Abstract
Zika virus (ZIKV) is emerging as a significant pathogen worldwide and may cause severe neurological disorders such as fetal microcephaly and Guillain-Barre syndrome. No drug or listed vaccines are currently available for preventing ZIKV infection. As a major target of neutralizing, ZIKV envelop (E) protein usually used for vaccine development. Nevertheless, the immunogenicity of ZIKV envelop (E) protein expressed by baculovirus display system has never been assessed. In this study, we reported a new strategy for surface display of ZIKV E protein by a recombinant baculovirus vector derived from Autographa californica multiple nuclear polyhedrosis virus (AcMNPV) and assessed its immunogenicity in mice. We produced recombinant fusion ZIKV E protein linked with signal peptide (SP) and transmembrane domain (TM) of AcMNPV GP64. The results showed that the recombinant protein was easy to produce by baculovirus display system. BALB/c mice immunized with this recombinant E protein developed ZIKV specific serum antibodies. The anti-E protein sera from the mice were able to effectively neutralize ZIKV in vitro. More importantly, AG6 (IFN-α/β and IFN-γ receptor deficient) mice immunized with recombinant E protein were protected against lethal ZIKV challenge. Together, these findings demonstrated that the recombinant E protein displayed by baculovirus can be conveniently prepared and displayed good immunogenicity in immunized mice. It is a promising practical approach for prompting the development of vaccine and related immunology research.
Electronic supplementary material
The online version of this article (10.1007/s12250-020-00238-x) contains supplementary material, which is available to authorized users.
Keywords: Zika virus (ZIKV), Envelope protein, Baculovirus, Immunogenicity, Neutralizing antibody, Viral challenge
Introduction
Zika virus (ZIKV) is an enveloped, positive single-stranded RNA virus and a member of genus Flavivirus of Flavividae family (Musso and Gubler 2016; Wikan and Smith 2016; Martinez-Sobrido and Almazán 2019). The entire genome of ZIKV is about 11 kb in length, encoding three structural proteins, envelope (E), pre-membrane/membrane (prM/M), and capsid (C) along with seven non-structural proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 (Enfissi et al. 2016; Kostyuchenko et al. 2016; Richner et al. 2017; Lin et al. 2018). The structural proteins play a significant role in viral entry, fusion and assembly. E protein is the primary target of neutralizing antibodies (Abbink et al. 2017; Dai et al. 2018; Saiz et al. 2018). Two genetic strains of ZIKV are identified, African and Asian/American, with only one serotype (Richner et al. 2017). ZIKV is transmitted by Aedes aegypti. Modes of its transmission can be sexual, mother-to-fetus, or transfusion via blood (D’Ortenzio et al. 2016; Morrison and Diamond 2017). Historically, it was considered as a modest public health concern, causing mild fever, maculopapular rash, and arthralgia in 20% of patients, with 80% of infections being asymptomatic (Dowall et al. 2016; Mlakar et al. 2016; Zhao et al. 2016). However, it had raised major public health concerns since 2015, when a large outbreak occurred in Brazil and rapidly spreaded to other countries in Central and South America (da Silva et al. 2018). ZIKV has become a clinically significant pathogen since its association with severe neurological diseases such as fetal microcephaly, Guillain–Barre syndrome and congenital joint-limb contractures (White et al. 2016; Dávila-Castrodad et al. 2018; de Araújo et al. 2018). Increasing evidence has indicated that ZIKV infection can also lead to infertility (Govero et al. 2016; Ma et al. 2017). There are an estimated 2.6 billion people at risk worldwide and 242 million people in China, are at risk of exposure to ZIKV infection (Bogoch et al. 2016). Clearly, ZIKV is a global pathogen occurring in Americas, Africa, and Asia, and poses a serious threat to global public health (Zhao et al. 2016).
The quest for development of vaccine against ZIKV has made spectacular progress over the past 4 years (Ghaffar et al. 2018). Several familiar vaccine candidates including live attenuated virus (LAV), DNA, purified inactivated virus (PIV), mRNA, and vectors vaccines are being tested on animal models or are already in clinical trials (Garg et al. 2018). However, there are some limitations about these candidates. LAV and DNA vaccines incur risks of viral activation or oncogenicity, especially in immunocompromised individuals. PIV and mRNA vaccines induce weak immunogenicity and are unstable. Among vector vaccines, adenovirus and measles virus are commonly used as vectors. However, pre-existing immune response aimed at these vectors in the host may weaken the immune effect of the vaccine (Saxena et al. 2013). Therefore, one of the biggest conundrums in research is to find a vaccine that is safe, effective in a vast majority and highly susceptible target populations, especially pregnant women and immunosuppressed patients (Garg et al. 2018; Ghaffar et al. 2018). Thereby, it becomes necessary to try another insect specific viral vector, baculovirus expression vector.
Recombinant baculovirus vectors derived from the Autographa californica multiple nuclear polyhedrosis virus (AcMNPV) are widely used to express foreign proteins in insect cells (Ono et al. 2018). These proteins are produced by inserting a gene fragment in frame between the signal peptide (SP) and the mature domain of the major surface glycoprotein 64 (GP64) nucleotide sequence of AcMNPV (Premanand et al. 2018). After expression in insect cells, these proteins are processed, modified and translocated to the plasma membrane, then displayed on the surface of the recombinant baculovirus with the GP64 (Kost et al. 2005). Proteins displayed in this manner could be highly accessible to immunorecognition when using viral particles as immunogens (Premanand et al. 2018). This method has been extended to develop pseudotype baculovirus as a potential recombinant protein delivery platform (Kaname et al. 2010).
Although ZIKV vaccine development has had a quick head start, further development may be hampered because of the decline in cases globally (Durbin and Wilder-Smith 2017). Currently, there are no authorized ZIKV vaccines and specific antivirals approved for clinical use, making prophylactic and effective vaccination the first and best approach to combat the disease (da Silva et al. 2018; Saiz et al. 2018). Furthermore, new outbreaks of ZIKV are unpredictable. Given that recombinant baculovirus is a promising recombinant protein expression vehicle (Wu et al. 2009), the primary aim of this study was to effectively display E protein on the baculovirus surface, with the hope that E protein would retain superior immunogenicity upon in vivo immunization.
Materials and Methods
Cells, Reagents, and Antibodies
Spodoptera frugiperda cell line Sf9 were grown at 27 °C in Grace’s insect media (Thermo Fisher Scientific, Waltham, MA, United States) supplemented with 10% fetal bovine serum (FBS, Life Technology, Australia). Recombinant baculovirus were propagated and titrated in Sf9 cells.
African green monkey kidney epithelial cells (Vero, ATCC CCL-81, American Type Culture Collection) were cultured at 37 °C with 5% CO2 in minimal essential medium (MEM, Thermo Fisher Scientific) supplemented with 10% FBS, 100 units/mL penicillin and 100 μg/mL streptomycin.
The following antibodies were used for Western blot analysis: ZIKV E protein monoclonal antibody (BioFront, BF-1176-56, Tallahassee, USA), Anti-baculovirus envelope GP64 protein (eBioscience, 14-6995-82, San Diego, USA) and HRP-conjugated secondary antibodies (Boster, Wuhan, China). FITC-conjugated Goat anti-mouse IgG (Proteintech, Wuhan, China) was used in immunofluorescence assays (IFA). Gold-conjugated Goat anti-mouse IgG (Boster, Wuhan, China), as the secondary antibody, was used in immuno-electron microscopy (IEM).
Complete Freund’s adjuvant and Incomplete Freund’s adjuvant (Sigma, USA) were used to immunize mice.
Virus Stock and Cell Culture
ZIKV strain SZ-WIV01 (GenBank: KU963796) was obtained from the Key Laboratory of Special Pathogens and Biosafety, Center for Emerging Infectious Diseases, Wuhan Institute of Virology, Chinese Academy of Sciences (Deng et al. 2016). The virus was grown in Vero cells. Supernatants containing viral particles were clarified by centrifugation at 2000 rpm at 4 °C before being stored at −80 °C. The viral stock titer was determined by plaque formation assay.
Construction of Recombinant Baculovirus
Recombinant baculovirus containing sequences coding for ZIKV E protein without transmembrane structure (TM) were constructed using the Bac-to-Bac baculovirus expression system (Invitrogen), which is based on site-specific transposon-mediated insertion of the foreign gene of interest from a donor plasmid pFastBac into a baculovirus genome called a bacmid in Escherichia coli. In brief, the genomic RNA of the ZIKV strain SZ-WIV01 was reversely transcribed using PrimeScriptTM RT reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions (Supplementary Table S1). The resultant cDNA was used as a template for the amplification of gene fragments coding for ZIKV E protein. The DNA fragment harbouring signal peptide (SP, 63 bp) of baculoviral envelope GP64, ZIKV E protein (1362 bp, 153 bp at the TM of itself was deleted) and transmembrane structure (TM, 90 bp) of GP64 were linked together by overlap PCR, the gene of recombinant protein is still 1515 bp (Supplementary Table S1). This construct was inserted into pFastBac vector at EcoR I/Hind III sites under the control of baculovirus polyhedrin (Ph) promoter, to obtain expression vector plasmids FastBac-E-GP64 (pFastBac-E-GP64) which could display recombinant protein EGP64 on the surface of baculovirus. The resultant construct pFastBac-E-GP64 was confirmed by enzyme digestion and DNA sequencing.
Generation of Recombinant Baculovirus
pFastBac-E-GP64 was used to generate the corresponding baculovirus. The procedures, including vector transformation, generation, selection and propagation of the recombinant baculovirus, were carried out according to the protocol of Bac-to-Bac baculovirus expression system (Invitrogen). In this regard, pFastBac-EGP64 was transformed into competent DH10Bac E. coli cells for generating recombinant bacmids. After selecting colonies through two rounds of blue/white selection, recombinant bacmids were isolated from white colonies. The recombinant DNA was then examined for the presence of the insert by PCR using E-specific and baculovirus GP64-specific primer pairs (Supplementary Table S1). The positive colonies were cultured to isolate the bacmid DNA.
Then, Sf9 cells were allowed to attach for 1 h. After attachment, recombinant bacmid DNA was transfected into 80% confluent Sf9 cells in 6-well plates to generate a corresponding recombinant baculovirus designated Bac-EGP64(EG). Transfected cells were incubated for 5 h at 27 °C and the transfection medium was replaced with fresh medium. After incubation for 72 h at 27 °C, the recombinant baculoviruses were harvested and purified by two-three rounds of plaque isolations. Individual recombinant viruses were subjected to biochemical analyses and measured titer by TCID50, then high titer stocks were utilized for infecting cells.
Biochemical Analyses
After 72 h post transfection, the Sf9 cells were harvested separately by centrifugation, then cells were lysed on ice for 10 min in RIPA lysis buffer (Beyotime) supplemented with PMSF (Beyotime) and protease inhibitor cocktail tablets (Roche). Following boiling at 95 °C for 10 min, the total protein in supernatant and whole cell lysates were respectively separated by 10% SDS-PAGE and then electro-transferred onto Immobilon-P PVDF membrane (Merck Millipore, Burlington, MA, US) in transfer buffer (30 mmol/L Tris, 200 mmol/L glycine, 20% (V/V) methanol) for 2.5 h at 4 °C. The membranes were blocked using 5% BSA dissolved in TBST (50 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.1% Tween 20, pH 7.4) for 1 h at 37 °C and then incubated with primary antibodies diluted with primary antibody dilution buffer (Beyotime) overnight at 4 °C. After washing with TBST 5 times for 7 min each, the membranes were incubated with HRP-conjugated secondary antibodies diluted with TBST with 0.5% BSA for 1 h at 37 °C. After washing for 5 times, the membranes were incubated with an Immobilon Western Chemiluminescent HRP substrate (Millipore) and subjected to a Bio-Rad Imaging System. The bands were analyzed by using Image Lab 4.0.1.
Meanwhile, Sf9 cells seeded in dishes (Nest Biotechnology, Wuxi, Jiangsu, China) were infected with Bac-EGP64(EG). After 72 h, cells were washed, fixed with 4% formaldehyde, and permeabilized with 2% Triton X-100. The fixed cells were blocked with PBS buffer containing 2% BSA (BioFroxx, Einhausen, Germany) and 5% normal goat serum (Beyotime Institute of Biotechnology, Nanjing, China) and then incubated with ZIKV E protein monoclonal antibody and anti-baculovirus GP64 protein (diluted 1:200) in PBS for 1 h at 37 °C. After washing 5 times with PBS, the cells were incubated with FITC-conjugated goat anti-mouse antibody at a 1:200 dilution for 1 h at 37 °C. After washing, cell nuclei were counterstained with Hoechst 33258 (Beyotime Institute of Biotechnology) and visualized on fluorescence microscope (Olympus Inverted Fluorescence 1X51 Microscope, Japan).
The Sf9 cells infected with EG were harvested and immobilized in glutaraldehyde after 72 h post infection (h.p.i.), then baculovirus particles were observed using a transmission electron microscope (TEM, Tecnai G2 20 TWIN, FEI company, USA).
Production and Purification of Recombinant Baculovirus
Sf9 cells were infected with Bac-EGP64 at a MOI of 5 for 72 h. The infected cells were harvested by centrifugation at 3000 rpm for 30 min followed by centrifugation at 10,000 rpm for 20 min at 4 °C. The supernatant was passed through a 0.45 μm filter to remove the debris and then concentrated using a 25% sucrose cushion at 27,000 rpm (SW32 rotor; Beckman) for 2 h at 4 °C. The pellets were resuspended in NTE buffer. For sucrose gradient sedimentation, the clarified lysates were layered onto 15%–65% sucrose gradients for ultracentrifugation at 35,000 rpm (SW41 rotor; Beckman) for 3 h. Then, 2 fractions were taken from top to bottom and were examined by Western blot. The recombinant E protein-enriched sucrose gradient fractions were resuspended in NTE buffer and subjected to 27,000 rpm (SW32 rotor; Beckman) for 2 h at 4 °C for desugaring. The pellets were resuspended in 1000 µL NTE buffer for subsequent TEM analysis and immunization. Then, the total protein concentration of EG was determined by BCA method. The E protein concentration in EG was determined by ELISA. The E protein of the known concentration expressed by E. coli was set as the standard substance and diluted serially. Both E proteins were coated on ELISA plates respectively, followed by the immune response of primary antibody, secondary antibody and chromogenic response, then the absorbance was detected. A standard curve was generated according to the absorbance of E protein of the known concentration, and the E protein quantity in EG was calculated.
Immune Electron Microscopy (IEM)
Purified recombinant baculovirus particles were adsorbed onto carbon-coated nickel grids (200 mesh) and blocked with 5% BSA for IEM. The primary antibodies were ZIKV E protein monoclonal antibody. The 12-nm colloidal gold-AffiniPure goat anti-mouse IgG (Boster, Wuhan, China) was used as the secondary antibody. Subsequently, the grids were stained negatively and examined with TEM.
Immunization of BALB/c Mice
Prior to immunization, recombinant Bac-EGP64(EG) (E protein was 15 μg/dose) were formulated separately with complete Freund’s adjuvant (1st immunization) or incomplete Freund’s adjuvant (2nd, 3rd immunization) by vortex at volume ratio 1:1. Non-recombinant baculovirus (WT) and PBS were formulated with the adjuvant and used as controls. Specific-pathogen-free, 6-week-old BALB/c female mice (n = 6 per group, Laboratory Animal Center, Wuhan Institute of Virology, Chinese Academy of Sciences) were intraperitoneally (i.p.) injected with EG, WT or PBS, respectively, at weeks 0, 2 and 4. Two weeks after the third injection, sera from immunized mice were collected by orbital venous plexus, the mice were sacrificed and splenocytes were separated.
Enzyme-Linked Immunosorbent Assay (ELISA)
Sera from immunized mice were heat inactivated at 56 °C for 30 min and then analyzed for EG antibodies as follows. Briefly, 96-well microtiter plates (Costar) were coated with E protein (produced by our lab) and incubated at 4 °C overnight. The plates were then blocked with PBS (pH 7.4) containing 5% skim milk at 37 °C for 1 h. Serially diluted mice sera was added to the wells and incubated at 37 °C for 1 h. Wells were rinsed 5 times with PBST (PBS containing 0.05% Tween-20), followed by a 30 min incubation with HRP-conjugated goat-anti-mouse IgG (Sigma) (1:10,000) at 37 °C. After five washings and a 15 min incubation with the substrate 3, 3′, 5, 5′- tetramethylbenzidine (TMB) solution at 37 °C in dark, reactions were terminated with a stop solution, and optical densities (ODs) were determined using a microplate reader set at 450 nm. Endpoint titres (EPTs) were defined as reciprocal sera dilutions that corresponded to 2 times the average OD values obtained with mock antigen.
Plaque Reduction Neutralisation Test (PRNT)
PRNT were performed to measure ZIKV neutralizing antibodies. In brief, monolayers of Vero cells were seeded in 12-well plates (Corning) and incubated at 37 °C in a CO2 incubator 1 day before viral infection. On day 2, heat-inactivated serum samples from immunized mice were serially diluted twofold and mixed with ZIKV to obtain approximately 100 plaque forming units (PFU) per 0.2 mL. The mixtures were incubated at 37 °C for 1.5 h and added onto confluent Vero cells in 12-well plates. During the 1.5 h incubation period, the plates were swirled every 15 min to ensure complete coverage of the cell monolayer, for even infection. After 1.5 h, 1.25% methyl cellulose overlay containing 2% FBS and 1% penicillin streptomycin was added to each well, and the plates were incubated at 37 °C for 4 days. Then, the plates were fixed with 4% formaldehyde and incubated at room temperature for 20 min. After removal of the fixative, the plates were stained with 0.5% crystal violet for 1 min. The number of plaques reported for each serum dilution was the average of the duplicate wells. PRNT50 titer was expressed as the reciprocal of the highest dilution of each serum sample that caused a 50% reduction in the plaque number relative to the control samples. Samples with titers of ≥ 10 were considered as seropositive.
Indirect Immunofluorescence Assays (IFA)
Vero cell monolayers were seeded in small dishes and inoculated for 90 min with the mixture of mice serum samples at 1:10 dilution and ZIKV (MOI = 0.005) that was premixed for 90 min at 37 °C. The cultured cells were respectively terminated at 2, 3 and 4 days post infection (dpi) and then detected by IFA as described as the above biochemical analysis section.
Elispot
The analysis of splenocytes from immunized mice for IFN-γ and IL-4 were performed by Enzyme-linked Immunospot Assay (ELISPOT) following the manufacturer’s instructions (DAKEWE, Shenzhen, China). Briefly, splenocytes (at 5 × 105 cells/well in 100 µL RPMI medium containing 1% FCS) from individual mice 2 weeks after three immunizations were added to a 48-well microplate coated with anti-mouse IFN-γ and IL-4 antibody. Inactivated ZIKV (8.35 × 107 PFU/mL) was diluted in complete RPMI-1640 medium at a volume of 10 µL/well and then added to wells. Phorbol 12-myristate 13-acetate (PMA) and ionomycin was set as the positive control and the medium only as the negative control. After incubation for 20 h at 37 °C, the plates were incubated with biotinylated anti-mouse IFN-γ or biotinylated anti-mouse IL-4 detection antibody, and then AP-conjugated streptavidin. AEC substrates were then added for color development. The plates were imaged and cytokine-secreting cell spots counted using an ELISPOT reader (Bioreader 4000; Bio-sys, Germany).
Immunization of AG6 Mice
The protective efficacy of recombinant vaccine was determined using active immunization/challenge assays. AG6 mice, a gift from Dr. Qibin Leng (Institute Pasteur of Shanghai, Chinese Academy of Sciences), were deficient in type I and II interferon (IFN-α/β/γ) receptors and bred under specific-pathogen-free conditions in the Animal Resource Center at the Wuhan Institute of Virology, Chinese Academy of Sciences. 4-week-old AG6 mice were injected i.p. with three doses of recombinant EG (E protein was 15 µg/dose), WT or PBS at 2-week interval. Two weeks after the third injection, the immunized mice were bled for the titration of neutralizing antibodies and then challenged through the i.p. route with 1 PFU of the ZIKV (SZ-WIV01 strain). After viral challenge, all mice were measured for viremia on day 3 after challenge and monitored daily for weight loss, survival, and assigned clinical scores for a period of 28 days. Clinical scores were graded as follows: 0, healthy; 1, reduced mobility; 2, limb weakness; 3, paralysis; and 4, death. On day 28 after challenge, all the mice were anesthetized and sacrificed. The viremia and neutralizing antibodies were respectively measured by plaque forming assay and PRNT as described previously.
Statistical Analysis
All experiments were reproducible and conducted in triplicates. The Student’s t test and analysis of variance (ANOVA) were used for the data analyses of all experiments. Differences were considered statistically significant when P value was less than 0.05 (P < 0.05). Statistical analyses were performed in GraphPad Prism 6.0 software.
Results
Construction of Recombinant Baculovirus, Bac-EGP64 (EG)
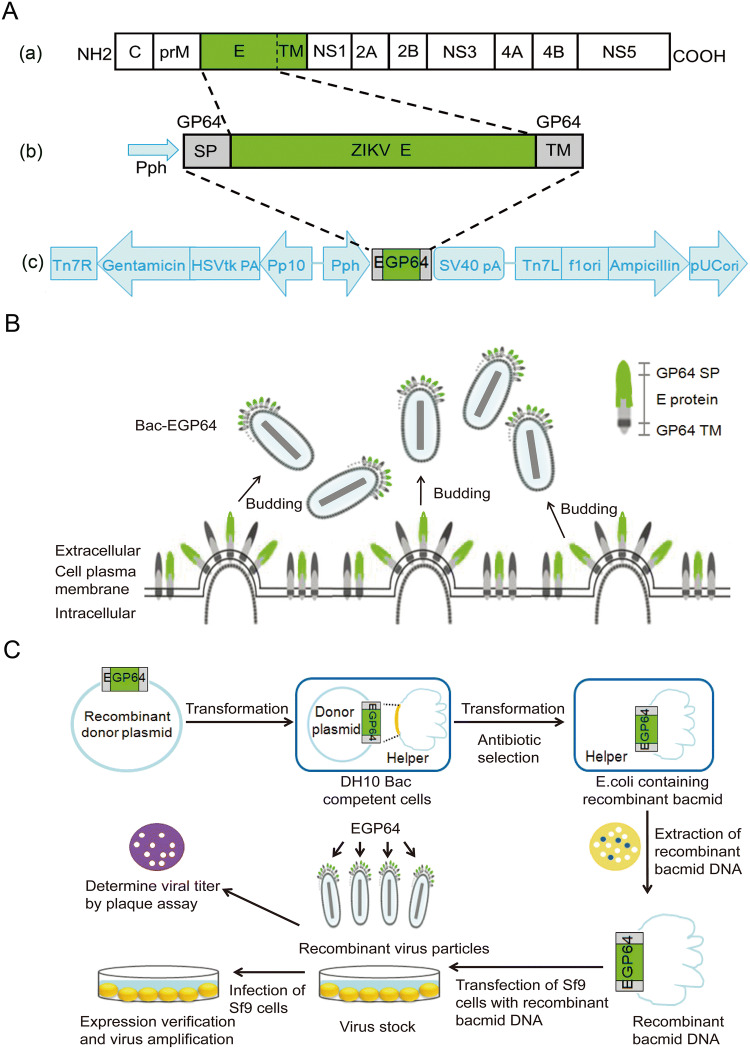
We constructed a recombinant baculovirus expressing E proteins of ZIKV as described earlier. E protein (TM deleted) genes of Zika strain SZ-WIV01 were linked with SP and TM of baculoviral envelope GP64 together and confirmed through sequence analysis. These constructs were cloned into AcMNPV bacmid under the control of the polyhedron (Ph) promoter, to generate the recombinant bacmid Bac-EGP64 (Fig. 1A). The strategy was that GP64 SP would facilitate the translocation of E protein to Sf9 cell plasma membrane and the expose of it on the outer surface. GP64 TM enables E protein to anchor on to the plasma membrane. So, the fusion protein containing GP64 SP, ZIKV E, and GP64 TM was primarily displayed on the surface of Sf9 cell membrane, then the baculovirus Bac-GP64 acquired the Sf9 cell membrane as its envelope after budding, ultimately leading to the fusion protein EG being displayed on the envelope of baculovirus (Fig. 1B). For the sake of brevity, the entire construction process from gene recombination to the production of recombinant baculovirus is shown in the flow chart (Fig. 1C).
Fig. 1.
Construction of recombinant baculovirus, Bac-EGP64. A-a Schematic representation of ZIKV polyprotein. Envelop (E) protein (aa. 1–504, green). A-b The strategy of fusion proteins. Recombinant EGP64 contained GP64 SP, ZIKV E, GP64 TM. A-c Representation of ZIKV E protein recombinant expression constructs. The E protein coding sequence was inserted between the signal peptide (SP) and transmembrane domain (TM) of AcMNPV bacmid envelope protein GP64 under the control of a polyhedrin promoter (PH), to ensure proper expression and transportation. B The perceived budding process of Bac-EGP64. The fusion protein EGP64 was displayed on the surface of Sf9 cell membrane and on the envelope of baculovirus following budding. C Flow chart of the construction process from cloning of ZIKV E gene into the pFastBac to the production of recombinant baculovirus.
Expression Confirmation and Generation of Bac-EGP64
Following transfection and infection, expression of the recombinant baculovirus, Bac-EGP64(EG), was confirmed by Western blot. That is to say, expression of the E and GP64 proteins in Sf9 cells infected by Bac-EGP64 and Bac-WT were detected by Western blot with anti E protein antibody and anti GP64 monoclonal antibody correspondingly. As shown in Fig. 2A(a), the protein band of EG group was located at 55 kDa as detected by anti-E antibody, it indicated that recombinant E protein was successfully expressed. As shown in Fig. 2A(b), the protein band of EG group was located at 64 kDa like GP64 of WT group, it indicated that the recombinant baculovirus could work normally and express unfused intact GP64. It was also worth noting that there was only one protein band in EG group (Fig. 2A(b)). The reason was that the epitope of GP64 monoclonal antibody was not located in the TM region of the fusion protein EGP64, and the antibody could not bind to EGP64 but to the unfused GP64. Thus, fused GP64 could be successfully expressed, except that it did not contain the antibody binding sites.
Fig. 2.
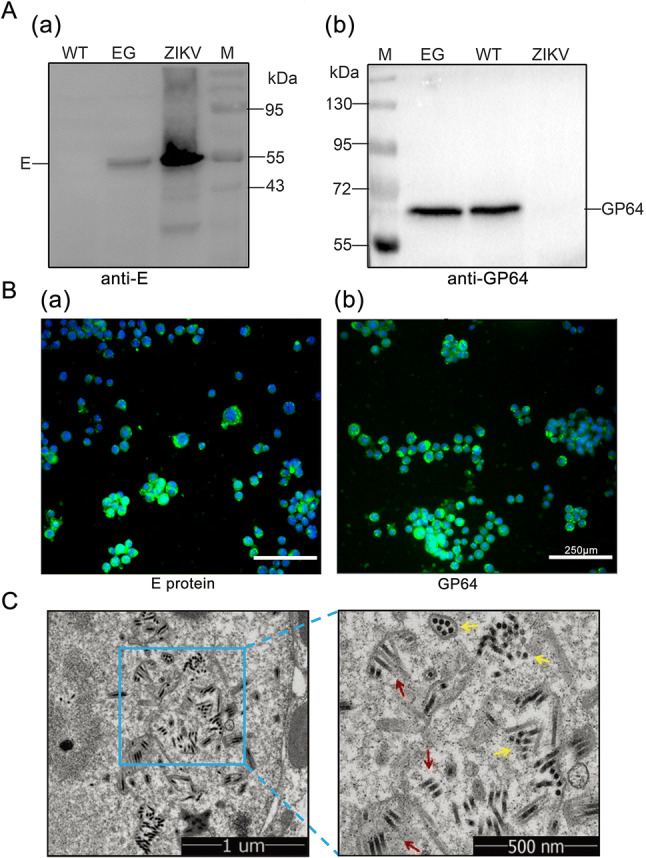
Expression confirmation and generation of Bac-EGP64. A Western blot analyses of E and GP64 protein, ZIKV was used as positive (a) and negative control (b) respectively. Bac-GP64 (WT) was used as a negative (a) and positive (b) control respectively. B Immunofluorescence assay for the E (a, anti-E) and GP64 (b, anti-GP64) proteins in Sf9 cells infected with Bac-EGP64 (Scale bar = 250μm). Green represents E or GP64 protein, blue represents nuclei (stained with Hoechst 33258). C Transmission electron micrographs of Sf9 cells infected with Bac-EGP64. Rod-like (red arrows) and sphere-like (yellow arrows) particles in Sf9 cells infected with Bac-EGP64. The right panels show enlarged views of the blue box regions in the corresponding left panels.
In addition, the expression of EG was also confirmed by indirect immunofluorescence assays. Sf9 cells infected by Bac-EGP64 displayed green fluorescence with anti E protein antibody (Fig. 2B(a)) and anti GP64 antibody (Fig. 2B(b)), indicating that Bac-EGP64 was successfully constructed to work normally and express the ZIKV E protein.
The Sf9 cells infected with Bac-EGP64 were harvested and immobilized in glutaraldehyde after 72 h.p.i., then observed using a TEM. As indicated in Fig. 2C, Sf9 cells presented with rod-shaped particles ranging from 30 to 60 nm in diameter and 250–300 nm in length, which were presumed to be Bac-EGP64.
Purification and Identification of Bac-EGP64
In order to further purify Bac-EGP64, infected Sf9 cell lysates were subjected to enrichment process with 15%–65% sucrose gradient ultracentrifugation. There were two resultant fractions (Fig. 3A). After desugarization, the two fractions were tested for E protein expression by Western blot, and the upper band was determined to be rich in Bac-EGP64 (Fig. 3B). Negative-stained purified Bac-EGP64 was analyzed by TEM and exhibited rough rod-like particles (Fig. 3C).
Fig. 3.
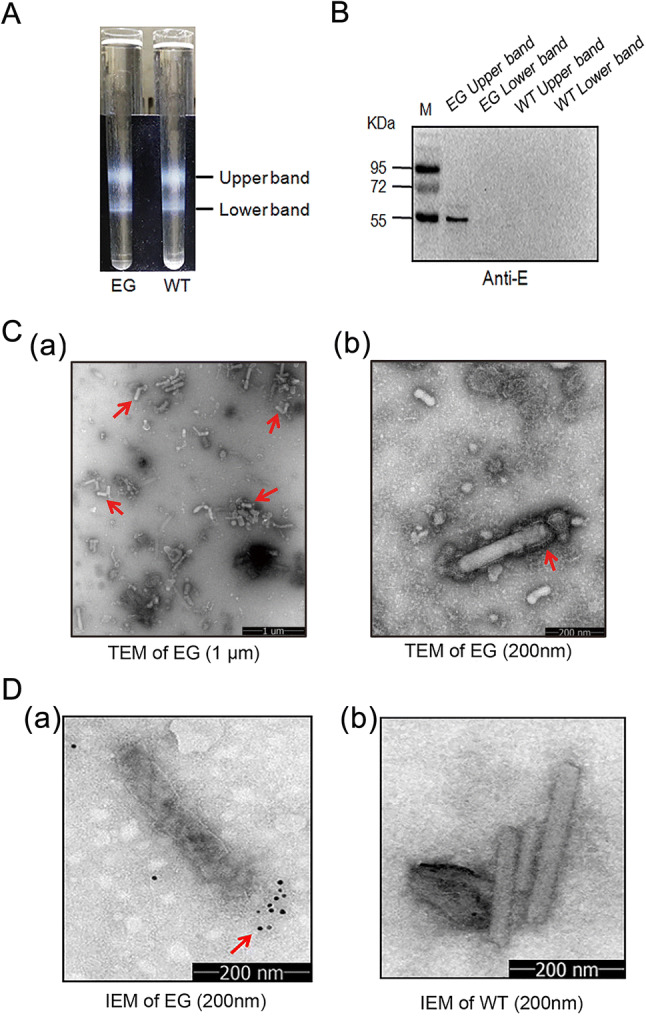
Characterization of E protein expressed by baculovirus. A Lysates of Sf9 cells infected by Bac-EGP64 (EG) were ultracentrifuged on a 15%–65% sucrose gradient, Bac-GP64 (WT) was used as a negative control. Two distinct bands were taken from top to bottom and assayed. B Western blot of a ZIKV E protein specific monoclonal antibody. C TEM of purified EG negatively stained with 1% phosphotungstic acid (a, scale bar = 1 μm; b, scale bar = 200 nm). D Immune Electron Microscopy (IEM) detection of purified baculovirus EG and WT by anti-E monoclonal antibodies. Gold particles are displayed on the surface of EG (a), and no gold particles are displayed on the surface of WT (b).
Immunogold electron microscope (IEM) using anti-E MAb further revealed immunogold particles surrounding one side of the rod-like Bac-EGP64 (Fig. 3D(a)), which were completely different with those of Bac-WT (Fig. 3D(b)), confirming the exposure of E protein on the outer surface of the baculoviral particle and the presence of E protein in extracellular particles in the medium. All these data also demonstrate that E protein can be expressed in Sf9 cells infected with Bac-EGP64 and purified from the lysates by sucrose gradient ultracentrifugation.
Bac-EGP64(EG) were Highly Immunogenic in Immunized BALB/c Mice
To assess the immunogenicity of Bac-EGP64 (EG), the antigens were formulated with adjuvant for BALB/c mice immunization study. Mice were administered EG (E protein was 15 μg/dose) intraperitoneally, at weeks 0, 2 and 4, and then serum samples were collected to measure antibody responses at week 6 (Fig. 4A). WT and PBS respectively mixed with adjuvant were set as the negative control. As shown in Fig. 4B, the mice immunized by EG developed significant anti-E protein specific IgG with geometric mean titer (GMT) of 110,000 at 6 weeks. In contrast, antisera from the PBS control group did not show significant reactivity in the E protein-capture ELISA at the lowest serum dilution (1:100). Similarly, the WT-immunized mice, produced lower serum antibodies with GMT of 7000 at 6 weeks. These data indicated that EG was capable of efficiently inducing specific antibody responses in immunized BALB/c mice.
Fig. 4.
Humoral and cellular immune responses elicited by Bac-EGP64(EG) in BALB/c mice. A BALB/c mice (n = 6) were immunized with three doses of Bac-EGP64 (EG), Bac-GP64 (WT) or PBS at weeks 0, 2 and 4. Blood samples were collected at weeks 6 (2 weeks after the third immunization). B Antibody responses were measured via ELISA. The antisera from mice were serially diluted and ZIKV E-specific IgG titres were detected by ELISA in which E protein was used as the capture antigen. The highest titres were defined as the last serial diluted specimens that gave positive readings. C Neutralizing titres of individual mouse antisera. Each symbol represents one mouse and the line indicates the GMT of the group. D IFA of E protein expression in Vero cells inoculated with the mixture of mice serum samples a 1:10 dilution and ZIKV (MOI = 0.005). Green represents E protein, and blue represents nuclei (stained with Hoechst 33258). The GFP fluorescence intensity in the ZIKV infected cells was quantified using ImageJ software by determining the mean gray value representing the average signal intensity. All the images were captured at 2, 3, and 4 dpi (Scale bar = 250 μm). Representative images of ZIKV infected Vero cells were shown (left panel). (E) IFN-γ-secreting splenocytes were measured via ELISPOT. (F) IL-4-secreting splenocytes were measured via ELISPOT. All data are presented as the mean ± SD of triplicate wells were shown. Asterisks represent significant differences: ns, not significant, P ≥ 0.05; *0.01 ≤ P < 0.05; **P < 0.01; ***P < 0.001;****P < 0.0001.
Plaque reduction neutralization assay indicated the presence of ZIKV neutralizing antibodies in the sera collected from the immunized mice. As shown in Fig. 4C, all the anti-EG sera could neutralize the ZIKV with PRNT50 values ranging from 29.15 to 67.61; in contrast, the antisera from the two control groups had no neutralization effect even at the lowest dilution (1:10). The geometric mean PRNT50 value of the anti-EG group was 47.76, indicating that the EG elicited anti-ZIKV neutralizing antibody response in the immunized mice.
Next, IFA of E protein expression in Vero cells inoculated with the mixture of mice sera and ZIKV were also used to evaluate the neutralizing capacity of sera. The green fluorescence of Vero cells showed a pronounced and wide distribution of E protein in WT group, PBS group and ZIKV control group at 2, 3, and 4 dpi; in contrast, there was less distribution of E protein in EG group (Fig. 4D). Additionally, the average signal intensity of GFP quantified using ImageJ software. As shown, the GFP signal in EG group is significantly decreased compared with other groups including WT, ZIKV and PBS groups (Fig. 4D). That was to say, the sera of EG immunized mice have the most significant neutralizing capacity to ZIKV.
Cellular immune responses induced by EG were evaluated by ELISPOT assay in splenocytes. As shown in Fig. 4E and 4F, the numbers of IFN-γ and IL-4-secreting splenocytes from EG immunized group and WT immunized group were significantly higher than PBS control group, whereas only baseline levels of IFN-γ and IL-4-secreting cells were detected in the PBS control group. These data indicated that baculovirus used as immunization vector could effectively elicit cellular responses in mice.
Immunization of AG6 Mice
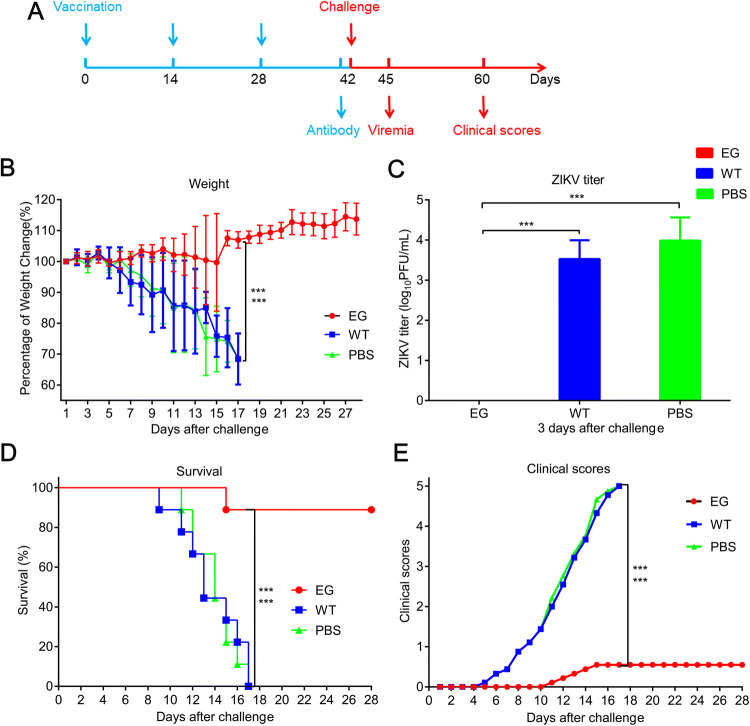
To evaluate the protective efficacy of recombinant EG, AG6 mice were intraperitoneally immunized with Bac-EGP64, WT or PBS control respectively, at weeks 0, 2, and 4. Two weeks later, the mice were challenged with a lethal dose of the ZIKV (Fig. 5A). The weight of WT group and PBS group dropped sharply compared with that of EG group, which was relatively stable and slightly increased at day 16 after challenge (Fig. 5B, P < 0.001). Because the pathogenicity and viral load in sera were usually positively correlated (Wang et al. 2015; Shan et al. 2017), viral titers in sera were measured by virus plaque assay. There was no detectable peripheral viremia in EG group at day 3 after challenge, in contrast, the mean viral load in WT group and PBS group reached up to 103.53 PFU/mL and 103.99 PFU/mL respectively (Fig. 5C), which ultimately resulted in disease manifestation and death of all mice in these two groups. In detail, 5 out of 6 mice in EG group were completely protected and survived without any obvious clinical manifestations through 28 days, only one mouse displayed weight loss, disease signs, and died on day 15 after challenge; in contrast, the mice in the other two groups began to exhibit disease signs, including weight loss, ruffled fur, hunched posture, decreased mobility, limb weakness, and hind limb paralysis, at day 5 after challenge (Fig. 5D, 5E, P < 0.001). These results indicate that EG could elicit protective immunity against lethal ZIKV infection in AG6 mice.
Fig. 5.
Protective efficacy of Bac-EGP64 in AG6 mice challenged by ZIKV. A AG6 mice (n = 6) were immunized with three doses of EG, WT or PBS at weeks 0, 2 and 4, then challenged by ZIKV at week 6 and blood samples were collected at day 3 after challenge. The challenged mice were monitored daily for 28 days for weight, clinical scores, viremia and survival. B Weight of the challenged mice. C Viremia detection of AG6 mice at day 3 after challenge. D Survival curves of challenged AG6 mice. Representative data of two independent experiments are shown. E Clinical scores of the challenged mice. Clinical scores were graded as follows: 0, healthy; 1, retardation and reduced mobility; 2, limb weakness; 3, tremors or imbalance; 4, paralysis; and 5, moribund or dead. ***P < 0.001.
Discussion
Zika virus (ZIKV) is a member of the Flaviviridae family (Musso and Gubler 2016; Wikan and Smith 2016; Martinez-Sobrido and Almazán 2019). It used to be an endemic and tropical pathogen. However, it has transmitted rapidly to many countries and regions around the world and was found to be primarily associated with fetal microcephaly, congenital Zika syndrome (CZS) and other neurological disorders such as Guillain-Barre syndrome in 2015–2016 (Cao-Lormeau et al. 2016; Martines et al. 2016). WHO declared a Public Health Emergency of International Concern in February 2016. Vaccine development by the different research communities was initiated and the advancements in this field were tremendous and unprecedented (Lecouturier et al. 2019).
In 4 years, 13 vaccine candidates have been evaluated in clinical phase I studies and 2 vaccine candidates have entered clinical phase II studies, including 5 DNA vaccines, 7 purified PIV vaccines, 1 LAV vaccine, 1 mRNA vaccine, and 1 vector vaccine (Garg et al. 2018). Although DNA vaccines provide many advantages such as ease of production and rapid adaptation to new and emerging infectious agents, they may not be durable and may integrate into chromosomes, which can lead to insertional mutagenesis and potential oncogenesis (Abbink et al. 2017; Richner et al. 2017; Garg et al. 2018). PIV is relatively safe, but it needs to improve its immunogenicity (Singh et al. 2018). Experience from vaccination studies with other flaviviruses such as yellow fever virus (YFV) and Japanese encephalitis virus (JEV) suggest that LAV for ZIKV are likely to be effective (Garg et al. 2018). However, LAV is a potential threat to immunocompromised individuals and pregnant women (Dai et al. 2016). mRNA vaccine has emerged as a promising new vaccine modality that can elicit potent immune responses (Pardi et al. 2017). In the long run, it is necessary to break the bottleneck restriction of instability and inefficiency of in vivo delivery of mRNA (Pardi et al. 2017). Subunit vaccines are non-infectious and safer to use, but require multiple doses and periodic boosts to produce protective antibodies. Virus vector vaccine is a non-pathogenic or the virulent virus has been removed from the vector. The target antigen gene fragments are inserted into the vector genome. The virus vector itself can enhance the immunogenicity of the vaccine, which can induce a strong cytotoxic T cell response. The foregoing vector vaccine takes the attenuated measles virus as the vector to express recombinant ZIKV protein, and it may trigger strong humoral and cellular immunity (Garg et al. 2018). But the drawback is that the pre-existing immune responses aimed at the vector in the host may weaken the subsequent immune response to the target antigen (Saxena et al. 2013). On this basis, it is a promising effort to choose non-mammalian specific viruses as vectors, such as baculovirus expression vector.
Baculovirus expression system permits target DNA to be inserted between the baculoviral envelope GP64 signal sequence and mature domain, anchoring the fusion protein in the envelope of the virus via the transmembrane (TM) region of GP64, thus displaying the foreign protein on the surface of viral particles. This may further augment its immunorecognition and immunoreactivity (Lindley et al. 2000; Premanand et al. 2018). This system exhibits several advantages: (1) it has a good biosafety profile in mice, (2) it can be readily manipulated for insertion of exogenous DNA fragments, (3) it has the capacity for rapid development and potential for low cost production, (4) high titres (> 109 PFU/mL) of virus can be easily reached in Sf9 cells, and (5) the recombinant protein expressed can be readily purified (Xu et al. 2011; Yamaji 2014; Premanand et al. 2018; Gupta et al. 2019). Due to its characteristic advantages of security, economy and convenience, the baculovirus expression system is regarded as the most versatile surface display system in eukaryotes (Zheng et al. 2018). To date, it has been commonly exploited to display proteins or peptides such as HA of influenza virus, VP1 of EV71, N and S proteins of SARS-like coronavirus (Bai et al. 2008; Baxter et al. 2011; Premanand et al. 2018).
The current trends of vaccines against ZIKV mainly concern the pre-membrane and/or envelope (E) proteins as the target epitopes for vaccine candidates due to the natural structure of the virus (Ghaffar et al. 2018). The E protein, encompassing the majority of the virion surface, is also the major envelope antigen and is closely related with pathogenicity and infection immunity of ZIKV (Musso and Gubler 2016; Wikan and Smith 2016; Lin et al. 2018; Martinez-Sobrido and Almazán 2019). Therefore, the principal aim of the present study was to construct a recombinant ZIKV E protein expressed by baculovirus and assess its immunogenicity and efficacy.
We selected most segment of E gene (only its TM removed) of Zika strain SZ-WIV01 for insertion, the two ends of spliced E gene were linked with GP64 signal peptide (SP) and transmembrane structure (TM) respectively, and then inserted into donor bacmid under the control of the polyhedrin promoter of donor plasmid (Fig. 1A). After the subsequent transformation, extraction, screening and transfection, Bac-EGP64 finally expressed and displayed recombinant ZIKV E protein in Sf9 cells (Fig. 1C). This construction strategy was likely to have at least two advantages: first, the recombinant E protein would be expressed effectively; and second, the recombinant E protein was likely to be displayed on the surface of viral particles by means of anchoring itself in the envelope of the virus via GP64 (Fig. 1B).
Next, animal experiments were performed to determine the immunogenicity of EG. The BALB/c mice were immunized with EG, then the serum samples were assayed by ELISA and PRNT. The mean specific antibody and neutralizing antibody titers were 1:110,000 and 1:47.76, respectively. It is known that most of the vaccines against ZIKV today focus on the induction of neutralizing antibody (nAb) responses because neutralizing antibodies can strongly bind to E proteins, and block viral attachment to host cells or prevent E proteins rearrangement before membrane fusion, and make it less infectious (Ghaffar et al. 2018; Yang et al. 2019). The neutralizing titer of 1:10 or more is accepted by World Health Organization as a evidence of protection against another flavivirus, JEV (Xu et al. 2011). In order to intuitively observe neutralizing effect of the antibody in vitro, IFA staining of Vero cells inoculated with the mixture of the serum and ZIKV was conducted. It were a wide viral distribution in WT group and PBS group; in contrast, it was not easy to detect viral distribution in EG group (Fig. 4D). Therefore, it is clear that EG is capable of eliciting the anti-ZIKV-specific neutralizing antibody response in BALB/c mice. That is to say, the E protein expressed by baculovirus is highly immunogenic.
In addition to humoral immunity, cellular immunity stimulated by EG was also evaluated. In general, IFN-γ and IL-4 are used to assess the function of TH1 and TH2 cells in cellular immunity, respectively (Bai et al. 2008; Ghaffar et al. 2018). In the present study, EG was able to induce IFN-γ (TH1) and IL-4 (TH2) cell response (Fig. 4E, 4F), which contributes to the strong cellular immune protection against virus infection. This might be partly explained why only immunocompromised mice such as AG6 mice susceptible to ZIKV could be used as infection models (Lazear et al. 2016). At the same time, there was a concern that the levels of IFN-γ and IL-4 in EG and WT group showed no significant difference (Fig. 4E, 4F). There were two possible reasons for this: (I) baculovirus itself has a strong immunogenicity, (II) ZIKV and baculovirus may have similar epitopes. For example, Thogoto virus (THOV) and Dhori virus (DHOV) Gp protein and baculovirus GP64 show a particularly similar domain structure (Peng et al. 2017). We are attempting to resolve this speculation by studying the crystal structure of proteins.
In our study, the protective efficacy of EG was assessed by challenge in AG6 mice that were deficient in the type I and type II IFN (IFN-α/β/γ) receptors and highly susceptible to ZIKV infection and display severe disease signs, including hind limb weakness, paralysis, and death, and provide a practical and convenient platform to determine ZIKV virulence and testing the efficacy of vaccines and antivirals (Aliota et al. 2016; Rossi et al. 2016; Morrison and Diamond 2017; Weger-Lucarelli et al. 2017; Li et al. 2018). We found that just 1 PFU of SZ-WIV01 by i.p. injection could sufficiently infect, induce severe clinic manifestations and then cause 100% mortality in control mice after challenge. This result was similar to those of ICR neonatal mice, all of which were dead after the challenge by 1 PFU of ZIKV (Qu et al. 2018). More importantly, 89% of mice immunized with EG survived (Fig. 5D). These data indicated that EG was efficient in protecting AG6 mice against lethal virus challenge. We further showed that viremia could not be detected in EG group at 3 days after challenge, in contrast, the viral titers could be detected in WT group and PBS group (Fig. 5C). These results suggest that neutralizing antibodies from AG6 mice immunized by EG may play an important role in the protection against ZIKV infection.
Furthermore, the titer of specific antibodies from BALB/c mice is very high, the titer of neutralizing antibodies and the challenge dose of AG6 mice were not as high as we originally expected. It is likely that proper folding, membrane association, and assembly of recombinant E protein still need the assistance of another membrane protein, such as pre-membrane (prM) and NS1 protein of ZIKV, although baculovirus expression system could in itself conduct post-translational modification of recombinant protein.
In conclusion, the present data demonstrate that the recombinant EG could be largely produced in Sf9 cells, induce humoral immune and cellular immune response in BALB/c mice and it impart protective immunity in AG6 mice from lethal ZIKV challenge. These results indicate good immunogenicity and protective efficacy of recombinant ZIKV E protein expressed in baculovirus display system and it may hold great promise as a novel platform for studying ZIKV vaccine. Our study, thus, demonstrates the potential of EG for preparing an effective ZIKV vaccine. At present, the lack of new Zika cases since 2018 has dampened the enthusiasm as well as urgency for the vaccine development (Garg et al. 2018). However, the risk of a fresh outbreak in the near or distant future cannot be ignored (Garg et al. 2018). In this scenario, development of a safe, effective and economical ZIKV vaccine aimed against a subsequent outbreak remains a real possibility and this exploration is endless.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by grants from the National Key R&D Program of China (2016YFD0500406), the National Natural Science Foundation of China (NSFC) (no. 81672036), and the Youth Innovation Promotion Association of CAS (2016302). We are grateful for Dr. Qibin Leng from the Institute Pasteur of Shanghai, Chinese Academy of Sciences, for providing AG6 mice. We also thank Xuefang An from Animal Resource Center of the Wuhan Institute of Virology, Chinese Academy of Sciences, for support in animal experiments. We would like to thank Pei Zhang and Du An-na from The Core Facility and Technical Support, Wuhan Institute of Virology, for their help with producing TEM micrographs.
Authors Contributions
ZZ conceived and designed the study. DL and YM developed the methodology, performed the experiments and drafted the manscript. XK, ZT and PL helped to conduct part of the experiments. CH analyzed the data by software and helped to draw the illustration. PL, YZ, YL and TW performed the formal analysis. DL, YM and JS contributed to the investigation. YM provided the visualization. HW supervised the experiment. ZZ finalized the manuscript and conducted the project administration. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This study was carried out in strict accordance with the recommendations of the national guidelines for the use of animals in scientific research ‘‘Regulations for the Administration of Affairs Concerning Experimental Animals’’. The protocol was also approved by the Animal Care Committee of Wuhan Institute of Virology (Permit Number: WIVA07201804). All efforts were made to minimize suffering.
Footnotes
Dan Luo and Yuanjiu Miao have contributed equally to this work.
References
- Abbink P, Larocca RA, Visitsunthorn K, Boyd M, De La Barrera RA, Gromowski GD, Kirilova M, Peterson R, Li Z, Nanayakkara O, Nityanandam R, Mercado NB, Borducchi EN, Chandrashekar A, Jetton D, Mojta S, Gandhi P, LeSuer J, Khatiwada S, Lewis MG, Modjarrad K, Jarman RG, Eckels KH, Thomas SJ, Michael NL, Barouch DH. Durability and correlates of vaccine protection against zika virus in rhesus monkeys. Sci Transl Med. 2017;9:eaao4163. doi: 10.1126/scitranslmed.aao4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliota MT, Caine EA, Walker EC, Larkin KE, Camacho E, Osorio JE. Characterization of lethal zika virus infection in ag129 mice. PLoS Negl Trop Dis. 2016;10:e0004682. doi: 10.1371/journal.pntd.0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Lu X, Meng J, Hu Q, Mao P, Lu B, Chen Z, Yuan Z, Wang H. Vaccination of mice with recombinant baculovirus expressing spike or nucleocapsid protein of sars-like coronavirus generates humoral and cellular immune responses. Mol Immunol. 2008;45:868–875. doi: 10.1016/j.molimm.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter R, Patriarca PA, Ensor K, Izikson R, Goldenthal KL, Cox MM. Evaluation of the safety, reactogenicity and immunogenicity of flublok® trivalent recombinant baculovirus-expressed hemagglutinin influenza vaccine administered intramuscularly to healthy adults 50–64 years of age. Vaccine. 2011;29:2272–2278. doi: 10.1016/j.vaccine.2011.01.039. [DOI] [PubMed] [Google Scholar]
- Bogoch II, Brady OJ, Kraemer MUG, German M, Creatore MI, Brent S, Watts AG, Hay SI, Kulkarni MA, Brownstein JS, Khan K. Potential for zika virus introduction and transmission in resource-limited countries in Africa and the Asia-pacific region: a modelling study. Lancet Infect Dis. 2016;16:1237–1245. doi: 10.1016/S1473-3099(16)30270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lormeau VM, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial AL, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra JC, Despres P, Fournier E, Mallet HP, Musso D, Fontanet A, Neil J, Ghawché F. Guillain-barré syndrome outbreak associated with zika virus infection in French polynesia: a case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S, Oliveira Silva Martins D, Jardim ACG. A review of the ongoing research on zika virus treatment. Viruses. 2018;10:255. doi: 10.3390/v10050255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Song J, Lu X, Deng YQ, Musyoki AM, Cheng H, Zhang Y, Yuan Y, Song H, Haywood J, Xiao H, Yan J, Shi Y, Qin CF, Qi J, Gao GF. Structures of the zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe. 2016;19:696–704. doi: 10.1016/j.chom.2016.04.013. [DOI] [PubMed] [Google Scholar]
- Dai S, Zhang T, Zhang Y, Wang H, Deng F. Zika virus baculovirus-expressed virus-like particles induce neutralizing antibodies in mice. Virol Sin. 2018;33:213–226. doi: 10.1007/s12250-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávila-Castrodad NM, Reyes-Bou Z, Correa-Rivas M, Pérez-Berenguer JL, Di-Pasquale L, García-García I. First autopsy of a newborn with congenital zika syndrome in puerto rico. Proc R Health Sci J. 2018;37:S81–s84. [PubMed] [Google Scholar]
- de Araújo TVB, Ximenes RAA, Miranda-Filho DB, Souza WV, Montarroyos UR, de Melo APL, Valongueiro S, de Albuquerque M, Braga C, Filho SPB, Cordeiro MT, Vazquez E, Cruz D, Henriques CMP, Bezerra LCA, Castanha P, Dhalia R, Marques-Júnior ETA, Martelli CMT, Rodrigues LC. Association between microcephaly, zika virus infection, and other risk factors in brazil: final report of a case-control study. Lancet Infect Dis. 2018;18:328–336. doi: 10.1016/S1473-3099(17)30727-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Liu S, Zhang Q, Xu M, Zhang H, Gu D, Shi L, He J, Xiao G, Zhang B. Isolation and characterization of zika virus imported to china using c6/36 mosquito cells. Virol Sin. 2016;31:176–179. doi: 10.1007/s12250-016-3778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ortenzio E, Matheron S, Yazdanpanah Y, de Lamballerie X, Hubert B, Piorkowski G, Maquart M, Descamps D, Damond F, Leparc-Goffart I. Evidence of sexual transmission of zika virus. N Engl J Med. 2016;374:2195–2198. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- Dowall SD, Graham VA, Rayner E, Atkinson B, Hall G, Watson RJ, Bosworth A, Bonney LC, Kitchen S, Hewson R. A susceptible mouse model for zika virus infection. PLoS Negl Trop Dis. 2016;10:e0004658. doi: 10.1371/journal.pntd.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin A, Wilder-Smith A. An update on zika vaccine developments. Expert Rev Vaccines. 2017;16:781–787. doi: 10.1080/14760584.2017.1345309. [DOI] [PubMed] [Google Scholar]
- Enfissi A, Codrington J, Roosblad J, Kazanji M, Rousset D. Zika virus genome from the americas. Lancet. 2016;387:227–228. doi: 10.1016/S0140-6736(16)00003-9. [DOI] [PubMed] [Google Scholar]
- Garg H, Mehmetoglu-Gurbuz T, Joshi A. Recent advances in zika virus vaccines. Viruses. 2018;10:631. doi: 10.3390/v10110631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffar KA, Ng LFP, Renia L. Fast tracks and roadblocks for zika vaccines. Vaccines (Basel) 2018;6:77. doi: 10.3390/vaccines6040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govero J, Esakky P, Scheaffer SM, Fernandez E, Drury A, Platt DJ, Gorman MJ, Richner JM, Caine EA, Salazar V, Moley KH, Diamond MS. Zika virus infection damages the testes in mice. Nature. 2016;540:438–442. doi: 10.1038/nature20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Tölzer C, Sari-Ak D, Fitzgerald DJ, Schaffitzel C, Berger I. Multibac: baculovirus-mediated multigene DNA cargo delivery in insect and mammalian cells. Viruses. 2019;11:198. doi: 10.3390/v11030198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaname Y, Tani H, Kataoka C, Shiokawa M, Taguwa S, Abe T, Moriishi K, Kinoshita T, Matsuura Y. Acquisition of complement resistance through incorporation of cd55/decay-accelerating factor into viral particles bearing baculovirus gp64. J Virol. 2010;84:3210–3219. doi: 10.1128/JVI.02519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost TA, Condreay JP, Jarvis DL. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol. 2005;23:567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Lim EX, Zhang S, Fibriansah G, Ng TS, Ooi JS, Shi J, Lok SM. Structure of the thermally stable zika virus. Nature. 2016;533:425–428. doi: 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. A mouse model of zika virus pathogenesis. Cell Host Microbe. 2016;19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecouturier V, Bernard MC, Berry C, Carayol S, Richier E, Boudet F, Heinrichs J. Immunogenicity and protection conferred by an optimized purified inactivated zika vaccine in mice. Vaccine. 2019;37:2679–2686. doi: 10.1016/j.vaccine.2019.04.013. [DOI] [PubMed] [Google Scholar]
- Li P, Ke X, Wang T, Tan Z, Luo D, Miao Y, Sun J, Zhang Y, Liu Y, Hu Q, Xu F, Wang H, Zheng Z. Zika virus attenuation by codon pair deoptimization induces sterilizing immunity in mouse models. J Virol. 2018;92:e00701-18. doi: 10.1128/JVI.00701-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Yip BS, Huang LM, Wu SC. Zika virus structural biology and progress in vaccine development. Biotechnol Adv. 2018;36:47–53. doi: 10.1016/j.biotechadv.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Lindley KM, Su JL, Hodges PK, Wisely GB, Bledsoe RK, Condreay JP, Winegar DA, Hutchins JT, Kost TA. Production of monoclonal antibodies using recombinant baculovirus displaying gp64-fusion proteins. J Immunol Methods. 2000;234:123–135. doi: 10.1016/s0022-1759(99)00133-7. [DOI] [PubMed] [Google Scholar]
- Ma W, Li S, Ma S, Jia L, Zhang F, Zhang Y, Zhang J, Wong G, Zhang S, Lu X, Liu M, Yan J, Li W, Qin C, Han D, Qin C, Wang N, Li X, Gao GF. Zika virus causes testis damage and leads to male infertility in mice. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.009. [DOI] [PubMed] [Google Scholar]
- Martines RB, Bhatnagar J, de Oliveira Ramos AM, Davi HP, Iglezias SD, Kanamura CT, Keating MK, Hale G, Silva-Flannery L, Muehlenbachs A, Ritter J, Gary J, Rollin D, Goldsmith CS, Reagan-Steiner S, Ermias Y, Suzuki T, Luz KG, de Oliveira WK, Lanciotti R, Lambert A, Shieh WJ, Zaki SR. Pathology of congenital zika syndrome in brazil: a case series. Lancet. 2016;388:898–904. doi: 10.1016/S0140-6736(16)30883-2. [DOI] [PubMed] [Google Scholar]
- Martinez-Sobrido L, Almazán F. New advances on zika virus research. Viruses. 2019;11:258. doi: 10.3390/v11030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, Vizjak A, Pižem J, Petrovec M, Avšič Županc T. Zika virus associated with microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Morrison TE, Diamond MS. Animal models of zika virus infection, pathogenesis, and immunity. J Virol. 2017;91:e00009-17. doi: 10.1128/JVI.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso D, Gubler DJ. Zika virus. Clin Microbiol Rev. 2016;29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono C, Okamoto T, Abe T, Matsuura Y. Baculovirus as a tool for gene delivery and gene therapy. Viruses. 2018;10:510. doi: 10.3390/v10090510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Pelc RS, Muramatsu H, Andersen H, DeMaso CR, Dowd KA, Sutherland LL, Scearce RM, Parks R, Wagner W, Granados A, Greenhouse J, Walker M, Willis E, Yu JS, McGee CE, Sempowski GD, Mui BL, Tam YK, Huang YJ, Vanlandingham D, Holmes VM, Balachandran H, Sahu S, Lifton M, Higgs S, Hensley SE, Madden TD, Hope MJ, Karikó K, Santra S, Graham BS, Lewis MG, Pierson TC, Haynes BF, Weissman D. Zika virus protection by a single low-dose nucleoside-modified mrna vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Zhang S, Cui Y, Shi Y, Gao GF, Qi J. Structures of human-infecting thogotovirus fusogens support a common ancestor with insect baculovirus. Proc Natl Acad Sci USA. 2017;114:E8905–e8912. doi: 10.1073/pnas.1706125114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premanand B, Zhong Wee P, Prabakaran M. Baculovirus surface display of immunogenic proteins for vaccine development. Viruses. 2018;10:298. doi: 10.3390/v10060298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu P, Zhang W, Li D, Zhang C, Liu Q, Zhang X, Wang X, Dai W, Xu Y, Leng Q, Zhong J, Jin X, Huang Z. Insect cell-produced recombinant protein subunit vaccines protect against zika virus infection. Antiviral Res. 2018;154:97–103. doi: 10.1016/j.antiviral.2018.04.010. [DOI] [PubMed] [Google Scholar]
- Richner JM, Himansu S, Dowd KA, Butler SL, Salazar V, Fox JM, Julander JG, Tang WW, Shresta S, Pierson TC, Ciaramella G, Diamond MS. Modified mrna vaccines protect against zika virus infection. Cell. 2017;168:1114–1125.e1110. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC. Characterization of a novel murine model to study zika virus. Am J Trop Med Hyg. 2016;94:1362–1369. doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz JC, Oya NJ, Blázquez AB, Escribano-Romero E, Martín-Acebes MA. Host-directed antivirals: a realistic alternative to fight zika virus. Viruses. 2018;10:453. doi: 10.3390/v10090453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M, Van TTH, Baird FJ, Coloe PJ, Smooker PM. Pre-existing immunity against vaccine vectors–friend or foe? Microbiology. 2013;159:1–11. doi: 10.1099/mic.0.049601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Muruato AE, Nunes BTD, Luo H, Xie X, Medeiros DBA, Wakamiya M, Tesh RB, Barrett AD, Wang T, Weaver SC, Vasconcelos PFC, Rossi SL, Shi PY. A live-attenuated zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat Med. 2017;23:763–767. doi: 10.1038/nm.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Dhama K, Khandia R, Munjal A, Karthik K, Tiwari R, Chakraborty S, Malik YS, Bueno-Marí R. Prevention and control strategies to counter zika virus, a special focus on intervention approaches against vector mosquitoes-current updates. Front Microbiol. 2018;9:87. doi: 10.3389/fmicb.2018.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang C, Tekes G, Mueller S, Paul A, Whelan SP, Wimmer E. Recoding of the vesicular stomatitis virus l gene by computer-aided design provides a live, attenuated vaccine candidate. mBio. 2015;6:e00237-15. doi: 10.1128/mBio.00237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger-Lucarelli J, Duggal NK, Bullard-Feibelman K, Veselinovic M, Romo H, Nguyen C, Rückert C, Brault AC, Bowen RA, Stenglein M, Geiss BJ, Ebel GD. Development and characterization of recombinant virus generated from a new world zika virus infectious clone. J Virol. 2017;91:e01765-16. doi: 10.1128/JVI.01765-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Wollebo HS, David Beckham J, Tyler KL, Khalili K. Zika virus: an emergent neuropathological agent. Ann Neurol. 2016;80:479–489. doi: 10.1002/ana.24748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikan N, Smith DR. Zika virus: history of a newly emerging arbovirus. Lancet Infect Dis. 2016;16:e119–e126. doi: 10.1016/S1473-3099(16)30010-X. [DOI] [PubMed] [Google Scholar]
- Wu Q, Fang L, Wu X, Li B, Luo R, Yu Z, Jin M, Chen H, Xiao S. A pseudotype baculovirus-mediated vaccine confers protective immunity against lethal challenge with h5n1 avian influenza virus in mice and chickens. Mol Immunol. 2009;46:2210–2217. doi: 10.1016/j.molimm.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Xu XG, Wang ZS, Zhang Q, Li ZC, Zhao HN, Li W, Tong DW, Liu HJ. Baculovirus surface display of e envelope glycoprotein of japanese encephalitis virus and its immunogenicity of the displayed proteins in mouse and swine models. Vaccine. 2011;29:636–643. doi: 10.1016/j.vaccine.2010.11.045. [DOI] [PubMed] [Google Scholar]
- Yamaji H. Suitability and perspectives on using recombinant insect cells for the production of virus-like particles. Appl Microbiol Biotechnol. 2014;98:1963–1970. doi: 10.1007/s00253-013-5474-9. [DOI] [PubMed] [Google Scholar]
- Yang C, Gong R, de Val N. Development of neutralizing antibodies against zika virus based on its envelope protein structure. Virol Sin. 2019;34:168–174. doi: 10.1007/s12250-019-00093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS, Fremont DH. Structural basis of zika virus-specific antibody protection. Cell. 2016;166:1016–1027. doi: 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Wang X, Ren F, Zou S, Feng M, Xu L, Yao L, Sun J. Construction of a highly efficient display system for baculovirus and its application on multigene co-display. Mol Genet Genomics. 2018;293:1265–1277. doi: 10.1007/s00438-018-1459-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.