Abstract
The formation of a tube-like structure is a basic step in the making of functional hearts in vertebrates and invertebrates and therefore, its understanding provides important information on heart development and function. In Drosophila, the cardiac tube originates from two bilateral rows of dorsally migrating cells. On meeting at the dorsal midline, coordinated changes in cell shape and adhesive properties transform the two sheets of cells into a linear tube. ECM and transmembrane proteins linked to the cytoskeleton play an important role during these dynamic processes. Here we characterize the requirement of Cbl-Associated Protein (CAP) in Drosophila heart formation. In embryos, CAP is expressed in late migrating cardioblasts and is located preferentially at their luminal and abluminal periphery. CAP mutations result in irregular cardioblast alignment and imprecisely controlled cardioblast numbers. Furthermore, CAP mutant embryos show a strongly reduced heart lumen and an aberrant shape of lumen forming cardioblasts. Analysis of double heterozygous animals reveals a genetic interaction of CAP with Integrin- and Talin-encoding genes. In post-embryonic stages, CAP closely colocalizes with Integrin near Z-bands and at cell–cell contact sites. CAP mutants exhibit a reduced contractility in larval hearts and show a locally disrupted morphology, which correlates with a reduced pumping efficiency. Our observations imply a function of CAP in linking Integrin signaling with the actin cytoskeleton. As a modulator of the cytoskeleton, CAP is involved in the establishment of proper cell shapes during cardioblast alignment and cardiac lumen formation in the Drosophila embryo. Furthermore, CAP is required for correct heart function throughout development.
Introduction
Significant insights in heart development and function at the molecular-genetic level were obtained by study of model organisms. The morphogenetic origin, structure and function of the Drosophila heart (dorsal vessel, DV) exhibits remarkable conservation with vertebrates [1–4] and has been instructive in revealing mechanisms of cardiomyopathies and aging [5–8]. The Drosophila heart derives from cardiac progenitors located in bilateral anlagen within the dorsal mesoderm. After their specification, cardioblasts (CBs) align as two lateral rows of migrating cells that eventually meet and line up at the dorsal midline ([9]; reviewed by [10–14]). Approaching the dorsal midline, the contralateral CBs polarize by reorganization of their extracellular matrix (ECM), transmembrane receptors and associated intracellular proteins, forming a leading edge and later distinct basal, junctional (J) and luminal (L) domains ([15–18]; see below). Contralateral CBs make first contact at their dorsal, cadherin-rich J domain. After dorsal sealing their cell shape changes from pear-like to crescent-like due to cytoskeletal remodeling [17]. By this event, the ventral sides of contralateral CBs come in close proximity, allowing establishment of cell contacts between their ventral J domains and a heart lumen forms between the non-adherent L domains [9, 19].
The wall of the DV is formed by 52 pairs of CBs, which are flanked by non-contractile, more loosely arranged pericardial cells that have nephrocytic functions. The posterior one-third of the DV, referred to as the heart, is distinguished from the aorta [20] by a wider lumen and by the differentiation of specialized CBs (ostial cells) into inflow tracts for the hemolymph. At its boundary to the aorta a valve, formed by another pair of special cells, inhibits retrograde flow from the aorta, the outflow tract of the heart [9, 10, 13]. Late in embryo development, the cardiomyoblasts organize a contractile apparatus in form of striated circular myofibrils. In the resulting cardiomyocytes (CMCs) these fibrils are embedded and anchored in the dorsal and ventral contact zone of contralateral cells, organized by the Integrin Adhesion Complex (IAC, see below). There, the myofibrils line up and become matched in register with the myofibrils of the contralateral cells [7]. In the larvae, the principal organization of the DV is maintained, but there is an eightfold hypertrophic growth of CMCs with a concomitant increase in the number of contractile myofibrils [7, 21]. During metamorphosis the most posterior cells of the heart undergo apoptosis. The aortal part takes over heart function, becomes widened and four pairs of ostia progenitor CMCs form new inflow tracts. In addition, three new intracardial valves are formed [22, 23]. In addition to the CMCs with a circular arrangement of myofibrils, a subset of heart tube-connected striated muscles (alary muscles) transdifferentiates into a layer of longitudinal myofibers associated with the ventral surface of the heart tube [21, 24].
Recent years have seen a significant progress in establishing molecular mechanisms that contribute to the heart assembly and lumen formation. The polarization of CBs is essential for correct heart tube morphogenesis [15–18]. During migration, the leading edge actively extends filopodia, where non-muscle Myosin II (Zipper), the Rho-GTPase Cdc42 and the formins dDAAM and Diaphanous are located [25]. This domain forms the dorsal J domain, the site of first contact between contralateral CBs. The dorsal and ventral J domains are characterized by the expression of adherens junction proteins β-Catenin (Armadillo) and E-cadherin (Shotgun). The non-adhesive L domain between the J domains displays the ECM protein Laminin and its receptor Integrin [16, 18, 26–28]. Integrin is an anchoring as well as signaling molecule that is required for adhesion and tension signaling to the cytoskeleton through links to the ECM established by the Integrin Adhesion Complex (see below). At this early stage Integrin is required for polarization and recruitment of Robo1/2 and its ligand, the signaling molecule Slit [29–31]. Slit and Robo in turn are essential for the formation of a non-adhesive luminal domain. The J and L domains maintain themselves by negative interactions and provide the cells with local information to regulate the adhesive properties and the cell shape changes required for tube and lumen formation [17, 18]. In addition, the Netrins together with their receptors Unc5 [32] or Frazzled [33] provide cues independent form the Slit/Robo pathway for DV tubulogenesis. Netrin signaling mediated by Frazzled facilitates cell shape changes and attachment of contralateral CBs [33].
Integrins can directly interact with actin, but normally this interaction is supported by adapter proteins like Talin, Vinculin and Paxillin [34–36]. These and other unknown adapter proteins together with Integrin-linked kinases form the Integrin Adhesion Complex (IAC). The IAC links and modulates integrin adhesion and signaling across the cell membrane to the cytoskeleton by regulating the affinity for ligands and recruiting numerous modifying proteins [37]. Talin (encoded by rhea) together with Integrin is an early marker of the L domain and supports Integrin in the localization of Slit and Robo [38]. Like Integrin, Talin is continuously required for heart growth to maintain the one to one apposition of myofibril ends between cardiomyocytes. The induced knock down of Integrin or Talin results in degeneration of cardiomyocyte contacts and a reduction in myofibril length and number [6, 7].
We report here the function of CAP in the development of the Drosophila DV. Drosophila CAP is an orthologue of the conserved mammalian CAP protein, an adapter protein of the CAV family (CAP/Ponsin, ArgBP2 and Vinexin; [39]). The CAV proteins regulate cell adhesion, cytoskeletal organization, and growth factor signaling. They contain a N-terminal SoHo (Sorbin homology) domain that was found to interact with Flotillin in lipid rafts in HEK293T cells in vitro [40]. Their three C-terminal SH3 domains mediate the interaction with multiple binding partners including signaling molecules and cytoskeletal proteins [41–44]. CAP was first described by its association with the c-Cbl protein, an E3 ubiquitin ligase involved in Insulin receptor signaling in mouse 3T3-L1 adipocytes [41]. In this context, CAP functions as an adapter to recruit c-Cbl to the insulin receptor where it becomes activated by phosphorylation [45]. Furthermore, CAP was shown to bind to dynamin and to play a role in receptor mediated endocytosis in mammalian cells [46]. A different role was attributed to CAP in the formation of stress fibers and focal adhesions in vitro [47]. This was consistent with its interaction with Focal Adhesion Kinase, Vinculin and Paxillin [39, 43, 44, 47]. Vertebrate CAP is present in costameres and intercalated discs, both in vitro and in mouse muscle sections [48] and is prominently expressed in the murine and zebrafish heart ([47]; S. Abdelilah-Seyfried, University of Potsdam, personal communication). In Drosophila, CAP was first described for its interactions with proteins of the Wnt signaling pathway [49, 50]. More recently, Bharadwaj and coworkers [51] reported that CAP regulates two actin-rich structures: muscle attachment sites (MAS), which connect somatic muscles to the body walls, and scolopale cells required for mechanosensation. They also found, that the CAP-interacting protein Vinculin is required for CAP recruitment to MAS and that vinculin mutants partially phenocopy CAP mutations.
We demonstrate that CAP is required lifelong for the development and function of the Drosophila heart, in cooperation with the IAC. In migrating cardioblasts, CAP loss of function results in irregular cardioblast alignment and variation in cardioblast numbers, indicating a role in the regulation of the coherence of migrating cells. CAP genetically interacts with scab (encoding the αPS3-Integrin subunit), and rhea (encoding Talin), and CAP mutant hearts show a strongly reduced heart lumen due to defects in cell shape. In larval, pupal and adult hearts, CAP colocalizes with Integrin near Z-bands and at cell contact sites of contralateral cardiomyocytes. Adult CAP-mutant hearts exhibit local defects in cell contacts and myofibrillar organization which result in strongly reduced heart function.
Materials and methods
Fly stocks and genetics
The control strain used for all experiments was w1118. The fly stocks scb2, rhea1, P{TRiP.HM05250}attP2 (UAS-CAP RNAi) and Df(2R)BSC281 were obtained from the Bloomington stock center. The homozygous lethal Df(2R)BSC281 (46D-47A) removes 3220 kbp DNA encoding CAP and 32 up- and downstream flanking protein coding genes. The FlyTrap GFP lines P{PTT-GB}ZASP52G00189 (ZASP52-GFP), P{PTT-un1}slsZCL2144 (Kettin-GFP; [52, 53]) and P{PTT-GA}CAPCA06924 (CAP-GFP; [52, 54]) were provided by L. Cooley (Yale University, USA). The handC3.1-GFP line [55] was provided by A. Paululat (University Osnabrück, Germany). For tissue-specific RNAi knockdown, the driver lines tinC+D-GAL4 and twi+how-GAL4, were generated by combining tinCΔ4-GAL4 [56] with tinD-GAL4 (=TMEN6-GAL4 #52D1; from J. Weiss, OHSU, Portland, USA; [57]) and 2xPE-twi-GAL4 [58] with how24B-GAL4 [59], respectively. The CAP42b and the CAP49e mutant fly strains and UAS-CAP-C [51] were kindly provided by A. Kolodkin (Johns Hopkins University, Baltimore, USA). All experiments were carried out at 25°C except the RNAi knockdown experiments that were carried out at 29°C.
Whole-mount in situ hybridizations
Whole-mount in situ hybridizations were performed as described by Lécuyer et al. [60]. Dig-labeled RNA sense and antisense probes were generated using the oligonucleotides CAP-fwd: ATGCCCAATAACCGCAATC and CAP-rev: TTTGGACTCGGTTCATTTTC. For probe detection Alkaline phophatase-coupled anti-Dig antibody (Roche Molecular Biochemicals) was used. Images were taken using Zeiss Axiophot microscope system with Nomarski optics.
Drosophila CAP antiserum
To generate a Myc-His fusion protein, the DNA encoding the short CAP isoform CAP-PE/N with three SH3 domains (NP_001137639), was cloned into the pMH vector [61]. Purified Myc-His-CAP-PE/N was injected into rabbits for polyclonal antibody production (BioGenes GmbH, Berlin). The CAP antisera were affinity purified using the recombinant CAP-E/N protein.
Immunoblotting
Protein extracts were isolated from 10 adult flies of the desired genotype. Shock frozen animals were homogenized in 50 μl 2× SDS sample buffer and boiled at 90°C for 10 min. Insoluble debris were removed by short centrifugation. For SDS-PAGE 10 μl of the resulting extracts per lane were used. The blot was blocked for 30 minutes with 10% dry milk powder (w/v) in phosphate buffered saline (PBS) containing 0.1% Triton X100 (PBT). Primary antibodies were diluted in 5% BSA in PBT and incubated overnight at 4°C. Antibodies used were rabbit anti-CAP (1:2.000, this study) and mouse anti-H2A (undiluted cell culture supernatant, monoclonal antibody Bx65; M. Frasch, H. Saumweber, unpublished). Alkaline phosphatase-coupled secondary antibodies (Dianova, Germany) were diluted 1:10.000 and phosphatase activity was visualized by colorimetric NBT/BCIP reaction.
Immunohistochemistry and microscopy
Embryos collected from grape juice plates were dechorionated in 50% DanKlorix (Colgate-Palmolive), fixed for 30 minutes in a 1:1 (v/v) mixture of 4% formaldehyde in PBS and n-heptane and devitellinized by shaking in a 1:1 (v/v) mixture of methanol and n-heptane. Dorsal vessels from adult flies were dissected in 1xPBS and fixed for 1h with 3.7% formaldehyde in 1xPBS. Staining of fixed embryos was as described by Knirr et al. [62]. If necessary Tyramide Signal Amplification (TSA, as indicated below) was performed using biotinylated secondary antibodies in combination with the Vectastain ABC Kit (Vector Laboratories) and fluorescent Tyramide reagent (PerkinElmer Inc.). Preparation of adult heart still inside an open abdomen was as described by Vogler and Ocorr [63]. Fixation and staining of such hearts still in their abdomina was according to Molina and Cripps [22] with the following modifications: Fixation in 3.7% formaldehyde was for 30–60 min at 4°C, blocking was for 30 min in PBT containing 5% BSA. Following overnight reaction with primary antibodies at 4°C, samples were washed 3x 15 min in PBT and binding of secondary antibody was for 2 h at room temperature. F-actin was visualized by staining fixed tissue using TRITC-coupled Phalloidin (Sigma, 0.3 μg/ml) in PBS for 1h at room temperature. Following three 15 min washes in PBT abdominal preparations were mounted in 85% glycerol, 4% propylgallate. The following primary antibodies were used: monoclonal antibodies for βPS1-Integrin/CF.6G11 (1:10), Rhea/Talin E16B (1:20, TSA) and Slit/C555.6D (1:10) were obtained from DSHB-NIH, University of Iowa, anti-GFP 3E6 (1:1000) was from Invitrogen. In addition the following polyclonal antibodies were used: rabbit anti-CAP (1:1000, this study), guinea pig anti-Doc2+3 (1:300, TSA; [64]) and rabbit anti-Mef2 (1:750; kindly provided by H.T.Nguyen, University of Erlangen-Nürnberg). Secondary antibodies were obtained from Invitrogen and used in a dilution of 1:1000. Data were taken using a Deltavision deconvolution microscope system (Applied precision, USA) and processed using softworx and imageJ software. The significance of changes in the number of Mef2+ or Doc+ cardioblasts was evaluated with a 2-sided t-test with equal variance. Differences with p-scores<0.05 were considered as significant (p<0.05 (*); p<0.01 (**); p<0.001 (***); S1 File).
Histological sections and electron microscopy
Late stage embryos of the desired genotype were prepared for TEM analysis essentially as described [65]. Following washing with 0.1 M cacodylate buffer, tissue was stained with 1% osmium tetroxide for 1 hour, dehydrated in a graded ethanol series and propylene oxide and embedded in Poly/BedR 812 (Polysciences, Inc., Eppelheim, Germany). Semi-thin cross sections of each 1 μm were made on a Leica Ultracut S ultramicrotome. Sections were stained with toluidine blue for 3 minutes at 70°C. Image acquisition was on a Zeiss Axiophot microscope system. Ultrathin sections were contrasted with uranyl acetate and lead citrate and examined with a FEI Morgagni electron microscope. Digital images were taken with a Morada CCD camera and the iTEM software (Olympus Soft Imaging Solutions GmbH, Münster, Germany).
Live-imaging of larval heart beat
Live imaging was performed using w1118 control or CAP49e mutant L3 larvae carrying the handC3.1-GFP reporter line [55]. Larvae were anesthetized for 3 minutes with Flynap® (Carolina Biological Supply Company) according to manufacturer´s instructions and immobilized on double-faced scotch tape on microscopic slides. Following 10 minutes recovery, larvae were imaged immediately on a Deltavision microscope system. Data were recorded as single optical sections with 10 frames per sec (fps) and processed using softworx software (Applied precision, USA). Data were further processed as avi files using imageJ software. From stills of these files the minimal (DCNmin) and maximal (DCNmax) distance of CMC nuclei close to the inner valve cells were measured for each 10 cycles of heart beat per animal. Average and standard deviation was determined for the control (n = 3) and the CAP49e mutant group of animals (n = 15). From these values percent of apparent fractional shortening (AFS) was determined according to: [(DCNmax–DCNmin)/DCNmax] x 100%. Following data recording the larvae were carefully removed from the tape and checked for vitality. Data were graphically displayed and significance of the data was evaluated by 2-sided t-test for unequal variances (type 3). Differences with p-scores<0.05 were considered as significant (p<0.05 (*); p<0.01 (**); p<0.001 (***); S1 File).
Dye angiography
For dye injections staged pharate adults (<90 h APF) were used. The preparation of flies, the injection process and the movie acquisition were described by Drechsler et al. [66]. Briefly, pharate adults were glued on a glass slide and the operculum was removed to allow recording of the dye accumulation. 1 μg/μl Uranin, a fluorescent tracer solution, was microinjected laterally into the abdomen of the animal. Dye accumulation in the head region was recorded for 180 sec by fluorescence stereomicroscopy with a digital camera at 10 frames/sec. Pixel intensities were measured within the region of interest (R.O.I.; see red outline in Fig 8b) in the fly head using ImageJ-Fiji software. The average from three consecutive frames (0.3 sec) was determined for each animal for a time interval of 180 sec. Background was removed from the data of each animal individually by subtracting the 0.3 sec interval value with the minimal value determined from all time points. The resulting values were graphically displayed for single animals and for the average of each genotype. The p-scores were determined by a two-sided t-test with unequal variance (type 3). Differences with p-scores<0.05 were considered as significant (p<0.05 (*); p<0.01 (**); p<0.001 (***); S1 File).
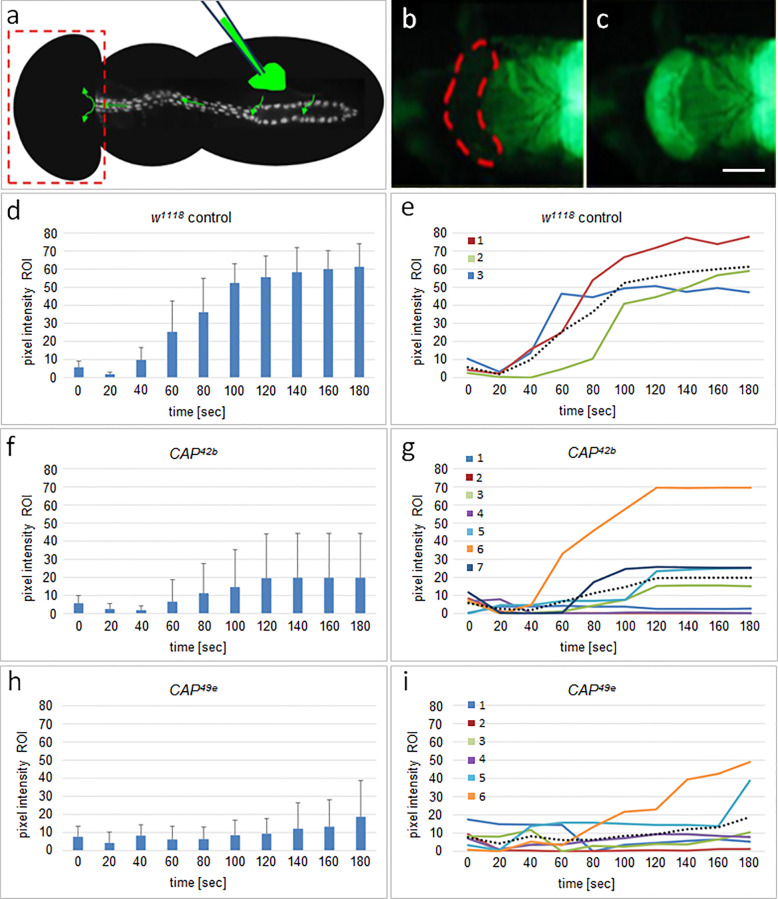
Fig 8. CAP is required for proper adult heart function.
(a) Illustration of the dye angiography assay to analyze adult heart function: Fluorescent dye is injected into the abdominal region of pharate adults as indicated, taken up by the heart, pumped anteriorly and delivered into the head region (boxed). Transport efficiency was quantified by integration of fluorescence intensity in the ROI of the head region (outlined in b) at different time points. (b) Snapshot of the head region of a w1118 control at time of injection. ROI is indicated by dashed red line. (c) Same region 180 seconds after injection. (d, f, h) Graphs showing the average pixel intensity in the R.O.I. with corresponding standard deviation at different time-points following injection for (d) w1118 controls (n = 3), and with significantly lower values for (f) homozygote CAP42b animals (n = 7, p = 0.0072; **) and (h) homozygote CAP49e animals (n = 6, p = 0.0039; **). The corresponding graphs in (e, g, i) show the time course of transport of single animals as differently color coded solid lines for w1118 (1–3), CAP42b (1–7) and CAP49e (1–6); dashed lines in these graphs represent the average for each genotype. See text and S1 File for details.
Results
Drosophila CAP is expressed in the dorsal vessel and is located at the periphery of cardioblasts
In a screen for new genes involved in Drosophila heart morphogenesis we found CAP to be expressed in the dorsal vessel (DV). Since CAP is conserved in evolution, and since the zebrafish and mouse CAP orthologues are highly expressed in the heart ([47]; S. Abdelilah-Seyfried, University Potsdam, unpublished), we decided to further explore the heart specific function of CAP in Drosophila.
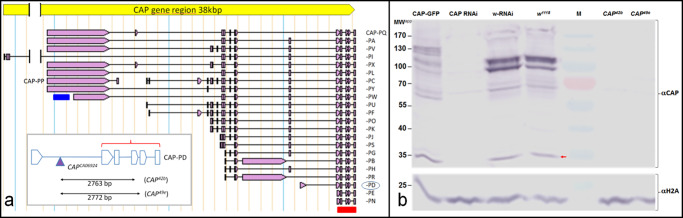
The CAP gene spans 38 kbp on chromosome 2R (46F9-47A1) encoding 23 alternate spliced transcripts that encode 20 unique polypeptides (Fig 1a; [67, 68]). To detect the CAP protein, we generated an antiserum against the short CAP isoform CAP-E/N, which consists of the three highly conserved SH3 domains (Fig 1a, Inset). According to the gene model this antiserum should detect all annotated CAP isoforms except CAP-PP (Fig 1a). On Western blots with cell extracts of adult flies (Fig 1b; for entire blot see S4 Fig) our CAP antiserum reacts with several (~17) polypeptides (Fig 1b, w1118 lane 4). This number corresponds to most of the 20 predicted CAP isoforms, as was reported previously for a similar CAP antiserum [51]. CAP42b and CAP49e mutant cell extracts show a weak nonspecific immunoreaction with CAP antiserum (Fig 1b, lanes 6 and 7). Extracts of animals with da-GAL4-induced UAS-CAP RNAi (see below) show a >90% reduction in signal intensity (Fig 1b, CAP RNAi lane 2 compare to. w1118, lane 4). Together, these results show that our antiserum is specific for CAP.
Fig 1. CAP gene structure and protein expression.
(a) Exon-intron structure of the annotated CAP proteins [68]. Note that all but the CAP-PP coding transcript contain 5 common 3´ exons. The bars indicate the location of probes used for in situ hybridization (blue) and the protein isoform CAP-PE/N used for CAP antigen expression (red), respectively. Inset: indicates length and position of the deletions CAP42b and CAP49e (double arrows) and the insertion site of CAPCA06924 (used to generate the theses deletions) in relation to isoform CAP-PD (also encircled in the list of all annotated CAP proteins) according to [51]. The deletions remove two of the three SH3 domains (drawn as pentagons) common to all CAP isoforms except CAP-PP. The red bracket indicates the exon-intron structure of CAP PE/N isoform that was used for immunization. b) Western blot reacted with either rabbit anti-CAP (top) or anti-H2A (loading control below). The type of the adult fly extracts loaded is as indicated on top and specified in the text: CAP-GFP (lane 1); w1118; UAS-CAP RNAi; da-GAL4 (lane 2); w1118; UAS-white RNAi; da-GAL4 (lane 3); w1118 control strain (lane 4); protein molecular weight marker (lane 5); CAP42b; hand-GFP (lane 6), CAP49e; hand-GFP (lane 7). Red arrow points to the position of CAP isoform PN/E.
Next, we studied CAP expression in the Drosophila dorsal vessel (Fig 2, S1 Fig). By whole mount in situ hybridization using an N-terminal probe we find strong CAP expression in the DV of the Drosophila embryo (Fig 2a). The first expression of CAP is detected in embryos at stage 11 in the garland cell primordium (S1a Fig). Starting with stage 13/14 CAP expression is detected in the heart progenitors and the visceral mesoderm (S1b Fig). Thereafter CAP expression in the heart increases up to stage 16 (S1c–S1e Fig). Until stage 16/17 CAP expression in the heart is restricted to cardioblasts (CBs).
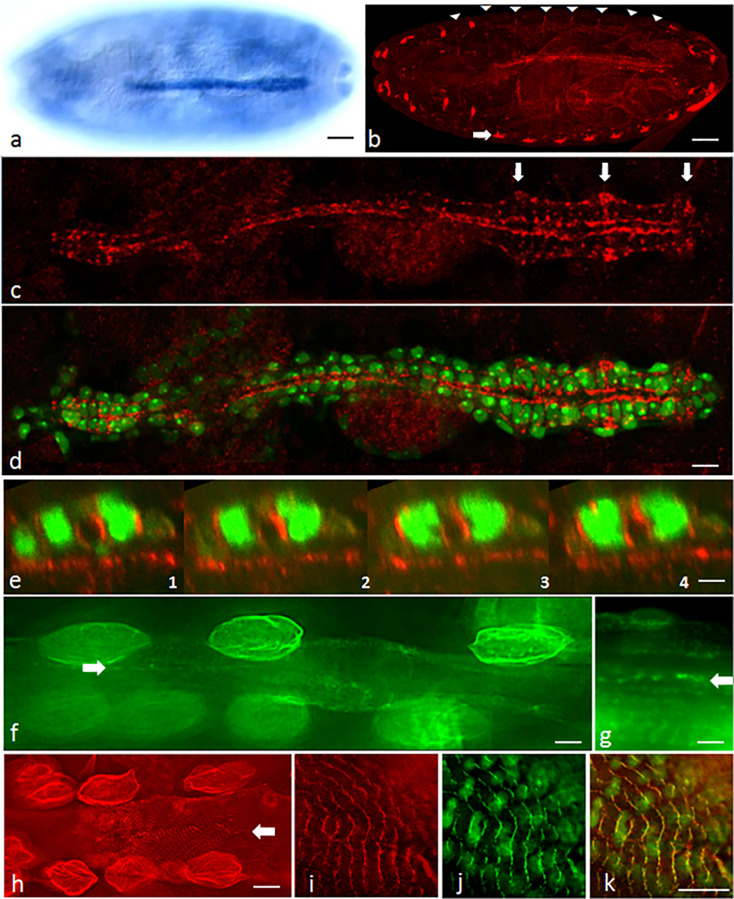
Fig 2. CAP is expressed in the Drosophila heart throughout development.
(a) Dorsal view of a stage 16 embryo following in situ hybridization using the 5´ CAP probe shown in Fig 1a. (b) Optical section of a whole mount stage 16 embryo following anti-CAP staining; white arrowheads: muscle attachment sites; open arrow: lateral pentascolopidial chordotonal organs; bar in (a, b): 35 μm; (c-e) Dorsal vessel of a stage 16 embryo stained for expression of CAP (red); hand-GFP signal green; (c) Dorsal view projections of anti-CAP staining only; arrow: prominent CAP staining near the ostia; (d) Same as (c) with additional hand-GFP signal. CAP is present in CMCs, but absent from pericardial cells, labeled by hand-GFP only; bar: 10 μm. (e) Consecutive Z-sections of the dorsal vessel; longitudinal distance between two consecutive sections: 20 μm; bar: 5 μm. (f, g) Optical sections of L3 larval heart expressing CAP-GFP (green); Note the presence of CAP at circular myofibrils and contact sites of contralateral CMCs (open arrow); strong staining is also observed in larval pericardial cells; bar (f): 10 μm; (g): 5 μm. (h) Optical sections of adult heart; CAP antibody staining (red) of circular and longitudinal myofibrils and at contacts of contralateral myocardial cells (open arrow). Pericardial cells are prominently stained; bar: 15 μm. (i-k) anti-CAP staining together with Kettin-GFP fluorescence of adult heart at higher magnification. (i) CAP (red) and (j) Kettin (green) colocalize near Z-bands on circular myofibrils (k, merge); bars (i-k): 8 μm.
In embryos we find CAP protein in the scolopale cells, a component of the chordotonal organ (Fig 2b, open arrow), the muscle attachment sites (MAS; Fig 2b, arrowheads) and in the DV, in agreement with data published previously [51]. CAP immunostaining of CBs is initially weak but increases during heart development. In stage 16, CAP is present in all CBs of the DV (Fig 2c), but not in the pericardial cells (Fig 2d). CAP is distributed in a granular pattern at the CB periphery. It is enriched at the luminal and abluminal side but is also present in between ipsilateral cells. On the luminal side CAP forms a rather continuous layer compared to the granular staining on the abluminal side. This difference is more obvious in the heart region. Locally, we observe abluminal accumulations of granular CAP staining near the ostial cells (Fig 2c, open arrows). In Z-sections, we see the same principal features of luminal and abluminal staining. Of note, besides the luminal location, CAP staining often reaches into the dorsal contact zones of paired contralateral cells (Fig 2e). In the larval and adult heart CAP is localized at cell contacts of contralateral cardioblasts and in the circular myofibrils of cardiomyocytes (Fig 2f–2h; S2d, S2g and S2j Fig). In adult hearts, in addition, it is localized in ventral longitudinal myofibers (S2a and S2j Fig). In both types of myofibrils CAP staining localizes near Z-bands, as evident by Kettin and ZASP colocalization (Fig 2i–2k; S2a–S2l Fig). CAP most closely colocalizes with Integrin at contacts of contralateral cardiomyocytes (CMCs; S2m–S2o Fig). The strong CAP/Integrin colocalization along the circular myofibrils in these cells suggests that CAP is localized in costameres (S2m–S2o Fig; see Discussion). In contrast to their embryonic precursors, larval and adult pericardial cells show a strong ubiquitous CAP distribution (Fig 2f and 2h; S2j, S2l, S2p and S2r Fig). Altogether our analysis shows that CAP is continuously expressed in cardiac cells throughout Drosophila development.
CAP mutants display abnormal cardioblast cell number and alignment
To investigate the role of CAP during heart morphogenesis we used the molecularly characterized P-element excision mutants CAP49e and CAP42b. Both mutants carry C-terminal deletions removing two of the three SH3-domain containing exons common to most annotated isoforms [51] except CAP-PP (see Fig 1a; inset). Western blot analysis and immunostaining indicates the absence of most CAP protein isoforms in these two alleles (Fig 1b). However, as mentioned, our antiserum is directed to the CAP C-terminus and therefore we cannot exclude the presence of isoform CAP-PP in these mutants (see Discussion). With this caveat in mind we studied the morphology of the DV of CAP mutants using anti-Mef2 antibodies, which stain all CB nuclei in combination with anti-Doc2+3 antibodies, which stain the ostial cell nuclei only (Fig 3).
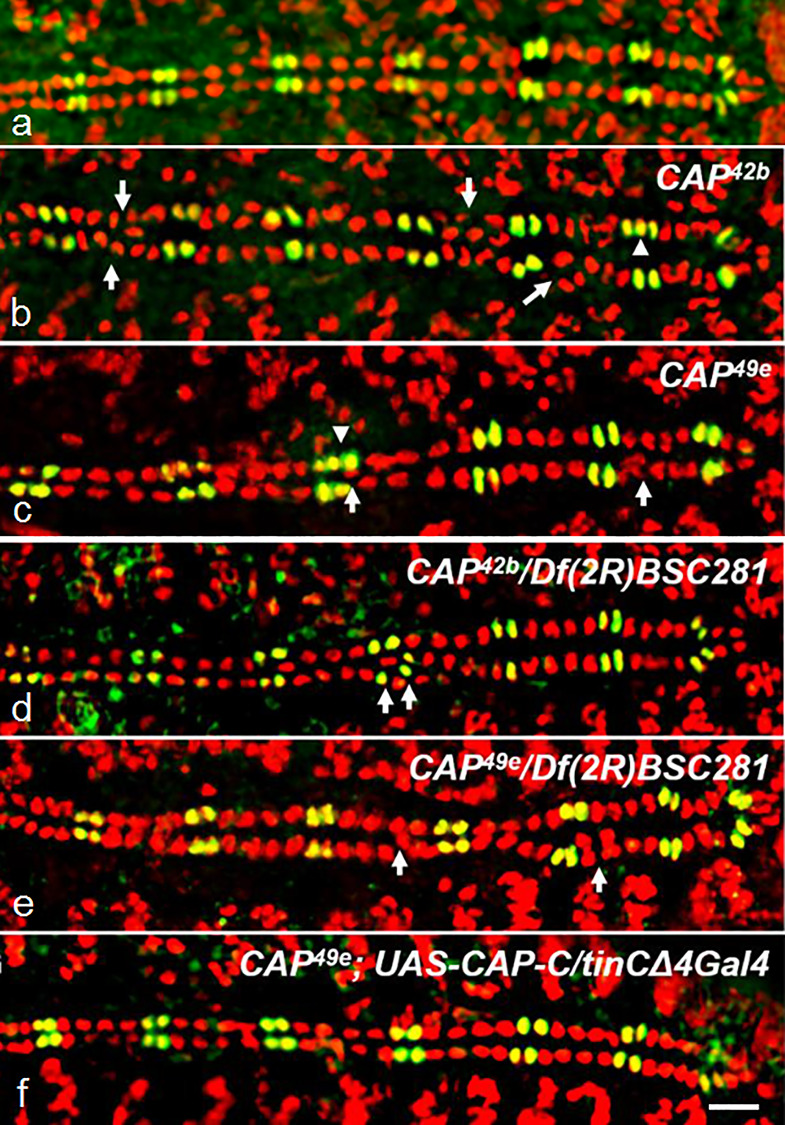
Fig 3. CAP mutants display abnormal cardioblast cell number and alignment.
Optical sections of stage 16 whole mount embryos costained for Doc2+3 (green) and Mef2 (red); anterior left; arrows: amplification and/or internalization of CBs; arrowheads: amplification of Doc+Mef2 positive CBs. (a) w1118 control showing the typical pattern of 2 Mef2+Doc CBs (yellow) and 4 Mef2-only CBs (red) per hemisegment; (b) CAP42b, (c) CAP49e, (d) CAP42b/Df(2R)BSC281, (e) CAP49e/Df(2R)BSC281: amplification and internalization of Mef2 only and Mef2+Doc CBs is observed. (f) CAP42b;UAS-CAP-C/tinCΔ4-GAL4 rescued embryo showing normal heart development (see Table 1); bar 10 μm.
In w1118 controls or heterozygous CAP mutants the CBs line up in two rows that pair exactly with their contralateral partners at the dorsal midline (Fig 3a). A segmental organization of the DV becomes apparent by molecular markers that is exactly matched between opposite CB rows. Wildtype type hemisegments contain two Doc/Svp-positive cells (stained for Mef2 and Doc) and four Tinman-positive cells (stained by anti-Mef2 only; Fig 3a). When homozygous, both CAP mutants show a DV phenotype characterized by irregularities in the alignment of CBs, blister formation, occlusion of CBs between the adjacent rows of cells and gain or loss of CBs. Instead of two contralateral CBs, often three cells are meeting at the dorsal midline, with one or more central CBs separating peripheral CBs (Fig 3b and 3c). In addition, CAP mutant DVs frequently contain additional CBs in several hemisegments and may have 6 to 8 Tinman-positive CBs per hemisegment (Fig 3b and 3c; Table 1). In a t-test of increased numbers of Mef2+ cells compared to w1118 the p-score for CAP42b is p = 7.00E-09 (***) and for CAP49e p = 2.98E-05 (***; S1 File). Occasionally we observed an increased number of Doc/Svp-positive CBs as well (Fig 3b and 3c). More rarely we also found hemisegments with less than 4 Tinman-positive CBs (arrows S3a Fig). The DV phenotype is already expressed during CB migration (S3a Fig).
Table 1. Quantification of the embryonic heart phenotype of CAP mutants, UAS-CAP RNAi, CAP mutant rescue and of CAP double heterozygous animals.
Hearts are categorized as defective if displaced or misaligned Mef2-stained CB nuclei where observed.
| Quantification of the CAP phenotype | |||
|---|---|---|---|
| Genotype | Heart defects | Mef expressing cardioblasts | Doc expressing cardioblasts |
| w1118 | 0% (n = 52) | 103.5 ± 1.2 (n = 22) | 28,2 ± 0.7 (n = 22) |
| CAP42b | 64% (n = 115) | 107.5 ± 1.2 (n = 27) | 28,4 ± 0.9 (n = 27) |
| CAP42b/Df(2R)BSC281 | 53% (n = 17) | 106.3 ± 1.9 (n = 10) | 28,4 ± 1.2 (n = 10) |
| CAP49e | 63% (n = 75) | 106.2 ± 2.6 (n = 29) | 28,6 ± 0.9 (n = 29) |
| CAP49e/Df(2R)BSC281 | 55% (n = 20) | 105.8 ± 2.9 (n = 8) | 28,9 ± 1.5 (n = 8) |
| Df(2R)BSC281 | 72% (n = 18) | n.d. | n.d. |
| UAS-CAP RNAi | 0% (n = 15) | 104.0 ± 0 (n = 15) | 28,2 ± 0.4 (n = 15) |
| UAS-CAP RNAi; tinC+D-GAL4 | 41% (n = 12) | 104.0 ± 0 (n = 12) | 28,0 ± 0.3 (n = 12) |
| UAS-CAP RNAi; twi+how-GAL4 | 64% (n = 14) | 104.0 ± 1.0 (n = 14) | 29,0 ± 1.0 (n = 14) |
| CAP49e;UAS-CAP-C/tinCΔ4Gal4 | 26% (n = 19) | 104.3 ± 1.2 (n = 8) | 28,3 ± 0.7 (n = 8) |
| CAP49e; rhea1 | 47% (n = 17) | 105.7 ± 2.0 (n = 6) | 28,2 ± 0.4 (n = 6) |
| CAP49e/scb2 | 48% (n = 29) | 104.2 ± 1.1 (n = 11) | 28,1 ± 0.3 (n = 11) |
The DV phenotype was observed in 63% of CAP49e and 64% of CAP42b embryos (Table 1). However, it is variable in expression. In the CAP mutant embryos with recognizable aberrations, the phenotype ranges from embryos in which only one segment is affected to clearly abnormal hearts which feature more irregularities. We did not further subdivide recognizable phenotypes according to their strength. On the other end, some of the subtle defects may not have been recognized and thus classified as wildtype-like (see Discussion). Both CAP mutations heterozygous over Df(2R)BSC281 show a similar DV phenotype (deviations in alignment and cell number) as the homozygous CAP mutants with a slightly reduced penetrance (Fig 3d and 3e; Table 1). However, in a t-test for the number of Mef positive CBs we see no significant difference between the CAP/Df(2R)BSC281 heterozygotes and the corresponding homozygous CAP mutants (p-scores for CAP42b/Df(2R)BSC281 are p = 0.1759 and for CAP49e/Df(2R)BSC281 p = 0.6491; S1 File). Homozygous Df(2R)BSC281 animals survive as stage 17 embryos and can unambiguously be identified by the absence of CAP or Nidogen (Ndg) staining (Ndg, is also removed by Df(2R)BSC281). 72% of Df(2R)BSC281 homozygotes (n = 18) show a DV alignment phenotype (Table 1). Probably the reduced penetrance in this case is caused by maternal effects of heterozygous female parents, however, this was not tested. To further corroborate the CAP DV phenotype we depleted CAP by RNAi knockdown. The UAS-CAP RNAi construct induced by the ubiquitous driver da-GAL4 strongly reduced CAP expression in adults (Fig 1b, lane 2) and induction of UAS-CAP RNAi generated a similar DV phenotype as described for CAP mutants. Following induction by tinC+D-GAL4 41% of the embryos show this phenotype. Induction by twi+how-Gal-4 produces a DV phenotype in 64% of randomly tested embryos. However, the number of Mef2+ CBs is closer to w1118 (S3 Fig; Table 1). The phenotypic effects of CAP49e and CAP42b are partially (~50%) suppressed by overexpressing the CAP-PC isoform via the CB -specific driver tinCΔ4-Gal-4 (Fig 3f, Table 1). Under rescue conditions (CAP49e;UAS-CAP-C/tinCΔ4Gal4) the number of Mef2+ CBs is closer to the w1118 than with CAP49e (p-score CAP49e;UAS-CAP-C/tinCΔ4Gal4 p = 0.0956 and CAP49e p = 2.9807E-05; S1 File).
CAP is required for heart lumen formation
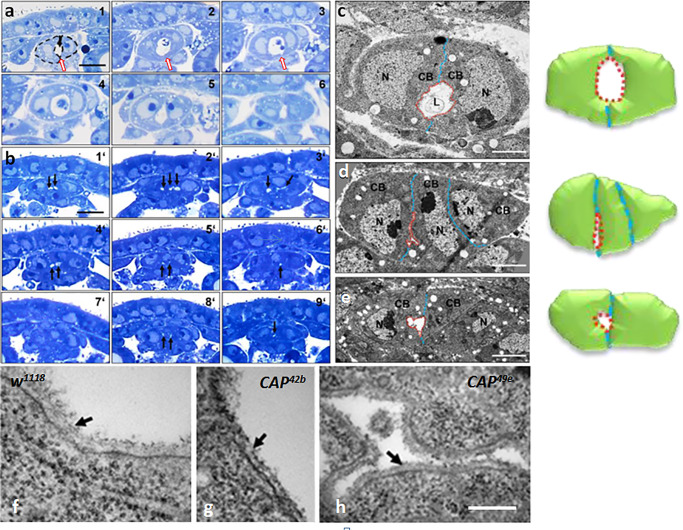
Next, we asked whether a normal heart lumen is formed in CAP mutants (Fig 4). To compare the ultrastructure of the CBs in normal and mutant DVs in terms of size, shape and lumen formation, we analyzed wild type and CAP mutant embryos on serial semi-thin cross sections by light microscopy (Fig 4a) and ultrathin sections by TEM microscopy (Fig 4b). In w1118 control animals the paired contralateral CBs are sealed at the dorsal and ventral junctional region and the intercellular space remaining between these connections establishes a DV lumen (Fig 4a, arrow). In contrast, about 60% of CAP mutant embryos reveal a defect in lumen formation. In both CAP49e and CAP42b mutants, the CBs meet each other normally at the dorsal midline, but form a slit-like lumen or several split lumina, both much smaller than in the control (Fig 4b; arrows). Serial semi-thin cross sections show that the lumen phenotype is consistent along the length of the aorta and the heart proper region (Fig 4a and 4b). The lumen defects are not exclusively connected to misplaced CBs since they also occur in areas with just two contralateral CBs (see Fig 4e). As evident in high resolution TEM, the typical crescent like shape of the CBs is distorted in CAP mutant animals. Especially the curved luminal domain seen in the controls seems to be disturbed. The cells have a more half moon-like appearance and enclose a slit-like lumen, as is emphasized in the corresponding cartoons (compare Fig 4c to 4d and 4e). Among the factors that affect CB shape and lumen formation are components of the basal membrane as well as molecules that guide CB polarization or organize membrane domains and their attachment to the cytoskeleton. According to our TEM data, CAP appears to be largely dispensable for basal membrane formation since an apparently normal basal membrane ensheaths the lumen of the DV in CAP mutants (Fig 4f–4h).
Fig 4. Heart lumen formation is disrupted in CAP mutants but basal membranes still form.
(a, b) Light microscopy of serial semi-thin sections through the dorsal vessel of stage 16 embryos stained by toluidine blue. (a) w1118 embryo (six different cut positions, a1-6). Two crescent shaped contralateral cardioblasts (dashed lines in A1) with light blue nuclei and darker blue cytoplasm encircle the white lumen. (b) In a CAP49e embryo (b1´-9´) the lumen (labeled by black arrows) is much smaller, often split in two or three minilumina or absent altogether (b7´); sometimes three CBs are involved; bar 10 μm. (c-e) Transmission EM of ultrathin sections of the heart region. (c) w1118 embryos; the luminal boundary between the two crescent shaped contralateral CBs is labeled red, their contact sites are labeled blue. (d) CAP49e; in the example shown three non-crescent formed CBs come together resulting in an extremely small lumen. (e) CAP42b; in the example shown only the left CB shows crescent formation. The right one does not, resulting in a significant reduction of the luminal space; bar: 2 μm. The cartoons to the right of c-e schematically illustrate the principal features shown of the TEM images. (f-h) Transmission EM of ultrathin sections of an embryo stage 16/17 showing CB luminal regions in controls and CAP mutants. (f) In the w1118 control the basal membrane at the luminal domain is marked (black arrow). An apparently normal basal membrane (arrows) is formed in (g) CAP42b and (h) CAP49e; bar in (h): 200 nm.
CAP is required for fine tuning the localization of Slit and Integrin at the luminal domain
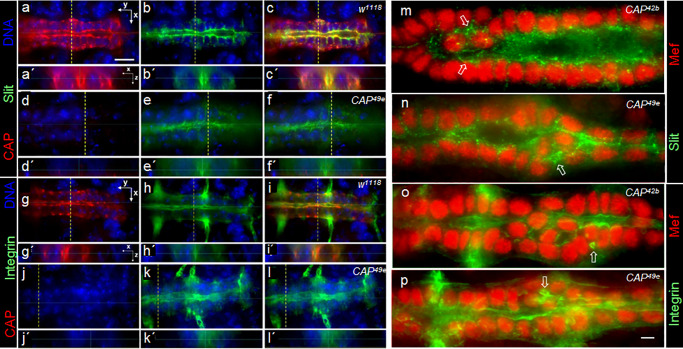
To find out whether CB polarization or the distribution of typical luminal marker proteins depends on CAP, we first analyzed the distribution of Slit and βPS1-Integrin in w1118 controls and CAP49e mutants (Fig 5). In controls the CAP protein is localized to the luminal, abluminal and lateral membranes (Fig 5a and 5g) and the luminal marker Slit is localized predominantly at the luminal domain of the CBs (Fig 5b). Luminal Slit is co-localized with CAP protein (Fig 5c). In CAP49e mutants no CAP protein can be detected (Fig 5d). Although Slit is still polarized and detected at the luminal domain of CAP49e CBs, its distribution is less distinct than in the control. As a consequence the boundaries towards the lumen become ill defined (Fig 5e and 5f). In Z-sections, the lumen, if visible at all, appears to be much smaller than in the controls (Fig 5e´ and 5f´). βPS1-Integrin in the control localizes to the luminal and abluminal membrane domains of CBs and is co-localized with CAP at these sites (Fig 5h and 5i). CAP49e mutants still display a polarized βPS1-Integrin localization, but similar to Slit, its distribution looks ill defined and the boundaries of the lumen are less well defined in comparison to the controls (Fig 5k and 5l). Co-staining of Slit and anti-βPS1-Integrin in the two CAP mutants with anti-Mef2 shows that the disruption of Slit and Integrin localization is stronger when associated with misaligned CBs. In such regions, Slit spreads into the basolateral space between peripheral CBs and around the central CBs as granular material that occasionally forms extra minilumina (Fig 5m and 5n). Similar spreading and minilumen formation associated with central CBs is also observed for ßPS1-Integrin (Fig 5o and 5p). This irregular organization often spreads along the heart tube from the misaligned CBs into immediate adjacent regions (Fig 5n and 5p).
Fig 5. CAP is required for establishment of a distinct luminal boundary.
Optical sections of the heart region of stage 16 whole mount embryos stained for CAP (red) and Slit (green) in (a-f) or CAP (red) and ßPS1-Integrin (green) in (g-l); DNA in blue. (a-c) w1118 control: CAP colocalizes with Slit at a clearly defined luminal boundary. (a´-c´) shows the heart lumen in Z-sections taken at the dashed lines in corresponding XY images. (d-f) CAP49e: The lumen is much smaller, with an ill-defined Slit stained boundary. (e´, f´) In Z-sections a lumen is hardly detectable. (g-i) w1118: CAP colocalizes with ßPS1-Integrin at the abluminal side and at a clearly defined luminal boundary. (j-l) CAP49e: The ßPS1-Integrin stained boundary towards the lumen looks ill defined. (k´, l´) A lumen is hardly detectable in Z-sections. anterior left; bar in (a): 10 μm. (m-p) Optical sections of the heart region stained by (m, n) Mef2 (red) and Slit antibodies (green) or (o, p) Mef2 (red) and ßPS1-Integrin antibodies (green). (m) CAP42b; note the ill-defined, granular Slit staining at the luminal cell boundary. On both sides of the central CBs minilumina are formed (arrows). (n) CAP49e: in the region of central CBs, Slit extensively spreads and encircles minilumina (arrow). (o) CAP42b: in the region of central CBs, the formation of a continuous lumen is obscured and ßPS1-Integrin is detected in extra luminal and miniluminal boundaries (arrow). (p) CAP49e: ill-defined ßPS1-Integrin distribution similar to (o); extra lumina form between peripheral and central CBs (arrow); bar in (p): 5 μm.
CAP genetically interacts with the integrin signaling components scab and rhea during heart development
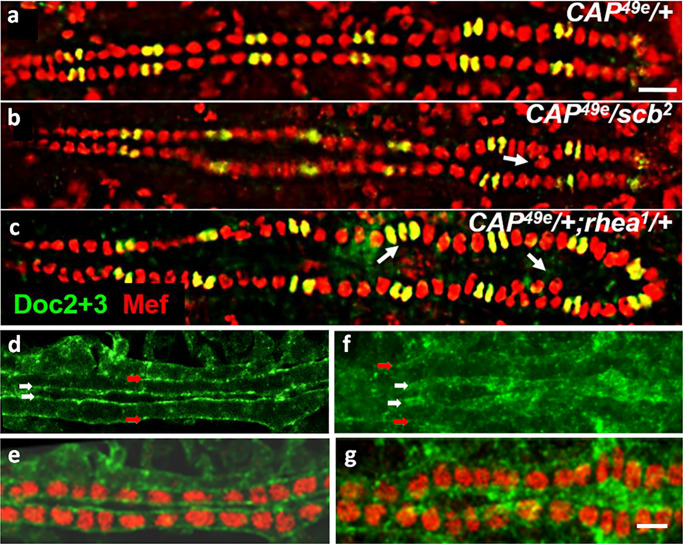
A recent study on CAP function implicates that CAP is acting downstream of Integrin signaling at muscle attachment sites and interacts with Vinculin [51]. Due to the co-localization of CAP and βPS1-Integrin in the Drosophila DV we asked whether CAP genetically interacts with Integrin signaling components during heart development (Fig 6). Previous studies reported that heterozygous scab and rhea mutants show normal DV development [18]. Heterozygous CAP49e mutants also have a normal DV morphology (Fig 6a). Interestingly, double heterozygous animals, with CAP49e either combined with scab2 or rhea1 show defects similar to those seen in CAP homozygous mutants (Fig 6). 48% of randomly selected scab2/CAP49e double heterozygous embryos show a DV phenotype (Fig 6b; Table 1). In the case of rhea1/CAP49e embryos 47% show a DV phenotype (Fig 6c; Table 1). This demonstrates that CAP genetically interacts with scab and rhea during heart development. As for Integrins, this observation is corroborated by our finding that Rhea/Talin localization is disrupted in a homozygous CAP mutant background. In control animals, Talin is localized at the luminal and abluminal periphery of CBs, demarcating a clearly defined cytoplasmic space (Fig 6d and 6e). In CAP49e mutant animals the luminal Talin staining is similarly ill defined as was observed for Slit and ßPS1-Integrin in CAP mutants (Fig 6f and 6g). On the abluminal domain Talin distribution is also altered and less distinct.
Fig 6. CAP genetically interacts with scab and rhea.
(a-c) CAP49e double heterozygote animals show the CAP phenotype. Optical sections of the heart region of stage 16 whole mount embryos costained with Mef2 (red) and Doc2+3 antisera (green). (a) CAP49e/+ control; (b) CAP49e/scab2 with an additional central cardioblast in the heart region (arrow); (c) CAP49e/+; rhea1/+ with 5 peripheral and 3 additional central cardioblasts and three ostial cells in the indicated heart regions. Anterior left; bar: 10 μm. (d-g) Optical sections of the dorsal vessel of stage 16 whole mount embryos costained with Mef2 (red) and Rhea/Talin (green) antibodies. (d) The w1118 control shows a distinct localization of Talin in the luminal and abluminal (open arrow) region with a clearly defined lumen (filled arrow). (e) same, costained with anti-Mef2. (f) In CAP49e the Talin distribution at the luminal boundary is less well defined than in the control (filled arrow) and in the abluminal domain of the dorsal vessel Talin staining is also less distinct (open arrows). (g) Same costained with anti-Mef2. There are no internal extra cardioblasts in this section. Anterior left; bar 10 μm.
CAP mutant larvae and adults show an impaired heart function
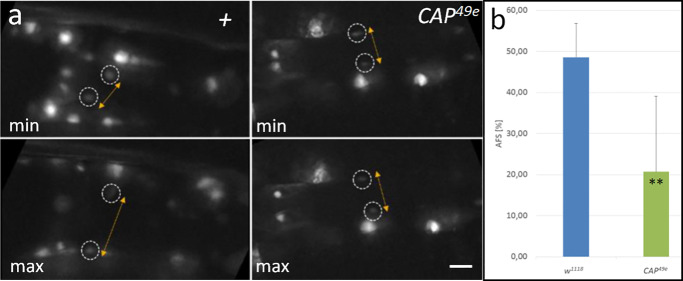
We argued, that the defects we have seen in CAP mutant embryos would affect the heart function later on. Therefore, we assayed the heart function in two different ways. First we checked for the heart contractility by performing live imaging of L3 larvae carrying the cardiac handC3.1-GFP reporter. Using this reporter, cycles of heart contraction and expansion can be observed and recorded in living larvae by the movement of CMC nuclei. We used the region immediately posterior to the internal valve to record the contraction cycles of the posterior heart as shown for the w1118 control (Fig 7a, S1 Movie). By measuring the distance of nuclei of contralateral CMCs from stills of the movies, the heart diameter at maximal extension, DCNmax (Fig 7a, top) and minimal extension DCNmin (Fig 7a, bottom) was determined and used to calculate the percentage of apparent fractional shortening (AFS% = [(DCNmax–DCNmin)/DCNmax] x 100%; for details see Materials and methods). In control animals (n = 3) we observe an average fractional shortening of 48.6 ± 8% (AFS ranging from 39.2 to 54.6%). By contrast, in CAP49e mutant larvae (n = 15) the CMC extension is significantly reduced (Fig 7b; S2 Movie), resulting in an AFS of 20.7 ± 18.3% for these animals (Fig 7c; ranging from 0.3 to 54.3%). Data analysis by t-test demonstrates that the difference between the controls and CAP mutants is statistically significant (p = 0.0044 (**); S1 File). Most of the mutant larvae display a clear reduction of the AFS with five out of 15 CAP49e larvae featuring less than 5% of the average of the controls and eight larvae feature intermediate AFS phenotypes (see S1 File for details). Three out of 15 CAP49e larvae (20%) exhibit an AFS in the range of the w1118 controls consistent with the incomplete penetrance of the phenotype in the DV of embryos.
Fig 7. CAP is required for function of larval hearts.
(a, b) Test for changes of apparent fractional shortening in L3 larvae. (a) hand-GFP controls (left panels, labeled +): stills of live imaging movies (see S1 Movie) are displayed and the distance of contralateral inner valve cell nuclei (encircled) at maximal extension (max) and minimal extension (min) is indicated by double arrows. (b) CAP49e (right panels): stills of live imaging movies (see S2 Movie). Due to reduced valve opening, phenotypically mutant CAP49e; hand-GFP larvae show only a small difference in nuclear distance between the two states; bar: 10 μm (c) The average and standard deviation of the apparent fractional shortening (AFS; see Materials and methods) for the hand-GFP control (n = 3) and hand-GFP; CAP49e mutants (n = 15) are displayed in the graph. The difference is significant (p = 0.0044¸**).
Our second assay examined the adult heart function after the heart remodeling at metamorphosis. For this assay we injected a fluorescent dye as a tracer into the abdomen of late pharate adults (Fig 8a; [66]). Due to active pumping by the heart, the dye accumulates with time in the head region of the flies, which can be observed as an increase of the fluorescence emission in a defined region of interest (ROI), as seen for a w1118 control in Fig 8b and 8c. By integration at defined time intervals and calculating the average pixel intensity, the accumulation of the tracer in the ROI is quantitated and displayed graphically for single flies as seen for three control flies (Fig 8e). The average and standard deviation for the group are plotted separately in Fig 8d. The same data is displayed for CAP42b and CAP49e mutants (Fig 8f–8i). Compared to the control at 180 sec, with an average pixel intensity of 61.3 (n = 3), the transport efficiency of CAP42b (average pixel intensity 19.8; n = 7) and CAP49e (average pixel intensity 18.8; n = 6) is strongly reduced. The differences between the w1118 control and the CAP mutants are statistically significant (p = 0.0072 for CAP42b (**) and p = 0.0039 (**) for CAP49e; see S1 File). However, considering single animals we observe differences in transport capability between tested animals. For instance, animals CAP42b #2 and #4 had almost lost activity, whereas CAP42b #3, #5, and #7 showed an intermediate phenotype, and CAP42b #6 behaved like wild type (Fig 8g). A similar variation in phenotypic strength is also seen with CAP49e (Fig 1i). This observation is in agreement with the variability of phenotypes observed in embryonic and larval CAP mutant hearts.
CAP mutants show local aberrations in the morphology of the adult heart tube
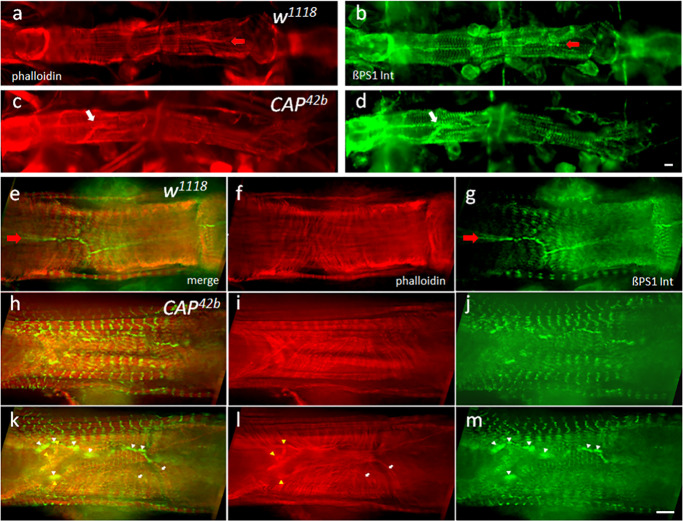
Normally CAP is colocalized with ßPS1-Integrin near Z-bands/costameres and cell contacts between contralateral cardiomyocytes (S2m–S2o Fig). Therefore we wondered whether the observed impaired heart function in CAP mutants results from changes in the adult heart structure, as a consequence of the disruption of Integrin adhesion complex (IAC; Fig 9). In contrast to the w1118 control, where the contralateral cardiomyocytes (CMCs) form a continuous spiral pattern of myofibrils stained for F-actin by phalloidin (Fig 9a) and a smooth line of cell attachment sites stained by Integrin (Fig 9b, open arrow), CAP mutant hearts often exhibit local structural aberrations with clusters of actin and Integrin elements (Fig 9c and 9d, white arrows). Optical sections of the w1118 control at higher magnification (Fig 9e–9g) reveal the regular arrangement CMCs with spiral myofibrils (Fig 9e and 9f) that line up at cell contact sites (Fig 9e and 9g) and are continued by matching myofibrils in the contralateral cell. A smoothly stained line of ßPS1-Integrin demarcates these cell contact sites between contralateral CMCs. Integrin is also found at the costameres of circular and longitudinal myofibrils (Fig 9g). In the CAP42b mutant (Fig 9h–9m) we do not observe obvious general defects in the Z-band architecture and costamere formation of myofibrils of CMCs. However, locally we detect aberrant, misaligned CMCs with a disrupted organization of βPS1-Integrin at cell contact sites (Fig 9j and 9m). In contrast to the controls, ßPS1-Integrin is found in such cells in clusters of increased Integrin accumulation (white arrowheads in Fig 9m), interrupted by sections that appear devoid of the protein. Interestingly, amorphous actin accumulations (Fig 9i and 9l) often form near such integrin clusters (yellow arrowheads in Fig 9l). Furthermore, CAP42b mutants feature aberrant CMCs in which the myofibrils fail to align in register and often do not attach at cell contact sites (Fig 9i and 9l). Instead they bend back into the cytoplasm or run for a while parallel to the long axis of the heart (white arrows in Fig 9l), or multiple myofibrils or amorphous actin fibers initiate from ßPS1-Integrin clusters in the mutant (yellow arrowheads in 9l). Together, the Integrin mislocalization correlates with a disturbed architecture of the circular myofibrils and a discontinuity in their arrangement across contralateral CMCs (Fig 9k). These local defects at cell contact sites could break the structural continuity between contralateral CMCs that is important to transmit coordinated contraction.
Fig 9. CAP mutants show local disruptions of adult heart morphology.
Stacks of optical sections of adult hearts costained by F-actin/phalloidin and ßPS1-Integrin antibodies. (a, b) low magnification view of a w1118 control heart stained for (a) F-actin by phalloidin (red) and (b) ßPS1-Integrin (green). (c, d) low magnification view of a CAP42b heart stained for (c) F-actin and (d) ßPS1-Integrin; note disruptions indicated by local accumulation of actin fibers and clusters of Integrin (white arrows in c and d, respectively); bar 8 μm. (e-m) higher magnification view of heart regions stained for F-actin (f, i, l) and anti-ßPS1-Integrin (g, j, m); (e, h, k) merge. (e-g) shows a projection of an image stack for the w1118 control with regularly arranged circular myofibrils (f; F-actin) that appear to smoothly continue across the line of contact sites of contralateral cells (open arrow in e and g; ßPS1-Integrin). (h-m) shows two consecutive projections of image stacks of a CAP42b heart with cytological defects: (i, l) Disorganized cell contacts indicated by unorganized local masses and clustering of actin fibers (yellow arrowheads in l). Myofibrils in this region often do not contact the cell contact site and/or fold back into the cell interior (white arrows in l); (j, m) discontinuous, patchy organization of ßPS1-Integrin at cell contact sites (white arrowheads in m). (k) Unorganized actin masses and Integrin patches often colocalize (white and yellow arrowheads). bar: 8 μm.
Discussion
As mentioned in the introduction, CAP has been reported to function in a variety of cell types. The data presented in our paper provide evidence that CAP plays a role in the robustness of Drosophila heart development and function. CAP mutant embryos show a DV phenotype characterized by local abnormalities in the alignment and numbers of CBs (often manifesting in luminal location of one or several CBs following closure of the dorsal vessel) as well as defects in lumen formation. Taken together with the protein localization data, this implies that CAP functions in processes near or within the membrane of cardiac cells. We suggest that the most important part of cardiac CAP function is related to the Integrin Adhesion Complex (IAC) for the following arguments. Apart from our observation that CAP mutants do not display breaks in the continuity of adjacent CBs, the DV phenotype is similar to that of homozygous slit, integrin, and talin mutants, double heterozygous mutant combinations among themselves or combinations with mutations in other Integrin-signaling components ([18, 27], and this work). This includes “clumps” [27] which we interpret as sites of misaligned CBs often forced into the DV lumen (see Fig 1H in [18]). In stage 15–16 embryos CAP colocalizes with ßPS1-Integrin (this work, Fig 5g–5i) and overlaps with Talin at the CB periphery [38]. Furthermore, we observed aberrant DV phenotypes, and hence a genetic interaction, in embryos double heterozygous for either CAP49e and the αPS3-Integrin-encoding scb2 or CAP49e and the Talin allele rhea1. The interaction with Scab may be cell non-autonomous. CAP/Vinculin interactions were documented in vitro [44] and more recently genetically in Drosophila muscle attachment sites in vivo [51]. We propose that CAP contributes to Integrin-actin cytoskeletal interaction to stably link the chain of CBs by cell adhesions and by adhesion to a common extracellular matrix (ECM). Local weakening or improper regulation of adhesiveness, possibly through a failure to transmit adhesion related forces or to coordinate organization of membrane microdomains, could result in mispositioning of migrating CBs relative to their neighbors.
The effect of the two CAP alleles used is not fully penetrant and is expressed as a series of intermediate states that differ in strength between individuals. Both alleles are small deletions removing two of the three SH3 domains common to all CAP isoforms except CAP-PP (Fig 1a inset; [51]). Bharadwaj and colleagues [51] suggest that both alleles likely result in a complete loss of CAP protein (amorphic type). We favor the idea that CAP42b and CAP49e are of non-amorphic type, consistent with the phenotypic effect observed. Although the deletions in both alleles affect the C-terminus of most isoforms, except CAP-PP, they leave their N-terminal portions intact, which, like CAP-PP, might have residual activity. In addition to CAP-PP, C-terminally truncated isoforms could be variably expressed in the CAP42b and CAP49e alleles and mediate a partial CAP function leading to variable mild, dosage-sensitive effects. A mild, dosage-sensitive antimorphic character would also explain why the rather subtle morphological phenotypes are not enhanced in hemizygous embryos with the CAP-deleting deficiency Df(2L)BSC281. Specific antisera are required to detect such putative truncated protein isoforms. Unfortunately, all available CAP antisera are directed to the conserved CAP C-terminus that is affected by the CAP excision mutants.
Currently, we can only speculate what causes the observed increase of CB numbers in CAP mutants. CB number has been shown to be positively and negatively regulated by the antagonistic actions of receptor tyrosine kinase (RTK/MAPK) and Notch signaling, respectively [69–73]. It is conceivable that CAP could influence signaling efficiency either through its proposed impact on cell adhesion or by interfering with the activity or distribution of certain signaling components (e.g. via its SH3 domains). Interestingly, the vertebrate CAP-homolog CAP/Ponsin negatively regulates MAPK signaling [44] by binding to components of the MAPK-pathway [74]. Alternatively, Integrin dependent adhesion, mediated by cell-cell or cell-extracellular matrix contacts, could serve as a mechanism to control cell division as has been reported in many mammalian cell types (reviewed in [75]). Changing the stability of the IAC by CAP mutation may interfere with such controls and trigger signals to initiate unscheduled cell division. This aspect of the cardiac CAP phenotype needs further investigation, although such investigations may be complicated by the subtleness of the phenotype and the complexity of CB number-determining signaling processes.
In the process of tube formation the CBs change their cell shape and cell surface properties. This requires a dramatic reorganization of their ECM, adhesion molecules and cytoskeletal organization [17]. CAP plays an important role in these processes since CAP mutants result in strong defects in lumen formation. CAP is most prominent at the luminal periphery of CBs where it colocalizes with ßPS1-Integrin, Talin and Slit, which are all essential for lumen formation. Integrin is the crucial player since it counteracts luminal spreading of Discs-large to the luminal domain and supports luminal location of Robo/Slit and the formation of a non-adhesive luminal ECM [18]. In CAP mutants the polarized Integrin distribution is maintained but the luminal domain of CBs stained for Integrin looks ill-defined and more expanded than in controls. This is similar to the aberrant broad distribution of ßPS1-Integrin at muscle attachment sites of somatic muscles in CAP mutant animals [51]. Interestingly, Talin is affected in a similar way. In contrast, CAP localization is not dramatically changed in zygotic rhea79 mutations; the observed changes were interpreted as secondary to the general perturbations in heart morphology by the rhea79 mutation [38]. In summary, this suggests a role for CAP in fine-tuning integrin-dependent adhesion that does not affect CB polarization and is upstream or independent of Talin-mediated functions at the luminal domain. Slit localization, though perturbed, is still observed at its original domains and an apparently normal basal membrane lining is still formed between contralateral CBs. Therefore, the adhesive properties of the luminal ECM might be unchanged, suggesting, that CAP mutants fail to establish a lumen not because of changes in adhesiveness, but owing to defects in the cytoskeletal architecture relevant for the formation of crescent shaped CBs. Due to defective Integrin assembly or its coupling to the cytoskeleton, CAP mutant CBs may be impaired in forming the functional connections between focal adhesions and the actin cytoskeleton that are required for the establishment of crescent cell shape. This is consistent with the observed role of CAP for the induction of stress fibers and focal adhesions in various mammalian cell types, and fits well with the proposed role for CAP and other mammalian SORBS family proteins in stiffness-sensing and force transmission [47, 76].
In the transition of the late embryo to the 1st instar larva and during pupation the dorsal vessel undergoes terminal differentiation to form the contractile larval and adult heart, respectively. This includes the establishment of robust links between the cytoskeleton and myofibrils and the cell contacts between contralateral CMCs. These contacts are structurally and functionally similar to intercalated discs in the vertebrate heart, which sense and transmit tension across the heart muscle [77]. Integrin and Talin play an important role in establishment and lifelong maintenance of CMC adhesion and their loss of function results in a severe retraction of myofibrils, losses of contacts between ipsilateral and contralateral myocytes and severely impaired function of the adult heart [6, 7]. CAP staining closely correlates with the Integrin localization at contact sites of contralateral CMCs and near Z-bands. Integrin is thought to contribute to the costamere structures that link the Z-bands to the ECM (reviewed in [35]), supported by proteins like ZASP that bind to and organize Z-bands and their Integrin connection [78]. The strong CAP-ßPS1-Integrin colocalization near Z-bands and in circular myofibrils suggests association of CAP with costameres, consistent with earlier observations made by Zhang and coworkers for the CAP homolog in mouse CMCs [48]. However, this has to be established by using more protein marker combinations at higher resolution.
Our physiological tests of CAP mutant larvae and adults uncovered animals with a reduced heart function. Apparent fractional shortening in larvae and pumping efficiency in adults were strongly affected in CAP mutants, although both parameters showed a similar incomplete penetrance as was observed for morphological defects in embryos. The physiological defects in heart function correlate with a locally defective morphology of adult CAP mutant hearts. A similar phenotype was found following Talin RNAi, when the knock down was limited to 2nd larval stage [7]. Also, hand-GAL4 driven knock down of Integrin-linked kinase ILK showed a similar phenotype [6]. Our data indicate that CAP is required for the maintenance and correct assembly of IACs at contact sites of contralateral CMCs and their connections to myofibrils. This function is in correspondence with its suggested role of CAP at muscle attachment sites of somatic muscles [51].
Overexpression of IAC components in Drosophila hearts results in premature aging and the age dependent increase in cardiac arrhythmias that could be prevented by reducing the gene dosage in heterozygous ilk and mys animals or by moderate cardiac specific knockdown of other IAC components [6]. Elimination of Talin1, one of two Talin genes in mice, also reduced the hypertrophic responses to increased cardiac load [79]. However, strong downregulation of Talin and other major components of the IAC cause serious defects, similar to dilated cardiomyopathy [6, 7]. Compared to other IAC proteins CAP depletion has rather mild effects on Drosophila heart structure and function. This and the incomplete penetrance of the phenotype classify CAP as a conserved modifier of IAC function that may provide a useful tool in the therapies of age dependent heart diseases.
Supporting information
Shown are lateral (a, c) or dorsal (b, d, e, f) views; anterior left. (a) Stage 11: first CAP expression in Garland cells (arrow). (b) Stage 13/14: CAP expression in the rows of cardioblasts (arrows; lower row out of focus). (c) Stage 14: cardioblasts (arrow) migrating dorsally. (d) Stage 15: the two rows of cardioblasts are approaching the dorsal midline (arrow). (e) Stage 16: the dorsally aligned cardioblast rows form the DV with a distinct posterior heart tube (arrow). (f) Stage 16 embryo hybridized with the 5´CAP sense probe: no signal; bar: 35 μm.
(PDF)
Colocalization of CAP using CAP antiserum combined with either Kettin-GFP expression, ZASP52-GFP expression or ßPS1-Integrin antibodies (a-f): Optical sections stained with CAP antiserum (a, d) with Kettin-GFP expression (b, e); merge (c, f). Focus to ventral longitudinal muscles (a-c) or to circular myofibrils (d-f). Bars: 5 μm. (g-l) Optical sections stained with CAP antiserum (g, j) with ZASP52-GFP expression (h, k); merge (i, l). Focus to circular myofibrils (g-i); heart at lower magnification focusing to ventral longitudinal muscles, to contact sites of contralateral CMCs (open arrows) and pericardial cells (j-l). Bars in (g-i): 5 μm, (j-l): 10 μm. (m-r) Optical sections stained with CAP antiserum (m, p), ßPS1-Integrin antibodies (n, q); merge (o, r). Focus to myofibrillar region and contact sites of contralateral CMCs (arrows; m-o); heart at lower magnification focusing to contact sites of contralateral CMCs (open arrows) and pericardial cells (p-r). Bars in (m-o): 6 μm, (p-r): 15 μm.
(PDF)
(a) Embryo with UAS-CAP RNAi driven by tinC+D-GAL4: One segment is disrupted by the loss of two Mef2+ CBs in the lower hemisegment (white arrows). Pairing with contralateral cells is delayed in the middle region. (b, c) Embryo with UAS-CAP RNAi driven by twi+how-GAL4 as inducer. (b) Note a group of 3 ostial cells in the heart region (3 arrows); (c) two central CBs in the posterior aorta (arrows). Bars: 10 μm.
(TIF)
Uncropped Western blot using adult whole fly extracts reacted with either rabbit anti-CAP (top) or anti-H2A (loading control below; see also Fig 1b). The type of extracts loaded is as follows: Lane 1: CAP-GFP, Lane 2: CAP RNAi (w1118; UAS-CAP RNAi; da-GAL4), Lane 3: white RNAi (w1118; UAS-white RNAi; da-GAL4), Lane 4: CAP42b [51], Lane 5: CAP49e [51], Lane 6: RNAi-control (w1118), Lane 7: protein molecular weight marker, Lane 8: CAP42b; hand-GFP, Lane 9: CAP49e; hand-GFP. The single 43 kD MWapp peptide stained in CAP mutant extracts (lanes 4 and 5; omitted in Fig 1b) is probably an artifact. It is not detected in CAP; hand-GFP mutant extracts (lanes 8, 9) or following RNAi (lane 2, 3). It is not or only marginally (overflow of adjacent lane?) present in control extracts (lanes 1, 6) and was not detected by Bharadwaj and coworkers (Fig 1D in [51]).
(DOCX)
Imaging was performed as described in materials and methods.
(RAR)
(RAR)
1. Evaluation of cardioblast numbers in DVs of different genotype: top line indicates the genotype of the embryos evaluated; second line lists the type of CBs evaluated and the third and following lines list the numbers of cells counted per embryo (compare Table 1). Line 32 displays the average for the CB numbers of the given genotype, followed by corresponding the standard deviation (S) in line 33 and the variance (S) in line 34. Line 35 lists the p-scores obtained by a 2-sided t-test with equal variance (type 2) in relation to w1118; line 36 the same in relation to CAP42b, line 37 in relation to CAP49e and line 38 in relation to CAP RNAi control without GAL4-driver (compare Table 1). 2. Evaluation of heart contraction cycle by live imaging. 2.1. lists the maximal (max) and minimal (min) distance of CMC nuclei close to the inner valve cells for hand-GFP;w1118 controls measured for each 10 cycles of heart beat per animal. The length determined for DCNmax (max) and DCNmin (min) are listed next to the avi file name and time point of the records in sec. Average and standard deviation for DCNmax and DCNmin were calculated. 2.2. Same is displayed for hand-GFP; CAP49e; 2.3. Evaluation of the data. Listed to the left are the values of AFS[%] calculated from the average DCNmax and DCNmin for each hand-GFP;CAP49e and hand-GFP; w1118 larva according to: [(DCNmax–DCNmin)/DCNmax] x 100%. From this the average and standard deviation for the whole group was determined for each genotype, listed in the table and used to calculate the variance. Then the p-scores were calculated using a 2-sided t-test with unequal variance (type 3). 3. Evaluation of heart transport activity of pharate adults determined by dye angiography. 3.1 Transport rate as average pixel intensity in ROI (w1118) lists the pixel values determined for three w1118 adults at different time points (for details see Materials and methods section) along with their average and standard deviation. 3.2 shows the same for 7 homozygous CAP42b animals and 3.3 the same for 6 CAP49e animals. These single fly values were graphically displayed in Fig 8e, 8g and 8i. 3.4 lists the average and the standard deviation of pixel intensities for each genotype that were graphically displayed in Fig 8d, 8f and 8h. From these data the p-scores to w1118 and significance were evaluated by a 2-sided t-test with unequal variance.
(PDF)
Acknowledgments
We thank Dr. L. Cooley (Yale University, USA), Dr. A. Paululat (University Osnabrück, Germany) and Dr. A. Kolodkin (John Hopkins University USA) for providing fly strains and Dr. H.T. Nguyen (University Erlangen-Nürnberg, Germany) for providing the Mef2 antiserum. Furthermore, we thank Dr. A. Paululat and Dr. M. Drechsler (University Osnabrück, Germany) for hosting J.J. in their lab and their support in dye angiography and we are grateful to Dr. B. Purfürst (Max Delbrück Center Berlin, Germany) for her support in TEM analysis and Dr. A. Seyfried (University Potsdam, Germany) for sharing unpublished information. Last but not least the many helpful discussions and suggestions made by the team of the Cytogenetics division, in particular by Dr. A. Klebes, Dr. A. Glotov and Dr. T. Zielke are also acknowledged.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was supported by funds of the MDC given to H.S. by the MDC (Max Delbrück Center, Berlin Buch; member of the Helmholtz foundation). J.J. obtained a PhD fellowship of the Transcard program by the MDC and a short term fellowship by the FAZIT foundation to finish her thesis (FAZIT Gemeinnützige Verlagsgesellschaft mbH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zaffran S, Frasch M. Early signals in cardiac development. Circulation research. 2002;91(6):457–69. 10.1161/01.res.0000034152.74523.a8 . [DOI] [PubMed] [Google Scholar]
- 2.Bier E, Bodmer R. Drosophila, an emerging model for cardiac disease. Gene. 2004;342(1):1–11. 10.1016/j.gene.2004.07.018 . [DOI] [PubMed] [Google Scholar]
- 3.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science (New York, NY. 2006;313(5795):1922–7. 10.1126/science.1132292 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad SM. Conserved signaling mechanisms in Drosophila heart development. Dev Dyn. 2017;246(9):641–56. 10.1002/dvdy.24530 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian L, Bodmer R. Probing the polygenic basis of cardiomyopathies in Drosophila. J Cell Mol Med. 2012;16(5):972–7. 10.1111/j.1582-4934.2012.01529.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimura M, Kumsta C, Kaushik G, Diop SB, Ding Y, Bisharat-Kernizan J, et al. A dual role for integrin-linked kinase and beta1-integrin in modulating cardiac aging. Aging Cell. 2014;13(3):431–40. 10.1111/acel.12193 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogatan S, Cevik D, Demidov V, Vanderploeg J, Panchbhaya A, Vitkin A, et al. Talin Is Required Continuously for Cardiomyocyte Remodeling during Heart Growth in Drosophila. PloS one. 2015;10(6):e0131238 10.1371/journal.pone.0131238 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu S, Han Z, Luo Y, Chen Y, Zeng Q, Wu X, et al. Molecular mechanisms of heart failure: insights from Drosophila. Heart Fail Rev. 2017;22(1):91–8. 10.1007/s10741-016-9590-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rugendorff A, Younossi-Hartenstein A, Hartenstein V. Embryonic origin and differentiation of the Drosophila heart. Roux Arch Dev Biol. 1994;203(5):266–80. 10.1007/BF00360522 . [DOI] [PubMed] [Google Scholar]
- 10.Bryantsev AL, Cripps RM. Cardiac gene regulatory networks in Drosophila. Biochim Biophys Acta. 2009;1789(4):343–53. Epub 2008/10/14. 10.1016/j.bbagrm.2008.09.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reim I, Frasch M. Genetic and genomic dissection of cardiogenesis in the Drosophila model. Pediatric cardiology. 2010;31(3):325–34. Epub 2009/12/25. 10.1007/s00246-009-9612-1 . [DOI] [PubMed] [Google Scholar]
- 12.Vogler G, Bodmer R. Cellular Mechanisms of Drosophila Heart Morphogenesis. J Cardiovasc Dev Dis. 2015;2(1):2–16. 10.3390/jcdd2010002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotstein B, Paululat A. On the Morphology of the Drosophila Heart. J Cardiovasc Dev Dis. 2016;3(2). 10.3390/jcdd3020015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes CJR, Jacobs JR. Dissecting the Role of the Extracellular Matrix in Heart Disease: Lessons from the Drosophila Genetic Model. Vet Sci. 2017;4(2). 10.3390/vetsci4020024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fremion F, Astier M, Zaffran S, Guillen A, Homburger V, Semeriva M. The heterotrimeric protein Go is required for the formation of heart epithelium in Drosophila. The Journal of cell biology. 1999;145(5):1063–76. 10.1083/jcb.145.5.1063 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haag TA, Haag NP, Lekven AC, Hartenstein V. The role of cell adhesion molecules in Drosophila heart morphogenesis: faint sausage, shotgun/DE-cadherin, and laminin A are required for discrete stages in heart development. Developmental biology. 1999;208(1):56–69. 10.1006/dbio.1998.9188 . [DOI] [PubMed] [Google Scholar]
- 17.Medioni C, Astier M, Zmojdzian M, Jagla K, Semeriva M. Genetic control of cell morphogenesis during Drosophila melanogaster cardiac tube formation. The Journal of cell biology. 2008;182(2):249–61. 10.1083/jcb.200801100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanderploeg J, Vazquez Paz LL, MacMullin A, Jacobs JR. Integrins are required for cardioblast polarisation in Drosophila. BMC Dev Biol. 2012;12:8 Epub 2012/02/23. 10.1186/1471-213X-12-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medioni C, Senatore S, Salmand PA, Lalevee N, Perrin L, Semeriva M. The fabulous destiny of the Drosophila heart. Current opinion in genetics & development. 2009;19(5):518–25. Epub 2009/09/01. 10.1016/j.gde.2009.07.004 . [DOI] [PubMed] [Google Scholar]
- 20.Rizki TM. The circulatory system and associated cells and tissues In: Ashburner M, Wright T. R. F., editor. The Genetics and Biology of Drosophila. New York, NY: Academic Press; 1978. p. 397–452. [Google Scholar]
- 21.Lehmacher C, Abeln B, Paululat A. The ultrastructure of Drosophila heart cells. Arthropod Struct Dev. 2012;41(5):459–74. Epub 2012/03/20. 10.1016/j.asd.2012.02.002 . [DOI] [PubMed] [Google Scholar]
- 22.Molina MR, Cripps RM. Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mechanisms of development. 2001;109(1):51–9. 10.1016/s0925-4773(01)00509-3 . [DOI] [PubMed] [Google Scholar]
- 23.Monier B, Astier M, Semeriva M, Perrin L. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development (Cambridge, England). 2005;132(23):5283–93. 10.1242/dev.02091 . [DOI] [PubMed] [Google Scholar]
- 24.Schaub C, März J, Reim I, Frasch M. Org-1-dependent lineage reprogramming generates the ventral longitudinal musculature of the Drosophila heart. Curr Biol. 2015;25(4):488–94. 10.1016/j.cub.2014.12.029 . [DOI] [PubMed] [Google Scholar]
- 25.Vogler G, Liu J, Iafe TW, Migh E, Mihaly J, Bodmer R. Cdc42 and formin activity control non-muscle myosin dynamics during Drosophila heart morphogenesis. The Journal of cell biology. 2014;206(7):909–22. 10.1083/jcb.201405075 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stark KA, Yee GH, Roote CE, Williams EL, Zusman S, Hynes RO. A novel alpha integrin subunit associates with betaPS and functions in tissue morphogenesis and movement during Drosophila development. Development (Cambridge, England). 1997;124(22):4583–94. . [DOI] [PubMed] [Google Scholar]
- 27.MacMullin A, Jacobs JR. Slit coordinates cardiac morphogenesis in Drosophila. Developmental biology. 2006;293(1):154–64. 10.1016/j.ydbio.2006.01.027 . [DOI] [PubMed] [Google Scholar]
- 28.Hollfelder D, Frasch M, Reim I. Distinct functions of the laminin beta LN domain and collagen IV during cardiac extracellular matrix formation and stabilization of alary muscle attachments revealed by EMS mutagenesis in Drosophila. BMC Dev Biol. 2014;14(1):26 Epub 2014/06/18. 10.1186/1471-213X-14-26 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian L, Liu J, Bodmer R. Slit and Robo control cardiac cell polarity and morphogenesis. Curr Biol. 2005;15(24):2271–8. 10.1016/j.cub.2005.10.037 . [DOI] [PubMed] [Google Scholar]
- 30.Santiago-Martinez E, Soplop NH, Kramer SG. Lateral positioning at the dorsal midline: Slit and Roundabout receptors guide Drosophila heart cell migration. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12441–6. 10.1073/pnas.0605284103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santiago-Martinez E, Soplop NH, Patel R, Kramer SG. Repulsion by Slit and Roundabout prevents Shotgun/E-cadherin-mediated cell adhesion during Drosophila heart tube lumen formation. The Journal of cell biology. 2008;182(2):241–8. 10.1083/jcb.200804120 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albrecht S, Altenhein B, Paululat A. The transmembrane receptor Uncoordinated5 (Unc5) is essential for heart lumen formation in Drosophila melanogaster. Developmental biology. 2011;350(1):89–100. Epub 2010/11/26. 10.1016/j.ydbio.2010.11.016 . [DOI] [PubMed] [Google Scholar]
- 33.Macabenta FD, Jensen AG, Cheng YS, Kramer JJ, Kramer SG. Frazzled/DCC facilitates cardiac cell outgrowth and attachment during Drosophila dorsal vessel formation. Developmental biology. 2013;380(2):233–42. Epub 2013/05/21. 10.1016/j.ydbio.2013.05.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das M, Ithychanda S, Qin J, Plow EF. Mechanisms of talin-dependent integrin signaling and crosstalk. Biochim Biophys Acta. 2014;1838(2):579–88. 10.1016/j.bbamem.2013.07.017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maartens AP, Brown NH. The many faces of cell adhesion during Drosophila muscle development. Developmental biology. 2015;401(1):62–74. 10.1016/j.ydbio.2014.12.038 . [DOI] [PubMed] [Google Scholar]
- 36.Maartens AP, Wellmann J, Wictome E, Klapholz B, Green H, Brown NH. Drosophila vinculin is more harmful when hyperactive than absent, and can circumvent integrin to form adhesion complexes. Journal of cell science. 2016;129(23):4354–65. 10.1242/jcs.189878 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphries JD, Chastney MR, Askari JA, Humphries MJ. Signal transduction via integrin adhesion complexes. Curr Opin Cell Biol. 2019;56:14–21. 10.1016/j.ceb.2018.08.004 . [DOI] [PubMed] [Google Scholar]
- 38.Vanderploeg J, Jacobs JR. Talin is required to position and expand the luminal domain of the Drosophila heart tube. Developmental biology. 2015;405(2):189–201. 10.1016/j.ydbio.2015.04.024 . [DOI] [PubMed] [Google Scholar]
- 39.Kioka N, Ueda K, Amachi T. Vinexin, CAP/ponsin, ArgBP2: a novel adaptor protein family regulating cytoskeletal organization and signal transduction. Cell Struct Funct. 2002;27(1):1–7. 10.1247/csf.27.1 . [DOI] [PubMed] [Google Scholar]
- 40.Kimura A, Baumann CA, Chiang SH, Saltiel AR. The sorbin homology domain: a motif for the targeting of proteins to lipid rafts. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(16):9098–103. 10.1073/pnas.151252898 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribon V, Printen JA, Hoffman NG, Kay BK, Saltiel AR. A novel, multifuntional c-Cbl binding protein in insulin receptor signaling in 3T3-L1 adipocytes. Molecular and cellular biology. 1998;18(2):872–9. 10.1128/mcb.18.2.872 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akamatsu M, Aota S, Suwa A, Ueda K, Amachi T, Yamada KM, et al. Vinexin forms a signaling complex with Sos and modulates epidermal growth factor-induced c-Jun N-terminal kinase/stress-activated protein kinase activities. The Journal of biological chemistry. 1999;274(50):35933–7. 10.1074/jbc.274.50.35933 . [DOI] [PubMed] [Google Scholar]
- 43.Mandai K, Nakanishi H, Satoh A, Takahashi K, Satoh K, Nishioka H, et al. Ponsin/SH3P12: an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. The Journal of cell biology. 1999;144(5):1001–17. 10.1083/jcb.144.5.1001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang M, Liu J, Cheng A, Deyoung SM, Chen X, Dold LH, et al. CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. The EMBO journal. 2006;25(22):5284–93. 10.1038/sj.emboj.7601406 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang SH, Baumann CA, Kanzaki M, Thurmond DC, Watson RT, Neudauer CL, et al. Insulin-stimulated GLUT4 translocation requires the CAP-dependent activation of TC10. Nature. 2001;410(6831):944–8. 10.1038/35073608 . [DOI] [PubMed] [Google Scholar]
- 46.Tosoni D, Cestra G. CAP (Cbl associated protein) regulates receptor-mediated endocytosis. FEBS Lett. 2009;583(2):293–300. 10.1016/j.febslet.2008.12.047 . [DOI] [PubMed] [Google Scholar]
- 47.Ribon V, Herrera R, Kay BK, Saltiel AR. A role for CAP, a novel, multifunctional Src homology 3 domain-containing protein in formation of actin stress fibers and focal adhesions. The Journal of biological chemistry. 1998;273(7):4073–80. 10.1074/jbc.273.7.4073 . [DOI] [PubMed] [Google Scholar]
- 48.Zhang M, Liu J, Cheng A, Deyoung SM, Saltiel AR. Identification of CAP as a costameric protein that interacts with filamin C. Mol Biol Cell. 2007;18(12):4731–40. 10.1091/mbc.e07-06-0628 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamazaki H, Nusse R. Identification of DCAP, a drosophila homolog of a glucose transport regulatory complex. Mechanisms of development. 2002;119(1):115–9. 10.1016/s0925-4773(02)00307-6 . [DOI] [PubMed] [Google Scholar]
- 50.Yamazaki H, Yanagawa S. Axin and the Axin/Arrow-binding protein DCAP mediate glucose-glycogen metabolism. Biochemical and biophysical research communications. 2003;304(2):229–35. 10.1016/s0006-291x(03)00582-5 . [DOI] [PubMed] [Google Scholar]
- 51.Bharadwaj R, Roy M, Ohyama T, Sivan-Loukianova E, Delannoy M, Lloyd TE, et al. Cbl-associated protein regulates assembly and function of two tension-sensing structures in Drosophila. Development (Cambridge, England). 2013;140(3):627–38. 10.1242/dev.085100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15050–5. 10.1073/pnas.261408198 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hudson AM, Petrella LN, Tanaka AJ, Cooley L. Mononuclear muscle cells in Drosophila ovaries revealed by GFP protein traps. Developmental biology. 2008;314(2):329–40. 10.1016/j.ydbio.2007.11.029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175(3):1505–31. 10.1534/genetics.106.065961 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sellin J, Albrecht S, Kölsch V, Paululat A. Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand. Gene Expr Patterns. 2006;6(4):360–75. 10.1016/j.modgep.2005.09.012 . [DOI] [PubMed] [Google Scholar]
- 56.Lo PC, Frasch M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mechanisms of development. 2001;104(1–2):49–60. Epub 2001/06/19. 10.1016/s0925-4773(01)00361-6 . [DOI] [PubMed] [Google Scholar]
- 57.Reim I, Frasch M. The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development (Cambridge, England). 2005;132(22):4911–25. 10.1242/dev.02077 . [DOI] [PubMed] [Google Scholar]
- 58.Baker R, Schubiger G. Autonomous and nonautonomous Notch functions for embryonic muscle and epidermis development in Drosophila. Development (Cambridge, England). 1996;122(2):617–26. Epub 1996/02/01. . [DOI] [PubMed] [Google Scholar]
- 59.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Cambridge, England). 1993;118(2):401–15. Epub 1993/06/01. . [DOI] [PubMed] [Google Scholar]
- 60.Lécuyer E, Parthasarathy N, Krause HM. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods in molecular biology (Clifton, NJ. 2008;420:289–302. 10.1007/978-1-59745-583-1_18 . [DOI] [PubMed] [Google Scholar]
- 61.Gorchakov AA, Kaltenhauser J, Eggert H, Saumweber H. [Construction of pMH, a convenient Escherichia coli protein expression vector]. Mol Biol (Mosk). 2004;38(4):713–6. . [PubMed] [Google Scholar]
- 62.Knirr S, Azpiazu N, Frasch M. The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development (Cambridge, England). 1999;126(20):4525–35. . [DOI] [PubMed] [Google Scholar]
- 63.Vogler G, Ocorr K. Visualizing the beating heart in Drosophila. J Vis Exp. 2009;(31). 10.3791/1425 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reim I, Lee HH, Frasch M. The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development (Cambridge, England). 2003;130(14):3187–204. 10.1242/dev.00548 . [DOI] [PubMed] [Google Scholar]
- 65.Soplop NH, Patel R, Kramer SG. Preparation of embryos for electron microscopy of the Drosophila embryonic heart tube. J Vis Exp. 2009;(34). 10.3791/1630 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drechsler M, Schmidt AC, Meyer H, Paululat A. The conserved ADAMTS-like protein lonely heart mediates matrix formation and cardiac tissue integrity. PLoS genetics. 2013;9(7):e1003616 Epub 2013/07/23. 10.1371/journal.pgen.1003616 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, et al. FlyBase 2.0: the next generation. Nucleic acids research. 2019;47(D1):D759–D65. 10.1093/nar/gky1003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.FlyBase FB2019_05, FBgn0033504 [Internet]. 2019 [cited Dec 03]. http://flybase.org/reports/FBgn0033504.
- 69.Hartenstein AY, Rugendorff A, Tepass U, Hartenstein V. The function of the neurogenic genes during epithelial development in the Drosophila embryo. Development (Cambridge, England). 1992;116(4):1203–20. . [DOI] [PubMed] [Google Scholar]
- 70.Carmena A, Buff E, Halfon MS, Gisselbrecht S, Jimenez F, Baylies MK, et al. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Developmental biology. 2002;244(2):226–42. Epub 2002/04/12. 10.1006/dbio.2002.0606 . [DOI] [PubMed] [Google Scholar]
- 71.Mandal L, Banerjee U, Hartenstein V. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nature genetics. 2004;36(9):1019–23. 10.1038/ng1404 . [DOI] [PubMed] [Google Scholar]
- 72.Grigorian M, Mandal L, Hakimi M, Ortiz I, Hartenstein V. The convergence of Notch and MAPK signaling specifies the blood progenitor fate in the Drosophila mesoderm. Developmental biology. 2011;353(1):105–18. Epub 2011/03/09. 10.1016/j.ydbio.2011.02.024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwarz B, Hollfelder D, Scharf K, Hartmann L, Reim I. Diversification of heart progenitor cells by EGF signaling and differential modulation of ETS protein activity. Elife. 2018;7:e32847 10.7554/eLife.32847 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roignot J, Soubeyran P. ArgBP2 and the SoHo family of adapter proteins in oncogenic diseases. Cell Adh Migr. 2009;3(2):167–70. 10.4161/cam.3.2.7576 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moreno-Layseca P, Streuli CH. Signalling pathways linking integrins with cell cycle progression. Matrix Biol. 2014;34:144–53. 10.1016/j.matbio.2013.10.011 . [DOI] [PubMed] [Google Scholar]
- 76.Ichikawa T, Kita M, Matsui TS, Nagasato AI, Araki T, Chiang SH, et al. Vinexin family (SORBS) proteins play different roles in stiffness-sensing and contractile force generation. Journal of cell science. 2017;130(20):3517–31. 10.1242/jcs.200691 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Israeli-Rosenberg S, Manso AM, Okada H, Ross RS. Integrins and integrin-associated proteins in the cardiac myocyte. Circulation research. 2014;114(3):572–86. 10.1161/CIRCRESAHA.114.301275 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jani K, Schock F. Zasp is required for the assembly of functional integrin adhesion sites. The Journal of cell biology. 2007;179(7):1583–97. 10.1083/jcb.200707045 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manso AM, Li R, Monkley SJ, Cruz NM, Ong S, Lao DH, et al. Talin1 has unique expression versus talin 2 in the heart and modifies the hypertrophic response to pressure overload. The Journal of biological chemistry. 2013;288(6):4252–64. 10.1074/jbc.M112.427484 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown are lateral (a, c) or dorsal (b, d, e, f) views; anterior left. (a) Stage 11: first CAP expression in Garland cells (arrow). (b) Stage 13/14: CAP expression in the rows of cardioblasts (arrows; lower row out of focus). (c) Stage 14: cardioblasts (arrow) migrating dorsally. (d) Stage 15: the two rows of cardioblasts are approaching the dorsal midline (arrow). (e) Stage 16: the dorsally aligned cardioblast rows form the DV with a distinct posterior heart tube (arrow). (f) Stage 16 embryo hybridized with the 5´CAP sense probe: no signal; bar: 35 μm.
(PDF)
Colocalization of CAP using CAP antiserum combined with either Kettin-GFP expression, ZASP52-GFP expression or ßPS1-Integrin antibodies (a-f): Optical sections stained with CAP antiserum (a, d) with Kettin-GFP expression (b, e); merge (c, f). Focus to ventral longitudinal muscles (a-c) or to circular myofibrils (d-f). Bars: 5 μm. (g-l) Optical sections stained with CAP antiserum (g, j) with ZASP52-GFP expression (h, k); merge (i, l). Focus to circular myofibrils (g-i); heart at lower magnification focusing to ventral longitudinal muscles, to contact sites of contralateral CMCs (open arrows) and pericardial cells (j-l). Bars in (g-i): 5 μm, (j-l): 10 μm. (m-r) Optical sections stained with CAP antiserum (m, p), ßPS1-Integrin antibodies (n, q); merge (o, r). Focus to myofibrillar region and contact sites of contralateral CMCs (arrows; m-o); heart at lower magnification focusing to contact sites of contralateral CMCs (open arrows) and pericardial cells (p-r). Bars in (m-o): 6 μm, (p-r): 15 μm.
(PDF)
(a) Embryo with UAS-CAP RNAi driven by tinC+D-GAL4: One segment is disrupted by the loss of two Mef2+ CBs in the lower hemisegment (white arrows). Pairing with contralateral cells is delayed in the middle region. (b, c) Embryo with UAS-CAP RNAi driven by twi+how-GAL4 as inducer. (b) Note a group of 3 ostial cells in the heart region (3 arrows); (c) two central CBs in the posterior aorta (arrows). Bars: 10 μm.
(TIF)
Uncropped Western blot using adult whole fly extracts reacted with either rabbit anti-CAP (top) or anti-H2A (loading control below; see also Fig 1b). The type of extracts loaded is as follows: Lane 1: CAP-GFP, Lane 2: CAP RNAi (w1118; UAS-CAP RNAi; da-GAL4), Lane 3: white RNAi (w1118; UAS-white RNAi; da-GAL4), Lane 4: CAP42b [51], Lane 5: CAP49e [51], Lane 6: RNAi-control (w1118), Lane 7: protein molecular weight marker, Lane 8: CAP42b; hand-GFP, Lane 9: CAP49e; hand-GFP. The single 43 kD MWapp peptide stained in CAP mutant extracts (lanes 4 and 5; omitted in Fig 1b) is probably an artifact. It is not detected in CAP; hand-GFP mutant extracts (lanes 8, 9) or following RNAi (lane 2, 3). It is not or only marginally (overflow of adjacent lane?) present in control extracts (lanes 1, 6) and was not detected by Bharadwaj and coworkers (Fig 1D in [51]).
(DOCX)
Imaging was performed as described in materials and methods.
(RAR)
(RAR)
1. Evaluation of cardioblast numbers in DVs of different genotype: top line indicates the genotype of the embryos evaluated; second line lists the type of CBs evaluated and the third and following lines list the numbers of cells counted per embryo (compare Table 1). Line 32 displays the average for the CB numbers of the given genotype, followed by corresponding the standard deviation (S) in line 33 and the variance (S) in line 34. Line 35 lists the p-scores obtained by a 2-sided t-test with equal variance (type 2) in relation to w1118; line 36 the same in relation to CAP42b, line 37 in relation to CAP49e and line 38 in relation to CAP RNAi control without GAL4-driver (compare Table 1). 2. Evaluation of heart contraction cycle by live imaging. 2.1. lists the maximal (max) and minimal (min) distance of CMC nuclei close to the inner valve cells for hand-GFP;w1118 controls measured for each 10 cycles of heart beat per animal. The length determined for DCNmax (max) and DCNmin (min) are listed next to the avi file name and time point of the records in sec. Average and standard deviation for DCNmax and DCNmin were calculated. 2.2. Same is displayed for hand-GFP; CAP49e; 2.3. Evaluation of the data. Listed to the left are the values of AFS[%] calculated from the average DCNmax and DCNmin for each hand-GFP;CAP49e and hand-GFP; w1118 larva according to: [(DCNmax–DCNmin)/DCNmax] x 100%. From this the average and standard deviation for the whole group was determined for each genotype, listed in the table and used to calculate the variance. Then the p-scores were calculated using a 2-sided t-test with unequal variance (type 3). 3. Evaluation of heart transport activity of pharate adults determined by dye angiography. 3.1 Transport rate as average pixel intensity in ROI (w1118) lists the pixel values determined for three w1118 adults at different time points (for details see Materials and methods section) along with their average and standard deviation. 3.2 shows the same for 7 homozygous CAP42b animals and 3.3 the same for 6 CAP49e animals. These single fly values were graphically displayed in Fig 8e, 8g and 8i. 3.4 lists the average and the standard deviation of pixel intensities for each genotype that were graphically displayed in Fig 8d, 8f and 8h. From these data the p-scores to w1118 and significance were evaluated by a 2-sided t-test with unequal variance.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.