Abstract
SARS-CoV-2 is a new virus responsible for an outbreak of respiratory illness known as COVID-19, which has spread to several countries around the world and a global effort is being undertaken to characterize the molecular features and evolutionary origins of this virus. In silico analysis of the transcription start sites, promoter regions, transcription factors and their binding sites, gene ontology, CpG islands for SARS-CoV-2 viral genome are a first step to understand the regulation mechanisms of gene expression and its association with genetic variations in the genomes. For this purpose, we first computationally surveyed all SARS-CoV-2 virus genes with the open reading frames from NCBI database and found eleven sequences to accomplish the mentioned features by using bioinformatics tools. Our analysis revealed that all (100%) of the SARS-CoV-2 virus genes have more than one TSS. By taking all TSSs with the highest predictive score we determined promoter regions and identified five common candidate motifs (MVI, MVII, MVIII, MVIV and MVV) of which MVI was found to be shared by all promoter regions of SARS-CoV-2 virus genes with the least E-value (3.8e-056, statistically highly significant). In our further analysis of MVI we showed MVI serve as binding sites for a single transcription factor (TF) family, EXPREG, involved in the regulatory mode of these genes. From EXPREG family four TFs that belongs to Cyclic AMP (cAMP) receptor protein (CRP) and Catabolite control protein A (CcpA) group mostly serve as transcriptional activator whereas two TFs that belong to LexA group always serve as transcriptional repressor in different kinds of cellular processes and molecular functions. Therefore, we unfolded SARS-CoV-2 viral genome to shed light on its gene expression regulation that could help to design and evaluate diagnostic tests, to track and trace the ongoing outbreak and to identify potential intervention options.
Keywords: COVID-19, CpG Island, Motif, Promoter, SARS-CoV-2, Transcription factor
Highlights
-
•
Computationally surveyed all SARS-CoV-2 genes with the open reading frames from NCBI database showed eleven gene sequences
-
•
Our analysis revealed that all (100%) of the SARS-CoV-2 genes have more than one TSS.
-
•
We determined promoter regions and identified five common candidate motifs of which MVI was found to be shared by all promoter regions of SARS-CoV-2 genes.
-
•
We showed MVI serve as binding sites for a single transcription factor family, EXPREG, involved in the regulatory mode of these genes.
1. Introduction
Emerging and reemerging pathogens are global threat for public health (Gao, 2018). Coronaviruses have been identified in human and several avian hosts (Ksiazek et al., 2003; Kuiken et al., 2003) as well as in various mammals, including camels, bats, masked palm civets, mice, dogs, and cats. Researchers showed with evidence as Severe Acute Respiratory Syndrome associated coronaviruses (SARS-CoV) could spread throughout the population of domestic and wild animals that come to market (Guan et al., 2003). It has been also reported for the potential risk of re-emergence of SARS-CoV that was circulating in bat populations and its cross-species transmission events leading to outbreaks in humans (Menachery et al., 2015). Accordingly, in late December 2019, several patients with pneumonic cases were found to be epidemiologically associated with the Huanan seafood market in Wuhan, in the Hubei province of China, where a number of non-aquatic animals such as birds and rabbits were also on sale (Lu et al., 2020). Because the disease, COVID-19, spread across China and beyond (Phan, 2020), WHO was obliged to declare as pandemic on March 11 and reported a total of confirmed 4,617,176 cases and 307,988 deaths until May 15, 22:41 GMT (WHO, 2020).
The Chinese public health, clinical, and scientific communities took prompt response to allow for timely recognition of the new virus, SARS-CoV-2, and shared the viral gene sequence to the world (Zhu et al., 2020). The genome of SARS-CoV-2 is a single-stranded positive-sense RNA of 30 kb (29,891 nucleotides) encoding 9860 amino acids. The genome is arranged in the order of 5′-replicase (orf1/ab)-structural proteins [Spike (S)-Envelope (E)-Membrane (M)-Nucleocapsid (N)] − 3′. The S, M, and E proteins together form the envelope of the virus. The M protein is the most abundant, mostly responsible for the shape of the envelope. The E protein is the smallest structural protein. The S and M proteins are also the transmembrane proteins that are involved in virus assembly during replication. N proteins remain associated with the RNA forming a nucleocapsid inside the envelope. It has been reported that although N protein is largely involved in processes related to the viral genome, it is also involved in other aspects of the virus replication cycle (assembly and budding) and the host cellular response to viral infection. Polymers of S proteins remain embedded in the envelope giving it a crown-like appearance, thus the name coronavirus (Chan et al., 2020; Sagar, 2020).
The first genetic sequence of the virus was completed in early January, and gave researchers an answer to the most basic questions about the disease; what pathogen is causing it? However, as the months go on, the diversity in the viral genome is increasing. At over 30,000 base pairs, the SARS-CoV-2 virus genome is thought to accumulate approximately one to two mutations a month. By following the mutation patterns of the virus as it spreads, researchers are attempting to track the mutation and determine how it is spreading. Though the virus is mutating, as yet this has not caused a significant change in its behavior, how it enters cells or how is transmitted (CDC, 2020; Phan, 2020; www.biotechniques.com/coronavirus-news, 2020) and how the gene expression is regulated. Therefore, this study was conducted to unfold SARS-CoV-2 virus genome to identify regulatory elements such as CpG islands, transcription factors (TFs) and their corresponding binding sites (TFBSs) involved in the regulation of its gene expression so that it provide baseline information for designing COVID-19 disease control strategy and for further detail molecular characterization of SARS-CoV-2 virus genome.
2. Methodology
2.1. Determination of transcription start sites and promoter regions for SARS-CoV-2 genes
SARS-CoV-2 virus genes (Wuhan seafood market pneumonia virus isolate sequences) were taken from NCBI genome browser on March 20, 2020. In this analysis all eleven SARS-CoV-2 gene coding sequences, available in NCBI database with the start codon at the beginning of the sequence and only functional genes (protein coding) were considered. To determine their respective transcription start sites (TSSs), 1 kb sequences upstream of the start codon were excised from each gene (Lenhard et al., 2012). All the TSSs of each of eleven SARS-CoV-2 virus genes were searched within this region by using the Neural Network Promoter Prediction (NNPP version 2.2) tool set with the minimum standard predictive score (between 0 and 1) cutoff value of 0.8 (Reese, 2001). This tool helps to locate the possible TSSs within the sequences upstream of the start codon where the RNA polymerases start their activity, transcription process. NNPP tool has ability to recognize precisely the position of a TSS for a given gene. For those regions containing more than one TSS, the one with the highest value of prediction score was considered to have trustable and accurate prediction. Therefore, as previously done for mice V1R gene promoter regions determination, SARS-CoV-2 virus gene promoter sequences were defined as 1 kb region upstream of each TSS (Michaloski et al., 2011).
2.2. Identification of common candidate motifs and transcription factors
Promoter sequences which were identified based on the criteria considered above from SARS-CoV-2 virus gene were analyzed using the MEME version 5.0.1 searches, via the web server hosted by the National Biomedical Computation Resource (http://meme.nbcr.net) (Bailey and Elkan, 1994) to look for common candidate motifs that serves for binding sites of transcription factors that regulate the expression of SARS-CoV-2 virus genes. MEME searches for statistically significant candidate motifs in the input sequence set. The MEME output are in the form of XML, text, MAST HTML, MAST XML, MAST text, HTML and shows the candidate motifs as local multiple alignments of the input promoter sequences. Briefly, MEME discovers novel, ungapped motifs (recurring, fixed-length patterns) in sequences submitted in it. A motif is an approximate sequence pattern that occurs repeatedly in a group of related sequences. MEME represents motifs as position-dependent letter-probability matrices that describe the probability of each possible letter at each position in the pattern. MEME takes as input a group of sequences and outputs as many motifs as requested. MEME uses statistical modeling techniques to automatically choose the best width, number of occurrences, and description for each motif (Bailey and Elkan, 1994). Buttons on the MEME HTML output allow one or all of the candidate motifs to be forwarded for further analysis, to better characterize the identified candidate motifs, by other web-based programs. In this case TOMTOM (Gupta et al., 2007) web server was used to search for sequences matching the identified motif for its respective TF. The output of TOMTOM includes LOGOS representing the alignment of the candidate motif and TF with the p-value and q-value (a measure of false discovery rate) of the match, and links back to the parent transcription database for more detailed information about it (Bailey et al., 2006; Bailey et al., 2009).
2.3. Gene ontology (GO) analysis
For further analysis for the gene ontology or gene function associated with the identified most common candidate motifs, we used GOMO (Gene Ontology for Motifs) version 5.0.1 searches, via the web server hosted by the National Biomedical Computation Resource (http://meme.nbcr.net) (Fabian et al., 2010). In general, GOMO scans all promoters using nucleotide motifs provided to determine if any motif is significantly associated with genes linked to one or more Genome Ontology (GO). The significant GO terms can suggest the biological roles of the motifs. GOMO assign roles or GO terms to DNA regulatory candidate motifs using comparative genomics. The GOMO output is HTML and shows the GO terms, GO name and Gene ID. Buttons on the GOMO HTML output allow one or all of the Gene ID to be forwarded for further analysis, to ascertain which molecular and biological functions are associated with the candidate motif. GO term associated with statistically highly significant motif was used to present our result.
2.4. Search for CpG islands
Two algorithms were used to search for CpG islands. First, the stringent search criteria, Takai and Jones algorithm: GC content ≥55%, ObsCpG/ExpCpG ≥0.65, and length ≥ 500 bp was used (Takai and Jones, 2002). For this purpose, the CpG island searcher program (CpGi130) available at web link http://dbcat.cgm.ntu.edu.tw/ was used. Secondly, the CLC Genomics Workbench ver. 3.6.5 (http://clcbio.com, CLC bio,Aarhus, Denmark) was used for searching the restriction enzyme MspI cutting sites (fragment sizes between 40 and 220 bps). Searching for MspI cutting sites is relevant for detection of CGIs, because studies using whole genome CpG island libraries prepared for different species revealed that, CpG islands are not randomly distributed but are concentrated in particular regions because CpG-rich regions are achieved by isolation of short fragments after MspI digestion that recognizes CCGG sites (Takamiya et al., 2006).
3. Results
3.1. Identification of transcription start sites (TSSs)
TSSs predicted for each of the eleven protein coding SARS-CoV-2 virus genes were summarized and presented in Table 1 . Accordingly, the prediction showed SARS-CoV-2 virus genes have 3, 4, 8, 10, 11, 12, 13, 14 and 21 TSSs. It was also revealed that the locations for more than half (54.54%) of the TSSs are below −500 bp relative to the start codon with the predictive score of 0.99, 1.00, 0.97 and 0.93 for 54.5%, 27.3%, 9.1% and 9.1%, respectively, of the TSSs.
Table 1.
Number and predictive score value for SARS-CoV-2 virus gene TSSs.
| Name/Gene ID | Corresponding promoter region name | Number of TSS identified | Predictive score at cutoff value of 0.8 | Location of the best TSS from start codon |
|---|---|---|---|---|
| orf1ab/43740578 | pro-43,740,578 | 3 | 0.92, 0.93, 0.88 | −77 |
| ORF8/43740577 | pro-43,740,577 | 13 | 0.84, 0.98, 0.90, 0.99, 0.85, 0.81, 0.89, 1.00, 0.93, 0.95, 0.99, 0.94, 0.96 | −376 |
| ORF10/43740576 | pro-43,740,576 | 4 | 0.90, 0.88, 0.82, 0.97 | −50 |
| N/43740575 | pro-43,740,575 | 14 | 0.99, 0.85, 0.81, 0.89, 1.00, 0.93, 0.95, 0.99, 0.94, 0.96, 0.81, 0.99, 0.83, 0.99 | −756 |
| ORF7b/43740574 | pro-43,740,574 | 10 | 0.84, 0.98, 0.90, 0.99, 0.85, 0.81, 0.89, 1.00, 0.93, 0.95, | −238 |
| ORF7a/43740573 | pro-43,740,573 | 12 | 0.99, 0.96, 0.84, 0.86, 0.91, 0.94, 0.84, 0.98, 0.90, 0.99, 0.85, 0.81 | −855 |
| ORF6/43740572 | pro-43,740,572 | 8 | 0.81, 0.99, 0.96, 0.84, 0.86, 0.91, 0.94, 0.84 | −663 |
| M/43740571 | pro-43,740,571 | 12 | 0.98, 0.88, 0.88, 0.93, 0.81, 0.90, 0.96, 0.96, 0.99, 0.98, 0.96, 0.81 | −422 |
| E/43740570 | pro-43,740,570 | 14 | 0.89, 0.97, 0.81, 0.98, 0.88, 0.88, 0.93, 0.81, 0.90, 0.96, 0.96, 0.99, 0.98, 0.96 | −144 |
| ORF3a/43740569 | pro-43,740,569 | 11 | 0.98, 0.97, 0.92, 0.84, 0.98, 0.82, 0.99, 0.90, 0.91, 0.83, 0.99 | −508 |
| S/43740568 | pro-43,740,568 | 21 | 0.95, 0.88, 0.94, 0.86, 0.81, 0.89, 0.98, 0.91, 0.86, 0.85, 0.99, 0.89, 0.94, 0.86, 0.99, 0.88, 0.82, 0.99, 0.99, 0.81, 0. 81 | −617 |
3.2. Common candidate motifs and associated transcription factors in the promoter regions of SARS-CoV-2 virus genes
Because one gene, orf1ab/43740578, doesn't has promoter sequence as it is located around the beginning (at 266 bp) of the genome coordinate, the study tried to identify the best candidate motifs for the remaining ten promoter sequences of SARS-CoV-2 virus gene. Accordingly, we revealed five common candidate motifs (MVI, MVII, MVIII, MVIV and MVV) that are shared by 80% (8/10) of the promoter input sequences (Table 2 ).
Table 2.
Identified common candidate motifs in SARS-CoV-2 virus gene promoter regions.
| Discovered motif | Number (%) of promoters containing each one of the motifs | E-value⁎ | Motif width | Total no. of binding sites |
|---|---|---|---|---|
| MVI | 7(87.5) | 3.8e-056 | 50 | 7 |
| MVII | 7(87.5) | 5.7e-056 | 50 | 7 |
| MVIII | 7(87.5) | 2.6e-052 | 50 | 7 |
| MVIV | 7(87.5) | 1.3e-051 | 50 | 7 |
| MVV | 7(87.5) | 3.6e-050 | 50 | 7 |
Probability of finding an equally well-conserved motif in random sequences.
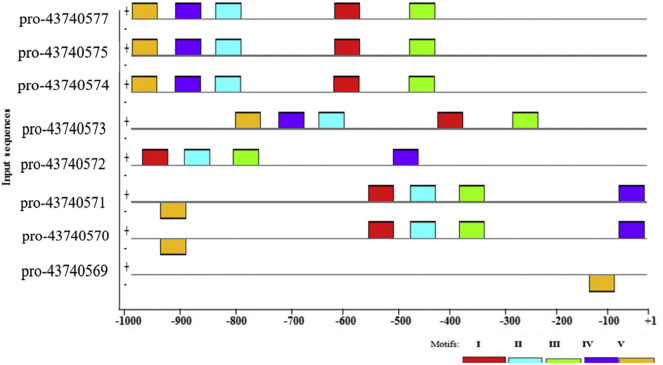
Fig. 1 presents the relative location and spatial distribution of these motifs in the promoter regions: majority of them are concentrated between −500 and -1 kb of the TSSs. The spatial position distribution of identified motifs seems to be consistent within the promoter sequences of SARS-CoV-2 virus genes. It is also interesting to notice that majority of the motifs are distributed on positive strands and only three on the negative strands of the input promoter sequences.
Fig. 1.
Block diagrams showing the relative positions of candidate motifs in different SARS-CoV-2 virus gene promoter sequences relative to TSSs. The nucleotide positions are indicated at the bottom ofthe graph from +1 (beginning of TSSs) to the upstream 1000 (−1000) bp.
To determine the motifs which are functionally important, motifs that shared by all promoter regions of SARS-CoV-2 virus gene with the least e-value (3.8e-056, statistically highly significant) was chosen. It has been reported that the motif which commonly present a large number of promoter region could provide significantly more information than others in mice (Michaloski et al., 2011). Accordingly, in the present analysis MVI was revealed as the common promoter motif for all (100%) SARS-CoV-2 virus genes that serve as binding sites for TFs involved in the expression regulation of these genes. Sequence logo for MVI generated by MEME is presented in Fig. 2 .
Fig. 2.
Sequence logos for the identified best motif (MVI) for SARS-CoV-2 virus genes promoter regions. The analysis was carried out using MEME Suite.
Furthermore, additional analyses were performed to get more insights on the MV1 motif of SARS-CoV-2 virus genes. MVI was compared to registered motifs in publically available databases such as Prokaryote DNA to see if they are similar to known regulatory motifs for transcription factor using the TOMTOM web application (Reese, 2001). TOMTOM provides LOGOS that represents the alignment of the motif with the candidate TF and also provides a numeric score for the match between them together with statistical significance. The output from TOMTOM also links the parent TF database for more detail information for activation, repression or dual regulatory effects on the matched motif. As a result, MVI matched with 10 out of 84 known motifs found in the databases. On the basis of their statistical significance values, all the 10 matched motifs were analyzed and presented in Table 3 . From the result of our analysis we revealed that all of them were found to be categorized under only one transcription factor family known as EXPREG that serves as best binding candidates for MVI motif. Analysis of the remaining four candidate motifs also revealed the same TF family. Further categorization of this TF family that contain ten individual TFs associated to MVI revealed that two, three and two of them belongs to CRP, CcpA and LexA group, respectively, whereas the remaining belonged to their individual group. Therefore, the MVI motif could also serve as a binding site for the EXPREG TF family in SARS-CoV-2 virus genes to regulate its expression as transcriptional activator or repressor or dual purpose.
Table 3.
The list of candidates from EXPREG transcription factors family which could bind to motif MVI.
| Candidate transcription factors | Statistical significance⁎ | Regulatory mode (%) |
|||
|---|---|---|---|---|---|
| Activation | Repression | Dual | Not specified | ||
| CRP (Y.pestis) | 4.18e+00 | 76.0 | 23.0 | 0.0 | |
| CRP (E.coli) | 7.34e+00 | 82.0 | 17.0 | 0.0 | 0.0 |
| CcpA (L.lactis) | 2.07e+00 | 69.0 | 30.0 | 0.0 | |
| CcpA (S.pneumoniae) | 9.82e+00 | 68.0 | 31.0 | 0.0 | 0.0 |
| CcpA (B.subtilis) | 9.68e+00 | 6.0 | 93.0 | 0.0 | 0.0 |
| Fur (P.syringae) | 1.50e+00 | 0.0 | 13.0 | 0.0 | 85.0 |
| LexA (S.meliloti) | 5.41e+00 | 0.0 | 100.0 | 0.0 | 0.0 |
| LexA (M.tuberculosis) | 8.23e+00 | 0.0 | 100.0 | 0.0 | 0.0 |
| GInR (S.erythraea) | 2.36e+00 | 35.0 | 38.0 | 0.0 | 26.0 |
| ArcA (S.oneidensis) | 9.62e+00 | 0.0 | 0.0 | 0.0 | 100.0 |
CcpA - Catabolite control protein A; ArcA - aerobic respiration response regulator; CRP - cAMP receptor protein; Fur - Ferric uptake regulation protein; GlnR - DNA-binding response OmpR family regulator; LexA - locus for X-ray sensitivity A.
Statistical significance for the binding of given transcription factors to MVI motif.
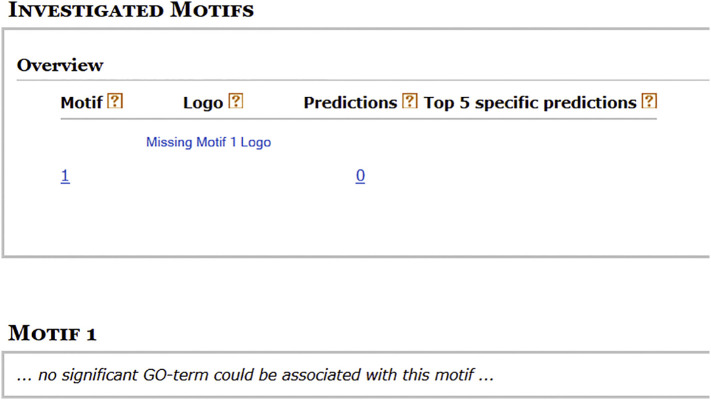
Further analysis was conducted to identify GO terms for the identified best motif of SARS-CoV-2 virus genes promoter sequences. But, no significant GO-term was found to be associated with MVI motif (Fig. 3 ).
Fig. 3.
GO-term associated with MVI motif. No gene ontology was identified.
3.3. Investigation for CpG islands (CGIs) in SARS-CoV-2 virus genes promoter regions
To further explore the regulatory elements that are involved in eleven SARS-CoV-2 virus genes, CpG islands were also investigated in both promoter and gene body regions using two algorithms. First, in silico analysis was conducted using Takai and Jones' algorithm (Takai and Jones, 2002) and found no CpG islands in both promoter and gene body regions. Similarly, a second alternative approach to search for the presence of CpG islands by in-silico digestion using restriction enzyme MspI revealed poor CpG islands in both promoter and gene body regions in SARS-CoV-2 virus genes (Table 4 ).
Table 4.
MspI cutting sites and fragment sizes for SARS-CoV-2 virus gene in promoter and gene body regions for eleven sequences.
| Region | Names of corresponding SARS-CoV-2 virus gene | Nucleotide positions of MspI sites | Fragment sizes (between 40 and 220 bps) |
|---|---|---|---|
| Promoter region | Pro-43,740,578 | No cut | – |
| Pro-43,740,577 | Single cut (at 235) | ||
| Pro-43,740,576 | No cut | ||
| Pro-43,740,575 | Single cut (at 235) | ||
| Pro-43,740,574 | Single cut (at 234) | ||
| Pro-43,740,573 | Single cut (at 435) | ||
| Pro-43,740,572 | Single cut (at 612) | ||
| Pro-43,740,571 | No cut | ||
| Pro-43,740,570 | No cut | ||
| Pro-43,740,569 | No cut | ||
| Pro-43,740,568 | Single cut (at 201) | 201 | |
| ORF8/43740577 | No cut | – | |
| ORF10/43740576 | No cut | – | |
| N/43740575 | No cut | – | |
| Gene body region | ORF7b/43740574 | No cut | – |
| ORF7a/43740573 | No cut | – | |
| ORF6/43740572 | No cut | – | |
| M/43740571 | Single cut (at 229) | – | |
| E/43740570 | No cut | – | |
| ORF3a/43740569 | Single cut (at 757) | 71 | |
| S/43740568 | Single cut (at 1423) | – | |
| orf1ab/43740578 | Multiple cut (at 3987, 12466, 12,933, 14,409, 15,693, 20,411) | – |
4. Discussion
Viral genome show a large diversity genome composition, structures, replication and transcription strategies with great implications in virus biology such as in virus–host interactions (Whelan, 2014). It has been reported that the accurate identification of TSSs and then promoter regions is an important step for in-silico understanding of the gene transcription regulation mechanisms. This is because promoter regions are found to share common subtle patterns or motifs that act as binding sites where TFs attach to facilitate or regulate transcription of genes (Mahdi and Rouchka, 2009). In this regard, our present prediction accuracy was ≥97% for 90.9% of the TSSs identified for SARS-CoV-2 virus genes. It has been shown that most genes have more than one TSS (Lenhard et al., 2012) which is in agreement with the present study where all (100%) of the SARS-CoV-2 virus genes analyzed have more than one TSS. On the other hand we identified five common candidate motifs of which MVI was found to be shared by all promoter regions of SARS-CoV-2 virus genes with the least E-value (3.8e-056, statistically highly significant). Unlike in higher organisms such as mouse and pig (Michaloski et al., 2011; Dinka and Thong Le, 2017), the spatial distributions for majority of these motifs in SARS-CoV-2 virus genes were found to be not closer to their TSSs.
It has been reported that regulatory proteins affect the general TFs of the pre-initiation complex, RNA polymerase II and its cofactors, histones and DNA polymerases and thereby modulate the rate of transcription initiation and elongation and replication initiation that likely affect the viral biology such as virus–host interactions (Bernard, 2013). In our analysis we found that not only MVI but also the remaining four candidate motifs serve as binding sites for a single transcription factor family, EXPREG, involved in the regulatory mode of SARS-CoV-2 virus genes. This is in agreement with the recent report by Ambrosini et al. (2020) where they confirmed by experiment that most of the best performing TFs are those coming from the same family. Detail further analysis of the ten TFs that belong to EXPREG family revealed that in more than 68% of the cases four TFs that belongs to Cyclic AMP (cAMP) receptor protein (CRP) and Catabolite control protein A (CcpA) group serve as transcriptional activator whereas two TFs that belong to LexA group always (100%) serve as transcriptional repressor. This is in agreement with the report of Akeo and his colleagues (2007) where CRP served as transcriptional activator for Thermus thermophilus bacterium. Furthermore, it has been reported that CRPs are global transcriptional regulators broadly distributed in bacteria playing diverse cellular roles such as carbohydrate metabolism, modulation of virulence gene expression and pathogenesis (Wolfgang et al., 2003; Zheng et al., 2004; Suh et al., 2002). As we tried to check the role of CRP TFs in UniProt protein database, they can act as an activator, repressor, co-activator or co-repressor in Escherichia coli and Yersinia pestis where similar function was revealed for CcpA transcription factor in Lactococcus lactis and Streptococcus pneumoniae.
In UniProt protein database LexA transcription factor is also named as LexA repressor functioning as a repressor of a number of genes in bacteria such as Rhizobium meliloti. LexA protein is the repressor, which, during normal bacterial growth down regulates its own expression and, in E. coli, the expression of at least 43 unlinked genes (Courcelle et al., 2001; Fernandez et al., 2000). Under normal growth conditions, LexA represses transcription of DNA damage-inducible genes by binding to an upstream DNA sequence (Dos Vultos et al., 2009). It has been reported that in E. coli the transcriptional repressor LexA controls a coordinated cellular response to DNA damage which is crucial for bacterial survival (Kamenšek et al., 2010; Smollett et al., 2012). In our analysis unlike for the other nine transcription factors, the role of ArcA transcription factor is not specified. But, further investigation about ArcA transcription factor from UniProt protein data base revealed it is a protein with two component signal transduction system controlling aerobic respiration response regulator with gene ontology (molecular function) of mainly serving as DNA-binding transcription activator activity in bacteria.
As we discussed above TFs that belong to EXPREG family are involved in the expression regulation of bacteria genes. The same might hold true for SARS-CoV-2 virus genes. Because our analysis is in silico, the role of the ten transcription factors identified in the current study need experimental validation by Electrophoretic mobility shift assay (EMSA) and other methods. Furthermore, the poor CpG islands we observed might suggest that SARS-CoV-2 virus gene expression regulation pattern is in tissue specific manner.
5. Conclusion
The main motivation of this work was to identify regulatory elements that can determine expression of SARS-CoV-2 virus genes. Accordingly, we revealed a panel of TFs that belong to a single transcription factor family, EXPREG, acting as activators and/or repressor of SARS-CoV-2 virus genes. Moreover, we predicted transcription start sites, promoter regions, transcription factor binding sites, CpG islands in SARS-CoV-2 viral genome that plays role in the process of gene expression regulation. These analyses would benefit for understanding the regulation mechanisms of gene expression, fill the gap of missing data and improve the process of knowledge discovery about SARS-CoV-2 virus genome by elucidating the molecular mechanisms behind the virus expression regulation. Robust and detail study about genomic features and their potential association with SARS-CoV-2 virus characteristics and virulence in humans need further attention.
Data availability
The datasets analyzed during the current study were taken from SARS-CoV-2 of NCBI Genome browser.
Declaration of Competing Statement
The authors declare that there is no conflict of interest.
References
- Akeo S., Satoshi K., Noriko N., Aiko K., Seiki K., Shigeyuki Y. Transcription activation mediated by a cyclic AMP receptor protein from Thermus thermophilus HB8. J. Microbiol. 2007;189(10):3891–3901. doi: 10.1128/JB.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G., Vorontsov I., Penzar D., Groux R., Fornes O., Nikolaeva D.D., Ballester B., Grau J., Grosse I., Makeev V., Kulakovskiy I., Bucher P. Insights gained from a comprehensive all-against-all transcription factor binding motif benchmarking study. Genome Biol. 2020;21 doi: 10.1186/s13059-020-01996-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey L.T., Elkan C. Proceedings of the Second International Conference on Intelligent Systems for MolecularBiology. AAAI Press; 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; pp. 28–36. [PubMed] [Google Scholar]
- Bailey L.T., Nadya W., Chris M., Wilfred W.L. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey L.T., Mikael B., Fabian A.B., Martin F., Charles E.G., Luca C., Jingyuan R., Wilfred W.L., William S.N. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H. Regulatory elements in the viral genome. Virol. 2013;445(1–2):197–204. doi: 10.1016/j.virol.2013.04.035. [DOI] [PubMed] [Google Scholar]
- CDC First Travel-Related Case of 2019 Novel Coronavirus Detected in United States. 2020. https://www.cdc.gov/media/releases/2020/p0121-novel-coronavirus-travel-case.html (Jan 21)
- Chan J.F., Kok K., Zhu Z., Chu H., To KK, Yuan S., Yuen K. Genomic characterization of the 2019 novel humanpathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Khodursky A., Peter B., Brown P.O., Hanawalt P.C. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinka H., Thong Le M. Analysis of pig Vomeronasal receptor type 1 (V1R) promoter region reveals a common promoter motif but poor CpG Islands. Anim. Biotech. 2017;29(4):293–300. doi: 10.1080/10495398.2017.1383915. [DOI] [PubMed] [Google Scholar]
- Dos Vultos T., Mestre O., Tonjum T., Gicquel B. DNA repair in mycobacterium tuberculosis revisited. FEMS Microbiol. Rev. 2009;33:471–487. doi: 10.1111/j.1574-6976.2009.00170.x. [DOI] [PubMed] [Google Scholar]
- Fabian A., Buske M.B., Denis C.B., Timothy L.B. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics. 2010;26:860–866. doi: 10.1093/bioinformatics/btq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D.H., Ogi A.R., Aoyagi T., Chafin S., Hayes D., Ohmori J.J., Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Gao G.F. From “a”IV to “Z”IKV: attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157–1159. doi: 10.1016/j.cell.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopolous J.A., Timothy B., William S.N. Quantifying similarity between motifs. Gen. Biol. 2007;8(2):R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenšek S., Podlesek Z., Gillor O., Zgur-Bertok D. Genes regulated by the Escherichia coli SOS repressor LexA exhibit heterogenous expression. BMC Microbiol. 2010;10:283. doi: 10.1186/1471-2180-10-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A.M., Schutten M. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B., Sandelin A., Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi R.N., Rouchka E.C. RBF-TSS: identification of transcription start site in human using radial basis functions network and oligonucleotide positional frequencies. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0004878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., Randell S.H., Lanzavecchia A., Marasco W.A., Shi Z.L., Baric R.S. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloski J.S., Galante P.A., Nagai M.H., Armelin C.L., Chien M.S., Matsunami H., Malnic B. Common promoter elements in odorant and vomeronasal receptor genes. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0029065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Gen. Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- Sagar A. Online Microbiology Notes. 2020. Structure and genome of SARS-CoV-2 (COVID-19) with diagram.https://microbenotes.com/structure-and-genome-of-sars-cov-2/ [Google Scholar]
- Smollett K.L., Kimberley M.S., Christina K., Kristine B., Roger S.B., Davis E.O. Global analysis of the regulon of the transcriptional repressor LexA, a key component of SOS response in mycobacterium tuberculosis. J. Biol. Chem. 2012;287(26):22004–22014. doi: 10.1074/jbc.M112.357715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh S.J., Runyen-Janecky L.J., Maleniak T.C., MacGregor C.H., Zielinski-Mozny N.A., Phibbs P.V., Jr., West S.E. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology. 2002;148:1561–1569. doi: 10.1099/00221287-148-5-1561. [DOI] [PubMed] [Google Scholar]
- Takai D., Jones P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamiya T., Hosobuchi S., Asai K., Nakamura E., Tomioka K., Kawase M., Kakutani T., Paterson A.H., Murakami Y., Okuizumi H. Restriction landmark genome scanning method using Isoschizomers (MspI/HpaI) for DNA MethylationAnalysis. Electrophoresis. 2006;27:2846–2856. doi: 10.1002/elps.200500776. [DOI] [PubMed] [Google Scholar]
- Whelan S. Viral replication strategies. In: Knipe D.M., Howley P.M., editors. Fields Virology. 6th ed. Vol. 2. Lippincott, Williams and Wilkins; Philadelphia, PA, USA: 2014. p. 2160. [Google Scholar]
- WHO Novel coronavirus – Republic of Korea (ex-China) 2020. http://www.who.int/csr/don/21-january-2020-novelcoronavirus-republic-of-korea-ex-china/en/ (Jan 21, 2020)
- Wolfgang M.C., Lee V.T., Gilmore M.E., Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Zheng D.C. Constantinidou, Hobman J.L., Minchin S.D. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. 2020 Jan 24. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study were taken from SARS-CoV-2 of NCBI Genome browser.