TO THE EDITOR:
Sickle cell disease (SCD) and thalassemia are the commonest forms of inherited anemia, affecting around 300 000 births per annum worldwide. They result in progressive end-organ damage mainly due to recurrent vaso-occlusive crises (in SCD) and iron overload. Allogeneic hemopoietic stem cell (HSC) transplantation has been the only curative option but generally is only suitable for a minority of young patients with severe disease due to the transplant-related mortality.1-3 Recently, it has become possible to use gene therapy and genome editing to modify HSCs ex vivo for autologous transplantation. Gene-therapy approaches are based on using lentiviral vectors to insert exogenous copies of β-globin (HBB)4 or to upregulate γ-globin (HBG) production.5 Several genome-editing strategies have been developed including induction of fetal hemoglobin,6 reduction of α-globin expression,7 and correction of the HBB gene in situ using homology-directed repair.8
A number of lines of evidence suggest that hemopoietic stem/progenitor cell (HSPC) function might be perturbed in SCD and thalassemia. SCD is an inflammatory condition,9 which results in chronic leukocytosis, particularly neutrophilia and monocytosis, and this chronic inflammatory state may perturb HSPC function.10,11 SCD is also associated with persistently elevated plasma erythropoietin12 and transferrin receptor13 levels. Murine models show that high levels of erythropoietin reprogram the transcriptome of HSPCs, supporting the development of committed erythroid progenitors.14 This results in an erythroid-lineage bias during differentiation and decreased myeloid output. Interestingly, murine models also show that the increased reactive oxygen species in SCD perturb HSC function.15
To create a platform for these advanced cellular therapies, we set out to perform an in-depth characterization of the cellular and molecular composition of the CD34+ HSPC in children with SCD and thalassemia.
CD34+ cells were magnetic-activated cell sorting–purified (Miltenyi Biotec) from pretransplant bone marrow harvests from 5 children with homozygous SCD (SCD1-5); 3 children with transfusion-dependent β-thalassemia major (BT1-3), and 4 healthy controls (see supplemental Table 1, available on the Blood Web site, for clinical details). Adult bone marrow was used as the control because the majority of data supporting the use of CD34 for HSC dosing in transplantation is derived from adult patients.16,17 Further fluorescence-activated cell sorted CD34+ cells were analyzed using a combination of single-cell RNA sequencing (scRNA-seq) on the Chromium platform (10X Genomics), multiparameter flow cytometry, and colony-forming assays (supplemental Methods).
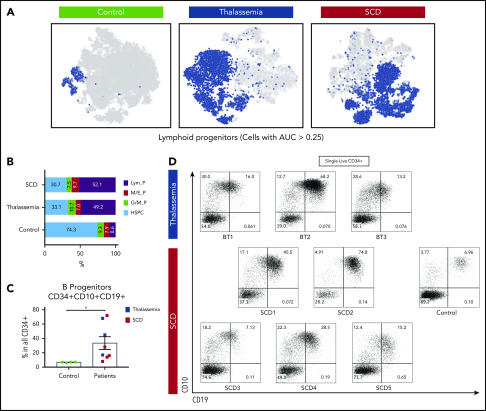
Analysis of scRNA-seq data using Seurat18 and AUcell19 (see supplemental Table 2 for quality control data) showed a marked expansion of CD34+ B-lymphoid progenitors (Figure 1A-B) in all 8 children with hemoglobinopathies compared with controls (supplemental Figures 1-6). Cells identified as B-lymphoid progenitors were enriched for expression of lymphoid genes20,21 including CD79A, CD79B, VPREB1, VPREB3, and EBF1 (supplemental Table 3; area under the curve 0.25 was used for all samples). As predicted, patients with SCD appear to show an increased proportion of cells with an erythroid gene–expression program although this did not reach statistical significance (Figure 1B; supplemental Table 3) (control, 7.86%; thalassemia, 6.97%; and SCD, 9.67%). No expansion of erythroid gene-expressing cells was seen in children with thalassemia, consistent with adherence to a rigorous regular transfusion protocol designed to suppress endogenous erythropoiesis in these patients (Figure 1A-B; supplemental Figures 1-5).
Figure 1.
Marked expansion of lymphoid-primed progenitors in the bone marrow CD34+ cells of children with SCDs and thalassemia. (A) Dimensionality reduction using t-distributed stochastic neighbor embedding (t-SNE) on pooled data from 4 controls (n = 29 086), 3 thalassemia (n = 11 034), and 5 SCD patients (n = 11 776); cells identified distinct cell populations while preserving intercluster relationships. Cell type was annotated based on the expression of reported marker genes using AUcell (see supplemental Table 3). (B) Components of cell types identified in single-cell sequencing data: lymphoid progenitors (Lym_P), megakaryocyte/erythroid progenitors (M/E_P), granulocyte/monocyte progenitors (G/M_P), and HSPCs. (C) Quantification of B-progenitor cells by flow cytometry in healthy donors and patients (SCD in red and thalassemia in blue). (D) Multiparameter flow cytometry plots showing the expansion of CD10+CD19+ cells in CD34+ cells derived from the bone marrow of patients with SCD and thalassemia. AUC, area under the curve.
Trajectory analysis was performed using an in-house R package,20 which allowed the expanded lymphoid progenitors to be subclassified into early B progenitors,22 expressing high levels of IL7R and IGHM, and late CD24-expressing B progenitors (supplemental Figure 5; supplemental Tables 1-3). These data were corroborated using multiparameter flow cytometry, which showed significant increases in the frequency of CD34+CD10+CD19+ cells in these patients (Figure 1C-D; supplemental Figures 7 and 8; supplemental Table 4). In some of the children (eg, BT2, SCD2), over 65% of all CD34+ cells were CD19+CD10+CD34+ B-progenitors (Figure 1C-D). There was significant variation between patients, which did not clearly correlate with age, genotype, or clinical factors (supplemental Figure 6B; supplemental Table 1). Previous studies show that children have higher numbers of CD10+CD19+ B cells in the bone marrow compared with adults but this rarely exceeds 20% of all cells23; however, there are no systematic studies regarding the proportion of CD34+ B-progenitor cells in the children. Our hypothesis is that a proinflammatory state in the children with hemoglobinopathies leads to these changes but it is possible that it relates to transfusion-related alloimmunization or it may be an age-related phenomenon.
scRNA-seq analysis also suggested a relative reduction in the frequency of multipotent HSPCs within the CD34+ compartment in all of the children with thalassemia and SCD (supplemental Figures 1 and 7-9). We carried out flow cytometric analysis, which confirmed a significant reduction in the proportion of CD38low\− cells as a proportion of total CD34+ cells in hemoglobinopathy patients compared with controls (Figure 2A-B; supplemental Figure 8). The proportion of HSCs (Lin−CD10−CD34+CD38low\−CD45RA−CD90+) and multipotent progenitors (MPPs) (Lin−CD10−CD34+CD38low\−CD45RA−CD90−) in the CD34+ compartment was also reduced in the hemoglobinopathy patients (Figure 2B), raising the possibility that such patients had a reduced frequency of editable long-term multipotent HSPCs.
Figure 2.
CD34-based HSPC estimation overestimates the HSC quantity in patients. (A) Flow plots show gating of Lin−CD10−CD34+CD38low\−CD45RA−CD90+ HSCs from a healthy donor and a child with SCD. (B) The proportion of phenotypic Lin−CD10−CD34+CD38low\−CD45RA−CD90+ HSCs in total CD34+ cells was reduced in patients (supplemental Figure 3B,D). This may be partially explained by the fact that she was the oldest and only postpubertal subject. (C) The proportion of phenotypic Lin−CD10−CD34+CD133+CD38low\−CD45RA−CD90+ HSCs in total CD133+ cells was not significantly different between patients and controls.
Despite this, when CD34+CD10+ lymphoid progenitors were excluded from the analysis by gating on the Lin−CD10−CD34+ compartment, the proportion of CD38low\− cells in the population was higher in SCD patients24 (median, 19.4% ± 6%) compared with controls (9.7% ± 0.55%) and thalassemia (7.7% ± 1.9%) (supplemental Figure 8B). Flow cytometric analysis of the Lin−CD10−CD34+CD38+ myeloid progenitor compartment showed no significant differences in granulocyte/monocyte progenitors, although common myeloid progenitors and megakaryocyte/erythroid progenitors were slightly decreased in the patients (supplemental Figure 8C,E). Colony-forming assays from sorted progenitor populations showed no significant differences between the patients and controls (supplemental Figure 9B), suggesting no functional differences between the progenitor populations.
The majority of gene-editing and gene-therapy preclinical and clinical trials use total CD34+ cells to determine cell dose for transplantation. The massively expanded B-progenitor compartment in the bone marrow of children with hemoglobinopathies will significantly underestimate the proportion of editable 3long-term multipotent HSPCs in autologous bone marrow samples. Consequently, we went on to investigate alternative strategies for quantifying HSPCs in these patients. First, we confirmed that the majority of CD34+ B-lymphoid progenitor cells were excluded when CD10 or CD19 were included in the lineage depletion cocktail (Figure 1B; supplemental Figure 8A). This was confirmed by paired chromium 10× analysis of lin−CD34+ cells with unselected CD34+ cells from a control sample (supplemental Figure 9A).
Our scRNA-seq data showed that CD133 (PROM1), a well-defined alternative to CD34 for quantification of HSPCs,21 is expressed by very few B-progenitors (supplemental Figure 10A-B). Addition of CD133 improved the reliability of flow cytometric measurement of HSC frequency. Gating on CD133+ cells removed CD34+ lymphoid progenitors and corrected the proportion of HSCs as quantified by CD133+Lin−CD10−CD34+CD38low\−CD45RA-CD90+ such that it was similar in both patients and controls (Figure 2C).
Our data show that the majority of children with SCD and BT have a marked increase in the proportion of B-progenitor cells, which do not have long-term repopulating capacity as measured by xenograft assays.25 As a result, an apparent rather than absolute reduction of HSCs is observed in the CD34+ compartment. CD34 is therefore a potentially unreliable marker as a surrogate for HSC frequency in these patients. Data from gene-therapy studies suggest that patients with hemoglobinopathies are often slow to engraft following transplantation with a gene-therapy product. In the first patient treated with lentiviral vector–mediated β-globin therapy, neutrophil engraftment occurred on day +384 and subsequent patients have also been slow to achieve neutrophil engraftment (median, 22 days; range, 17-29 days).16 We hypothesize that this may be in part due to the large numbers of CD34+ B-lymphoid cells causing the number of HSCs to be overestimated for a given dose of CD34+ cells per kilogram.
In conclusion, we have shown that, compared with healthy adults, a high percentage of CD34+ cells in bone marrow harvests taken from children with SCD and thalassemia are B-lymphoid progenitor cells. This reduces the proportion of other stem and progenitor populations within the CD34 compartment, making it unreliable to use CD34 alone as a biomarker for quantifying HSCs in these patients. This could potentially be circumvented at least partially either by the use of lineage depletion or the addition of CD133 to exclude B progenitor cells. Further work is required to delineate the mechanisms behind this phenomenon, particularly because there are limited studies defining how the CD34+ stem and progenitor compartment changes with age. This finding has important implications for transplantation, gene therapy, and genome editing for these diseases.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
This work was supported by a Medical Research Council (MRC) Discovery Award led by Doug Higgs (“Developing an initiative in stem cell editing for human genetic diseases” [MC_PC_15069]). J.O.J.D. and P.H. were also supported by an MRC Clinician Scientist Award (MRC Clinician Scientist Fellowship ref. MR/R008108) (J.O.J.D.).
Footnotes
The sequencing data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE133181).
Authorship
Contribution: P.H. designed, performed, and analyzed experiments, performed bioinformatics analyses, and wrote the manuscript; G.W., B.P., and A.R. contributed to experimental analyses; K.C. performed fluorescence-activated cell sorting; N.A. contributed to the single-cell experiment; Y.H. supplied patients’ details and assisted with obtaining consent; and J.O.J.D., C.N., J.d.l.F., N.R., S.M.W., S.T., and I.R. conceived and supervised the project, designed and analyzed experiments, and contributed to the writing of the manuscript.
Conflict-of-interest disclosure: J.O.J.D. is a cofounder of Nucleome Therapeutics Ltd to which he provides consultancy. The remaining authors declare no competing financial interests.
Correspondence: James O. J. Davies, MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, University of Oxford, John Radcliffe Hospital, Headington, Oxford, OX3 9DS, United Kingdom; e-mail: james.davies@ndcls.ox.ac.uk.
REFERENCES
- 1.Bolaños-Meade J, Brodsky RA. Blood and marrow transplantation for sickle cell disease: is less more? Blood Rev. 2014;28(6):243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talano JA, Cairo MS. Hematopoietic stem cell transplantation for sickle cell disease: state of the science. Eur J Haematol. 2015;94(5):391-399. [DOI] [PubMed] [Google Scholar]
- 3.Walters MC, De Castro LM, Sullivan KM, et al. Indications and results of HLA-identical sibling hematopoietic cell transplantation for sickle cell disease. Biol Blood Marrow Transplant. 2016;22(2):207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ribeil J-A, Hacein-Bey-Abina S, Payen E, et al. Gene therapy in a patient with sickle cell disease. N Engl J Med. 2017;376(9):848-855. [DOI] [PubMed] [Google Scholar]
- 5.Bauer DE, Kamran SC, Lessard S, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253-257.24115442 [Google Scholar]
- 6.Lin MI, Paik E, Mishra B, et al. CRISPR/Cas9 genome editing to treat sickle cell disease and B-thalassemia: re-creating genetic variants to upregulate fetal hemoglobin appear well-tolerated, effective and durable [abstract]. Blood. 2017;130(suppl 1):284. [Google Scholar]
- 7.Mettananda S, Fisher CA, Hay D, et al. Editing an α-globin enhancer in primary human hematopoietic stem cells as a treatment for β-thalassemia. Nat Commun. 2017;8(1):424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dever DP, Bak RO, Reinisch A, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539(7629):384-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoppe CC. Inflammatory mediators of endothelial injury in sickle cell disease. Hematol Oncol Clin North Am. 2014;28(2):265-286. [DOI] [PubMed] [Google Scholar]
- 10.Nogueira-Pedro A, Dias CC, Regina H, et al. Nitric oxide-induced murine hematopoietic stem cell fate involves multiple signaling proteins, gene expression, and redox modulation. Stem Cells. 2014;32(11):2949-2960. [DOI] [PubMed] [Google Scholar]
- 11.Pietras EM, Mirantes-Barbeito C, Fong S, et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol. 2016;18(6):607-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croizat H, Nagel RL. Circulating cytokines response and the level of erythropoiesis in sickle cell anemia. Am J Hematol. 1999;60(2):105-115. [DOI] [PubMed] [Google Scholar]
- 13.Lulla RR, Thompson AA, Liem RI. Elevated soluble transferrin receptor levels reflect increased erythropoietic drive rather than iron deficiency in pediatric sickle cell disease. Pediatr Blood Cancer. 2010;55(1):141-144. [DOI] [PubMed] [Google Scholar]
- 14.Grover A, Mancini E, Moore S, et al. Erythropoietin guides multipotent hematopoietic progenitor cells toward an erythroid fate. J Exp Med. 2014;211(2):181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javazon EH, Radhi M, Gangadharan B, Perry J, Archer DR. Hematopoietic stem cell function in a murine model of sickle cell disease. Anemia. 2012;2012:387385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanter J, Walters MC, Hsieh MM, et al. Interim results from a phase 1/2 clinical study of lentiglobin gene therapy for severe sickle cell disease [abstract]. Blood. 2016;128(22):1176. [Google Scholar]
- 17.Gorin NC. Bone marrow harvesting for HSCT In: Carreras E, Dufour C, Mohty M, Kröger N, eds. The EBMT Handbook: Hematopoietic Stem Cell Transplantation and Cellular Therapies, Cham, Switzerland: Springer International Publishing; 2019:109-115. [PubMed] [Google Scholar]
- 18.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Psaila B, Wang G, Meira AR, et al. Single-cell analyses reveal aberrant pathways for megakaryocyte-biased hematopoiesis in myelofibrosis and identify mutant clone-specific targets [published online ahead of print 20 May 2019]. bioRxiv. doi:10.1101/642819. [Google Scholar]
- 21.Hua P, Kronsteiner B, van der Garde M, et al. Single-cell assessment of transcriptome alterations induced by Scriptaid in early differentiated human haematopoietic progenitors during ex vivo expansion. Sci Rep. 2019;9(1):5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bendall SC, Davis KL, Amir AD, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157(3):714-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreoni C, Rigal D, Bonnard M, Bernaud J. Phenotypic analysis of a large number of normal human bone marrow sample by flow cytometry. Blut. 1990;61(5):271-277. [DOI] [PubMed] [Google Scholar]
- 24.Luck L, Zeng L, Hiti AL, Weinberg KI, Malik P. Human CD34(+) and CD34(+)CD38(-) hematopoietic progenitors in sickle cell disease differ phenotypically and functionally from normal and suggest distinct subpopulations that generate F cells. Exp Hematol. 2004;32(5):483-493. [DOI] [PubMed] [Google Scholar]
- 25.Byrne S, Elliott N, Rice S, et al. Discovery of a CD10-negative B-progenitor in human fetal life identifies unique ontogeny-related developmental programs. Blood. 2019;134(13):1059-1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.