Since the initial outbreak of COVID-19 and identification of a novel enveloped RNA betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), reports suggest the virus might be evolving, albeit at a lower rate than influenza viruses.1, 2, 3 Nevertheless, in light of the rapid and pandemic-scale spread of COVID-19, mutations in SARS-CoV-2 raise new diagnostic challenges, including the redesign of the oligonucleotide sequences in use in RT-qPCR assays to circumvent potential primer–sample mismatches.
We report an analysis of all high-coverage SARS-CoV-2 genome sequences (1825 in total) deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database4 as of March 30, 2020. We aligned the sequences against the reference sequence obtained from the Wuhan seafood market pneumonia virus isolate, Wuhan-Hu-1 (NC_045512). Subsequently, we annotated in the alignments the binding sites of 33 oligonucleotides developed by different centres and shared by WHO5 for use in the RT-qPCR detection of SARS-CoV-2 from human samples. We then calculated the nucleotide diversity (π)6 in the binding region of each oligonucleotide (figure ).
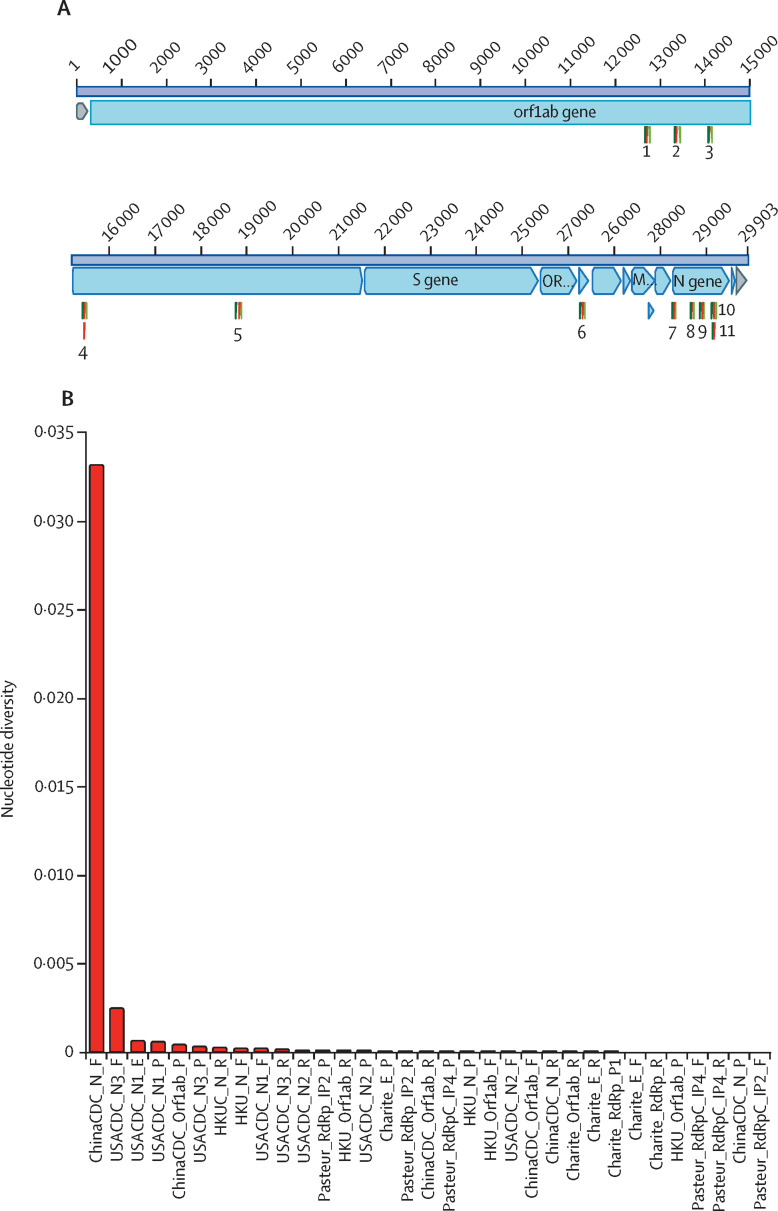
Figure.
Genomic position and nucleotide diversity in the binding sites of oligonucleotides in use in RT-qPCR diagnostic assays across 1825 SARS-CoV-2 genomes isolated from humans in different countries
(A) Graphical representation of a SARS-CoV-2 genome (29 903 nucleotides, NC_045512), showing genes (blue) and oligonucleotide primer binding sites (forward primers in dark green, reverse primers in light green, and double-dye oligonucleotide probes in red). (1) Pasteur_RdRp_IP2, (2) ChinaCDC_Orf1ab, (3) Pasteur_RdRp_IP4, (4) Charité_RdRp, (5) HKU_Orf1ab, (6) Charité_E, (7) USACDC_N1, (8) USACDC_N3, (9) ChinaCDC_N, (10) HKU_N, and (11) USACDC_N2.5 (B) Nucleotide diversity (π) in the binding sites of the oligonucleotides in use in RT-qPCR diagnostic assays across 1825 SARS-CoV-2 genomes isolated from humans in different countries. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
The analysis revealed that 79% (26 of 33) of the primer binding sites used in the RT-qPCR assays were mutated in at least one genome. The substitution of three nucleotides (GGG substituted to AAC) at the beginning of the binding site of the forward primer designed by the Chinese National Institute for Viral Disease Control and Prevention in the gene encoding for the nucleocapsid phosphoprotein was of relevance. The AAC variant was in 14% (258 of 1825) of the genomes isolated and sequenced in 24 different countries (Australia, Belgium, Brazil, Chile, China, Czech Republic, Denmark, England, Finland, France, Germany, Greece, Hungary, Iceland, Italy, Mexico, the Netherlands, Peru, Portugal, Spain, Switzerland, the USA, Vietnam, and Wales).
Notwithstanding the possibility of sequencing errors, the fact that some variants were consistently found in different sequencing experiments at independent test sites suggests that these genetic variants are true variants. Moreover, the observation that at least one of the previously designed primers is now likely to be ineffective at detecting up to 14% of the virus variants in circulation strengthens the need to continue optimising the oligonucleotides in use in assays being developed. Oligonucleotide optimisation will be facilitated by global sharing of SARS-CoV-2 genomes and the frequently updated reports on sequence analysis that are available on the GISAID4 website.
Acknowledgments
We declare no competing interests. We acknowldege all authors originating and submitting the sequences used in this Correspondence (appendix).
Supplementary Material
References
- 1.Lai A, Bergna A, Acciarri C, Galli M, Zehender G. Early phylogenetic estimate of the effective reproduction number of SARS-CoV-2. J Med Virol. 2020 doi: 10.1002/jmv.25723. published online Feb 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, Wang W, Zhao X, et al. Transmission dynamics and evolutionary history of 2019-nCoV. J Med Virol. 2020;92:501–511. doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shu Y, McCauley J. GISAID: Global Initiative on Sharing All Influenza Data—from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO World Health Organization (WHO) list of in-house-developed molecular assays for SARS-CoV-2 detection. 2020. https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf
- 6.Tajima F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.