Abstract
Klebsiella pneumoniae carbapenemase (KPC) have become a major therapeutic challenge because of its increasingly fast dissemination throughout the world. Accurate detection of KPC is essential for optimal treatment. The Clinical and Laboratory Standards Institutes (CLSI) for fast detection of KPC producers currently recommend Modified Hodge Test (MHT) and Carba NP test. MHT can directly detect carbapenemase production in Enterobacteriaceae isolates. The current study was conducted to evaluate the capacity of MHT with two carbapenem disks for accurate detection of KPC. MHT was performed according to guidelines of CLSI to identify isolates with carbapenem resistance. In doing so, two substrates of MHT were assigned into two groups for examination: meropenem and ertapenem groups. A total of 96 non-repetitive clinical isolates of Klebsiella pneumoniae were tested. The presence of the blaKPC gene in each MHT-positive isolate was examined by PCR. A total of 54 isolates exhibited reduced susceptibility or resistance to carbapenems. Sensitivity of MHT with two carbapenem disks was similar. Specificity of the MHT with meropenem disk was 64% and with ertapenem disk was 53%. Detection of KPC by MHT with meropenem disk was found to be more effective than with ertapenem disk. Based on our results, the presence of KPC does not in itself influence the categorization of resistance. Therefore, the use of MHT with ertapenem disk for the rapid detection of KPC among K. pneumoniae for infection control should not be recommended.
Key words: Klebsiella pneumoniae, carbapenem disks, detection method, Modified Hodge Test, KPC
Introduction
Klebsiella pneumoniae causes serious hospital-acquired infections of the urinary tract, respiratory tract, surgical sites, and the bloodstream and can cause severe diseases, such as pneumonia, sepsis, and bacteremia (Maroncle et al. 2002; Paterson 2006).The resistance of K. pneumoniae has significantly increased with the rampant use of beta-lactam antibiotics, such as carbapenems. Carbapenems are often the last treatment option for infections caused by these multidrug-resistant bacteria. Therefore, the major concern is the development of resistance against carbapenems (Nordmann et al. 2011; Barwa and Shaaban 2017).
Klebsiella pneumoniae carbapenemase (KPC) is a class-A β-lactamase, and the most active family of carbapenemases. The development of antibiotic resistance by class-A Ambler enzymes, such as KPC, generally leads to increased cessation in the treatment of infections (Hashemi et al. 2014; Bachman et al. 2015; Barwa and Shaaban 2017). The strains possessing the blaKPC gene have spread worldwide; this has resulted in increased concern for healthcare services worldwide (Woodford et al. 2010). Detection of carbapenemases in Enterobacteriaceae is essential to control the development of resistance in this family, particularly in K. pneumoniae isolates. The identification of K. pneumoniae isolates producing KPC has become a major concern for clinicians (Djahmi et al. 2014).
In recent years, various phenotypic confirmatory tests to detect the presence of carbapenemase enzymes in Enterobacteriaceae have been evaluated by testing the growth of such organisms in CHROMagar KPC medium, Chrom ID ESBL medium, Supercarba medium and other tests, i.e., Neo sensitabs (Samra et al. 2008; Nordmann et al. 2011; Aliskan et al. 2012; Hansen et al. 2012).
CLSI has recommended the use of MHT and Carba NP tests for carbapenemase detection in Enterobacteriaceae. Because of variations in sensitivity and specificity for detection of KPC producers, the identification of such bacteria through these tests is difficult (Vasoo et al. 2013; Shinde et al. 2017). MHT has acceptable sensitivity. It is easy to perform, inexpensive and feasible for practically all-clinical laboratories; however, it lacks specificity (Anderson et al. 2007; Pasteran et al. 2009; Pasteran et al. 2010). MHT was suggested for detection of carbapenemase-producers based on their in vivo production of carbapenemases (Galani et al. 2008; Centers for Disease and Prevention 2009; Miriagou et al. 2010).
The purpose of this study was to evaluate the effectiveness of MHT as a phenotypic confirmatory test with 2 different substrates for KPC screening. Additionally, PCR was performed for the detection of blaKPC gene.
Experimental
Materials and Methods
Bacterial collection and Ribotyping analysis. K. pneumoniae isolates were collected from 96 consecutive hospital inpatients and/or outpatients admitted to the AL Zahra hospital in Esfahan, Iran between February and June 2016.
PCR amplification was used for PCR-ribotyping based on internal transcribed spacer (ITS). The procedure implemented was as follows. Template DNA for the PCR was prepared from overnight culture of K. pneumoniae on nutrient agar (Scharlo, Spain). Five colonies from the overnight culture on TSA medium (Scharlo, Spain) were suspended in 100 μl distilled water. The boiling lysis method was used for DNA extraction. Cell debris was centrifuged at 13684 RCF for three minutes. Supernatants were used as the source of template DNA for amplification. Strain identifications were performed by analysis of the ITS. PCR-ribotyping was performed using PCR Master Mix 2X (Thermo Scientific, Germany) and specific primers (Liu et al. 2008).
Antimicrobial susceptibility testing. The disk diffusion methods using meropenem, ertapenem disks (Rosco, Denmark) were conducted on the basis of CLSI recommendations. Escherichia coli ATCC 25922 and K. pneumoniae 700603 were used as reference strains for susceptibility testing (CLSI).
Phenotypic confirmation test. In our study all isolates were subjected to MHT, according to the following procedure. Ertapenem disks (10 μg) (Rosco, Denmark), and meropenem disks (10 μg) (Rosco, Denmark) were used for MHT. The MHT indicator organism, E. coli ATCC 25922, was suspended in Mueller-Hinton broth (Scharlo, Spain) to obtain a suspension with turbidity of a 0.5 McFarland standard. The suspension was diluted 1:10 and plated on Mueller-Hinton agar (Scharlo, Spain). The carbapenem disk was placed in the centre of plates, and the isolates were streaked from the margin to the central disk by sterile swab. Two isolates were tested per plate. The plates were then incubated at 35°C for 18–20 hours. The production of a clover leaf-like indentation of the E. coli ATCC 25922 growth indicated a positive result for MHT (CLSI).
Genotypic confirmation test. All isolates were subjected to PCR to check for the presence of the blaKPC gene. K. pneumoniae ATCC BAA-1705 and K. pneumoniae ATCC BAA 1706 were used as quality control strains. A commercial DNA plasmid extraction kit was used (IBRC, Iran) to purify and characterize the plasmid DNA derived from the isolates. The primers used for amplification of blaKPC gene were designed in this study. The forward and reverse primers were KPC-F: ATGTCACTGTATCGCCGTCT, and KPC-R: GCTGTGCTTGTCATCCTTGT (Fazabiotech, Iran), respectively. The primers were used at 1 μmol concentration. The amplification product was expected to be 819 bp in length.
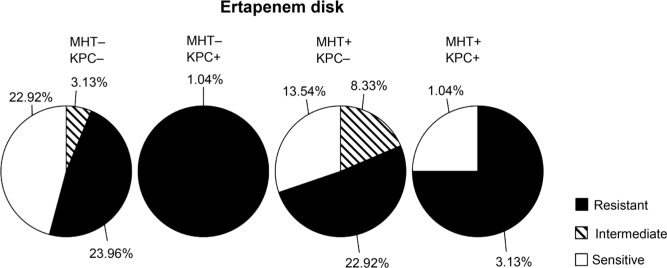
Fig. 1.
Result of antimicrobial susceptibility of positive MHT isolates by ertapenem disk. Antimicrobial susceptibility of ertapenem disk and its patterns compared with existence of the blaKPC gene. Results show that 25% isolates were MHT-negative and resistant to ertapenem disk.
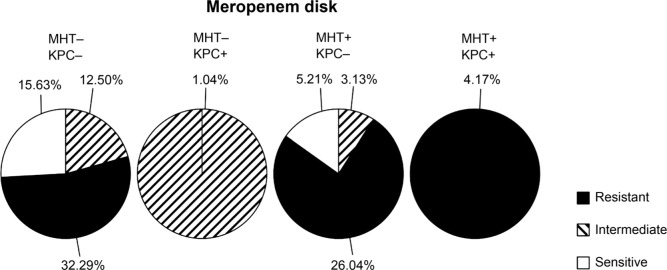
Fig. 2.
Result of antimicrobial susceptibility of positive MHT isolates by meropenem disk. Antimicrobial susceptibility of meropenem disk and its patterns compared with existence of the blaKPC gene. Results show that 32.29% isolates were MHT-negative and resistant to meropenem disk.
PCR reactions were performed using a thermocycler with the following conditions: initial denaturing at 95°C for 5 min, followed by 35 cycles of 60 sec of denaturation at 95°C, annealing at 53°C for 30 sec, elongation at 72°C for 90 sec, and a final extension at 72°C for 10 min.
Results
Bacterial isolation. A total of 96 K. pneumoniae were isolated from 96 different patients of whom 15.6% were outpatient and 84.4% were admitted to internal medicine wards. Clinical specimens included 30 urine, 16 wound swabs, one peritoneal-fluid, 6 blood samples, 34 respiratory secretions, and 9 other specimens.
Positive MHT patterns. 37 isolates were classified as carbapenem-resistant using MHT with the meropenem disk, and 47 with the ertapenem disk. A total of 54 isolates (some isolates showed positive result with the use of both disks) were classified as carbapenemase-producer.
Antimicrobial susceptibility patterns. The antimicrobial susceptibility test for two carbapenem antibiotics revealed that 49 isolates (51.04%) were resistant to ertapenem, and 60 isolates (62.5%) were resistant to meropenem.
Antimicrobial susceptibility patterns based on MHT results. 25 percent of the isolates were resistant to ertapenem, whereas they showed negative results by MHT with ertapenem disk (Table I). Only 29 (30.2%) of the MHT-positive isolates with meropenem disk were resistant to meropenem (Table II).
Table I.
| Sensitive | Intermediate | Resistant | ||
|---|---|---|---|---|
| MHT | Negative | 22.9% | 3.1% | 25.0% |
| Positive | 14.6% | 8.3% | 26.0% | |
The interpretation was performed based in CLSI guideline (M100-S25). Antimicrobial susceptibility tests were determined using disk diffusion methodology.
MHT positive and negative results were interpreted using the CLSI guideline(M100-S25).
Table II.
| Sensitive | Intermediate | Resistant | ||
|---|---|---|---|---|
| MHT | Negative | 15.6% | 13.5% | 32.3% |
| Positive | 5.2% | 3.1% | 30.2% | |
The interpretation was performed based in CLSI guideline (M100-S25). Antimicrobial susceptibility tests were determined using disk diffusion methodology.
MHT positive and negative results were interpreted using the CLSI guideline (M100-S25).
Molecular analysis of blaKPC gene. Five of the 54 isolates, which were carbapenem-resistant and possessed the blaKPC gene, were detected using PCR. Three of these 5 isolates exhibited MHT-positive phenotype with both disks, one with meropenem disk, and one with ertapenem.
Antimicrobial susceptibility of blaKPC gene. The disk diffusion method was interpreted for KPC-producing K. pneumoniae isolates as follows: four isolates (4.17%) were resistant to carbapenem disks, two isolates were sensitive to ertapenem disk, and one isolate (1.04%) was intermediate to meropenem disk.
Discussion
The rapid detection of carbapenemase-producing strains in clinical samples is critically important to provide appropriate treatment. As a phenotypic test, MHT is widely used for first-line detection of carbapenem resistance in clinical laboratories (Carvalhaes et al. 2010; Cury et al. 2012; Chande et al. 2013). In the current study, we evaluated the efficiency of MHT test with two different carbapenem disks as substrate for identification of KPC enzyme and compared it using PCR.
In this examination, 54 K. pneumoniae isolates were MHT-positive, while only five of them were carriers of blaKPC gene. Our finding seems to be consistent with those other studies that obtained false-positive results for MHT. Therefore, the use of MHT alone should not be recommended to confirm the presence of carbapenemase produced by Enterobacteriaceae (Bayramoglu et al. 2016)
Varying sensitivity and specificity of MHT were found in different studies (Doyle et al. 2012; Lari et al. 2014; Shinde et al. 2017; Sun et al. 2017). One study revealed that MHT with ertapenem disk could be utilized for the detection of carbapenemases in isolates that showed intermediate or sensitive zone diameter on disk diffusion (Amjad et al. 2011). Another study demonstrated that the MHT with ertapenem disk had positive predictive significance of KPC detection in Enterobacteriaceae (Cury et al. 2012).
Our results revealed that the sensitivity of MHT test with meropenem and ertapenem disks was equal (80%). Hence, using two carbapenem disks as a substrate for MHT led to excellent sensitivity for the detecting of KPC producers. The specificity of MHT with ertapenem and meropenem disks was found to be low: 53% and 64%, respectively. However, the specificity of ertapenem disk was much lower than the meropenem disk.
MHT cannot be considered as a good indicator for the detection of KPC producers because of its low specificity. The meropenem disk was more effective than ertapenem disk as a substrate for MHT. Nevertheless, MHT results alone are not sufficient to predict carbapenem resistance. One limitation of the study is that we did not detect other carbapenemase producers. MHT showed carbapenemase activity other than carbapenemase production. The resistance could be related to some other mechanisms. PCR yielded sufficient results for detection of KPC-producing isolates. We recommended that improvement in MHT for the screening of KPC producers with high specificity is required for accurate detection.
Acknowledgements
The authors would like to thank Isfahan science and technology town for this work. No funds have been provided for this study.
Footnotes
Ethical approval
The study was approved by the Ethics Committee of the Alzahra Hospital (Letter number A/120). All microbiological samples were taken as part of standards care procedures. No written informed consent was necessary for this type of study.
Literature
- Aliskan HE, Colakoglu S, Turunc T, Demiroglu YZ. 2012. Evaluation of the chromid esbl agar for the detection of esbl-positive Enterobacteriaceae and vancomycin-resistant enterococcus isolates from urine cultures. Mikrobiyol Bul. 46(1):17–25. [PubMed] [Google Scholar]
- Amjad A, Mirza I, Abbasi S, Farwa U, Malik N, Zia F. 2011. Modified hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol. 3(4):189–193. [PMC free article] [PubMed] [Google Scholar]
- Anderson KF, Lonsway DR, Rasheed JK, Biddle J, Jensen B, McDougal LK, Carey RB, Thompson A, Stocker S, Limbago B et al.. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol. 45(8): 2723–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman MA, Breen P, Deornellas V, Mu Q, Zhao L, Wu W, Cavalcoli JD, Mobley HL. 2015. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. MBio. 6(3):e00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barwa R, Shaaban M. 2017. Molecular characterization of Klebsiella pneumoniae clinical isolates with elevated resistance to carbapenems. Open Microbiol J. 11:152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayramoglu G, Ulucam G, Gencoglu Ozgur C, Kilic AO, Aydin F. 2016. Comparison of the modified hodge test and the Carba NP test for detection of carbapenemases in Enterobacteriaceae isolates. Mikrobiyol Bul. 50(1):1–10. [DOI] [PubMed] [Google Scholar]
- Carvalhaes CG, Picao RC, Nicoletti AG, Xavier DE, Gales AC. 2010. Cloverleaf test (modified hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: Be aware of false positive results. J Antimicrob Chemother. 65(2):249–251. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention 2009. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb Mortal Wkly Rep. 58(10):256–260. [PubMed] [Google Scholar]
- Chande C, Veer P, Chivate A, Joshi SG, Chowdhary A. 2013. What should be the criteria for application of modified Hodge test for carbapenemases in Klebsiella pneumoniae? Indian J Med Microbiol. 31(3):318–319. [DOI] [PubMed] [Google Scholar]
- CLSI CaLSI 2016. Performance standards for antimicrobial susceptibility testing; twenty-sixth informational supplement. Wayne, PA. [Google Scholar]
- Cury AP, Andreazzi D, Maffucci M, Caiaffa-Junior HH, Rossi F. 2012. The modified Hodge test is a useful tool for ruling out Klebsiella pneumoniae carbapenemase. Clinics (Sao Paulo). 67(12): 1427–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djahmi N, Dunyach-Remy C, Pantel A, Dekhil M, Sotto A, Lavigne JP. 2014. Epidemiology of carbapenemase-producing Enterobacteriaceae and Acinetobacter baumannii in Mediterranean countries. Biomed Res Int. 2014:305784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. 2012. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 50(12):3877–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I, Rekatsina PD, Hatzaki D, Plachouras D, Souli M, Giamarellou H. 2008. Evaluation of different laboratory tests for the detection of metallo-beta-lactamase production in Enterobacteriaceae. J Antimicrob Chemother. 61(3):548–553. [DOI] [PubMed] [Google Scholar]
- Hansen F, Hammerum AM, Skov RL, Giske CG, Sundsfjord A, Samuelsen O. 2012. Evaluation of rosco neo-sensitabs for phenotypic detection and subgrouping of esbl-, ampc- and carbapenemase-producing Enterobacteriaceae. APMIS. 120(9):724–732. [DOI] [PubMed] [Google Scholar]
- Hashemi A, Fallah F, Erfanimanesh S, Hamedani P, Alimehr S, Goudarzi H. 2014. Detection of beta-lactamases and outer membrane porins among Klebsiella pneumoniae strains isolated in Iran. Scientifica (Cairo). 2014:726179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lari AR, Azimi L, Rahbar M, Alaghehbandan R, Sattarzadeh-Tabrizi M. 2014. First report of Klebsiella pneumoniae carbapenemase-producing Pseudomonas aeruginosa isolated from burn patients in Iran: Phenotypic and genotypic methods. GMS Hyg Infect Control. 9(1):Doc06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu C, Zheng W, Zhang X, Yu J, Gao Q, Hou Y, Huang X. 2008. PCR detection of Klebsiella pneumoniae in infant formula based on 16s-23s internal transcribed spacer. Int J Food Microbiol. 125(3):230–235. [DOI] [PubMed] [Google Scholar]
- Maroncle N, Balestrino D, Rich C, Forestier C. 2002. Identification of Klebsiella pneumoniae genes involved in intestinal colonization and adhesion using signature-tagged mutagenesis. Infect Immun. 70(8):4729–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miriagou V, Cornaglia G, Edelstein M, Galani I, Giske CG, Gniadkowski M, Malamou-Lada E, Martinez-Martinez L, Navarro F, Nordmann P et al.. 2010. Acquired carbapenemases in gram-negative bacterial pathogens: Detection and surveillance issues. Clin Microbiol Infect. 16(2):112–122. [DOI] [PubMed] [Google Scholar]
- Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 17(10): 1791–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteran F, Mendez T, Guerriero L, Rapoport M, Corso A. 2009. Sensitive screening tests for suspected class a carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol. 47(6):1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteran F, Mendez T, Rapoport M, Guerriero L, Corso A. 2010. Controlling false-positive results obtained with the Hodge and Masuda assays for detection of class a carbapenemase in species of Enterobacteriaceae by incorporating boronic acid. J Clin Microbiol. 48(4):1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson DL. 2006. Resistance in gram-negative bacteria: Enterobacteriaceae. Am J Infect Control. 34(5 Suppl 1):S20–28; discussion S64–73. [DOI] [PubMed] [Google Scholar]
- Samra Z, Bahar J, Madar-Shapiro L, Aziz N, Israel S, Bishara J. 2008. Evaluation of Chromagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 46(9):3110–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde S, Gupta R, Raut SS, Nataraj G, Mehta PR. 2017. Carba NP as a simpler, rapid, cost-effective, and a more sensitive alternative to other phenotypic tests for detection of carbapenem resistance in routine diagnostic laboratories. J Lab Physicians. 9(2):100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Xu X, Yan J, Zhang L. 2017. Evaluation of six phenotypic methods for the detection of carbapenemases in Gram-negative bacteria with characterized resistance mechanisms. Ann Lab Med. 37(4):305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasoo S, Cunningham SA, Kohner PC, Simner PJ, Mandrekar JN, Lolans K, Hayden MK, Patel R. 2013. Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase-producing Gram-negative bacilli. J Clin Microbiol. 51(9):3097–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N, Eastaway AT, Ford M, Leanord A, Keane C, Quayle RM, Steer JA, Zhang J, Livermore DM. 2010. Comparison of BD Phoenix, Vitek 2, and Microscan automated systems for detection and inference of mechanisms responsible for carbapenem resistance in Enterobacteriaceae. J Clin Microbiol. 48(8):2999–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]