Abstract
Background:
Attention-deficit/hyperactivity disorder (ADHD) is a neurodevelopmental disorder which is consistently associated with lower levels of educational attainment. A recent large genome-wide association study (GWAS) identified common gene variants associated with ADHD, but most of the genetic architecture remains unknown.
Methods:
We analyzed independent GWAS summary statistics for ADHD (19,099 cases and 34,194 controls), educational attainment (EDU) (n = 842,499) and general intelligence (INT) (n = 269,867) using a conditional/conjunctional false discovery rate (condFDR/conjFDR) statistical framework that increases power of discovery by conditioning the FDR on overlapping associations. The genetic variants identified were characterized in terms of function, expression and biological processes.
Results:
We identified 58 LD-independent ADHD-associated loci (condFDR < 0.01), of which 30 are shared between ADHD and EDU or INT (conjFDR < 0.01), and 46 are novel risk loci for ADHD.
Conclusions:
These results expand on previous genetic and epidemiological studies and support the hypothesis of a shared genetic basis between these phenotypes. Although the clinical utility of the identified loci remains to be determined, they can be used as resources to guide future studies aiming to disentangle the complex etiologies of ADHD, educational attainment and general intelligence.
Keywords: Cognition, pleiotropy, genetic overlap, mental health, psychiatric disorder, ADHD
Introduction
Attention-deficit/hyperactivity disorder (ADHD) is a neurodevelopmental psychiatric disorder that affects approximately 5% of children and 2.5% of adults globally, with an estimated heritability of 0.7 to 0.8 (1, 2). Epidemiological and clinical studies implicate genetic and environmental factors in the etiology of the disorder, many of which affect the structure and functional capacity of brain networks involved in behavior and cognition (1, 2). As a result, ADHD is consistently associated with lower levels of educational attainment (3, 4) and children with ADHD experience cognitive problems such as increased risk of learning disabilities and communication disorders (3, 5–7).
Despite the high heritability, significant ADHD-associated risk loci were only recently identified using the genome-wide association study (GWAS) methodology (8). In addition, success was also obtained using a conditional/conjunctional false discovery rate (condFDR/conjFDR) method (9). This method exploits the shared genetic background of phenotypes to boost association signals in a phenotype of interest by employing genome-wide association data from one or more secondary phenotypes. By combining an educational attainment GWAS in more than 300,000 individuals (10) with an initial moderately powered GWAS of ADHD (11), Shadrin et al. identified five novel loci for ADHD risk and provided evidence for shared genetic basis between ADHD and educational attainment (9).
After the publication of that study, two larger GWASs for general intelligence (n=269,867) (12), and educational attainment (n > 1.1 million) (13) uncovered multiple novel loci associated with these phenotypes. Furthermore, GWAS summary statistics for a substantially larger ADHD cohort are now also available (8). Following in the steps of Shadrin et al. (9), we therefore aimed to apply the condFDR/conjFDR approach to these new GWAS summary statistics in order to identify additional novel loci associated with ADHD and shared between ADHD and educational attainment or general intelligence. In addition, we performed positional and functional annotation of significant ADHD-associated variants to explore their potential biological context.
Methods and Materials
GWAS Samples
GWAS summary statistics for ADHD were obtained from the Psychiatric Genomics Consortium (PGC) (8). The summary statistics for general intelligence (INT) were obtained from the meta-analysis of 14 independent cohorts (12). For our analyses of educational attainment (EDU) we used summary statistics generated from meta-analysis of data from the Social Science Genetic Association Consortium (13) and 23andMe (10). The meta-analysis was performed using an inverse-weighted fixed effects model implemented in the software METAL (http://csg.sph.umich.edu//abecasis/Metal/) (14). All participants in the GWAS samples were of European origin. A summary of these GWAS samples is shown in Table 1. More detailed descriptions are available in the Supplementary Methods and original publications (8, 12, 13).
Table 1.
GWAS summary statistics characteristics
| Sample | Sample Size (N)* | Sample Size inlcuded (N)** | Age Group | Ref |
|---|---|---|---|---|
| PGC1,2 | 53 293 (19 099 ADHD, 34 194 CON) |
53 293 | Adult and Children | (8) |
| INT1 | 269 867 | 269 867 | Adult and Children | (12) |
| EDU1 | 1 131 881 | 842 499 | Adult | (13) |
| deCODE2 | - | 348 561 (10 217 ADHD, 338 344 CON) |
Adult and Children | - |
| EAGLE2 | 17 666 | 17 666 | Children | (23) |
Sample size of the cohort in the referenced study
Sample size of the cohort included and analyzed in this study.
GWAS summary statistics used for condFDR/conjFDR analyses.
GWAS summary statistics used for sign concordance evaluation.
INT, general intelligence. EDU, educational attainment. ADHD, cases. CON, controls.
Statistical Analyses
To assess cross-phenotype polygenic enrichment we generated conditional QQ-plots, conditioning ADHD on EDU or INT and vice versa. QQ-plots depict the quantiles of the observed p-values on the y-axis against the theoretical quantiles under no association on the x-axis. In the case of no association a QQ-plot follows a straight line, but deflects from this null line when some form of systematic association is present. Conditional QQ-plots depict the differential enrichment between pre-specified strata of single nucleotide polymorphisms (SNPs). Points on the QQ-plot are weighted according to LD structure, using n=200 iterations of random pruning at LD threshold r2=0.1. We focused on the SNP p-values of trait 1 (ADHD), and defined strata based on trait 2 (EDU or INT). More specifically we plotted the SNP p-values of trait 1 conditional on different strength of association with trait 2 (i.e. –log10 p-values > 1, 2, or 3). This enables us to determine if conditioning on a secondary trait leads to stronger association in the primary trait of interest. A stronger enrichment together with increased evidence for association with the secondary trait can be an indicator of a shared polygenic architecture between the two traits. To further support this, we estimated the genetic correlation between ADHD and EDU or INT using LD score regression (15–17).
To identify shared loci between ADHD and EDU or INT we employed the condFDR/conjFDR method (18, 19). The condFDR method utilizes genetic association summary statistics from a trait of interest (ADHD) together with those of a conditional trait (EDU or INT) to estimate the posterior probability that a SNP has no association with the primary trait, given that the p-values for that SNP in both the primary and conditional traits are lower than the observed p-values. This method increases the power to identify loci associated with the primary trait by leveraging associations with conditional traits, thereby re-ranking SNPs compared to the original GWAS p-value ranking. The conjFDR statistic is defined as the maximum of the two mutual condFDR values and is a conservative estimate of the posterior probability that a SNP has no association with either trait, given that the p-values for that SNP in both the primary and conditional traits are lower than the observed p-values. The conjFDR method thus allows the identification of loci associated with both traits. A conservative FDR level of 0.01 per pair-wise comparison was set for condFDR/conjFDR, corresponding to 1 false positive per 100 reported associations. More details can be found in the original and subsequent publications (9, 18–22), and Supplementary Methods.
Evaluation of Detected Loci in Two Independent ADHD Cohorts
To assess the robustness of the condFDR/conjFDR results we examined the most significant SNPs in the identified loci in the association summary statistics from a case-control ADHD cohort from deCODE Genetics and the GWAS on ADHD symptoms conducted by the EAGLE consortium (23) (Table 1). Additional details are provided in the Supplementary Methods. Sign concordance tests were performed to compare the effect directions for the identified SNPs between the PGC-GWAS (8) and the deCODE and EAGLE samples, respectively. Fisher’s exact tests were used to determine if the number of concordant SNPs was significantly greater (p < 0.025 = 0.05/2 cohorts) than expected by chance in each comparison.
In Silico Analyses of Significant Variants
Positional and functional annotation of significantly associated SNPs was performed using ANNOVAR (24), implemented in FUMA (25). To evaluate the potential biological context of significantly associated genetic variants identified through condFDR/conjFDR analyses we queried for known expression quantitative trait loci (eQTLs) in brain tissue using the GTEx portal (http://gtexportal.org), the Braineac database (http://www.braineac.org) and the CommonMind Consortium knowledge portal (https://www.synapse.org/#!Svnapse:syn2759792). In addition, we checked age-dependent variations of expression for the genes associated with identified eQTL SNPs using the Human Brain Transcriptome database (http://hbatlas.org) (26).
Results
Genetic Overlap and Correlation
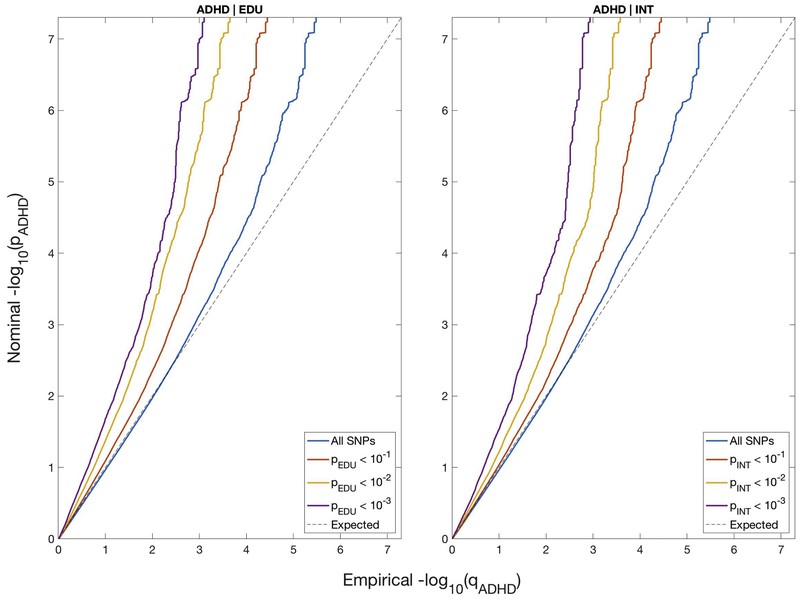
The conditional QQ-plots show strong enrichment for ADHD given EDU or INT (Figure 1). The blue lines are drawn using the genome-wide summary statistics for ADHD, including all SNPs regardless of their association with EDU or INT. An increasingly leftward deflection from the dashed line of no association is observed with stronger associations with EDU or INT. Furthermore, we note the symmetry of the observed enrichment and show the conditional QQ-plots for EDU or INT given ADHD in Figure S1.
Figure 1.
Conditional QQ-plots of nominal vs empirical –log10 p-values (corrected for inflation) in ADHD below the standard genome-wide association study threshold of p < 5.0 × 10−8 as a function of significance of association with educational attainment (EDU) or general intelligence (INT) at the level of –log10 p-values of 1, 2, or 3, corresponding to p = 0.10, p = 0.01 and p = 0.001, respectively. The dashed lines indicate the null hypothesis.
We used partitioned LD score regression to assess the statistical significance of enrichment for each QQ-plot stratum (15). After adjusting for multiple testing (two conditional traits and three strata) we identified significant enrichment for ADHD given EDU or INT for all three strata (Table S1). For ADHD given EDU, the enrichment parameters ranged from 2.877 (–log10pval >1) to 4.916 (–log10pval >2) and 8.093 (–log10pval >3), while for ADHD given INT the enrichment parameters ranged from 2.586 (–log10pval >1) to 5.046 (–log10pval >2) and 6.866 (–log10pval >3). Significant enrichment parameters for EDU or INT given ADHD for all three strata were also identified (Table S1). Moreover, LD score regression analyses also showed significant negative genetic correlation between ADHD and EDU (rg −0.520, SE 0.025, p = 1.333 × 10−93) and between ADHD and INT (rg −0.366, SE 0.030, p = 1.023 × 10−34), respectively.
ADHD-Associated Loci and Related Genes
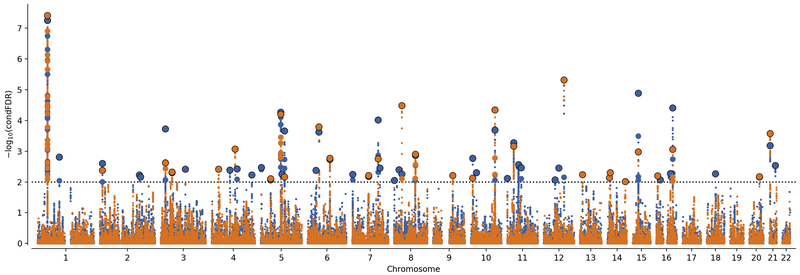
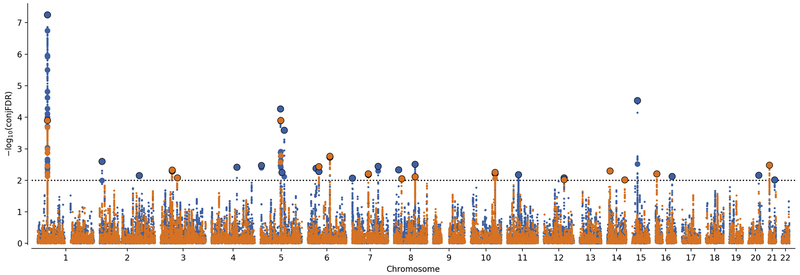
Using condFDR we identified 48 (Table S2) and 31 (Table S3) LD-independent loci to be significantly (condFDR < 0.01) associated with ADHD after conditioning on association with EDU and INT, respectively. To provide a map of shared loci between ADHD and EDU and INT we performed conjFDR analyses. We thereby identified 24 shared loci between ADHD and EDU (conjFDR < 0.01), of which seven are novel to both ADHD and EDU (Table S4). Similarly, we identified 15 loci shared by ADHD and INT (conjFDR < 0.01), of which four are novel to both phenotypes (Table S5). Manhattan plots from condFDR and conjFDR analyses are presented in Figures 2 and 3, respectively.
Figure 2.
Conditional False Discovery Rate (condFDR) Manhattan Plot of Conditional –log10(FDR) Values. ADHD conditioned on educational attainment (EDU) (ADHD|EDU) is shown in blue and ADHD conditioned on general intelligence (ADHD|INT) is shown in orange. Linkage disequilibrium (LD) independent single nucleotide polymorphisms (SNPs) with conditional –log10(FDR) higher than 2.0 (horizontal dotted line) (ie, condFDR < 0.01) are shown with large points. A black line around the large points indicates the most significant SNP in a locus.
Figure 3.
Conjunctional False Discovery Rate (conjFDR) Manhattan Plot of Conjunctional –log10(FDR) Values. ADHD and Educational Attainment (EDU) (ADHD & EDU) is shown in blue, and ADHD and General Cognitive Ability (ADHD & INT) is shown in orange. Linkage disequilibrium (LD) independent single nucleotide polymorphisms (SNPs) with conjunctional –log10(FDR) higher than 2.0 (horizontal dotted line) (ie, conjFDR < 0.01) are shown with large points. A black line around the large points indicates the most significant SNP in a locus.
Combining the results of the aforementioned analyses yields a list of 58 LD-independent loci associated with ADHD (Table 2, Table S6), by condFDR and conjFDR analyses with EDU or INT. Thirty of these loci are shared between ADHD and EDU or INT. The majority of the 58 loci showed discordant direction of effect between ADHD and EDU (52 loci, 23 shared loci (ADHD&EDU conjFDR < 0.01)), and ADHD and INT (51 loci, 15 shared loci (ADHD&INT conjFDR < 0.01)), respectively. Nine loci showed concordant direction of effect between ADHD and EDU or ADHD and INT (3 shared loci (ADHD&EDU or ADHD&INT conjFDR < 0.01)), of which three loci were concordant between all three phenotypes. Twelve of these loci were significantly associated with ADHD in the previous GWAS (8) (Table S6), and three loci were previously reported for ADHD by leveraging polygenic overlap with educational attainment (9) (Table S6). All SNPs with conjFDR < 0.1 (ADHD&EDU and ADHD&INT) and r2 ≧ 0.6 with a representative SNP are shown in Table S7. Gene-set analysis of the genes implicated by the SNPs within the 58 loci (Table 2, Table S6 and Table S7) revealed no significantly enriched biological processes, cellular components or molecular functions.
Table 2.
The 58 LD-independent loci associated with ADHD, by condFDR and conjFDR analyses with educational attainment (EDU) or general intelligence (INT).
| Locus | Chr | Lead SNP | A1/A2 | Nearest Gene | Functional category | ADHD|EDU condFDR | ADHD&EDU conjFDR | ADHD|INT condFDR | ADHD&INT conjFDR | P-value ADHD | P-value EDU | P-value INT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | rs112984125 | G/A | KDM4A-AS1:ST3GAL3 | ncRNA_intronic | 5.622E-08 | 5.622E-08 | 3.898E-08 | 5.068E-04 | 1.08E-12 | 2.11E-23 | 8.61E-08 |
| 2 | 1 | rs2391734 | G/T | RNU1-130P | intergenic | 1.553E-03 | 1.923E-01 | 2.766E-02 | 1.000E+00 | 6.70E-08 | 7.81E-03 | 8.60E-01 |
| 3* | 2 | rs55748262 | G/A | PDIA6 | intergenic | 2.492E-03 | 2.492E-03 | 4.150E-03 | 4.614E-02 | 2.94E-06 | 8.45E-14 | 3.00E-04 |
| 4* | 2 | rs2676507 | G/A | RAPGEF4 | intronic | 5.955E-03 | 6.942E-03 | 2.948E-01 | 9.305E-01 | 1.09E-05 | 1.23E-06 | 5.98E-01 |
| 5* | 2 | rs79699670 | G/A | RBM45 | intronic | 6.818E-03 | 9.253E-02 | 4.494E-02 | 5.049E-01 | 2.36E-06 | 1.08E-03 | 6.23E-02 |
| 6 | 2 | rs13023832 | G/A | SPAG16:AC107218.3 | ncRNA_intronic | 9.270E-03 | 9.997E-01 | ND | ND | 9.33E-08 | 1.37E-01 | ND |
| 7 | 3 | rs4858241 | T/G | RNU6-815P | intergenic | 1.892E-04 | 1.051E-01 | 2.410E-03 | 6.905E-01 | 8.17E-09 | 1.89E-03 | 2.29E-01 |
| 8* | 3 | rs12493769 | A/G | SNRK:ANO10 | intronic | 5.161E-02 | 2.576E-01 | 9.620E-03 | 2.764E-02 | 2.09E-05 | 1.53E-02 | 1.19E-04 |
| 9* | 3 | rs28535523 | C/T | UBA7 | intronic | 4.986E-03 | 4.986E-03 | 4.721E-03 | 4.721E-03 | 8.25E-06 | 1.38E-20 | 2.65E-07 |
| 10* | 3 | rs6789751 | T/C | FOXP1 | intronic | 1.097E-02 | 1.097E-02 | 9.86E-03 | 9.86E-03 | 2.92E-05 | 1.84E-09 | 1.78E-06 |
| 11* | 3 | rs11710737 | A/G | BBX | intronic | 3.842E-03 | 1.514E-02 | 1.243E-02 | 1.132E-01 | 5.55E-06 | 1.89E-05 | 1.55E-03 |
| 12* | 3 | rs7634587 | A/G | BBX | intronic | 1.789E-02 | 2.708E-01 | 9.560E-03 | 1.755E-01 | 2.20E-06 | 2.14E-02 | 3.76E-03 |
| 13 | 4 | rs28522755 | A/G | PCDH7 | intergenic | 3.141E-02 | 8.895E-01 | 3.819E-03 | 3.442E-01 | 1.53E-07 | 7.56E-01 | 1.88E-02 |
| 14* | 4 | rs1484144 | T/C | LINC01088:NAA11 | ncRNA_intronic | 4.105E-03 | 5.372E-02 | 1.472E-01 | 9.259E-01 | 1.98E-06 | 1.61E-04 | 5.89E-01 |
| 15* | 4 | rs227372 | T/C | MANBA | intronic | 2.099E-02 | 8.693E-01 | 8.547E-04 | 1.508E-01 | 8.43E-08 | 6.00E-01 | 2.74E-03 |
| 16* | 4 | rs72678859 | C/T | RP11-255I10.1 | intergenic | 3.789E-03 | 3.789E-03 | 8.591E-02 | 5.542E-01 | 5.43E-06 | 1.94E-08 | 8.89E-02 |
| 17* | 4 | rs62338074 | T/C | GPM6A | intronic | 5.921E-03 | 2.166E-02 | 9.039E-02 | 5.079E-01 | 8.47E-06 | 2.51E-05 | 6.36E-02 |
| 18* | 5 | rs13163845 | T/C | CTD-2029E14.1 | intergenic | 3.337E-03 | 3.337E-03 | 2.509E-02 | 2.650E-01 | 4.51E-06 | 2.10E-08 | 9.58E-03 |
| 19* | 5 | rs13176429 | T/C | ZNF131 | intronic | 8.511E-03 | 2.851E-01 | 7.845E-03 | 3.211E-01 | 5.03E-07 | 2.45E-02 | 1.55E-02 |
| 20 | 5 | rs4916723 | A/C | LINC00461 | ncRNA_intronic | 5.352E-05 | 5.352E-05 | 3.122E-03 | 1.25E-04 | 1.81E-08 | 2.32E-13 | 1.44E-01 |
| 21* | 5 | rs7733142 | C/A | FAM172A | intronic | 5.508E-03 | 5.508E-03 | 2.019E-01 | 7.348E-01 | 9.62E-06 | 3.41E-08 | 2.93E-01 |
| 22* | 5 | rs12658032 | A/G | RP11-6N13.1 | ncRNA_intronic | 2.202E-04 | 2.550E-04 | 6.925E-03 | 5.183E-01 | 1.15E-07 | 3.05E-10 | 6.86E-02 |
| 23* | 6 | rs57349798 | G/A | RP1-153P14.8 | ncRNA_intronic | 4.164E-03 | 4.164E-03 | 6.048E-02 | 4.389E-01 | 6.27E-06 | 7.60E-10 | 3.86E-02 |
| 24* | 6 | rs141547796 | G/A | RP1-28O17.1 | intergenic | 2.398E-04 | 5.223E-03 | 1.622E-04 | 4.285E-03 | 9.64E-08 | 1.40E-05 | 3.44E-06 |
| 25* | 6 | rs4839923 | G/A | RP11-436D23.1 | ncRNA_intronic | 1.827E-03 | 1.827E-03 | 1.699E-03 | 1.699E-03 | 1.90E-06 | 2.46E-15 | 1.12E-08 |
| 26* | 7 | rs61409925 | G/A | MAD1L1 | intronic | 5.653E-03 | 8.469E-03 | 1.990E-01 | 9.762E-01 | 1.90E-05 | 2.31E-09 | 6.97E-01 |
| 27* | 7 | rs1978102 | C/T | CALN1 | intronic | 6.510E-03 | 6.510E-03 | 6.159E-03 | 6.159E-03 | 1.25E-05 | 1.58E-18 | 1.74E-15 |
| 28 | 7 | rs9969232 | G/A | FOXP2 | intronic | 9.586E-05 | 3.707E-03 | 1.474E-02 | 9.336E-01 | 3.87E-08 | 1.65E-06 | 6.05E-01 |
| 29* | 7 | rs3757541 | A/G | CADPS2 | intronic | 3.486E-03 | 2.106E-01 | 4.881E-02 | 9.886E-01 | 2.04E-07 | 5.72E-03 | 7.26E-01 |
| 30* | 8 | rs1532744 | A/G | ERICH1-AS1 | ncRNA_intronic | 8.935E-03 | 1.401E-01 | 1.562E-02 | 2.662E-01 | 2.07E-06 | 2.46E-03 | 9.68E-03 |
| 31* | 8 | rs4383968 | T/C | LINC00681 | ncRNA_intronic | 9.360E-03 | 9.299E-02 | 1.989E-01 | 7.216E-01 | 1.01E-05 | 1.34E-04 | 2.72E-01 |
| 32* | 8 | rs4739249 | A/C | AC009695.1 | intergenic | 3.966E-03 | 4.623E-03 | 8.431E-02 | 5.389E-01 | 5.83E-06 | 9.46E-07 | 7.96E-02 |
| 33 | 8 | rs74760947 | A/G | RP1-84O15.2 | intergenic | 5.563E-03 | 8.077E-01 | 3.271E-05 | 8.829E-03 | 1.39E-08 | 5.31E-01 | 1.38E-05 |
| 34 | 8 | rs10956838 | A/C | RP11-700E23.2 | intergenic | 1.365E-03 | 4.134E-03 | 1.260E-03 | 7.59E-03 | 1.28E-06 | 4.23E-07 | 2.63E-05 |
| 35* | 9 | rs295268 | T/C | GKAP1 | intronic | 1.955E-02 | 1.141E-01 | 6.215E-03 | 2.049E-02 | 1.27E-05 | 2.14E-03 | 6.91E-05 |
| 36* | 10 | rs3928823 | G/A | RP11-575N15.1 | intergenic | 1.855E-03 | 4.685E-02 | 7.396E-03 | 2.672E-01 | 6.66E-07 | 6.47E-05 | 9.76E-03 |
| 37* | 10 | rs220370 | T/C | KIAA1217 | intronic | 4.988E-03 | 1.471E-02 | 3.053E-01 | 1.000E+00 | 8.25E-06 | 1.13E-05 | 7.72E-01 |
| 38* | 10 | rs10786831 | T/G | SORCS3 | intronic | 5.254E-04 | 6.115E-03 | 4.544E-05 | 5.55E-03 | 1.08E-05 | 1.14E-06 | 9.51E-05 |
| 39* | 11 | rs28633403 | G/A | RPLP2 | downstream | 7.556E-03 | 2.747E-01 | 2.874E-02 | 6.374E-01 | 4.46E-07 | 1.23E-02 | 1.62E-01 |
| 40* | 11 | rs4275621 | A/G | RP11-960D24.1 | intergenic | 5.273E-04 | 2.707E-02 | 3.406E-03 | 2.781E-01 | 2.03E-07 | 3.90E-05 | 1.08E-02 |
| 41* | 11 | rs11040490 | T/G | RP11-707M1.1 | ncRNA_intronic | 3.433E-03 | 6.601E-03 | 1.216E-01 | 7.683E-01 | 1.28E-05 | 1.62E-06 | 3.41E-01 |
| 42* | 11 | rs1791794 | A/G | DAGLA | intergenic | 3.407E-03 | 3.320E-02 | 4.177E-02 | 4.803E-01 | 2.45E-06 | 4.78E-05 | 5.21E-02 |
| 43* | 12 | rs7953911 | T/C | KCNH3 | intronic | 8.496E-03 | 1.932E-02 | 8.343E-02 | 3.954E-01 | 1.66E-05 | 2.14E-05 | 2.79E-02 |
| 44* | 12 | rs10400419 | T/C | HMGA2 | intergenic | 3.522E-03 | 2.438E-02 | 6.402E-02 | 5.414E-01 | 3.41E-06 | 6.76E-05 | 8.10E-02 |
| 45 | 12 | rs1427829 | A/G | RP11-1109F11.3 | upstream | 6.962E-06 | 8.223E-03 | 4.818E-06 | 9.36E-03 | 1.35E-09 | 3.64E-06 | 3.04E-05 |
| 46* | 13 | rs66931513 | A/G | WDR95P | intergenic | 1.034E-02 | 4.857E-02 | 5.795E-03 | 2.177E-02 | 1.09E-05 | 1.29E-04 | 7.73E-05 |
| 47* | 14 | rs140802584 | A/G | CTD-2384A14.1:RP11-148E17.1 | ncRNA_intronic | 1.579E-02 | 7.490E-02 | 7.258E-03 | 2.284E-02 | 1.49E-05 | 1.22E-03 | 8.42E-05 |
| 48* | 14 | rs2300861 | C/T | AKAP6 | intronic | 1.316E-02 | 8.374E-02 | 5.016E-03 | 5.016E-03 | 9.04E-06 | 9.18E-04 | 1.85E-12 |
| 49* | 14 | rs12435486 | G/A | RP11-61O1.1 | ncRNA_intronic | 2.998E-02 | 1.251E-01 | 9.675E-03 | 9.675E-03 | 2.64E-05 | 1.44E-03 | 3.20E-07 |
| 50 | 15 | rs8039398 | T/C | SEMA6D | intronic | 1.314E-05 | 3.721E-05 | 1.061E-03 | 6.761E-01 | 2.99E-09 | 3.75E-11 | 2.09E-01 |
| 51* | 16 | rs11861310 | C/T | RP11-420N3.2 | ncRNA_intronic | 1.629E-02 | 8.850E-02 | 6.236E-03 | 6.236E-03 | 1.28E-05 | 5.66E-04 | 2.27E-06 |
| 52* | 16 | rs1428102 | G/A | RPL7P47 | upstream: downstream | 8.555E-03 | 1.043E-01 | 1.363E-01 | 7.843E-01 | 2.98E-06 | 9.10E-04 | 3.65E-01 |
| 53* | 16 | rs1369918 | G/A | CDH8 | intronic | 5.347E-03 | 3.742E-02 | 2.385E-02 | 2.534E-01 | 4.51E-06 | 1.92E-04 | 8.58E-03 |
| 54 | 16 | rs212178 | G/A | AC004158.2 | ncRNA_intronic | 3.914E-05 | 8.266E-03 | 8.681E-04 | 4.177E-01 | 1.20E-08 | 3.37E-06 | 3.30E-02 |
| 55* | 18 | rs4144756 | G/A | RP11-188I24.1 | intergenic | 5.377E-03 | 3.489E-01 | 4.708E-02 | 1.000E+00 | 1.46E-07 | 2.08E-02 | 9.11E-01 |
| 56* | 20 | rs2024568 | T/C | RPL13P2 | intergenic | 7.068E-03 | 7.068E-03 | 6.679E-03 | 2.102E-02 | 1.42E-05 | 3.37E-09 | 7.28E-05 |
| 57* | 21 | rs992936 | T/C | NEK4P1 | intergenic | 6.537E-04 | 3.910E-02 | 2.679E-04 | 3.602E-03 | 1.78E-07 | 1.45E-04 | 2.50E-06 |
| 58* | 21 | rs2898433 | C/T | YRDCP3 | intergenic | 2.915E-03 | 9.528E-03 | 1.860E-01 | 8.972E-01 | 1.04E-05 | 4.36E-06 | 5.37E-01 |
The most strongly associated SNPs in novel genomic loci associated with ADHD at condFDR/conjFDR < 0.01 (bold typeset) with EDU or INT after merging regions < 250 kb apart into a single locus. The table presents chromosomal position (Chr), nearest gene and functional category. Conditional FDR (condFDR) values are reported when ADHD is conditioned on EDU (ADHD|EDU) and when ADHD is conditioned on INT (ADHD|INT). The inverse condFDR results (EDU|ADHD and INT|ADHD) are not shown. The conjunctional FDR (conjFDR) columns report the maximum condFDR value, from each pair of condFDR analyses, for each SNP. FDR values in bold typeset are significant FDR < 0.01. P-values from the original summary statistics on ADHD (8), EDU (13) and INT (12) are also reported. P-values in bold typeset are significant p < 5.0E-8. For more details see Supplementary Table 6.
Indicates novel ADHD loci, defined as those not associated with ADHD in the original GWAS (8).
ND, not determined.
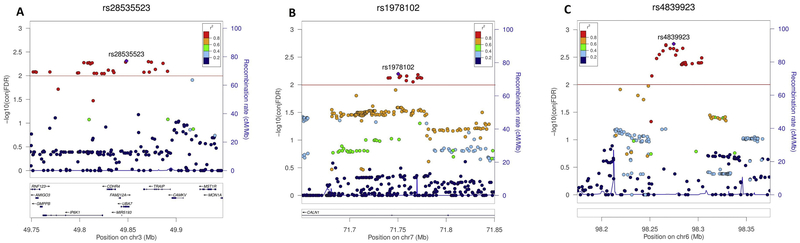
Four LD-independent loci are shared between ADHD, EDU and INT (conjFDR < 0.01), and are represented by SNPs rs112984125, rs28535523, rs4839923, rs1978102 (Table 2, Table S6). The intronic ST3GAL3 rs112984125 was previously associated with ADHD risk (8, 9), and showed the most significant association with ADHD (ADHD|INT condFDR = 3.999 × 10−8) in this study. The remaining three loci are novel for ADHD, however they were all previously significantly associated with EDU (13), and rs1978102 and rs4839923 were also previously significantly associated with INT (12). Both rs28535523 and rs1978102 are intronic variants within the UBA7 gene on chromosome 3p21.31 (Figure 4A) and CALN1 gene on chromosome 7q11.22 (Figure 4B), respectively. No protein-coding genes were identified to be in the region represented by intergenic SNP rs4839923 on chromosome 6q16.1 (Figure 4C).
Figure 4.
Genetic context for three novel loci associated with ADHD, and shared between ADHD, educational attainment (EDU) and general intelligence (INT) in conjunctional false discovery rate (conjFDR). The SNPs –log10(conjFDR) values are shown on the left y-axes. In each sub-plot the representative SNP for the locus (strongest association with ADHD) is shown in the purple square. The color of the remaining markers reflects the degree of linkage disequilibrium (LD) with the representative SNP measured as r2 coefficient. The recombination rate is plotted in blue and it’s value is indicated on the right y-axes. The red line indicates the FDR threshold (conjFDR < 0.01). Surrounding of the strongest association in conjFDR analysis: (A) rs28535523 (conjFDR = 4.376 × 10−3), (B) rs1978102 (conjFDR = 5.789 × 10−3) and (C) rs4839923 (conjFDR = 1.479 × 10−3). Figures are generated with LocusZoom (47).
Evaluation of Detected Loci in Two Independent ADHD Cohorts
Of the 58 LD-independent loci identified through condFDR/conjFDR analyses, the lead SNPs within 44 loci showed the same direction of effect in the PGC (8) and EAGLE (23) GWASs (significantly more than expected by chance, p = 0.007) (Table S6). This was consistent for the previously identified loci (10/12; p = 0.193) (8), novel loci identified in this study (34/46; p = 0.031) and when only considering shared (conjFDR < 0.01) loci (22/30; p = 0.063) (Table S6). When comparing the effect direction for lead SNPs within the 58 loci between the PGC and deCODE ADHD cohorts, 29 showed the same effect direction (p = 1.000) (Table S6). Similar results were observed when considering lead SNPs in previously identified loci (4/12; n.s.) (8) and novel loci identified in this study (24/46; n.s.) (Table S6). A slightly improved concordance rate was observed for shared (conjFDr < 0.05) loci (18/30; n.s.).
In Silico Identification of Variant Effects on Transcription
In order to determine if the SNPs identified by condFDR/conjFDR are associated with gene expression in brain tissues we evaluated the brain regions within the GTEx database with all 58 representative SNPs from Table S6. Nineteen SNPs were identified as potential eQTLs, predicted to alter the expression of 22 genes, in GTEx brain regions (Table S8). In order to validate these findings we further evaluated these 19 eQTL SNPs in the Braineac database and CMC knowledge portal. Five of these 19 SNPs were also identified as eQTLs for 10 genes in brain regions in the Braineac database (Table S9), while 10 of the 19 SNPs were identified as eQTLs for 23 genes in the dorsolateral prefrontal cortex in the CMC knowledge portal (Table S10). The most significant eQTLs were observed between rs28633403 and PIDD1 in the cerebellum in the GTEx database (p = 2.63 × 10−17) and between rs28633403 and NS3BP in the thalamus in the Braineac database (p = 4.00 × 10−9). The rs28633403 SNP was also observed as an eQTL for PNPLA2 in the frontal cortex in the GTEx (p = 4.55 × 10−6) and Braineac databases (p = 1.20 × 10−4) and in the dorsolateral prefrontal cortex in the CMC knowledge portal (FDR < 0.01). According to Human Brain Transcriptome data (26), 19 genes identified from evaluation of the GTEx, Braineac and CMC databases (Tables S8 - S10) have apparent expression in different brain regions during development and adulthood (Figure S2).
Discussion
This study identified 58 ADHD-associated loci by leveraging genetic overlap between ADHD, EDU and INT, of which 30 are shared between ADHD, EDU and INT (Table 2, Table S6). Of these loci, 46 are novel risk loci for ADHD (Table 2, Table S6). These results suggest shared polygenic architecture between educational attainment, general intelligence and ADHD, which may further our understanding of the relationship between these phenotypes observed in epidemiological studies (1, 2).
We identified polygenic overlap between ADHD and both EDU and INT, as illustrated by the increasingly significant enrichment in ADHD when conditioning on EDU or INT (Figure 1, Table S1). The majority of the identified shared loci show discordant effects on ADHD and EDU or INT (Table S6). These findings are consistent with the phenotypic relationship whereby risk alleles for ADHD are associated with lower educational attainment and reduced general intelligence scores, and the significant negative genetic correlations between both ADHD and EDU (rg −0.520, SE 0.025, p = 1.333 × 10–93), as well as ADHD and INT (rg −0.366, SE 0.030, p = 1.023 × 10–34). An advantage of the condFDR/conjFDR method is to discover loci with both similar and opposite effects. Interestingly, nine loci show concordant effect directions for ADHD and EDU or INT (Table S6), three of which show concordant effect directions between all three phenotypes. The majority of these concordant loci are represented by intergenic SNPs, however, for two of the loci the nearest genes include PCDH7 and CADPS2 (Table S6). The PCDH7 and CADPS2 genes are implicated in epilepsy (27), autism spectrum disorder and learning disability (28), respectively. The PCDH7 protein is also known to bind to phosphatase 1α within dendritic spines where it may play a role in learning and memory (29). Further investigation of these concordant loci is warranted since this may help to explain some of the heterogeneity seen among patients with ADHD. These results add further support to the hypothesis of a shared complex genetic basis underlying ADHD, educational attainment and general intelligence.
Only four of the significant ADHD-associated risk loci identified in this study were implicated by the conjFDR analysis with both EDU and INT (Table 2, Table S6). The most significant SNPs for these regions are rs112984125, rs28535523, rs4839923 and rs1978102. Three of these loci (lead SNPs: rs28535523, rs4839923 and rs1978102) are novel for ADHD risk, although they were previously implicated in EDU (13) and INT (12), and may therefore provide new insights into the underlying mechanisms of the disorder. The intronic ST3GAL3 rs112984125 showed the most significant association with ADHD (ADHD|INT condFDR = 3.999 × 10–8) in this study, and was previously implicated through the most recent ADHD GWAS (8) and the condFDR/conjFDR method employed in this study (9).
The rs28535523 SNP is located on chromosome 3p21.31 and is intronic to the UBA7 gene (Figure 4A). A nonsense mutation located at chr3:49848458, 44 bp away from rs28535523 and within this risk locus (Table S6), was previously associated with mild cognitive disability (30). Furthermore, rs28535523 was identified as an eQTL for the AMT gene in both the GTEx and Braineac databases (Tables S8 and S9). The AMT gene has previously been implicated in autism spectrum disorder (31)
The intronic CALN1 rs1978102 SNP is located on chromosome 7q11.22 (Figure 4B). Although there is no evidence previously implicating this gene in ADHD etiology, deletions in a region containing the AUTS2, WBSCR17 and CALN1 genes were associated with a syndromic form of intellectual disability (32). Furthermore, this locus was also identified as a risk locus for schizophrenia after conditioning on educational attainment using the same method described in this study (33).
The fourth shared ADHD-risk locus identified is rs4839923 on chromosome 6q16.1 (Figure 4C). No protein-coding genes were identified in this region; however, this SNP is intronic to a long non-coding RNA (lncRNA) RP11–436D23.1. lncRNAs have been implicated in a number of neurological and psychiatric disorders (34), including fragile X mental retardation (35), schizophrenia (36, 37) and autism spectrum disorder (38), highlighting the need to better characterize their role in other brain-related phenotypes such as ADHD.
Although only identified by condFDR, as a novel risk locus for ADHD, rs28633403 was the most significant eQTL identified in the GTEx database with PIDD1 (Table S8) and in the Braineac database with NS3BP (Table S9), respectively. This SNP was also the only SNP identified as an eQTL for the same gene (PNPLA2) within the same brain tissue (frontal cortex) in all databases (Table S8–10). The PIDD1 gene was previously associated with ADHD risk by gene-wise association (8), and the PNPLA2 gene was implicated in ADHD risk after being identified within a gene set significantly enriched in ADHD copy number variations (39). These results highlight potential mechanisms through which this locus may influence ADHD risk.
In addition to the loci mentioned above, all 12 ADHD-risk loci identified in the most recent GWAS were maintained (8), and three of the five ADHD-risk loci previously identified using this condFDR/conjFDR methodology were replicated (9). The two non-replicated loci, on chromosome 1p36.12 and 2p24, were also not identified in the most recent GWAS (8). Furthermore, the 1p36.12 locus was represented by only a single SNP (rs17414302) with no LD-linked SNPs in the direct vicinity highlighting the potential of a false positive (9). These results demonstrate the sensitivity of the condFDR/conjFDR methodology to the quality and power of the GWASs employed for these analyses. As such, the condFDR/conjFDR method shares some of the limitations and strengths of GWASs in that sample size limits the power to detect associations and that identified associations require replication. As the sample sizes and ensuing power of GWASs increase so too does the power of this method to identify cross-phenotype polygenic enrichment.
The most significant SNPs identified by condFDR/conjFDR analyses were evaluated in two independent ADHD cohorts, a case-control cohort (deCODE) and a GWAS on ADHD symptoms (EAGLE) (23). Lead SNPs within 44 of 58 loci identified in our study showed consistent direction of effect between the PGC-GWAS (8) and the EAGLE GWAS (23) (Table S6). This concordance rate is similar to that reported for the genome-wide significant loci in the PGC-GWAS (10/12 sign concordance) (8) a and when considering the novel loci identified in this study (34/46 sign concordance). ADHD diagnosis and continuous measures of ADHD, including symptom scores, have been shown to share substantial genetic background (± 90%) (8). Furthermore, polygenic risk scores calculated from associations with ADHD diagnosis have also been shown to predict variability in ADHD symptoms (40). The consistent direction of effects found here may therefore be considered as a validation of our findings. Lead SNPs for 29/58 loci were concordant between the PGC-GWAS (8) and deCODE case-control cohort. A similar difference in effect concordance, between these two cohorts, has been previously reported (8). These differences may be due to the difference in ascertainment of ADHD affected individuals in the deCODE cohort compared to the PGC and EAGLE cohorts (Table 1). These results highlight the need for large well-powered independent cohorts to replicate identified genetic loci.
Despite the focus on representative (most significant) SNPs within the identified loci, as always with GWAS, it must be considered that these SNPs may be in LD with other causal SNPs. Further studies are required to identify truly causal variants with biological relevance that may explain the cross-phenotype polygenic enrichment observed between ADHD, educational attainment and general intelligence. Furthermore, we do not know in what way the alleles identified here confer risk to ADHD and influence cognitive performance. Some of the overlapping gene loci may be driven by the natural occurrence of ADHD in the general population from which the EDU and INT samples were recruited. Although this is likely a small fraction (~ 2%) (1, 2), some of the identified shared genetic architecture could be driven by this effect. It is also possible that the identified shared loci might influence a common cognitive sub-phenotypic trait affecting both ADHD risk and cognitive performance such as attention, or that the loci might affect more basic neurobiological mechanisms that contribute to both higher-level phenotypes. Although no significantly enriched biological processes, cellular components or molecular functions were identified in this study, a number of the identified genes were previously implicated in the genetic overlap between schizophrenia and intelligence (FOXP1, CALN1, SORCS3 and AKAP6) (20, 21), and bipolar disorder and intelligence (CDH8 and RP11-436D23.1) (21). These findings are suggestive of a common genetic architecture underlying the relationship between psychiatric disorders and cognitive performance, in line with identified common-variant correlations (41). Similar biological processes to those identified for the shared loci between schizophrenia and intelligence, related to neurodevelopment, synaptic integrity, and neurotransmission (21), may therefore also play a role in the shared genetic component of ADHD and intelligence. Discovery of additional ADHD-risk loci is required to increase the statistical power of gene-set analysis to better understand the underlying neurobiological mechanisms.
ADHD medications are effective at reducing core ADHD symptoms (42, 43), however they are also known to improve academic performance (44, 45). This provides further evidence suggestive of overlapping biological mechanisms between cognitive performance and ADHD, in line with the current findings of shared polygenic architecture. However, despite the discovery of several novel ADHD-risk loci, and the implication of a number of novel genes, these results are not yet of clinical relevance for treatment of individual patients. Future studies are required to unravel and understand the complex underlying genetic architecture of ADHD, and how it overlaps with cognitive phenotypes, to reach the level of clinical utility.
Previous analysis of the PGC ADHD and EDU datasets using the condFDR/conjFDR method highlighted the sample overlap (WTCCC58C cohort) (46) between these datasets, which may potentially inflate the condFDR/conjFDR results. This overlap, however, is very limited, and amounts to approximately 2800 ADHD control samples (8) that were also included in the EDU GWAS (13). To the best of our knowledge no ADHD cases were shared between any of the datasets used in these analyses.
Since all of the GWAS summary statistics analyzed in this study were generated from cohorts of European ancestry, as was the case for the original GWAS, the results may not be generalizable to non-European populations. In addition, the difference in prevalence of ADHD in children and adults (1, 2) suggests that age specific factors may interact with genetic risk factors. However, currently available GWAS data does not allow for analyses of potential age-genotype interactions.
In conclusion, we have demonstrated shared polygenic architecture between ADHD and both EDU and INT. We leveraged this genetic overlap to identify 46 novel risk loci for ADHD, four of which are associated with ADHD risk, educational attainment and general intelligence. Interestingly, using the condFDR/conjFDR method we identified nine loci with concordant effects on ADHD and EDU or INT, contrasting the genome-wide genetic correlation findings between these phenotypes. These results expand on previous genetic and epidemiological studies to further support the hypothesis of a shared genetic basis between these phenotypes. Although the clinical utility of the identified risk loci remains to be determined, they can be used as resources to guide future studies aiming to disentangle the complex etiologies of ADHD, educational attainment and general intelligence.
Supplementary Material
KEY RESOURCES TABLE
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Deposited Data; Public Database | PGC-ADHD GWAS summary statistics | 30478444; | ||
| Deposited Data; Public Database | Intelligence GWAS summary statistics | PMID: 29942086 | ||
| Deposited Data; Public Database | Educational attainment GWAS summsary statis | PMID: 30038396 | ||
| Deposited Data; Public Database | ADHD symptoms GWAS summsary statistics | PMID: 27663945 | ||
| Software; Algorithm | Conditional/ConjunctionalFDR scripts | https://github.com/precimed/pleiofdr |
Acknowledgements
Funding: NIH (NS057198, EB00790); the Research Council of Norway (229129, 213837, 223273, 226971); the South-East Norway Regional Health Authority (2013–123); KG Jebsen Foundation (SKGJ-2011–36). The authors thank the Psychiatric Genetics Consortium (PGC), the Social Science Genetic Association Consortium (SSGAC) and the Complex Traits Genetics (CTG) Consortium for access to GWAS data. The authors thank Thomas Bjella, of the Oslo University Hospital & Institute of Clinical Medicine, for support with the database. We thank the research participants and employees of 23andMe, Inc. for their contribution to this study.
Disclosures
Dr. Andreassen reports grants from Research Council of Norway, grants from KG Jebsen Stiftelsen, grants from South-East Norway Health Authority, during the conduct of the study; personal fees from Lundbeck, outside the submitted work. Dr. Dale reports that he is a Founder of and holds equity in CorTechs Labs, Inc., and serves on its Scientific Advisory Board. He is a member of the Scientific Advisory Board of Human Longevity, Inc., and receives funding through research grants with General Electric Healthcare. The terms of these arrangements have been reviewed by and approved by UCSD in accordance with its conflict of interest policies. Dr. Steen reports grants from NIH, grants from Research Council of Norway, grants from South-East Norway Regional Health Authority, grants from KG Jebsen Foundation, during the conduct of the study. Dr. Fan reports personal fees from Multi-Modal Imaging Service, outside the submitted work. Dr. Haavik reports personal fees from Eli-Lilly, personal fees from HB Pharma, personal fees from Medice, personal fees from Biocodex, personal fees from Shire, outside the submitted work. G.B.W., O.O.G., H.S. and K.S. are employees of deCODE genetics/Amgen. All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Faraone SV, Larsson H (2018): Genetics of attention deficit hyperactivity disorder. Mol Psychiatry. doi: 10.1038/s41380-018-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franke B, Michelini G, Asherson P, Banaschewski T, Bilbow A, Buitelaar JK, et al. (2018): Live fast, die young? A review on the developmental trajectories of ADHD across the lifespan. Eur Neuropsychopharmacol. 28: 1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claesdotter E, Cervin M, Åkerlund S, Råstam M, Lindvall M (2018): The effects of ADHD on cognitive performance. Nord J Psychiatry. 72: 158–163. [DOI] [PubMed] [Google Scholar]
- 4.Voigt RG, Katusic SK, Colligan RC, Killian JM, Weaver AL, Barbaresi WJ (2017): Academic Achievement in Adults with a History of Childhood Attention-Deficit/Hyperactivity Disorder: A Population-Based Prospective Study. J Dev Behav Pediatr. 38: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strine TW, Lesesne CA, Okoro CA, McGuire LC, Chapman DP, Balluz LS, Mokdad AH (2006): Emotional and behavioral difficulties and impairments in everyday functioning among children with a history of attention-deficit/hyperactivity disorder. Prev Chronic Dis. 3: A52. [PMC free article] [PubMed] [Google Scholar]
- 6.Czamara D, Tiesler CMT, Kohlböck G, Berdel D, Hoffmann B, Bauer C-P, et al. (2013): Children with ADHD symptoms have a higher risk for reading, spelling and math difficulties in the GINIplus and LISAplus cohort studies. PLoS ONE. 8: e63859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korrel H, Mueller KL, Silk T, Anderson V, Sciberras E (2017): Research Review: Language problems in children with Attention-Deficit Hyperactivity Disorder - a systematic meta-analytic review. J Child Psychol Psychiatry. 58: 640–654. [DOI] [PubMed] [Google Scholar]
- 8.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. (2019): Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 51: 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shadrin AA, Smeland OB, Zayats T, Schork AJ, Frei O, Bettella F, et al. (2018): Novel Loci Associated With Attention-Deficit/Hyperactivity Disorder Are Revealed by Leveraging Polygenic Overlap With Educational Attainment. J Am Acad Child Adolesc Psychiatry. 57: 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, et al. (2016): Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 533: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch K- P, et al. (2010): Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 49: 884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. (2018): Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 50: 912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. (2018): Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 50: 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer CJ, Li Y, Abecasis GR (2010): METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, et al. (2015): Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 47: 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulik-Sullivan BK, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, et al. (2015): An atlas of genetic correlations across human diseases and traits. Nat Genet. 47: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. (2015): LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 47: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, et al. (2013): Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 92: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreassen OA, Thompson WK, Dale AM (2013): Boosting the Power of Schizophrenia Genetics by Leveraging New Statistical Tools. Schizophr Bull. 40: 13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smeland OB, Frei O, Kauppi K, Hill WD, Li W, Wang Y, et al. (2017): Identification of Genetic Loci Jointly Influencing Schizophrenia Risk and the Cognitive Traits of Verbal-Numerical Reasoning, Reaction Time, and General Cognitive Function. JAMA Psychiatry. 74: 1065–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smeland OB, Bahrami S, Frei O, Shadrin A, O’Connell K, Savage J, et al. (2019): Genome wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry. doi: 10.1038/s41380-018-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smeland OB, Frei O, Shadrin A, O’Connell K, Fan C-C, Bahrami S, et al. (2019): Discovery of shared genomic loci using the conditional false discovery rate approach. Hum Genet. doi: 10.1007/s00439-019-02060-2. [DOI] [PubMed] [Google Scholar]
- 23.Middeldorp CM, Hammerschlag AR, Ouwens KG, Groen-Blokhuis MM, Pourcain BS, Greven CU, et al. (2016): A Genome-Wide Association Meta-Analysis of Attention-Deficit/Hyperactivity Disorder Symptoms in Population-Based Pediatric Cohorts. J Am Acad Child Adolesc Psychiatry. 55: 896–905. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Li M, Hakonarson H (2010): ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe K, Taskesen E, Bochoven A van, Posthuma D (2017): Functional mapping and annotation of genetic associations with FUMA. Nature Communications. 8: 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. (2011): Spatio-temporal transcriptome of the human brain. Nature. 478: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manford M (2017): Recent advances in epilepsy. J Neurol. 264: 1811–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonora E, Graziano C, Minopoli F, Bacchelli E, Magini P, Diquigiovanni C, et al. (2014): Maternally inherited genetic variants of CADPS2 are present in autism spectrum disorders and intellectual disability patients. EMBO Mol Med. 6: 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braithwaite SP, Stock JB, Lombroso PJ, Nairn AC (2012): Protein phosphatases and Alzheimer’s disease. Prog Mol Biol Transl Sci. 106: 343–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam M, Trampush JW, Yu J, Knowles E, Davies G, Liewald DC, et al. (2017): Large-scale Cognitive GWAS Meta-Analysis Reveals Tissue-Specific Neural Expression and Potential Nootropic Drug Targets. Cell Rep. 21: 2597–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, et al. (2013): Using whole-exome sequencing to identify inherited causes of autism. Neuron. 77: 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beunders G, Voorhoeve E, Golzio C, Pardo LM, Rosenfeld JA, Talkowski ME, et al. (2013): Exonic Deletions in AUTS2 Cause a Syndromic Form of Intellectual Disability and Suggest a Critical Role for the C Terminus. Am J Hum Genet. 92: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Hellard S, Wang Y, Witoelar A, Zuber V, Bettella F, Hugdahl K, et al. (2017): Identification of Gene Loci That Overlap Between Schizophrenia and Educational Attainment. Schizophr Bull. 43: 654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blokhin I, Khorkova O, Hsiao J, Wahlestedt C (2018): Developments in lncRNA drug discovery: where are we heading? Expert Opin Drug Discov. 1–13. [DOI] [PubMed] [Google Scholar]
- 35.Peschansky VJ, Pastori C, Zeier Z, Wentzel K, Velmeshev D, Magistri M, et al. (2016): The long non-coding RNA FMR4 promotes proliferation of human neural precursor cells and epigenetic regulation of gene expression in trans. Mol Cell Neurosci. 74: 49–57. [DOI] [PubMed] [Google Scholar]
- 36.Gibbons A, Udawela M, Dean B (2018): Non-Coding RNA as Novel Players in the Pathophysiology of Schizophrenia. Noncoding RNA. 4. doi: 10.3390/ncrna4020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian T, Wei Z, Chang X, Liu Y, Gur RE, Sleiman PMA, Hakonarson H (2018): The Long Noncoding RNA Landscape in Amygdala Tissues from Schizophrenia Patients. EBioMedicine. 34: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Zhao X, Ju W, Flory M, Zhong J, Jiang S, et al. (2015): Genome-wide differential expression of synaptic long noncoding RNAs in autism spectrum disorder. Transl Psychiatry. 5: e660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos-Quiroga J-A, Sánchez-Mora C, Casas M, Garcia-Martínez I, Bosch R, Nogueira M, et al. (2014): Genome-wide copy number variation analysis in adult attention-deficit and hyperactivity disorder. J Psychiatr Res. 49: 60–67. [DOI] [PubMed] [Google Scholar]
- 40.Groen-Blokhuis MM, Middeldorp CM, Kan K-J, Abdellaoui A, van Beijsterveldt CEM, Ehli EA, et al. (2014): Attention-deficit/hyperactivity disorder polygenic risk scores predict attention problems in a population-based sample of children. J Am Acad Child Adolesc Psychiatry. 53: 1123–1129. e6. [DOI] [PubMed] [Google Scholar]
- 41.Anttila V, Bulik-Sullivan B, Finucane HK, Walters R, Bras J, Duncan L, et al. (2017): Analysis of shared heritability in common disorders of the brain. bioRxiv. 048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaughan BS, March JS, Kratochvil CJ (2012): The evidence-based pharmacological treatment of paediatric ADHD. Int J Neuropsychopharmacol. 15: 27–39. [DOI] [PubMed] [Google Scholar]
- 43.Caye A, Swanson JM, Coghill D, Rohde LA (2019): Treatment strategies for ADHD: an evidence-based guide to select optimal treatment. Mol Psychiatry. 24: 390–408. [DOI] [PubMed] [Google Scholar]
- 44.Baweja R, Mattison RE, Waxmonsky JG (2015): Impact of Attention-Deficit Hyperactivity Disorder on School Performance: What are the Effects of Medication? Paediatr Drugs. 17: 459–477. [DOI] [PubMed] [Google Scholar]
- 45.Keilow M, Holm A, Fallesen P (2018): Medical treatment of Attention Deficit/Hyperactivity Disorder (ADHD) and children’s academic performance. PLoS ONE. 13: e0207905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Power C, Elliott J (2006): Cohort profile: 1958 British birth cohort (National Child Development Study). Int J Epidemiol. 35: 34–41. [DOI] [PubMed] [Google Scholar]
- 47.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. (2010): LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.