect membrane properties [3,4]. A downward shift in temperature from an optimal reduces membrane fluidity, referred to as rigidification [3], which compromises its functionality and limits cell growth. To maintain physiological homeostasis, cells employ interconnected networks of synthesis and turnover of major lipid classes that allow membrane biophysical properties to be suited as needed [5,6]. A lower environmental temperature brings about a change in membrane lipid composition, characterized by an increased abundance of phospholipids with shorter chain lengths and/or unsaturated fatty acids [7,8]. How changes in membrane fluidity are perceived, and how the abundance and composition of different lipid classes is properly balanced, remain largely unknown.
In the yeast Saccharomyces cerevisiae, phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] is the most abundant species of plasma membrane phosphoinositide (PPIn), a term that refers to the phosphorylated derivatives of phosphatidylinositol, PI [9]. A schematic representation of the PPIn pathway is shown in Fig. 1. PI(4,5)P2 interacts with membrane-associated proteins to regulate important cellular processes [10,11], and plays a central role in cellular signaling, either as a precursor of second messengers or a regulator of signaling proteins such as small GTPases [12,13]. Increasing evidence also suggests that PI(4,5)P2 is an important regulator of lipid metabolism in response to thermal stress. PI(4,5)P2 levels at the plasma membrane rise in heat-stressed yeast cells, a signal that activate the cell wall integrity (CWI) pathway [14,15]. This regulation appears essential for lipid homeostasis since the mutants in Slt2, the MAPK of the pathway, show severe defects in lipid metabolism [16,17]. Furthermore, a link between elements of the CWI pathway and the synthesis of very long-chain fatty acids, which are required for the production of sphingolipids (SLs), has also been reported [18].
Fig. 1.
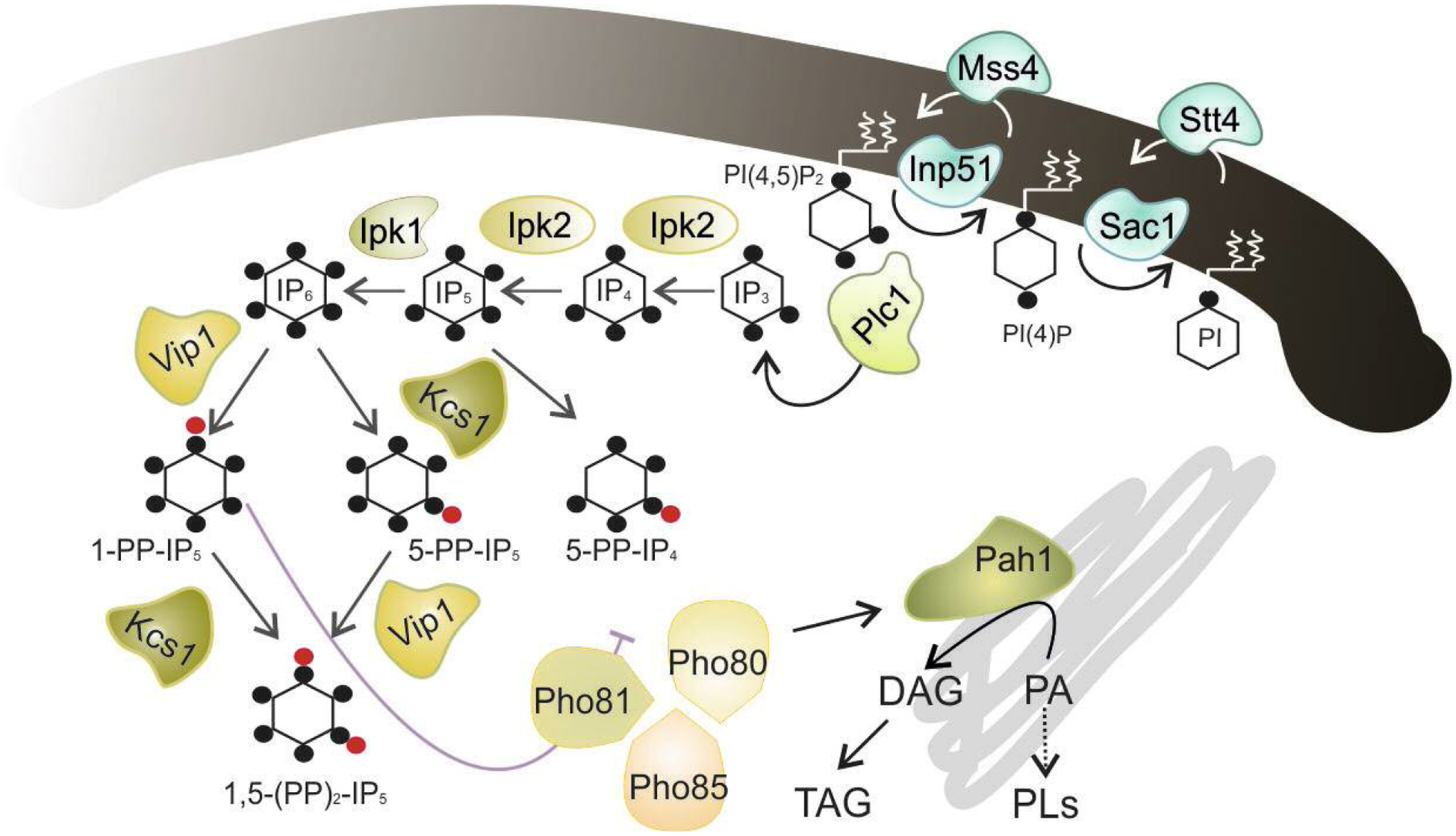
Schematic representation of the PPIn, IPs and DPIPs synthesis and its interacton with the Pho80-Pho81-Pho85 complex. The metabolic steps and enzymes involved in the synthesis and degradation of PI(4,5)P2 are shown (see [10,12,13,35,36,37] for representative reviews). The hydrolysis of PI(4,5)P2 by Plc1 generates IP3, which is sequentially phosphorylated to form in the last steps the DPIP 1-PP-IP5 (1-IP7) through the action of the IP6 kinase Vip1 [PP-IP5 kinase or IP7 kinase in mammals]. The 1-IP7 isomer acts as an inhibitor of the cyclin-regulated kinase complex Pho81-Pho80-Pho85 [37,38], which responds to phosphate availability [39], and regulates, among others, the abundance and activity of the phosphatidate phosphatase Pah1 [41], a key enzyme in the synthesis of NLs and PLs [42]. The black dot indicates a single phosphate group. The red dot represents a high-energy phosphate or pyrophosphate. For more details see text.
Enrichment of PI(4,5)P2 in the plasma membrane of heat-shocked cells may simultaneously activate additional SLs-regulatory networks. Evidence links SLs homeostasis to the activity of the yeast orthologs of mammalian Akt and SGK, Ypk1/2 [19–21]. Ypk1, and likely its paralog Ypk2, inhibits the activity of transmembrane protein Orm2, a negative regulator of serine palmitoyltransferase (SPT), which catalyzes the first step in the de novo SLs biosynthesis pathway [22,23]. In addition, Ypk1/2 phosphorylates ceramide synthase to stimulate the synthesis of complex SLs [24]. Activation of Ypk1/2 is mediated by PDK orthologs Pkh1/2 and the TORC2 kinase complex [19,25,26] which is, in turn, regulated by the PI(4,5)P2-binding proteins Slm1/2, which target Ypk1/2 to the plasma membrane [21,27,28]. A previous connection between the PI(4,5)P2 pathway and Slm1/2 in regulating the content of inositol phosphorylceramides (IPCs) has been previously established [29]. Consistently with this, heat-induced membrane stress increases the phosphorylation of Ypk by TORC2, an event that diminishes in the stt4ts and mss4ts mutants, which display low PI(4,5)P2 levels due to impaired PI- and PI(4)P-kinase activity (Fig. 1), respectively [30]. Then, Ypk kinase transmits the heat-stress signal to the SLs biosynthesis pathway via Orm phosphorylation [31], which leads to the rapid and transient accumulation of long-chain bases (LCBs) and ceramides (Cer) [32,33]. Whether PI(4,5)P2 signaling regulates the TORC2-Pkh1-Ypk1 module to adjust the SLs profile in response to a downward shift in temperature remains unknown.
In addition to its role in membrane recruitment and the regulation of cellular signaling proteins, PI(4,5)P2 serves as a substrate for phospholipase C (PLC), which generates second messengers, diacylglycerol (DAG) and inositol-1,4,5-trisphosphate (IP3) [12]. Then, IP3 is sequentially phosphorylated by several kinases (see Fig. 1) to form an array of more polar inositol phosphates (IPs) and diphosphoinositol phosphates (DPIPs), also known as inositol pyrophosphates [34]. IPs and DPIPs species have proven critical for development and nuclear signaling [35,36]. In yeast cells, the level of the DPIP isomer 1-IP7, which acts as an inhibitor of the cyclin-regulated kinase complex Pho80-Pho85 [37,38], increases in response to phosphate limitation. This regulation allows the nuclear import of the transcriptional activator Pho4, which results in the induction of phosphate metabolism genes. Although the exact mechanism by which phosphate-limitation triggers increased 1-IP7 levels is unknown, the metabolism of PI(4,5)P2 could play a role. High levels of PI(4,5)P2 obtained by the knock-out of the PPIn-5-phosphatase INP51 gene (also named SJL1) feed the DPIP-pathway and stimulate the production of 1-IP7 as it is suggested by the effect that the inp51 mutation has on Pho85-dependent read-outs [4].
The Pho85 kinase is a member of the yeast cyclin dependent kinase (CDK) family, which interacts with at least 10 different cyclins that determine its functionality and specificity [39]. In addition of its function in phosphate regulation, Pho85 is known to have a pleiotropic role on different aspects of the metabolism, including amino acid biosynthesis, carbohydrate storage and lipid metabolism [40]. Interestingly, Pho85 regulates, together with other kinases, the activity of Pah1 [41], the yeast phosphatidate phosphatase [42] that catalyzes the conversion of phosphatidate (PA) into DAG, the substrate used by yeast cells to produce triacylglycerol (TAG), and phosphatidylcholine (PC) and phosphatidylethanolamine (PE) via the Kennedy pathway. Whether Pho85 is regulated by 1-IP7 in response to cold, and whether this affects the homeostasis of neutral lipids (LNs) and PLs remains unclear.
The role of Pho85 in regulating lipid metabolism could also imply additional targets and involve different lipid classes. Pho85 influences the expression of genes containing in their promoters ICRE (inositol/choline-responsive element=UASINO) sequences, like INO1 [43], by phosphorylating the transcriptional repressor Opi1 [44]. Pho85 also phosphorylates the calcineurin-regulated transcription factor Crz1 [45], which functions, among others, in the stress response to elevated temperature [46]. In addition, there is evidence that calcineurin signaling downregulates cellular processes controlled by TORC2, including the synthesis of complex SLs [47], likely through the regulation of Slm1/2 [21]. Previous work has also shown that Pho85 functions in the phosphorylation and degradation of Lcb4, the major yeast sphingoid Long Chain Bases [LCB]-kinase [48]. Overall, these data support a regulatory role of Pho85 in the yeast SLs metabolism. Whether the SLs homeostasis is triggered in response to cold and whether this adaptation is connected to the activity of Pho85 remains to be elucidated.
Here, we examined the main cold-induced regulatory events that control lipid homeostasis in yeast cells. Our results provide evidence that PI(4,5)P2 modulates the activity of essential signaling effectors in response to cold and that Pho85 is a central regulator of lipid homeostasis. Its activity connects different lipid biosynthesis pathways to endow yeast cells with the optimal lipid profile to face with thermal stress.
2. Materials and methods
2.1. Strains, plasmids, media and culture conditions
The yeast strains, plasmids and oligonucleotides used in this study are listed in supplementary Tables S1, S2 and S3, respectively. Ypk1 and Orm2 C-terminal tagging with the 3xHA epitope was carried out by PCR-based gene tagging using plasmids pFA6a-3HA-His3MX6 [49] as a template and the appropriate target-gene specific plasmid pairs (Table S2). Previously described standard methods were used for YPD and SD media preparation [50]. SCD contained 2% glucose as a carbon source, and the appropriate amino acid drop-out mixture (Formedium, England). For the cold stress experiments, the cells grown to the mid-log phase at 30°C (OD600 ~ 0.5) were transferred to a 15°C-pre-chilled water bath, and were maintained under shaking conditions for 1–3 h, as indicated. In some experiments, cells were grown in high phosphate medium [0.67% yeast nitrogen base without amino acids and phosphate (Formedium), 0.15% KH2PO4 and 2.0% glucose in 0.025 M Na+citrate/citric acid buffer (pH 4.7)] supplemented with the appropriate drop-out mixture (Formedium). Phosphate-free medium was prepared by replacing KH2PO4 by KCl [51] in the high-phosphate medium. Stock solutions of 2 mM Myriocin (Myr) and 25 mM phytosphingosine (PhS) were prepared in ethanol, sampled in small volumes and stored at −20°C until use at the indicated concentrations.
2.2. Preparation of protein extracts and the Western blot analysis
Protein extracts were prepared, and proteins were separated by SDS-PAGE and analyzed by a Western blot as formerly described [4]. Protein extracts from HA-tagged Orm2 cells were obtained by using a NaOH-based protocol as previously described [52]. For the phosphate-affinity electrophoresis, protein samples were resolved in the SDS-PAGE gels that contained 6% acrylamide, 10 μM Phos-tag™ affinity reagent (Wako Chemicals USA, Inc.) and 20 μM Mn2+. The proteins tagged with 3xHA and 13xMyc (Pah1-Myc) were visualized by using an anti-HA rabbit polyclonal (1:2,000; cat# sc-805; Santa Cruz Biotechnology, Santa Cruz, CA) and an anti-Myc (1:2000; cat# sc-40, Santa Cruz Biotechnology) antibody, respectively. Mouse monoclonal Glucose 6 Phosphate Dehydrogenase (G6Pdh) antibody (1:3,000; cat# sc-47724) and rabbit antiPma1 (1:10,000; kindly provided by R. Serrano), were used as loading control of cytosolic and membrane-associated proteins, respectively. The secondary antibody used was horseradish peroxidase-conjugated goat anti-rabbit (1:2,000; cat# 7074; Cell Signaling, Danvers, MA, USA) or rabbit anti-mouse (1:5,000, cat# P0260; Dako, Carpinteria, CA). Blots were done and images were captured as described elsewhere [4].
2.3. Fluorescence microscopy
Cells were grown as described above in SCD medium at 30°C (control). One-ml samples were spun for 10 s, the supernatant was decanted, and cells were resuspended in the residual liquid. Aliquots of 4 μl were directly observed under a Zeiss 510 Meta Confocal microscope with a 63× Plan-Apochromat 1.4 NA Oil DIC objective lens (Zeiss). Images were acquired at identical exposures and processed identically by using Photoshop CS5.1 (Adobe System).
2.4. PI(4,5)P2 quantification
PI(4,5)P2 was extracted from cell pellets ( ~ 0.5 g) of the CEN.PK2–1C wild-type strain, separated of other PPIn species by TLC and quantified by GC/MS as described [53]. Data are the mean (± SD) of at least three independent experiments.
2.5. SLs extraction and mass spectrometry analysis
The cells (~ 100 OD600 units) grown in SCD (OD600 ~ 0.5 – 1.0) at 30°C (control) and transferred to 15°C for 3 h were suspended in 1.5 ml of Mandala extraction buffer [54] and lipid standards (C17-SpH, C17-DhS, SpH-1P and C8-PhS; Avanti Polar Lipids, Alabaster, AL) and glass beads (1.0 g; acid washed, 0.4-mm diameter) were added. Then, the mixture was vortexed 3 times for 1 min each time and incubated at 60°C for lipid preparation as described [55,56].
For LC-MS, lipid samples were dissolved in 150 μl of ammonium formate (1 mM) with 0.2% formic acid in methanol and separated in a Thermo Accela HPLC system (San Jose, CA) coupled to a Peeke Scientific Spectra C8 (Redwood city, CA) HPLC column (150 × 3 mm) and the HESI source of a Thermo TSQ Quantum Ultra triple quadrupole mass spectrometer (San Jose, CA, USA). LCBs and their phosphorylated forms (LCBPs) were analyzed, and the mass spectrometer was operated as described [56]. Samples were processed by the Thermo Xcalibur 2.2 Quan Browser software and exported to Excel for reporting results. The levels of the different lipid species in each sample were normalized to the units of processed OD600. All the data were calculated from at least two biological replicates (± SD).
2.6. NLs and PLs analysis
SCD-grown cells at 30°C (Control; OD600 ~ 0.5) were transferred to 15°C for 3 h, collected by centrifugation (3,000 × g, 2 min, 4°C), washed twice with ultrapure water (MilliQ RO 10 Plus; Millipore, Bedford, MA) and kept at −80°C until the extraction was carried out. Then, the cell pellet was suspended in ultrapure water, glass beads (1.0 g; acid washed, 0.4-mm diameter) were added, the mixture was vortexed 3 times for 1 min each time, centrifuged and the supernatant was recovered. Finally, yeast samples were diluted (~ 20 OD600 units/ml), and subjected to basic lipidomic analysis.
Quantitative lipid analysis was carried out by the lipidomic service of Lipotype (Lipotype GmbH, Dresden, Germany), which use a high throughput mass-spectrometry-based shotgun lipidomics platform. Lipids were extracted using chloroform and methanol [57]. Samples were spiked with lipid-class specific internal standards prior to extraction. After drying and resuspending in MS acquisition mixture, lipid extracts were subjected to mass spectrometric analysis. Mass spectra were acquired on a hybrid quadrupole/Orbitrap mass spectrometer equipped with an automated nano flow electrospray ion source in both positive and negative ion mode. Lipid identification using LipotypeXplorer [58] was performed on unprocessed (*.raw format) mass spectra. For MS-only mode, lipid identification was based on the molecular masses of the intact molecules. MSMS mode included the collision induced fragmentation of lipid molecules and lipid identification was based on both the intact masses and the masses of the fragments. Prior to normalization and further statistical analysis, lipid identifications were filtered according to mass accuracy, occupation threshold, noise and background. Lists of identified lipids and their intensities were stored in a database optimized for the particular structure inherent to lipidomic datasets. Intensity of lipid class-specific internal standards was used for lipid quantification. The amounts of individual lipid molecules (species) of a given lipid class were normalized as the mol%. Likewise, the quantities of the lipid species containing the same number of double bonds or the same number of carbon atoms in the hydrocarbon moiety are summed and these values are normalized to the total amount of the given lipid class. Additional details concerning sample handling and processing, reagents, equipment, procedures and data visualization tools can be found at https://www.lipotype.com/. Data represent the mean value (±SD) of three independent biological replicates.
2.7. Gene expression analysis
Quantitative RT-PCR (qPCR) experiments were carried out to determine the changes in mRNA levels. Total RNA and cDNA were prepared as described in Gomar-Alba et al. [59]. qPCR was performed in a DNA Engine Peltier Thermal Cycler (Bio Rad) using the SYBR Premix Ex Taq Tli RNaseH Plus Green with ROX (Takara, Shiga, Japan) and specific oligonucleotides (Table S2) as previously described [60]. Relative quantification of gene expression was determined using the comparative Ct (threshold cycle number) method analysis [61]. Fold change values were calculated as the 2^(−ΔdCt), where dCt = Ct[Target]-Ct[Housekeeping], and ΔdCt = (ΔExperimental condition)-(ΔControl). Samples were run in triplicate and normalized to ACT1 mRNA as housekeeping gene. Each graph is representative of at least three independent experiments.
2.8. The β-galactosidase assay
β-galactosidase activity was determined at 30°C using cell extracts and the substrate ONPG as previously described [62]. One unit was defined as the amount of enzyme that is able to convert 1 nmol of ONPG per min under the assay conditions. The values provided represent the mean (± SD) of at least three independent experiments.
2.9. Determination of acid phosphatase activity
The phosphatase activity was assayed according to the method described by Neef and Kladde [63] with slight modifications. Briefly, yeast cells were washed twice with water, and resuspended in 0.1 M sodium acetate buffer, pH 4.0 at 4°C. The reaction was started by mixing 250 μl of cell suspension, previously incubated at 30°C for 5 min, and 250 μl of 20 mM p-nitrophenylphosphate (Sigma). After 10 min at 30°C, the reaction was stopped by adding 125 μl of 1 M Na2CO3 and quantified by measuring the absorbance at 420 nm. Activities are reported in units per OD600 [(OD420)/(OD600 × volume [in milliliters] of cells assayed × 10 min)], and are the mean (± SD) of at least three independent experiments.
2.10. Statistical analysis
Sample averages were compared using a Student s t-test with the Excel software (Microsoft). p< 0.01 (**, ##) and p< 0.05 (*,#) were considered statistically significant.
3. Results
3.1. Cold exposure reduces plasma membrane PI(4,5)P2 abundance
We examined how PI(4,5)P2 levels respond to a downward shift in temperature. PPIn species of 30°C-grown and cold-shocked cells of the CEN.PK2–1C wild-type strain were extracted, separated by TLC and the amount of unlabeled PI(4,5)P2 was quantified by GC/MS as described [53]. As it is shown, a cold treatment reduced significantly (p< 0.05) the PI(4,5)P2 content (Fig. 2A), an opposite change to that reported for heat-stressed yeast cells [14,15].
Fig. 2.
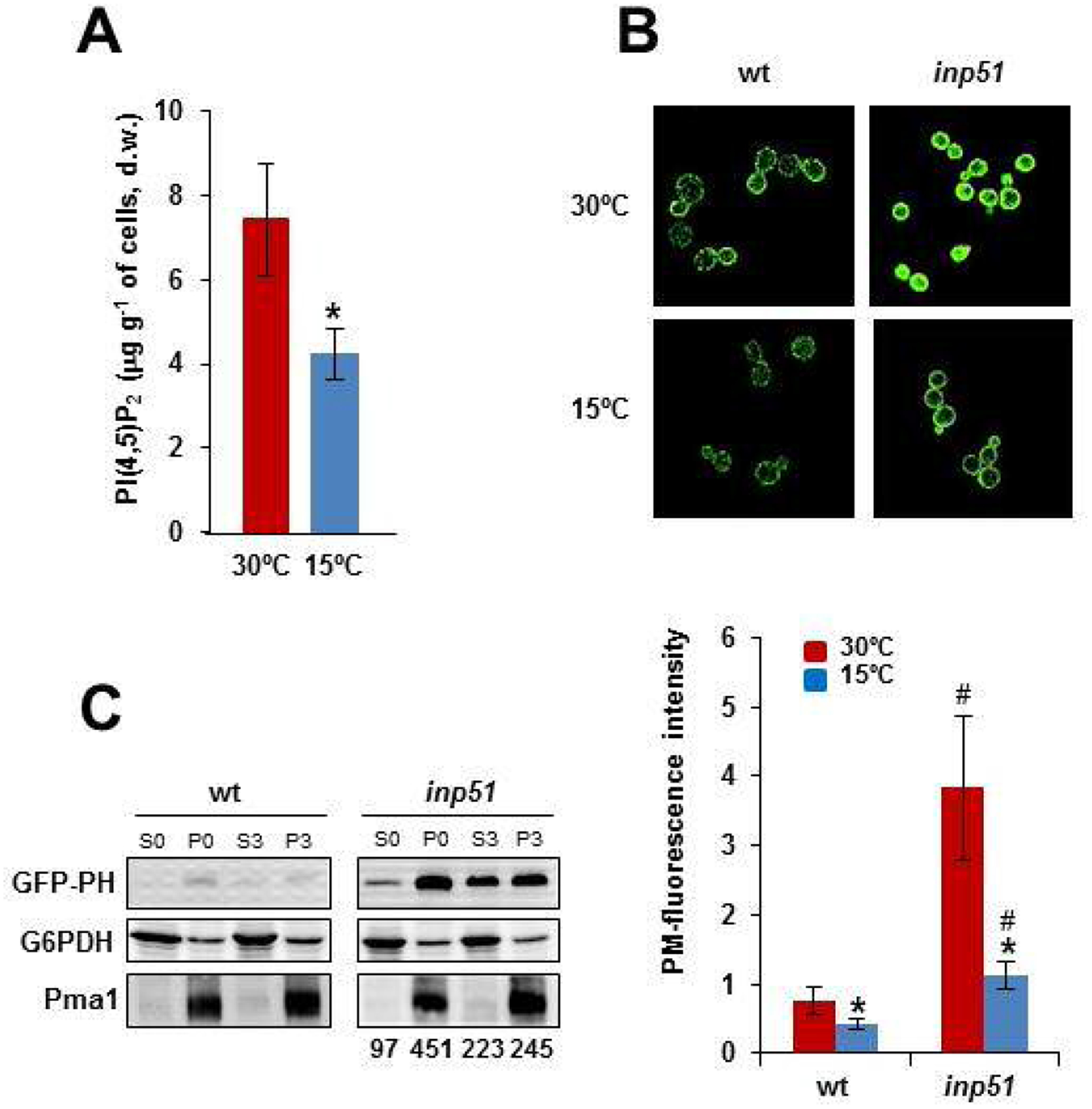
Plasma membrane PI(4,5)P2 levels lower in the cold-shocked cells. A) Cells of the wild-type CEN.PK2–1C strain were grown to the mid-logarithmic phase in SCD medium at 30°C. An aliquot was withdrawn for the analysis and the rest of the culture was shifted to 15°C for 3 h. Samples were analyzed for PI(4,5)P2 levels by GC/MS. The values represent the mean (± SD) of at least three independent experiments (*; p< 0.05). B) The plasmid pRS414–7×2-PHO5-GFP-hPLC (see Table S3) which encodes a GFP-tagged PH domain was used to visualize the cellular location of yeast PI(4,5)P2. Cells of the CEN.PK2–1C wild-type (wt) and the corresponding inp51 mutant strain were grown to the mid-logarithmic phase at 30°C. An aliquot was withdrawn for the analysis and the rest of the culture was shifted to 15°C for 3 h. Cells were concentrated and visualized as described in the Materials and Methods section. The cells photographed at 30°C and 15°C are shown as an example. Quantification of plasma membrane fluorescence was done with Image J and expressed as arbitrary units. Data are the mean (± SD) of at least three independent experiments in which 100 cells were processed per sample. The 15°C samples were significantly different compared with their respective control at 30°C (*; p< 0.05). The differences between the wt and the inp51 samples at the same temperature, 30°C or 15°C, were also statistically significant (#; p< 0.05). C) The GFP-PH levels from the protein extracts of the mentioned strains were analyzed by Western blot. Protein extracts were centrifuged at 17,900 × g for 10 min, and the corresponding pellet (P, membrane-enriched fraction) and supernatant (S, soluble fraction) fractions were separated by SDS-PAGE and immunoblotted with anti-GFP antibody. The cells from 30°C-grown cells (S0, P0) or transferred (3 h) to 15°C (S3, P3) were compared. The amount of G6Pdh and Pma1 were tested as loading control of cytosolic and membrane-associated proteins. Relative quantification of GFP signals in the inp51 mutant was done with Image J and is expressed as arbitrary units. The intensity of wild-type signals was under the detection limit. For more details see the Materials and Methods section. A representative experiment is shown.
Then, a genetically encoded fluorescent probe consisting of a GFP-tagged version of the PLC-δ PH domain (GFP-PH) was used to further analyze the PI(4,5)P2 dynamics in response to cold. Previous studies showed this probe binds specifically to PI(4,5)P2 [64]. Consistent with this, the GFP-signal in cells of the wild-type was mainly localized in the plasma membrane (Fig. 2B). We also observed that cold reduced the abundance of PI(4,5)P2 signals at the plasma membrane (Fig. 2B). This change was also observed in cells lacking the PI(4,5)P2 phosphatase Inp51 (Fig. 2B), which has been reported to contain higher levels of this PPIn as compared with the wild-type [4,10,14], a circumstance that was confirmed in our study (Fig. 2B).
To further verify the cold-instigated changes in PI(4,5)P2, we analyzed by Western blot the distribution of the GFP-PH probe in membrane-enriched (P) and cytosolic (S) protein fractions of 30°C-grown control (P0 and S0) and cold-exposed cells (S3 and P3). As shown in Fig. 1C, we detected a clear redistribution of the GFP-PH construct to the cytosol in response to cold, although the signals could only be quantified in samples of the inp51 mutant (Fig 2C), where the total cellular content of GFP-PH (S+P) was higher at either 30°C or 15°C (Fig. 2C). We interpret that the higher expression of the probe in the mutant is consequence of the reduced activity of the Pho85 kinase in the inp51 strain [4], since the expression of the GFP-PH construct is directed by the promoter of PHO5, a gene downregulated by Pho85. Overall, our results show a decrease in the PI(4,5)P2 accumulation in the plasma membrane of cold-shocked cells.
3.2. 5-PP-IP4 and 1-IP7 readouts vary in response to cold
Taking into account that some human PLCγ2 variants and Arabidopsis PLC are irreversibly activated by cold exposure [65,66], we wonder if the cold-induced clearance of PI(4,5)P2 observed at the yeast plasma membrane is caused by the increased activity of Plc1 at low temperature. Such Plc1 activation could be revealed by an increase in DPIPs levels, as occurs in the inp51 mutant [4]. However, DPIPs are traditionally evaluated by using radioactive experimental approaches [67] that require a highly controlled research environment and sophisticated equipment to extract and purify radioactive derivatives. In addition, the acidic conditions often used for their analysis appear to cause changes in the profile of DPIPs, in particular of IP7 and IP8 [68]. As an alternative, here we take advantage of the regulatory effects exerted by some DPIPs. First, we measured the expression of INO1, the first enzyme in the inositol biosynthesis pathway [69], which responds positively to the content of 5-PP-IP4 [70], the pyro-phosphorylation product of IP5 by Kcs1 (Fig. 1). As shown in Fig. 3A, transfer of yeast wild-type cells to 15°C caused a strong induction of an INO1-LacZ reporter. Similar results were observed when the expression of INO1 was assayed by qPCR (data not shown). However, blocking DPIPs production by knock-out of PLC1 (Fig. 1) abrogated the upregulation of the reporter gene (Fig. 3A).
Fig. 3.
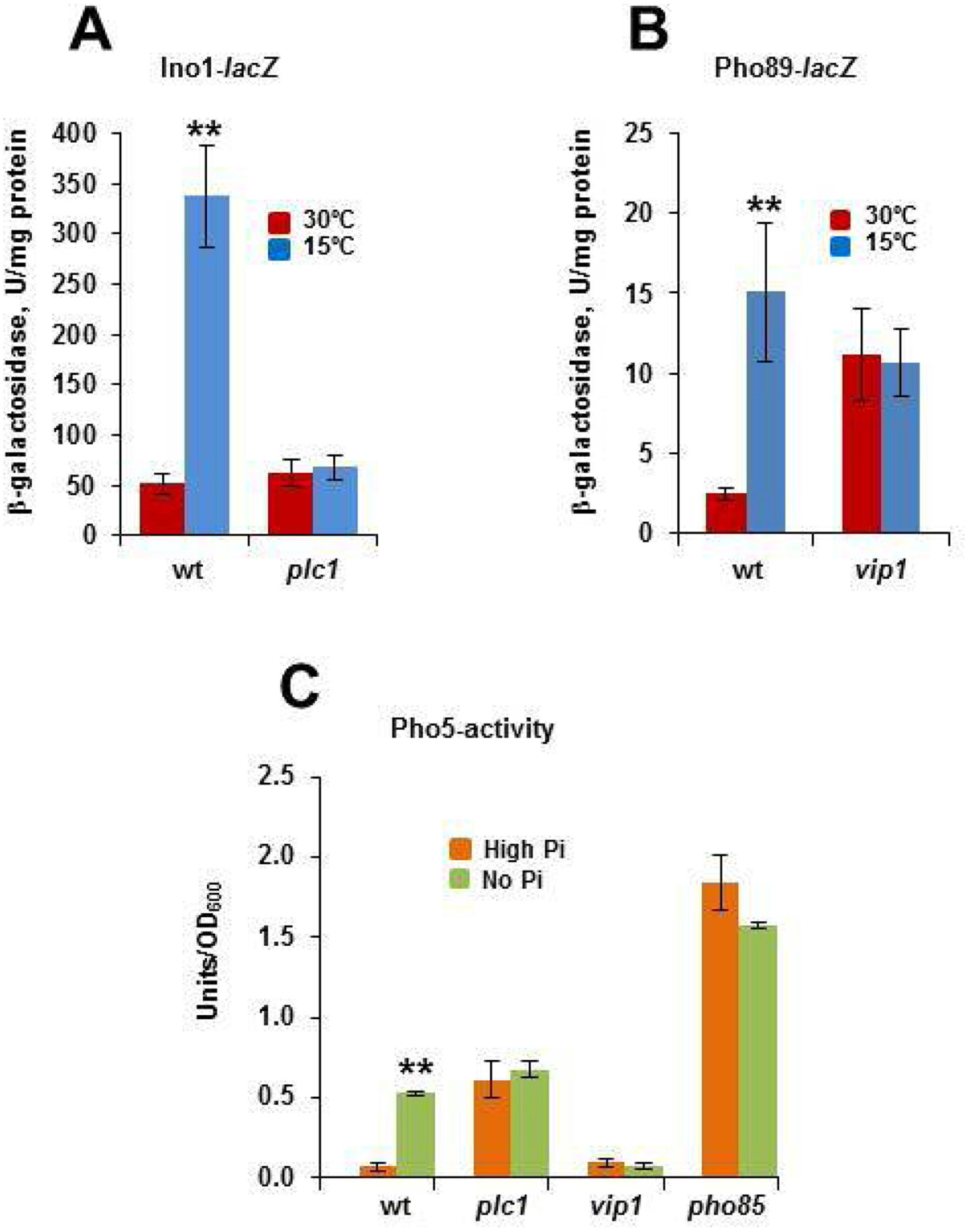
DPIPs and Pho85 exert regulatory functions in response to cold. A) Cells of the CEN.PK2–1C wild-type (wt) strain and its corresponding plc1 mutant were grown as described above (Fig. 2A), and the expression of INO1 was analyzed by using an INO1-lacZ reporter as indicated in the Materials and methods section. Data represent the mean value (± SD) of three independent experiments (**; p< 0.01). B) The 1-IP7 content in the cells of the wild-type and vip1 mutant strain was estimated by using a PHO89::lacZ reporter. Cells were grown at 30°C in SCD and then transferred to 15°C for 3 h. Data represent the mean value (± SD) of three independent experiments (**; p< 0.01). C) The activity of the repressible acid phosphatase Pho5 (Units/OD600) was measured before and after transfer for 3 h of yeast cells from high-phosphate to phosphate-free medium. Cells of the wild-type, plc1, vip1 and pho85 mutants were analyzed. Data represent the mean value (± SD) of three independent experiments (**; p< 0.01).
Then, we assayed the expression of PHO89 by using a PHO89-lacZ reporter. As mentioned above, high levels of 1-IP7 regulate positively the expression of several PHO-genes, including PHO5, PHO11, PHO84 and the high-affinity phosphate transporter-encoding gene PHO89 [37]. As shown in Fig. 3B, the expression of the reporter gene increased 6-fold in cold-transferred cells of the wild-type strain. On the contrary, no effect could be detected in cells of the vip1 mutant, which are unable to synthesize 1-IP7 (Fig. 3B). As expected, cold induction of PHO89 is not observed in a strain lacking Plc1, which as vip1 is unable to produce 1-IP7 (Supplementary Fig. S1). That the effect of 1-IP7 on the cold activation of PHO89 occurs through the inhibition of Pho85 kinase is suggested by the constitutive expression of the reporter in the pho85 mutant (Fig. S1). Quite remarkably, the cold-induced regulatory effects were replicated in cells starved for phosphate. As shown in Fig. 3C, transfer of wild-type cells from high-phosphate to starvation medium stimulated the synthesis of the repressible acid phosphatase Pho5. As expected, lack of Pho85 gave rise to high levels of Pho5 activity even in high-phosphate conditions, while the PLC1 mutation stimulated the synthesis of the phosphatase to intermediate levels (Fig. 3C), a result that has been previously reported [71]. Finally, either plc1 or vip1 cells were insensitive to phosphate starvation. We conclude that cold-signals are transmitted from PI(4,5)P2 to DPIP species, among them 5-PP-IP4 and 1-IP7, which exert regulatory effects.
3.3. PI(4,5)P2 effects on membrane fluidity and cold tolerance require a functional Pho85 protein
The above results suggested a link between PI(4,5)P2 levels and DPIPs species that may target Pho85 in mediating the cold-signaling. To further confirm this relationship, we investigated whether the phenotype of inp51 mutant cells, is suppressed by the lack of Pho85 activity. As earlier reported, the INP51 mutation endows yeast cells with cold tolerance [4,72], a feature that fully depends on a functional Pho85 kinase (Fig. 4A). Lack of Inp51 also increased tolerance of yeast cells to the toxicity of fatty acid palmitoleate (C16:1), which causes hyper-fluidity of the membrane [73], myriocin, which inhibits the activity of serine palmitoyl transferase, SPT [74], the first enzyme in the sphingolipid biosynthesis pathway [22,75], and phytosphingosine (PhS). Remarkably, all of these phenotypes were suppressed by the combined knock-out of INP51 and PHO85, while the single pho85 mutant did not display an apparent phenotype under these conditions (Fig. 4A).
Fig. 4.
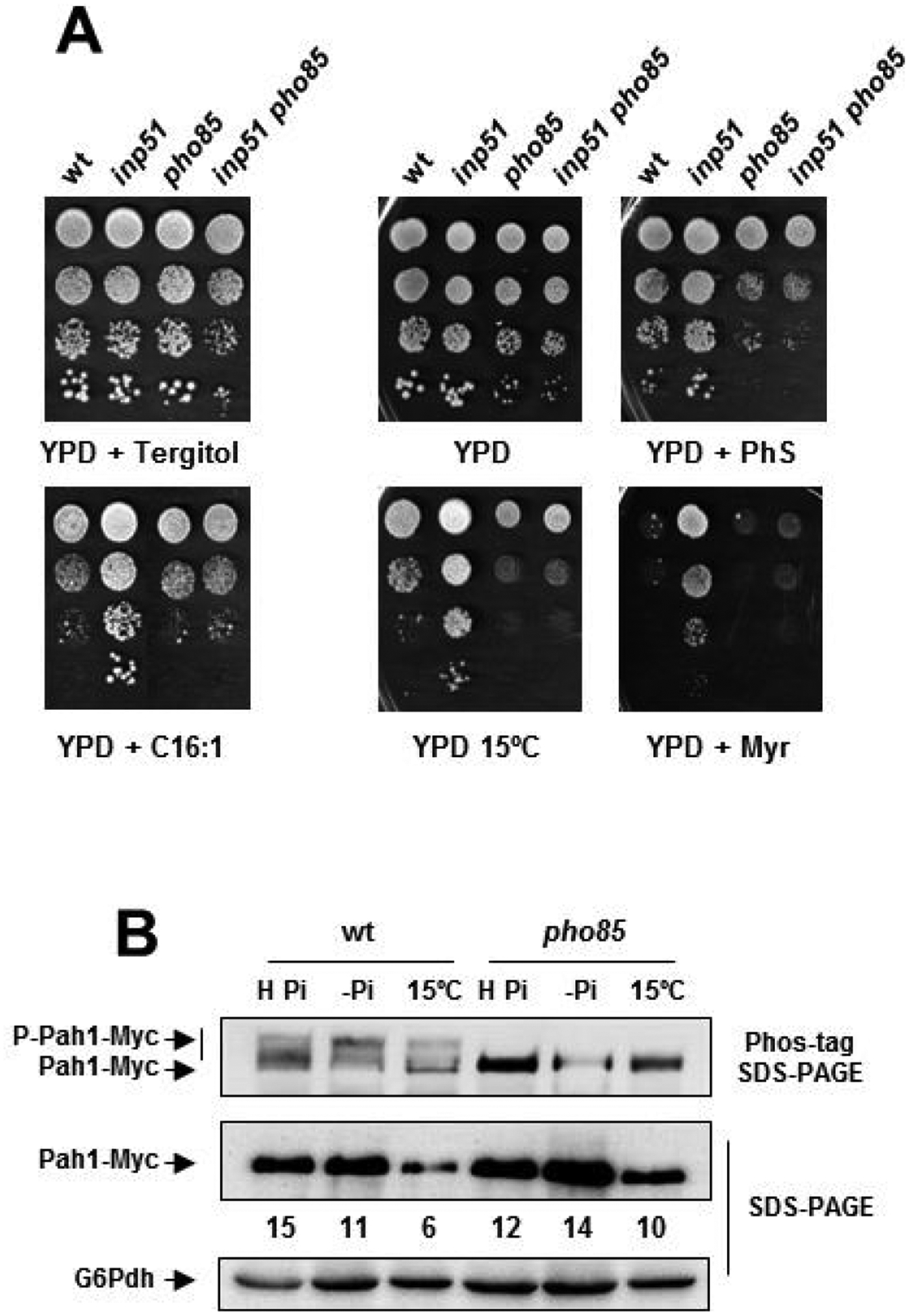
Pah1 abundance and main inp51 phenotypes depend on Pho85. A) Cells of the CEN.PK2–1C wild-type (wt) strain and its corresponding mutants inp51, pho85 and inp51 pho85 were examined for growth in YPD lacking or containing 1.2 μM myriocin (YPD + Myr), 25 μM phytosphingosine (YPD + PhS), or exposed to 15°C (YPD 15°C). Toxicity of palmitoleic acid (C16:1) was tested on YPD-0.05% tergitol medium (YPD + Tergitol) supplemented with 750 μM of C16:1 (YPD + C16:1). Overnight YPD-grown cultures were adjusted to OD600 ~ 0.5, diluted (1–10−3), spotted (3 μl) onto the mentioned media, and incubated at 30°C for 2–5 days. B) The protein extracts from Pah1-Myc tagged cells of the CEN.PK2–1C wild-type strain and its corresponding pho85 mutant were analyzed by Western blot. The 30°C-incubated cell cultures in High-phosphate medium (H Pi; OD600 ~ 0.5) were transferred to phosphate starvation medium for 3 h (-Pi) or to 15°C for the same period (15°C). Antibodies against Myc and G6Pdh (loading control) were used. A representative experiment is shown.
3.4. Pah1 abundance lowers in response to cold
The above results indicated that Pho85 could be a hub element in the cold signaling mechanism. To further support this idea, we examined the effect of cold on different Pho85 read-outs. First, we focused our attention on Pah1, the yeast phosphatidate phosphatase, a well-known target of Pho85 [41]. Pah1 suffers multiple phosphorylations by this and additional kinases, among them Cdc28, PKA and PKC [76,77]. In addition, cross-talk phosphorylations by different kinases have been described [78], making more complex the biochemical regulation of the enzyme. Because of this, we designed an experiment in which nutrient conditions were strictly controlled. High-phosphate, phosphate-starved and cold-shocked cells of the wild-type and pho85 mutant strains containing a chromosomal copy of Pah1 tagged with 13xMyc were lysed and analyzed by Western blot. As Fig. 4B shows, analysis of wild-type and pho85 mutant protein extracts by regular SDS-PAGE detected a major band corresponding to Pah1. Abundance of Pah1 decreased in both strains by phosphate starvation and, especially by cold treatment (Fig. 4B). Nevertheless, the analysis by Phos-tag™ affinity SDS-PAGE, which reduces the mobility of phosphorylated isoforms, provided additional information. As shown in Fig. 4B, the single SDS-PAGE band in wild-type samples was resolved by the Phos-tag™ system in at least two additional bands of lower mobility. These bands were absent or under the detection limit in pho85 samples, confirming that the kinase participates in the posttranslational modification of Pah1, as previously reported [41]. Phosphate-starvation seemed to increase the intensity of the lowest mobility band suggesting a hyperphosphorylation. However, we were unable to show noticeable changes in the profile of Pah1 bands upon cold transfer (Fig. 4B).
3.5. A downward shift in temperature stimulates the remodeling of NLs and PLs
We investigated if the cold-signals controlling the INO expression extend to other lipid metabolism genes, and how the cold regulation affects the biosynthesis and turnover of NLs and PLs. In addition to INO1, almost all PLs biosynthetic genes contain UASINO sequences [79]. Among them, CHO1, which encodes the phosphatidylserine synthase, CDS1, the yeast CDP-diacylglycerol synthase and PSD1, encoding the phosphatidylserine decarboxylase, were analyzed. Supplementary Fig. S2 shows the fold-change of mRNA levels measured by qPCR at 30 and 15°C. As can be seen, a downward shift in temperature had a positive effect on the transcription of these genes.
Then, the profiles of the NLs and PLs of the 30°C and 15°C-transferred yeast cells were compared. A cold transfer altered the relative abundance of lipid classes and subclasses (Fig. 5A). In particular, the relative content of NLs, except for DAG, lowered in the cold-exposed cells, while that of PLs increased, with PC, PE and PI representing the most important changes. On the contrary, PS was the unique individual PLs whose relative content was found to decrease in response to low temperature (Fig. 5A). A similar trend has been reported when the exponentially-growing cells of a pah1 mutant were compared with its corresponding wild-type [80]. Finally, total carbon length and unsaturation degree of PLs were found to decrease and increase respectively, in response to cold treatment (Fig. 5B), a result that confirms previous reports [7,8]. Supplementary Tables S4–S12 show the relative content at 30°C and 15°C of all NLs and PLs species identified in this study. Overall, our results strongly agree with a role of the PPIn and DPIP pathways in controlling the activity of Pho85, the Pah1 content and the expression of UASINO-containing genes, which contribute to remodeling the profile of PLs and TAG at low temperature.
Fig. 5.
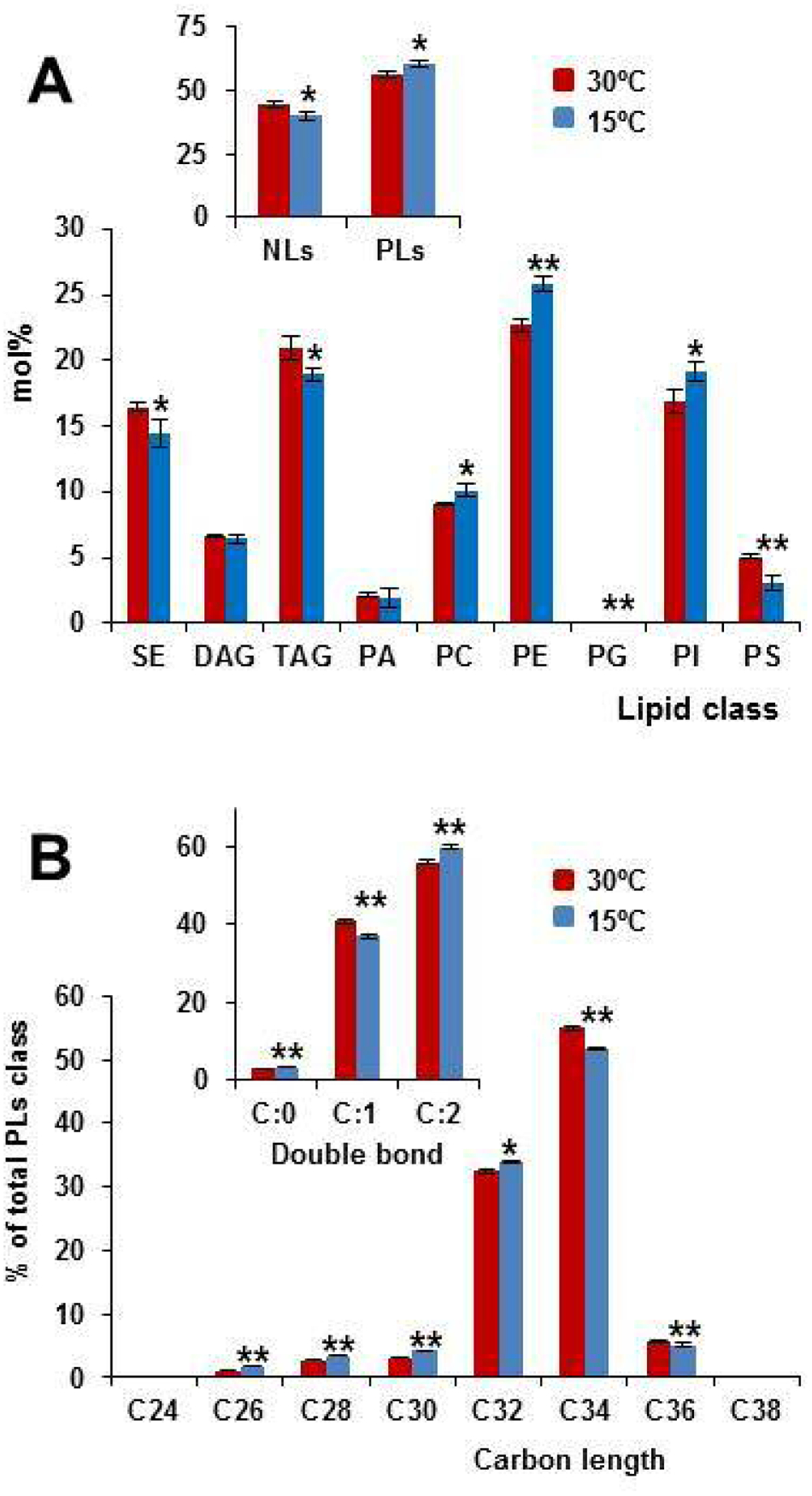
Cold exposure affects lipid composition. A) Lipid profiles of wild-type cells in SCD were analyzed for NLs and PLs classes. 30°C-grown (OD600 ~ 0.5) and 15°C-exposed (3 h) cells were analyzed by mass-spectrometry-based shotgun lipidomics. The amount of NLs, PLs, and their corresponding classes were normalized to the total lipid content and expressed as the mol%. B) The quantities of the PLs species containing the same number of double bonds or the same number of carbon atoms in the hydrocarbon moiety are summed and these values were normalized to the total amount of PLs and expressed as mol%. * (p< 0.05) and ** (p< 0.01) denote significant differences between 30°C and 15°C samples. Data represent the mean value (± SD) of three independent biological replicates.
3.6. Ypk1 and Orm2 are regulated in response to cold
We examined additional connections between PI(4,5)P2 and Pho85 in the context of lipid homeostasis. A previous systematic lipidomic analysis identified kinase and phosphatase mediated phospho-signaling pathways as playing a role in sphingolipid metabolism [17]. Both kcs1 and pho85 cells were found to exhibit a similar profile of SLs characterized by reduced Cer and IPC content. On the other hand, PI(4,5)P2 plays an important role in recruiting effector proteins to the plasma membrane and regulating the TORC2-Pkh1-Ypk1 signaling circuit that controls, among others, the synthesis of LCBs, the precursors of Cer and complex SLs (see Fig. 6A). Thus, we inspected the effect of cold in the phosphorylation status of Ypk1 and Orm2.
Fig. 6.
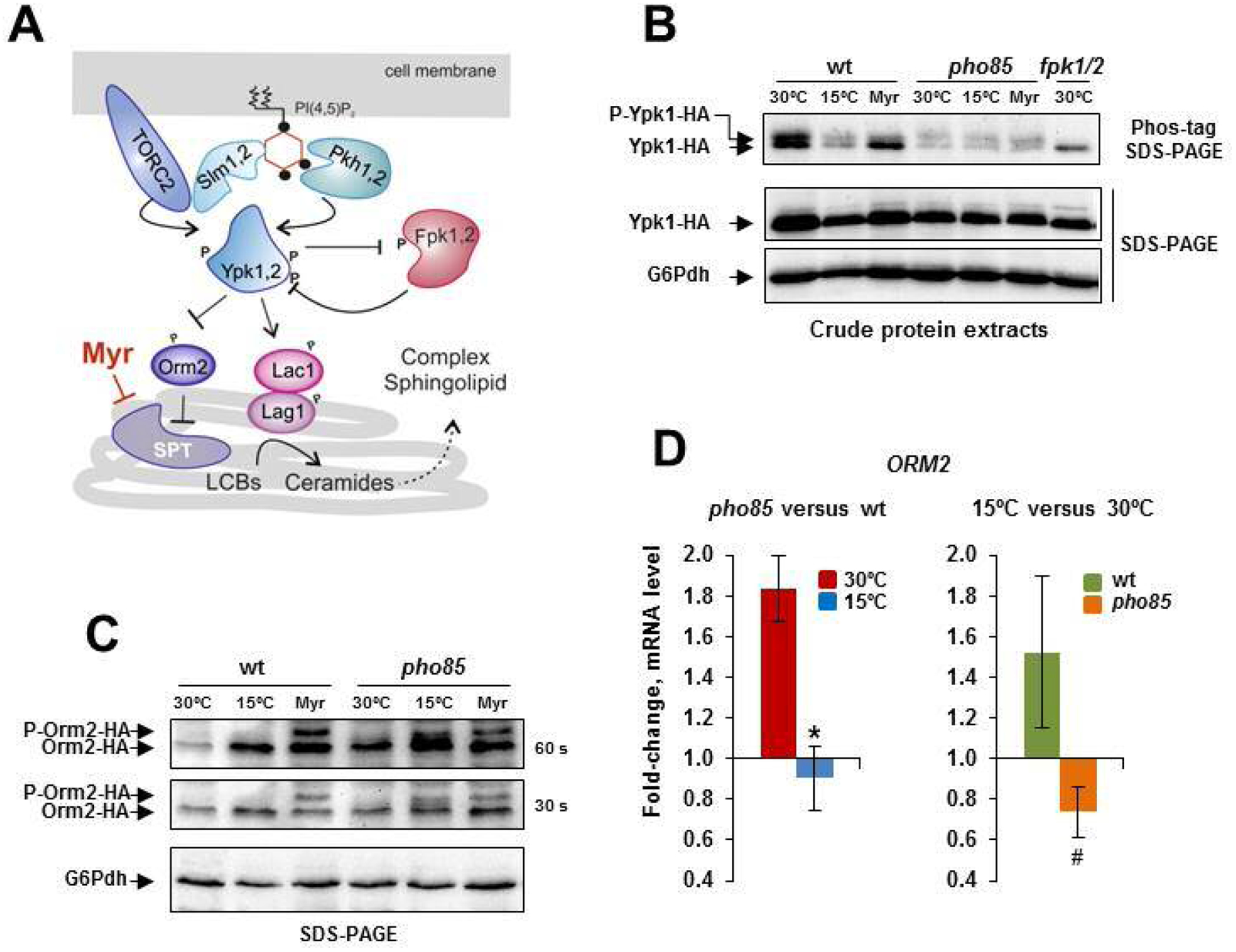
Ypk1 and Orm2 are cold-regulated through control protein level. A) Schematic representation of the TORC2-Pkh1-Ypk1 signaling module. At the plasma membrane, the interaction of PI(4,5)P2 and the PH-like domain-containing proteins Slm1/2 facilitates the targeting of Ypk1/2 to the plasma membrane and its later phosphorylation by the TORC2 complex and kinases Pkh1/2 [21,28], which also interact with PI(4,5)P2. Activated Ypk1/2 stimulates the production of SLs by phosphorylating SPT inhibitor Orm2 [22,23] and ceramide synthase components Lag1 and Lac1 [24]. A black dot indicates a phosphate group. For more details, see the text. B) The protein crude extracts from the Ypk1-HA tagged cells of the CEN.PK2–1C wild-type strain and pho85 mutant were analyzed by Phos-tag affinity SDS-PAGE (upper panel) or regular SDS-PAGE (lower panel). Cells grown at 30°C in SCD medium, transferred to 15°C for 3 h (15°C) or exposed to 2 μM myriocin for 30 min (Myr) were examined. Protein extracts from 30°C-grown cells of the fpk1 fpk2 double mutant (fpk1/2) were used as controls. Images show only a part of the gels where Ypk1-HA isoforms were visualized with an anti-HA antibody. The slower migrating bands, which most likely correspond to the phosphorylated isoforms by Fpk1/2 (P-Ypk1-HA), are indicated. The band indicated by the arrow might be unspecific/phosphorylated and unphosphorylated Ypk1 isoforms (Ypk1-HA). The G6Pdh level was used as a loading control for crude extracts. A representative experiment is shown. C) Protein extracts from the Orm2-HA tagged cells of the wild-type and pho85 mutant strain were obtained by NaOH-treatment and analyzed by regular SDS-PAGE. Cultures at 30°C, transferred to 15°C for 3 h (15°C) or treated with 2 μM myriocin for the same time (Myr) were tested. The myriocin treatment stimulates the phosphorylation of Orm2 (P-Orm2-HA) by Ypk1 [31]. Membranes exposed for 30 and 60 s are shown. The G6Pdh level was used as a loading control for crude extracts. A representative experiment is shown. D) The ORM2 mRNA levels in cells of the CEN.PK2–1C wild-type (wt) strain and its corresponding pho85 mutant were analyzed by qPCR. Expression differences between wt and pho85 samples at 30 and 15°C (left panel) or between control (30°C) and cold-treated (15°C, 3 h) samples for the wt and pho85 strain (right panel) are shown as fold-change. Data represent the mean (± SD) of at least three independent experiments. Statistically significant (p< 0.05) differences are denoted (*, #).
Fig. 6B shows the electrophoretic profile (analyzed by Phos-tag affinity SDS-PAGE) of Ypk1-HA from 30°C-grown, cold-shocked or myriocin (Myr)-treated wild-type and pho85 cell samples. As mentioned above, the fungal natural product myriocin depress the sphingolipid biosynthesis [74] by inhibiting the activity of SPT [81], which leads to a compensatory response through the TORC2-Pkh-Ypk1 signaling module (Fig. 6A). As control, protein extracts from a double fpk1/2 mutant were also analyzed. As opposite to TORC2/Pkh1/2, the protein kinases Fpk1/2 downregulate Ypk1 in response to increased levels of complex SLs [82,83]. In addition, the Ypk1 isoform phosphorylated by Fpk1/2 is the only one readily detected in yeast cell extracts by SDS-PAGE [82]. Using Phospho-tag methodology with anti-HA antibody, we were able to detect two bands in Ypk1-HA cells extracts (Fig. 6B). The fact that the upper band is reduced by myriocin treatment [82], and completely absent in the fpk1/2 mutant indicates that this band is indeed consequence of the phosphorylation of Ypk1 by Fpk1/2 kinases. Cold exposure caused minor changes, if any, in the profile of Ypk1-HA (Fig. 6B; Phos-tag), but decreased its abundance (Fig. 6B). Remarkably, in the pho85 mutant the reduced Ypk1 content found in cold-treated wild-type cells was also observed in 30°C-grown control and myriocin-treated cells. This indicates that Pho85 functions in regulating positively Ypk1 abundance in response to optimal growth temperature, and suggests that inactivation of Pho85 may be on the basis of the downregulation of Ypk1 content upon cold exposure.
Then, we tested the abundance and phosphorylation state of Orm2, which provides the majority of Orm-inhibitory activity in yeast cells [75]. Because of the low Orm2 content in yeast cells, a method that use NaOH to enhance the permeability of yeast cell wall prior to protein extraction with sample buffer, was employed [52], and the proteins were analyzed by SDS-PAGE (Fig. 6C). As shown, a single weak band was visible when protein extracts from 30°C-grown wild-type cells were visualized with anti-HA antibody. In contrast with the regulation of Ypk1 abundance showed above, cold exposure increased the Orm2-HA content (Fig. 6C). Moreover, no change in the electrophoretic profile of the protein could be observed. Compared with this, myriocin-treated control cells displayed an additional strong lower mobility band, which likely corresponds to phosphorylated species (Fig. 6C). Orm2 phosphorylation in response to inhibition of SLs production has been associated to increased Ypk1 kinase activity [23]. We also observed that the response to myriocin was not altered in a pho85 mutant (Fig. 6C). However, the Orm2 content was clearly higher in 30°C-grown cells of the mutant as compared with the wild-type. In addition, cold-exposed pho85 cells showed weak slower mobility species (Fig. 6C; 30s-exposed membrane). Consistent with these results, the mRNA level of ORM2 was higher in pho85 than in wild-type cells at 30°C (Fig. 6D). In addition, ORM2 expression was stimulated in cold-exposed cells of the wild-type, while hardly varied in those of the mutant (Fig. 6D). We conclude that Ypk1 and Orm2 are coordinately regulated in response to cold in a Pho85-dependent manner. This regulation is mainly exerted through control of protein level, although we cannot discard additional mechanisms and effectors.
3.7. Pho85 controls cellular LCBs and LCBPs abundance and it is involved in their regulation at low temperature
The finding that Ypk1 and Orm2 content are down and upregulated, respectively, suggested that SLs synthesis is repressed under cold conditions. Previous results have shown that LCBs are decreased when ORM2 is overexpressed [84]. In addition, lowering Ypk1 content should reduce the flux of SLs from LCBs to Cer, thus adjusting the synthesis and demand of LCBs (see Fig. 6A). Indeed, Ypk1, among other actions, stimulates the synthesis of Cer by phosphorylating ceramide synthase [24]. To confirm the cold-regulation of the SLs synthesis, we examined the levels of the LCBs dihydrosphingosine (DhS) and phytosphingosine (PhS) from 30°C-grown cells or cold-shocked at 15°C for 3 h (Fig. 7A). In wild-type cells, a shift to 15°C resulted in decreased content of LCBs, in particular of PhS, the major LCB species in yeast [85]. Compared with this, deletion of PHO85 reduced the PhS levels at 30°C by around 55%, while no significant change could be detected in response to cold (Fig. 7A).
Fig. 7.
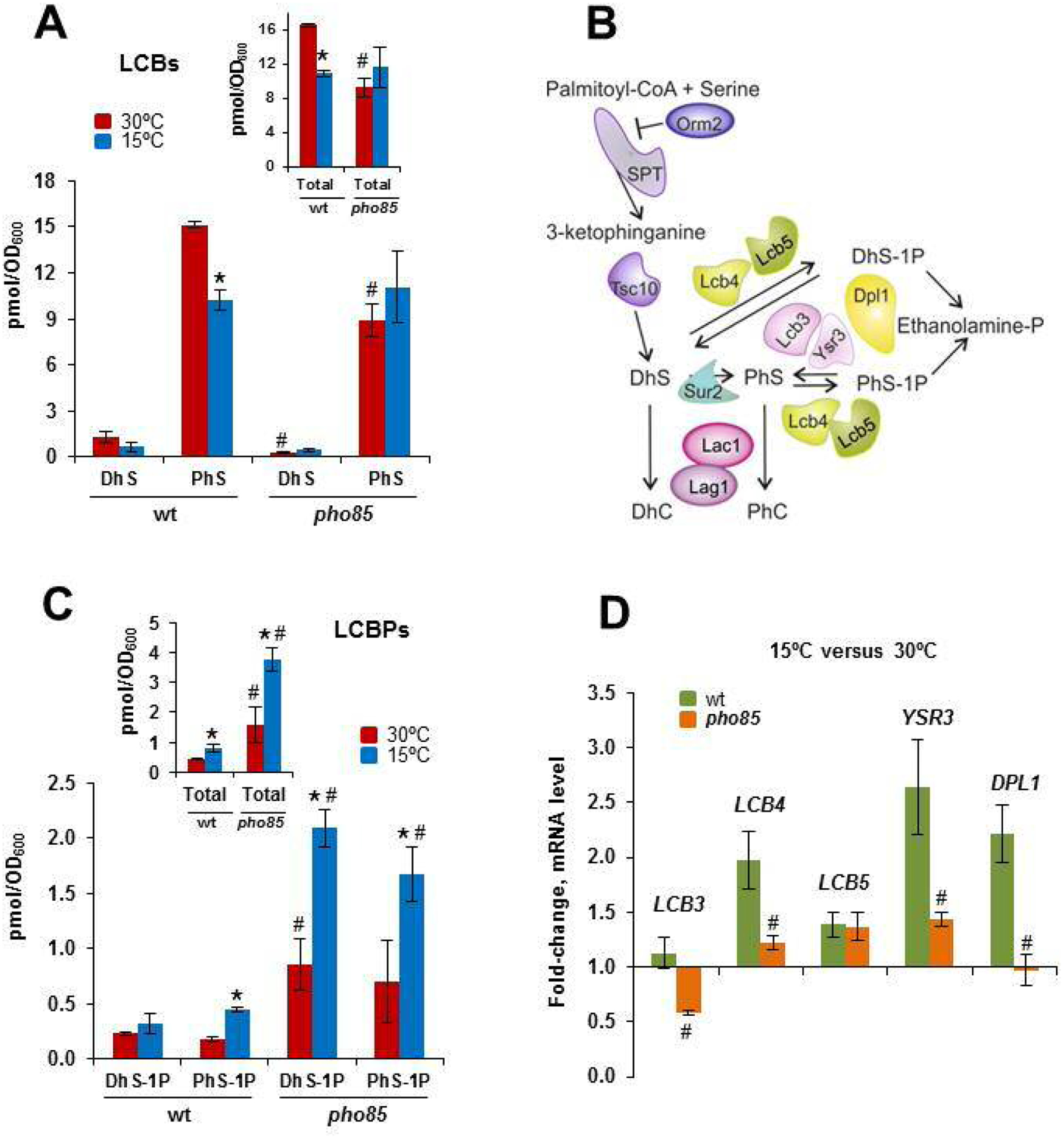
A downward shift in temperature downregulates the synthesis of LCBs and stimulates its phosphorylation. A) The cells of the CEN.PK2–1C wild-type and corresponding pho85 mutant strain were grown in SCD medium at 30°C until the mid-log phase (OD600 ~ 0.5) and were then transferred to 15°C for 3 h. Lipids were extracted, and the LCBs level was analyzed by mass spectrometry as described in the Materials and methods section. Data are the mean (± SD) of two independent biological replicates. Statistically significant differences (p< 0.05) between 30°C- and 15°C-samples (*), and between the wt and pho85 strains at the same temperature (#) are indicated. B) Schematic representation showing the initial steps of the SLs biosynthesis from the rate-limiting step catalyzed by the serine palmitoyltransferase (SPT) complex to ceramide synthase, and the LCBs-to-glycerophospholipid metabolic pathway. Orm2 acts as an inhibitor of SPT. LCBs kinases (Lcb4, Lcb5) use DhS and PhS, the precursors of DhC and PhC, to form their corresponding phosphorylated forms DhS-1P and PhS-1P. Details about each step, and the enzymes, effectors and regulators involved, can be found in the text and recent reviews [85,90–92,97]. C) Cells of the indicated strains were grown as indicated above and analyzed for LCBPs content. Statistically significant differences (p< 0.05) between 30°C- and 15°C-samples (*), and between the wt and pho85 strains at the same temperature (#) are indicated. D) The mRNA levels of the indicated genes in cells of the CEN.PK2–1C wild-type (wt) strain and its corresponding pho85 mutant were analyzed by qPCR. The expression changes in response to a cold-transfer from 30 to 15°C for 3 h are shown as fold-change for each strain. Data represent the mean (± SD) of at least three independent experiments. Statistically significant differences between wt and pho85 samples are denoted (#; p< 0.05).
Then, we inspected the content of LCBPs, dihydrosphingosine-1-phosphate (DhS-1P) and phytosphingosine-1-phosphate (PhS-1P). LCBs can be phosphorylated by two kinases, Lcb4 and Lcb5, to form LCBPs, which can be, in turn, dephosphorylated by Ysr3 and Lcb3, the main yeast LCBPs phosphatases, or irreversibly degraded by the LCBPs lyase Dpl1 (Fig. 7B). Thus, phosphorylation of LCBs represents an additional mechanism to that of Orm2-SPT to control the flux into the SLs pathway. Consistent with this role, a downward shift in temperature stimulated the accumulation of both DhS-1P and PhS-1P in the wild-type strain (Fig. 7C). Hence LCBs and LCBPs decrease and increase, respectively, in response to cold transfer. This opposite behavior is remarkably similar to that previously observed for the cells that express a temperature-sensitive TORC2 allele, torc2ts [47] or an ATP-analog-sensitive allele Ypk1-AS [86]. Regarding the pho85 mutant, samples from 30°C-grown cells exhibited much higher LCBPs levels as compared with those of the wild-type. Moreover, the LCBPs level in pho85 cells increased in response to cold and this variation was even more intense than that observed for the wild-type (Fig. 7C). Interestingly, the ratio PhS-1P/DhS-1P raised in cold-exposed cells of the wild-type from 0.78 ± 0.09 at 30°C to 1.48 ± 0.35 at 15°C (p< 0.05), while did not vary in the pho85 mutant, 0.80 ± 0.22 and 0.80 ± 0.06, at 30 and 15°C, respectively. This result could reflect a certain substrate preference of LCBs-kinases and/or LCBPs-phosphatases and their distinct cold-regulation in wild-type and pho85 mutant cells.
To further confirm this idea, we analyzed by qPCR the mRNA levels of genes involved in the metabolism of LCBs to glycerophospholipids (Fig. 7B). As shown in Fig. 7D, the expression of the five genes analyzed increased in cold-exposed cells of the wild-type strain. The change was especially intense for the major kinase LCB4 and especially for the LCBPs-lyase encoding gene DPL1, while LCB3, which represents the most important source of phosphatase activity, was not induced, accounting thus for the accumulation of LCBPs at low temperature. Comparing with this, LCB4, LCB5 and DPL1 hardly varied in cold-exposed cells of the pho85 mutant. In addition, the phosphatase activity encoded by LCB3 was repressed. This is consistent with the exacerbated LCBPs content found in the mutant strain (Fig. 7C). We conclude that Pho85 is involved in regulating not only the de novo synthesis of LCBs, but also their turnover via their conversion to glycerophospholipids.
4. Discussion
Here we provide direct evidence that a drop in temperature reduces PI(4,5)P2 levels and increases the availability of water-soluble DPIPs. These observations suggest that PI(4,5)P2 functions as a second messenger and that the PPIn pathway is a candidate to play a role in the thermosensing mechanism/s of S. cerevisiae. Further evidence supports this idea; 1) PPIn kinases and phosphatases, as well as PLC, reside in the plasma membrane or interact with it [15]; 2) heat-stressed yeast cells show the opposite dynamics of PI(4,5)P2 levels [14]; 3) the enzymes of this pathway, in particular PLC, appear to display special sensitivity to temperature changes and are required for molecular responses to thermal stress [15,29,30]. Some human PLCγ2 variants appear to display unique thermodynamic features and are irreversibly activated by cold exposure [65]. Likewise, Arabidopsis PLC activity increases in minutes after cold exposure [66]; 4) the knock-out and ectopic expression of the PPIn kinases- and phosphatases-encoding genes, or PLC1, has physiological consequences in cold acclimation and survival [4].
The finding that the PI(4,5)P2 level decreases in response to cold suggested that this variation could alter the content of intermediates of the DPIP-pathway and trigger regulatory mechanisms mediated by, among others, Pho85. It has been previously suggested that poly-phosphate metabolism is connected to that of DPIPs [87], and recent evidence shows that several yeast SPX domains, which are found in proteins involved in phosphate metabolism [88], bind to both IP6 and IP7 [89]. S. cerevisiae possesses 10 proteins that contain an SPX domain, including Pho81 and Pho4 [88], which thus explain the previously described inhibitory effect of 1-IP7 on the kinase complex Pho81-Pho80-Pho85 [37,38]. Consistent with this, cold-shocked cells displayed increased expression of a PHO89-lacZ reporter, indicating increased levels of 1-IP7 and downregulation of Pho85 at low temperature. Cold also caused a strong transcriptional induction of UASINO sequences containing genes like INO1, or the PLs biosynthetic genes CHO1, CDS1 and PSD1. Expression of INO1 depends on the level of 5-PP-IP4 [70], an intermediate of the DPIPs-pathway that is produced from IP5 by the kinase Kcs1 [36]. Remarkably, kcs1 mutant cells exhibit inositol auxotrophy, a defect that is suppressed by deletion of INP51 [4], more likely by increasing the flux through the DPIPs-pathway. Additional data support that cold increases the flux through the DPIPs-pathway and regulates the activity of Pho85. 1) Blocking the synthesis of DPIPs by knock-out of PLC1 makes the expression of INO1 and PHO89-lacZ insensitive to low temperature; 2) Cold induction of PHO89-lacZ is not observed in cells of the vip1 mutant, which are unable to produce 1-IP7; 3) Relevant phenotypic features of the inp51 mutant, including C16:1 toxicity and cold tolerance, are suppressed by mutation of PHO85; and 4) a downward shift in temperature increases and reduces the content of PLs and TAG, respectively, and alters the relative abundance of individual PLs classes, all of them typical features of cells displaying reduced Pah1 activity [80], a well-known target of Pho85 [78]. We conclude that cold increases the abundance of DPIPs, which has molecular consequences in remodeling the profile of NLs and PLs.
The central role of Pho85 as a master regulator of lipid homeostasis was further established by analyzing the effects of cold on the electrophoretic profile of Ypk1 and Orm2, which receives signals from the TORC2-Pkh1 module, the main pathway in controlling SLs homeostasis [90–92]. To our surprise, neither Ypk1 nor Orm2 showed apparent changes in their phosphorylation state in response to cold. Instead, Ypk1 abundance lowers, while Orm2 content increases, indicating that the main mechanism for regulating SLs synthesis under this condition is through control protein level. Increasing Orm2 abundance contributes to the inhibition of the SPT complex [84,92], explaining thus the reduced synthesis of LCBs at low temperature. On the other hand, Ypk1 activity stimulates the synthesis of Cer by phosphorylating ceramide synthase [24], and thus, lowering Ypk1 content may be aimed to coupling the demand of LCBs to their synthesis. Compared with this, PHO85 disruption leads to reduced and increased content of Ypk1 and Orm2, respectively, at 30°C, indicating that inhibition of the effector Pho85 protein kinase may be part of the SLs homeostasis mechanism at low temperature. In agreement with this, the LCBs content at 30°C is decreased in pho85 mutant cells, a result that fits well with previous systematic lipidomic analysis [17], while no changes could be observed in response to cold.
The regulation of Ypk1 and Orm2 protein level could be mediated by the signaling of the Ca2+/calmodulin-calcineurin pathway, which appears to depress cellular processes controlled by TORC2, including the synthesis of SLs [92], likely through the regulation of Slm1/2 [29,93]. This regulation could be controlled, at least in part, by the activity of Pho85, which has been reported to phosphorylate the calcineurin-regulated transcription factor Crz1 [45], and the PI(4,5)P2 levels. Indeed, the plasma membrane association of Ypk1, a prerequisite for its activation by phosphorylation, depends directly or indirectly on the activity of the PI(4,5)P2-binding proteins Slm1/2 [21,29,92,93]. In agreement with this model, heat-induced membrane stress increases the phosphorylation of Ypk1/2 by TORC2, an event that diminishes in the stt4ts and mss4ts mutants, which display low PI(4,5)P2 levels [30]. However, we have no evidence of this signaling mechanisms working in response to cold. Additional experiments are required to further investigate the role of PI(4,5)P2 and Pho85 in the regulation of Ypk1/Orm2 abundance and the implication of the Ca2+/calmodulin-calcineurin pathway in response to cold.
Quite remarkably, cold exposure and loss of Pho85 stimulated LCBPs accumulation, suggesting that the phosphorylation of LCBs contributes in a Pho85-dependent manner to regulate the flux toward Cer under this stress condition. Yeast mutants that lack the LCBP-phosphatase Lcb3 display smaller amounts of Cer than the corresponding wild-type [94]. In addition, the increased LCBPs content could supply, through the Dpl1-dependent reaction, phosphoethanolamine for the enhanced synthesis of PLs at low temperature. Our study also provided evidence that higher levels of LCBPs by pho85 mutation are driven, at least in part, by a defect in the cold regulation of genes involved in the metabolism of LCBs to glycerophospholipids. Indeed, upregulation of LCB4 and LCB5, or DPL1 in cold-shocked cells of the wild-type was missing by loss of Pho85. Finally, the cold-induced accumulation of LCBPs may perform alternative functions. A previous work by Gallego et al. [95] showed that both PI(4,5)P2 and DhS-1P can simultaneously bind the PH-like domain of Slm1. Although the specific effects that result from this binding are unclear, it has been suggested that they could affect the activity of Slm1 [95]. Thus, the increased accumulation of LCBPs could provide an additional regulatory input to the TORC2-Pkh1 module. Further work is required to demonstrate this signaling interplay and its consequences on the regulation of the metabolism of SLs in response to cold stress.
The adaptation and survival of S. cerevisiae to membrane stress requires the deep remodeling of the lipid repertoire. A downward shift in temperature decreases membrane fluidity and permeability, and homeostasis mechanisms are expected to operate to sustain membrane functionality. In this scenario, the finding that cold exposure reduces SLs abundance in favor of more PLs has a physiological meaning. Complex sphingolipids with long saturated hydrocarbon chains exhibit a higher affinity for sterols, and together with them, promote the formation of a thicker, more compact and less permeable plasma membrane than that provided by a lipid matrix which contains high proportions of unsaturated phospholipids [96]. The finding that Pho85 connects different lipid synthesis pathways uncovers a mechanism of how lipid metabolism is coordinated to tailor cell membrane composition and properties to environment temperature changes. More work is required to acknowledge the implications and usefulness of these findings in applied aspects.
Supplementary Material
Fig. S1. The expression of PHO89 does not vary in cold-shocked cells of the plc1 and pho85 mutants.
Fig. S2. Cold stimulates the expression of UASINO sequences containing genes.
Table S1. The Saccharomyces cerevisiae strains used in this study.
Table S2. The oligonucleotides used in this study.
Table S3. The plasmids used in this study.
Table S4. Composition, total carbon length and total double bond of TAG molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S5. Composition, total carbon length and total double bond of SE molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S6. Composition, total carbon length and total double bond of DAG molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S7. Composition, total carbon length and total double bond of PA molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S8. Composition, total carbon length and total double bond of PC molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S9. Composition, total carbon length and total double bond of PE molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S10. Composition, total carbon length and total double bond of PG molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S11. Composition, total carbon length and total double bond of PI molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S12. Composition, total carbon length and total double bond of PS molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Fig. S1. The expression of PHO89 does not vary in cold-shocked cells of the plc1 and pho85 mutants. PHO89::lacZ transformants of the CEN.PK2–1C mutants plc1 and pho85 were grown at 30°C in SCD, transferred to 15°C for 3 h, and the β-galactosidase activity of cellular extracts was analyzed as indicated in the Materials and methods section. Data represent the mean value (± SD) of three independent experiments. Activity differences between plc1 and pho85 samples were statistically significant at both 30 and 15°C (##; p< 0.01).
Fig. S2. Cold stimulates the expression of UASINO sequences containing genes. A) Schematic representation of pathways in which the phospholipid biosynthetic genes CHO1, which encodes the phosphatidylserine synthase, CDS1, the yeast CDP-diacylglycerol synthase and PSD1, encoding the phosphatidylserine decarboxylase, are involved. Details are given in the text and reviews [79]. B) Cells of the CEN.PK2–1C wild-type (wt) strain were grown to the mid-logarithmic phase in SCD medium at 30°C. An aliquot was withdrawn for the analysis and the rest of the culture was shifted to 15°C for 1 h or 3 h. Samples were analyzed for total mRNA levels of the mentioned genes by qPCR as indicated in the Materials and methods section. Expression differences between control (30°C) and cold-treated (15°C) samples are represented as the fold-change (*; p<0.05) (**; p<0.01). Data represent the mean (± SD) of at least three independent experiments.
Acknowledgments
We would like to thank J. Ariño (Universitat Autònoma de Barcelona), T. Levine (UCL Institute of Ophthalmology, London), R. Serrano (Universitat Politècnica de València) and Y. Kozutsumi (Kyoto University) for providing us with antibodies and plasmids. We acknowledge the help of A. Rella, M.A. Munshi and C. Lazzarini (Stony Brook University) with lipid extractions and equipment use. This research has been supported by the Comisión Interministerial de Ciencia y Tecnología Project (BIO2015-71059-R) from the Spanish Ministry of Science, Innovation and Universities (MICINN/FEDER) and the National Institutes of Health (AI116420 and AI125770 to M.D.P.). I.C.-S. was supported by a pre-doctoral fellowship as part of the “FPI” Program [MICINN].
Abbreviations
- Cer
ceramides
- CDK
cyclin dependent kinase
- CWI
Cell Wall Integrity
- DAG
diacylglycerol
- DhC
dihydroceramide
- DhS
dihydrosphingosine
- DhS-1P
dihydrosphingosine-1-phosphate
- DPIP
diphosphoinositol phosphate
- ICRE
inositol/cholineresponsive element
- LCBs
long-chain bases
- LCBPs
phosphorylated long-chain bases
- IPC
inositol phosphorylceramide
- IPs
inositol phosphates
- IP3
inositol-1,4,5-trisphosphate
- MAPK
mitogen-activated protein kinase
- NLs
neutral lipids
- PA
phosphatidate
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PhC
phytoceramide
- PhS
phytosphingosine
- PhS-1P
phytosphingosine-1-phosphate
- PI
phosphatidylinositol
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PLC
phospholipase C
- PLs
phospholipids
- PPIn
phosphoinositide
- PS
phosphatidylserine
- SLs
sphingolipids
- 5-PP-IP4
5-diphospho-myo-inositol 1,3,4,6-tetrakisphosphate
- 1-PP-IP5/1-IP7
1-diphosphomyo-inositol 2,3,4,5,6-pentakisphosphate
- SE
steryl ester
- SPT
serine palmitoyltransferase
Footnotes
Conflict of interest
Dr. Maurizio Del Poeta is a Co-Founder and Chief Scientific Officer [CSO] of MicroRid Technologies Inc. All other authors have no conflict of interest.
References
- [1].Simons K, Sampaio JL (2011) Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol 3: a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dowhan W (2017) Understanding phospholipid function: Why are there so many lipids? J Biol Chem 292: 10755–10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Los DA, Mironov KS, Allakhverdiev SI (2013) Regulatory role of membrane fluidity in gene expression and physiological functions. Photosynth Res 116: 489–509. [DOI] [PubMed] [Google Scholar]
- [4].Córcoles-Sáez I, Hernández ML, Martínez-Rivas JM, Prieto JA, Randez-Gil F (2016) Characterization of the S. cerevisiae inp51 mutant links phosphatidylinositol 4,5-bisphosphate levels with lipid content, membrane fluidity and cold growth. Biochim Biophys Acta 1861: 213–226. [DOI] [PubMed] [Google Scholar]
- [5].Gaspar ML, Hofbauer HF, Kohlwein SD, Henry SA (2011) Coordination of storage lipid synthesis and membrane biogenesis: evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis J Biol Chem 286: 1696–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rajakumari S, Rajasekharan R, Daum G (2010) Triacylglycerol lipolysis is linked to sphingolipid and phospholipid metabolism of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1801: 1314–1322. [DOI] [PubMed] [Google Scholar]
- [7].Rodríguez-Vargas S, Sánchez-García A, Martínez-Rivas JM, Prieto JA, Randez-Gil F (2007) Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl Environ Microbiol 73: 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Martin CE, Oh CS, Jiang Y (2007) Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim Biophys Acta 1771: 271–285. [DOI] [PubMed] [Google Scholar]
- [9].Shah ZH, Jones DR, Sommer L, Foulger R, Bultsma Y, D’Santos C, Divecha N (2013) Nuclear phosphoinositides and their impact on nuclear functions. FEBS J 280: 6295–6310. [DOI] [PubMed] [Google Scholar]
- [10].Michell RH, (2011) Inositol and its derivatives: their evolution and functions. Adv Enzyme Regul 51: 84–90. [DOI] [PubMed] [Google Scholar]
- [11].Hammond GR, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF (2012) PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity Science 337: 727–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Delage E, Puyaubert J, Zachowski A, Ruelland E (2013) Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: convergences and divergences among eukaryotic kingdoms. Prog Lipid Res 52: 1–14. [DOI] [PubMed] [Google Scholar]
- [13].de Craene JO, Bertazzi DL, Bär S, Friant S (2017) Phosphoinositides, major actors in membrane trafficking and lipid signaling pathways. Int J Mol Sci 18: pii: E634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Morales-Johansson H, Jenoe P, Cooke FT, Hall MN (2004) Negative regulation of phosphatidylinositol 4,5-bisphosphate levels by the INP51-associated proteins TAX4 and IRS4. J Biol Chem 279: 39604–39610. [DOI] [PubMed] [Google Scholar]
- [15].Audhya A, Emr SD (2002) Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade Dev Cell 2: 593–605. [DOI] [PubMed] [Google Scholar]
- [16].Nunez LR, Jesch SA, Gaspar ML, Almaguer C, Villa-Garcia M, Ruiz-Noriega M, Patton-Vogt J, Henry SA (2008) Cell wall integrity MAPK pathway is essential for lipid homeostasis J Biol Chem 283: 34204–34217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].da Silveira Dos Santos AX, Riezman I, Aguilera-Romero MA, David F, Piccolis M, Loewith R, Schaad O, Riezman H (2014) Systematic lipidomic analysis of yeast protein kinase and phosphatase mutants reveals novel insights into regulation of lipid homeostasis. Mol Biol Cell 25: 3234–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Olson DK, Fröhlich F, Farese RV Jr, Walther TC (2015) Rom2-dependent phosphorylation of Elo2 controls the abundance of very long-chain fatty acids. J Biol Chem 290: 4238–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y (2005) Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol 25: 7239–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26: 663–674. [DOI] [PubMed] [Google Scholar]
- [21].Niles BJ, Mogri H, Hill A, Vlahakis A, Powers T (2012) Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc Natl Acad Sci USA 109: 1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463: 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J (2011) Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 108: 19222–19227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Muir A, Ramachandran S, Roelants FM, Timmons G, Thorner J (2014) TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. Elife doi: 10.7554/eLife.03779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Casamayor A, Torrance PD, Kobayashi T, Thorner J, Alessi DR (1999) Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol 9: 186–197. [DOI] [PubMed] [Google Scholar]
- [26].Roelants FM, Torrance PD, Bezman N, Thorner J (2002) Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol Biol Cell 13: 3005–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Roelants FM, Torrance PD, Thorner J (2004) Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology 150: 3289–3304. [DOI] [PubMed] [Google Scholar]
- [28].Berchtold D, Piccolis M, Chiaruttini N, Riezman I, Riezman H, Roux A, Walther TC, Loewith R (2012) Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol 14: 542–547. [DOI] [PubMed] [Google Scholar]
- [29].Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD (2006) The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol Cell Biol 26: 5861–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Omnus DJ, Manford AG, Bader JM, Emr SD, Stefan CJ (2016) Phosphoinositide kinase signaling controls ER-PM cross-talk. Mol Biol Cell 27: 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sun Y, Miao Y, Yamane Y, Zhang C, Shokat KM, Takematsu H, Kozutsumi Y, Drubin DG (2012) Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol Biol Cell 23: 2388–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dickson RC, Nagiec EE, Skrzypek M, Tillman P, Wells GB, Lester RL (1997) Sphingolipids are potential heat stress signals in Saccharomyces. J Biol Chem 272: 30196–30200. [DOI] [PubMed] [Google Scholar]
- [33].Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun Y (1997) Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J Biol Chem 272: 32566–32572. [DOI] [PubMed] [Google Scholar]
- [34].Wundenberg T, Mayr GW (2012) Synthesis and biological actions of diphosphoinositol phosphates (inositol pyrophosphates), regulators of cell homeostasis. Biol Chem 393: 979–998. [DOI] [PubMed] [Google Scholar]
- [35].Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657. [DOI] [PubMed] [Google Scholar]
- [36].Tsui MM, York JD (2010) Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv Enzyme Regul 50: 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee YS, Mulugu S, York JD, O’Shea EK (2007) Regulation of a cyclin-CDKCDK inhibitor complex by inositol pyrophosphates. Science 316: 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee YS, Huang K, Quiocho FA, O’Shea EK (2008) Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol 4: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM (2014) Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 38: 254–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Choi J, Rajagopal A, Xu YF, Rabinowitz JD, O’Shea EK (2017) A systematic genetic screen for genes involved in sensing inorganic phosphate availability in Saccharomyces cerevisiae. PLoS One 17: e0176085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Choi HS, Su WM, Han GS, Plote D, Xu Z, Carman GM (2012) Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J Biol Chem 287: 11290–11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pascual F, Carman GM (2013) Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim Biophys Acta 1831: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nishizawa M, Katou Y, Shirahige K, Toh-e A (2004) Yeast Pho85 kinase is required for proper gene expression during the diauxic shift. Yeast 21: 903–918. [DOI] [PubMed] [Google Scholar]
- [44].Kliewe F, Engelhardt M, Aref R, Schüller HJ. (2017) Promoter recruitment of corepressors Sin3 and Cyc8 by activator proteins of the yeast Saccharomyces cerevisiae. Curr Genet 63: 739–750. [DOI] [PubMed] [Google Scholar]
- [45].Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, Boone C, Andrews B (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21: 319–330. [DOI] [PubMed] [Google Scholar]
- [46].Cyert MS (2001) Genetic analysis of calmodulin and its targets in Saccharomyces cerevisiae. Annu Rev Genet 35: 647–672. [DOI] [PubMed] [Google Scholar]
- [47].Aronova S, Wedaman K, Aronov PA, Fontes K, Ramos K, Hammock BD, Powers T (2008) Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab 7: 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Iwaki S, Kihara A, Sano T, Igarashi Y (2005) Phosphorylation by Pho85 cyclin-dependent kinase acts as a signal for the down-regulation of the yeast sphingoid long-chain base kinase Lcb4 during the stationary phase. J Biol Chem 280: 6520–6527. [DOI] [PubMed] [Google Scholar]
- [49].Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- [50].Guthrie C, Fink GR (1991) Guide to yeast genetics and molecular biology. Methods Enzymol 194: 21–37. [PubMed] [Google Scholar]
- [51].To-E A, Ueda Y, Kakimoto SI, Oshima Y (1973) Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J Bacteriol 113: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Quilis I, Taberner FJ, Martínez-Garay CA, Alepuz P, Igual JC (2019) Karyopherin Msn5 is involved in a novel mechanism controlling the cellular level of cell cycle regulators Cln2 and Swi5. Cell Cycle 18: 580–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].König S, Hoffmann M, Mosblech A, Heilmann I (2008) Determination of content and fatty acid composition of unlabeled phosphoinositide species by thin-layer chromatography and gas chromatography. Anal Biochem 378: 197–201. [DOI] [PubMed] [Google Scholar]
- [54].Mandala SM, Thornton RA, Frommer BR, Curotto JE, Rozdilsky W, Kurtz MB, Giacobbe RA, Bills GF, Cabello MA, Martín I, Peláez F, Harris GH (1995) The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J Antibiot (Tokyo) 48: 349–356. [DOI] [PubMed] [Google Scholar]
- [55].Singh A, Qureshi A, Del Poeta M (2011) Quantitation of cellular components in Cryptococcus neoformans for system biology analysis. Methods Mol Biol 734: 317–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Singh A, Wang H, Silva LC, Na C, Prieto M, Futerman AH, Luberto C, Del Poeta M (2012) Methylation of glycosylated sphingolipid modulates membrane lipid topography and pathogenicity of Cryptococcus neoformans. Cell Microbiol 14: 500–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K (2012) Flexibility of a eukaryotic lipidome--insights from yeast lipidomics. PLoS One 7: e35063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A (2011) A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol 12: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gomar-Alba M, Méndez E, Quilis I, Bañó MC, Igual JC (2017) Whi7 is an unstable cell-cycle repressor of the Start transcriptional program. Nat Commun 8: 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jiménez-Martí E, Zuzuarregui A, Ridaura I, Lozano N, del Olmo M (2009) Genetic manipulation of HSP26 and YHR087W stress genes may improve fermentative behaviour in wine yeasts under vinification conditions. Int J Food Microbiol 130: 122–130. [DOI] [PubMed] [Google Scholar]
- [61].Schmittgen T, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- [62].Torres-Quiroz F, García-Marqués S, Coria R, Randez-Gil F, Prieto JA (2010) The activity of yeast Hog1 MAPK is required during endoplasmic reticulum stress induced by tunicamycin exposure. J Biol Chem 285: 20088–20096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Neef DW, Kladde MP (2003) Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol Cell Biol 23: 3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lemmon MA, Ferguson KM, O’Brien R, Sigler PB, Schlessinger J (1995) Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci USA 92: 10472–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Schade A, Walliser C, Wist M, Haas J, Vatter P, Kraus JM, Filingeri D, Havenith G, Kestler HA, Milner JD, Gierschik P (2016) Cool-temperature-mediated activation of phospholipase C-γ2 in the human hereditary disease PLAID. Cell Signal 28: 1237–1251. [DOI] [PubMed] [Google Scholar]
- [66].Ruelland E, Cantrel C, Gawer M, Kader JC, Zachowski A (2002) Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol 130: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wilson MSC, Saiardi A (2017) Importance of Radioactive Labelling to Elucidate Inositol Polyphosphate Signalling. Top Curr Chem (Cham) 375:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Losito O, Szijgyarto Z, Resnick AC, Saiardi A (2009) Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS One 4:e5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Donahue TF, Henry SA (1981) myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J Biol Chem 256: 7077–7085. [PubMed] [Google Scholar]
- [70].Ye C, Bandara WM, Greenberg ML (2013) Regulation of inositol metabolism is fine-tuned by inositol pyrophosphates in Saccharomyces cerevisiae. J Biol Chem 288: 24898–24908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Auesukaree C, Tochio H, Shirakawa M, Kaneko Y, Harashima S (2005) Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J Biol Chem 280: 25127–25133. [DOI] [PubMed] [Google Scholar]
- [72].Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD (1998) INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem 273: 11852–11861. [DOI] [PubMed] [Google Scholar]
- [73].Lockshon D, Olsen CP, Brett CL, Chertov A, Merz AJ, Lorenz DA, Van Gilst MR, Kennedy BK (2012) Rho signaling participates in membrane fluidity homeostasis. PLoS One 7: e45049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T (1995) Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun 211: 396–403. [DOI] [PubMed] [Google Scholar]
- [75].Han S, Lone MA, Schneiter R, Chang A (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci USA 107: 5851–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Choi HS, Su WM, Morgan JM, Han GS, Xu Z, Karanasios E, Siniossoglou S, Carman GM (2011) Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), and SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J Biol Chem 286: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Su WM, Han GS, Casciano J, Carman GM (2012) Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J Biol Chem 287: 33364–33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Su WM, Han GS, Carman GM (2014) Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites. J Biol Chem 289: 34699–34708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Henry SA, Kohlwein SD, Carman GM (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190: 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Fakas S, Qiu Y, Dixon JL, Han GS, Ruggles KV, Garbarino J, Sturley SL, Carman GM (2011) Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J Biol Chem 286: 29074–29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wadsworth JM, Clarke DJ, McMahon SA, Lowther JP, Beattie AE, Langridge-Smith PR, Broughton HB, Dunn TM, Naismith JH, Campopiano DJ. (2013) The chemical basis of serine palmitoyltransferase inhibition by myriocin. J Am Chem Soc 135: 14276–14285. [DOI] [PubMed] [Google Scholar]
- [82].Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J (2010) A protein kinase network regulates the function of aminophospholipid flippases. Proc Natl Acad Sci USA 107: 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Yamane-Sando Y, Shimobayashi E, Shimobayashi M, Kozutsumi Y, Oka S, Takematsu H (2014) Fpk1/2 kinases regulate cellular sphingoid long-chain base abundance and alter cellular resistance to LCB elevation or depletion. Microbiologyopen 3: 196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Liu M, Huang C, Polu SR, Schneiter R, Chang A (2012) Regulation of sphingolipid synthesis through Orm1 and Orm2 in yeast. J Cell Sci 125: 2428–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Dickson RC (2010) Roles for sphingolipids in Saccharomyces cerevisiae. Adv Exp Med Biol 688: 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Niles BJ, Joslin AC, Fresques T, Powers T (2014) TOR complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell Rep 6: 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Livermore TM, Azevedo C, Kolozsvari B, Wilson MS, Saiardi A (2016) Phosphate, inositol and polyphosphates. Biochem Soc Trans 44: 253–259. [DOI] [PubMed] [Google Scholar]
- [88].Azevedo C, Saiardi A (2017) Eukaryotic phosphate homeostasis: The inositol pyrophosphate perspective. Trends Biochem Sci 42: 219–231. [DOI] [PubMed] [Google Scholar]
- [89].Wild R, Gerasimaite R, Jung JY, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M, Mayer A (2016) Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352: 986–990. [DOI] [PubMed] [Google Scholar]
- [90].Teixeira V, Costa V (2016) Unraveling the role of the Target of Rapamycin signaling in sphingolipid metabolism. Prog Lipid Res 61: 109–133. [DOI] [PubMed] [Google Scholar]
- [91].Roelants FM, Leskoske KL, Martinez Marshall MN, Locke MN, Thorner J (2017) The TORC2-Dependent Signaling Network in the Yeast Saccharomyces cerevisiae. Biomolecules 7: pii: E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gururaj C, Federman RS, Chang A (2013) Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J Biol Chem 288: 20453–20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Mulet JM, Martin DE, Loewith R, Hall MN (2006) Mutual antagonism of target of rapamycin and calcineurin signaling. J Biol Chem 281: 33000–33007. [DOI] [PubMed] [Google Scholar]
- [94].Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, Menzeleev R, Spiegel S (1998) Sphingoid base 1-phosphate phosphatase: A key regulator of sphingolipid metabolism and stress response. Proc Natl Acad Sci USA 95: 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gallego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, Matetzki C, Aguilar-Gurrieri C, Beltran-Alvarez P, Bonn S, Fernández-Tornero C, Jensen LJ, Kuhn M, Trott J, Rybin V, Müller CW, Bork P, Kaksonen M, Russell RB, Gavin AC (2010) A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol Syst Biol 6: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Lester RL, Withers BR, Schultz MA, Dickson RC (2013) Iron, glucose and intrinsic factors alter sphingolipid composition as yeast cells enter stationary phase. Biochim Biophys Acta 1831: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Megyeri M, Riezman H, Schuldiner M, Futerman AH (2016) Making Sense of the Yeast Sphingolipid Pathway. J Mol Biol 428: 4765–4775. [DOI] [PubMed] [Google Scholar]
- [98].Nakano K, Yamamoto T, Kishimoto T, Noji T, Tanaka K (2008) Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol Biol Cell 19: 1783–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wach A, Brachat A, Pöhlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The expression of PHO89 does not vary in cold-shocked cells of the plc1 and pho85 mutants.
Fig. S2. Cold stimulates the expression of UASINO sequences containing genes.
Table S1. The Saccharomyces cerevisiae strains used in this study.
Table S2. The oligonucleotides used in this study.
Table S3. The plasmids used in this study.
Table S4. Composition, total carbon length and total double bond of TAG molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S5. Composition, total carbon length and total double bond of SE molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S6. Composition, total carbon length and total double bond of DAG molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S7. Composition, total carbon length and total double bond of PA molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S8. Composition, total carbon length and total double bond of PC molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S9. Composition, total carbon length and total double bond of PE molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S10. Composition, total carbon length and total double bond of PG molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S11. Composition, total carbon length and total double bond of PI molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Table S12. Composition, total carbon length and total double bond of PS molecular species found in S. cerevisiae wild-type cells grown at 30°C and cold-shocked at 15°C for 3 h.
Fig. S1. The expression of PHO89 does not vary in cold-shocked cells of the plc1 and pho85 mutants. PHO89::lacZ transformants of the CEN.PK2–1C mutants plc1 and pho85 were grown at 30°C in SCD, transferred to 15°C for 3 h, and the β-galactosidase activity of cellular extracts was analyzed as indicated in the Materials and methods section. Data represent the mean value (± SD) of three independent experiments. Activity differences between plc1 and pho85 samples were statistically significant at both 30 and 15°C (##; p< 0.01).
Fig. S2. Cold stimulates the expression of UASINO sequences containing genes. A) Schematic representation of pathways in which the phospholipid biosynthetic genes CHO1, which encodes the phosphatidylserine synthase, CDS1, the yeast CDP-diacylglycerol synthase and PSD1, encoding the phosphatidylserine decarboxylase, are involved. Details are given in the text and reviews [79]. B) Cells of the CEN.PK2–1C wild-type (wt) strain were grown to the mid-logarithmic phase in SCD medium at 30°C. An aliquot was withdrawn for the analysis and the rest of the culture was shifted to 15°C for 1 h or 3 h. Samples were analyzed for total mRNA levels of the mentioned genes by qPCR as indicated in the Materials and methods section. Expression differences between control (30°C) and cold-treated (15°C) samples are represented as the fold-change (*; p<0.05) (**; p<0.01). Data represent the mean (± SD) of at least three independent experiments.


