Significance
G-quadruplex (G4) structures in functionally important genomic regions regulate multiple biological processes in cells. This study demonstrates a genome-wide correlation between the occurrence of endogenous oxidative base damage, activation of BER, and formation of G4 structures. Unbiased mapping of AP sites, APE1 binding, and G4 structures across the genome reveal a distinct distribution of AP sites and APE1 binding, predominantly in G4 sequences. Furthermore, APE1 plays an essential role in regulating the formation of G4 structures and G4-mediated gene expression. Our findings unravel a paradigm-shifting concept that endogenous oxidized DNA base damage and binding of APE1 in key regulatory regions in the genome have acquired a novel function in regulating the formation of G4 structures that controls multiple biological processes.
Keywords: endogenous damage, 8-oxoguanine, G-quadruplex structures, APE1, base excision repair
Abstract
Formation of G-quadruplex (G4) DNA structures in key regulatory regions in the genome has emerged as a secondary structure-based epigenetic mechanism for regulating multiple biological processes including transcription, replication, and telomere maintenance. G4 formation (folding), stabilization, and unfolding must be regulated to coordinate G4-mediated biological functions; however, how cells regulate the spatiotemporal formation of G4 structures in the genome is largely unknown. Here, we demonstrate that endogenous oxidized guanine bases in G4 sequences and the subsequent activation of the base excision repair (BER) pathway drive the spatiotemporal formation of G4 structures in the genome. Genome-wide mapping of occurrence of Apurinic/apyrimidinic (AP) site damage, binding of BER proteins, and G4 structures revealed that oxidized base-derived AP site damage and binding of OGG1 and APE1 are predominant in G4 sequences. Loss of APE1 abrogated G4 structure formation in cells, which suggests an essential role of APE1 in regulating the formation of G4 structures in the genome. Binding of APE1 to G4 sequences promotes G4 folding, and acetylation of APE1, which enhances its residence time, stabilizes G4 structures in cells. APE1 subsequently facilitates transcription factor loading to the promoter, providing mechanistic insight into the role of APE1 in G4-mediated gene expression. Our study unravels a role of endogenous oxidized DNA bases and APE1 in controlling the formation of higher-order DNA secondary structures to regulate transcription beyond its well-established role in safeguarding the genomic integrity.
G-quadruplexes (G4) are noncanonical tetrahelical nucleic acid structures that arise from the self-stacking of two or more guanine quartets into a planar array of four guanine residues coordinated through Hoogsteen hydrogen bonding (1, 2). Numerous in vitro biochemical and structural analyses have established that both DNA and RNA sequences with a G4 consensus motif (G≥3 N1–7 G≥3 N1–7 G≥3 N1–7 G≥3) can form G4 structures (3). The formation of G4 DNA structures in the genome has emerged as an epigenetic mechanism for regulating transcription, replication, translation, and telomere maintenance (4). Dysregulation of formation (folding) or unfolding of G4 has been implicated in transcriptional dysregulation, telomere defects, replication stress, genomic instability, and many human diseases, including cancer and neurodegeneration (5, 6). The recent genome-wide mapping of G4s in human cells using high-throughput chromatin immunoprecipitation sequencing (ChIP-Seq) was a breakthrough in establishing the regulatory role of G4s in vivo (4, 7). Mapping revealed that G4 structures are overrepresented in key regulatory regions like gene promoters, 5′ and 3′ untranslated regions, and telomeric regions, which indicates a positive selective pressure for retention of these motifs at specific sites in the genome to regulate multiple biological processes. In vitro, many G4 DNA structures are thermodynamically more stable than double-stranded DNA (8). However, G4 formation (folding), stabilization, and unfolding must be regulated to coordinate biological processes. Several proteins (DNA or RNA helicases) that resolve G4 structures have been characterized (9); however, the mechanisms underlying the spatiotemporal formation and stabilization of G4 structures in the genome are largely unknown.
Guanine (G) bases in potential G-quadruplex−forming sequences (PQS) have the lowest redox potential and are susceptible to the formation of 8-oxoguanine (8-oxoG), a prevalent endogenous oxidized DNA base damage in the genome (10, 11). The 8-oxoG DNA glycosylase (OGG1) initiates the repair of 8-oxoG via the evolutionarily conserved DNA base excision repair (BER) pathway. OGG1 removes the oxidized base to generate an Apurinic/apyrimidinic (AP/abasic) site (12), and human AP endonuclease 1 (APE1) is subsequently recruited to the AP sites for repair through the BER pathway (13). APE1, a key enzyme in the BER pathway, is a multifaceted protein involved in telomere maintenance, transcription regulation, and antibody genesis, which highlights the role of BER beyond its genome maintenance function (14–16). However, the molecular and functional connection of endogenous DNA damage and APE1 with the G4 structures remains largely unclear. Here, we conduct an unbiased genome-wide mapping of G4 structures, AP sites, and binding of APE1 and OGG1 proteins, and provide direct evidence that the occurrence of endogenous oxidized DNA base-derived AP site damage is nonrandom and is predominant in PQS sequences. High-resolution microscopy of G4 dynamics in cells revealed that oxidized DNA base damage and the associated repair complexes play a critical role in the spatiotemporal regulation of G4 structures. Loss of either stable AP site binding/coordination or acetylation of APE1 results in the abrogation of G4 structures and dysregulation of G4-mediated gene expression. Using in vitro biophysical and cell biological assays, we provide evidence that AP site damage and binding of APE1 to a PQS promotes G4 formation and facilitates transcription factor (TF) loading to regulate gene expression. Overall, our study comprehensively elucidates the role of endogenous oxidized DNA base damage and APE1 in controlling the formation of G4 structures in the genome to regulate transcription and other biological processes.
Results
Genome-wide Mapping of Endogenous AP Site Damage and Binding of Repair Proteins.
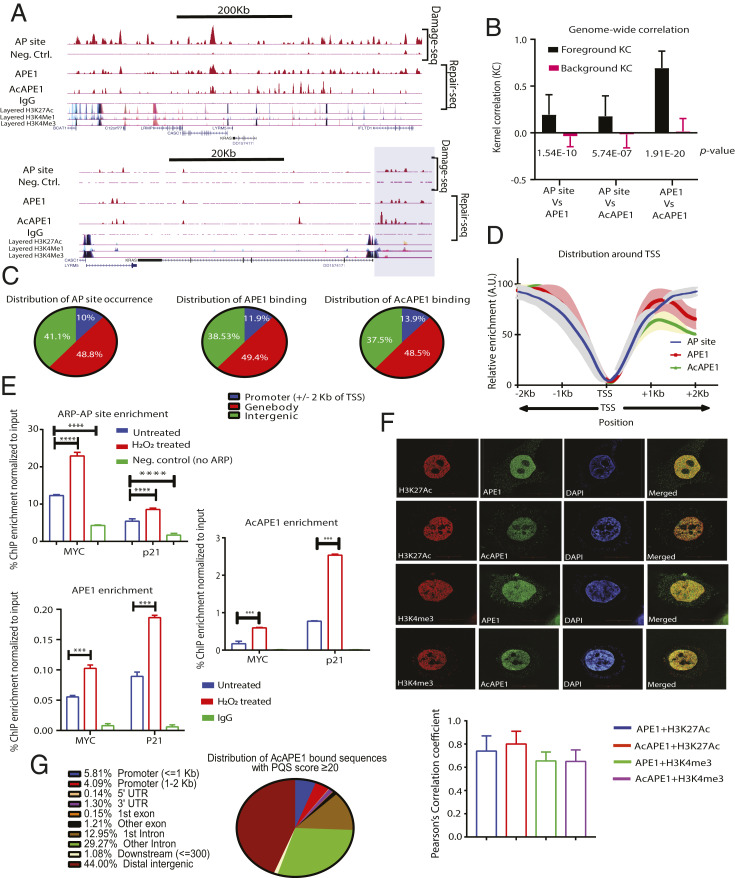
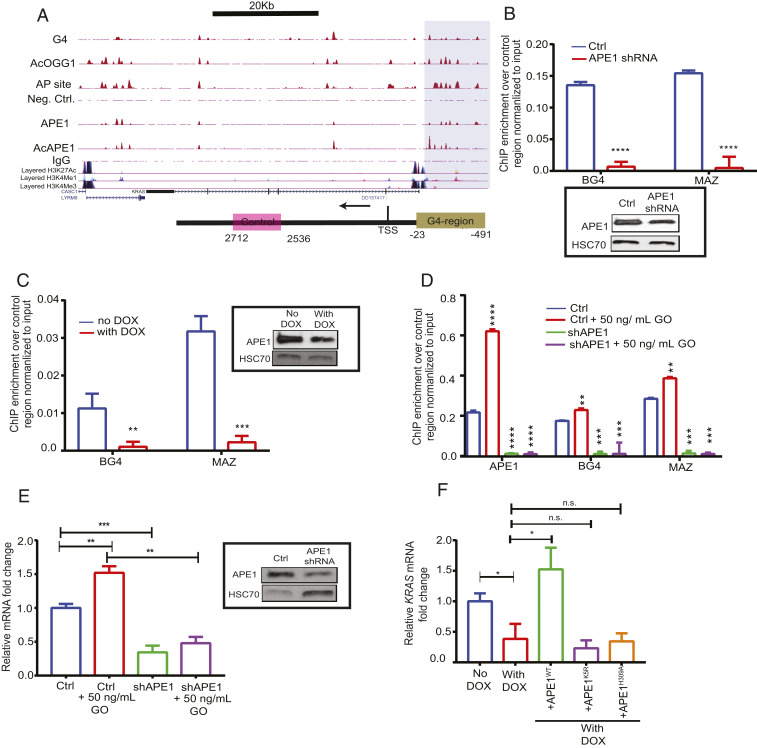
AP sites are the most prevalent type of endogenous DNA damage in cells and are generated spontaneously or after cleavage of modified bases, including oxidized G in the BER pathway (17). To determine the genome-wide occurrence of AP site damage, we developed a technique to map AP sites (AP-seq) in the genome, which employs a biotin-labeled aldehyde-reactive probe that reacts explicitly with an AP site in DNA (18, 19). By pull-down of biotin-tagged AP site DNA with streptavidin followed by sequencing, we could map AP sites in the genome at a ∼300-base pair (bp) resolution (see SI Appendix, Fig. S1 A, Left and Materials and Methods for details). We found a statistically significant (P < 0.001) occurrence of AP site damage in specific regions in the genome of lung adenocarcinoma A549 and colon cancer HCT116 cells (Fig. 1A and SI Appendix, Fig. S1B). We also mapped genome-wide occupancy of APE1, the primary enzyme responsible for repairing AP sites, and acetylated APE1 (AcAPE1), which is acetylated at AP site damage in chromatin (20), in A549 and HCT116 cells by ChIP-Seq analysis using α-APE1 and α-AcAPE1 antibodies (Abs) (repair-seq; Fig. 1A and SI Appendix, Fig. S1 A, Right). The disappearance of AcAPE1 peaks in HCT116 cells expressing APE1-specific short hairpin RNA (shRNA) compared to isogenic wild-type (WT) HCT116 cells confirms the specificity of APE1 binding (SI Appendix, Fig. S1B). To analyze the genome-wide correlation between AP sites (AP-seq), APE1, and AcAPE1, we used the StereoGene method to estimate Kernel correlation (KC), which provides correlation as a function of genomic position (21). Genome-wide correlation analysis between AP sites and APE1 binding revealed a statistically significant, positive KC (KC = 0.2, P = 10−10) and Spearman’s correlation (r = 0.84) (Fig. 1B and SI Appendix, Fig. S1C), which suggests that generation of AP sites and binding of APE1 and AcAPE1 are nonrandom and predominantly occur in specific regions in the genome. Analysis of endogenous AP site damage and AcAPE1 binding distribution relative to annotated genomic features revealed a predominant occurrence (∼60%) in gene bodies (exon and intron) and gene promoter regions (2,000 bp upstream [−] and downstream [+]; ±2 kilobases [kb]) of the transcription start site (TSS) (Fig. 1C and SI Appendix, Fig. S1D). Interestingly, we observed a significant fraction (∼10%) of AP site damage and APE1 and AcAPE1 binding in promoter regions, although promoters represent a very small portion of the human genome. Analysis of distribution across ±2 kb of TSS revealed that AP site damage and binding of APE1 and AcAPE1 are significantly enriched upstream and downstream of TSS (Fig. 1D). Furthermore, genome-wide karyogram mapping revealed clusters of APE1 and AcAPE1 in gene-rich regions (SI Appendix, Fig. S1 E and F). The AP-seq and AcAPE1 ChIP-Seq data were validated by real-time ChIP-PCR analysis of the MYC and P21 gene promoters with or without induction of oxidative DNA damage (Fig. 1E). Consistent with our ChIP-Seq data, superresolution (110 nm) structured illumination microscopy (SIM) revealed that AcAPE1 localizes to specific regions in the genome that bear the active enhancer marker H3K27ac and active promoter marker H3K4me3 (Fig. 1F). Collectively, these results demonstrate that endogenous AP site damage is not randomly distributed in the genome but is predominant at defined gene transcription regulatory regions.
Fig. 1.
Genome-wide mapping of endogenous AP site damage and binding of APE1. (A) (Top) A representative region of chromosome 12 showing the distribution of AP site damage, APE1, and AcAPE1 in A549 cells. (Bottom) A focused region of the KRAS gene. The shaded box highlights the KRAS promoter region cooccurrence of AP site damage, APE1, and AcAPE1. Cells not treated with aldehyde reactive probe (ARP) (Neg. Ctrl.) or immunoprecipitation with IgG were used as controls. Layered H3K27Ac, H3K4me1 (active enhancer), and H3K4me3 (active promoter) marks of seven cell lines from ENCODE are shown below. (B) Genome-wide positional correlations between AP sites and APE1 or AcAPE1 binding were estimated by KC values. The bar diagram shows the average KC for foreground and background distributions. The P values for the differences between these distributions are shown. (C) Analyses of distribution of endogenous AP site damage, APE1, and AcAPE1 binding relative to promoters, gene bodies, and intergenic regions in A549 cells. (D) Metaprofiles of the relative enrichment of APE1, AcOGG1, and AcAPE1 with respect to ±2,000 bp of the TSSs of ∼28,000 protein-coding genes. Shaded regions represent the 0.95 CIs. (E) Validation of APE1 and AcAPE1 ChIP-Seq and AP-seq data by real-time ChIP-PCR analysis with respect to P21 and MYC promoter regions in A549 cells. Hydrogen peroxide (H2O2) was used as a positive control for inducing oxidative DNA damages. Cells not treated with ARP were used as Neg. Ctrl. for AP site enrichment. IgG was used as a control for APE1 and AcAPE1 enrichment. An unpaired Student’s t test comparing untreated vs. Neg. Ctrl. and untreated vs. H2O2-treated samples was used to determine P values (****P < 0.0001, ***P < 0.001, error bars denote +SD). (F) Three-dimensional (3D) SIM images show colocalization of APE1 and AcAPE1 with H3K27Ac (active enhancer) and H3K4Me3 (active promoter) in A549 cells counterstained with DAPI. (Magnification: 63×.) Pearson coefficient was calculated (n = 10 cells) as an indicator of colocalization frequency. (G) Genome-wide distribution of AcAPE1-bound sequences with a PQS score of ≥20 in indicated annotated genomic regions in A549 cells.
Genome-wide Mapping of Oxidative Base Damage and G4 Structure Formation.
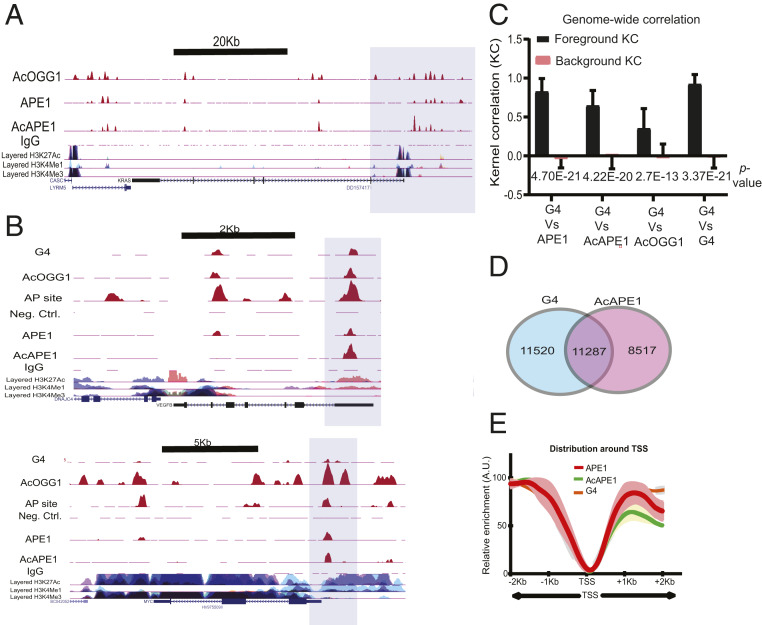
Analysis of AcAPE1-bound regions showed enrichment of G-rich sequences, which significantly overlapped with PQS in the genome. We used QGRS mapper (a web-based server for G-quadruplex prediction) (22) and found 72% of AcAPE1-bound sequences had a PQS score greater than 20 (PQS score 20 is considered significant). Upon overlap with genome-wide PQS map (23) (with cutoff of 20), AcAPE1-bound sequences were found to be predominantly enriched in promoter and gene regulatory regions (Fig. 1G). Since G bases in PQSs are more susceptible to form 8-oxoG, and repair of 8-oxoG is initiated by OGG1 (10), we mapped the genome-wide binding of OGG1 using our previously generated acetylated OGG1 (AcOGG1) antibody (24) (Fig. 2A). The AcOGG1 ChIP-Seq showed a significant positive Spearman’s correlation (r > 0.8) with APE1 and AcAPE1 binding (SI Appendix, Fig. S2A). To examine the association between AP site damage, binding of repair proteins, and G4 formation, we mapped the genome-wide occurrence of G4s using a G4-specific antibody (BG4) (25). As previously observed, G4 structures were enriched in promoters, 5′ untranslated regions (UTR), and gene bodies (Fig. 2B) (4). We observed genome-wide statistically significant, positive KCs and Spearman’s correlations between G4 and APE1, AcAPE1, or AcOGG1 (Fig. 2C and SI Appendix, Fig. S2A), which indicates a genome-wide relationship between oxidized base damage, binding of OGG1 and APE1, and G4 structures (Fig. 2 B and C).
Fig. 2.
Genome-wide mapping of binding of OGG1 and APE1 repair proteins and G4 structures. (A) The shaded region highlights the overlap of AcOGG1, APE1, and AcAPE1 binding in the KRAS promoter region in A549 cells. (B) Cooccurrence of G4 structures, AP site damage, and binding of AcOGG1, APE1, and AcAPE1 in VEGF and MYC promoter regions (shaded region) which were previously shown to form G4. (C) Genome-wide positional correlations between G4 structures and APE1, AcAPE1, and AcOGG1 were estimated by KC. The bar graph shows the average KC for foreground and background distributions. P values for the difference between foreground and background KC are shown. (D) Venn diagram represents overlapping G4 and AcAPE1 enriched sequences that have a PQS score of ≥20. (E) Metaprofiles of the relative enrichment of G4, APE1, and AcAPE1 with respect to ±2,000 bp of the TSSs of ∼28,000 protein-coding genes in A549 cells. Shaded regions represent the 0.95 CIs.
Alignment of genomic sequences (434,272) with a PQS score of 20 and our G4 ChIP-Seq reads revealed that 5.25% (22,807) of PQSs formed G4s in A549 cells (23), and 49.5% of G4 enriched sequences overlap with AcAPE1-bound sequences (Fig. 2D and Dataset S1). Intriguingly, G4 distribution (with 95% confidence level) across ±2 kb of TSS showed similar patterns to APE1 and AcAPE1 (Fig. 2E). Random shuffling of G4, APE1, or AcAPE1 reads shows no distinct pattern across ±2 kb of TSS (SI Appendix, Fig. S2B).
Many have reasoned that gene promoters experience negative superhelicity during transcription, which converts the duplex DNA to a G-quadruplex on the purine-rich strand (26). We inhibited either transcription initiation or elongation with Triptolide and Actinomycin D, respectively (27), and performed APE1, AcAPE1, AcOGG1, and G4 ChIP-Seq. Interestingly, we found no significant alteration of the APE1 or G4 ChIP-Seq profiles after inhibition of transcription (SI Appendix, Fig. S2C). Furthermore, MYC and P21 promoter-directed real-time ChIP-PCR after Actinomycin D or Triptolide treatment showed no change in APE1 binding or G4 formation (SI Appendix, Fig. S2 D–G). These observations indicate that the occurrence of AP sites or G4 formation is not an indirect consequence of transcription. However, we found a significant reduction of enrichment of AcAPE1 and AcOGG1, which indicates that acetylation of these proteins is dependent on active transcription (SI Appendix, Fig. S2 C–G). Together, our data reveal a genome-wide correlation between AP site damage, binding of AcOGG1 and APE1, and formation of G4 structures, which suggests a link between endogenous damage, activation of BER, and G4 formation in vivo.
Stable AP Site Binding and Acetylation of APE1 Play a Crucial Role in the Formation of G4 Structures in Cells.
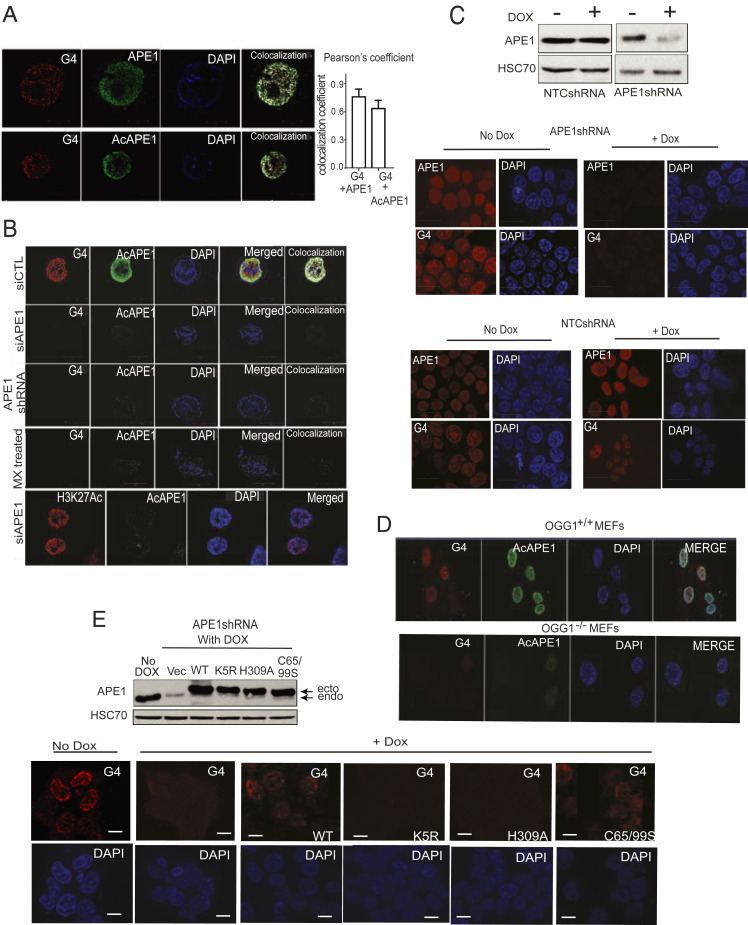
G4 structures were visualized in human cells by confocal and SIM microscopy using the G4 DNA-specific antibody 1H6. We found the formation of G4 foci in the nucleus of several cell lines, including primary lung fibroblast IMR90, A549, HCT116, and mouse embryonic fibroblasts (MEFs) (Fig. 3 A and B and SI Appendix, Fig. S3A). G4 foci detected by 1H6 was sensitive to DNase treatment, but not RNase A treatment (SI Appendix, Fig. S3B), which confirmed the specificity of 1H6 to DNA G4 structures. We observed a high colocalization frequency (r = 0.78) of G4 structures and APE1 or AcAPE1 staining (Fig. 3A). Down-regulation of APE1 levels by transient expression of APE1 small interfering RNA (siRNA) or stable expression of APE1 shRNA in HCT116 cells abolished the formation of G4 foci compared to isogenic control HCT116 cells, which indicates the critical importance of APE1 in regulating the formation or stabilization of G4 structures in the genome (Fig. 3B and SI Appendix, Fig. S3C). We further confirmed this finding by expressing two independent APE1 shRNA under a Doxycycline (Dox)-inducible promoter (Fig. 3C) and observed a significant reduction in G4 foci formation upon Dox treatment (Fig. 3C and SI Appendix, Fig. S3 D and E). As a negative control, we stained for histone H3K27Ac and c-Jun in APE1 down-regulated cells and observed no change in staining, which further supports that APE1 down-regulation specifically reduced G4 staining (Fig. 3B and SI Appendix, Fig. S4A). To determine whether binding of APE1 to AP site damage is essential for the formation of G4 structures, we treated HCT116 cells with methoxyamine (MX), a small molecule which binds to AP sites and competitively inhibits binding of APE1 to AP sites both in vitro and in cells. We found that pretreatment of cells with MX significantly inhibited the formation of G4 structures (Fig. 3B). Furthermore, we assessed whether OGG1, which initiates the repair of 8-oxoG by generating an AP site, plays a role in the formation of G4 by comparing G4 staining between WT (OGG1+/+) MEF and OGG1−/− MEF cells (28). Formation of G4 foci was significantly reduced in OGG1−/− MEF (Fig. 3D). We also tested whether deletion of the ECD gene, a non-BER protein, in adeno-Cre expressing ECDfl/fl MEFs (29) affected G4 staining, and found deletion did not alter G4 foci formation (SI Appendix, Fig. S4 B–D). Altogether, these data demonstrate that the absence of either OGG1 or APE1 abolished the formation of most genomic G4 in cells, which suggests their critical role in G4 formation. Interestingly, although down-regulation of APE1 nearly abolished G4 staining with the 1H6 antibody, ChIP-Seq data revealed APE1 bound to only 50% of G4 enriched sequences (Fig. 2D). To resolve the apparent disagreement between the two results, we conducted G4 ChIP-Seq in APE1 down-regulated cells to determine the differential G4 peak formation in the absence of APE1. Our analysis revealed a significant reduction of G4 enriched peaks upon APE1 knockdown in A549 cells (Fig. 4A). Reduced BG4 ChIP-Seq peaks between control and APE1 knockdown cells were determined using THOR peak caller based on log2 fold change of 0.5, adjusted P value of 0.05 (30). Distribution of reduced G4 peaks relative to annotated genomic features revealed a predominant occurrence in the promoter and gene body regions in the genome (SI Appendix, Fig. S5A). Interestingly, there were some G4 peaks that rather increased in some regions of the genome in the absence of APE1, which suggests that APE1 regulates and stabilizes the formation of most, but not all G4 structures in the genome.
Fig. 3.
DNA repair function and acetylation of APE1 play a crucial in the formation of stable G4 structures in the genome. (A) The 3D SIM images of A549 cells immunostained with α-1H6, α-APE1, and α-AcAPE1 Abs and counterstained with DAPI. (Magnification: 63×.) Pearson coefficient was calculated (n = 10 cells) as an indicator of colocalization frequency. (B) HCT116 cells transiently transfected with control siRNA (siCTL) or APE1 siRNA (siAPE1), HCT116 cells constitutively expressing APE1 shRNA, and HCT116 cells treated with MX (50 mM) for 30 min were immunostained with α-1H6 and α-AcAPE1. HCT116siAPE1 cells were immunostained with α-H3K27Ac and α-AcAPE1 as a control. (Magnification: 63×.) (C) HEK-293T cells expressing nontargeting control shRNA (NTCshRNA) or APE1-specific shRNA (APE1shRNA) under a Dox-inducible promoter (Bottom) were treated (Left) without or (Right) with Dox (2 µg/mL) for 3 days and immunostained with α-1H6 and α-AcAPE1. (Top) APE1 level in these cell extracts was examined by Western blot analysis with α-APE1 and α-HSC70 (loading control). (Scale bars: 10 uM; magnification: 40×.) (D) MEF cells established from OGG1-null mice (OGG1−/− MEF) and WT mice (OGG1+/+ MEF) were immunostained with α-1H6 and visualized by confocal microscopy. (E) (Bottom) HEK-293T cells expressing APE1shRNA were treated with Dox (2 µg/mL) for 2 days and then transfected with FLAG-tagged WT, acetylation-defective K5R, repair-defective H309A, or redox-defective C65,99S APE1 plasmid constructs for 24 h. Cells were then immunostained with α-1H6 and α-AcAPE1 and visualized by confocal microscopy. (Top) APE1 levels in these cell extracts were examined by Western blot analysis with α-APE1 and α-HSC70 (loading control). (Scale bars: 10 uM; magnification: 40×.)
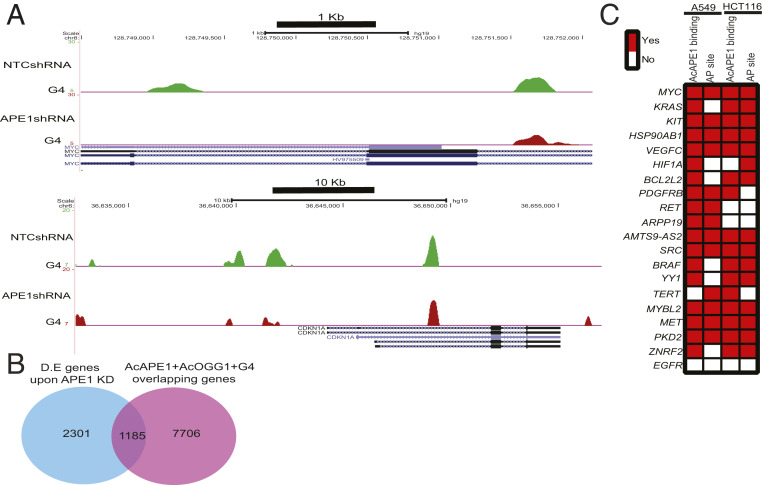
Fig. 4.
Genome-wide G4 regulation by APE1. (A) A549 cells expressing NTCshRNA or APE1shRNA under a Dox-inducible promoter were treated with Dox (2 µg/mL) for 3 days, and ChIP was performed with α-BG4. Representative genome browser views of MYC and P21 genes showing G4 profiles in control and APE1 KD A549 cells. (B) RNA sequencing analysis of RNA isolated from A549 cells expressing NTCshRNA or APE1shRNA treated with Dox (2 µg/mL) for 3 days. Overlapping between differentially expressed (D.E.) genes and genes with overlapping enrichment of AcOGG1, AcAPE1, and G4. The data are represented as a Venn diagram. (C) The occurrence of AP sites and AcAPE1 binding in G4 containing oncogene promoters.
APE1 is a multifunctional protein with a DNA repair function (AP endonuclease activity) and a transcription regulatory Ref-1 function (31). Furthermore, several of our previous studies had shown that APE1 is acetylated at multiple Lys (Lys6, 7, 27, 31, and 32) residues after binding to AP site damage in chromatin, and AcAPE1 modulates the expression of genes via functioning as a transcriptional coactivator or corepressor (20, 32). To elucidate which function of APE1 (DNA repair, Ref-1, or acetylation) is important for regulating G4 formation in cells, we ectopically expressed WT, active site mutant H309A (33), Ref-1 function-deficient Cys65S/Cys99S (31), or acetylation-defective K5R (Lysine 6, 7, 27, 31, and 32 mutated to arginine) (20) APE1 in Dox-inducible APE1 down-regulated cells. Our data show that expression of WT APE1 or Ref-1 mutant Cys65/Cys99 APE1 restored the formation of G4 structures in Dox-inducible APE1 down-regulated cells; however, active site mutant H309A and acetylation-defective K5R APE1 mutants failed to restore the formation of G4 foci in cells (Fig. 3E). H309 residue in active site pocket of APE1 is found to be critical for formation of hydrogen bonding with phosphate oxygen of AP site and coordinating the catalysis (34–36). Thus, this result suggests that disruption of AP site binding and the formation of a stable preincision complex of APE1 abrogates G4 formation. All together, both the AP site binding and acetylation of APE1 play a crucial role in the formation of genomic G4 structures in cells.
APE1 Modulates G4-Mediated Expression of Genes.
To identify genes that are regulated by APE1 and G4 structures, we performed RNA-Seq analysis with control and Dox-inducible APE1 knock-down (APE1 KD) A549 cells and compared differentially expressed (log2 ≥ twofold change, adjusted p ) genes with our A549 AcAPE1, AcOGG1, and G4 ChIP-Seq and AP-seq data. We found that 33% of differentially expressed genes have overlapping peaks of AP site damage, AcAPE1, AcOGG1, and G4 structures (Fig. 4B and Dataset S2). The role of promoter G4 structures in regulating the expression of protooncogenes, such as MYC, KRAS, and BCL-2, has been well characterized in multiple studies (37–39). We found that these oncogene G4 promoters have overlapping endogenous AP site damage and AcAPE1 occupancy in A549 and HCT116 cells (Fig. 4C). Thus, to understand the role of APE1 in G4-mediated gene transcription, we used oncogenes KRAS and MYC as a model in our study. Promoter-directed ChIP-qPCR showed significant enrichment of APE1, AcAPE1, and G4 in the previously reported KRAS G4 promoter region (37), but not in the control non-G4 sequence region (Fig. 5A). To establish whether APE1 is involved in folding of the KRAS promoter G4, we performed ChIP-qPCR in WT and APE1 KD cells. The qPCR amplification revealed a significant reduction of G4 enrichment on the KRAS promoter compared to the negative control region in APE1 KD cells (Fig. 5 B and C). To investigate the importance of APE1 acetylation on G4 structure formation, we analyzed the correlation of the genome-wide occupancy of p300 [the acetyltransferase responsible for APE1 acetylation (40)], AcAPE1, and G4 and observed a positive Spearman’s correlation (>0.4) (SI Appendix, Fig. S5B). Moreover, inhibition of APE1 acetylation by adenovirus E1A 12S (E1A inhibits HAT function of p300) protein overexpression abrogated G4 enrichment in the KRAS promoter (SI Appendix, Fig. S5C), which indicates that AcAPE1 is important for G4 folding. A recent study has shown that specific G oxidation and binding of MAZ TF to the KRAS G4 promoter sequence regulate KRAS expression (37). ChIP-qPCR analysis in control, APE1 KD, or E1A overexpressing cells showed decreased MAZ occupancy on the KRAS G4 promoter in the absence of APE1 or AcAPE1 (Fig. 5 B and C and SI Appendix, Fig. S5C). Furthermore, induction of oxidative DNA damage by glucose oxidase (GO) increased enrichment of G4, APE1, and MAZ on the KRAS G4 promoter in WT cells but not in APE1 KD HCTT16 cells (Fig. 5D). Induction of oxidative damage by GO treatment increased KRAS gene expression in WT cells but not in APE1 KD cells (Fig. 5E). Importantly, APE1 KD attenuates both basal and oxidative stress-induced KRAS expression (Fig. 5E). Ectopic expression of WT APE1 in APE1 KD cells restored KRAS gene expression; however, acetylation-defective K5R APE1 or active site mutant H309A APE1 could not restore KRAS gene expression (Fig. 5F), which shows that both the AP site stable binding and acetylation of APE1 are necessary for modulating KRAS gene expression.
Fig. 5.
APE1 modulates G4-mediated expression of genes. (A). (Top) Overlapping enrichment of G4 structures, AP sites, AcOGG1, APE1, and AcAPE1 in the KRAS gene in A549 cells. The shaded region highlights the established KRAS G4 promoter region which shows overlapping AP site damage, G4, and binding of AcOGG1, APE1, and AcAPE1. (Bottom) The shaded region of the schematic highlights the KRAS G4 promoter region and pink box denotes non−G4-forming region. (B–D) Primers were utilized to amplify the G4 region (−23 to −491) and a non-G4 control region (+2,536 to +2,712) to examine the enrichment of G4, MAZ, and APE1 in the G4 region over the non-G4 control region by real-time ChIP-PCR. (B) Enrichment of G4 and MAZ in HCT116 control and constitutively expressing APE1 shRNA cells. (C) Enrichment of G4 and MAZ in HCT116 cells expressing Dox-inducible NTCshRNA and APE1shRNA. (Inset) APE1 levels in these cell extracts were examined by Western blot analysis with α-APE1 and α-HSC70 (loading control). (D) Enrichment of APE1, BG4, and MAZ in HCT116 control and APE1 shRNA cells treated with 50 ng/mL GO for 30 min. (E) Relative KRAS gene expression (normalized to GAPDH) in control (ctrl) and APE1 knockdown cells with or without GO treatment was measured by real-time PCR. (Inset) The APE1 level in these cell extracts was examined by Western blot analysis with α-APE1 and α-HSC70 (as loading control); mRNA, messenger RNA. (F) HCT116 cells expressing APE1shRNA were treated with Dox (2 µg/mL) for 2 days and then transfected with FLAG-tagged WT, or acetylation-defective K5R (Lysine 6, 7, 27, 31, and 32 to arginine), or repair-defective H309A APE1 plasmid constructs for 24 h. RT-PCR was performed to measure KRAS expression. The P value was calculated using unpaired Student’s t tests (****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, n.s. (nonsignificant) = P > 0.05). Error bars denote +SD.
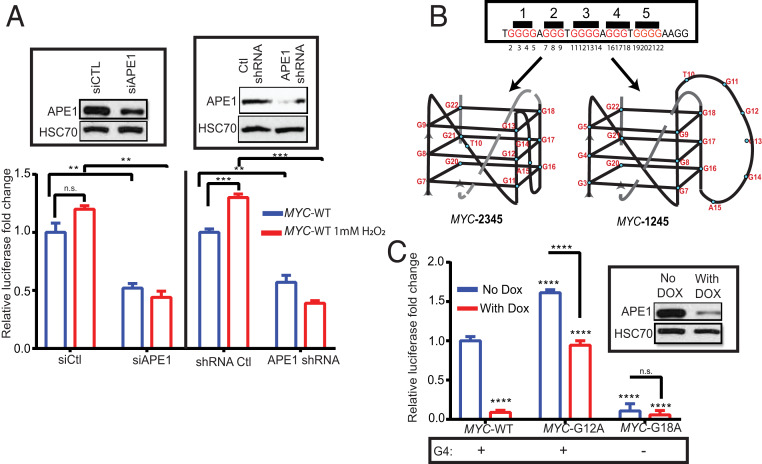
Our ChIP-Seq data showed enrichment of APE1, AcAPE1, and G4 structures in the previously reported MYC G4 promoter, which was also validated by ChIP-qPCR (41) (Figs. 2B and 1E, and SI Appendix, Fig. S5D). To examine the role of APE1 in regulating G4-mediated gene expression, we utilized a promoter luciferase reporter with the WT c-MYC (MYC-WT) G4 sequence in the upstream promoter region of a firefly luciferase coding gene. The expression of c-MYC firefly luciferase was normalized to the relative expression of the renilla luciferase gene with a non-G4 promoter sequence (pRL-TK). We found that induction of oxidative damage by hydrogen peroxide activated MYC-WT luciferase expression in WT cells but not in APE1 KD cells (Fig. 6A). Importantly, APE1 KD attenuated both basal and oxidative stress-induced MYC-WT luciferase activity (Fig. 6A). The c-MYC promoter PQS sequence has five G tracks and was shown to form two alternative G4 structures in vitro utilizing four G tracks. The c-MYC promoter G4 can form using the second, third, fourth, and fifth G tracks (MYC-2345 G4; predominant form) or the first, second, fourth, and fifth G tracks (MYC-1245 G4) (Fig. 6B) (41). To confirm whether the effect of APE1 on gene expression is mediated through the G4 sequence, we introduced two separate mutations in the c-MYC G4 sequence of the promoter luciferase reporter: 1) MYC-G12A (G to A mutation of the 12th position of c-MYC G4), which can only form MYC-1245 G4, and 2) MYC-G18A (G to A mutation of the 18th position of c-MYC G4), which cannot form a G4 structure. Our results demonstrate that MYC-G12A has increased luciferase activity relative to MYC-WT (Fig. 6C), which suggests that the MYC-1245 conformation is involved in the induction of promoter activity compared to MYC-2345 G4, which was previously shown to negatively regulate MYC gene expression (42). MYC-G18A, which cannot form a G4, had significantly decreased luciferase activity relative to MYC-WT (Fig. 6C). Down-regulation of APE1 in cells reduced (∼fourfold) MYC-WT and (∼twofold) MYC-G12A luciferase expression, but did not significantly affect MYC-G18A luciferase expression (Fig. 6C). Similarly, activation of BCL-2 G4 promoter luciferase expression upon oxidative damage was found to be dependent on APE1 (SI Appendix, Fig. S5E) (38). Overall, our results suggest that APE1 modulates G4-mediated gene expression, such as MYC or KRAS, via promoting the formation of G4 structures and facilitating TF loading.
Fig. 6.
APE1 regulates G4-mediated Myc gene expression. (A) WT MYC G4 promoter luciferase reporter (MYC-WT) and pRL-TK−renilla luciferase were cotransfected in HCT116 cells transiently expressing siCtl or siAPE1 or constitutively expressing APE1shRNA. Forty-eight hours after transfection, cells were treated with 1 mM H2O2 for 1 h, and luciferase activity was measured and normalized with renilla luciferase. (Insets) The level of APE1 in these cell extracts was examined by Western blot analysis with α-APE1 and α-HSC70. (B) The sequence of the G4-forming Nuclease Hypersensitive element III1 (NHEIII1) element of the c-MYC gene. Bases are numbered according to the sequence of the NHEIII1. Two alternative G4 folding patterns (MYC-1245 and MYC-2345) of NHEIII1 are shown. (C) MYC–WT promoter luciferase reporters or MYC harboring a G to A mutation of the 12th base (MYC-G12A) and 18th base (MYC-G18A) were transfected in Dox-inducible HCT116APE1shRNA cells. Firefly luciferase activity was measured and normalized with renilla luciferase activity. (Inset) The level of APE1 knockdown in these cell extracts was examined by Western blot analysis with α-APE1 and α-HSC70. The P value was determined using an unpaired Student’s t test (****P < 0.0001, ***P < 0.001, **P < 0.01, n.s. (nonsignificant) = P > 0.05). Error bars denote +SD.
Binding of APE1 to AP Site Damage in PQS Promotes the G4 Folding, and Acetylation of APE1 Enhances Residence Time.
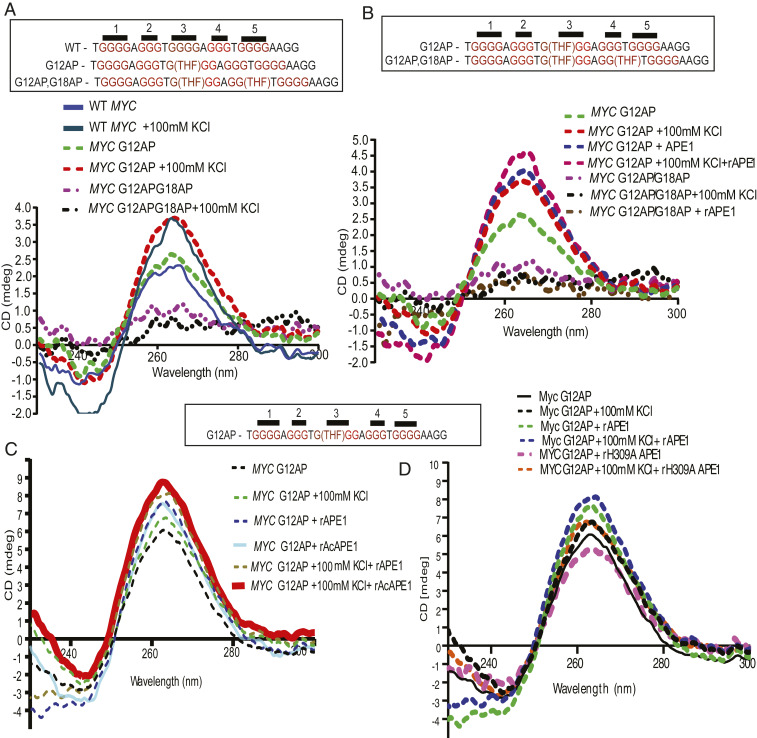
Finally, we investigated the mechanistic connection of AP site damage and binding of APE1 in a PQS on the stable formation of G4 structures using a 28-mer c-MYC promoter G4 (WT MYC) oligo. To test the effects of AP site damage in G4 sequences, we used MYC promoter G4 oligos containing AP site analog (tetrahydrofuran) at G12 (MYC G12AP) or two AP site analogs at G12 and G18 (MYC G12AP/G18AP) (Fig. 7A). Circular dichroism (CD) spectroscopy was performed to determine the secondary structure of DNA (43). The presence of a strong positive peak at 265 nm with a weak negative signal at 240 nm is indicative of a parallel G4 structure. We found that WT MYC and MYC G12AP G4 formation increased in the presence of KCl (Fig. 7A). The MYC G12AP/G18AP oligo with AP sites in both the third and fourth G tracks served as a negative control, as it was unable to form a G4 structure in the presence of KCl. To test whether APE1 can induce the formation of stable G4s, we incubated MYC G12AP and G12AP/G18AP oligos with recombinant APE1 (rAPE1). The addition of APE1 stimulated the folding of the MYC G12AP oligo to a G4 structure even in the absence of KCl, which suggests that APE1 promotes the formation of G4 folding in vitro (Fig. 7B). In contrast, addition of APE1 to MYC G12AP/G18AP oligo, which cannot form G4, showed no effect on CD signals. We also observed that in vitro AcAPE1 also enhanced the formation of G4 (Fig. 7C). To examine whether stable coordination/binding with AP site or catalytic activity of APE1 is required for promoting G4 folding in vitro, we incubated WT APE1 or H309A or N212A APE1 mutants with MYC G12AP oligo. X-ray crystallography and molecular modeling studies of APE1 bound to substrate AP site DNA and in vitro activity assays suggest that H309 stabilizes the preincision complex by forming a hydrogen bond with a nonbridging phosphate oxygen atom of the AP site to orient and polarize the phosphate backbone for nucleophilic attack mediated by N212A (33, 35, 36, 44, 45). Therefore, H309 in active pocket primarily plays a role in stabilization of APE1–AP site preincision complex, while N212 is crucial for AP site cleavage or catalysis. Our CD results demonstrate that, while both WT APE1 and catalytically inactive N212A APE1 mutant can promote the MYC G12AP folding, H309A mutant was unable to stimulate the G4 folding (Fig. 7D and SI Appendix, Fig. S6A), suggesting that formation of a stable APE1–AP site preincision complex, rather than AP site cleavage or catalytic activity of APE1, is required for promoting G4 formation in vitro. Interestingly, we found that, while APE1 has very little effect on WT MYC G4 folding, APE1 can stimulate its folding in the presence of KCl (SI Appendix, Fig. S6B), which indicates APE1 can stabilize a preformed G4 structure in vitro. We compared the binding affinity of APE1 to AP site containing MYC duplex (DS) and preformed G4 MYC promoter oligos by electrophoretic mobility-shift assay (EMSA) assay. We found that rAPE1 has an equal binding affinity for an AP site when present in MYC duplex or G4 (Fig. 8A); however, APE1 could not stably bind to a MYC G4 oligo without an AP site (SI Appendix, Fig. S6C). Interestingly, our EMSA data show that the binding ability of H309A and N212A mutant APE1 proteins to an AP site containing DNA oligo is comparable to that of WT APE1 protein (SI Appendix, Fig. S6D).
Fig. 7.
Binding of APE1 to an AP site in a PQS promotes G4 folding. (A) The 28-mer c-MYC G4-forming oligonucleotide sequences with or without an AP site analog (THF) are shown. CD spectra of WT-MYC, MYC G12AP, and MYC G12APG18AP at 20 °C in the presence or absence of 100 mM KCl. The ordinate indicates the ellipticity signal expressed in millidegrees. (B) CD spectra of MYC G12AP and MYC G12APG18AP at 20 °C in the presence of 100 mM KCl or 1 µg of rAPE1, or both. (C and D) CD spectra of MYC G12AP at 20 °C in the presence of 100 mM KCl alone or in combination with 1 µg of recombinant (C) APE1 or AcAPE1 or (D) APE1 or H309A mutant APE1 proteins.
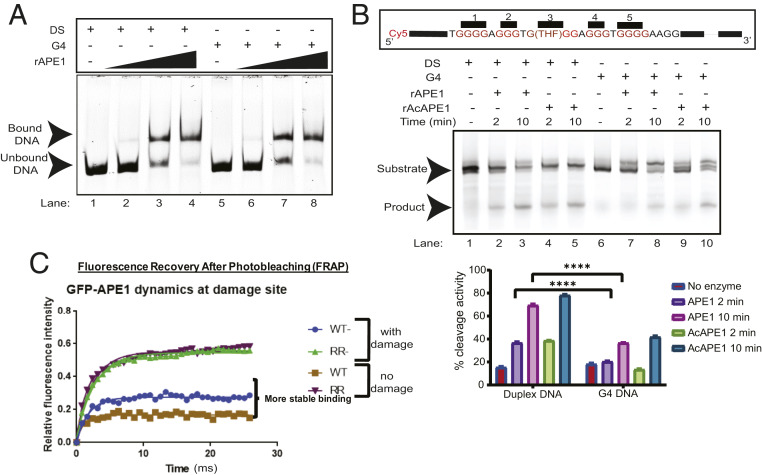
Fig. 8.
Acetylation of APE1 enhances residence time. (A) Electrophoretic mobility shift assay was performed to analyze the binding of rAPE1 to a Cy5-labeled DS or G4 oligonucleotide substrates. Substrates were incubated with 6.4, 64, and 64 ng of rAPE1. (B) (Top) Comparison of rAPE1 and rAcAPE1 endonuclease activity on a Cy5-labeled 75-mer DS and G4 substrate. Formation of a 37-mer cleaved product is shown at 2- and 10-min time points after incubation. (Bottom) Quantitations of the cleaved product from three independent experiments are shown. (C) FRAP analysis of GFP-APE1WT and GFP-APE1K6R/K7R expressing cells was performed before and after DNA damage for 30 min. Each point represents the average of n = 15 cells. Experiments were repeated three times and consistently showed similar mobility differences.
A previous study observed that APE1 could bind to AP sites within a G4 structure, but the cleavage rate is attenuated (46). Consistent with this, we also found that rAPE1 cleaves an AP site less efficiently when it is present in a G4 (Fig. 8B). We recently showed that APE1 is acetylated after binding to an AP site in chromatin (20). Our current study suggests that both stable interaction or coordination with AP site and the acetylation of APE1 are essential for G4 (Fig. 3E) formation in cells; therefore, we examined the effect of acetylation on endonuclease activity of APE1 on a G4 substrate. Recombinant AcAPE1 (rAcAPE1) had similar activity compared to unmodified rAPE1 when the AP site was present in a folded G4 structure (Fig. 8B and SI Appendix, Fig. S6E). It was previously shown that APE1 remains bound to the cleaved AP site and coordinates recruitment of the downstream BER enzyme polymerase β (47). To test whether acetylation of APE1 increases residence time at AP site damage in chromatin, we measured the mobility of WT APE1 GFP and RR-APE1 (lysine 6 and 7 acetylation-defective) Lysine 6 and 7 acetylation-defective RR-APE1 GFP at damage sites by performing fluorescence recovery after photobleaching (FRAP) assay with or without induction of AP site damage in cells. We found that WT APE1 GFP protein has a higher residence time at damage sites and a less mobile fraction compared to RR-APE1 GFP in control or damage-treated cells (Fig. 8C). The FRAP results suggest that acetylation may impede the complete repair of an AP site and increase the residence time of APE1 on a G4 structure to coordinate transcriptional activation or repression via regulating loading of TFs to promoters.
Discussion
Oxidative DNA damage is conventionally viewed as detrimental to cellular processes. However, mounting evidence supports the interplay between oxidative stress signaling, formation of 8-oxoG and AP sites, binding of cognate repair protein OGG1 and APE1 in promoters, and transcriptional activation or repression of mammalian genes (48–50). Studies have shown that oxidized DNA base damage has a strong positive correlation with elevated oncogene and proinflammatory gene expression (51, 52). Of note is that many genes with a G4 promoter are known to be regulated by oxidative stress (53). Recent studies have demonstrated that 8-oxoG or AP sites in G4-forming promoter sequences of VEGF, BCL-2, and KRAS induced up-regulation of the respective genes (37, 38, 54). In this study, using genome-wide ChIP-Seq analyses, cell-based assays, and in vitro biochemical analyses, we have provided a mechanistic framework linking oxidized DNA base-derived AP sites, binding of APE1 to G4 formation/stability, and the control of gene expression. We demonstrate that the occurrence of endogenous oxidized base-derived AP site damage and APE1 binding are not random and are predominant in G4 sequences. Furthermore, our study reveals an essential role of APE1 in the formation of G4s in the genome to regulate gene expression.
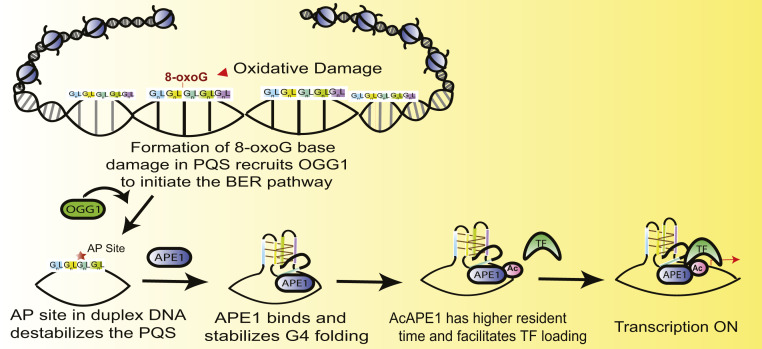
The observations that loss of OGG1, APE1, or AcAPE1 abrogates G4 formation raise questions about the biological and mechanistic roles of these proteins and their acetylation in the formation of G4s. We propose that cellular oxidants oxidize guanine base in PQSs, which recruit OGG1 to initiate the BER pathway (Fig. 9). OGG1 cleaves an 8-oxoG to generate an AP site and remains bound to the AP site (55). OGG1 is then acetylated by histone acetyltransferase p300, a protein commonly found in promoters/enhancers and gene bodies due to association with TFs and RNA Pol II. We previously demonstrated that the acetylation of OGG1 enhances its catalytic turnover by reducing its affinity for AP sites in DNA (24). The generation of an AP site significantly impacts the thermal stability of duplex DNA, unlike 8-oxoG paired with C (56); therefore, we propose that the generation of an AP site in a PQS after excision of an 8-oxoG by OGG1 destabilizes and opens up the duplex (57, 58). Subsequent binding and stable coordination of APE1 with AP site in PQS promotes the formation and stabilization of a G-quadruplex structure with the AP site containing G track looped out. APE1 bound to an AP site is then acetylated by p300, which reduces its dissociation from AP sites (59). AcAPE1 bound to a G4 structure is then primed to stimulate the loading of activator or repressor TFs. Several lines of evidence support this model. We have shown a genome-wide overlap of AP site damage, APE1/AcAPE1, AcOGG1, and G4 (Figs. 1 A and B and 2 A–C). Furthermore, we observed a positive correlation between the occupancy of p300, AcAPE1, and G4 (SI Appendix, Fig. S5B). Our data show that loss of OGG1, APE1, AcAPE1, or stable APE1–AP site coordination, abrogates G4 formation (Figs. 3 and 5 B–D and SI Appendix, Figs. S3 C–E and S5C). Additionally, knockdown of APE1 or inhibition of APE1 acetylation reduced MAZ TF loading on the KRAS G4 promoter (Fig. 5 B–D and SI Appendix, Fig. S5C). Our finding that APE1 is required for MAZ TF loading to the KRAS G4 promoter is further supported by several previous studies which have reported the interaction and cooccupancy of APE1 with TFs, such as STAT3, HIF-1α, AP-1, nuclear factor (NF)-κB, and HDAC1, at promoter regions (40, 59–61). The higher mobility or lower residence time of acetylation-defective APE1 (APE1K6R/K7R) in chromatin compared to WT APE1 indicates that acetylation of APE1 delays dissociation from an AP site (Fig. 8C). Furthermore, AcAPE1-bound regions significantly overlap with G4-forming regions that bear active enhancer and promoter histone marks (Fig. 1F). Finally, our in vitro CD experiments show that rAPE1 stimulates the formation/folding of MYC G4 (Fig. 7B).
Fig. 9.
Schematic representation linking oxidized G base damage (8-oxoG), AP site formation, and binding of APE1 in a PQS sequence in the genome to promote G4 structure formation. Oxidation of G in G4 motif sequences (GnLGnLGnLGnLGn; where n ≥ 3 and L is a loop region containing any nucleotide) initiates the BER pathway by recruiting OGG1. OGG1 cleaves 8-oxoG to generate an AP site, which destabilizes and opens up the duplex. Subsequent binding of APE1 to the AP sites in PQS promotes the formation and stabilization of a G-quadruplex structure with the AP site containing G track looped out. APE1 is then acetylated by p300, which enhances its residence time and delays its dissociation from AP sites. AcAPE1 bound to a G4 structure is then primed to stimulate the loading of TFs to regulate gene expression.
Our in vitro CD data demonstrate that stabilization of APE1–AP site preincision complex but not the cleavage of AP site in the MYC PQS oligo is required for promoting the formation of G4 structures in vitro (Fig. 7 B and D and SI Appendix, Fig. S6A); however, we do not know the exact molecular mechanism by which APE1 mediates the folding and stabilization of G4 structures. Structural characterization of APE1 bound to substrate AP site DNA suggests that initial binding of APE1 to AP site DNA induces DNA bending which causes AP site eversion into the enzyme active site pocket (34). Upon AP site eversion, H309 forms hydrogen bonding with AP sites and stabilizes the APE1–AP site preincision complex (33–36); therefore, it is likely that H309 is a critical residue in promoting G4 structure mediated by APE1-induced DNA conformational changes. Supporting this idea, we demonstrated that H309A mutant which cannot bind/stabilize AP sites in enzyme active site pocket or by MX which blocks APE1 binding to AP sites abrogates the formation of G4 structures in cells (Fig. 3 B and E). Although less efficiently, APE1 can cleave AP site in G4 but remains bound to cleaved AP site; APE1 is then acetylated by p300, which further reduces the dissociation of APE1 from G4 structures and likely prevents loading of G4 resolving helicases. This is congruent with previous studies which have shown that APE1 can interact with and inhibit the activity of WRN, a member of RecQ family human helicases reported to resolve G4 DNA in human telomeres (62). Finally, deacetylation of APE1 by SIRT1 promotes the dissociation of APE1 from G4 structures, and, subsequently, 3′ to 5′ DNA helicases such as WRN or BLM are recruited to resolve the G4 structures. After resolution of G4 structures, downstream BER proteins can then repair the APE1-generated DNA nick and restore the G4 sequence. Earlier studies from our laboratory and others have shown that APE1 acetylation/deacetylation cycles by p300 and SIRT1 regulate endogenous damage repair via BER pathway (63, 64). Nonetheless, it is imperative that, in cells, both AP site binding and its AP site cleavage activity are equally important for regulating the spatiotemporal formation and stability of G4 and to prevent mutation in G4 DNA sequences. Further studies are necessary to address this hypothesis.
Down-regulation of APE1 abolished the formation of G4 structures in cells, as evident from the loss of G4-specific 1H6 antibody staining in immunofluorescence (Fig. 3 B and C); however, G4 ChIP-Seq in APE1 down-regulated cells revealed that a fraction of the G4 structures can still form in the genome. The apparent disagreement between the two results can be explained by the differences in the experimental approach. Immunofluorescence imaging with the G4-specific 1H6 antibody likely does not have sufficient resolution or sensitivity to detect all G4 structures in the genome. Since PQSs have diverse sequence motifs, and G4 structures can exist in multiple conformations (e.g., parallel, antiparallel, with varying loop lengths) with variable degrees of stability, APE1 may stabilize specific G4 structure conformations. Additional experiments and detailed PQS motif analyses are necessary to explore this area.
The impact of oxidative stress on multiple biological processes is well documented (52, 54, 65). During the onset of reactive oxygen species-associated inflammation, OGG1 and APE1 were shown to augment proinflammatory gene expression by facilitating TF binding to promoters (52). We propose that G4 sequences act as a sensor for oxidative stress. Due to low oxidation potential, G bases of G4 sequences are more susceptible to oxidize and form 8-oxoG damage. Initiation of BER in G4 sequences could serve as an intermediate step in a signal transduction cascade that regulates G4-mediated gene expression and impacts multiple biological processes, including cell proliferation and innate immune response (65). The idea that G4s can function as a sensor of oxidative stress is supported by human genome sequence analyses which have shown that promoter G4s are highly conserved in comparison to other genomic regions (66).
Although this study shows that G4 structure formation is coupled with endogenous oxidative DNA damage and subsequent activation of BER, the source of site-specific G oxidation in PQS promoter sequences remains a question. Random oxidation of G bases is too erratic to constitute the mechanism. We have observed a reproducible enrichment of AP sites and binding of BER proteins in specific gene promoter and gene bodies in multiple independent experiments in different cell lines. This discovery raises the intriguing question about whether endogenous oxidative or AP site damage occurs in a targeted or site-specific manner. Although a few recent studies have shown a region-specific distribution of DNA damage (67, 68), additional high-resolution (single-base) mapping of base damage in the genome is warranted. Previous studies have indicated the presence of a target-specific DNA base damage mechanism in cells. For example, eliciting targeted DNA base damage appears to be a common first step in hormone-induced activation of many genes. Perillo et al. (69) have shown that estrogen-induced activation of BCL-2 is controlled by LSD1 flavin-dependent demethylation of H3K9me2 in the BCL-2 promoter. Interestingly, the flavin-dependent mechanism by which LSD1 demethylates H3K9me2 generates H2O2. Local oxidation arising from LSD1 demethylation was shown to produce oxidized G bases in the BCL-2 promoter, which contains a PQS (38). Perillo et al. demonstrated that LSD1-mediated oxidation and OGG1 recruitment in the BCL-2 promoter was essential for BCL-2 gene activation. Pan et al. (49) have also shown that oxidation of G in NF-κB binding sites promotes NF-κB binding and stimulates transcription. Further studies are necessary to address how an indiscriminate oxidant like H2O2 liberated by LSD1 generates specific G oxidation in PQS sequences to induce G4 formation.
G4 structures are often formed in the 3′ overhang regions of telomere sequences (70) and can serve regulatory roles by protecting telomere cap structures (4). Madlener et al. (71) showed that the absence of either APE1 endonuclease function or acetylation of APE1 results in telomere shortening and fusion and the formation of micronuclei. Telomere defects in the absence of APE1 acetylation or endonuclease activity could be a result of the destabilization of telomere G4 structures. G4-induced replication stress, DNA damage, and genomic instability are linked with many cancers (72). Studies have found that G4-forming sequences are enriched at translocation breakpoints (73). We propose that activation of BER upon endogenous oxidative DNA damage not only repairs damaged bases but also regulates the formation and stability of G4 structures in the genome to coordinate multiple biological processes. Our study introduces a perspective in region-specific endogenous oxidative damage and activation of BER in the regulation of G4 to coordinate multiple cellular processes. This function of endogenous damage and the BER machinery defines a role beyond the well-characterized role as a safeguard for maintaining genomic integrity.
Materials and Methods
Detailed materials and methods are described in SI Appendix, Materials and Methods, including cell lines, plasmids, reagents, immunofluorescence analysis, Western blot analysis, ChIP method and ChIP-Seq and statistical analysis, RNA-Seq and qRT-PCR techniques, Luciferase assay, CD, EMSA, and AP endonuclease activity assay protocols.
Data Availability.
The sequence data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (GSE142284). All materials generated (such as cell lines) are available to readers upon request.
Supplementary Material
Acknowledgments
We are grateful to Dr. Istvan Boldogh, for providing WT and OGG1-null MEF cells, and to Dr. Jyoti Dash, for MYC-G12A luciferase construct. This work was supported by NIH/NCI (National Cancer Institute) (Grants R01 CA148941 and R03 CA235214) to K.K.B., and S.R. is supported by NCI/NIH Fellowship F99/K00 CA223064. The University of Nebraska Medical Center (UNMC) Advanced Confocal Microscopy Core facility is funded by the Nebraska Research Initiative and Fred and Pamela Buffet Cancer Center support Grant P30 CA036727.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (GSE142284). All ChIP-Seq and RNA-Seq data will be available to readers upon manuscript publication.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912355117/-/DCSupplemental.
References
- 1.Bochman M. L., Paeschke K., Zakian V. A., DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gellert M., Lipsett M. N., Davies D. R., Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 48, 2013–2018 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burge S., Parkinson G. N., Hazel P., Todd A. K., Neidle S., Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 34, 5402–5415 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hänsel-Hertsch R., et al. , G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 48, 1267–1272 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Maizels N., G4-associated human diseases. EMBO Rep. 16, 910–922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhodes D., Lipps H. J., G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 43, 8627–8637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hänsel-Hertsch R., Di Antonio M., Balasubramanian S., DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 18, 279–284 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Lane A. N., Chaires J. B., Gray R. D., Trent J. O., Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 36, 5482–5515 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauer M., Paeschke K., G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 45, 1173–1182 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Cadet J., Wagner J. R., Shafirovich V., Geacintov N. E., One-electron oxidation reactions of purine and pyrimidine bases in cellular DNA. Int. J. Radiat. Biol. 90, 423–432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleming A. M., Burrows C. J., 8-Oxo-7,8-dihydro-2′-deoxyguanosine and abasic site tandem lesions are oxidation prone yielding hydantoin products that strongly destabilize duplex DNA. Org. Biomol. Chem. 15, 8341–8353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson D. M. 3rd, Bohr V. A., The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst.) 6, 544–559 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Li M., Wilson D. M. 3rd, Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox Signal. 20, 678–707 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frossi B., et al. , Endonuclease and redox activities of human apurinic/apyrimidinic endonuclease 1 have distinctive and essential functions in IgA class switch recombination. J. Biol. Chem. 294, 5198–5207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tell G., Quadrifoglio F., Tiribelli C., Kelley M. R., The many functions of APE1/Ref-1: Not only a DNA repair enzyme. Antioxid. Redox Signal. 11, 601–620 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izumi T., et al. , Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl. Acad. Sci. U.S.A. 102, 5739–5743 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindahl T., Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Atamna H., Cheung I., Ames B. N., A method for detecting abasic sites in living cells: Age-dependent changes in base excision repair. Proc. Natl. Acad. Sci. U.S.A. 97, 686–691 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubo K., Ide H., Wallace S. S., Kow Y. W., A novel, sensitive, and specific assay for abasic sites, the most commonly produced DNA lesion. Biochemistry 31, 3703–3708 (1992). [DOI] [PubMed] [Google Scholar]
- 20.Roychoudhury S., et al. , Human apurinic/apyrimidinic endonuclease (APE1) is acetylated at DNA damage sites in chromatin, and acetylation modulates its DNA repair activity. Mol. Cell. Biol. 37, e00401-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stavrovskaya E. D., et al. , StereoGene: Rapid estimation of genome-wide correlation of continuous or interval feature data. Bioinformatics 33, 3158–3165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikin O., D’Antonio L., Bagga P. S., QGRS Mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 34, W676–W682 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsico G., et al. , Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 47, 3862–3874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhakat K. K., Mokkapati S. K., Boldogh I., Hazra T. K., Mitra S., Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell. Biol. 26, 1654–1665 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hänsel-Hertsch R., Spiegel J., Marsico G., Tannahill D., Balasubramanian S., Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 13, 551–564 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Brooks T. A., Hurley L. H., Targeting MYC expression through G-quadruplexes. Genes Cancer 1, 641–649 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bensaude O., Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2, 103–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bacsi A., Chodaczek G., Hazra T. K., Konkel D., Boldogh I., Increased ROS generation in subsets of OGG1 knockout fibroblast cells. Mech. Ageing Dev. 128, 637–649 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J. H., et al. , Role of mammalian Ecdysoneless in cell cycle regulation. J. Biol. Chem. 284, 26402–26410 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allhoff M., Seré K., F Pires J., Zenke M., G Costa I., Differential peak calling of ChIP-seq signals with replicates with THOR. Nucleic Acids Res. 44, e153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley M. R., Georgiadis M. M., Fishel M. L., APE1/Ref-1 role in redox signaling: Translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Curr. Mol. Pharmacol. 5, 36–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chattopadhyay R., et al. , Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol. Cell. Biol. 28, 7066–7080 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsutakawa S. E., et al. , Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J. Biol. Chem. 288, 8445–8455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mol C. D., Izumi T., Mitra S., Tainer J. A., DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403, 451–456 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Batebi H., Dragelj J., Imhof P., Role of AP-endonuclease (Ape1) active site residues in stabilization of the reactant enzyme-DNA complex. Proteins 86, 439–453 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Lowry D. F., et al. , Investigation of the role of the histidine-aspartate pair in the human exonuclease III-like abasic endonuclease, Ape1. J. Mol. Biol. 329, 311–322 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Cogoi S., Ferino A., Miglietta G., Pedersen E. B., Xodo L. E., The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: Implications on transcription. Nucleic Acids Res. 46, 661–676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agrawal P., Lin C., Mathad R. I., Carver M., Yang D., The major G-quadruplex formed in the human BCL-2 proximal promoter adopts a parallel structure with a 13-nt loop in K+ solution. J. Am. Chem. Soc. 136, 1750–1753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleming A. M., Zhou J., Wallace S. S., Burrows C. J., A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent. Sci. 1, 226–233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sengupta S., Mantha A. K., Mitra S., Bhakat K. K., Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1. Oncogene 30, 482–493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beckett J., Burns J., Broxson C., Tornaletti S., Spontaneous DNA lesions modulate DNA structural transitions occurring at nuclease hypersensitive element III(1) of the human c-myc proto-oncogene. Biochemistry 51, 5257–5268 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Siddiqui-Jain A., Grand C. L., Bearss D. J., Hurley L. H., Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varizhuk A., et al. , The expanding repertoire of G4 DNA structures. Biochimie 135, 54–62 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Masuda Y., Bennett R. A., Demple B., Dynamics of the interaction of human apurinic endonuclease (Ape1) with its substrate and product. J. Biol. Chem. 273, 30352–30359 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Kim W.-C., et al. , Characterization of the endoribonuclease active site of human apurinic/apyrimidinic endonuclease 1. J. Mol. Biol. 411, 960–971 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broxson C., Hayner J. N., Beckett J., Bloom L. B., Tornaletti S., Human AP endonuclease inefficiently removes abasic sites within G4 structures compared to duplex DNA. Nucleic Acids Res. 42, 7708–7719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson S., Kunkel T. A., Passing the baton in base excision repair. Nat. Struct. Biol. 7, 176–178 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Clark D. W., Phang T., Edwards M. G., Geraci M. W., Gillespie M. N., Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radic. Biol. Med. 53, 51–59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan L., et al. , Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor κB-driven gene expression. J. Biol. Chem. 291, 25553–25566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fleming A. M., Ding Y., Burrows C. J., Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U.S.A. 114, 2604–2609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leone A., Roca M. S., Ciardiello C., Costantini S., Budillon A., Oxidative stress gene expression profile correlates with cancer patient poor prognosis: Identification of crucial pathways might select novel therapeutic approaches. Oxid. Med. Cell. Longev. 2017, 2597581 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ba X., et al. , 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J. Immunol. 192, 2384–2394 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fedeles B. I., G-quadruplex-forming promoter sequences enable transcriptional activation in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 114, 2788–2790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pastukh V., et al. , An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell. Mol. Physiol. 309, L1367–L1375 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill J. W., Hazra T. K., Izumi T., Mitra S., Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: Potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 29, 430–438 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J., Fleming A. M., Averill A. M., Burrows C. J., Wallace S. S., The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 43, 7171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Esposito V., et al. , Effects of abasic sites on structural, thermodynamic and kinetic properties of quadruplex structures. Nucleic Acids Res. 38, 2069–2080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minetti C. A., et al. , Impact of bistrand abasic sites and proximate orientation on DNA global structure and duplex energetics. Biopolymers 109, e23098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhakat K. K., Izumi T., Yang S. H., Hazra T. K., Mitra S., Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 22, 6299–6309 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhakat K. K., Mantha A. K., Mitra S., Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid. Redox Signal. 11, 621–638 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ray S., Lee C., Hou T., Bhakat K. K., Brasier A. R., Regulation of signal transducer and activator of transcription 3 enhanceosome formation by apurinic/apyrimidinic endonuclease 1 in hepatic acute phase response. Mol. Endocrinol. 24, 391–401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahn B., Harrigan J. A., Indig F. E., Wilson D. M. 3rd, Bohr V. A., Regulation of WRN helicase activity in human base excision repair. J. Biol. Chem. 279, 53465–53474 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Yamamori T., et al. , SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 38, 832–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sengupta S., et al. , Elevated level of acetylation of APE1 in tumor cells modulates DNA damage repair. Oncotarget 7, 75197–75209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aguilera-Aguirre L., et al. , Whole transcriptome analysis reveals an 8-oxoguanine DNA glycosylase-1-driven DNA repair-dependent gene expression linked to essential biological processes. Free Radic. Biol. Med. 81, 107–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakken S., Rognes T., Hovig E., The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 37, 5749–5756 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poetsch A. R., Boulton S. J., Luscombe N. M., Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis. Genome Biol. 19, 215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mao P., et al. , Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity. Genome Res. 27, 1674–1684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perillo B., et al. , DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319, 202–206 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Fouquerel E., et al. , Oxidative guanine base damage regulates human telomerase activity. Nat. Struct. Mol. Biol. 23, 1092–1100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Madlener S., et al. , Essential role for mammalian apurinic/apyrimidinic (AP) endonuclease Ape1/Ref-1 in telomere maintenance. Proc. Natl. Acad. Sci. U.S.A. 110, 17844–17849 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y., et al. , G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 10, 943 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bacolla A., Ye Z., Ahmed Z., Tainer J. A., Cancer mutational burden is shaped by G4 DNA, replication stress and mitochondrial dysfunction. Prog. Biophys. Mol. Biol. 147, 47–61 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (GSE142284). All materials generated (such as cell lines) are available to readers upon request.